ABSTRACT
Yersinia pestis, the causative agent of plague, evolved from the closely related pathogen Yersinia pseudotuberculosis. During its emergence, Y. pestis is believed to have acquired its unique pathogenic characteristics through numerous gene gains/losses, genomic rearrangements, and single nucleotide polymorphism (SNP) changes. One such SNP creates a single amino acid variation in the DNA binding domain of PhoP, the response regulator in the PhoP/PhoQ two-component system. Y. pseudotuberculosis and the basal human-avirulent strains of Y. pestis harbor glycines at position 215 of PhoP, whereas the modern human-virulent strains (e.g., KIM and CO92) harbor serines at this residue. Since PhoP plays multiple roles in the adaptation of Y. pestis to stressful host conditions, we tested whether this amino acid substitution affects PhoP activity or the ability of Y. pestis to survive in host environments. Compared to the parental KIM6+ strain carrying the modern allele of phoP (phoP-S215), a derivative carrying the basal allele (phoP-G215) exhibited slightly defective growth under a low-Mg2+ condition and decreased transcription of a PhoP target gene, ugd, as well as an ∼8-fold increase in the susceptibility to the antimicrobial peptide polymyxin B. The phoP-G215 strain showed no apparent defect in flea colonization, although a phoP-null mutant showed decreased flea infectivity in competition experiments. Our results suggest that the amino acid variation at position 215 of PhoP causes subtle changes in the PhoP activity and raise the possibility that the change in this residue have contributed to the evolution of increased virulence in Y. pestis.
IMPORTANCE Y. pestis acquired a single nucleotide polymorphism (SNP) in phoP when the highly human-virulent strains diverged from less virulent basal strains, resulting in an amino acid substitution in the DNA binding domain of the PhoP response regulator. We show that Y. pestis carrying the modern phoP allele has an increased ability to induce the PhoP-regulated ugd gene and resist antimicrobial peptides compared to an isogenic strain carrying the basal allele. Given the important roles PhoP plays in host adaptation, the results raise an intriguing possibility that this amino acid substitution contributed to the evolution of increased virulence in Y. pestis. Additionally, we present the first evidence that phoP confers a survival fitness advantage to Y. pestis inside the flea midgut.
KEYWORDS: PhoP, Yersinia pestis, evolution, transcription
INTRODUCTION
Yersinia pestis is a Gram-negative bacterium that is the causative agent of plague. It has been responsible for at least three human pandemics—the Justinian plague (1), Black Death (2), and modern plague—and has claimed lives of as many as 200 million people throughout human history (3). The infectious life cycle of Y. pestis is complex, alternating between two hosts: mammals (mainly rodents) and insect vectors (fleas) (4). The flea acquires Y. pestis by taking a blood meal from an infected mammal. The ingested bacteria can form a biofilm inside the flea gut and block the flea foregut and are eventually regurgitated into a new mammalian host during a fleabite to cause bubonic plague (4). Y. pestis also can be transmitted through an aerosol route to cause pneumonic plague (3). Consequently, Y. pestis encounters a wide range of host environments during its life cycle and must adapt to each to achieve successful propagation.
The PhoP/PhoQ two-component system plays a critical role in the adaptation of many Gram-negative bacteria to stressful host environments as well as in pathogenesis (5). In a typical prokaryotic two-component system, an environmental stimulus is detected by the sensor kinase, initiating a phosphorelay that results in the phosphorylation and the activation of the response regulator. The activated response regulator binds to the promoters of the target genes, ultimately leading to coordinated changes in global gene expression profiles (5). The PhoP/PhoQ system consists of the sensor kinase PhoQ and the response regulator PhoP and has been shown to detect several stimuli, including low-Mg2+ conditions (6), acidic pH (7), and the presence of cationic antimicrobial peptides (8). In Y. pestis, phoP is required for growth in low-Mg2+ environments, for survival inside macrophages, and for resistance to antimicrobial peptides (9–12). Loss of phoP function in Y. pestis results in an approximately 2- to 75-fold increase in the 50% lethal dose (LD50) and an increased time to death in murine bubonic plague models, suggesting that phoP plays a key role in virulence (11, 13). In addition, Y. pestis strains lacking phoP are reported to have reduced abilities to form biofilms in a flow cell system at 25°C and to block fleas in vivo, indicating that PhoP/PhoQ is also important for developing a transmissible infection in the flea (14). PhoP activity affects the expression of ∼200 to 700 genes according to microarray studies, and at least 30 genes, including ugd and the seven-gene pmr operon, are known to be direct targets of PhoP (15–19). The pmrHFIJKLM and ugd genes encode enzymes that are necessary for the addition of the sugar 4-amino-4-deoxy-l-arabinose (aminoarabinose) to the lipid A moiety of Y. pestis lipopolysaccharide (LPS) (16). The aminoarabinose modification reduces the net negative charge on the outer surface of the bacterium, leading to an increased resistance to the cationic antimicrobial peptides (9, 16, 20).
The phylogenetic analyses of various Y. pestis and Y. pseudotuberculosis strains have indicated that Y. pestis is a clone of Y. pseudotuberculosis that evolved as recently as 5,000 to 79,000 years ago (21–24). Despite their close relationships, Y. pestis and Y. pseudotuberculosis have remarkably distinct ecologies; while Y. pseudotuberculosis is a soil- and waterborne pathogen that normally causes self-limiting gastrointestinal disease, Y. pestis is a highly virulent insect vector-borne pathogen (25). A comparison of Y. pestis and Y. pseudotuberculosis genomes suggests that Y. pestis acquired its unique pathogenic characteristics through numerous changes in its genome, including the acquisition of two plasmids (pMT1 and pPCP1), genomic rearrangements, massive gene inactivation, and gene gains (25, 26). Small genetic changes, including single nucleotide polymorphisms (SNPs), have also contributed to the adaptation of Y. pestis to the insect and mammalian hosts. For example, Zimbler et al. showed that the plasminogen activator protease Pla acquired a single amino acid substitution during Y. pestis evolution, and this substitution enhanced the ability of Y. pestis to disseminate within its mammalian host (27). Similarly, Sun et al. showed that in addition to the gain of the murine toxin gene ymt on the pMT1 plasmid, the stepwise acquisition of inactivating mutations in three genes controlling biofilm formation enhanced the transmissibility of Y. pestis by its flea vector (28). The comparative examination of ancient human Y. pestis genomes from archaeological sites suggests that the pandemic strains of Y. pestis arose from less virulent non-flea-borne strains capable of infecting humans and that the small changes in the functions of key genes may have led to the increased virulence and transmissibility of Y. pestis (23). However, the specific genetic changes that transformed Y. pestis into a highly virulent human pathogen remain elusive.
Y. pestis strains are classified into several phylogenetic branches on the basis of comprehensive genomic analyses (21, 22, 29). A basal/ancestral branch (termed 0) is rooted in Y. pseudotuberculosis and includes all human-avirulent Y. pestis (Pestoides and subsp. microtus) strains. Branches 1 and 2, which contain most of the modern human-pathogenic strains sampled worldwide, as well as the newly discovered branches 3 and 4, all diverged from the basal branch 0 (21, 22, 29). In particular, the East Smithfield strain (ESS) that was isolated from the skeletal remains of Black Death victims is placed near the base of branch 1, very close to the polytomy from which branches 1 through 4 diverged (2, 29). In contrast, the Y. pestis subsp. microtus 91001 strain belongs to the basal branch 0 and is considered one of the closest relatives of the branch 1 and 2 strains that are avirulent to humans (29, 30). Therefore, genetic differences between these two strains may represent changes that played key roles in enhancing the virulence or host specificity of Y. pestis during the emergence of the modern human-virulent strains (2). A comparison of the genomic sequences between Y. pestis subsp. microtus 91001 and ESS revealed SNPs in 113 genes. Notably, some of the SNPs are found in genes implicated in the virulence or the host adaptation of Y. pestis, including hmsT (biofilm formation), iucD (iron acquisition), ail (host cell adhesion and serum resistance), and phoP (2). The SNP located in phoP creates a single amino acid change at position 215 of the encoded PhoP protein; Y. pestis subsp. microtus 91001 encodes glycine at this residue, while serine is coded for in ESS (2). Given the known roles of PhoP in host adaptation, we hypothesized that the phoP allele encoding serine might have conferred increased fitness in the flea and/or mammalian host environments and thus have been positively selected during Y. pestis evolution. Herein, we tested this possibility by introducing an S-to-G amino acid substitution in PhoP of the modern Y. pestis strain KIM6+ and assessed its effects on protein function. We show that the S-to-G substitution indeed results in the differential regulation of the transcription of a PhoP target gene and in antimicrobial peptide susceptibility.
RESULTS
Comparison of the Y. pestis ESS and Y. pestis subsp. microtus 91001 genomic sequences revealed a G/A nucleotide polymorphism inside phoP, which generates a serine/glycine single amino acid change at position 215 of the encoded PhoP response regulator (2). A further comparison of the phoP sequences from other Yersinia strains showed that glycine (G) is encoded in Y. pseudotuberculosis and other basal strains of Y. pestis, such as Angola and Pestoides, while serine (S) is shared by all the modern strains, including KIM and CO92 (Fig. 1A). Hence, it was concluded that this SNP was acquired on the basal branch (branch 0) of the Y. pestis phylogenetic tree, before the polytomy where branches 1 to 4 diverged (29). The modeling of PhoP in Y. pestis and of a related response regulator protein PmrA in Salmonella predicts that this residue is located in the DNA binding domain of the protein, within the flexible loop that is likely to contact the target DNA (Fig. 1B) (10, 31). Therefore, we hypothesized that the amino acid change at this position may alter the activity of PhoP.
FIG 1.
A nonsynonymous SNP in phoP creates a G-to-S single amino acid substitution in PhoP between Y. pestis subsp. microtus and East smithfield strain (ESS). (A) The codon change caused by the SNP in phoP and the resulting amino acid substitution are listed. Yersinia strains are classified based on the amino acid residue (glycine or serine) at position 215 of the PhoP protein. (B) Amino acid sequences and predicted secondary structure of PhoP proteins from Y. pestis ESS and Y. pestis subsp. microtus 91001. The secondary structure prediction is based on the previously published work by Grabenstein et al. (10) and Perez and Groisman (33). The black strips indicate the predicted α-helices, and the gray arrows indicate the predicted β-sheets. The location of the G-to-S amino acid substitution is indicated in a shaded box.
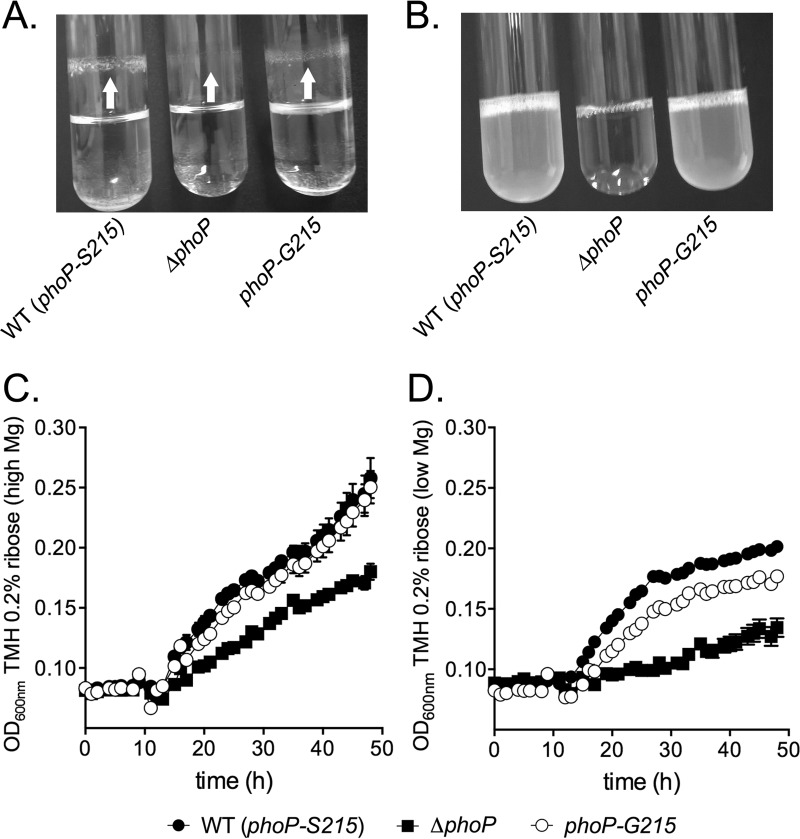
To assess the effects of this amino acid variation on PhoP function, we introduced the S-to-G substitution in PhoP of the KIM6+ strain to represent the basal allele of phoP. The resulting strain, KIM6+ phoP-G215, was cultured in TMH medium containing 20 μM MgCl2 (a phoP-activating low-Mg2+condition), and its growth was compared to that of the parental wild-type (phoP-S215) strain and the isogenic KIM6+ ΔphoP strain (Table 1). At 21°C, all the strains were able to grow and formed tightly packed biofilms at the liquid-air interface of the glass tubes. The thicknesses of the biofilms were visibly different among the three strains, with the wild-type strain forming the most robust aggregates and the ΔphoP strain forming the thinnest biofilm. The biofilm of the phoP-G215 strain showed an intermediate appearance (Fig. 2A). At 37°C, the wild-type and phoP-G215 strains were able to grow normally, while the growth of the ΔphoP strain was inhibited. No biofilm formation was observed in any of the strains at this temperature (Fig. 2B).
TABLE 1.
Yersinia and Salmonella strains used in this study
| Strain | Relevant characteristics | Reference |
|---|---|---|
| Y. pestis KIM6+ | Molecular grouping 2.MED, pCD1− | 48 |
| Y. pestis KIM6+ ΔphoP | KIM6+ phoPΔ127–429 | 15 |
| Y. pestis KIM6+ phoP-G215 | KIM6+ phoP-G215 | This work |
| Y. pestis KIM6+/pGFP | KIM6+/p67GFP3.1 Ampr Carbr | 49 |
| Salmonella Typhimurium | Salmonella enterica serovar Typhimurium ATCC 14028 | 50 |
FIG 2.
Growth characteristics of Y. pestis strains. (A and B) Y. pestis KIM6+ (wild type [WT]) or the indicated KIM6+-derived strains were cultured overnight in TMH medium containing 20 μM Mg2+ (phoP-activating low-Mg2+ condition) in glass tubes at 21°C (A) or 37°C (B), and their gross growth phenotypes were compared. The arrows indicate the biofilms formed on the surfaces of the glass tubes. The biofilms appear to have formed above the liquid-air interface, because the liquid was reaching higher levels in the tubes during incubation in a rotary shaker. The pictures are representatives of three independent experiments. (C and D) Growth curve analysis was performed at ambient room temperature (∼23°C) using TMH supplemented with 0.2% ribose as the carbon source. The indicated Y. pestis KIM6+-derived strains were cultured successively over two nights, first in brain heart infusion medium and then in TMH ribose containing 20 mM Mg2+. On day three, the strains were diluted to 1:1,000 in TMH ribose with 20 mM Mg2+ (high Mg2+) (C) or TMH ribose with 10 μM Mg2+ (low Mg2+) (D) and growth was recorded on a Bioscreen C (Growth Curves, USA) for 48 h. Data represent the average from three biological replicates.
To see if the variable biofilm formation in the strains with different phoP alleles was due to differences in growth or in biofilm production, a growth curve analysis was performed at ∼23°C in TMH medium supplemented with 0.2% ribose as the carbon source. In the high-Mg2+ TMH medium containing 20 mM MgCl2, the growth rates of wild-type and phoP-G215 strains were almost identical, while the ΔphoP strain showed a slightly reduced growth rate. However, in the low-Mg2+ TMH medium containing 10 μM MgCl2, the phoP-G215 strain showed slightly reduced growth compared to that of the wild-type strain. The growth of the ΔphoP strain was highly defective as expected, since PhoP function is essential for bacterial survival under the low-magnesium conditions (10). These results suggested that the subtle difference in biofilm formation at this temperature is likely due to the reduced growth rate of the phoP-G215 bacteria and that the S-to-G substitution in PhoP may have caused a small change in the functionality of the protein.
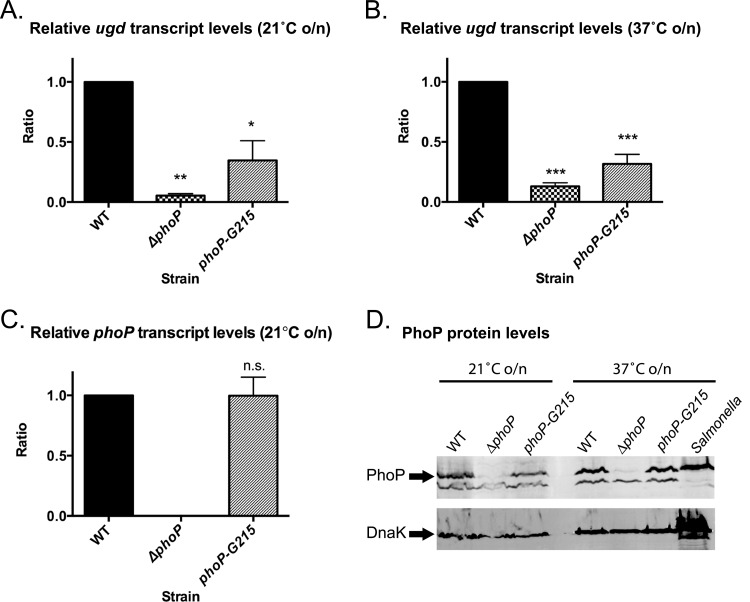
To determine if the amino acid change in PhoP has any effect on the activity of the protein, we compared the expression levels of a PhoP target gene, ugd, in the phoP variant strains. RNA samples were collected from the overnight cultures of Y. pestis strains grown at different temperatures in low-Mg2+ (20 μM MgCl2) TMH medium, and the ugd transcript levels were determined by quantitative reverse transcription-PCR (RT-qPCR). The results were normalized against 16S rRNA levels. An approximately 2-fold decrease in the ugd transcript level was observed in the phoP-G215 strain compared to that in the wild type (phoP-S215), and even less transcript was detected in the ΔphoP strain (Fig. 3A and B). Similar results were obtained when ugd expression levels were determined in logarithmically growing cultures (data not shown). The expression levels of other PhoP targets such as pmrH and mgtC showed similar trends, although the results were not statistically significant (data not shown). The transcript levels of phoP itself were not significantly different between the wild-type and the phoP-G215 strains (Fig. 3C), and the immunoblot of the bacterial cell lysates showed similar PhoP protein levels between these strains (Fig. 3D). Therefore, we concluded that the S-to-G change does not alter the expression level or the stability of PhoP but decreases the ability of PhoP to induce the transcription of its target, ugd.
FIG 3.
ugd transcript levels are decreased in Y. pestis KIM6+ phoP-G215, while phoP transcript or PhoP protein levels are unaffected. (A to C) mRNA levels of the PhoP-regulated gene ugd at 21°C (A) and 37°C (B), as well as phoP at 21°C (C), were determined in the indicated Y. pestis strains by RT-qPCR. The strains were grown overnight (o/n) at indicated temperatures in low-Mg2+ (20 μM MgCl2) TMH medium. The results were normalized against the 16S rRNA expression levels. Relative levels of expression (with the wild-type levels set at 1) are shown. Data represent the averages from at least three biological replicates. Error bars represent standard errors of the means (SEMs). The asterisks indicate significant differences compared to KIM6+ WT (*, P < 0.05; ***, P < 0.001) and “n.s.” denotes not significant statistically (P > 0.05) as determined by one-way ANOVA with Tukey's multiple-comparison tests. (D) PhoP protein levels of the indicated strains were compared by immunoblotting. Approximately 2 × 108 bacteria grown in low-Mg2+ TMH medium were lysed in 1× Laemmli sample buffer and loaded in each lane. DnaK protein levels were used as a loading control, and Salmonella Typhimurium was used as a positive control for the anti-PhoP antibody. At least three independent experiments were performed and the images of a representative blot are shown.
The decreased induction of the PhoP target gene ugd in the KIM6+ phoP-G215 strain is consistent with the idea that the PhoP-G215 protein has a decreased activity as a transcriptional regulator compared to that of the wild-type PhoP. To test if PhoP-G215 has a reduced ability to bind the target DNA sequence, electrophoretic mobility shift assays (EMSAs) were performed. Increasing amounts of purified PhoP proteins (either PhoP-S215 or PhoP-G215) were incubated with biotin-labeled PCR-amplified DNA fragments spanning the promoter regions of ugd and pmrH, and the ability of the protein to cause a gel shift was determined. As shown in Fig. 4, similar amounts of PhoP-S215 and PhoP-G215 proteins were required to retard the mobility of the ugd and pmrH promoter fragments. Similar gel shift patterns were observed when the PhoP proteins were preincubated with acetyl phosphate to produce phosphorylated PhoP (data not shown).
FIG 4.
WT PhoP and PhoP-G215 proteins have similar affinities to ugd and pmr promoters, as determined by electrophoretic mobility shift assays. Electrophoretic mobility shift assays (EMSAs) were performed using 1 nM biotin-labeled DNA fragments spanning the promoter regions of ugd (A) and pmrH (B) and increasing amounts (0 to 2 μM) of purified WT PhoP or PhoP-G215 proteins as indicated. The results shown are representative of three independent experiments.
The PhoP-regulated ugd and pmrHFIJKLM genes promote the resistance of Y. pestis to antimicrobial peptides by adding aminoarabinose to the lipid A portion of LPS (16). Since the S-to-G substitution in PhoP affects the transcription levels of ugd, we next determined the susceptibility of the phoP variant strains to the antimicrobial peptide polymyxin B. The strains were incubated in low-Mg2+ (20 μM MgCl2) TMH medium containing various concentrations of polymyxin B, and the MICs were determined. At 37°C, both the wild-type and the phoP-G215 strains were highly susceptible to polymyxin B. However, at 21°C and 28°C, the MICs of the phoP-G215 strain were approximately 4-fold lower than those of the wild type, showing that the phoP-G215 strain has an increased susceptibility to polymyxin B (Table 2). The control strain lacking phoP was completely susceptible to polymyxin B at all temperatures (Table 2).
TABLE 2.
Y. pestis phoP G215 mutant strain is more susceptible to antimicrobial peptide polymyxin Ba
| Strain | Polymyxin B MIC (μM) at: |
||
|---|---|---|---|
| 21°C | 28°C | 37°C | |
| KIM6+ WT | 7.81 | 31.25 | 0.49 |
| KIM6+ ΔphoP | 0.49 | 0.98 | ND |
| KIM6+ phoP-G215 | 0.98 | 3.91 | 0.49 |
Bacteria were grown for 20 h in low-Mg2+ TMH medium containing various concentrations of polymyxin B (0.12 to 125 μM) at the indicated temperatures, and the MICs were determined as the lowest concentration at which no bacterial growth was seen. At least three independent experiments were performed, and the results of one representative experiment are shown.
The increased susceptibility to antimicrobial peptides could influence the ability of Y. pestis to survive inside insect or mammalians hosts. As mammalian macrophages are known to produce cationic antimicrobial peptides such as CRAMPs to limit the intracellular growth of bacterial pathogens (32), we first tested whether the bacterial survival inside macrophages is affected in the phoP-G215 strain. We infected murine bone marrow-derived macrophages with the phoP-G215 and control strains that were pregrown at 28°C in a rich heart infusion (HI) medium and enumerated the intracellular bacteria at various times postinfection. No significant difference was detected between the wild-type and phoP-G215 strains in their ability to survive inside macrophages (data not shown).
We next compared the ability of the phoP variant strains to colonize the flea midgut in coinfection experiments. The flea vector Xenopsylla cheopis was fed with the 1:1 mixture of wild-type and the mutant bacteria in a blood meal, and the percentage of each strain colonizing the midgut was determined at various times postinfection. When the wild-type (phoP-S215) and the phoP-G215 strains were coinfected, similar numbers of bacteria from each strain were recovered from the flea midgut at 7 days postinfection, showing that neither strain had a significant competitive advantage over the other (Fig. 5). In contrast, wild-type bacteria dominated when the wild-type and the ΔphoP strains were coinfected, suggesting that phoP function may be necessary for the successful survival and colonization of the bacteria in the flea midgut (Fig. 5).
FIG 5.
KIM6+ ΔphoP mutant is outcompeted by the wild type during flea coinfection, while KIM6+ phoP-G215 can colonize the flea equally well as the wild type. Cohorts of Xenopsylla cheopis fleas were fed on a blood meal containing a 1:1 mixture of wild-type (KIM6+ harboring an ampicillin/carbenicillin resistance cassette on plasmid pGFP [KIM6+/pGFP]) and the indicated phoP mutant or wild-type KIM6+ Y. pestis strains. At days 0 (T0) and 7 (T7) postinfection, Y. pestis cells colonizing the flea midgut were enumerated and the percentages of each strain colonizing the midgut were determined. For each coinfection experiment at a given time point, CFU was determined for 20 to 25 fleas that had fed on infected blood. Data points showing that 0 CFU were recovered from a flea were removed from the analysis (there was no apparent difference among three phoP variant strains in the number of fleas that gave 0 CFU.) Data were plotted as the percentage of the KIM6+/pGFP recovered from each coinfected flea. Error bars represent means ± standard deviations (SDs) from the percentage CFU data. Bars indicate the two groups that were assessed for statistical differences.
DISCUSSION
The aim of this study was to compare two naturally occurring alleles of phoP in Y. pestis that result in the S-to-G single amino acid substitution at position 215 of the PhoP response regulator. We found that this amino acid substitution has small but reproducible effects on PhoP functions. Compared to the parental KIM6+ strain carrying the modern allele (phoP-S215), a derivative of KIM6+ carrying the basal allele (phoP-G215) exhibited slightly defective growth under a low-Mg2+ condition and decreased transcription of a PhoP target gene, ugd, as well as an ∼8-fold increase in the susceptibility to the antimicrobial peptide polymyxin B. The increased susceptibility to polymyxin B is likely due to the decreased aminoarabinose modification of lipid A, since ugd encodes an enzyme required for this process (16). Given the known roles of Y. pestis PhoP in growth under low-Mg2+ conditions and resistance against antimicrobial peptides (9–12), our results suggest that the S-to-G amino acid substitution has led to the slight reduction in the activity of PhoP protein.
Residue 215 of the PhoP protein is predicted to be located within the DNA binding domain that is likely to contact the target DNA (10, 31). A similar single amino acid variation exists in the equivalent residue (amino acid 211) of a related response regulator, PmrA, in Salmonella (31). The PmrA protein from Salmonella enterica serovar Paratyphi B has a glutamate residue at position 211 (PmrA E211), whereas most other S. enterica strains have glycines at this position (PmrA G211). The purified PmrA E211 protein was shown to bind DNA less efficiently than PmrA G211 and have a decreased ability to activate PmrA-dependent genes, including the pmrCAB operon itself (31). It was proposed that this is partly because a negatively charged glutamate at position 211 does not bind strongly to DNA, which is also negatively charged (31). Similarly, the decreased ugd expression in KIM6+ phoP-G215 may be caused by the lower binding affinity of PhoP-G215 to the ugd promoter, as the polar serine residue may have a stronger affinity for DNA than the nonpolar glycine. Our EMSAs did not reveal an observable difference in the abilities of PhoP-S215 and PhoP-G215 proteins to bind the promoters of ugd or pmrH. The change in the DNA affinity due to this single amino acid substitution may be too small to be detected by EMSA under the conditions tested. This possibility is consistent with our finding that the changes in the expression levels of PhoP target genes are also small between the phoP-S215 and phoP-G215 strains. Alternatively, the amino acid substitution could affect the transcription of PhoP-regulated genes more indirectly in vivo, involving other factors. For example, it could influence the phosphorylation status of PhoP, thereby affecting the activity of the transcription factor. In Salmonella, the G211E substitution in PmrA changes not only the DNA affinity but also the levels of phosphorylated PmrA in vivo, likely due to the altered efficiency of the PmrD protein to protect PmrA from dephosphorylation (31). The amino acid substitution in PhoP could also alter its interaction with RNA polymerase via a conformational change, potentially affecting the transcription of the downstream genes (33). More sensitive and quantitative assays for the DNA affinity and/or assays that can detect the PhoP/DNA interaction in vivo (e.g., chromatin coimmunoprecipitation) might reveal the mechanism for differential gene regulation by PhoP-S215 and PhoP-G215 proteins.
The reduced ugd expression in the phoP-G215 strain suggests that the expression levels of other PhoP-regulated genes also may be altered. It would be interesting to see if the S-to-G amino acid substitution results in a uniform decrease in the expression of all the PhoP-induced genes or the alteration of the repertoire of genes activated by this response regulator. Interestingly, the levels of phoP RNA as well as PhoP protein were unchanged in the phoP-G215 strain even though the autoregulation of PhoP has been reported in both Salmonella and Y. pestis (34, 35). Further studies should reveal the extent of the effects the S215G amino acid substitution causes on the PhoP regulon.
The resistance to cationic antimicrobial peptides is important for many pathogens, since both insect and mammalian hosts produce a variety of them to combat infectious agents (36). Our finding that the phoP-G215 strain is more susceptible to polymyxin B than the phoP-S215 strain raises a possibility that the acquisition of the phoP-S215 allele contributed to the increased virulence of the modern Y. pestis strains through increased resistance to antimicrobial peptides. Consistent with this idea, Anisimov et al. reported previously that Y. pestis subsp. microtus bv. caucasica, a biovar that is virulent in mice but not in guinea pigs or humans, also carries the phoP-G215 allele and shows increased susceptibility to polymyxin B at 25 to 28°C (37, 38). However, so far, we have been unable to establish that the strains carrying phoP-S215 have increased fitness in the host environments. In our coinfection experiment using the flea host, the wild-type (S215) strain did not exhibit any competitive advantage over the phoP-G215 strain in colonizing the flea midgut. We also did not detect a difference between the S215 and G215 strains in their ability to survive inside murine macrophages, although our experiment did not use the bacteria grown in low-Mg2+ TMH medium (data not shown). Also unknown is the effect of this amino acid variation in the virulence of Y. pestis in mammalian hosts. Previous infection studies using mouse bubonic plague models showed that even the complete loss of PhoP function results in only mild attenuation (11, 13). Therefore, the single amino acid change in PhoP may not cause a dramatic attenuation in these models. Further studies are needed to determine if this allelic variation causes any changes in the ability of Y. pestis to infect and colonize mammalian hosts or to be transmitted between hosts.
In previous single-strain infection studies, phoP was shown to play roles in biofilm formation and the blocking of the proventriculus in fleas but not in the infectivity of the bacteria, since a ΔphoP strain exhibited bacterial loads and infection rates in the flea midgut similar to those of the wild type (14). However, our coinfection studies showed that a phoP deletion strain is outcompeted by the wild-type strain in the flea midgut. To our knowledge, this is the first study that directly indicated a potential role of phoP in the survival of Y. pestis within the flea digestive tract. Our results are consistent with the finding in a Drosophila melanogaster model for the Y. pestis colonization of insect vectors that the loss of phoP results in decreased colonization of the midgut of fly larvae (39). Although the mechanism by which phoP promotes bacterial survival in the fleas is unclear, it may be a subtle role that is not easily detected in a single-strain infection assay. It has been demonstrated that the Y. pestis strain lacking ugd is not defective in either flea infection or blockage, suggesting that the lipid A modification is not involved in these processes (14). However, this was determined in single-strain infection assays and not coinfection assays. A recent study reported that Y. pestis lacking other genes in the lipid A modification pathway (galU and arnB) is defective in colonizing the flea midgut (40). In addition, the aforementioned Drosophila model of Y. pestis colonization showed that the decreased colonization of fly larvae by the phoP mutant strain can be rescued by the inactivation of the host imd, the gene that drives the production of antimicrobial peptides (39). Therefore, we cannot rule out the possibility that the lipid A modification and the resistance to antimicrobial peptide play some role in Y. pestis survival in the flea. Alternatively, the flea colonization by Y. pestis may depend on phoP-dependent pathways other than for lipid A modification, such as those for metabolism (19, 41) or the formation of a cohesive biofilm (14).
Recent studies have shown that several SNPs and other small genetic changes have played critical roles in the emergence of modern Y. pestis strains during the evolution of the pathogen (27, 28). However, it is difficult to determine whether a given genetic change/polymorphism has been fixed in the modern Y. pestis genomes because it confers a selective advantage (i.e., positive selection) or it is simply the result of neutral genetic drift. Indeed, it has been argued that most Y. pestis SNPs fall into the latter category (29). Accordingly, the presence of the phoP-S215 allele in modern Y. pestis strains could have little to do with any putative selective advantage. Nevertheless, our observation that the phoP-S215 allele renders the bacteria more resistant to polymyxin B is consistent with the idea that this allele confers a slight fitness advantage in hostile host environments and that the acquisition of this allele might have contributed to the increased virulence of the modern Y. pestis strains. The evolution of bacteria can be driven by changes in the gene regulatory network due to the altered functions of transcription regulators, because such changes can affect the ecological niche in which the pathogen can thrive (33). Therefore, it is possible that the subtle changes in PhoP can influence the adaptability or the specificity of Y. pestis to its hosts through the modification of its regulon.
In addition to phoP, a comparison of the genome sequences of Y. pestis subsp. microtus 91001 and Y. pestis ESS revealed a number of SNPs in genes implicated in virulence, such as hmsT, iucD, and ail (2). Interestingly, Y. pestis bv. caucasica, which carries the same ail allele as Y. pestis subsp. microtus 91001, is reported to be less resistant to human serum than strain KIM, suggesting that this SNP may also contribute to the increased virulence of modern Y. pestis to humans (37). Perhaps the SNP in phoP is one of many small genetic changes that yielded subtle effects on the pathogenic characteristics of Y. pestis, and it is the combinatorial effects of multiple mutations that have led to more noticeable differences in pathogen virulence.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The Y. pestis strains used in this study (Table 1) were derived from strain KIM (molecular group 2.MED [21]). All Y. pestis strains used did not contain the pCD1 plasmid, and are therefore avirulent and exempt from tier 1 select agent guidelines. Unless otherwise indicated, Y. pestis strains were cultivated at 28°C on heart infusion ([HI] Difco) plates, and cultures were grown in HI broth or the chemically defined TMH medium (42) containing 20 μM magnesium chloride (low-Mg2+ TMH) with aeration at 21°C (the room temperature of the Bliska lab), 28°C, or 37°C. The growth medium for Y. pestis was supplemented with kanamycin at 25 μg/ml or chloramphenicol at 10 μg/ml when appropriate. Y. pestis cells from the 21°C cultures were collected by scraping off the biofilm that had formed on the surface of the glass tube at the air-liquid interface, as there were not enough bacteria in the liquid suspension. The Escherichia coli strain S17-1 used for plasmid and mutant construction was grown as described previously (10). The Salmonella enterica serovar Typhimurium strain ATCC 14028 was a gift from M. Starnbach (Table 1). It was grown on Luria-Bertani (LB) agar plates at 37°C and cultured in LB or low-Mg2+ TMH medium with aeration at 37°C. To isolate logarithmically growing bacteria, the overnight cultures were subcultured in fresh medium and bacteria were grown with aeration for 4 h at indicated temperatures.
Construction of KIM6+ phoP-G215 mutants.
The phoP open reading frame was amplified from KIM6+ by PCR using primers phoP-F2 and phoP-R (Table 3), digested with BamHI and NotI, and ligated into the pSB890 suicide plasmid (43) that also had been cut with BamHI and NotI. The resulting plasmid was subjected to site-directed mutagenesis to introduce a serine-to-glycine substitution at position 215 of PhoP protein, using primers phoP-mut1 and phoP-mut2 (Table 3) and a QuikChange site-directed mutagenesis kit (Stratagene/Agilent Technologies) according to the manufacturer's instructions. The mutagenesis also introduced a silent mutation that creates an additional MscI restriction site in the phoP-G215 allele for easy recognition. The suicide plasmid carrying phoP-G215 was mated into wild-type KIM6+, and the KIM6+ phoP-G215 strain was created by allelic exchange as described previously (10).
TABLE 3.
List of primers used in this study
| Primer | Sequence (5′→3′) | Reference |
|---|---|---|
| phoP-F2a | TCTAGCGGCCGCGTGCTCAAGACGGTATAGCACC | |
| phoP-Ra | TTAGGATCCAGAGGTCGCGGCGTAGGTATTG | |
| phoP-mut1b | AAGTCATTACGACTATTCGTGGCCAGGGATATCGTTTTGAC | |
| phoP-mut2b | GTCAAAACGATATCCCTGGCCACGAATAGTCGTAATGACTT | |
| phoP-qRTF | TGTTGCGTCACCATCTGACA | |
| phoP-qRTR | TCACCGGGCAAACCAAGAT | |
| ugd-RTF | GCTCCGTTGGTCAAAGAAAA | |
| ugd-RTR | CCGTCTTCATCAGGAGGTGT | |
| yr005 (16S rRNA)_f | TGAACCCAGATGGGATTAGC | 51 |
| yr005 (16S rRNA)_r | CGCTTTACGCCCAGTAATTC | 51 |
| mgtC-RTF | GCACAGATTGTTTCGGGAAT | |
| mgtC-RTR | GACCACAAAGCACACCAATG | |
| pmrH-RTF | CATGCGATCGCTGTATGTTC | |
| pmrH-RTR | CACGTCTGTGATGGCGTAAT | |
| ugd-EMSA-F2 (CO92-YPO2174EMSA-F) | biotin(TEG)-GCTTAACAATGGTGTCCC | 17 |
| ugd-EMSA-R2 (CO92-YPO2174EMSA-R) | biotin(TEG)-ACTCCCAGTGATTATCGG | 17 |
| pmrH-EMSA-F1 | biotin(TEG)-GCGTTAAATCCAACTCATTG | |
| pmrH-EMSA-R1 | biotin(TEG)-GCGATCGCATGTTTGCAAC |
Underlined letters represent specific restriction sites added to the primer.
Letters in boldface font represent specific nucleotide changes that were introduced by the site-directed mutagenesis, and the underlined letters show the MscI restriction site that has been introduced by the change.
Growth curves.
Growth analysis was performed at ambient room temperature ∼23°C in the Vadyvaloo lab) using TMH medium supplemented with 0.2% ribose as the carbon source. Ribose was added because a transcriptional analysis of Y. pestis from blocked fleas suggests that Y. pestis specifically takes up pentose sugars, including ribose, during development in the flea digestive tract (41). The strains were cultured successively over two nights, first in brain heart infusion (BHI) medium and then in TMH ribose containing 20 mM Mg2+ (high Mg2+). On day three, the strains were diluted 1:100 in high-Mg2+ TMH ribose medium or TMH ribose containing 10 μM Mg2+ (low Mg2+), and growth without shaking was recorded on a Bioscreen C (Growth Curves, USA) for 48 h. Growth curves were plotted using GraphPad Prism 5.
Polymyxin B MIC assay.
The MIC of polymyxin B for Y. pestis was determined as previously described (44), except for the following modifications. Polymyxin B (Sigma) was serially diluted 2-fold into low-Mg2+ (20 μM Mg2+) TMH medium at concentrations ranging from 250 to 0.24 μM, and 100 μl of the dilution was transferred to the wells of the 96-well plate. Overnight cultures of Y. pestis were grown at 21°C, 28°C, or 37°C in low-Mg2+ TMH medium and diluted in fresh low-Mg2+ TMH medium, and 100 μl of the dilution was added to the wells containing a TMH-polymyxin B solution at a final concentration of ∼1 × 106 CFU/ml. The plates were incubated at 21°C, 28°C, or 37°C, respectively, for 20 h with shaking in a C24 shaker (New Brunswick Scientific) at 200 rpm. The bacterial growth in each was examined visually and compared to that of a control grown in the medium with no polymyxin B.
RT-qPCR.
Relative transcription levels of phoP and ugd and other PhoP target genes were determined by quantitative reverse transcription-PCR (RT-qPCR). The primers used for RT-qPCR were designed using the Primer3 Plus program (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) (45) and are listed in Table 3. Total bacterial RNA was stabilized and isolated from overnight cultures or logarithmically growing cultures of Y. pestis grown in low-Mg2+ (20 μM MgCl2) TMH medium at the indicated temperatures (21°C, 28°C, or 37°C). To obtain logarithmically growing cultures, the overnight cultures were diluted in low-Mg2+ TMH medium to an optical density at 600 nm (OD600) of ∼0.1, and incubation was continued at the same temperatures for 4 h with aeration. For the stabilization of RNA, 2 to 5 ml of each culture (∼1 × 109 cells) was centrifuged, and the cell pellet was mixed immediately with 1 ml of RNAprotect Bacteria reagent (Qiagen) and incubated at room temperature for 5 min. The cells were then harvested by centrifugation at 4,000 × g for 10 min. The cell pellets were used immediately for RNA isolation or stored at −80°C until use. Total RNA was isolated using a Qiagen RNeasy Mini kit, and the on-column digestion of genomic DNA was performed using RNase-free DNase (Qiagen) as described previously (15).
cDNA synthesis was performed with 0.6 μg of total RNA and 300 ng of random hexamers (Invitrogen), using SuperScript III reverse transcriptase according to the manufacturer's instructions, except that the reaction was carried out for ∼8 h at 42°C. Each of the 25-μl qPCR mixtures contained 2.0 μl of a 1:20 dilution of cDNA, 0.3 μM gene-specific primers, and 12.5 μl of 2× QuantiTect SYBR green master mix (Qiagen). The reaction was performed according to the manufacturer's instructions and monitored using an ABI Prism Applied Biosystems 7300 SDS real-time PCR machine. The expression level for each gene was calculated using standard curves, and the results were normalized to Y. pestis 16S rRNA. RT-qPCR was performed on at least three independent RNA samples.
Detection of PhoP protein by immunoblotting.
Approximately 2 × 108 bacteria (as determined by the OD600) grown in low-Mg2+ (20 μM MgCl2) TMH medium were lysed in 1× Laemmli sample buffer and loaded on a 12% SDS-PAGE gel. The blots were probed with a 1:2,000 dilution of a rabbit anti-Salmonella PhoP antibody (a gift from E. Groisman) in Tris-buffered saline–0.05% Tween 20 (TBST) containing 1% casein for 2 h at room temperature. The Salmonella enterica serovar Typhimurium strain ATCC 14028 (Table 1) was run as a positive control for the anti-PhoP antibody. As a loading control, a mouse anti-DnaK monoclonal antibody (Enzo) was used at a 1:500 dilution. Secondary antibodies, an IRDye680-conjugated anti-IgG rabbit antibody and an IRDye800-conjugated anti-IgG mouse antibody (LI-COR), were used at dilutions of 1:5,000 and 1:10,000, respectively. The infrared signal from each band was detected and quantified using an infrared imaging system (Odyssey; LI-COR).
Purification of His-tagged PhoP and its derivative mutant.
His-tagged Y. pestis PhoP proteins were expressed in E. coli BL21(DE) containing either pT7.7-PhoP yersinia (33) (a gift from E. Groisman) or pT7.7-PhoP-G215 yersinia expression vectors. pT7.7-PhoP-G215 yersinia was created from pT7.7-PhoP yersinia carrying wild-type PhoP (PhoP-S215) by site-directed mutagenesis, using phoP-mut1/phoP-mut2 primers (Table 3) and a QuikChange site-directed mutagenesis kit (Stratagene/Agilent Technologies). Overnight cultures of these expression strains were diluted to an OD600 of 0.1 in 50 to 150 ml LB medium supplemented with 100 μg/ml ampicillin and grown at 37°C until reaching an OD600 of ∼0.6. Then, isopropyl-1-thio-B-d-galacopyranoside (IPTG) was added to a final concentration of 100 μM and incubated at 30°C for an additional 4 h. The cells were harvested and suspended in lysis buffer (50 mM Tris [pH 8.0], 100 mM NaCl, 10 mM imidazole) and disrupted by sonication. The insoluble fraction was pelleted and removed by centrifugation at 14,000 rpm for 20 min. PhoP protein was purified using a His-Bind purification kit (Novagen) according to the manufacturer's instructions, except that the lysis buffer described above was used instead of the binding buffer provided by the kit. The purified protein was concentrated and the buffer was exchanged into the storage buffer (50 mM Tris [pH 7.6], 100 mM NaCl, 10% glycerol, 10 mM MgCl2, 0.1 mM EDTA, 1 mM dithiothreitol [DTT]) using Amicon Ultra centrifugal filter units (molecular weight cutoff [MWCO] 10; Millipore).
EMSAs.
Biotin-labeled DNA fragments spanning the promoter regions of PhoP target genes ugd and pmrH were generated by PCR using Y. pestis KIM6+ genomic DNA as a template. The upstream sequence of ugd containing the consensus PhoP box was amplified using primers ugd-EMSA-F2 (CO92-YPO2174EMSA-F) and ugd-EMSA-R2 (CO92-YPO2174EMSA-F) (17) (Table 3), and that of pmrH was amplified using primers pmrH-EMSA-F1 and pmrH-EMSA-R1 (Table 3), resulting in 405-bp and 386-bp fragments, respectively. The PCR products were gel purified with the QIAquick PCR purification kit (Qiagen). The biotin-labeled DNA fragment and the purified PhoP protein were mixed with binding buffer in a total volume of 20 μl and incubated at room temperature for 20 min. The binding reaction mixture contained 20 fmol (1 nM) of the labeled DNA target, various amounts (0 to 40 pmol or 0 to 2 μM) of PhoP protein, 25 mM Tris-HCl (pH 7.6), 50 mM NaCl, 50 μg/ml poly(dI-dC), 5 mM MgCl2, 50 μg/ml bovine serum albumin [BSA], 0.05 mM EDTA, 0.5 mM DTT, and 5% glycerol. Samples were separated on a 6% nondenaturing polyacrylamide gel in 0.5× Tris-borate-EDTA (TBE) buffer at 4°C and transferred to a nitrocellulose membrane (Thermo Fisher). The biotin-labeled DNA fragments were detected using a LightShift chemiluminescent EMSA kit (Thermo Fisher) according to the manufacturer's instructions. To phosphorylate PhoP proteins, purified PhoP was incubated with 20 mM fresh acetyl phosphate (Sigma) for 2.5 h (33) at room temperature or 37°C prior to the EMSA.
Flea coinfection experiments.
For flea coinfections, the KIM6+/pGFP strain carrying a carbenicillin/ampicillin antibiotic selective marker was coinfected with either the KIM6+ wild-type, phoP mutant, or phoP-G215 strains. Bacteria were grown in BHI broth overnight, first at 28°C with aeration and then at 37°C without aeration. Bacteria were harvested by centrifugation. The two strains were mixed together in a 1:1 ratio and used to seed prewarmed mouse blood (Bioreclamation) to a concentration of ∼5 × 108 cells/ml per strain. A cohort of Xenopsylla cheopis fleas was allowed to feed on the infected blood using a previously described artificial feeding chamber (46). Fleas that ingested a blood meal were maintained at 21°C and 75% relative humidity and fed twice a week on uninfected mice. The total Y. pestis bacterial load per flea was determined from 20 to 25 infected fleas at 0 and 7 days postinfection by plating on Yersinia selective agar base (YSAB) to determine CFU. Simultaneously, plating was carried out on YSAB plus 100 μg/ml carbenicillin to select for the KIM6+/pGFP strain. The percentage of KIM6+/pGFP cells was calculated from the CFU counts. For each experiment, the CFU was determined only on the fleas that were microscopically confirmed to contain red blood in their midguts at time zero. The data points were removed from the calculation of the average CFU and the graph if 0 CFU were recovered. In situations where a slightly higher CFU count was obtained in the plate containing antibiotic than in the one without antibiotic (i.e., >100% KIM6+/pGFP values indicating that fleas contained KIM6+/pGFP only), the data points were plotted at 100%.
Macrophage infection and analysis of bacterial survival.
Murine bone marrow-derived macrophages (BMMs) were cultured and infected with Y. pestis as described previously (10, 47). The bacterial survival inside macrophages was quantified by CFU assays at 0, 0.25, 1.5, 4, and 24 h postinfection as described previously (47), with the following modifications. BMMs were seeded in 24-well tissue culture plates at a concentration of 1.5 × 105 cells/well and infected with Y. pestis at a multiplicity of infection (MOI) of 5. The infection was performed with Y. pestis grown overnight at 28°C in HI broth. For CFU assays, the results were log transformed prior to statistical analysis.
Statistical analyses of the data.
When appropriate, statistical tests were performed using a one-way analysis of variance (ANOVA) with Tukey's multiple-comparison test (Prism version 4.0c for Macintosh; GraphPad Software, San Diego, CA). For flea coinfections, a one-way ANOVA with Bonferroni's multiple-comparison test was applied to the data (GraphPad Prism version 5).
Ethics statement.
Animal work (the flea infection study) was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols (04001 and 04524) were approved by the Committee on the Ethics of Animal Experiments at Washington State University.
ACKNOWLEDGMENTS
We thank Eduardo Groisman and Jinki Yeom for providing anti-Salmonella PhoP antibody and pT7.7-his-PhoP expression plasmid and for their advice on immunoblotting. We also thank Michael Starnbach for providing Salmonella enterica serovar Typhimurium 14028, Athena Lemon for performing the Y. pestis growth curve assays, and Caitlin Unkenholz for help with purification of PhoP proteins.
REFERENCES
- 1.Wagner DM, Klunk J, Harbeck M, Devault A, Waglechner N, Sahl JW, Enk J, Birdsell DN, Kuch M, Lumibao C, Poinar D, Pearson T, Fourment M, Golding B, Riehm JM, Earn DJ, Dewitte S, Rouillard JM, Grupe G, Wiechmann I, Bliska JB, Keim PS, Scholz HC, Holmes EC, Poinar H. 2014. Yersinia pestis and the plague of Justinian 541–543 AD: a genomic analysis. Lancet Infect Dis 14:319–326. doi: 10.1016/S1473-3099(13)70323-2. [DOI] [PubMed] [Google Scholar]
- 2.Bos KI, Schuenemann VJ, Golding GB, Burbano HA, Waglechner N, Coombes BK, McPhee JB, DeWitte SN, Meyer M, Schmedes S, Wood J, Earn DJ, Herring DA, Bauer P, Poinar HN, Krause J. 2011. A draft genome of Yersinia pestis from victims of the Black Death. Nature 478:506–510. doi: 10.1038/nature10549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perry RD, Fetherston JD. 1997. Yersinia pestis—etiologic agent of plague. Clin Microbiol Rev 10:35–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chouikha I, Hinnebusch BJ. 2012. Yersinia–flea interactions and the evolution of the arthropod-borne transmission route of plague. Curr Opin Microbiol 15:239–246. doi: 10.1016/j.mib.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Groisman EA. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J Bacteriol 183:1835–1842. doi: 10.1128/JB.183.6.1835-1842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.García Véscovi E, Soncini FC, Groisman EA. 1996. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84:165–174. doi: 10.1016/S0092-8674(00)81003-X. [DOI] [PubMed] [Google Scholar]
- 7.Prost LR, Daley ME, Le Sage V, Bader MW, Le Moual H, Klevit RE, Miller SI. 2007. Activation of the bacterial sensor kinase PhoQ by acidic pH. Mol Cell 26:165–174. doi: 10.1016/j.molcel.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, Miller SI. 2005. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122:461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 9.Rebeil R, Ernst RK, Gowen BB, Miller SI, Hinnebusch BJ. 2004. Variation in lipid A structure in the pathogenic yersiniae. Mol Microbiol 52:1363–1373. doi: 10.1111/j.1365-2958.2004.04059.x. [DOI] [PubMed] [Google Scholar]
- 10.Grabenstein JP, Marceau M, Pujol C, Simonet M, Bliska JB. 2004. The response regulator PhoP of Yersinia pseudotuberculosis is important for replication in macrophages and for virulence. Infect Immun 72:4973–4984. doi: 10.1128/IAI.72.9.4973-4984.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oyston PC, Dorrell N, Williams K, Li SR, Green M, Titball RW, Wren BW. 2000. The response regulator PhoP is important for survival under conditions of macrophage-induced stress and virulence in Yersinia pestis. Infect Immun 68:3419–3425. doi: 10.1128/IAI.68.6.3419-3425.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hitchen PG, Prior JL, Oyston PC, Panico M, Wren BW, Titball RW, Morris HR, Dell A. 2002. Structural characterization of lipo-oligosaccharide (LOS) from Yersinia pestis: regulation of LOS structure by the PhoPQ system. Mol Microbiol 44:1637–1650. doi: 10.1046/j.1365-2958.2002.02990.x. [DOI] [PubMed] [Google Scholar]
- 13.Bozue J, Mou S, Moody KL, Cote CK, Trevino S, Fritz D, Worsham P. 2011. The role of the phoPQ operon in the pathogenesis of the fully virulent CO92 strain of Yersinia pestis and the IP32953 strain of Yersinia pseudotuberculosis. Microb Pathog 50:314–321. doi: 10.1016/j.micpath.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Rebeil R, Jarrett CO, Driver JD, Ernst RK, Oyston PC, Hinnebusch BJ. 2013. Induction of the Yersinia pestis PhoP-PhoQ regulatory system in the flea and its role in producing a transmissible infection. J Bacteriol 195:1920–1930. doi: 10.1128/JB.02000-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grabenstein JP, Fukuto HS, Palmer LE, Bliska JB. 2006. Characterization of phagosome trafficking and identification of PhoP-regulated genes important for survival of Yersinia pestis in macrophages. Infect Immun 74:3727–3741. doi: 10.1128/IAI.00255-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winfield MD, Latifi T, Groisman EA. 2005. Transcriptional regulation of the 4-amino-4-deoxy-l-arabinose biosynthetic genes in Yersinia pestis. J Biol Chem 280:14765–14772. doi: 10.1074/jbc.M413900200. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Gao H, Qin L, Li B, Han Y, Guo Z, Song Y, Zhai J, Du Z, Wang X, Zhou D, Yang R. 2008. Identification and characterization of PhoP regulon members in Yersinia pestis biovar Microtus. BMC Genomics 9:143. doi: 10.1186/1471-2164-9-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou D, Han Y, Qin L, Chen Z, Qiu J, Song Y, Li B, Wang J, Guo Z, Du Z, Wang X, Yang R. 2005. Transcriptome analysis of the Mg2+-responsive PhoP regulator in Yersinia pestis. FEMS Microbiol Lett 250:85–95. doi: 10.1016/j.femsle.2005.06.053. [DOI] [PubMed] [Google Scholar]
- 19.Vadyvaloo V, Viall AK, Jarrett CO, Hinz AK, Sturdevant DE, Joseph Hinnebusch B. 2015. Role of the PhoP-PhoQ gene regulatory system in adaptation of Yersinia pestis to environmental stress in the flea digestive tract. Microbiology 161:1198–1210. doi: 10.1099/mic.0.000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knirel YA, Dentovskaya SV, Bystrova OV, Kocharova NA, Senchenkova SN, Shaikhutdinova RZ, Titareva GM, Bakhteeva IV, Lindner B, Pier GB, Anisimov AP. 2007. Relationship of the lipopolysaccharide structure of Yersinia pestis to resistance to antimicrobial factors. Adv Exp Med Biol 603:88–96. doi: 10.1007/978-0-387-72124-8_7. [DOI] [PubMed] [Google Scholar]
- 21.Achtman M, Morelli G, Zhu P, Wirth T, Diehl I, Kusecek B, Vogler AJ, Wagner DM, Allender CJ, Easterday WR, Chenal-Francisque V, Worsham P, Thomson NR, Parkhill J, Lindler LE, Carniel E, Keim P. 2004. Microevolution and history of the plague bacillus, Yersinia pestis. Proc Natl Acad Sci U S A 101:17837–17842. doi: 10.1073/pnas.0408026101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morelli G, Song Y, Mazzoni CJ, Eppinger M, Roumagnac P, Wagner DM, Feldkamp M, Kusecek B, Vogler AJ, Li Y, Cui Y, Thomson NR, Jombart T, Leblois R, Lichtner P, Rahalison L, Petersen JM, Balloux F, Keim P, Wirth T, Ravel J, Yang R, Carniel E, Achtman M. 2010. Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nat Genet 42:1140–1143. doi: 10.1038/ng.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rasmussen S, Allentoft ME, Nielsen K, Orlando L, Sikora M, Sjogren KG, Pedersen AG, Schubert M, Van Dam A, Kapel CM, Nielsen HB, Brunak S, Avetisyan P, Epimakhov A, Khalyapin MV, Gnuni A, Kriiska A, Lasak I, Metspalu M, Moiseyev V, Gromov A, Pokutta D, Saag L, Varul L, Yepiskoposyan L, Sicheritz-Ponten T, Foley RA, Lahr MM, Nielsen R, Kristiansen K, Willerslev E. 2015. Early divergent strains of Yersinia pestis in Eurasia 5,000 years ago. Cell 163:571–582. doi: 10.1016/j.cell.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Achtman M, Zurth K, Morelli G, Torrea G, Guiyoule A, Carniel E. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc Natl Acad Sci U S A 96:14043–14048. doi: 10.1073/pnas.96.24.14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wren BW. 2003. The yersiniae—a model genus to study the rapid evolution of bacterial pathogens. Nat Rev Microbiol 1:55–64. doi: 10.1038/nrmicro730. [DOI] [PubMed] [Google Scholar]
- 26.Chain PS, Carniel E, Larimer FW, Lamerdin J, Stoutland PO, Regala WM, Georgescu AM, Vergez LM, Land ML, Motin VL, Brubaker RR, Fowler J, Hinnebusch J, Marceau M, Medigue C, Simonet M, Chenal-Francisque V, Souza B, Dacheux D, Elliott JM, Derbise A, Hauser LJ, Garcia E. 2004. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc Natl Acad Sci U S A 101:13826–13831. doi: 10.1073/pnas.0404012101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zimbler DL, Schroeder JA, Eddy JL, Lathem WW. 2015. Early emergence of Yersinia pestis as a severe respiratory pathogen. Nat Commun 6:7487. doi: 10.1038/ncomms8487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun YC, Jarrett CO, Bosio CF, Hinnebusch BJ. 2014. Retracing the evolutionary path that led to flea-borne transmission of Yersinia pestis. Cell Host Microbe 15:578–586. doi: 10.1016/j.chom.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui Y, Yu C, Yan Y, Li D, Li Y, Jombart T, Weinert LA, Wang Z, Guo Z, Xu L, Zhang Y, Zheng H, Qin N, Xiao X, Wu M, Wang X, Zhou D, Qi Z, Du Z, Wu H, Yang X, Cao H, Wang H, Wang J, Yao S, Rakin A, Li Y, Falush D, Balloux F, Achtman M, Song Y, Wang J, Yang R. 2013. Historical variations in mutation rate in an epidemic pathogen, Yersinia pestis. Proc Natl Acad Sci U S A 110:577–582. doi: 10.1073/pnas.1205750110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song Y, Tong Z, Wang J, Wang L, Guo Z, Han Y, Zhang J, Pei D, Zhou D, Qin H, Pang X, Han Y, Zhai J, Li M, Cui B, Qi Z, Jin L, Dai R, Chen F, Li S, Ye C, Du Z, Lin W, Wang J, Yu J, Yang H, Wang J, Huang P, Yang R. 2004. Complete genome sequence of Yersinia pestis strain 91001, an isolate avirulent to humans. DNA Res 11:179–197. doi: 10.1093/dnares/11.3.179. [DOI] [PubMed] [Google Scholar]
- 31.Chen HD, Jewett MW, Groisman EA. 2012. An allele of an ancestral transcription factor dependent on a horizontally acquired gene product. PLoS Genet 8:e1003060. doi: 10.1371/journal.pgen.1003060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenberger CM, Gallo RL, Finlay BB. 2004. Interplay between antibacterial effectors: a macrophage antimicrobial peptide impairs intracellular Salmonella replication. Proc Natl Acad Sci U S A 101:2422–2427. doi: 10.1073/pnas.0304455101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perez JC, Groisman EA. 2009. Transcription factor function and promoter architecture govern the evolution of bacterial regulons. Proc Natl Acad Sci U S A 106:4319–4324. doi: 10.1073/pnas.0810343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soncini FC, Vescovi EG, Groisman EA. 1995. Transcriptional autoregulation of the Salmonella typhimurium phoPQ operon. J Bacteriol 177:4364–4371. doi: 10.1128/jb.177.15.4364-4371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Wang L, Han Y, Yan Y, Tan Y, Zhou L, Cui Y, Du Z, Wang X, Bi Y, Yang H, Song Y, Zhang P, Zhou D, Yang R. 2013. Autoregulation of PhoP/PhoQ and positive regulation of the cyclic AMP receptor protein-cyclic AMP complex by PhoP in Yersinia pestis. J Bacteriol 195:1022–1030. doi: 10.1128/JB.01530-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hancock RE, Lehrer R. 1998. Cationic peptides: a new source of antibiotics. Trends Biotechnol 16:82–88. doi: 10.1016/S0167-7799(97)01156-6. [DOI] [PubMed] [Google Scholar]
- 37.Anisimov AP, Dentovskaya SV, Titareva GM, Bakhteeva IV, Shaikhutdinova RZ, Balakhonov SV, Lindner B, Kocharova NA, Senchenkova SN, Holst O, Pier GB, Knirel YA. 2005. Intraspecies and temperature-dependent variations in susceptibility of Yersinia pestis to the bactericidal action of serum and to polymyxin B. Infect Immun 73:7324–7331. doi: 10.1128/IAI.73.11.7324-7331.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kislichkina AA, Bogun AG, Kadnikova LA, Maiskaya NV, Platonov ME, Anisimov NV, Galkina EV, Dentovskaya SV, Anisimov AP. 2015. Nineteen whole-genome assemblies of Yersinia pestis subsp. microtus, including representatives of biovars caucasica, talassica, hissarica, altaica, xilingolensis, and ulegeica. Genome Announc 3:e01342-. doi: 10.1128/genomeA.01342-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Earl SC, Rogers MT, Keen J, Bland DM, Houppert AS, Miller C, Temple I, Anderson DM, Marketon MM. 2015. Resistance to innate immunity contributes to colonization of the insect gut by Yersinia pestis. PLoS One 10:e0133318. doi: 10.1371/journal.pone.0133318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aoyagi KL, Brooks BD, Bearden SW, Montenieri JA, Gage KL, Fisher MA. 2015. LPS modification promotes maintenance of Yersinia pestis in fleas. Microbiology 161:628–638. doi: 10.1099/mic.0.000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vadyvaloo V, Jarrett C, Sturdevant DE, Sebbane F, Hinnebusch BJ. 2010. Transit through the flea vector induces a pretransmission innate immunity resistance phenotype in Yersinia pestis. PLoS Pathog 6:e1000783. doi: 10.1371/journal.ppat.1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Straley SC, Bowmer WS. 1986. Virulence genes regulated at the transcriptional level by Ca2+ in Yersinia pestis include structural genes for outer membrane proteins. Infect Immun 51:445–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palmer LE, Hobbie S, Galan JE, Bliska JB. 1998. YopJ of Yersinia pseudotuberculosis is required for the inhibition of macrophage TNF-alpha production and downregulation of the MAP kinases p38 and JNK. Mol Microbiol 27:953–965. doi: 10.1046/j.1365-2958.1998.00740.x. [DOI] [PubMed] [Google Scholar]
- 44.Klein KA, Fukuto HS, Pelletier M, Romanov G, Grabenstein JP, Palmer LE, Ernst R, Bliska JB. 2012. A transposon site hybridization screen identifies galU and wecBC as important for survival of Yersinia pestis in murine macrophages. J Bacteriol 194:653–662. doi: 10.1128/JB.06237-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rozen S, Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386. [DOI] [PubMed] [Google Scholar]
- 46.Hinnebusch BJ, Perry RD, Schwan TG. 1996. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science 273:367–370. doi: 10.1126/science.273.5273.367. [DOI] [PubMed] [Google Scholar]
- 47.Pujol C, Bliska JB. 2003. The ability to replicate in macrophages is conserved between Yersinia pestis and Yersinia pseudotuberculosis. Infect Immun 71:5892–5899. doi: 10.1128/IAI.71.10.5892-5899.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Une T, Brubaker RR. 1984. In vivo comparison of avirulent Vwa− and Pgm− or Pstr phenotypes of yersiniae. Infect Immun 43:895–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pujol C, Grabenstein JP, Perry RD, Bliska JB. 2005. Replication of Yersinia pestis in interferon gamma-activated macrophages requires ripA, a gene encoded in the pigmentation locus. Proc Natl Acad Sci U S A 102:12909–12914. doi: 10.1073/pnas.0502849102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jarvik T, Smillie C, Groisman EA, Ochman H. 2010. Short-term signatures of evolutionary change in the Salmonella enterica serovar Typhimurium 14028 genome. J Bacteriol 192:560–567. doi: 10.1128/JB.01233-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fukuto HS, Svetlanov A, Palmer LE, Karzai AW, Bliska JB. 2010. Global gene expression profiling of Yersinia pestis replicating inside macrophages reveals the roles of a putative stress-induced operon in regulating type III secretion and intracellular cell division. Infect Immun 78:3700–3715. doi: 10.1128/IAI.00062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]