ABSTRACT
Roger W. Hendrix was at the forefront of bacteriophage biology for nearly 50 years and was central to our understanding of both viral capsid assembly and phage genomic diversity and evolution. Roger's warm and gentle demeanor belied a razor-sharp mind and warmed him to numerous highly productive collaborations that amplified his scientific impact. Roger was always completely open with scientific ideas while at the same time quietly agitating with a stream of new ways of thinking about problems and nudging our communities to search for innovative solutions: a gentle but highly effective provocateur.
KEYWORDS: bacteriophages
INTRODUCTION
Absquatulate: we're not quite sure what it means either, but Roger Hendrix would frequently utter similar murmurings as greetings or as goodbyes or for no particular reason at all, other than perhaps to amuse himself and anyone within earshot. But these wordplay utterances were always creative, amusing, entertaining, nonthreatening, and generally delightful and were typically interspersed with short recitations of Ogden Nash verse (frequently “The Llama”). All of Roger's colleagues greatly enjoyed his company, and for good reason.
Roger Hendrix was a molecular musician. He excelled at all things scientific and musical from an early age, playing saxophone in his high school jazz band and leading the school mathematics team in Walnut Creek, CA. His love of both science and music lasted a lifetime; Roger ardently believed that each enriched the other, and it is hard to argue otherwise. Roger played numerous instruments, was a longtime clarinetist with the University of Pittsburgh Orchestra, and was an active participant in the Renaissance and Baroque Society. He also made a rare appearance to play the shawm as a guest with the Pittsburgh Symphony Orchestra (Fig. 1). None of this interfered with his scientific pursuits and his love of bacteriophages; it just spurred him on.
FIG 1.
Roger Hendrix and Susan Godfrey playing a pair of krumhorns. Image provided by Dick Burgess.
Roger loved machines, and his basement was full of lathes, routers, saws, and other impressive instrumentation. Bacteriophages, the ultimate nanomachines, were thus a perfect match for his curiosity. Phages first entered Roger's world when, after graduating from the California Institute of Technology (Cal Tech), he began pursuit of a Ph.D. in 1965 with James Watson at Harvard University, where he was the first to use SDS-polyacrylamide gel electrophoresis (SDS-PAGE) to identify the phage lambda-encoded proteins as bands in an electrophoresis gel. This may sound trivial today, but such experiments were technically difficult at the time, and his results were important. Before his work, only plaque-forming units (virions with unknown actual protein components), lysozyme, and an encoded nuclease could be assayed among the many lambda gene products. Lambda represented a central system in which the mechanisms of gene regulation (repressors, activators, transcription antiterminators) were being worked out at the time, and Roger's thesis work gave a sense of reality and a much more detailed picture of phage lambda's molecular life cycle. Before that, it was, in a sense, a rather esoteric “genetic house of cards.” Jim Watson recalls him as “the most decent [of all his graduate students]—never uttering a nasty word or comment and always helping anyone in need of help or encouragement” and described his Ph.D. thesis (published only as a dense chapter in a 1971 phage lambda book [1]) as a “massive assault on the proteins made following lambda infection.” Fittingly, Roger would go on to jointly edit the hugely influential second book, “Lambda II,” in 1983 (2).
Roger moved on in 1970 to a postdoctoral position with Dale Kaiser in the Biochemistry Department at Stanford University, where he continued his interests in phage biology but focused now on virion structure and assembly and the roles of host proteins in this process. He was supposed to write several additional major papers on his thesis work during his postdoc at Stanford, but he was having so much fun doing science (as well as hiking and climbing [Fig. 2]) that they never got written. He was actually a spectacularly good writer but had to overcome a large activation energy problem. This meant that, through the course of his entire career, when he did publish a paper, it was meaty, important, and without experimental flaws; the reader always knew that what Roger said in print was thoroughly reliable. Roger took a faculty position at the University of Pittsburgh in 1973, and phage virion assembly, together with phage gene expression, genomics, diversity, and evolution, would engage Roger for his next 44 years. As Jim Pipas (a long-time faculty colleague in Roger's department) notes, Roger was “soft-spoken, with a razor-sharp mind.” We also note that Roger especially enjoyed the collegial camaraderie and scientific give and take of a productive collaboration, so many of his projects involved major longtime collaborations, and we felt privileged to be part of some of them. Indeed, his complete openness with ideas was unique and a major boost to our whole scientific community.
FIG 2.

Roger Hendrix, 1971: Roger, ropes, and camera, adventuring on Mount Conness, Yosemite National Park, on a Stanford Biochemistry Department outing. Image provided by Sherwood Casjens.
PROTEIN CHAPERONES
His kind and respectful demeanor meant that even as a postdoc Roger did not look down on lowly grad students, and our similar scientific interests nudged one of us (S. R. Casjens—then a graduate student) into collaborating with him on the analysis of the protein components in what are now known to be phage lambda virion precursor procapsids (at the time, they were just “head-related” structures and were usually thought of as off-pathway dead-end particles). We were able to deduce the pathway for the assembly of the lambda head, including characterizing the protein cleavages that accompany virion assembly. We also showed that a protein, such as the lambda terminase, can be required for assembly of a virus even though it is not part of the final virion (3, 4). This information was soon used by Kaiser and Masuda (5) to devise the in vitro lambda DNA packaging system that was very important in the early development of cloning of DNA fragments into the lambda genome. At that time, Roger also collaborated with Costa Georgopoulos to determine the role of the first known Escherichia coli protein chaperone, GroE, in phage assembly (6, 7).
At the University of Pittsburgh, Roger's career took off with important early publications in which he was the first to try to come to grips with the possible biological roles of the (now ubiquitous) protein symmetry mismatches that occur in phage virions (specifically the 12-fold/5-fold portal-coat protein mismatch) (8). He and collaborator Isao Katsura demonstrated templated length determination of a macromolecular protein structure when they showed that the length of the lambda gene H protein (which he dubbed the “tape measure” protein) determines the length of the lambda tail (9). In addition, Roger and his first graduate student, Lapchee Tsui, were among the very first to clone and express a targeted gene (groEL) using the phage λ-gt cloning vector developed in Ron Davis's laboratory (10). In 1979, he published the observations that GroEL is a seven-member ring with a central cavity and that it had weak ATPase activity (11). This discovery of 7-fold symmetry was extremely unusual and surprising at the time, and of course he would regale everyone he met with the importance of everything in the world that had to do with the number 7—from the seven deadly sins to the seven notes in a scale, the seven colors in the rainbow, the seven continents, the seven days in a week, the seven wonders of the world, and the Seven Dwarfs. As with many new ideas, this discovery was independently made at the same time by Costa Georgopoulos with Barbara and Tom Hohn (12, 13). But in his typical noncombative style, Roger collaborated with Costa Georgopoulos, Sean Hemmingsen, and John Ellis to determine the sequence of the groEL gene and one of its eukaryotic homologues, the RubisCO binding protein of plants (14) (cited nearly 1,300 times [Google Scholar, January 2018]).
CHOREOGRAPHY
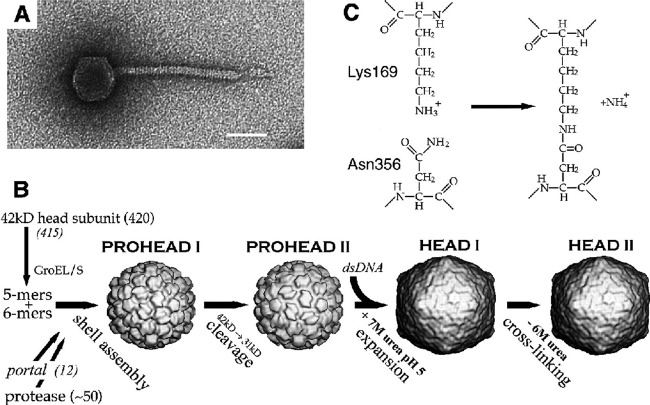
In or around 1987, Roger learned about a coliphage, HK97, isolated by T. S. Dhillon from a sample collected at a pig farm in Hong Kong. His interest in HK97 was tweaked by the fact that it was similar to but had a longer tail than lambda and thus was predicted to have a different length of tape measure protein. However, when he and student Melanie Popa began to investigate, they observed that the protein profile of whole HK97 virions separated by SDS-PAGE lacked any intense band that might correspond to the major capsid protein. Instead, several protein species barely migrated into the separating gel while an abundance of stained material remained in the stacking gel. Rather than concluding that “this phage's weird major capsid protein does not run normally in gels” and forgetting about it as most would no doubt have done, they looked deeper and discovered wholesale covalent cross-linking of the coat proteins in the phage HK97 capsid (15). This feature is now known to be fairly common among phage major head proteins. Roger and Bob Duda, a postdoc from Fred Eiserling's laboratory at the University of California, Los Angeles (UCLA), who joined Roger's laboratory in 1988, showed that the isopeptide cross-links between lysine and asparagine side chains form by an autocatalytic mechanism and that the coat protein rings thus formed are topologically interlocked to form a “molecular chain mail” (Fig. 3) (16–19). At the same time, Roger developed long-term, amazingly productive collaborations with members of the laboratories of Alasdair Steven at NIH and Jack Johnson at Scripps Institute that were designed to reveal the structures of the HK97 virion and its precursors using cryo-electron microscopy (cryo-EM) and X-ray crystallography, as well as to reveal the transitions between these structures. This work resulted in the following ground-breaking findings. (i) The atomic structure of the HK97 head revealed for the first time the protein fold of a tailed phage major capsid protein (Fig. 3) (16) (cited over 500 times [Google Scholar, January 2018]), which is now known to be the same in all tailed phages and herpesviruses. The abundance of tailed phages on Earth makes this one of the most abundant protein folds on our planet. (ii) Determination of the structures of a number of intermediates in the head assembly pathway yielded a wonderful vision of the process that Roger referred to as a protein “ballet” (20, 21) (Fig. 4). (iii) Comparison of early and late intermediate particles in the HK97 head assembly pathway illuminated the mechanism by which such proteins shells are first assembled into less-stable and (usually) smaller shells which then mature into more-stable shells by virtue of a major capsid protein conformational change (chain mail cross-linking happens during the latter stages of this process) (22–24). At the same time, Roger's laboratory pursued the process of assembly from monomer subunits to the finished virion by genetic and biochemical means (25–28). This amazing body of work made the assembly pathway of the icosahedral HK97 head shell from monomer subunits to the completed virion by far the best-characterized, best-understood, and most scientifically influential shell assembly pathway for any double-stranded DNA (dsDNA) virus.
FIG 3.
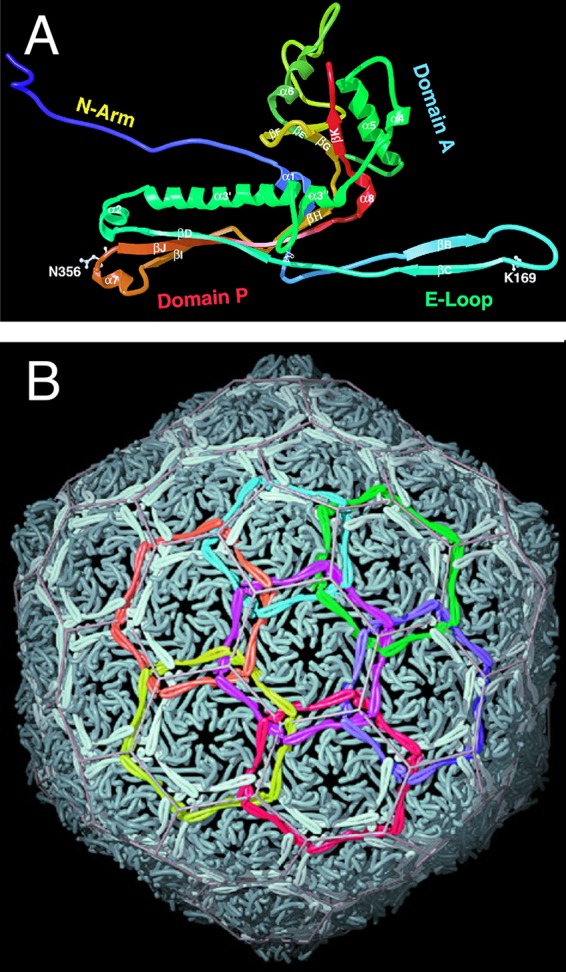
HK97 major capsid protein fold and “chain mail.” (A) Ribbon diagram of the HK97 major capsid protein with cross-linked amino acids K169 and N356 indicated. (B) Chain mail arrangement of interlocked covalent rings of six major capsid proteins in HK97 virions. (Republished from reference 16 with permission of the publisher.)
FIG 4.
The capsid assembly pathway of phage HK97. (A) Negatively stained electron micrograph of an HK97 virion. Bar, 50 nm. (B) HK97 head assembly and maturation pathway. (C) The HK97 major capsid protein cross-linking reaction. (Republished from reference 16 with permission of the publisher.)
SHIFTING FRAMES
Roger also loved examining phage nucleotide sequences for features of interest. In about 1991, he noticed that a lambda tail assembly gene that he was interested in, gene T, had unusual features; namely, it had no recognizable translation start site and its open reading frame overlapped the immediately upstream gene G. After careful analysis of the sequence, he deduced that the downstream T gene might be expressed only through a programmed translational frameshift from the upstream gene. With student Margaret Levin and in collaboration with one of us (S. R. Casjens), he went on to prove that this is in fact the case (the “T protein” is expressed only as a C-terminal fusion to the G protein) and to show exactly where this frameshift takes place. At that time, this was very surprising because such programmed frameshifts had been discovered only recently in retroviruses and there were only two specialized examples in phages, one of which was nonessential (29, 30) and the other of which has remained unique to phage T4 to this day (31). With graduate student Jun Xu, Roger went on to show that, in fact, it is probable that all phages with long noncontractile tails have a pair of tail genes that are expressed in this way, in spite of the proteins being so divergent in amino acid sequence that they show no evidence of homology (32). More recently, Bob Duda and Jun Xu in Roger's laboratory have shown that the G and G-T fusion proteins, neither of which is present in the completed virion, are both critical for tail shaft assembly. They presented a beautiful model that explains how these two proteins function as chaperones for assembly of the major tail shaft subunit, the V protein, along the H protein tape measure (33).
ALL THE WORLD'S A PHAGE
Roger's interests in phage evolution started early with conjecture on why the lambda virion assembly genes are arranged as they are (34, 35), and this approach continued to infuse his genomic analyses through the rest of his career (36–38). In 1989, Roger decided to sequence the complete HK97 genome. Although the prospect of sequencing an ∼50-kbp genome was still quite daunting at that time, Roger figured that if “the new kid” on the faculty (G. F. Hatfull) in his department was sequencing a mycobacteriophage genome, it could not be all that difficult. A graduate student, Tony Youlton, dove into the project, and by the early 1990s the sequenced genomes of mycobacteriophage L5 and HK97 were completed (39, 40); only a few dsDNA tailed phage genome sequences were available at that time, including those of lambda, T7, and ϕ29 (41–43). Comparing these genomes was very illuminating, revealing shared organizational features but little or no sequence commonality. This spurred a commentary published in Seminars in Virology coauthored by Roger and us laying the groundwork for thinking about how phages are constructed and how they evolved (44).
As DNA sequencing methodologies began to advance in the 1990s, sequencing of dsDNA tailed phages became less challenging, and the genomes of phages HK022, N4, N15, ES18, Bxb1, TM4, and D29 were sequenced in Pittsburgh, as was that of Streptomyces phage ØC31 in work spearheaded by Maggie Smith in Nottingham, United Kingdom. In 1995, the first complete genome sequence of a bacterial genome was reported (45) and revealed a cryptic prophage encoding the HindIII restriction system, followed in 1998 by the genome of Mycobacterium tuberculosis, which contains two prophage-like elements (46). Comparisons of these phage and prophage sequences provided several key insights, notably, that phage genomes are highly mosaic and that genes are shared through horizontal genetic exchange but with the phages having different levels of access to the large common gene pool (47). We also found that the otherwise constant lambdoid phage virion assembly gene clusters sometimes harbor novel non-assembly-related gene expression modules that are inserted between the universally present genes. Bob Duda called these “extra” genes “morons“ to indicate the fact that there is more DNA when one is present in the genome than when it is not present; Roger loved the term, which has caught on and is now in common use in phage genome comparisons (33, 37). Furthermore, with his math prowess, Roger was quick to figure out that tailed phages are enormously abundant in nature, with an estimated total abundance of 1031, making them the majority of all biological entities (38, 47). He dubbed the idea of a common tailed phage gene pool the “All the World's a Phage” hypothesis, with a delightful Shakespearian allusion that he took full advantage of. The paper describing this was, sadly, rejected by a widely read American scientific journal but was accepted for publication in the Proceedings of the National Academy of Sciences of the United States of America (PNAS). The stand-alone title was not permitted but was allowed as a second clause, with the full title being “Evolutionary relationships among diverse bacteriophages and prophages: all the world‘s a phage” (but only after fighting a PNAS editorial urge to print it as “All the world is a phage”!). The paper has been cited over 800 times (Google Scholar, January 2018) and is one of Roger's most highly cited and influential publications. An e-mail that Roger sent in October 2012 captures his spirit: “Not that we pay attention to such things, and certainly not that we value crass numbers over transcendent artistic beauty and literary elegance, but the ERADBAP/ATWAP paper has just broken 200 citations.”
THE RATCHET
Roger remained actively engaged in phage genomics after publication of the “All the world's a phage” paper but was increasingly interested in what he referred to as “jumbo” phages, which he broadly defined as those with genomes over 200 kbp long (48, 49). Jumbo phages not only provided a multitude of genomic puzzles but also have larger capsids and thus fully integrated Roger's interests in virion assembly and genome organization. The entry point was the phage G of Megabacterium isolated in 1968 (50) and characterized by Phil Serwer, who showed that the virion chromosome length was approximately 670 kbp (51–53). The phage G genome was sequenced in Pittsburgh and was shown to have a unique sequence of 498 kbp, suggesting that it has substantial terminal redundancy and is packaged into a massive capsid with a triangulation value of 52 (49).
Ron Davis noted that during discussions, Roger would often come up with a whole new way of thinking about something. Incorporating the genome sequences and cryo-EM reconstructions (in collaboration with James Conway and colleagues) of other jumbo phages, Roger proposed his “ratchet hypothesis” to account for the enormous variation among viral capsid sizes and symmetries (49). The hypothesis posits that, given the commonalities of DNA packaging densities, larger phage genomes must be packaged into larger capsids. If a phage genome acquires additional genes by horizontal exchange, then it becomes too large to package unless other genes are deleted. However, if a capsid gene mutation occurs that promotes formation of a larger capsid (assembling larger numbers of capsid protein subunits into alternative icosahedral symmetries but retaining a common protein fold), then it will require a larger genome; this could be accomplished by acquisition of genes by horizontal transfer, duplication of its current genes, or expansion of its terminal redundancy. This facilitates the acquisition of genes that are advantageous for phage success, but once the genome is enlarged, it is difficult for the capsid to revert to a smaller size. Phage sizes thus get ratcheted up unidirectionally, counterbalanced presumably by the increased metabolic costs of DNA replication and virion protein expression (49).
JUST PLAIN FUN
Roger was as adept at making science fun as he was at doing scientific experiments. Several examples are as follows. In his typical whimsical style (he once calculated that a single ribosome would take about 1015 years to translate a molecule of the mass of a chocolate doughnut), he discovered that if every phage magically transformed into a beetle, there would be a layer of beetles that was something like 50,000 km thick on Earth (at a time when the number of phages was thought to be 10-fold less than the current 1031 estimate) (54) and that if all of Earth's tailed phage particles were joined end to end, the line formed would be 200 million light years long (Fig. 5).
FIG 5.

All the world's a phage. Roger calculated that if the Earth's total of 1031 phage particles were joined end to end, they would stretch for a total of 200 million light years, as indicated in Roger's illustration of this line of phages passing the Eta Carinae nebula (a mere 7,500 light years from Earth).
In 1992 Roger became excited about an observation made by Haggård et al. (55) that noted that two of the reading frames in the original lambda genome sequence could be combined by a single (mental) frameshift to form a single gene that is similar to the T4 long tail fiber gene. Roger and Bob Duda immediately obtained the original lambda lysogen and showed that it did indeed produce virions with T4-like long tail fibers and that these fibers increased the rate of adsorption of lambda virions to the surface of E. coli. Thus, the original true wild-type phage lambda (dubbed “Ur-lambda”) had such fibers (Fig. 6), but they are dispensable in the laboratory. They were likely selected against when a large plaque (called lambda PaPa because it was a cross between strains from Paris and Pasadena) was picked very early on to maximize lambda's progeny phage yield and tractability in the laboratory, and all studies since the mid-1950s (56) have used the fiberless phage as the “wild type.” The resulting paper that Roger published in Science was titled “Bacteriophage lambda PaPa: not the mother of all lambda phages” (57). Although the paper does point out that lambda adsorption to cells is likely more complex in the wild than in the laboratory, this finding has had relatively little impact on the progress of understanding phages in general, but it is certainly fun to understand the details of the history of such an important player in the development of molecular genetics as we know it today.
FIG 6.
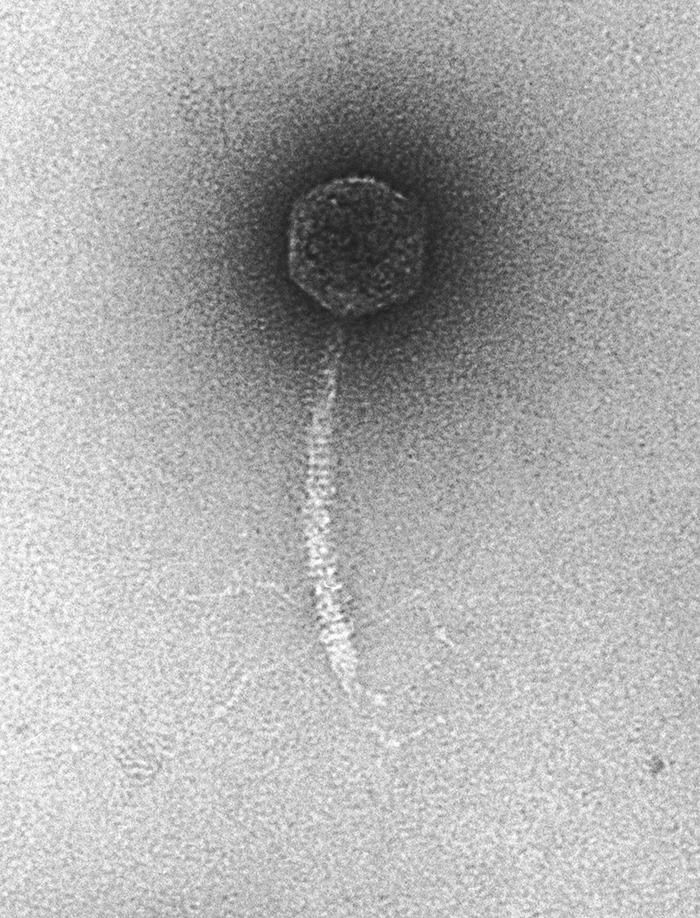
The Ur-λ virion. Negative stain electron microscopy shows that the original λ isolate had long side tail fibers, which have been lacking in all derivatives used since the mid-1950s. (Courtesy of Bob Duda, reproduced with permission.)
In line with his natural gregariousness and eagerness to share ideas, Roger and his wife Susan Godfrey would from time to time host small scientific conferences at their summer home on Vinalhaven Island, ME. These included the “Intergalactic Phage Meetings” on phage evolution and on phage head shell assembly, and the participants would be representative of Roger's currently most urgent interests. The fact that colleagues flocked there from around the world speaks volumes for the respect they had for Roger. Those of us lucky enough to attend one or more of these enjoyed exceptional scientific give and take of ideas and some wonderful fresh-caught-lobster meals as well.
FINAL THOUGHTS
On 15 August 2017, the world lost an important and creative scientist and wonderful person, Roger W. Hendrix. He had a zest for life and science that endeared him to all who knew him, and he will be greatly missed. It is rare that we can hold up the example of a colleague as a role model for all young scientists, but if ever there were one, it is Roger. His intellect was considerable, but it was the way that he chose to deploy it that bestowed on it the greatest impact: by sharing it freely and openly with faculty colleagues and students alike. No one ever got tired of talking to Roger. In the all-too-competitive environment of academic science, collegiality is sometimes given lip service but is rarely practiced to its fullest extent. Roger showed us that the essence of success lies within all that he embodied: a joie de vivre, a sense of adventure, a passion for the arts as well as the sciences, a sense of humor, mischievousness, and a humility that avoided the overinflation of self-importance. Go forth and absquatulate.
ACKNOWLEDGMENTS
We thank the following for the helpful insights and comments on the manuscript: Shankar Adhya, Dennis Bamford, James Conway, Alan Davidson, Bob Duda, Costa Georgopoulos, Susan Godfrey, Jack Johnson, Jeffrey Lawrence, Craig Peebles, Jim Pipas, Jeff Roberts, Forest Rohwer, Lucia Rothman-Denes, Alasdair Steven, and Ry Young.
Biographies

Sherwood R. Casjens is a professor in the Division of Microbiology and Immunology of the Department of Pathology at the University of Utah School of Medicine. Sherwood was a graduate student in Dale Kaiser's laboratory at Stanford University when Roger Hendrix came there as a postdoctoral student in 1970. They immediately became close friends and remained so ever since. Their common interests in phage virion assembly and gene expression, diversity, and evolution gave rise to long-term collaborations, joint grants, and coauthorship of numerous publications.

Graham F. Hatfull is a professor in the Department of Biological Sciences at the University of Pittsburgh. Graham was recruited as an assistant professor in 1988, when Roger Hendrix was chair of the department. Roger and Graham shared interests in phage genomics and evolution, and they collaborated and had fun on numerous projects. Together, they received several joint grants and coauthored a number of publications.
REFERENCES
- 1.Hershey AD. (ed). 1971. The bacteriophage lambda. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 2.Hendrix RW, Roberts JW, Stahl FW, Weisberg RA. 1983. Lambda II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 3.Hendrix RW, Casjens SR. 1974. Protein cleavage in bacteriophage lambda tail assembly. Virology 61:156–159. doi: 10.1016/0042-6822(74)90250-5. [DOI] [PubMed] [Google Scholar]
- 4.Hendrix RW, Casjens SR. 1975. Assembly of bacteriophage lambda heads: protein processing and its genetic control in petit lambda assembly. J Mol Biol 91:187–199. doi: 10.1016/0022-2836(75)90159-X. [DOI] [PubMed] [Google Scholar]
- 5.Kaiser D, Masuda T. 1973. In vitro assembly of bacteriophage lambda heads. Proc Natl Acad Sci U S A 70:260–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Georgopoulos CP, Hendrix RW, Kaiser AD, Wood WB. 1972. Role of the host cell in bacteriophage morphogenesis: effects of a bacterial mutation on T4 head assembly. Nat New Biol 239:38–41. doi: 10.1038/newbio239038a0. [DOI] [PubMed] [Google Scholar]
- 7.Georgopoulos CP, Hendrix RW, Casjens SR, Kaiser AD. 1973. Host participation in bacteriophage lambda head assembly. J Mol Biol 76:45–60. doi: 10.1016/0022-2836(73)90080-6. [DOI] [PubMed] [Google Scholar]
- 8.Hendrix RW. 1978. Symmetry mismatch and DNA packaging in large bacteriophages. Proc Natl Acad Sci U S A 75:4779–4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katsura I, Hendrix RW. 1984. Length determination in bacteriophage lambda tails. Cell 39:691–698. doi: 10.1016/0092-8674(84)90476-8. [DOI] [PubMed] [Google Scholar]
- 10.Hendrix RW, Tsui L. 1978. Role of the host in virus assembly: cloning of the Escherichia coli groE gene and identification of its protein product. Proc Natl Acad Sci U S A 75:136–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hendrix RW. 1979. Purification and properties of groE, a host protein involved in bacteriophage assembly. J Mol Biol 129:375–392. doi: 10.1016/0022-2836(79)90502-3. [DOI] [PubMed] [Google Scholar]
- 12.Georgopoulos CP, Hohn B. 1978. Identification of a host protein necessary for bacteriophage morphogenesis (the groE gene product). Proc Natl Acad Sci U S A 75:131–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hohn T, Hohn B, Engel A, Wurtz M, Smith PR. 1979. Isolation and characterization of the host protein groE involved in bacteriophage lambda assembly. J Mol Biol 129:359–373. doi: 10.1016/0022-2836(79)90501-1. [DOI] [PubMed] [Google Scholar]
- 14.Hemmingsen SM, Woolford C, van der Vies SM, Tilly K, Dennis DT, Georgopoulos CP, Hendrix RW, Ellis RJ. 1988. Homologous plant and bacterial proteins chaperone oligomeric protein assembly. Nature 333:330–334. doi: 10.1038/333330a0. [DOI] [PubMed] [Google Scholar]
- 15.Popa MP, McKelvey TA, Hempel J, Hendrix RW. 1991. Bacteriophage HK97 structure: wholesale covalent cross-linking between the major head shell subunits. J Virol 65:3227–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE. 2000. Topologically linked protein rings in the bacteriophage HK97 capsid. Science 289:2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- 17.Gan L, Conway JF, Firek BA, Cheng N, Hendrix RW, Steven AC, Johnson JE, Duda RL. 2004. Control of crosslinking by quaternary structure changes during bacteriophage HK97 maturation. Mol Cell 14:559–569. doi: 10.1016/j.molcel.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 18.Tso D, Peebles CL, Maurer JB, Duda RL, Hendrix RW. 2017. On the catalytic mechanism of bacteriophage HK97 capsid crosslinking. Virology 506:84–91. doi: 10.1016/j.virol.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duda RL. 1998. Protein chainmail: catenated protein in viral capsids. Cell 94:55–60. doi: 10.1016/S0092-8674(00)81221-0. [DOI] [PubMed] [Google Scholar]
- 20.Wikoff WR, Conway JF, Tang J, Lee KK, Gan L, Cheng N, Duda RL, Hendrix RW, Steven AC, Johnson JE. 2006. Time-resolved molecular dynamics of bacteriophage HK97 capsid maturation interpreted by electron cryo-microscopy and X-ray crystallography. J Struct Biol 153:300–306. doi: 10.1016/j.jsb.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 21.Hendrix RW, Duda RL. 1998. Bacteriophage HK97 head assembly: a protein ballet. Adv Virus Res 50:235–288. doi: 10.1016/S0065-3527(08)60810-6. [DOI] [PubMed] [Google Scholar]
- 22.Lee KK, Gan L, Tsuruta H, Moyer C, Conway JF, Duda RL, Hendrix RW, Steven AC, Johnson JE. 2008. Virus capsid expansion driven by the capture of mobile surface loops. Structure 16:1491–1502. doi: 10.1016/j.str.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang RK, Khayat R, Lee KK, Gertsman I, Duda RL, Hendrix RW, Johnson JE. 2011. The Prohead-I structure of bacteriophage HK97: implications for scaffold-mediated control of particle assembly and maturation. J Mol Biol 408:541–554. doi: 10.1016/j.jmb.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cardone G, Duda RL, Cheng N, You L, Conway JF, Hendrix RW, Steven AC. 2014. Metastable intermediates as stepping stones on the maturation pathways of viral capsids. mBio 5:e02067. doi: 10.1128/mBio.02067-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hasek ML, Maurer JB, Hendrix RW, Duda RL. 2017. Flexible connectors between capsomer subunits that regulate capsid assembly. J Mol Biol 429:2474–2489. doi: 10.1016/j.jmb.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duda RL, Oh B, Hendrix RW. 2013. Functional domains of the HK97 capsid maturation protease and the mechanisms of protein encapsidation. J Mol Biol 425:2765–2781. doi: 10.1016/j.jmb.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dierkes LE, Peebles CL, Firek BA, Hendrix RW, Duda RL. 2009. Mutational analysis of a conserved glutamic acid required for self-catalyzed cross-linking of bacteriophage HK97 capsids. J Virol 83:2088–2098. doi: 10.1128/JVI.02000-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oh B, Moyer CL, Hendrix RW, Duda RL. 2014. The delta domain of the HK97 major capsid protein is essential for assembly. Virology 456–457:171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Condron BG, Atkins JF, Gesteland RF. 1991. Frameshifting in gene 10 of bacteriophage T7. J Bacteriol 173:6998–7003. doi: 10.1128/jb.173.21.6998-7003.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sipley J, Stassi D, Dunn J, Goldman E. 1991. Analysis of bacteriophage T7 gene 10A and frameshifted 10B proteins. Gene Expr 1:127–136. [PMC free article] [PubMed] [Google Scholar]
- 31.Huang WM, Ao SZ, Casjens S, Orlandi R, Zeikus R, Weiss R, Winge D, Fang M. 1988. A persistent untranslated sequence within bacteriophage T4 DNA topoisomerase gene 60. Science 239:1005–1012. doi: 10.1126/science.2830666. [DOI] [PubMed] [Google Scholar]
- 32.Xu J, Hendrix RW, Duda RL. 2004. Conserved translational frameshift in dsDNA bacteriophage tail assembly genes. Mol Cell 16:11–21. doi: 10.1016/j.molcel.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 33.Xu J, Hendrix RW, Duda RL. 2014. Chaperone-protein interactions that mediate assembly of the bacteriophage lambda tail to the correct length. J Mol Biol 426:1004–1018. doi: 10.1016/j.jmb.2013.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Casjens SR, Hendrix RW. 1988. Control mechanisms in dsDNA bacteriophage assembly, p 1–91. In Calendar R. (ed), The bacteriophages. Plenum Press, New York, NY. [Google Scholar]
- 35.Casjens S, Hendrix R. 1974. Comments on the arrangement of the morphogenetic genes of bacteriophage lambda. J Mol Biol 90:20–25. doi: 10.1016/0022-2836(74)90253-8. [DOI] [PubMed] [Google Scholar]
- 36.Hendrix RW, Lawrence JG, Hatfull GF, Casjens S. 2000. The origins and ongoing evolution of viruses. Trends Microbiol 8:504–508. doi: 10.1016/S0966-842X(00)01863-1. [DOI] [PubMed] [Google Scholar]
- 37.Hendrix RW, Hatfull GF, Smith MC. 2003. Bacteriophages with tails: chasing their origins and evolution. Res Microbiol 154:253–257. doi: 10.1016/S0923-2508(03)00068-8. [DOI] [PubMed] [Google Scholar]
- 38.Hendrix RW. 2002. Bacteriophages: evolution of the majority. Theor Popul Biol 61:471–480. doi: 10.1006/tpbi.2002.1590. [DOI] [PubMed] [Google Scholar]
- 39.Hatfull GF, Sarkis GJ. 1993. DNA sequence, structure and gene expression of mycobacteriophage L5: a phage system for mycobacterial genetics. Mol Microbiol 7:395–405. doi: 10.1111/j.1365-2958.1993.tb01131.x. [DOI] [PubMed] [Google Scholar]
- 40.Juhala RJ, Ford ME, Duda RL, Youlton A, Hatfull GF, Hendrix RW. 2000. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J Mol Biol 299:27–51. doi: 10.1006/jmbi.2000.3729. [DOI] [PubMed] [Google Scholar]
- 41.Dunn JJ, Studier FW. 1983. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol 166:477–535. doi: 10.1016/S0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- 42.Sanger F, Coulson AR, Hong GF, Hill DF, Petersen GB. 1982. Nucleotide sequence of bacteriophage lambda DNA. J Mol Biol 162:729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- 43.Vlcek C, Paces V. 1986. Nucleotide sequence of the late region of Bacillus phage ø29 completes the 19,285-bp sequence of ø29 genome. Comparison with the homologous sequence of phage PZA. Gene 46:215–225. [DOI] [PubMed] [Google Scholar]
- 44.Casjens S, Hatfull GF, Hendrix R. 1992. Evolution of dsDNA tailed-bacteriophage genomes. Semin Virol 3:383–397. [Google Scholar]
- 45.Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM, McKenney K, Sutton G, FitzHugh W, Fields C, Gocayne JD, Scott J, Shirley R, Liu L-I, Glodek A, Kelley JM, Weidman JF, Phillips CA, Spriggs T, Hedblom E, Cotto MD, Utterback TR, Hanna MC, Nguyen DT, Saudek DM, Brandon RC, Fine LD, Fritchman JL, Fuhrmann JL, Geoghagen NSM, Gnehm CL, McDonald LA, Small KV, Fraser CM, Smith HO, Venter JC. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 46.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE III, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell BG. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 47.Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc Natl Acad Sci U S A 96:2192–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hendrix RW. 2009. Jumbo bacteriophages. Curr Top Microbiol Immunol 328:229–240. [DOI] [PubMed] [Google Scholar]
- 49.Hua J, Huet A, Lopez CA, Toropova K, Pope WH, Duda RL, Hendrix RW, Conway JF. 2017. Capsids and genomes of jumbo-sized bacteriophages reveal the evolutionary reach of the HK97 fold. mBio 8:e01579-17. doi: 10.1128/mBio.01579-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hutson MS, Holzwarth G, Duke T, Viovy J-L. 1995. Two-dimensional motion of DNA during 120° pulsed-field gel electrophoresis. I. Effect of molecular weight. Biopolymers 35:297–306. doi: 10.1002/bip.360350305. [DOI] [Google Scholar]
- 51.Sun M, Serwer P. 1997. The conformation of DNA packaged in bacteriophage G. Biophys J 72:958–963. doi: 10.1016/S0006-3495(97)78730-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hutson MS, Holzwarth G, Duke Viovy J-L. 1995. Two-dimensional motion of DNA during 120 pulsed-field gel electrophoresis. I. Effect of molecular weight. Biopolymers 35:297–306. [Google Scholar]
- 53.Serwer P, Hayes SJ, Thomas JA, Hardies SC. 2007. Propagating the missing bacteriophages: a large bacteriophage in a new class. Virol J 4:21. doi: 10.1186/1743-422X-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brüssow H, Hendrix RW. 2002. Phage genomics: small is beautiful. Cell 108:13–16. doi: 10.1016/S0092-8674(01)00637-7. [DOI] [PubMed] [Google Scholar]
- 55.Haggård-Ljungquist E, Halling C, Calendar R. 1992. DNA sequences of the tail fiber genes of bacteriophage P2: evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. J Bacteriol 174:1462–1477. doi: 10.1128/jb.174.5.1462-1477.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dove WF. 1969. Strains of phage lambda in current use. Virology 38:349–351. doi: 10.1016/0042-6822(69)90378-X. [DOI] [PubMed] [Google Scholar]
- 57.Hendrix RW, Duda RL. 1992. Bacteriophage lambda PaPa: not the mother of all lambda phages. Science 258:1145–1148. doi: 10.1126/science.1439823. [DOI] [PubMed] [Google Scholar]