Abstract
To elucidate the mechanism of internodal elongation in rice (Oryza sativa L.), we analyzed genes encoding xyloglucan endotransglycosylase (XET), a cell wall-loosening enzyme essential for cell elongation. Four rice XET-related (XTR) genes, OsXTR1, OsXTR2, OsXTR3, and OsXTR4, were isolated and their expression patterns in rice plants determined. The expression of the four XTR genes showed different patterns of organ specificity and responses to several plant hormones. OsXTR1 and OsXTR3 were up-regulated by gibberellin and brassinosteroids, whereas OsXTR2 and OsXTR4 showed no clear response to these hormones. Expression of the four XTR genes was also investigated in elongating internodes at different developmental stages. OsXTR1 and OsXTR3 were preferentially expressed in the elongating zone of internodes, while OsXTR2 and OsXTR4 were expressed in nodes and in the divisional and elongating zones of internodes. In three genetic mutants with abnormal heights, the expression of OsXTR1 and OsXTR3 correlated with the height of the mutants, whereas no such correlation was observed for OsXTR2 and OsXTR4. Based on these observations, we discuss the roles that OsXTR1 and OsXTR3 may play in internodal elongation in rice.
Internodal elongation in rice (Oryza sativa L.) is a specific developmental phenomenon that accompanies panicle development. This process involves cell division followed by differentiation, with dividing cells forming a secondary meristematic tissue, and subsequent cell elongation leading to dramatic lengthening of the internode. Therefore, the mechanism of internodal elongation includes control of both cell division and elongation. It has been pointed out that there is a close relationship between internodal elongation and phase change in the shoot apical meristem from vegetative to reproductive (Kawahara et al., 1968). This observation indicates that a regulatory mechanism exists which responds to the phase change in the shoot apical meristem and induces internodal elongation.
Because the length and strength of the rice culm are agronomically important traits, a large number of dwarf mutants deficient in internodal elongation have been isolated and characterized in an effort to further our understanding of this process (Kamijima et al., 1996). Some of these mutants have been tested for response to plant growth regulators such as gibberellin (GA) and auxin (Murakami, 1972; Mitsunaga et al., 1993), revealing that these hormones (especially GA) promote internodal elongation by enhancing cell division and/or elongation in the internode.
During morphogenesis at any developmental stage, all plant cells require modifications of the structure of their cell wall. The cell wall is the main factor that determines cell shape, and cell wall reconstruction makes possible its modification during cell elongation. According to a cell wall model (McQueen-Mason, 1996; Cosgrove, 1997), the primary cell wall consists of three co-extensive polymer networks: the cellulose-xyloglucan framework, pectin, and structural protein. It is considered that structural changes in these networks are regulated by enzymatic modification, and therefore wall-modifying enzymes would be expected to play an important role in regulating the plasticity of the cell wall.
Xyloglucan endotransglycosylase (XET) internally cleaves xyloglucan polymers and ligates the newly generated reducing ends to other xyloglucan chains (Nishitani and Tominaga, 1991; Smith and Fry, 1991; Fry et al., 1992; Nishitani and Tominaga, 1992). Because xyloglucan mediates the cross-linking of cellulose microfibrils in the plant cell wall (McCann et al., 1990; Hayashi et al., 1994), XET must have a role for cell wall plasticity, resulting in cell elongation. In various species, including both dicot and monocot plants, XET is encoded by a multigene family and, therefore, it is suggested that the expression of the individual XET genes is differentially regulated at various developmental stages and by diverse environmental stimuli (for review, see Nishitani, 1997). Arabidopsis, for example, contains a complex gene family consisting of more than 20 XET genes that are differentially regulated by several environmental stimuli, suggesting a recruitment of distinct XET genes that may control the properties of cell walls during development (Xu et al., 1996). It is reasonable to expect that, in rice internodal elongation, a specific subset of XET genes is also regulated by particular hormonal effects or environmental stimuli.
We report here the identification of four rice XET-related (XTR) genes, OsXTR1, OsXTR2, OsXTR3, and OsXTR4, and the characterization of their developmental and hormonal regulation. The role of the XTR genes in rice internodal elongation is discussed.
MATERIALS AND METHODS
Plant Materials
Wild-type rice (Oryza sativa L. cv Thai-Chung No. 65 [T65]) and three rice mutants (Akibare Dwarf, Waito C, and Awaodori) were grown in a greenhouse under the conditions of natural temperature and daylight in April. Three-week-old plants were used for total RNA extraction for expression analysis of XTR genes and for the treatments with plant hormones. Various organs from 11- to 13-week-old plants were harvested for RNA extraction. For extraction of RNA from elongating culms, 3- to 5-cm internodes of 7- to 8-week-old plants were sectioned (approximately 5 mm thick). The sections were divided into four groups: the node, the divisional zone of the internode immediately above the node, the elongating zone, and the elongated zone.
Hormone Treatments
Whole 3-week-old plants were treated by immersion in distilled water, 1 μm brassinolide, or 30 μm GA (GA3), grown in continuous light at 30°C for 12, 24, or 48 h, then rapidly frozen in liquid nitrogen and used for RNA extraction.
Cloning and Sequencing of XTR Genes
Rice XTR genes were initially identified among rice expressed sequence tag (EST) clones. Three independent clones were identified based on their nucleotide sequence similarity to Arabidopsis XET-related genes (EXT, XTR3, and XTR2) and were termed OsXTR1, OsXTR2, and OsXTR3, respectively. A fourth clone, OsXTR4, with similarity to conserved regions of the Bacillus licheniformis β-1,3-1,4-glucanase gene, was also identified. The entire nucleotide sequence of OsXTR1 was determined by sequencing the EST clone (accession no. D41305). Full-length cDNAs corresponding to OsXTR2 and OsXTR4 were isolated from a rice cDNA library (Sentoku et al., 1999) and sequenced. Because a full-length OsXTR3 cDNA was not obtained from the cDNA library, we screened a genomic library. One clone that contained a sequence identical to the partial OsXTR3 cDNA was isolated and used to determine the 5′ sequence of OsXTR3.
DNA and RNA Gel-Blot Analyses
To determine whether the various XTR clones cross-hybridized, 1 μg of plasmid DNA from each cDNA clone was digested with a suitable restriction enzyme, electrophoresed in a 1% (w/v) agarose gel, and transferred to a nylon membranes (Hybond N+, Amersham, Buckinghamshire, UK) under alkaline conditions. Southern hybridization was performed using the entire sequence of each XTR clone as probes under stringent conditions in hybridization solution containing sodium phosphate at 65°C, as described by Church and Gilbert (1984).
Total RNA from whole plants or from different organs was extracted with aurin tricarboxylic acid (González et al., 1980). Ten micrograms of RNA per lane was electrophoresed in a 1% (w/v) agarose/formaldehyde gel and transferred to nylon membrane. For quantitative comparison of mRNA levels among different XTR genes in each experiment, the activity of radiolabeled probes of full-length cDNAs was equalized and a different blot was performed for each gene by standardizing the RNA for each lane. Northern hybridization was performed under stringent conditions in Denhardt's solution at 65°C.
RESULTS
Cloning of Rice XET-Related Genes
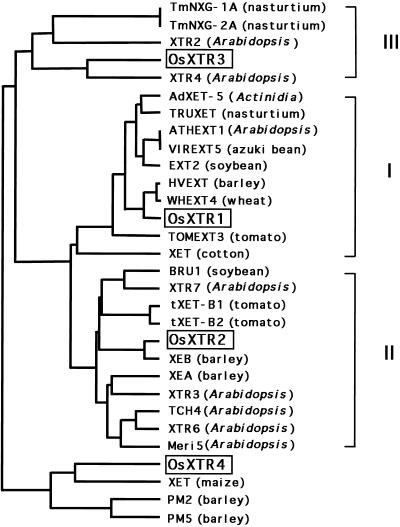
A number of XET-related partial sequences were identified by BLAST searching of the EST database. Using the EST clones kindly provided by the Rice Genome Project as probes, we also isolated other clones from cDNA and genomic libraries. As XET-related proteins are categorized into three groups based on phylogenetic analysis (Nishitani, 1997), we attempted to isolate clones representing each group. Based on partial sequences around their conserved regions, we selected four clones (OsXTR1–4) that fell into the three groups, plus a novel, unique group (see below). We determined the entire sequences of the clones. Figure 1A shows an alignment of the deduced amino acid sequences of OsXTR1, OsXTR2, OsXTR3, and OsXTR4. The inferred proteins share between 33.9% and 58.9% identity in their amino acid sequences (Fig. 1B). The alignment of the rice XTR proteins with the reported XETs from various plants revealed that OsXTR1, OsXTR2, and OsXTR3 could be grouped into subfamilies I, II, and III, respectively. OsXTR4 formed a new XET group, which was quite distant from the other subfamilies.
Figure 1.
Comparison of the products of four rice XET-related genes. A, Alignment of the deduced amino acid sequences of OsXTR1, OsXTR2, OsXTR3, and OsXTR4. Boxes indicate identical residues. Bar a, Catalytic active site shared with B. licheniformis β-glucanase. Bar b, Proposed N-linked glycosylation sites. Shaded boxes show the conserved Cys residues. B, Percent amino acid identity between the XTR gene products.
OsXTR1 and OsXTR2 contained a conserved DEIDFEFLG sequence that was the same as the sequence around the active site of the B. licheniformis β-glucanase (Borriss et al., 1990). Such conservation between the plant and bacterial proteins indicates that this region is critical for the cleavage of β-(1-4) glycosyl linkages (de Silva et al., 1993; Okazawa et al., 1993). OsXTR3 contained a similar sequence with a single conservative amino acid substitution (Ile to Leu). The XET-related proteins in subfamily III commonly have the same substitution at this position. The conserved sequence was also present in OsXTR4, but with two amino acid substitutions. In this protein, the first Asp is replaced by Asn and Ile is replaced by Phe. Despite these differences in the putative active site of OsXTR4, we believe that OsXTR4 is an XTR gene for the following reasons. First, the three putative catalytic residues (the first and second Glu and the second Asp [Planas et al., 1992; Juncosa et al., 1994]) are conserved in OsXTR4 (Fig. 1A, bar a). OsXTR4 has several novel sequences that are commonly observed among XET-related proteins, e.g. a potential site of N-linked glycosylation (N-X-T) on the C-terminal side of the conserved sequence (Fig. 1A, bar b), and three Cys residues in the C-terminal portion, which are considered to form disulfide bridges (Fig. 1A, shaded boxes). Additionally, phylogenic analysis (Fig. 2) indicates that the full-length sequence of the OsXTR4 protein has a high degree of similarity to the product of a maize XET gene (Saab and Sachs, 1995) and barley XET genes (PM2 and PM5, Schünmann et al., 1997).
Figure 2.
Phylogenetic tree of XET-related gene products. The similarity of previously reported XET-related gene products was calculated using the UPGMA program. Full-length protein sequences were used for the comparison. GenBank accession nos. are: TmNXG-1A, X68254; TmNXG-2A, X68255; XTR2, U43487; XTR4, U43486; AdXET-5, L49762; TRUXET1G, L43094; ATHEXT1, D16454; VIREXT5, D16458; EXT2 (soybean), D16455; HVEXT, X91659; WHEXT4, D16457; TOMEXT3, D16456; XET (cotton), D88413; BRU1, L22162; XTR7, U43489; tXET-B1, X82685; tXET-B2, X82684; XEB, X93175; XEA, X93174; XTR3, U43485; TCH4, U27609; XTR6, U43488; meri5, D63508; XET (maize), U15781; PM2, X91660; and PM5, X93173.
Organ-Specific Expression of Rice XTR Genes
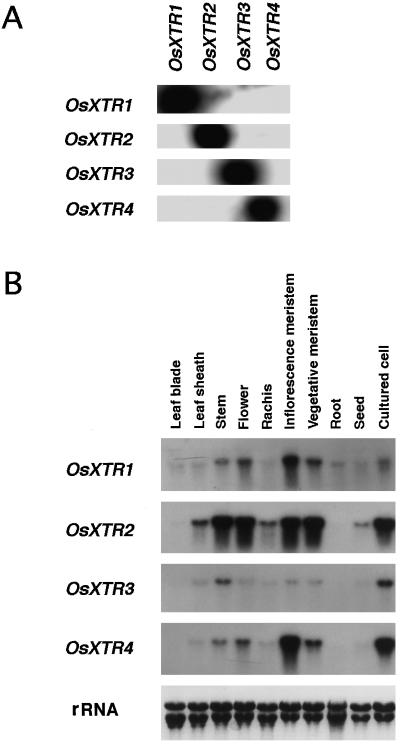
It has been shown that different members of the XET gene family are specifically regulated by various physiological and environmental stimuli (Xu et al., 1996). For this reason, we determined the expression patterns and hormone responses of the four rice XTR genes. For this analysis, we first prepared specific probes for each of the genes. The specificity of each probe was verified by cross-hybridization under high-stringency conditions (Fig. 3A).
Figure 3.
A, DNA gel-blot analysis to verify the specificity of the various XTR gene-specific probes. Entire cDNA clones were used as probes. Hybridization was carried out under stringent conditions in solution containing sodium phosphate at 65°C. B, Organ-specific expression of rice XET-related genes. Each lane contained 10 μg of total RNA isolated from the indicated organ. Hybridization was carried out under stringent conditions with Denhardt's solution at 65°C.
Differences in the patterns of organ-specific RNA accumulation were seen among the four XTR genes (Fig. 3B). The most significant difference was their level of expression, although the total expression pattern throughout an organ was similar (except for OsXTR3). In meristematic tissues such as vegetative and inflorescence meristems and in cultured cells, all of the XTR genes were expressed, but at different levels. The four genes were also expressed in elongating stems. Very low or no XET-related RNA accumulation was seen in leaf blades or roots. A low level of OsXTR2 RNA expression was seen in germinating seeds. Expression of OsXTR2 at a much higher level than the other XTR genes was also seen in other organs that we tested. The organ specificity of OsXTR1 RNA expression was similar to that of OsXTR4. OsXTR3 showed a restricted expression pattern, with expression seen mainly in elongating stems and cultured cells.
Hormonal Regulation of XTR Gene Expression
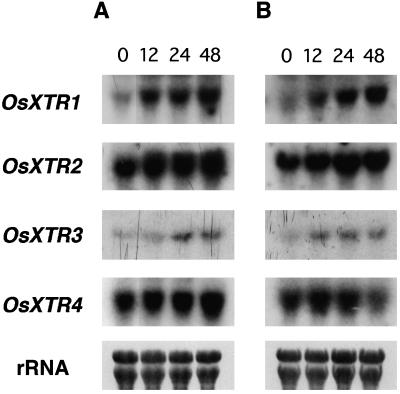
It has been demonstrated that XET genes are regulated by particular hormonal stimuli. For example, BRU1, a soybean XET gene, was originally identified as a gene regulated by brassinosteroids (Zurek and Clouse, 1994), and TCH4, an Arabidopsis XET gene, is also up-regulated by auxin and brassinosteroids (Xu et al., 1995). To determine the hormonal responses of the rice XTR genes, we treated rice plants with a range of plant hormones, including indole-3-acetic acid, 2,4-dichlorophenoxyacetic acid, GA, kinetin, brassinosteroids, and abscisic acid, at various concentrations (0.1, 1, and 10 μm for each hormone). Changes in XTR RNA expression were only seen in plants treated with GA or brassinosteroids; no changes were observed in plants treated with any concentration of indole-3-acetic acid, 2,4-dichlorophenoxyacetic acid, kinetin, or abscisic acid for 12, 24, or 48 h (data not shown). Upon treatment with GA, the levels of expression of OsXTR1 and OsXTR3 increased with increasing duration of treatment (Fig. 4A), whereas expression of OsXTR2 and OsXTR4 showed only a slight increase. The expression levels of OsXTR1 and OsXTR3 were also increased by treatment with brassinosteroids, whereas OsXTR2 showed little or no change and OsXTR4 showed a slight decrease after brassinosteroid treatment for 48 h (Fig. 4B).
Figure 4.
Induction of expression of XET-related genes by GA (A) and brassinosteroids (B). Lanes contain 10 μg of total RNA from whole rice plants grown for 3 weeks. Hormone treatments were performed by immersing the plants in 30 μm GA3 (A) or 1 μm brassinolide (B) for 12, 24, or 48 h.
Expression Patterns of XTR Genes in Elongating Rice Stems
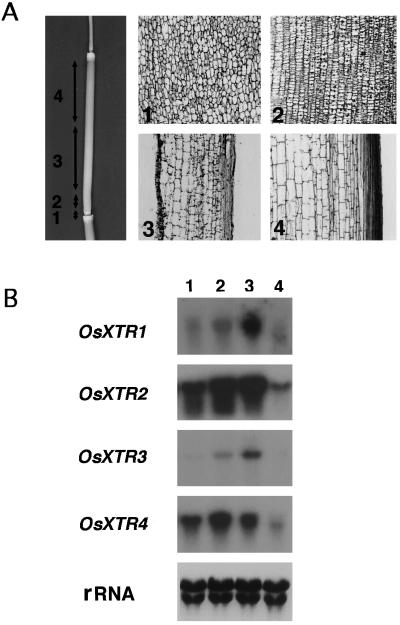
The rice culm is composed of nodes and internodes, which are the consequence of cell proliferation and elongation. Nodes represent the segmental region between two internodes, and consist of a population of small, isodiametric cells undergoing continuous cell division in random directions (Fig. 5A, 1). The basal part of the internode immediately above the node is termed the divisional zone and is defined as the intercalary meristem in which cells undergo continuous longitudinal divisions. As a result of active longitudinal division, specifically in the upper direction without cell elongation, small, flat cells line up (Fig. 5A, 2). These cells begin to elongate preferentially toward the upper direction. Therefore, the upper region of the divisional zone (Fig. 5A, 3) consists of cells with the greatest elongation activity. The uppermost region of the internode consists of longitudinally oriented cells in which elongation is complete (Fig. 5A, 4).
Figure 5.
A, Photographs of external shape of elongating stem and sections from each developmental region: 1, node; 2, divisional zone at the basal region of internode; 3, elongating zone of internode; and 4, elongated zone of internode. B, Differential expression of XET-related genes in elongating stems. Each lane contains 10 μg of total RNA from the regions designated in A.
To characterize the differential expression of the XTR genes in developing internodes, we divided the culm into four sections: node, divisional zone, elongating zone, and elongated zone of the internode (Fig. 5A), based on the developmental stages described above. XTR RNA expression was examined in each of the four sections (Fig. 5B). Expression of OsXTR1 and OsXTR3 was quite low in nodes, increased with cell development in internodes, and highest in the elongation zone. Little or no expression of these two genes was observed in the elongated zone of the internode (Fig. 5B). This preferential expression of OsXTR1 and OsXTR3 in the internode suggests that these XTR genes play an important role in cell elongation in the internode. In contrast to OsXTR1 and OsXTR3, OsXTR2 and OsXTR4 were expressed not only in the division and elongation zones, but also in the node, where cells divide.
Expression of XTR Genes in Rice Mutants with Abnormal Heights
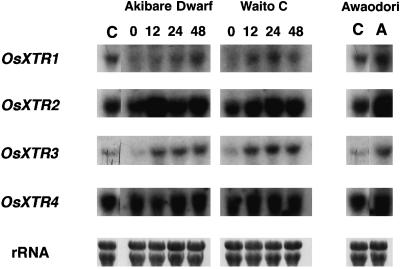
XTR RNA expression was investigated in rice mutants with abnormal heights. Akibare Dwarf and Waito C are allelic dwarf mutants that are responsive to GA. The heights of the mature mutant plants are approximately 30% and 50%, respectively, of that of wild-type plants. RNA gel-blot analysis demonstrated that the levels of OsXTR1 and OsXTR3 mRNA in the mutants were lower than in wild-type plants, whereas the levels of OsXTR2 and OsXTR4 mRNA were approximately equal to those in wild-type plants (Fig. 6). The expression of OsXTR1 and OsXTR3 in the mutants was induced to exceed wild-type levels by treatment with GA for 12 to 48 h. Awaodori is a GA-insensitive overgrowth mutant whose stem constantly elongates to 2 to 3 times the length of the wild type. As shown in Figure 6, the level of expression of all four of the XTR genes was higher than that of wild-type plants. The expression of the XTR genes did not change after treatment with GA in this mutant (data not shown).
Figure 6.
Expression of XET-related genes in genetic mutants exhibiting abnormal heights. Total RNA from two dwarf mutants, Akibare Dwarf and Waito C, and the overgrowth mutant Awaodori (lane A, right) was isolated. Ten micrograms of RNA was electrophoresed in each lane. Lanes C, RNA expression of XTR genes in wild-type plants. XTR RNA expression was restored in the two dwarf mutants by treatment with 30 μm GA3 for 12, 24, or 48 h (indicated by numbers at top).
DISCUSSION
The four rice XET-related genes isolated in this study were classified into four different subfamilies in the previously reported phylogenetic tree of XET and XET-related genes. Three subfamilies (Fig. 2, I, II, and III) are found in a wide range of flowering plants and the evolution of monocotyledonous plants therefore probably occurred after the divergence of these three subfamilies. Thus, the diversification of the structure of the XET genes of each subfamily may reflect a unique functional assignment of the different subfamilies that is essential throughout flowering plants.
OsXTR4 is an XET-related gene that falls outside the three established subfamilies, suggesting that it may have some peculiar functional features. Another XET-related gene that does not fit within the three established subfamilies has been reported in barley (Schünmann et al., 1997) and shows a unique expression pattern during leaf development. Whether these similar genes outside of the three subfamilies encode functional XET enzymes should be tested in the future: it is very possible that they encode XET or XET-related enzymes, since their products retain several characteristic features commonly observed among all XET enzymes.
The rice XTR genes exhibited different expression patterns in terms of organ specificity, response to GA and brassinosteroids, stage specificity during internodal elongation, and expression in several mutant backgrounds with abnormal heights. These results indicate that OsXTR1 and OsXTR3 may both play a role in internodal elongation in rice. The evidence for this is that the two genes are preferentially expressed in the elongation-active region of the internode (Fig. 5B).
Based on their organ specificity of expression, the functional differentiation of the four XTR genes can be summarized as follows: OsXTR1 and OsXTR4 exhibit similar organ specificity, but their expression in culms differ. OsXTR1 is expressed in both the elongating and divisional zones of internodes, with a higher level of expression in the elongating zone (Fig. 5B). In contrast, OsXTR4 is expressed in the divisional zone of internodes and in nodes. This expression pattern suggests that OsXTR4 acts mainly in the rearrangement of the cell wall in division-active cells, whereas OsXTR1 acts mainly on elongating cells in internodes. OsXTR2 is expressed at a much higher level than the other rice XTR genes, and is expressed in most organs, with the exception of well-developed leaf blades and roots. In the elongating internode, OsXTR2 is expressed in all four developmental zones. From these results, OsXTR2 is constitutively active in most organs, suggesting that it may have a role for reconstruction of the cell wall in cell elongation and division throughout rice development. In the case of OsXTR3, the level of RNA expression is lower than in other genes. Furthermore, OsXTR3 RNA expression shows a strict organ specificity in elongating stems, being notably higher in the internode elongation zone than in the divisional zone.
RNA expression of OsXTR1 and OsXTR3 is increased by treatment with either GA or brassinosteroids, suggesting that these two XTR genes act in altering the structure of the cell wall in response to these two hormones. It has been reported that GA has a large effect on internodal elongation, especially activating cell division and cell elongation (Kamijima, 1981). Additionally, XET activity and mRNA level of XET-related genes are regulated by GA to induce leaf elongation (Smith et al., 1996; Schünmann et al., 1997). In agreement with this, we have shown here that OsXTR1 and OsXTR3 are activated by GA and are involved in internodal elongation. Furthermore, brassinosteroids have recently been shown to regulate stem elongation in various plants, and several XET-related genes such as TCH4 (Xu et al., 1995) and BRU1 (Zurek and Clouse, 1994) have been reported to be regulated by brassinosteroids. It has been suggested that brassinosteroids are essential for plant development and play an important role in the control of cell elongation (Kauschmann et al., 1996; Azpiroz et al., 1998). Brassinosteroid-insensitive rice mutants have been identified, and exhibit a dwarf phenotype with repressed internodal elongation (C. Yamamuro and M. Matsuoka, personal communication). Brassinosteroids may regulate the expression of brassinosteroid-dependent XTR genes such as OsXTR1 and OsXTR3, at a specific developmental stage of internodal elongation.
It has been shown that d18, the mutant allele of Akibare Dwarf and Waito C, shows a high level of endogenous GA20 and a low level of GA1, and therefore the product of d18 may catalyze 3β-hydroxylation of GA20 to GA1 (Kobayashi et al., 1989). The expression of the XTR genes in these two dwarf mutants indicates that inhibition of GA biosynthesis is accompanied by preferential suppression of OsXTR1 and OsXTR3 expression. This result suggests the specific response of OsXTR1 and OsXTR3 to GA and the correlation of these genes with internodal elongation. These results suggest that OsXTR1 and OsXTR3 may be indirectly regulated by GA.
The overgrowth mutant Awaodori exhibited up-regulation of OsXTR1 and OsXTR3 compared with the wild type. The phenotype of this mutant is believed to be caused by a defect in a suppressive gene in the GA signal transduction pathway, which causes a continuous GA response without GA (Peng et al., 1997). This result also supports the possible regulation of OsXTR1 and OsXTR3 by GA.
In the d18 dwarf mutants, there was a synergistic relationship between the effects of GA and brassinosteroids on the expression of XTR genes. Although the expression of OsXTR1 and OsXTR3 increased after treatment with either exogenous GA or brassinosteroids in wild-type plants, the expression of OsXTR1 and OsXTR3 in the mutants did not increase after the treatment with brassinosteroids (data not shown). This suggests that the presence of GA is essential for the expression of these genes and that their induction by brassinosteroids occurs only if GA is also present. On the other hand, a GA-insensitive mutant, lkb of pea, has been shown to be deficient in brassinosteroid biosynthesis (Nomura et al., 1997). This suggests that brassinosteroids are essential for GA sensitivity and also indicates that cross-talk may take place between the brassinosteroid and GA signal transduction pathways. Further analysis of the precise expression patterns of the XTR genes using GA, and brassinosteroid biosynthesis, and response mutants may permit the elucidation of the synergistic effects of GA and brassinosteroids.
LITERATURE CITED
- Azpiroz R, Wu Y, LoCascio JC, Feldmann KA. An Arabidopsis brassinosteroid-dependent mutant is blocked in cell elongation. Plant Cell. 1998;10:219–230. doi: 10.1105/tpc.10.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borriss R, Buettner K, Maentsaelae P. Structure of the β-1,3-1,4-glucanase gene of Bacillus macerans: homologies to other β-glucanases. Mol Gen Genet. 1990;222:278–283. doi: 10.1007/BF00633829. [DOI] [PubMed] [Google Scholar]
- Church G, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. Assembly and enlargement of the primary cell wall in plants. Annu Rev Cell Dev Biol. 1997;13:171–201. doi: 10.1146/annurev.cellbio.13.1.171. [DOI] [PubMed] [Google Scholar]
- de Silva J, Jarman CD, Arrowsmith S, Stronach MS, Chengappa S, Sidebottom C, Reid JSG. Molecular characterization of a xyloglucan-specific endo-(1-4)-β-d-glucanase (xyloglucan endotransglycosylase) from nasturtium seeds. Plant J. 1993;3:701–711. [PubMed] [Google Scholar]
- Fry SC, Smith RC, Renwick KF, Martin DJ, Hodge SK, Matthews KJ. Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochem J. 1992;282:821–828. doi: 10.1042/bj2820821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González RG, Haxo RS, Schleich T. Mechanism of action of polymeric aurintricarboxylic acid, a potent inhibitor of protein-nucleic acid interactions. Biochemistry. 1980;19:4299–4303. doi: 10.1021/bi00559a023. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Ogawa K, Mitsuishi Y. Characterization of the adsorption of xyloglucan to cellulose. Plant Cell Physiol. 1994;35:1199–1205. [PubMed] [Google Scholar]
- Juncosa M, Pons J, Dot T, Querol E, Planas A. Identification of active site carboxylic residues on Bacillus licheniformis 1,3-1,4-β-d-glucan 4-glucanylhydrolase by site-directed mutagenesis. J Biol Chem. 1994;269:14530–14535. [PubMed] [Google Scholar]
- Kamijima O. Consideration on the mechanism of expression of dwarf genes in rice plants. II. The actions of dwarf genes on cell division and cell elongation in parenchyma of internode. Jpn J Breed. 1981;31:302–315. [Google Scholar]
- Kamijima O, Tanisaka T, Kinoshita T. Gene symbols for dwarfness. Rice Genet Newslett. 1996;13:19–25. [Google Scholar]
- Kauschmann A, Jessop A, Koncz C, Szekeres M, Willmitzer L, Altmann T. Genetic evidence for an essential role of brassinosteroids in plant development. Plant J. 1996;9:701–713. [Google Scholar]
- Kawahara H, Chonan N, Wada K. Studies on morphogenesis in rice plants. 3. Interrelation of the growth among laves, panicle and internodes, and a histological observation on the meristem of culm. Proc Crop Sci Soc Jpn. 1968;37:372–383. [Google Scholar]
- Kobayashi M, Sakurai A, Saka H, Takahashi N. Quantitative analysis of endogenous gibberellins in normal and dwarf cultivars of rice. Plant Cell Physiol. 1989;30:963–969. [Google Scholar]
- McCann MC, Wells B, Roberts K. Direct visualization of cross-links in the primary plant cell wall. J Cell Sci. 1990;96:323–334. [Google Scholar]
- McQueen-Mason S. Plant cell walls and the control of growth. Biochem Soc Trans. 1996;25:204–214. doi: 10.1042/bst0250204. [DOI] [PubMed] [Google Scholar]
- Mitsunaga S, Tashiro J, Yamaguchi J. Identification and characterization of gibberellin-insensitive mutants selected from among dwarf mutants of rice. Theor Appl Genet. 1993;87:705–712. doi: 10.1007/BF00222896. [DOI] [PubMed] [Google Scholar]
- Murakami Y. Dwarfing genes in rice and their relation to gibberellin biosynthesis. In: Carr DJ, editor. Plant Growth Substances, 1970. Berlin: Springer-Verlag; 1972. pp. 166–174. [Google Scholar]
- Nishitani K. The role of endoxyloglucan transferase in the organization of plant cell wall. Int Rev Cytol. 1997;173:3–52. doi: 10.1016/s0074-7696(08)62477-8. [DOI] [PubMed] [Google Scholar]
- Nishitani K, Tominaga R. In vitro molecular weight increase in xyloglucans by an apoplastic enzyme preparation from epicotyl of Vigna angularis. Physiol Plant. 1991;82:490–497. [Google Scholar]
- Nishitani K, Tominaga R. Endo-xyloglucan transferase, a novel class of glycosyltransferase that catalyzes transfer of a segment of xyloglucan molecule to another xyloglucan molecule. J Biol Chem. 1992;267:21058–21064. [PubMed] [Google Scholar]
- Nomura T, Nakayama M, Reid JB, Takeuchi Y, Yokota T. Blockage of brassinosteroid biosynthesis and sensitivity causes dwarfism in Pisum sativum. Plant Physiol. 1997;133:31–37. doi: 10.1104/pp.113.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazawa K, Sato Y, Nakagawa T, Asada K, Kato I, Tomita E, Nishitani K. Molecular cloning and cDNA sequencing of endoxyloglucan transferase, a novel class of glycosyltransferase that mediates molecular grafting between matrix polysaccharides in plant cell walls. J Biol Chem. 1993;268:25364–25368. [PubMed] [Google Scholar]
- Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP. The Arabidopsis GAIgene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 1997;11:3194–3205. doi: 10.1101/gad.11.23.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas A, Juncosa M, Lloberas J, Querol E. Essential catalytic role of Glu134 in endo-β-1,3-4-d-glucan 4-glucanohydrolase from B. licheniformisas determined by site-directed mutagenesis. FEBS Lett. 1992;308:141–145. doi: 10.1016/0014-5793(92)81262-k. [DOI] [PubMed] [Google Scholar]
- Saab IN, Sachs MM. Complete cDNA and genomic sequence encoding a flooding-responsive gene from maize (Zea maysL.) homologous to xyloglucan endotransglycosylase. Plant Physiol. 1995;108:439–440. doi: 10.1104/pp.108.1.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schünmann PHD, Smith RC, Lång V, Matthews PR, Chandler PM. Expression of XET-related genes and its relation to elongation in leaves of barley (Hordeum vulgareL.) Plant Cell Environ. 1997;20:1439–1450. [Google Scholar]
- Sentoku N, Sato Y, Kitano H, Ito Y, Kurata N, Matsuoka M. Regional expression of the rice KN1-type homeobox gene family during embryo, shoot, and flower development. Plant Cell. 1999;11:1651–1663. doi: 10.1105/tpc.11.9.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RC, Fry SC. Endotransglycosylation of xyloglucans in plant cell suspension cultures. Biochem J. 1991;279:529–535. doi: 10.1042/bj2790529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RC, Matthews PR, Schunmann PHD, Chandler PM. The regulation of leaf elongation and xyloglucan endotransglycosylase by gibberellin in ‘Himalaya’ barley (Hordeum vulgareL.) J Exp Bot. 1996;47:1395–1404. [Google Scholar]
- Xu W, Campbell P, Vargheese AV, Braam J. The Arabidopsis XET-related gene family: environmental and hormonal regulation of expression. Plant J. 1996;9:879–889. doi: 10.1046/j.1365-313x.1996.9060879.x. [DOI] [PubMed] [Google Scholar]
- Xu W, Purugganan MM, Poliensky DH, Antosiewicz DM, Fry SC, Braam J. Arabidopsis TCH4, regulated by hormones and the environment, encodes a xyloglucan endotransglycosylase. Plant Cell. 1995;7:1555–1567. doi: 10.1105/tpc.7.10.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurek DM, Clouse SD. Molecular cloning and characterization of a brassinosteroid-regulated gene from elongating soybean (Glycine maxL.) epicotyls. Plant Physiol. 1994;104:161–170. doi: 10.1104/pp.104.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]