ABSTRACT
The phototropic bacterium Synechocystis sp. strain PCC 6803 is able to adapt its morphology in order to survive in a wide range of harsh environments. Under conditions of high salinity, planktonic cells formed cell aggregates in culture. Further observations using crystal violet staining, confocal laser scanning microscopy, and field emission-scanning electron microscopy confirmed that these aggregates were Synechocystis biofilms. Polyamines have been implicated in playing a role in biofilm formation, and during salt stress the content of spermidine, the major polyamine in Synechocystis, was reduced. Two putative arginine decarboxylases, Adc1 and Adc2, in Synechocystis were heterologously expressed in Escherichia coli and purified. Adc2 had high arginine decarboxylase activity, whereas Adc1 was much less active. Disruption of the adc genes in Synechocystis resulted in decreased spermidine content and formation of biofilms even under nonstress conditions. Based on the characterization of the adc mutants, Adc2 was the major arginine decarboxylase whose activity led to inhibition of biofilm formation, and Adc1 contributed only minimally to the process of polyamine synthesis. Taken together, in Synechocystis the shift from planktonic lifestyle to biofilm formation was correlated with a decrease in intracellular polyamine content, which is the inverse relationship of what was previously reported in heterotroph bacteria.
IMPORTANCE There are many reports concerning biofilm formation in heterotrophic bacteria. In contrast, studies on biofilm formation in cyanobacteria are scarce. Here, we report on the induction of biofilm formation by salt stress in the model phototrophic bacterium Synechocystis sp. strain PCC 6803. Two arginine decarboxylases (Adc1 and Adc2) possess function in the polyamine synthesis pathway. Inactivation of the adc1 and adc2 genes leads to biofilm formation even in the absence of salt. The shift from planktonic culture to biofilm formation is regulated by a decrease in spermidine content in Synechocystis. This negative correlation between biofilm formation and polyamine content, which is the opposite of the relationship reported in other bacteria, is important not only in autotrophic but also in heterotrophic bacteria.
KEYWORDS: arginine decarboxylase, biofilm, polyamine, cyanobacteria, stress response
INTRODUCTION
Microorganisms have developed adaptation mechanisms to survive environmental stress conditions, such as changes in temperature, pH, or extracellular osmotic pressure. Most bacteria can exist in either a planktonic single-cell state or in an immobilized adhesive multicellular state known as a biofilm. Biofilm formation is tightly linked to stress responses (1). Within the biofilm, cells are embedded in a matrix. This extracellular matrix is composed of proteins, exopolysaccharides, and DNA (2) and acts as a shield, protecting bacteria within the biofilm (3). Synechocystis sp. strain PCC 6803 is a unicellular photosynthetic prokaryote that can survive a wide range of environmental conditions and therefore can be used as a model for the study of molecular response mechanisms to various stresses (4). Several membrane transport systems in Synechocystis function in the maintenance of intracellular ion homeostasis (5, 6). Prolonged exposure to a high-salinity environment induces morphological changes as well as the production of extracellular components. Exopolysaccharide production is considered an adaptive response that enhances the tolerance of Synechocystis to the toxic effects of salt and heavy metals (7, 8). The amount and composition of these exopolysaccharides are strongly influenced by the concentration of NaCl in the growth medium (7).
Polyamines (putrescine, spermidine, and spermine) are involved in the modulation of cell growth and differentiation according to the proposed model of the polyamine modulon (9, 10). Genes that are part of the polyamine modulon are characterized by enhanced expression at the level of translation in the presence of polyamines. Polyamines are organic polycationic compounds that are synthesized in cells and functionally different from essential cations like K+ and Mg2+. In Escherichia coli, polyamines are found predominantly as polyamine-RNA complexes where the binding of the polyamines leads to structural changes of the RNA. These structural changes result in enhanced expression of a set of genes encoding proteins related to cell proliferation and viability (9, 11). Most eukaryotes synthesize putrescine from ornithine by an ornithine decarboxylase; however, bacteria and plants possess another pathway for putrescine biosynthesis from arginine through the action of arginine decarboxylase (Adc) (12). The product of that reaction, agmatine, is subsequently converted to putrescine, spermidine, and spermine via a series of steps in the polyamine synthetic pathway. Synechocystis contains two putative adc genes and the presence of the corresponding mRNAs has been reported, but it does not seem to possess a gene encoding ornithine decarboxylase (13–15). In several other bacteria, biofilm formation accompanies changes in polyamine concentrations (16). In general, reduction of the amount of polyamines through inactivation of genes encoding relevant biosynthetic enzymes decreases biofilm formation in Bacillus subtilis (12) and E. coli (17).
In this study, we focused on the effects of salt stress on biofilm formation in Synechocystis using crystal violet staining, confocal laser scanning microscopy (CLSM), and field emission-scanning electron microscopy (FE-SEM). We analyzed the polyamine content of Synechocystis cells and the enzymatic properties of two putative arginine decarboxylases (Adc1 and Adc2) predicted to be responsible for the initial step of polyamine biosynthesis. Mutants defective in either or both of the adc genes were generated, and their capacity to form biofilms was analyzed to explore the relationship between salt stress, polyamine content, and biofilm formation in Synechocystis.
RESULTS
Salt stress-induced biofilm formation in Synechocystis.
During cultivation of Synechocystis, an aggregation of cells at the bottom of the flask that increased under high-salinity conditions was observed. After crystal violet staining to visualize adherent cells, strong purple staining was observed in the flask of a culture supplemented with 500 mM NaCl but not in the flask of a culture grown without additional NaCl (Fig. 1A). This strongly suggested that biofilm formation was affected by the salinity condition. The amount of biofilm produced by Synechocystis was quantified with a microtiter plate assay based on crystal violet staining (see Materials and Methods). The biofilm cell density increased with increasing NaCl concentration, 1.74-fold (250 mM NaCl) and 2.65-fold (500 mM NaCl), compared to cell density in medium without added NaCl (Fig. 1B). In contrast, KCl or sorbitol had relatively small effects on biofilm production. To further explore the NaCl-induced biofilm production, the morphology of Synechocystis cells was observed under high-salinity conditions using both confocal laser scanning microscopy (CLSM) and field emission-scanning electron microscopy (FE-SEM). Detection of autofluorescence from Synechocystis cells by CLSM (18, 19) confirmed that high-salt concentrations induced biofilm formation on a glass surface (Fig. 1C). This was consistent with the results of the crystal violet assays (Fig. 1A and B). FE-SEM imaging made it possible to visualize the three-dimensional architecture of the Synechocystis biofilm in detail (Fig. 1D). Under control conditions, single spherical cells appeared on the polycarbonate membrane. In NaCl-supplemented medium, an increased number of adhered cells remained after washing of the membrane.
FIG 1.
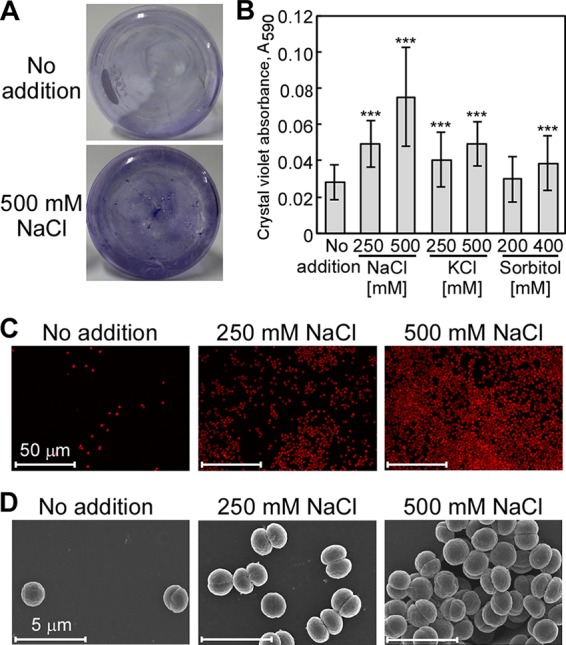
Salt stress-induced attachment of Synechocystis on glass. (A) Synechocystis wild-type culture. Synechocystis cells were grown in BG11 medium without (upper) or with (bottom) 500 mM NaCl. (B) Effect of NaCl, KCl, or sorbitol on biofilm formation in cells grown in 96-well polystyrene plates. The values shown are the absorbances (OD590) of crystal violet extracted from stained biofilms remaining in the wells after removal of the culture. Each column represents the mean ± standard deviation (SD) of data obtained from 77 to 80 individual cultures. Significant differences between no addition and each condition were analyzed by Student's t test (***, P < 0.001). (C) Autofluorescence of Synechocystis cells imaged by CLSM was used as a measure of biofilm formation. Higher autofluorescence was seen in cells grown in medium with added NaCl (250 mM and 500 mM). CLSM images shown are representative images of experiments performed in triplicate. (D) FE-SEM images of cells within the biofilms formed under the same culture conditions as those described for panel C.
Salt stress decreased polyamine content in Synechocystis.
Polyamine accumulation is known to be closely correlated with salt tolerance and cell growth in Synechocystis (13, 15, 20). The polyamine content in Synechocystis was analyzed under normal or salt stress conditions. In the controls, spermidine had the highest concentration (19.3 ± 8.1 nmol mg−1 protein−1) among the investigated polyamines, while agmatine (0.033 ± 0.040 nmol mg−1 protein−1), putrescine (0.093 ± 0.141 nmol mg−1 protein−1), and spermine (0.011 ± 0.008 nmol mg−1 protein−1) accumulated to much lower concentrations (Fig. 2). The fact that spermidine was the most abundant polyamine in nonstressed Synechocystis was consistent with the results by Jantaro et al. (20). Compared to cultures without added NaCl, the spermidine content decreased by 33.9% in 250 mM NaCl-supplemented medium and by 54.0% in 500 mM NaCl-supplemented medium. A decrease in concentration was also found for putrescine (by 59.1%) and spermine (by 43.7%) in cultures with 500 mM NaCl. In contrast, the agmatine concentration increased during salt stress. These results showed that spermidine was the major polyamine present in Synechocystis and that salt stress led to an overall decline in the content of polyamines.
FIG 2.
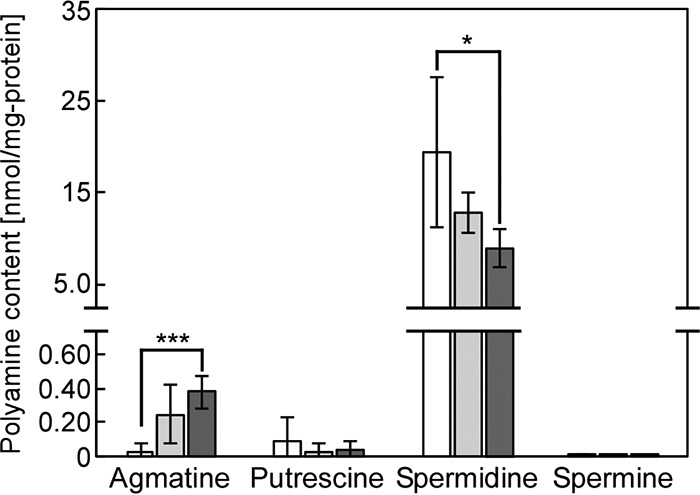
Changes in polyamine content in Synechocystis during salt stress. Agmatine, putrescine, spermidine, and spermine contents were determined after addition of NaCl to the medium (white bars, no NaCl added; light gray bars, 250 mM NaCl; dark gray bars, 500 mM NaCl). Cells were grown for 5 days in the medium indicated. Each value corresponds to the average ± SD (n = 3 or 4). Significant differences between no addition and each added NaCl concentration were analyzed by Student's t test (*, P < 0.05; ***, P < 0.001).
Adc1 and Adc2 catalyzed arginine decarboxylation in Synechocystis.
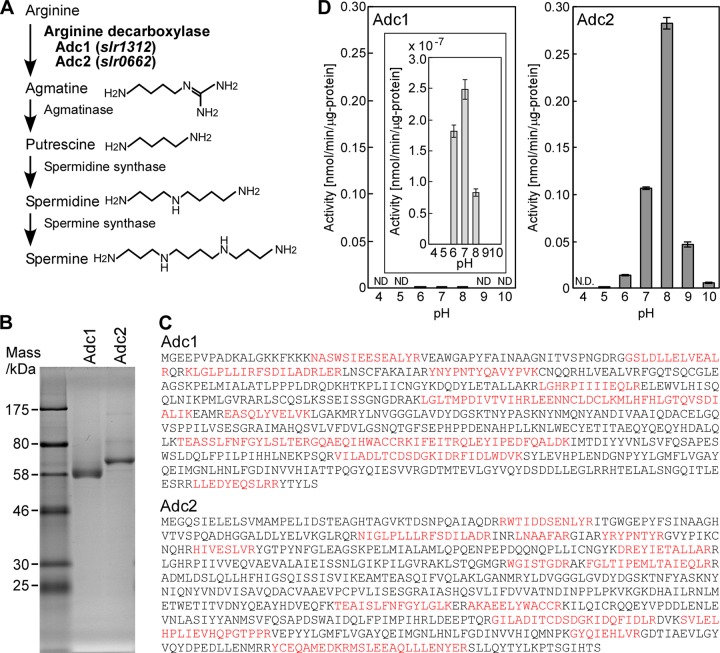
To evaluate the relation of polyamines to biofilm formation in Synechocystis, we characterized the activity of polyamine synthetic enzymes (Fig. 3A). According to the annotation of the Synechocystis genome, Synechocystis contains two genes encoding putative arginine decarboxylases, adc1 and adc2. These enzymes catalyze the conversion of arginine to agmatine, which is the initial step in the polyamine synthesis pathway (Fig. 3A) (14, 15, 20, 21). Histidine-tagged Adc1 and Adc2 were heterologously expressed in E. coli and purified to homogeneity (Fig. 3B). Their apparent mass was lower than expected from their deduced amino acid sequences (expected masses were 74.5 kDa for Adc1 and 78.2 kDa for Adc2). The results suggested the occurrence of N-terminal or C-terminal degradation of Adc1 and Adc2 during purification. Since the histidine tag required for the purification was fused to the N-terminal end of both proteins, any change in length of the purified proteins would have to result from processing of the C-terminal end. To elucidate the observed deviation in protein mass, the purified proteins were digested with trypsin and the resultant peptides analyzed by liquid chromatography mass spectrometry. The amino acid sequence of several peptides was determined as described in Materials and Methods (Fig. 3C). Interestingly, the analyzed purified peptides covered almost the entire protein sequence of Adc1 and Adc2. This led us to conclude that the actual molecular mass of purified Adc1 and Adc2 is larger than the apparent molecular mass shown in Fig. 3B. We subsequently tested the enzymatic activity of purified Adc1 and Adc2 (Fig. 3D). Purified Adc2 protein showed high arginine decarboxylase activity compared to that of purified Adc1. The pH profiles of both Adcs showed typical bell-shaped curves, with an apparent pH optimum of pH 7.0 for Adc1 and pH 8.0 for Adc2 (Fig. 3D). The results suggested that Adc2 purified from E. coli was responsible for the majority of the arginine decarboxylase activity, while Adc1 showed only negligible arginine decarboxylase activity.
FIG 3.
Enzymatic activities of Adc1 and Adc2. (A) Putative pathway of polyamine synthesis from arginine in Synechocystis. The enzymes for the biosynthesis of polyamines from arginine in Synechocystis are indicated (13, 14, 23) according to the information available from CyanoBase (http://genome.microbedb.jp/cyanobase). (B) Histidine-tagged Adc1 and Adc2 were expressed in E. coli, affinity purified, separated by SDS-PAGE on a 10% polyacrylamide gel, and stained with Coomassie brilliant blue R-250. (C) The amino acid sequences of tryptic peptides of Adc1 (top) and Adc2 (bottom) isolated from E. coli are highlighted in red. (D) Measurement of arginine decarboxylase activity of Adc1 and Adc2 at various pH values. Each data point corresponds to the average ± SD (n = 3). ND stands for not detected.
Effect of NaCl addition on expression and enzymatic activity of Adc1 and Adc2.
When Synechocystis was subjected to NaCl stress, the putrescine content decreased but the agmatine concentration increased (Fig. 2). This indicates a rise in the production of arginine decarboxylase and/or downregulation of the amount of agmatinase, which catalyzes the conversion of agmatine to putrescine (Fig. 3A). To determine whether adc1 and adc2 were induced or repressed, we performed a promoter-lacZ assay in Synechocystis. Based on the Synechocystis genomic sequence, the putative promoter sequences of adc1 (180 bp) and adc2 (414 bp) each were put in front of lacZ in an expression plasmid. The promoter-lacZ constructs were then introduced into Synechocystis. High-salt application (500 mM NaCl) did not significantly change the β-galactosidase activity in Synechocystis (Fig. 4A). Since the promoter regions have not been definitively determined, we cannot exclude the possibility that the promoter sequences selected for the constructs were not completely accurate. To corroborate the data, we also performed semiquantitative RT-PCR (Fig. 4B). When 500 mM NaCl was added to the medium, the expression of adc1 and adc2 was not significantly changed compared to that of the nonstressed cells, which is consistent with the promoter-lacZ data shown in Fig. 4A. Since adc1 and adc2 were constitutively expressed, we investigated whether the arginine decarboxylase activity of Adc1 and Adc2 was directly affected by addition of NaCl to the assay. For those assays, Adc1 and Adc2 heterologously expressed in E. coli were purified using the same buffer as that for Fig. 3 but without NaCl. To determine whether NaCl had a direct effect on the activities of Adc1 and Adc2, their relative enzyme activities were measured in the presence of 250 mM or 500 mM NaCl and compared to their activities in reaction buffer without NaCl (Fig. 4C). Adc2 exhibited 14-fold and 13-fold increases in activity in buffer containing 250 mM NaCl and 500 mM NaCl, respectively. Although the absolute activity of Adc1 was extremely low, addition of NaCl to the buffer also increased the activity of Adc1 by 2.9-fold with 250 mM NaCl and 2.2-fold with 500 mM NaCl. The NaCl stimulation of the activities of Adc1 and Adc2 might account for the increase in agmatine concentration in Synechocystis under high-salt conditions (Fig. 2), whereas the production of agmatinase might be inhibited.
FIG 4.
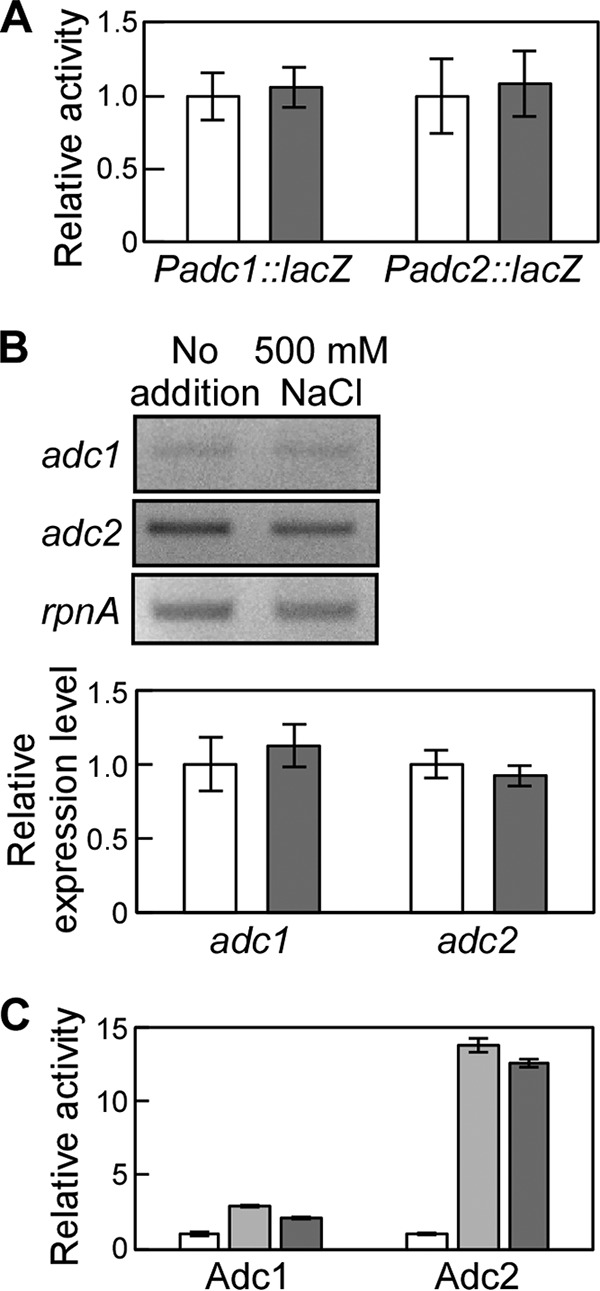
Expression and enzymatic activity of Adc1 and Adc2 during salt stress. (A) adc1 and adc2 promoter activities were measured by determining the β-galactosidase activity of the corresponding reporter strain during salt stress (white bars, no NaCl added; gray bars, 500 mM NaCl). Each value corresponds to means ± SD (n = 3). No significant differences (P > 0.05) were found by Student's t test. (B) Semiquantitative RT-PCR to determine adc1 and adc2 expression in Synechocystis during salt stress. (Top) The RT-PCR products of adc1, adc2, and rpnA amplified from total RNA were separated by agarose gel electrophoresis. (Bottom) The band intensity of adc1 and adc2 was determined and related to the band intensity of rpnA to calculate the relative expression level. Each value corresponds to means ± SD (n = 3). No significant differences (P > 0.05) were found by Student's t test. White bars, no NaCl added; gray bars, 500 mM NaCl. (C) Relative arginine decarboxylase activity of Adc1 and Adc2 in the presence of 250 mM (light gray bars) or 500 mM (dark gray bars) NaCl with respect to the activity measured in the buffer without NaCl (white bars). Each data point corresponds to the average ± SD (n = 3).
Inactivation of adc reduced spermidine content in Synechocystis.
To evaluate the enzymatic activity and physiological function of Adc1 and Adc2 with respect to cell aggregation in Synechocystis, adc1 and adc2 deletion mutants were generated in Synechocystis by insertion of antibiotic resistance gene cassettes into the genome sequences (Fig. 5A). We obtained null mutants of adc1 (adc1 knockout strain ko1) and a double null mutant of adc1 and adc2 (ko1ko2) (Fig. 5B, C, and E). Complete deletion of the adc2 gene in the wild type was not achieved (adc2 knockdown strain kd2) (Fig. 5D), although the amount of PCR product amplified from the original adc2 gene in kd2 was decreased compared with that of the wild type (data not shown). Note that the genome of Synechocystis consists of approximately 12 copies of its chromosome (22). Although we could not obtain an adc2 null mutant, simultaneous disruption of adc1 and adc2 allowed growth of Synechocystis. We also generated two strains in the ko1ko2 background overexpressing either adc1 (ko1ko2/1ox) or adc2 (ko1ko2/2ox) (Fig. 5F).
FIG 5.
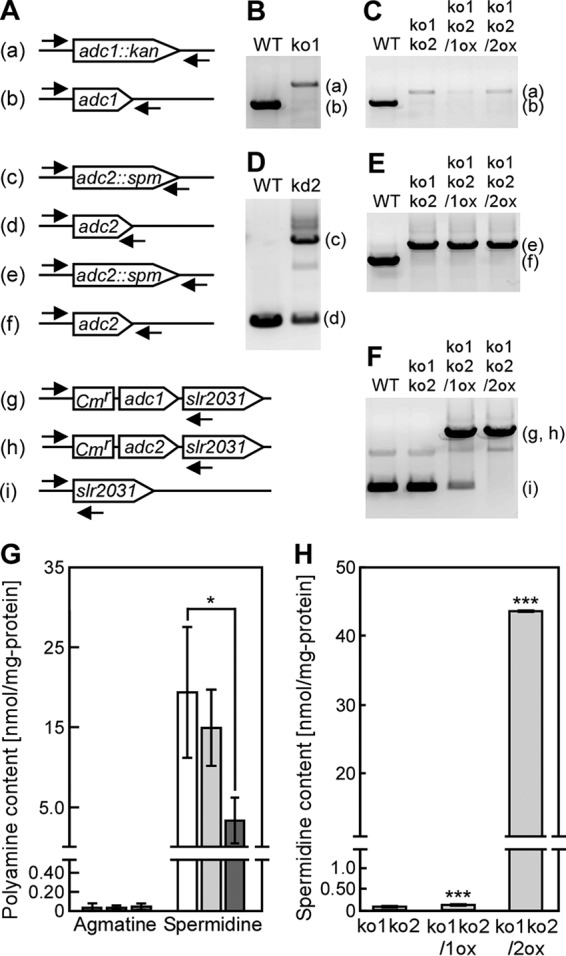
Spermidine contents in Synechocystis mutants. (A to F) The arrows indicate the positions and orientation of the primers for PCR to confirm the successful disruption or reintroduction of adc1 and adc2 in the Synechocystis genome. The letters on the left correspond to the PCR bands in panels B to F. The overexpression cassette of adc1 (g) or adc2 (h) was integrated at a neutral site, the slr2031 gene. Disruption of the genes in the insertion mutants ko1, kd2, and ko1ko2, obtained by repeated homogeneity segregation, was determined by the presence of a PCR product of the expected length. Note that for adc2 no complete disruption could be obtained (see the text for details). (G) Agmatine and spermidine content of the wild-type (WT) (white bars), ko1 (light gray bars), and ko2 (dark gray bars) strains were measured in cells grown for 5 days in medium without added NaCl. The spermidine content data for the wild type are the same as those shown in Fig. 2. Each value corresponds to the mean ± SD (n = 4). Significant differences between the WT and each mutant were analyzed by Student's t test (*, P < 0.05). No significant differences (P > 0.05) were found in agmatine content by Student's t test. (H) The spermidine contents of ko1ko2, ko1ko2/1ox, and ko1ko2/2ox strains were measured in cells grown for 7 days in medium without added NaCl. Each value corresponds to the mean ± SD (n = 3). Significant differences between ko1ko2 and ko1ko2/1ox or ko1ko2/2ox were analyzed by Student's t test (***, P < 0.001).
We measured the content of polyamines in Synechocystis mutants grown in BG11 medium and evaluated the arginine decarboxylase activity of Adc1 and Adc2 in Synechocystis. The mutants contained very low concentrations of putrescine and spermine compared to the amounts of spermidine (data not shown), which were similar to the concentrations found in the wild type (Fig. 2). The agmatine concentrations in wild-type, ko1, and kd2 strains were very low, and no significant differences were observed. On the other hand, the spermidine concentration in kd2 (3.47 ± 2.90 nmol/mg) was approximately 18% of that found in the wild type (19.3 ± 8.15 nmol/mg) (Fig. 5G). Loss of function of both adc1 and adc2 (ko1ko2) led to a strong reduction of spermidine content (0.07 ± 0.01 nmol/mg). In order to be able to relate the in vitro data showing low arginine decarboxylase activity of Adc1 and high activity of Adc2 (Fig. 3) to the in vivo situation, we determined the spermidine content in the ko1ko2/1ox and ko1ko2/2ox strains (Fig. 5H). ko1ko2/1ox showed only a small increase in spermidine content compared to the ko1ko2 strain, whereas ko1ko2/2ox had much higher levels of spermidine. This, together with the data on in vitro arginine decarboxylase activity of Adc1 and Adc2 (Fig. 3), supported the interpretation that Adc2 functions as the main arginine decarboxylase in Synechocystis.
Considering the very low arginine decarboxylase activity of Adc1 and the high activity of Adc2 (Fig. 3), we speculated that other unidentified cellular components are required for enzymatic activity of Adc1 and Adc2 in Synechocystis. To supply these putative cellular components, crude extracts from ko1ko2, ko1ko2/1ox, and ko1ko2/2ox were prepared (Fig. 6A and B) and used in enzyme assays. Extracts of ko1ko2/1ox and ko1ko2/2ox contained similar amounts of Adc1 and Adc2, respectively (Fig. 6A). Arginine decarboxylase activity of Adc1 remained very low, and there was no significant increase in spermidine production. On the other hand, Adc2 showed high arginine decarboxylase activity and a corresponding strong increase in spermidine production compared to that of ko1ko2 (Fig. 6C and D). Overall the observed enzyme activities were similar to the results shown in Fig. 3D. The fact that deletion of adc1 in ko1 led to a decrease in spermidine content (Fig. 5G) indicates that Adc1 has in vivo arginine decarboxylase activity and that its physiological role may be to enhance the enzyme activity of Adc2.
FIG 6.
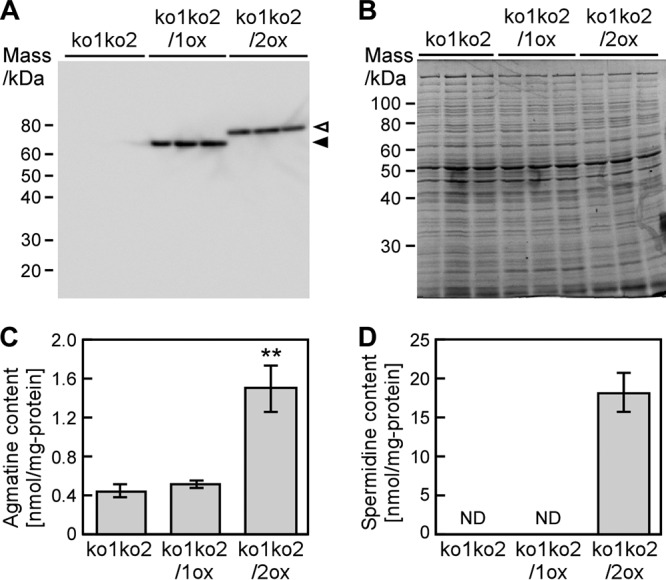
Effect of cellular components from Synechocystis on Adc1- or Adc2-mediated arginine decarboxylase activity and spermidine production. (A) Detection of Adc1 and Adc2 expressed in Synechocystis mutants by Western blotting. Arrowheads point to the bands corresponding to histidine-tagged Adc1 (filled arrowhead) and Adc2 (open arrowhead). (B) Total protein isolated from Synechocystis stained with Coomassie brilliant blue R-250. Each lane in panels A and B corresponds to one biological replicate. (C and D) Enzymatic activity measurements. Arginine was added to extracts of ko1ko2, ko1ko2/1ox, and ko1ko2/2ox. Amounts of agmatine (C) and spermidine (D) were determined after 1 h. Each value corresponds to the mean ± SD (n = 3). Significant differences between ko1ko2 and ko1ko2/1ox or ko1ko2/2ox were analyzed by Student's t test (**, P < 0.01). ND stands for not detected.
Synechocystis adc mutants showed biofilm formation without salt stress.
To analyze the effect of polyamine content on biofilm formation of the mutant cells, we compared cell aggregation and growth of the mutants with the wild type without added NaCl (Fig. 7). The morphology of adhered cells of ko1, kd2, and ko1ko2 strains and the wild type was observed by CLSM and FE-SEM (Fig. 7A and B). The adc mutants formed aggregation-like biofilms even without added NaCl, whereas the wild type scarcely showed biofilm formation under these conditions. The number of aggregated cells increased after addition of 500 mM NaCl. To confirm these observed differences between the mutants and the wild type, cell growth and amount of biofilm formation were estimated (Fig. 7C). Under nonstress conditions, the growth rate of the ko1ko2 strain was slightly decreased, and this was complemented by expression of adc2 (ko1ko2/2ox) (Fig. 7C). An increased number of ko1ko2 cells in biofilm were left on the test surface after the wash treatment compared to that for the wild type (Fig. 7C). The amount of biofilm in adc1-expressing cells (ko1ko2/1ox) was similar to that in the ko1ko2 strain, whereas a decrease in biofilm formation was observed in ko1ko2/2ox (Fig. 7C). These results indicated that the amount of biofilm formation and the amount of spermidine were in inverse correlation.
FIG 7.
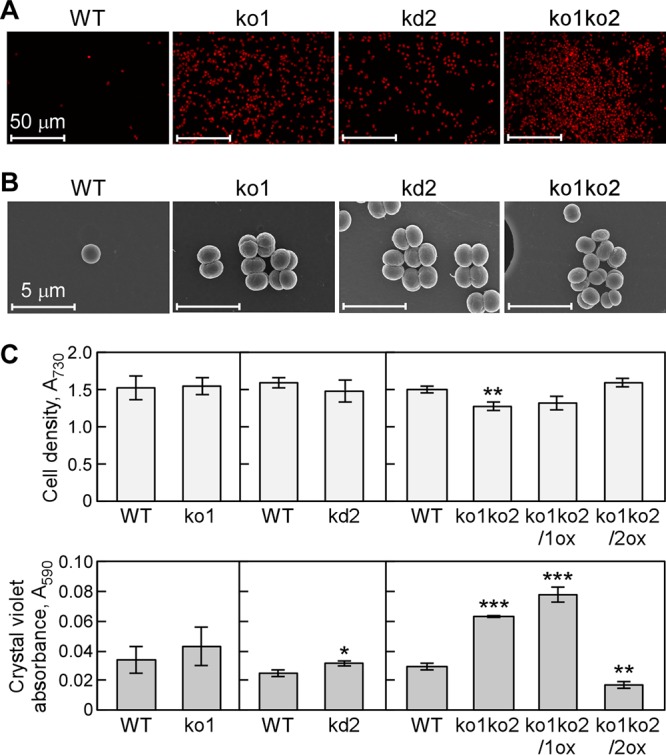
Effect of inactivation of adc1 and adc2 on growth and biofilm formation in Synechocystis. (A and B) Representative microscopic images obtained by CLSM (A) or FE-SEM (B) show biofilm formation of ko1, kd2, and ko1ko2 strains in medium after 5 days not seen with the wild type. Sample preparation was essentially the same as that described for Fig. 1C and D. The experiments were replicated three times and representative images are shown. (C) Growth curves of Synechocystis wild-type, ko1, kd2, ko1ko2, ko1ko2/1ox, and ko1ko2/2ox strains cultivated in BG11 medium without NaCl addition in 96-well polystyrene plates. (Top) The cell density was measured as the OD730. (Bottom) The amount of cells attached to the walls of the plates was quantified by crystal violet staining as described in the legend to Fig. 1. Each value corresponds to the mean ± SD (n = 3). Significant differences between the wild type (WT) and each mutant were analyzed by Student's t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
DISCUSSION
This study showed that in Synechocystis, salinity stress stimulated the formation of biofilm, which coincided with a reduction in the content of the most abundant intracellular polyamine, spermidine. The products of two genes predicted to encode key enzymes in the spermidine biosynthetic pathway, adc1 and adc2, were shown to possess arginine decarboxylase activity, and Adc2 was essential for the synthesis of spermidine in Synechocystis. Adc1 had extremely low enzymatic activity. Disruption of both of the adc genes led to biofilm formation under nonstress conditions. A reverse correlation between intracellular polyamine content and biofilm formation was observed in Synechocystis, and therefore Adc1 and Adc2 support the lifestyle of the cells through their enzymatic and regulatory function (Fig. 8).
FIG 8.
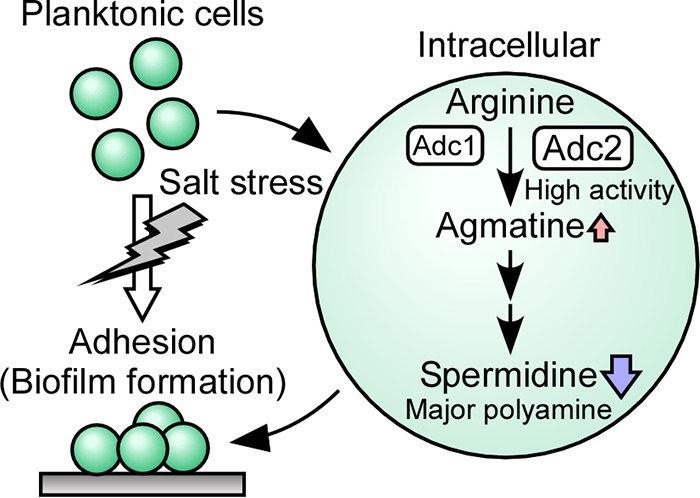
Morphological changes in a Synechocystis culture. During salt stress a change in life style occurs, leading planktonic Synechocystis cells to form cell aggregations in a biofilm. The conversion of arginine to agmatine, which is the initial step in the polyamine synthesis pathway in Synechocystis, is performed by two arginine decarboxylases, Adc1 and Adc2. Spermidine contents decline when the biofilm is formed.
Harsh environmental stress threatens the survival of bacteria. Under stress conditions, it becomes impossible for bacteria to proliferate as usual, and therefore in order to cope with salt stress bacteria undergo morphological changes. Available data from heterotrophic bacteria show that biofilm formation is correlated with the stress response and that in a biofilm cells are protected from physical and chemical damage in a range of environments (1–3). Biofilm formation constitutes a drastic morphological change from a planktonic existence to immobilized cells. For example, exopolysaccharide production occurs upon addition of NaCl, CoCl2, and CdSO4 and leads to increased tolerance against salt and heavy metal stress (7, 8). The morphological change results from a change in the metabolism of Synechocystis. For the purpose of this study, we focused on the effect of polyamine content on biofilm formation in Synechocystis, because polyamines are known modulators of tolerance to salt stress in other organisms (17) that are thought to enhance the cellular response to salt stress. The polyamines in Synechocystis and a possible synthetic pathway have been described (13, 14, 20, 23). In Synechocystis spermidine was the most abundant polyamine, while putrescine and spermine were present in smaller amounts (Fig. 2). These results were consistent with previous results by Jantaro et al. (20) but different from the situation in E. coli, where putrescine was the most abundant polyamine (24, 25).
Polyamines function as essential compounds for cell proliferation and differentiation in prokaryotes and eukaryotes (1, 2). Pioneering work on the cellular regulatory mechanism of polyamines in E. coli indicates that the majority of polyamines exist as polyamine-RNA complexes that alter RNA conformation, resulting in enhanced protein synthesis (10, 26). Furthermore, in E. coli, synthesis of the response regulators (UvrY and CpxR) of two-component systems as well as of the ribosome recycling factor was enhanced at the level of translation by polyamines functioning as part of the polyamine modulon (17). In animal cells, regulation by polyamine binding to specific RNAs was also found to be involved in protein synthesis associated with cell growth (26, 27). These findings indicate that polyamines function as positive growth factors. This is in agreement with our results in this study; the observed decrease in polyamine content in Synechocystis (Fig. 2) likely promoted the shift from more prolific planktonic cell division to cell aggregation, a state of decreased growth. This shift was induced by the addition of NaCl to the medium in Synechocystis (Fig. 1C).
We further investigated the effect of polyamines on biofilm formation in Synechocystis. A search of the genome sequence of Synechocystis sp. strain PCC 6803 indicated that an l-arginine decarboxylase (Adc) pathway exists in Synechocystis. Adc enzyme activity constitutes the initial step in the polyamine biosynthesis pathway in Synechocystis (Fig. 3A) (13–15, 20, 21, 28). A multiple-sequence alignment of Adc1 (slr1312) and Adc2 (slr0662) confirmed high similarity (77 to 79% amino acid sequence similarity) with Adcs from other organisms (SpeA from E. coli and Yersinia pestis and AtADC1 and AtADC2 from Arabidopsis thaliana). Adc1 shares 62% identity or 90% similarity with Adc2. Purified Adc1 and Adc2 showed clear differences in enzyme activity and pH dependency (Fig. 3D). Adc2 had high enzyme activity, whereas Adc1 did not. The results from the disruption mutants confirmed the dominant role of Adc2 in polyamine biosynthesis in Synechocystis (Fig. 6). The low activity of Adc1 (Fig. 3 and 6) indicates a different physiological role for Adc1 in vivo. In fact, Adc enzymes in other bacteria function as one of the components leading to the acquisition of extreme acid resistance at low external pH (29–31). Spermine also contributes to protection of photosystem II against photoinhibition conditions in spinach (32). Interestingly, a gene encoding a putative D1 protein, a component of photosystem II in the thylakoid membrane, is positioned in front of adc1 and in the same orientation in the Synechocystis genome. It is therefore possible that Adc1 and/or Adc2 also participate in other physiological processes besides salt stress adaptation. Another possible role of Adc1 is to directly regulate the catalytic activity of Adc2. The arginine decarboxylases from Methanococcus jannaschii (33) and Paramecium bursaria chlorella virus (34) form trimers and dimers, respectively. Assuming that in Synechocystis Adc1 and Adc2 also form multimeric structures, the presence of Adc1 in a heteroprotein complex might alter the overall activity provided by Adc2.
To constitutively reduce intracellular polyamine levels in Synechocystis, we generated disruption mutants of adc1 and adc2 (Fig. 5). The double mutant contained only negligible levels of spermidine (Fig. 5), strongly suggesting that both Adc1 and Adc2 are key enzymes for polyamine synthesis in Synechocystis. Interestingly, while the levels of three different polyamines decreased during salt stress in the wild type, agmatine accumulated even at 250 mM NaCl (Fig. 2). The expression of adc1 and adc2 was not altered by NaCl addition to the medium (Fig. 4); however, their arginine decarboxylase activity increased when NaCl was added to the reaction (Fig. 4C). At the same time it is possible that agmatinase production, which is required for the conversion of agmatine to putrescine, is inhibited by salt stress. This is a likely factor in the reduction of polyamine production in response to salt stress. The adc deletion strains showed increased biofilm formation even under nonstress conditions (Fig. 7). These results show that spermidine production was negatively correlated with biofilm formation in Synechocystis. A positive relationship between polyamine content and biofilm formation had been reported primarily in several heterotrophic bacteria, namely, Y. pestis, B. subtilis, Vibrio cholerae, and E. coli (12, 16, 17, 35). In Y. pestis, a mutant defective in the two main putrescine biosynthetic enzymes, arginine decarboxylase (SpeA) and ornithine decarboxylase (SpeC), shows a loss of biofilm formation (35). However, a limited number of examples for negative correlation were described: in Shewanella oneidensis, mutants deficient in ornithine decarboxylase (SpeF) show enhanced adherence and biofilm formation (36), and in V. cholerae, addition of spermine and spermidine inhibits biofilm formation (37, 38). A recent report also shows an increase of biofilm formation in response to spermidine depletion in Agrobacterium tumefaciens (39). The relationship between polyamine content and biofilm formation therefore varies among bacterial species. In plants, the amount of intracellular polyamines also changes in response to environmental challenges. In A. thaliana and rice, accumulation of polyamines helps to alleviate high-salinity stress (40, 41). However, there are also reports from plants where salt stress leads to a decrease of polyamines similar to the situation in Synechocystis. Accumulation of polyamines decreases in cucumber roots treated for 24 h with 200 mM NaCl (42). In tomato, the initial response to salt stress includes accumulation of polyamines followed by accumulation of Na+ in the cells, causing a disturbance of the ionic balance, which then results in a rapid decrease in the concentration of polyamines (43, 44). Like plants, cyanobacteria have the capability to convert atmospheric CO2 directly to organic compounds. Considering these similarities between cyanobacteria and plants, the way polyamines contribute to biofilm formation in phototropic bacteria like Synechocystis is more similar to the role of polyamines in sensing and responding to environmental changes found in plants.
The reason for the difference between the observed decrease of polyamines in a 5-day-old culture reported here (Fig. 2) and the moderate increase of polyamines reported by Jantaro et al. (20) in a 10-day-old culture is unclear. It may be due to differences in experimental conditions and cell status. Polyamines in planktonic cells might be produced when the cells are subjected to salt stress and hyperosmotic shock. However, once the cells have shifted from planktonic cell growth to biofilm formation during prolonged culture, our data showed a decrease in polyamine content. Biofilms also degrade and cells return to a planktonic state as a normal part of their life cycle (2). The intensity of stress, the physiological state of the cells, and polyamine synthesis may form a more complicated interrelationship in Synechocystis, influencing its lifestyle. Additional work will be needed to elucidate the exact mechanism of how polyamines induce biofilm formation.
MATERIALS AND METHODS
Cells and growth conditions.
Synechocystis sp. strain PCC 6803 wild-type and mutant cells were grown at 29°C in BG11 medium containing 20 mM TES-KOH (pH 8.0) (45). Continuous illumination was provided by fluorescent lamps (50 μmol of photons m−2 s−1; 400 to 700 nm).
Crystal violet assay.
Synechocystis wild-type and mutant cells cultured in liquid BG11 medium were diluted in BG11 medium with or without added NaCl, KCl, or sorbitol to an optical density at 730 nm (OD730) of 0.05 and grown for 7 days. The medium was removed, and flasks were washed gently with water. After drying the flasks, biofilm cells were stained with 0.1% crystal violet for 15 min and then washed three times with distilled water before staining was assessed. For the microtiter plate assay, biofilm formation was estimated by staining cells attached to microtiter plate wells with crystal violet (46). Diluted cultures (0.15 ml) were incubated in a 96-well polystyrene microtiter plate (TPP tissue culture plate; TPP Techno Plastic Products AG, Switzerland) at 29°C for 24 to 192 h, and cell density was measured as OD730. After discarding the medium in the wells, the wells were rinsed three times with water. Crystal violet solution (1%) was added to the microtiter plate wells. After incubation for 15 min, the staining solution was discarded and the wells rinsed with water. The crystal violet bound to the biofilm was extracted with 70% ethanol. The liquid was transferred to a new microtiter plate, and the absorbance (590 nm) was measured using a plate reader (Epoch microplate spectrophotometer; BioTek Instruments, Inc., Winooski, VT).
Analysis of CLSM images.
Synechocystis wild-type and mutant cells cultured in liquid BG11 medium for 7 days were diluted with fresh BG11 medium with or without added NaCl to an OD730 of 0.05. Diluted cultures (1 ml) were incubated in the presence of a coverslip (1 by 1 cm), functioning as a base plate, in the wells of a 12-well polystyrene tissue culture plate at 29°C for 5 days. The coverslips were then washed three times with BG11 and mounted on microscope slides (Vector Laboratories). Biofilm formation was monitored under a CLSM (Zeiss LSM5 Pa; Carl Zeiss, Germany) with a 63× objective (Plan-Apochromat 63×/1.4-numeric-aperture oil immersion lens; Carl Zeiss, Germany). A 543-nm HeNe laser and an LP560 emission filter were used to excite and detect the stained cells. CLSM images were obtained from three independent controls or treated biofilms and processed using Zen 2009 image software (Carl Zeiss, Germany).
FE-SEM analysis.
Synechocystis wild-type and mutant cells cultured in liquid BG11 medium for 7 days were diluted in BG11 medium with or without added NaCl to an OD730 of 0.05. The diluted cells (1 ml) were allowed to grow on polycarbonate membrane inserts (Nunc polycarbonate membrane inserts in multidishes; Thermo Fisher Scientific Inc., Waltham, MA) in 12-well polystyrene tissue culture plates at 29°C for 5 days. The polycarbonate membranes were then washed with 0.1 M phosphate buffer and fixed in 1.25% glutaraldehyde for 12 h at 4°C. Afterwards the membranes were washed with 0.1 M phosphate buffer and dehydrated through an ethanol series (from 0% to 100%). The fixed polycarbonate membranes were immersed in t-butyl alcohol, frozen at −20°C, and placed in a vacuum evaporator (FD-1200; EYELA, Tokyo, Japan). Finally, the polycarbonate membranes were sputter coated with osmium (POC-3 osmium plasma coater; Meiwafosis Co., Japan) and examined under an FE-SEM (S-4800; Hitachi, Japan).
HPLC detection of polyamines.
Putrescine, spermidine, spermine, and agmatine were measured by high-performance liquid chromatography (HPLC) using a TSK gel IEX215 column (4 by 160 mm; Toyo Soda) as described previously (47). Briefly, polyamines were extracted from cell lysate with 5% trichloroacetic acid (TCA), and after centrifugation at 27,000 × g for 15 min the supernatant was used for HPLC analysis. The protein content of the pellet was determined by the Lowry method.
Expression and purification of Adc1 and Adc2.
The genes encoding arginine decarboxylases (slr1312 and slr0662) were amplified by PCR from Synechocystis sp. strain PCC 6803 genomic DNA as the template with a set of primers for adc1 (slr1312) (5′-GGAATTCCATATGGGGGAAGAACCTGTGCCGGC-3′ and 5′-GGAGGATCCTCAACTTAGATAGGTGTAGCGCCG-3′) or adc2 (slr0662) (5′-GGAATTCCATATGGAAGGGCAGTCAATCGAACTAG-3′ and 5′-GGAGGATCCTTAGCTGGTGTGGATGCCTGAAGTGG-3′). The PCR products were digested with NdeI and BamHI and ligated into the corresponding sites of plasmid pET16b (Merck Millipore, MA). The DNA sequences of slr1312 and slr0662 were verified by DNA sequencing. The resultant plasmids were designated pET16b-adcl and pET16b-adc2. The E. coli BL21(DE3) strain (Merck Millipore, MA) containing the plasmids was grown in Luria-Bertani (LB) medium containing 100 μg/ml ampicillin overnight at 37°C. An aliquot of the culture was inoculated (100-fold dilution) into 500 ml fresh LB medium containing 100 μg/ml ampicillin. When the culture reached an OD600 of 0.6, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.4 mM and the culture was grown at 20°C for another 20 h. The cells were harvested by centrifugation (5,200 × g) for 15 min. The cells were disrupted in buffer A (50 mM Tris-HCl [pH 8], 150 mM NaCl, 10% glycerol, 40 μM pyridoxal-5′-phosphate [PLP], 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF]) by high-pressure lysis in a French press (EmulsiFlex-B15; Avestin, Inc., Ottawa, Canada). After centrifugation at 190,000 × g for 30 min, the supernatant was incubated with 500 μl of Talon metal affinity resin (TaKaRa Bio Inc., Shiga, Japan) at 4°C for 4 h. The resin was packed into columns, and then the columns each were washed with a total of 50 ml of buffer A. Adc1 or Adc2 was eluted by a brief centrifugation with 900 μl of cold elution buffer (50 mM Tris-HCl [pH 8], 150 mM NaCl, 10% glycerol, 40 μM PLP, 1 mM DTT, 1 mM PMSF, and 0.3 M imidazole). For those experiments performed to analyze the effect of NaCl on the enzymatic activity of Adc1 and Adc2, NaCl was omitted from buffer A during purification.
β-Galactosidase assay.
The 180-bp and 414-bp regions upstream from the start codons of adc1 and adc2 (Padc1 and Padc2) were amplified by PCR from Synechocystis sp. strain PCC 6803 genomic DNA and ligated into the AflII-NdeI sites of pNS1::lacZ+Cmrd (6). Using the resultant plasmid, Padc1::lacZ or Padc2::lacZ was integrated into the TS1 region in the Synechocystis genome. The strains containing Padc1::lacZ or Padc2::lacZ were cultured until they reached an OD730 of 0.8 to 1.2 and then transferred to fresh BG11 medium with or without 500 mM NaCl and incubated for 24 h. LacZ activity was determined according to the method of Miller (48). The assay solution consisted of 100 mM sodium phosphate buffer (pH 7; 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol, 0.002% SDS, 2% chloroform, and 0.08% o-nitrophenyl-β-d-galactoside).
Semiquantitative RT-PCR.
Synechocystis wild-type cells cultured in liquid BG11 medium was diluted to an OD730 of 0.05 in BG11 medium with or without added 500 mM NaCl and grown for 3 days. Total RNA was extracted with hot acidic phenol (49). The cDNAs for adc1, adc2, and rpnA were synthesized using the ReverTra Ace quantitative PCR reverse transcription master mix with genomic DNA Remover (Toyobo, Japan). The resultant cDNAs were then amplified by PCR with a set of primers for adc1 (5′-ACCGTCAGGACAAGCATACC-3′ and 5′-AATGTCCGAAACTTGCGTTCC-3′), adc2 (5′-TACGGGCCAAACTGAGTACC-3′ and 5′-ACAGGACAGGGCACTTCAG-3′), or rpnA (5′-GGACTACCCAAAACACTGC-3′ and 5′-CAATAATCCCAGCTTGGCT-3′). The numbers of PCR cycles used for adc1, adc2, and rpnA were 24, 24, and 22. PCR products were subjected to 2% agarose gel electrophoresis. The band intensities were evaluated by ImageJ (https://imagej.nih.gov/ij/).
Inactivation of adc1 and adc2 in Synechocystis sp. strain PCC 6803.
Disruption mutations of adc1 (slr1312) and adc2 (slr0662) were generated by insertion of a kanamycin resistance gene into adc1 and a spectinomycin resistance gene into adc2, respectively (Fig. 5A) (6). The mutant strains ko1, kd2, and ko1ko2 were then selected by successive streaking to homogeneity segregation in BG11 medium containing kanamycin (20 μg/ml) and/or spectinomycin (20 μg/ml) at 29°C. Disruption of adc1 and adc2 was confirmed by PCR using forward primer 5′-ATTGCACTCTCCTTCAATGGG-3′ and reverse primer 5′-AGTCTCAACTCTATCACGGC-3′ for adc1 and forward primer 5′-AGTCTCAACTCTATCACGGC-3′ and reverse primer 5′-ATTTCCGGGATGGTTAAACC-3′ for adc2, with genomic DNA prepared from the wild type and mutants as the template (Fig. 4A).
Reintroduction of adc1 and adc2 into Synechocystis mutants.
For reintroduction of adc1 or adc2 into the ko1ko2 double knockout strains, adc1 or adc2 was amplified with a set of primers 5′-ACGATAAGGATCATATGGGGGAAGAACCTGTG-3′ and 5′-TGAGGTTAACAGATCTTCAACTTAGATAGGTGTAGCGC-3′ for adc1 and 5′-ACGATAAGGATCATATGGAAGGGCAGTCAATCG-3′ and 5′-TGAGGTTAACAGATCTTTAGCTGGTGTGGATGCC-3′ for adc2 from genomic DNA. The PCR-amplified products were ligated into the NdeI-BglII sites located downstream of the trc promoter in the expression plasmid provided by Masahiro Ikeuchi (50). The resultant plasmids encoded either Adc1 or Adc2 with a histidine tag fused to the N terminus. The plasmid was then used to integrate adc1 or adc2 into the slr2031 region in the Synechocystis genome. Disruption of adc1 and adc2 was confirmed by PCR using the forward primer 5′-ATTGCACTCTCCTTCAATGGG-3′ and the reverse primer 5′-AGTCTCAACTCTATCACGGC-3′ for adc1 and the forward primer 5′-GAGTGGAGTACGGTTTGAAGC-3′ and the reverse primer 5′-CAGTTCCTGCCCCATAAGC-3′ for adc2. Reintegration of either adc1 or adc2 was confirmed by PCR using 5′-CCATGGGGAAGTTTGCTGG-3′ and 5′-CTTGGCGCAATTGACGGTC-3′. The resultant strains were named ko1ko2/1ox (adc1 reintroduced) and ko1ko2/2ox (adc2 reintroduced).
Assay of arginine decarboxylase activity.
Adc1 or Adc2 protein purified from E. coli was mixed with 50 mM citrate-phosphate buffer (pH 4.0 to 8.0) or 50 mM glycine-sodium hydroxide buffer (pH 9.0 to 10.0) containing 10 mM l-arginine–HCl, 150 mM NaCl, 10% glycerol, 40 μM PLP, and 1 mM DTT and incubated at 37°C after 28 h of incubation (for Adc1) or 30 min of incubation (for Adc2). To investigate the effect of NaCl on Adc1 and Adc2, the purified Adc1 or Adc2 was mixed with Tris-HCl (pH 7.0 for Adc1 and pH 8.0 for Adc2) containing 10 mM l-arginine-HCl, NaCl (0 mM, 250 mM, or 500 mM), 10% glycerol, 40 μM PLP, and 1 mM DTT and incubated at 37°C for 28 h or 1 h, respectively. Five microliters of 100% TCA was added to terminate the reaction. After centrifugation (15,000 × g) for 5 min, the concentration of agmatine in the supernatant was determined by HPLC on a TSK gel IEX215 column (4 by 160 mm; Toyo Soda) as described previously (25). To determine the arginine decarboxylase activity in ko1ko2, ko1ko2/1ox, and ko1ko2/2ox strains, crude extracts were prepared in extraction buffer (50 mM Tris-HCl [pH 8.4], 0.5 mM PLP, 0.1 mM EDTA, 5 mM DTT). The reaction was initiated by addition of arginine (final concentration, 20 mM) to the crude extract (150 μg of protein). After incubation for 1 h, the reaction was stopped by addition of TCA and the reaction mixture was immediately incubated at 100°C for 3 min. After centrifugation at 15,000 × g for 15 min, the concentration of agmatine in the supernatant was determined by HPLC on a TSK gel IEX215 column.
Protein sequence analysis of Adc1 and Adc2.
Proteins were extracted from E. coli, separated by SDS-PAGE, and blotted onto polyvinylidene difluoride (PVDF) membrane. Proteins on the PVDF membrane were stained for 1 min with 0.1% (wt/vol) Coomassie brilliant blue G-250 in 50% (vol/vol) methanol and destained with 10% (vol/vol) acetic acid containing 50% (vol/vol) methanol to remove background staining. After washing with distilled water, the membrane was dried, the protein spots were excised, and the amino acid sequence was analyzed by a gas-phase Edman sequencer which sequentially cleaves N-terminal amino acids from the protein and analyzes the resulting phenylthiohydantoin (PTH)-amino acid residues by HPLC (PPSQ-53A; Shimadzu, Kyoto, Japan).
Immunodetection.
Crude protein extracts (5 μg) from Synechocystis strains ko1ko2, ko1ko2/1ox, and ko1ko2/2ox were separated by SDS-PAGE on a 10% polyacrylamide gel and blotted onto PVDF membrane or stained with Coomassie brilliant blue R-250. Immunodetection was performed with ECL-Plus Western blotting detection reagents (GE Healthcare, Little Chalfont, England) using the monoclonal antibody anti-His tag MAb-HRP-DirecT (MBL, Nagoya, Japan).
ACKNOWLEDGMENTS
We thank Soichi Furukawa and Yasushi Morinaga (Nihon University) for their comments on the crystal violet assay and Anke Reinders (University of Minnesota) for critical reading of the manuscript. We are grateful to Kiyoshi Onai (Kyoto University), Masahiro Ishiura (Nagoya University), and Masahiro Ikeuchi (University of Tokyo) for kindly providing the vector pNS1::lacZ+Cmrd and the Synechocystis expression vector.
This work was supported by Grants-in-Aid for Scientific Research (15H02226, 16H06558, and 16H04906 to N.U. and 16K18670 to K.K.) from the Ministry of Education, Culture, Sports, Science, and Technology.
REFERENCES
- 1.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 2.McDougald D, Rice SA, Barraud N, Steinberg PD, Kjelleberg S. 2012. Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat Rev Microbiol 10:39–50. doi: 10.1038/nrmicro2695. [DOI] [PubMed] [Google Scholar]
- 3.Candela T, Maes E, Garenaux E, Rombouts Y, Krzewinski F, Gohar M, Guerardel Y. 2011. Environmental and biofilm-dependent changes in a Bacillus cereus secondary cell wall polysaccharide. J Biol Chem 286:31250–31262. doi: 10.1074/jbc.M111.249821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Los DA, Murata N. 1999. Responses to cold shock in cyanobacteria. J Mol Microbiol Biotechnol 1:221–230. [PubMed] [Google Scholar]
- 5.Akai M, Onai K, Kusano M, Sato M, Redestig H, Toyooka K, Morishita M, Miyake H, Hazama A, Checchetto V, Szabo I, Matsuoka K, Saito K, Yasui M, Ishiura M, Uozumi N. 2011. Plasma membrane aquaporin AqpZ protein is essential for glucose metabolism during photomixotrophic growth of Synechocystis sp. PCC. 6803. J Biol Chem 286:25224–25235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nanatani K, Shijuku T, Takano Y, Zulkifli L, Yamazaki T, Tominaga A, Souma S, Onai K, Morishita M, Ishiura M, Hagemann M, Suzuki I, Maruyama H, Arai F, Uozumi N. 2015. Comparative analysis of kdp and ktr mutants reveals distinct roles of the potassium transporters in the model cyanobacterium Synechocystis sp. strain PCC. 6803. J Bacteriol 197:676–687. doi: 10.1128/JB.02276-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jittawuttipoka T, Planchon M, Spalla O, Benzerara K, Guyot F, Cassier-Chauvat C, Chauvat F. 2013. Multidisciplinary evidences that Synechocystis PCC6803 exopolysaccharides operate in cell sedimentation and protection against salt and metal stresses. PLoS One 8:e55564. doi: 10.1371/journal.pone.0055564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ozturk S, Aslim B. 2010. Modification of exopolysaccharide composition and production by three cyanobacterial isolates under salt stress. Environ Sci Pollut Res Int 17:595–602. doi: 10.1007/s11356-009-0233-2. [DOI] [PubMed] [Google Scholar]
- 9.Yoshida M, Kashiwagi K, Shigemasa A, Taniguchi S, Yamamoto K, Makinoshima H, Ishihama A, Igarashi K. 2004. A unifying model for the role of polyamines in bacterial cell growth, the polyamine modulon. J Biol Chem 279:46008–46013. doi: 10.1074/jbc.M404393200. [DOI] [PubMed] [Google Scholar]
- 10.Igarashi K, Kashiwagi K. 2006. Polyamine modulon in Escherichia coli: genes involved in the stimulation of cell growth by polyamines. J Biochem 139:11–16. doi: 10.1093/jb/mvj020. [DOI] [PubMed] [Google Scholar]
- 11.Valdes-Santiago L, Ruiz-Herrera J. 2014. Stress and polyamine metabolism in fungi. Front Chem 2:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burrell M, Hanfrey CC, Murray EJ, Stanley-Wall NR, Michael AJ. 2010. Evolution and multiplicity of arginine decarboxylases in polyamine biosynthesis and essential role in Bacillus subtilis biofilm formation. J Biol Chem 285:39224–39238. doi: 10.1074/jbc.M110.163154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jantaro S, Kidron H, Chesnel D, Incharoensakdi A, Mulo P, Salminen T, Maenpaa P. 2006. Structural modeling and environmental regulation of arginine decarboxylase in Synechocystis sp. PCC. 6803. Arch Microbiol 184:397–406. [DOI] [PubMed] [Google Scholar]
- 14.Schriek S, Ruckert C, Staiger D, Pistorius EK, Michel KP. 2007. Bioinformatic evaluation of l-arginine catabolic pathways in 24 cyanobacteria and transcriptional analysis of genes encoding enzymes of l-arginine catabolism in the cyanobacterium Synechocystis sp. PCC. 6803. BMC Genomics 8:437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pothipongsa A, Jantaro S, Incharoensakdi A. 2012. Polyamines induced by osmotic stress protect Synechocystis sp. PCC 6803 cells and arginine decarboxylase transcripts against UV-B radiation. Appl Biochem Biotechnol 168:1476–1488. [DOI] [PubMed] [Google Scholar]
- 16.Karatan E, Michael AJ. 2013. A wider role for polyamines in biofilm formation. Biotechnol Lett 35:1715–1717. doi: 10.1007/s10529-013-1286-3. [DOI] [PubMed] [Google Scholar]
- 17.Sakamoto A, Terui Y, Yamamoto T, Kasahara T, Nakamura M, Tomitori H, Yamamoto K, Ishihama A, Michael AJ, Igarashi K, Kashiwagi K. 2012. Enhanced biofilm formation and/or cell viability by polyamines through stimulation of response regulators UvrY and CpxR in the two-component signal transducing systems, and ribosome recycling factor. Int J Biochem Cell Biol 44:1877–1886. doi: 10.1016/j.biocel.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 18.Liu BR, Huang YW, Lee HJ. 2013. Mechanistic studies of intracellular delivery of proteins by cell-penetrating peptides in cyanobacteria. BMC Microbiol 13:57. doi: 10.1186/1471-2180-13-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisher ML, Allen R, Luo Y, Curtiss R. 2013. Export of extracellular polysaccharides modulates adherence of the cyanobacterium Synechocystis. PLoS One 8:e74514. doi: 10.1371/journal.pone.0074514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jantaro S, Maenpaa P, Mulo P, Incharoensakdi A. 2003. Content and biosynthesis of polyamines in salt and osmotically stressed cells of Synechocystis sp. PCC. 6803. FEMS Microbiol Lett 228:129–135. doi: 10.1016/S0378-1097(03)00747-X. [DOI] [PubMed] [Google Scholar]
- 21.Quintero MJ, Muro-Pastor AM, Herrero A, Flores E. 2000. Arginine catabolism in the cyanobacterium Synechocystis sp. strain PCC 6803 involves the urea cycle and arginase pathway. J Bacteriol 182:1008–1015. doi: 10.1128/JB.182.4.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Labarre J, Chauvat F, Thuriaux P. 1989. Insertional mutagenesis by random cloning of antibiotic resistance genes into the genome of the cyanobacterium Synechocystis strain PCC. 6803. J Bacteriol 171:3449–3457. doi: 10.1128/jb.171.6.3449-3457.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Incharoensakdi A, Jantaro S, Raksajit W, Mäenpää P. 2010. Polyamines in cyanobacteria: biosynthesis, transport and abiotic stress response, p 23–32. In Ḿendez-Vilas A (ed), Current research, technology and education topics in applied microbiology and microbial biotechnology. Formatex Research Center, Badajoz, Spain. [Google Scholar]
- 24.Munro GF, Hercules K, Morgan J, Sauerbier W. 1972. Dependence of the putrescine content of Escherichia coli on the osmotic strength of the medium. J Biol Chem 247:1272–1280. [PubMed] [Google Scholar]
- 25.Kashiwagi K, Igarashi K. 1988. Adjustment of polyamine contents in Escherichia coli. J Bacteriol 170:3131–3135. doi: 10.1128/jb.170.7.3131-3135.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Igarashi K, Kashiwagi K. 2010. Modulation of cellular function by polyamines. Int J Biochem Cell Biol 42:39–51. doi: 10.1016/j.biocel.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 27.Mandal S, Mandal A, Johansson HE, Orjalo AV, Park MH. 2013. Depletion of cellular polyamines, spermidine and spermine, causes a total arrest in translation and growth in mammalian cells. Proc Natl Acad Sci U S A 110:2169–2174. doi: 10.1073/pnas.1219002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamura Y, Kaneko T, Hirosawa M, Miyajima N, Tabata S. 1998. CyanoBase, a www database containing the complete nucleotide sequence of the genome of Synechocystis sp. strain PCC6803. Nucleic Acids Res 26:63–67. doi: 10.1093/nar/26.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richard H, Foster JW. 2004. Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J Bacteriol 186:6032–6041. doi: 10.1128/JB.186.18.6032-6041.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alvarez-Ordonez A, Fernandez A, Bernardo A, Lopez M. 2010. Arginine and lysine decarboxylases and the acid tolerance response of Salmonella Typhimurium. Int J Food Microbiol 136:278–282. doi: 10.1016/j.ijfoodmicro.2009.09.024. [DOI] [PubMed] [Google Scholar]
- 31.Iyer R, Iverson TM, Accardi A, Miller C. 2002. A biological role for prokaryotic ClC chloride channels. Nature 419:715–718. doi: 10.1038/nature01000. [DOI] [PubMed] [Google Scholar]
- 32.Hamdani S, Gauthier A, Msilini N, Carpentier R. 2011. Positive charges of polyamines protect PSII in isolated thylakoid membranes during photoinhibitory conditions. Plant Cell Physiol 52:866–873. doi: 10.1093/pcp/pcr040. [DOI] [PubMed] [Google Scholar]
- 33.Tolbert WD, Graham DE, White RH, Ealick SE. 2003. Pyruvoyl-dependent arginine decarboxylase from Methanococcus jannaschii: crystal structures of the self-cleaved and S53A proenzyme forms. Structure 11:285–294. doi: 10.1016/S0969-2126(03)00026-1. [DOI] [PubMed] [Google Scholar]
- 34.Shah R, Akella R, Goldsmith EJ, Phillips MA. 2007. X-ray structure of Paramecium bursaria Chlorella virus arginine decarboxylase: insight into the structural basis for substrate specificity. Biochemistry 46:2831–2841. doi: 10.1021/bi6023447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel CN, Wortham BW, Lines JL, Fetherston JD, Perry RD, Oliveira MA. 2006. Polyamines are essential for the formation of plague biofilm. J Bacteriol 188:2355–2363. doi: 10.1128/JB.188.7.2355-2363.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding Y, Peng N, Du Y, Ji L, Cao B. 2014. Disruption of putrescine biosynthesis in Shewanella oneidensis enhances biofilm cohesiveness and performance in Cr(VI) immobilization. Appl Environ Microbiol 80:1498–1506. doi: 10.1128/AEM.03461-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sobe RC, Bond WG, Wotanis CK, Zayner JP, Burriss MA, Fernandez N, Bruger EL, Waters CM, Neufeld HS, Karatan E. 2017. Spermine inhibits Vibrio cholerae biofilm formation through the NspS-MbaA polyamine signaling system. J Biol Chem 292:17025–17036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karatan E, Duncan TR, Watnick PI. 2005. NspS, a predicted polyamine sensor, mediates activation of Vibrio cholerae biofilm formation by norspermidine. J Bacteriol 187:7434–7443. doi: 10.1128/JB.187.21.7434-7443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Kim SH, Natarajan R, Heindl JE, Bruger EL, Waters CM, Michael AJ, Fuqua C. 2016. Spermidine inversely influences surface interactions and planktonic growth in Agrobacterium tumefaciens. J Bacteriol 198:2682–2691. doi: 10.1128/JB.00265-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamaguchi K, Takahashi Y, Berberich T, Imai A, Miyazaki A, Takahashi T, Michael A, Kusano T. 2006. The polyamine spermine protects against high salt stress in Arabidopsis thaliana. FEBS Lett 580:6783–6788. doi: 10.1016/j.febslet.2006.10.078. [DOI] [PubMed] [Google Scholar]
- 41.Krishnamurthy R, Bhagwat KA. 1989. Polyamines as modulators of salt tolerance in rice cultivars. Plant Physiol 91:500–504. doi: 10.1104/pp.91.2.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Janicka-Russak M, Kabala K, Mlodzinska E, Klobus G. 2010. The role of polyamines in the regulation of the plasma membrane and the tonoplast proton pumps under salt stress. J Plant Physiol 167:261–269. doi: 10.1016/j.jplph.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 43.SantaCruz A, Acosta M, Perez Alfocea F, Bolarin MC. 1997. Changes in free polyamine levels induced by salt stress in leaves of cultivated and wild tomato species. Physiologia Plantarum 101:341–346. doi: 10.1111/j.1399-3054.1997.tb01006.x. [DOI] [Google Scholar]
- 44.Santa-Cruz A, Estan MT, Rus A, Bolarin MC, Acosta M. 1997. Effects of NaCl and mannitol iso-osmotic stresses on the free polyamine levels in leaf discs of tomato species differing in salt tolerance. J Plant Physiol 151:754–758. [Google Scholar]
- 45.Matsuda N, Kobayashi H, Katoh H, Ogawa T, Futatsugi L, Nakamura T, Bakker EP, Uozumi N. 2004. Na+-dependent K+ uptake ktr system from the cyanobacterium Synechocystis sp PCC 6803 and its role in the early phases of cell adaptation to hyperosmotic shock. J Biol Chem 279:54952–54962. doi: 10.1074/jbc.M407268200. [DOI] [PubMed] [Google Scholar]
- 46.Narisawa N, Furukawa S, Ogihara H, Yamasaki M. 2005. Estimation of the biofilm formation of Escherichia coli K-12 by the cell number. J Biosci Bioeng 99:78–80. doi: 10.1263/jbb.99.78. [DOI] [PubMed] [Google Scholar]
- 47.Igarashi K, Kashiwagi K, Hamasaki H, Miura A, Kakegawa T, Hirose S, Matsuzaki S. 1986. Formation of a compensatory polyamine by Escherichia coli polyamine-requiring mutants during growth in the absence of polyamines. J Bacteriol 166:128–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 49.Kera K, Takahashi S, Sutoh T, Koyama T, Nakayama T. 2012. Identification and characterization of a cis,trans-mixed heptaprenyl diphosphate synthase from Arabidopsis thaliana. FEBS J 279:3813–3827. doi: 10.1111/j.1742-4658.2012.08742.x. [DOI] [PubMed] [Google Scholar]
- 50.Yoshihara S, Katayama M, Geng X, Ikeuchi M. 2004. Cyanobacterial phytochrome-like PixJ1 holoprotein shows novel reversible photoconversion between blue- and green-absorbing forms. Plant Cell Physiol 45:1729–1737. doi: 10.1093/pcp/pch214. [DOI] [PubMed] [Google Scholar]