Diffuse gliomas are classified according to the 2016 World Health Organization (WHO) Classification of Tumors of the Central Nervous System,1 which combines histological and molecular features. Diagnosis requires the assessment of mutations in the isocitrate dehydrogenase genes (IDH1 and IDH2), key genetic alterations characterizing gliomas with favorable outcome.2
Because IDH1 and IDH2 are highly similar enzymes, the WHO classification, as most of the current studies, combines these mutations into the same molecular group; however, it is unclear whether these tumors share the same characteristics.
We analyzed data from 1517 patients included in the French POLA Network to investigate differences between IDH1- and IDH2-mutant gliomas.
Inclusion criteria were the written consent of the patient for clinical data collection and genetic analysis and sufficient material for molecular studies allowing classification according to the 2016 WHO classification.
IDH1-R132H mutational status was evaluated using automated immunohistochemistry in all cases (n = 1517). Direct sequencing3 was performed in 978 cases and demonstrated IDH mutation in 573 cases (this includes confirmation of IDH1-R132H mutation in 468 cases, other IDH1 mutations in 61 cases, and IDH2 mutation in 44 cases). The 1p/19q codeletion status was determined based on single nucleotide polymorphism arrays, comparative genomic hybridization arrays, and/or microsatellite marker analysis.3 The following data were recorded: age, sex, follow-up, and MRI features (tumor location, extension, contrast enhancement, edema). All statistical analysis was done using IBM SPSS statistics software version 23. Chi-square test was used to compare qualitative variables. Continuous variables were compared using the Mann–Whitney U-test, and the Kaplan–Meier method was used to estimate survival distributions.
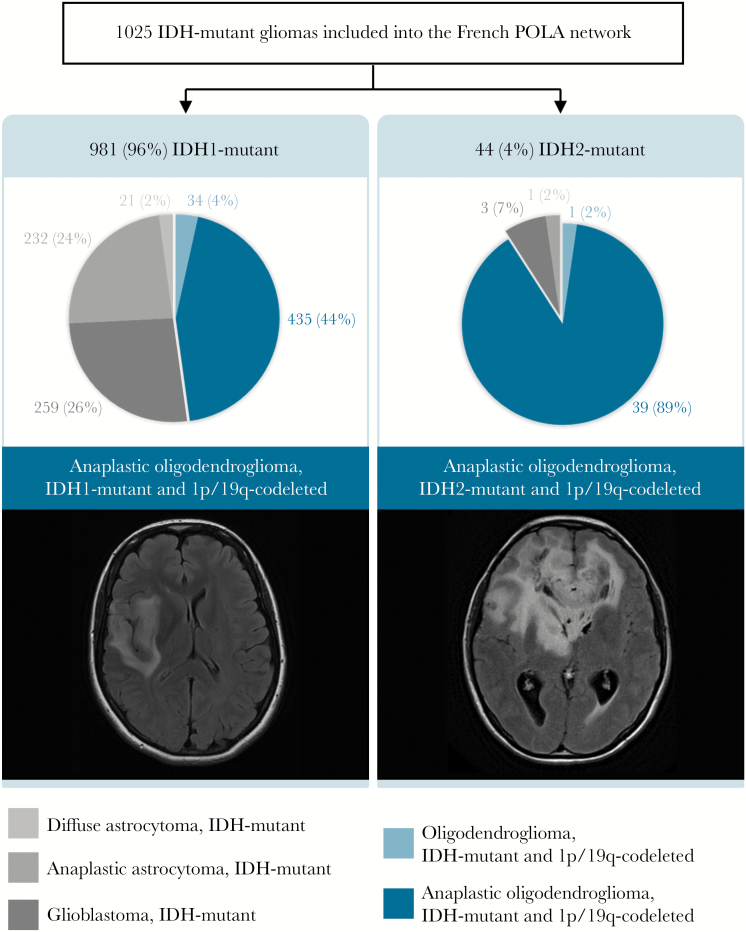
Among the 1517 patients, 1025 had an IDH-mutant tumor: 96% were IDH1-mutant and 4% IDH2-mutant. Integrated diagnoses are summarized in Fig. 1. The frequency of 1p/19q codeletion was higher in the IDH2-mutant group compared with the IDH1-mutant group (91% vs 48%, P < 0.001).
Fig. 1.
Integrated diagnosis of the 1025 IDH-mutant cases of the French POLA cohort according to the updated fourth WHO classification and representative fluid attenuated inversion recovery MRI axial sections among the subgroups of anaplastic oligodendroglioma, IDH-mutant, and 1p/19q codeleted.
Wang and coworkers previously reported higher frequency of 1p/19q codeletion in IDH2-mutant gliomas (9/18 samples) compared with IDH1-mutant.4 The percentage of each category in our study does not reflect the normal distribution of glioma because of the inclusion criteria in the POLA Network (ie, high-grade glioma with oligodendroglial component). However, the higher proportion of 1p/19q codeleted glioma in the IDH2-mutant group cannot be attributed to the inclusion criteria.
Because the main population of glioma associated with IDH2 mutation was 1p/19q codeleted anaplastic oligodendroglioma, we focused on this subgroup to search for differences compared with IDH1 mutation. Among these patients (n = 474), we did not observe any difference in terms of age, sex, progression-free survival, or overall survival between IDH1- and IDH2-mutant tumors. However, IDH2-mutant anaplastic oligodendrogliomas presented more frequently with multilobar extension (56% of the IDH2-mutant vs 35% of the IDH1-mutant, P = 0.015) and edema (79% vs 57%, P = 0.02). Furthermore, bifrontal location with corpus callosum involvement was more frequent in IDH2-mutant compared with IDH1-mutant tumors (41% vs 16%, P < 0.001).
IDH mutation is supposed to be one of the first “hits” of gliomagenesis,5 resulting in production of an oncometabolite, D-2-hydroxyglutarate (D-2HG), which impacts the α-ketoglutarate–dependent dioxygenase functions. Previous studies demonstrated that the potential for IDH-mutant enzymes to produce D-2HG depends on the mutation type.6 Based on our observations, we could hypothesize that the higher D-2HG accumulation induced by IDH2 mutation may lead to a phenotype that is favorable to 1p/19q chromosomal loss. It may also impact distinct cellular pathways that promote a more invasive phenotype. Whether IDH1 or IDH2 mutations impact distinct glial precursor cells with differential invasive properties remains to be elucidated.
In conclusion, our results illustrate that IDH2-mutant gliomas are commonly associated with 1p/19q codeletion. Most of IDH2-mutant anaplastic oligodendroglioma 1p/19q codeleted are multilobar. Understanding the genomic events involved in these specificities may represent a step forward for therapeutic development.
Funding
This work was funded by the French Institut National du Cancer (INCa, POLA Network) and is part of the national program Cartes d′Identité des Tumeurs (CIT) (http://cit.ligue-cancer.net/), which is funded and developed by the Ligue nationale contre le cancer. This work was supported by grants from the Institut National du Cancer (grant INCa-DGOS-Inserm 6038; SIRIC-Marseille).
Acknowledgments
We thank the ARTC-Sud patients’ association (Asso ciation pour le Recherche sur les Tumeurs Cérébrales) and the Cancéropôle PACA.
Conflict of interest statement. The responsible authors disclosed no conflict of interest.
POLA Network: Amiens (C. Desenclos, H. Sevestre), Angers (P. Menei, A. Rousseau), Annecy (T. Cruel, S. Lopez), Besançon (M-I. Mihai, A. Petit), Bicêtre (C. Adam, F. Parker), Brest (P. Dam-Hieu, I. Quintin-Roué), Bordeaux (S. Eimer, H. Loiseau), Caen (L. Bekaert, F. Chapon), Clamart (D. Ricard) Clermont-Ferrand (C. Godfraind, T. Khallil), Clichy (D. Cazals-Hatem, T. Faillot), Colmar (C. Gaultier, M. C. Tortel), Cornebarrieu (I. Carpiuc, P. Richard), Créteil (W. Lahiani), Dijon (H. Aubriot-Lorton, F. Ghiringhelli), Lille (C. A. Maurage, C. Ramirez), Limoges (E. M. Gueye, F. Labrousse), Marseille (O. Chinot), Montpellier (L. Bauchet, V. Rigau), Nancy (P. Beauchesne, G. Gauchotte), Nantes (M. Campone, D. Loussouarn), Nice (D. Fontaine, F. Vandenbos-Burel), Nimes (A. Le Floch, P. Roger) Orléans (C. Blechet, M. Fesneau), Paris (A. Carpentier, J. Y. Delattre [POLA Network national coordinator], S. Elouadhani-Hamdi, M. Polivka), Poitiers (D. Larrieu-Ciron, S. Milin), Reims (P. Colin, M. D. Diebold), Rennes (D. Chiforeanu, E. Vauleon), Rouen (O. Langlois, A. Laquerriere), Saint-Etienne (F. Forest, M. J. Motso-Fotso), Saint-Pierre de la Réunion (M. Andraud, G. Runavot), Strasbourg (B. Lhermitte, G. Noel), Suresnes (S. Gaillard, C. Villa), Toulon (N. Desse), Tours (C. Rousselot-Denis, I. Zemmoura), Toulouse (E. Cohen-Moyal, E. Uro-Coste), Villejuif (F. Dhermain)
Contributor Information
POLA Network:
C Desenclos, H Sevestre, P Menei, A Rousseau, T Cruel, S Lopez, M-I Mihai, A Petit, C Adam, F Parker, P Dam-Hieu, I Quintin-Roué, S Eimer, H Loiseau, L Bekaert, F Chapon, D Ricard, C Godfraind, T Khallil, D Cazals-Hatem, T Faillot, C Gaultier, M C Tortel, I Carpiuc, P Richard, W Lahiani, H Aubriot-Lorton, F Ghiringhelli, C A Maurage, C Ramirez, E M Gueye, F Labrousse, O Chinot, L Bauchet, V Rigau, P Beauchesne, G Gauchotte, M Campone, D Loussouarn, D Fontaine, F Vandenbos-Burel, A Le Floch, P Roger, C Blechet, M Fesneau, A Carpentier, J Y Delattre, S Elouadhani-Hamdi, M Polivka, D Larrieu-Ciron, S Milin, P Colin, M D Diebold, D Chiforeanu, E Vauleon, O Langlois, A Laquerriere, F Forest, M J Motso-Fotso, M Andraud, G Runavot, B Lhermitte, G Noel, S Gaillard, C Villa, N Desse, C Rousselot-Denis, I Zemmoura, E Cohen-Moyal, E Uro-Coste, and F Dhermain
References
- 1. Louis DN, Perry A, Reifenberger G et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 2. Yan H, Parsons DW, Jin G et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tabouret E, Nguyen AT, Dehais C et al. ; For POLA Network Prognostic impact of the 2016 WHO classification of diffuse gliomas in the French POLA cohort. Acta Neuropathol. 2016;132(4):625–634. [DOI] [PubMed] [Google Scholar]
- 4. Wang HY, Tang K, Liang TY et al. The comparison of clinical and biological characteristics between IDH1 and IDH2 mutations in gliomas. J Exp Clin Cancer Res. 2016;35:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174(4):1149–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ward PS, Lu C, Cross JR et al. The potential for isocitrate dehydrogenase mutations to produce 2-hydroxyglutarate depends on allele specificity and subcellular compartmentalization. J Biol Chem. 2013;288(6):3804–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]