Abstract
Gliomas are a heterogeneous group of brain tumors with distinct biological and clinical properties. Despite advances in surgical techniques and clinical regimens, treatment of high-grade glioma remains challenging and carries dismal rates of therapeutic success and overall survival. Challenges include the molecular complexity of gliomas, as well as inconsistencies in histopathological grading, resulting in an inaccurate prediction of disease progression and failure in the use of standard therapy. The updated 2016 World Health Organization (WHO) classification of tumors of the central nervous system reflects a refinement of tumor diagnostics by integrating the genotypic and phenotypic features, thereby narrowing the defined subgroups. The new classification recommends molecular diagnosis of isocitrate dehydrogenase (IDH) mutational status in gliomas. IDH-mutant gliomas manifest the cytosine-phosphate-guanine (CpG) island methylator phenotype (G-CIMP). Notably, the recent identification of clinically relevant subsets of G-CIMP tumors (G-CIMP-high and G-CIMP-low) provides a further refinement in glioma classification that is independent of grade and histology. This scheme may be useful for predicting patient outcome and may be translated into effective therapeutic strategies tailored to each patient. In this review, we highlight the evolution of our understanding of the G-CIMP subsets and how recent advances in characterizing the genome and epigenome of gliomas may influence future basic and translational research.
Keywords: CpG island methylator phenotype (CIMP), DNA methylation, G-CIMP (glioma-CIMP), glioma progression, IDH mutation, molecular subtypes
Gliomas: An Overview
Gliomas are a heterogeneous group of brain tumors with distinct biological and clinical properties.1,2 Classically, glioma subtypes and grading were exclusively defined by histological features. However, this classification strategy does not reflect the heterogeneity of the tumor, is prone to subjectivity and discordance among neuropathologists, falls short of predicting disease course, and cannot reliably guide treatment.2 Therefore, multiple research efforts have sought to identify molecular signatures that define more discrete glioma subgroups and have a greater impact and relevance in the clinical setting.1–5 Corroborating the importance of molecular markers in the cancer classification, researchers observed that 1 in 10 cancer patients would be better classified by molecular taxonomy, rather than by the current system based on the primary tissue of origin and stage of disease.6
The 2016 update to the World Health Organization (WHO) classification of tumors of the CNS represents a shift in tumor diagnostics by integrating molecular and phenotypic features into the classification of tumors and thereby narrowing the defined subgroups.1 Among the genetic alterations associated with diffuse gliomas, isocitrate dehydrogenase (IDH) mutation, histone H3 K27M status, and the integrity status of chromosomes 1 and 19 (1p and 19q) are now integrated with the traditional histology/grade-based glioma classification.1,7 Gliomas harboring mutations in IDH1/2, as well as codeletion of the 1p and 19q chromosome arms (1p/19q), have shown favorable prognostic and/or predictive values in relation to their counterparts (IDH-wildtype or 1p/19q intact [also known as “non-codel”]).8–12 A number of epigenomic markers also have shown prognostic and/or predictive values.13 Patients harboring gliomas that carry O6-methylguanine-DNA methyltransferase (MGMT) promoter DNA methylation demonstrate increased overall survival (OS) and time to progression of the disease after treatment with temozolomide (TMZ) or radiation.14–17MGMT promoter DNA methylation status is used to guide therapeutic management for anaplastic gliomas with wildtype IDH1/2 and in glioblastoma (GBM) of the elderly.14–17 Another important milestone highlighting the clinical importance of epigenetic signatures in gliomas was the discovery of the glioma cytosine-phosphate-guanine (CpG) island methylator phenotype (G-CIMP).18 Patients carrying G-CIMP (G-CIMP+) tumors have shown a better prognosis than those not carrying that phenotype (G-CIMP−). G-CIMP+ tumors were closely related to IDH mutation and nearly all IDH-mutant gliomas were G-CIMP+ and had a favorable prognosis.18,19 However, comprehensive DNA methylation profiling in a large cohort of glioma patients revealed that not all IDH-mutant/G-CIMP+ tumors had the same prognosis.7 Based on the extent of global DNA methylation, this study uncovered 2 subsets of IDH-mutant/G-CIMP+ gliomas, one that presented a low degree of DNA methylation and poorer outcome (G-CIMP-low) and another subset depicting high DNA methylation and good OS as usually described for IDH-mutant/G-CIMP+ (G-CIMP-high).7,18,19 G-CIMP+ subsets (-low and -high) have shown distinct biological features and clinical implications that will be further explored in this review. Following a brief introduction to the role of epigenomics in cancer and in neuro-oncology, we will detail the evolution of our understanding that led to the identification of the G-CIMP subsets. We will also describe how recent advances in DNA methylation-based biomarkers that characterize gliomas may influence future basic and translational research. Our review will conclude with current and upcoming clinical applications, focusing on the potential translational importance of G-CIMP subsets.
Epigenetic Modifications in Cancer
Within the nucleus of a cell, the 3 billion base pairs of the human DNA sequence are stored and organized as chromatin, which is made up of repeating units called nucleosomes. Nucleosomes are formed by octamers of histone proteins that are prone to chemical modifications at distinct amino acid residues, such as methylation and acetylation. Several epigenetic mechanisms may operate in synchrony to modify the packaging of the genome, including DNA methylation, histone modifications, nucleosome remodeling, small and long noncoding RNA, protein–DNA interactions via chromatin modifying transcription factors, and 3-D chromatin architecture.20 The resulting epigenomic landscape determines the accessibility of regulatory elements, thereby modulating the transcriptional regulation given by transcription factors.20 Epigenetic mechanisms are implicated in both physiological and pathological events, such as tissue specificity and carcinogenesis, respectively.20 In the process of malignant transformation, gene inactivating mutations have been shown to control the epigenomic landscape, implying a crosstalk between the genome and the epigenome.21 In the present review, we will focus on the first epigenetic modification to be linked to cancer and probably the most extensively studied epigenetic modification in mammals—DNA methylation.22
DNA Methylation
DNA methylation status results from the action of methylating or demethylating enzymes. DNA methylation is the covalent transfer of methyl groups to the 5ʹ position of the cytosine ring, primarily at dinucleotide CpGs, resulting in 5-methylcytosine (5mC). DNA methyltransferases (DNMTs), known as the DNA methylation “writer” enzymes, catalyze the transfer of the methyl group to 5ʹ cytosine.23 On the other hand, demethylation of 5mC is promoted by ten-eleven translocation (TET)1/2 methylcytosine dioxygenases, known as the DNA methylation “erasers,” to generate 5-hydroxymethylcytosine (5hmC).24 Some stretches of DNA contain frequent CpG sites, defined as CpG islands (CGIs), which are preferentially located at the 5ʹ end of genes overlapping gene promoters.25 CpG sites are also found in gene bodies and in other regions, named CpG shores (2 kb regions flanking CGIs), CpG shelves (>2 kb regions flanking CpG shores), and open sea regions (>4 kb to the nearest CGIs) relative to their proximity to CGIs.26 Intergenic regions that are enriched for CpG but located in open seas may encompass distal genomic regulatory elements, such as enhancers, silencers, and insulators. These transcriptional regulatory elements contain recognition sites for DNA-binding transcription factors, which function either to enhance or repress transcription. Specifically, enhancers are elements able to activate target genes from distal locations independently of their orientation,27 while insulators are boundary elements that possess the ability to block or insulate the signals of either enhancers or silencers.28 The interplay between protein:DNA complexes defines the spatial organization of the human genome. As a result, chromatin loops are formed, mediated by insulator proteins, such as the CCCTC-binding factor (CTCF), facilitating or blocking enhancer–promoter interactions. However, epigenomic alterations, such as the gain of DNA methylation at the binding site of a regulatory element, could disrupt those interactions,29 while removing site-specific DNA methylation has shown to reverse the process, thereby providing opportunities for potential clinical therapy.30
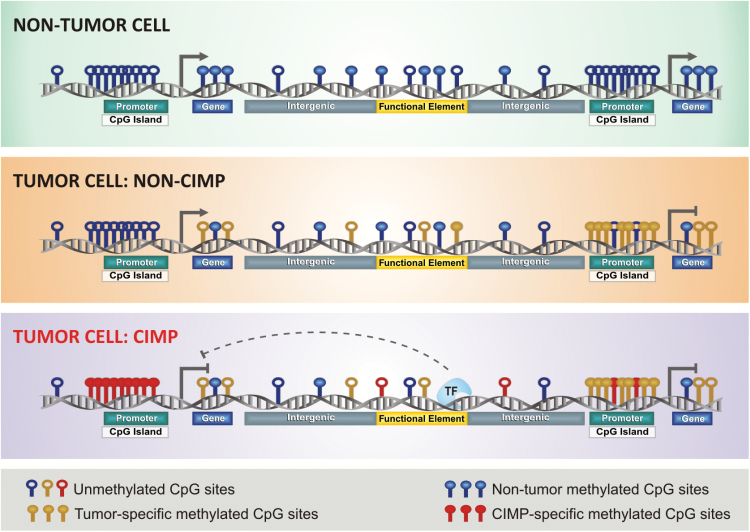
Unlike the CpG sites dispersed throughout the genome that are usually methylated, CGIs located at promoter regions are generally unmethylated. In physiological conditions, CGI methylation usually occurs as a mechanism of gene repression in specific regions, such as the inactive X-chromosome, imprinted genes, and germline cell-specific genes.31 In cancer, DNA methylation becomes aberrant and is mostly characterized by focal hypermethylation around the promoters of genes and gene bodies and global hypomethylation among nonpromoter elements32,33 (Fig. 1). Promoter hypermethylation is an important mechanism of epigenetic silencing of tumor suppressor genes.34,35 DNA methylation in nonpromoter regions is proposed to play a major role in intratumoral expression heterogeneity.36 In contrast, global hypomethylation largely affects the intergenic and intronic regions of the genome, which may also result in chromosomal instability. In addition to affecting the gene expression and chromosomal status, aberrant DNA methylation also modulates isoform expression and facilitates mutational events in the adult stem and progenitor cells.13,33,35
Fig. 1.
CIMP subtypes in human cancer. This illustration depicts aberrant DNA methylation changes at specific genomic loci in normal and tumor cells, especially in CIMP tumors. Each DNA strand represents one individual methylome. Methylated CpG sites in normal state are represented in blue, non-CIMP tumor DNA methylation gain in yellow, and aberrant DNA hypermethylation in CIMP tumors in red (modified from Weisenberger38).
CIMP: CpG Island Methylator Phenotype
Classically, CIMP is defined by genome-wide hypermethylation of CGIs (Fig. 1). Tumors carrying this phenotype were first described and validated in the context of colorectal cancers, also known as colorectal CIMP.37,38 Since then, CIMP has been described in several tumors, including gliomas.18,39 Interestingly, the CIMP+ tumor subset exhibits distinct epidemiological, clinicopathological, and molecular features in relation to its CIMP− counterpart. CIMP may have prognostic significance (both favorable and unfavorable) in terms of the OS or predictive value of therapeutic response in certain tumors.39–41 Despite tissue-specific differences, evidence suggests that CIMP may be a universal feature across different tumors. Addressing this issue, researchers applied a genome-wide unbiased and unsupervised hierarchical clustering of cancer-specific methylated CGI genes across 15 tumor types; however, gliomas were not included in the cohort.42 These researchers observed a set of 89 discriminative loci that allowed the segregation of 12 tumor types into CIMP+ and CIMP− subgroups according to the highest or lowest average levels of DNA methylation, respectively.42
Glioma Epigenomic Molecular Signatures
G-CIMP: Glioma-CpG Island Methylator Phenotype
The glioma-CIMP (G-CIMP) subtype was first described by Noushmehr et al in GBM and was further validated in lower-grade glioma (LGG).18,19 The G-CIMP+ subtype occurred frequently in LGG specimens, whereas in GBM it was mostly associated with secondary or recurrent (treated) tumors harboring mutations of the IDH gene.18,19 By integrating the DNA methylation data with 4 gene expression clusters previously described in GBM by Verhaak et al (ie, proneural, neural, classical, and mesenchymal),43,44 the authors found that G-CIMP+ subtypes were highly enriched among the proneural subtypes and in younger patients, compared with G-CIMP− tumors. Later, the G-CIMP subtype was also described in pediatric glioma patients.45 Importantly, G-CIMP+ was shown to be closely associated with IDH-mutant gliomas in several studies.18,19
By performing a large multiplatform genomic analysis across 1122 lower- and high-grade primary adult gliomas, our team, as part of Ceccarelli et al, uncovered 7 discrete subtypes with distinct biological features and clinical outcomes.7 This DNA methylation-based classification refined and recapitulated previous glioma stratification subgroups based on IDH mutation and 1p/19q codeletion status. The subgroups were mainly driven by IDH1/2 mutation status and classified as (i) IDH-wildtype, enriched for GBM; these were further segregated as classic-like, mesenchymal-like, LGm6-GBM, and pilocytic astrocytoma (PA)-like subgroups, and (ii) IDH-mutants, enriched for LGG and further clustered as 1p/19q codel and non-codel. The IDH-mutant non-codel cluster was further refined into 2 distinct subgroups, based on the extent of DNA methylation, G-CIMP-low (6% of IDH-mutant) and G-CIMP-high (55% of IDH-mutant), which were determined by a low or high degree of DNA methylation, respectively.7
Prognostic Value of G-CIMP+ and Its Subsets
G-CIMP DNA methylation showed relevant prognostic value, independently of the known OS predictors in adult diffuse glioma, such as grade and age.18 The favorable prognostic value of G-CIMP+ in both LGGs and GBMs has been reported in many other studies.7,18,19,45–47 Notably, IDH-mutant G-CIMP+ GBM presented favorable survival and molecular similarities to LGG. On the other hand, LGG carrying IDH-wildtype G-CIMP− subsets presented molecular and clinical behavior similar to GBM.7,19,45,46 The refinement of the G-CIMP+ stratification revealed that not all IDH-mutant G-CIMP+ tumors had the same prognosis. The G-CIMP-low subset had the poorest OS among the IDH-mutant gliomas (median survival G-CIMP-low = 2.7 y, G-CIMP-high = 7.2 y, Cox regression P < 0.001; and vs codel = 7.9 y, P < 0.001) resembling the behavior observed in the IDH-wildtype gliomas (median survival 1.2 y).7
Relationship Between G-CIMP+ Subsets and Established Prognostic and/or Predictive Molecular Biomarkers
IDH mutations, codeletion of 1p/19q, MGMT promoter methylation, and G-CIMP+ are all independent favorable prognostic biomarkers.48 However, combining some of them has been shown to improve their individual prognostic value.7,18,48 For instance, patients harboring a triple combination of 1p/19q codeletion, IDH mutation, and MGMT methylation had significantly better OS than those carrying the MGMT methylation biomarker alone.49 In addition, some of them (codeletion of 1p/19q and MGMT promoter methylation) have also been established as predictive biomarkers and have been used in decision making for chemotherapy and/or radiation treatment.50,51 The relationship between G-CIMP and the aforementioned biomarkers will be explored in the following sections.
IDH Mutation
One of the most relevant clinical discoveries in neuro-oncology involves the role of IDH1 mutations as a prognostic marker and potential as a drug target in glioma.1,7,10,19,45,52–54 The R132H IDH1 mutation, an amino acid substitution at a single arginine residue in the active site of the enzyme, is highly prevalent in grade II and III gliomas and appears in secondary GBMs, which develop from lower-grade tumors.10 Although considerably less common, mutations in IDH2, the mitochondrial homolog of the cytosolic IDH1, have also been identified in gliomas.55
Due to the close relationship between IDH mutations and G-CIMP+, it was suggested that the G-CIMP+ prognostic value was mainly due to its relation to IDH mutations.52 The mechanisms associated with favorable prognosis in IDH-mutant G-CIMP+ tumors are still under investigation.13,18 However, a meta-analysis of a G-CIMP gene list with prior gene expression analyses suggested that G-CIMP+ tumors may be less aggressive because of the silencing of key mesenchymal genes.18
The predictive value of IDH mutations for treatment response is still controversial4,50,51,56; however, a phase III clinical trial reported that IDH-mutant anaplastic gliomas benefited from an early combination of procarbazine, lomustine, and vincristine.51 The predictive value of G-CIMP+ and its subsets is still unknown.
1p/19q
The 1p/19q codel has an established favorable prognostic and predictive value in gliomas47,50 and is found exclusively in IDH-mutant tumors.57 The interaction between 1p/19q status and G-CIMP+ has also been reported.12,58 The IDH-mutant G-CIMP+ codel tumors, for example, were associated with a better OS than the IDH-mutant G-CIMP+ non-codel (mean survival G-CIMP+ codel = 9.9 y and G-CIMP+ non-codel = 4.4 y).58 In our pan-glioma cohort, 9.1% of the IDH-mutant G-CIMP+ non-codel subgroup was represented by the G-CIMP-low subtype, with the remainder represented by the G-CIMP-high subtype.7 As described, among the IDH-mutant G-CIMP+ non-codel tumors, the G-CIMP-high subset had a similar survival to the IDH-mutant codel tumors.7
MGMT Promoter Methylation
The epigenetic silencing of MGMT by methylation of its promoter has an established prognostic and predictive value in gliomas and is used in therapeutic decision planning.14–17 The MGMT enzyme repairs the DNA damage caused by alkylating agents, such as TMZ, used to treat patients with GBM, playing a key role in tumor cell resistance to the cytotoxic effect of alkylating agents.
MGMT promoter methylation is associated with IDH mutation and G-CIMP status, regardless of glioma grade.7,11,59–62 In fact, the prognostic versus predictive value of MGMT promoter methylation was found to be at least partly dependent on the context of IDH mutation and G-CIMP status. In IDH-wildtype gliomas, also considered G-CIMP− tumors, MGMT promoter methylation was found to be a predictive marker of a favorable response to alkylating agent chemotherapy.16,50 In contrast, in gliomas harboring IDH mutation, including G-CIMP+ cases, it was reported that MGMT promoter methylation is a prognostic indicator for better survival regardless of treatment with radiotherapy and alkylating agent chemotherapy or with radiotherapy only, but it did not predict the response to treatment.16,50
From our published data,7 we estimated that among the IDH-mutant cohort of The Cancer Genome Atlas (TCGA), 91.8% of glioma specimens presented with MGMT promoter methylation compared with 40.0% among IDH-wildtype tumors (P < 0.001). Notably, G-CIMP-low specimens presented a lower proportion of MGMT promoter methylation (68.0%) compared with G-CIMP-high specimens (88.8%, P = 0.008, unpublished data). These data suggest that G-CIMP+ subsets (-low or -high), as well as IDH1/2 mutation, should be considered when determining the predictive significance of MGMT promoter methylation in gliomas in revealing the benefit from alkylating chemotherapy.
Potential Drivers Associated with G-CIMP
Putative driver mechanisms underpinning the aberrant CpG methylation that occurs in CIMP tumors are still under surveillance in gliomas. Several potential cancer-specific mutated driver genes include IDH1/2 and H3F3A.18,19,45,63 Hotspot mutations in IDH1/2 genes provoke IDH to display a neomorphic enzymatic activity that leads to the reduction of α-ketoglutarate (α-KG) to 2-hydroxyglutarate (2-HG), an oncometabolite that functions as a competitive inhibitor of α-KG.54,64,65 The accumulation of 2-HG impairs the activity of α-KG–dependent dioxygenases, such as histone and DNA demethylases (eg, TET enzymes), leading to global hypermethylation (CIMP), as well as the impairment of cell differentiation.19,64,66 Another consequence of IDH mutations is the alteration of the intracellular levels of the coenzyme NAD+.67 NAD+ has a key role in intracellular signaling pathways implicated in cancer cell growth. It was shown that IDH-mutant cells had a reduced expression of nicotinate phosphoribosyl transferase 1 (NAPRT1), a rate-limiting enzyme within the NAD+ salvage system. Reduced expression of NAPRT1 led to lower basal NAD+ levels, rendering the IDH-mutant cell more vulnerable to death.67,68 Furthermore, IDH mutations also result in the methylation of histones that contribute to DNA methylation by themselves.69 Histone H3F3A mutations were also reported to drive the global aberrant pattern of DNA methylation in a subset of pediatric GBM by as yet unknown mechanisms.45 Interestingly, in a subset of GBM, the authors found recurrent and mutually exclusive mutations either in IDH1 or H3F3A (affecting amino acids K27 or G34, respectively) with distinct clinical, genomic, and epigenomic features.45
Recent large-scale genome-wide association studies provided evidence for a defined germline variant located on chromosome 9p21.3, which was found to be enriched among G-CIMP tumors,70 and a variant mapped to an intronic region located on chromosome 2q33.3, 50K base pairs from IDH1.71 Despite the lack of functional experiments, these findings, in combination with known somatic alterations, offer potential insights into the role of genetic variants in the biology and etiology of G-CIMP tumor development and possibly progression.
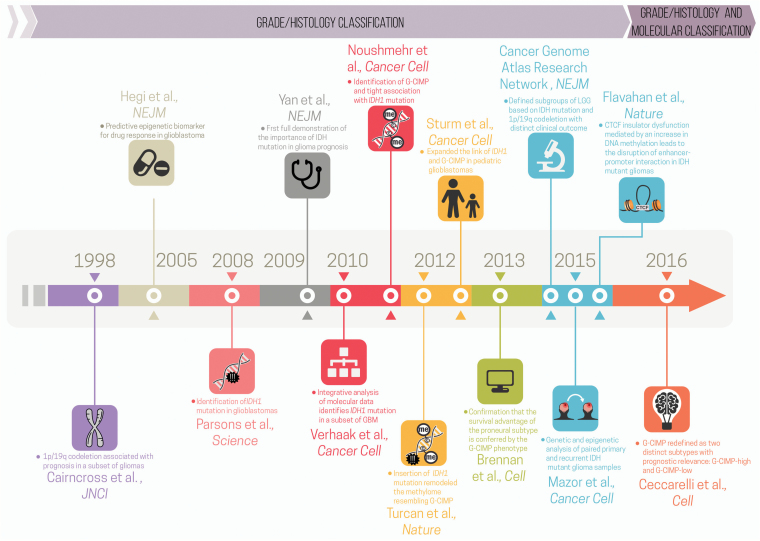
Fig. 2 summarizes the major milestones in integrating genomics and epigenomics data to uncover glioma molecular and clinical phenotypes that led to the characterization (either directly or indirectly) of G-CIMP subsets.
Fig. 2.
Timeline of major milestones in integrating genomics and epigenomics data that directly or indirectly uncovered glioma molecular and clinical phenotypes associated with G-CIMP subsets. Each milestone is indicated by marker papers that reported key molecular findings with clinical implications, along with a bullet summarizing their contribution. The timeline is broken up by the 2016 WHO publication (before and after) (modified from the original copy-free design by Freepik).
G-CIMP+ Subsets and Glioma Progression/Recurrence
WHO grades II and III IDH-mutant non-codel (astrocytic) gliomas habitually recur and unpredictably undergo malignant transformation to highly aggressive and treatment-resistant grade IV GBM.1,72 Remarkably, the analysis of a small cohort of primary and recurrent matched tumor specimens, composed of LGG and GBM, revealed that some G-CIMP-high tumors exhibited a demethylated pattern after relapse that was similar to those observed in the G-CIMP-low gliomas, suggesting a progression from the G-CIMP-high to the G-CIMP-low subset.7 Unpublished work from our own laboratory may provide further refinement of the epigenetic shift from G-CIMP-high to G-CIMP-low upon tumor recurrence in TCGA and non-TCGA specimens.73 Interestingly, in that cohort, G-CIMP-low at recurrence appeared in 12% of all gliomas and shared epigenomic features resembling IDH-wildtype primary GBM.73 Genome-wide decreases in DNA methylation levels associated with progression have also been reported by other studies.74,75
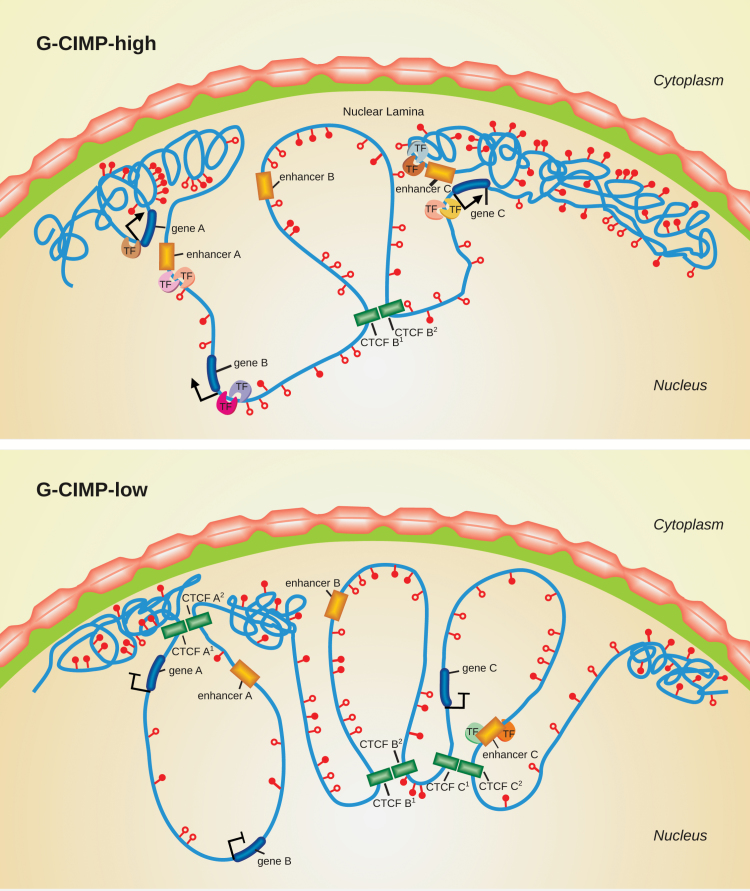
The hypothesis that G-CIMP-high tumors may relapse as G-CIMP-low gliomas suggests that variations in DNA methylation could be a key determinant of the mechanisms that drive glioma progression. Notably, the majority of CpG sites that underwent significant DNA demethylation in G-CIMP-low recurrent tumors were primarily found within intergenic (open sea) regions7 (Fig. 1). CTCF, a methylation-sensitive insulator protein, has an important role in stabilizing enhancer–gene interactions in intergenic regions, as mentioned previously in this review. In IDH-mutant gliomas, it was shown that hypermethylation at CTCF binding sites reduced CTCF binding. The consequent loss of insulation led to aberrant enhancer–gene interactions that ultimately resulted in the upregulation of a glioma oncogene.76 Since the discovery of IDH-mutant glioma subtypes, our group began to investigate the potential role of DNA methylation status at a single base pair resolution using whole-genome bisulfite sequencing. Confirming previous data,7 our unpublished findings revealed that, compared with G-CIMP-high (as well as with nontumoral brain specimens), G-CIMP-low presented DNA hypomethylation at some CTCF binding sites. Collectively, these findings may offer potential support for the hypothesis that the loss of DNA methylation at CTCF binding sites will also influence the chromatin architecture, which is mediated by the disruption of insulator binding. This phenomenon will, in turn, dysregulate nearby genes (Fig. 3) (Sabedot TS et al, unpublished data). However, further studies are needed to confirm this hypothesis. We also found that the hypomethylated intergenic regions were enriched for the oligodendrocyte transcription factor and sex determining region Y-box (Sox)–family binding motifs,7 which have been described as neurodevelopmental transcription factors essential for GBM propagation.77 Sox-family genes are transcription factors that are also involved in the induction and maintenance of stem cell pluripotency,78 promote self-renewal of neural stem cells in the nervous system,79 and regulate the plasticity between glioma stem cell and non–stem cell states within brain tumors.80 Accordingly, G-CIMP-low tumors displayed abnormalities in cell cycle pathway genes, such as cyclin-dependent kinase 4 (CDK4) and cyclin-dependent kinase inhibitor 2A (CDKN2A), supporting the association between stem cell signaling pathways and tumor proliferation in gliomas. Therefore, loss of CpG methylation at these functional genomic elements, known to be associated with normal development and pluripotency, defines a possible mechanism of glioma progression.7,73
Fig. 3.
Chromatin changes in the progression of G-CIMP-high to G-CIMP-low tumors. This illustration shows a model of chromatin reorganization during the progression from G-CIMP-high to G-CIMP-low. G-CIMP-low (lower panel) shows a loss of DNA methylation at specific loci causing disruption of CTCF binding sites, reorganization of chromatin, and dysregulation of gene expression.
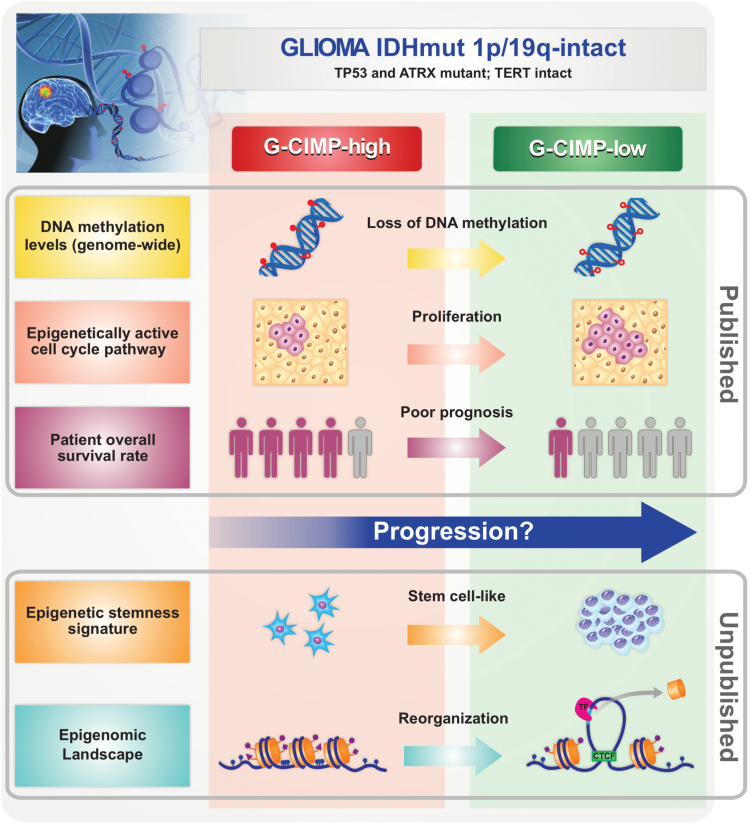
In addition, Bai et al found that key developmental transcription factors that are regulated by polycomb repressive complex 2 in human stem cells became hypermethylated during glioma progression, resulting in gene silencing, which could ultimately cause GBM cells to enter a continuous state of stem cell–like self-renewal.75 Mazor et al reported specific hypomethylation events that may contribute to increased cell proliferation upon progression from LGGs to GBMs.74 Recently, we derived a metric to measure the degree of de-differentiation of tumors based on DNA methylation and found a strong association between a high undifferentiated score and glioma molecular subtypes associated with worse clinical outcomes (ie, G-CIMP-low, classic-like, mesenchymal-like, LGm6-GBM, and PA-like). Interestingly, G-CIMP-low patients showed higher undifferentiated scores compared with G-CIMP-high patients, resembling those found in primary IDH-wildtype tumors (Malta TM et al, unpublished). This finding may suggest that a stem cell–like phenotype may be involved in unfavorable clinical outcomes and reinforce the importance of epigenetic alterations occurring in intergenic regions that may disrupt important regulatory elements affecting oncogenesis and tumor progression (Fig. 4).
Fig. 4.
Overview of major discoveries that define G-CIMP-high and G-CIMP-low glioma subsets. G-CIMP-high and G-CIMP-low tumors share the following genomic alterations: IDH-mutant–1p/19q intact, telomerase reverse transcriptase promoter wildtype, and alpha thalassemia/mental retardation syndrome X-linked and tumor protein 53 mutant. However, the G-CIMP-low subset defined a subgroup of IDH-mutant 1p/19q intact gliomas associated with DNA demethylation. Changes in chromatin architecture led to an imbalance between the insulators and enhancers and the consequent activation of cell-cycle related genes, the increase in stemness features, and poor clinical outcome compared with G-CIMP-high gliomas (cartoon representation, not to scale).
G-CIMP Detection
The diagnosis of G-CIMP was reported by Noushmehr et al.18 This diagnosis was based on an epigenetic biomarker panel consisting of 7 hypermethylated loci (ANKRD43, HFE, MAL, LGALS3, FAS-1, FAS-2, and RHO-F) and 1 hypomethylated locus, DOCK5, validated in silico. A sample was considered G-CIMP+ if at least 6 genes displayed a combination of DOCK5 DNA hypomethylation and/or hypermethylation of the remaining genes in the panel. The utilization of the MethyLight assay to detect the aforementioned biomarkers showed perfect concordance with the results obtained by array platforms and may be suitable for clinical utilization.18
Later, G-CIMP-high and -low subsets were defined and validated in silico by a panel of 163 DNA-methylated probe signatures using methylation arrays.7 Our group developed a concise DNA methylation biomarker panel, derived from those probes, consisting of 14 methylated probes allowing the identification of the 7 glioma subgroups that were previously reported7 and validated in silico (unpublished data). Eight of those probes distinguished between G-CIMP+ subsets (-low and -high) in the context of IDH-mutant glioma specimens; however, their detection by more readily available assays warrants investigation.
Epigenomic- and Chromatin-Targeted Therapies
Epigenetic mechanisms are heritable and potentially actionable, making them an attractive target for the treatment of diseases, including cancer.81 Many of the epigenome-targeting drugs have been demonstrated to be beneficial and safe in hematological malignancies.82,83 However, studies in solid tumors are limited.53,81,84 Epigenetic drugs approved or in clinical trials have been thoroughly explored in recently published reviews.53,81,85
In gliomas, few trials from this class have been completed.82,83,85,86 To our knowledge, there are no clinical trials specifically addressing G-CIMP+ subtypes. However, preclinical and clinical studies using drugs targeting DNA methylation directly, or the mechanisms involved in G-CIMP, are under way. Hypomethylating agents such as the inhibitors of DNMT, of histone deacetylase (HDAC), and of the bromodomain and extraterminal motif proteins (eg, BRD4 inhibitor) represent epigenetic drugs with broad actions.81
DNMT inhibitors or DNA demethylating agents (eg, 5-azacytidine, 5-aza-2-deoxycytidine-decitabine) promote dose-dependent global demethylation activity by depleting or degrading DNA methyltransferases. This may reverse aberrant expression of genes related to nearly all pathways involved in cancer initiation and progression (ie, tumor suppressor genes, oncogenes, genes associated with stemness or pluripotency, apoptosis, cell-cycle, and immune response).87,88 Moreover, DNMT inhibitors were also shown to reverse tumor immune evasion through viral infection mimicry and upregulation of viral defense pathways.89–91 For instance, in an IDH1-mutant glioma xenograft, low doses of the demethylating agent decitabine restored the activity of DNMT1 and, consequently, reversed DNA methylation marks in promoters of differentiating genes. This change resulted in the loss of stemlike features and in the arrest of glioma growth in that model.92 Although preclinical studies using these drugs in gliomas seem promising, a corresponding clinical trial using decitabine on adult glioma has not been reported. Considering the potential adverse impact of loss of methylation in the progression or recurrence of gliomas, reported previously,73,74 it seems reasonable to take G-CIMP subsets into account during clinical trial design.
Targeting IDH-mutant tumors, including G-CIMP+ gliomas, with the IDH-mutant enzyme-inhibiting agents is currently under investigation in clinical trials.53,67,81 The mechanistic intent of these drugs involves the metabolic pathways altered in IDH-mutant tumors (ie, decrease of 2-HG in hematological or salvage of NAD+ in solid tumors).67 However, a preclinical study has not identified a decrease or epigenetic change in IDH-mutant cancer initiating cells exposed to an IDH-R132 inhibitor drug, despite marked reduction in 2-HG.67 Tateishi et al postulated that the activity of this class of drugs may be limited to a subset of tumors. Moreover, NAD+ metabolic depletion was recently shown to be an attractive therapeutic target in IDH1-mutant cells vulnerable to a cytotoxic response when exposed to a nicotinamide phosphoribosyl transferase (NAMPT) inhibitor. The same response was not seen in IDH1-wildtype cell lines67; however, a later preclinical model demonstrated that NAMPT inhibitors enhanced the cytotoxic effects of TMZ in GBM cells.93
Immune modulation is also affected by epigenomic alterations and has been shown to be an attractive target for pharmacotherapy in cancer.94 The identification/targeting of disease-specific neoantigens has demonstrated success in other diseases, renewing excitement for immunotherapy in glioma. An immunogenic epitope vaccine targeting mutant IDH1 in glioma with promising preclinical work is now in phase I clinical trials.95,96 Several other clinical trials are under way in glioma, including phase III targeted immunotherapy trials.97 The use of immune checkpoint inhibitors targeting programmed cell death protein 1 (PD1) and/or its ligand 1 (PD-L1) has also shown activity in several cancer types and is currently being tested in GBM clinical trials. PD-L1 promoter methylation and lower PD-L1 expression in IDH-mutant gliomas compared with IDH-wildtype point to an epigenetic regulation of PD-L1 and indicate that patients harboring IDH-mutant gliomas may not benefit from monotherapy with drugs targeting the blocking of the PD1/PD-L1 pathway.98
Coadministration of epigenomic agents (eg, DNA-demethylating agents, HDAC inhibitors) has demonstrated improved efficacy of immunotherapy in many tumor types by increasing the tumoral immune response, enhancing the expression of immune blockade checkpoints, and reducing cellular adaptation that leads to drug resistance.99 Given the potential epigenetic regulation of PD-L1 expression, patients harboring IDH-mutant (G-CIMP+) tumors may benefit from combined treatment modalities, including demethylating agents and PD1/PD-L1 inhibitors. The relationship between the G-CIMP subsets and response to immunotherapy, either as a predictive marker or epigenetic therapy target, is largely unknown and worthy of further investigation.
Final Remarks and Perspectives
Advances in glioma research have highlighted the significance of epigenomic alterations. The incorporation of genetic markers into the traditional WHO histopathological classification of CNS tumors reflects widespread adoption of the latest scientific and clinical advances in molecular neuro-oncology into clinical practice.1 Moreover, evidence suggests that comprehensive analysis, such as unsupervised glioma subtyping based on gene expression or on G-CIMP status, rather than individual molecular markers, may improve prognostic and predictive outcomes.100 Currently, the guidelines for the detection of established prognostic biomarkers are based on individual marker assays—for example, immunohistochemistry or DNA sequencing for mutations in IDH1/2 and H3F3A genes, fluorescent in situ hybridization or microsatellite PCR-based loss of heterozygosity analyses for codeletion of chromosomal arms 1p and 19q, and real- time methylation-specific PCR for MGMT promoter methylation.61 However, the advent of genome-wide analysis technologies has enabled the concurrent detection of DNA methylation patterns and genomic and copy number alterations in tumor specimens.61 Remarkably, the identification of a DNA methylation–based classifier of glioma specimens enabled researchers to capture already known prognostic markers, such as IDH mutation and 1p/19q codeletion.7 In addition, it uniquely enabled the identification of a subset of tumors within the IDH-mutant/G-CIMP+ group (ie, G-CIMP-low) that presented a prognostic disadvantage in relation to their counterparts (IDH-mutant G-CIMP-high non-codels and codels). In addition to refining glioma classification, the identification of G-CIMP+ subsets also shed light on the role of the demethylation of specific genes in the pathogenesis of glioma progression, independently of grade or IDH status.7
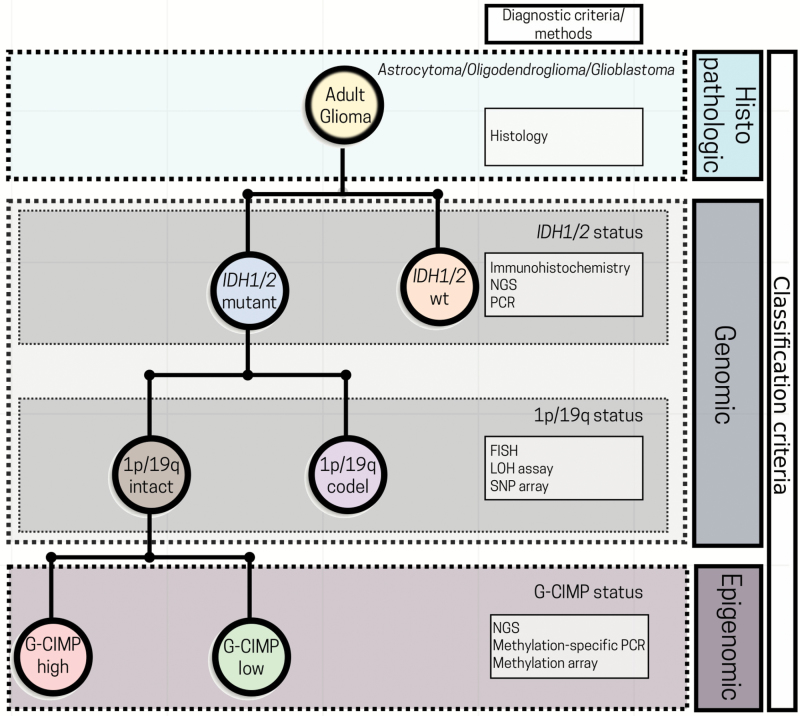
In summary, the detection of G-CIMP+ and its subsets (-high and -low) builds on the 2016 WHO molecular effort to refine glioma classification (Fig. 5) and provides insight into disease course and treatment opportunity.73 The prognostic significance of G-CIMP+ across all glioma types has been confirmed in many studies.7,18,19,45–47 It is possible that, as for other established therapeutic predictive markers in gliomas, such as 1p/19q codeletion and MGMT promoter methylation, G-CIMP+ subsets will be used for patient counseling and be part of algorithms used for clinical trial design and for therapeutic decisions.4 However, the utility of these classifiers and biomarkers in planning treatment strategies and designing clinical trials has not been fully validated to date.
Fig. 5.
Perspectives in incorporating G-CIMP subsets into the current 2016 WHO glioma classification algorithm. A simplified diagram for glioma classification based on histological, genetic, and epigenetic features. The incorporation of G-CIMP subsets further refined glioma classification. NGS: next-generation sequencing; PCR: polymerase chain reaction; FISH: fluorescent in situ hybridization; LOH: loss of heterozygosity; SNP: single-nucleotide polymorphism; wt: wildtype (modified from the original copy-free design by Freepik).
Funding
Support for this work was provided by an institutional Grant Henry Ford Hospital. Grants 14/02245-3, 15/07925-5, 16/01389-7, 16/06488-3, 16/01975-3, 16/15485-8, 16/12329-5, 16/10436-9, and 16/11039-3 Sao Paulo Research Foundation (FAPESP). Grant 14/08321-3 Sao Paulo Research Foundation (FAPESP) and Coordination of Improvement of Higher Education Personnel (CAPES). Grant 164061/2015-0 Brazilian National Council for Scientific and Technological Development (CNPq).
Acknowledgments
The authors thank Sandra Navarro at the Regional Blood Bank of Ribeirao Preto for carefully assembling the figures and OMICs laboratory members for their helpful discussion and contribution. We thank Michelle Felicella and Chunhai (Charlie) Hao for insightful discussion about glioma classification. We would also like to thank Laila Poisson, Ana deCarvalho,Tobias Walbert and Susan MacPhee-Gray for critical review of the manuscript.
Conflict of interest statement. The authors declare no conflict of interest.
References
- 1. Louis DN, Perry A, Reifenberger G et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 2. Masui K, Mischel PS, Reifenberger G. Molecular classification of gliomas. Handb Clin Neurol. 2016;134:97–120. [DOI] [PubMed] [Google Scholar]
- 3. Siegal T. Clinical relevance of prognostic and predictive molecular markers in gliomas. Adv Tech Stand Neurosurg 2016;(43):91–108. [DOI] [PubMed] [Google Scholar]
- 4. Taylor JW, Chi AS, Cahill DP. Tailored therapy in diffuse gliomas: using molecular classifiers to optimize clinical management. Oncology (Williston Park). 2013;27(6):504–514. [PubMed] [Google Scholar]
- 5. Gittleman H, Lim D, Kattan MW et al. An independently validated nomogram for individualized estimation of survival among patients with newly diagnosed glioblastoma: NRG Oncology RTOG 0525 and 0825. Neuro Oncol 2016;19(5):669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hoadley KA, Yau C, Wolf DM et al. ; Cancer Genome Atlas Research Network Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158(4):929–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ceccarelli M, Barthel FP, Malta TM et al. ; TCGA Research Network Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. van den Bent MJ, Looijenga LH, Langenberg K et al. Chromosomal anomalies in oligodendroglial tumors are correlated with clinical features. Cancer. 2003;97(5):1276–1284. [DOI] [PubMed] [Google Scholar]
- 9. Cairncross JG, Ueki K, Zlatescu MC et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst. 1998;90(19):1473–1479. [DOI] [PubMed] [Google Scholar]
- 10. Parsons DW, Jones S, Zhang X et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eckel-Passow JE, Lachance DH, Molinaro AM et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372(26):2499–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brat DJ, Verhaak RG, Aldape KD et al. ; Cancer Genome Atlas Research Network Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med 2015;372(26):2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. LeBlanc VG, Marra MA. DNA methylation in adult diffuse gliomas. Brief Funct Genomics. 2016;15(6):491–500. [DOI] [PubMed] [Google Scholar]
- 14. Esteller M, Garcia-Foncillas J, Andion E et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343(19):1350–1354. [DOI] [PubMed] [Google Scholar]
- 15. Hegi ME, Diserens AC, Godard S et al. Clinical trial substantiates the predictive value of O-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin Cancer Res. 2004;10(6):1871–1874. [DOI] [PubMed] [Google Scholar]
- 16. Hegi ME, Diserens AC, Gorlia T et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. [DOI] [PubMed] [Google Scholar]
- 17. Rivera AL, Pelloski CE, Gilbert MR et al. MGMT promoter methylation is predictive of response to radiotherapy and prognostic in the absence of adjuvant alkylating chemotherapy for glioblastoma. Neuro Oncol. 2010;12(2):116–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Noushmehr H, Weisenberger DJ, Diefes K et al. ; Cancer Genome Atlas Research Network Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Turcan S, Rohle D, Goenka A et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31(1):27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shen H, Laird PW. Interplay between the cancer genome and epigenome. Cell. 2013;153(1):38–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4(2):143–153. [DOI] [PubMed] [Google Scholar]
- 23. Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. [DOI] [PubMed] [Google Scholar]
- 24. Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25(23):2436–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196(2):261–282. [DOI] [PubMed] [Google Scholar]
- 26. Sandoval J, Heyn H, Moran S et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6(6):692–702. [DOI] [PubMed] [Google Scholar]
- 27. Blackwood EM, Kadonaga JT. Going the distance: a current view of enhancer action. Science. 1998;281(5373):60–63. [DOI] [PubMed] [Google Scholar]
- 28. Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 2006;7(9):703–713. [DOI] [PubMed] [Google Scholar]
- 29. Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405(6785):486–489. [DOI] [PubMed] [Google Scholar]
- 30. Liu XS, Wu H, Ji X et al. Editing DNA methylation in the mammalian genome. Cell. 2016;167(1):233–247.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. De Carvalho DD, You JS, Jones PA. DNA methylation and cellular reprogramming. Trends Cell Biol. 2010;20(10):609–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee ST, Wiemels JL. Genome-wide CpG island methylation and intergenic demethylation propensities vary among different tumor sites. Nucleic Acids Res. 2016;44(3):1105–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Baylin SB, Esteller M, Rountree MR, Bachman KE, Schuebel K, Herman JG. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum Mol Genet. 2001;10(7):687–692. [DOI] [PubMed] [Google Scholar]
- 34. Berdasco M, Esteller M. Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Dev Cell. 2010;19(5):698–711. [DOI] [PubMed] [Google Scholar]
- 35. Esteller M. Epigenetic gene silencing in cancer: the DNA hypermethylome. Hum Mol Genet 2007;16(1):R50-9. [DOI] [PubMed] [Google Scholar]
- 36. Hansen KD, Timp W, Bravo HC et al. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43(8):768–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96(15):8681–8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Weisenberger DJ. Characterizing DNA methylation alterations from The Cancer Genome Atlas. J Clin Invest. 2014;124(1):17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miller BF, Sánchez-Vega F, Elnitski L. The emergence of pan-cancer CIMP and its elusive interpretation. Biomolecules 2016;6(4):pii: E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Suzuki H, Yamamoto E, Maruyama R, Niinuma T, Kai M. Biological significance of the CpG island methylator phenotype. Biochem Biophys Res Commun 2014;455(1–2):35–42. [DOI] [PubMed] [Google Scholar]
- 41. Witte T, Plass C, Gerhauser C. Pan-cancer patterns of DNA methylation. Genome Med. 2014;6(8):66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sánchez-Vega F, Gotea V, Margolin G, Elnitski L. Pan-cancer stratification of solid human epithelial tumors and cancer cell lines reveals commonalities and tissue-specific features of the CpG island methylator phenotype. Epigenetics Chromatin. 2015;8:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Verhaak RG, Hoadley KA, Purdom E et al. ; Cancer Genome Atlas Research Network Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Phillips HS, Kharbanda S, Chen R et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. [DOI] [PubMed] [Google Scholar]
- 45. Sturm D, Witt H, Hovestadt V et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22(4):425–437. [DOI] [PubMed] [Google Scholar]
- 46. Brennan CW, Verhaak RG, McKenna A et al. ; TCGA Research Network The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wiestler B, Capper D, Sill M et al. Integrated DNA methylation and copy-number profiling identify three clinically and biologically relevant groups of anaplastic glioma. Acta Neuropathol. 2014;128(4):561–571. [DOI] [PubMed] [Google Scholar]
- 48. Mur P, Rodríguez de Lope Á, Díaz-Crespo FJ et al. Impact on prognosis of the regional distribution of MGMT methylation with respect to the CpG island methylator phenotype and age in glioma patients. J Neurooncol. 2015;122(3):441–450. [DOI] [PubMed] [Google Scholar]
- 49. Leu S, von Felten S, Frank S et al. IDH/MGMT-driven molecular classification of low-grade glioma is a strong predictor for long-term survival. Neuro Oncol. 2013;15(4):469–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wick W, Meisner C, Hentschel B et al. Prognostic or predictive value of MGMT promoter methylation in gliomas depends on IDH1 mutation. Neurology. 2013;81(17):1515–1522. [DOI] [PubMed] [Google Scholar]
- 51. Cairncross JG, Wang M, Jenkins RB et al. Benefit from procarbazine, lomustine, and vincristine in oligodendroglial tumors is associated with mutation of IDH. J Clin Oncol. 2014;32(8):783–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cohen AL, Holmen SL, Colman H. IDH1 and IDH2 mutations in gliomas. Curr Neurol Neurosci Rep. 2013;13(5):345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mondesir J, Willekens C, Touat M, de Botton S. IDH1 and IDH2 mutations as novel therapeutic targets: current perspectives. J Blood Med. 2016;7:171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yan H, Parsons DW, Jin G et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hartmann C, Meyer J, Balss J et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1010 diffuse gliomas. Acta Neuropathol. 2009;118(4):469–474. [DOI] [PubMed] [Google Scholar]
- 56. Wiestler B, Claus R, Hartlieb SA et al. ; Neuro-oncology Working Group (NOA) of the German Cancer Society Malignant astrocytomas of elderly patients lack favorable molecular markers: an analysis of the NOA-08 study collective. Neuro Oncol. 2013;15(8):1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Labussière M, Idbaih A, Wang XW et al. All the 1p19q codeleted gliomas are mutated on IDH1 or IDH2. Neurology. 2010;74(23):1886–1890. [DOI] [PubMed] [Google Scholar]
- 58. Mur P, Mollejo M, Ruano Y et al. Codeletion of 1p and 19q determines distinct gene methylation and expression profiles in IDH-mutated oligodendroglial tumors. Acta Neuropathol. 2013;126(2):277–289. [DOI] [PubMed] [Google Scholar]
- 59. Bady P, Sciuscio D, Diserens AC et al. MGMT methylation analysis of glioblastoma on the Infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status. Acta Neuropathol. 2012;124(4):547–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. van den Bent MJ, Gravendeel LA, Gorlia T et al. A hypermethylated phenotype is a better predictor of survival than MGMT methylation in anaplastic oligodendroglial brain tumors: a report from EORTC study 26951. Clin Cancer Res. 2011;17(22):7148–7155. [DOI] [PubMed] [Google Scholar]
- 61. Wiestler B, Capper D, Hovestadt V et al. Assessing CpG island methylator phenotype, 1p/19q codeletion, and MGMT promoter methylation from epigenome-wide data in the biomarker cohort of the NOA-04 trial. Neuro Oncol. 2014;16(12):1630–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wick W, Roth P, Hartmann C et al. ; Neurooncology Working Group (NOA) of the German Cancer Society Long-term analysis of the NOA-04 randomized phase III trial of sequential radiochemotherapy of anaplastic glioma with PCV or temozolomide. Neuro Oncol. 2016;18(11):1529–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Duncan CG, Barwick BG, Jin G et al. A heterozygous IDH1R132H/WT mutation induces genome-wide alterations in DNA methylation. Genome Res. 2012;22(12):2339–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Dang L, White DW, Gross S et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ward PS, Patel J, Wise DR et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17(3):225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Xu W, Yang H, Liu Y et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 2011;19(1):17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tateishi K, Wakimoto H, Iafrate AJ et al. Extreme vulnerability of IDH1 mutant cancers to NAD+ depletion. Cancer Cell. 2015;28(6):773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chiarugi A, Dölle C, Felici R, Ziegler M. The NAD metabolome—a key determinant of cancer cell biology. Nat Rev Cancer. 2012;12(11):741–752. [DOI] [PubMed] [Google Scholar]
- 69. Lu C, Ward PS, Kapoor GS et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483(7390):474–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Dahlin AM, Wibom C, Ghasimi S, Brännström T, Andersson U, Melin B. Relation between established glioma risk variants and DNA methylation in the tumor. PLoS One. 2016;11(10):e0163067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Melin BS, Barnholtz-Sloan JS, Wrensch MR et al. ; GliomaScan Consortium Genome-wide association study of glioma subtypes identifies specific differences in genetic susceptibility to glioblastoma and non-glioblastoma tumors. Nat Genet. 2017;49(5):789–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sanai N, Chang S, Berger MS. Low-grade gliomas in adults. J Neurosurg. 2011;115(5):948–965. [DOI] [PubMed] [Google Scholar]
- 73. de Souza CF, Sabedot TS, Malta TM et al. Distinct epigenetic shift in a subset of glioma CpG island methylator phenotype (G-CIMP) during tumor recurrence. bioRxiv 156646. doi:10.1101/156646
- 74. Mazor T, Pankov A, Johnson BE et al. DNA methylation and somatic mutations converge on the cell cycle and define similar evolutionary histories in brain tumors. Cancer Cell. 2015;28(3):307–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bai H, Harmancı AS, Erson-Omay EZ et al. Integrated genomic characterization of IDH1-mutant glioma malignant progression. Nat Genet. 2016;48(1):59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Flavahan WA, Drier Y, Liau BB et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529(7584):110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Suvà ML, Rheinbay E, Gillespie SM et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157(3):580–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sarkar A, Hochedlinger K. The Sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 2013;12(1):15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39(5):749–765. [DOI] [PubMed] [Google Scholar]
- 80. Berezovsky AD, Poisson LM, Cherba D et al. Sox2 promotes malignancy in glioblastoma by regulating plasticity and astrocytic differentiation. Neoplasia. 2014;16(3):193–206, 206.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Jones PA, Issa JP, Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet. 2016;17(10):630–641. [DOI] [PubMed] [Google Scholar]
- 82. Chen R, Cohen AL, Colman H. Targeted therapeutics in patients with high-grade gliomas: past, present, and future. Curr Treat Options Oncol. 2016;17(8):42. [DOI] [PubMed] [Google Scholar]
- 83. Azad N, Zahnow CA, Rudin CM, Baylin SB. The future of epigenetic therapy in solid tumours—lessons from the past. Nat Rev Clin Oncol. 2013;10(5):256–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Reifenberger G, Wirsching HG, Knobbe-Thomsen CB, Weller M. Advances in the molecular genetics of gliomas—implications for classification and therapy. Nat Rev Clin Oncol. 2017;14(7):434–452. [DOI] [PubMed] [Google Scholar]
- 85. Maio M, Covre A, Fratta E et al. Molecular pathways: at the crossroads of cancer epigenetics and immunotherapy. Clin Cancer Res. 2015;21(18):4040–4047. [DOI] [PubMed] [Google Scholar]
- 86. Nervi C, De Marinis E, Codacci-Pisanelli G. Epigenetic treatment of solid tumours: a review of clinical trials. Clin Epigenetics. 2015;7:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Mund C, Brueckner B, Lyko F. Reactivation of epigenetically silenced genes by DNA methyltransferase inhibitors: basic concepts and clinical applications. Epigenetics. 2006;1(1):7–13. [DOI] [PubMed] [Google Scholar]
- 88. Chiappinelli KB, Zahnow CA, Ahuja N, Baylin SB. Combining epigenetic and immunotherapy to combat cancer. Cancer Res. 2016;76(7):1683–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Roulois D, Yau HL, De Carvalho DD. Pharmacological DNA demethylation: implications for cancer immunotherapy. Oncoimmunology. 2016;5(3):e1090077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Licht JD. DNA methylation inhibitors in cancer therapy: the immunity dimension. Cell. 2015;162(5):938–939. [DOI] [PubMed] [Google Scholar]
- 91. Wrangle J, Wang W, Koch A et al. Alterations of immune response of non-small cell lung cancer with azacytidine. Oncotarget. 2013;4(11):2067–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Turcan S, Fabius AW, Borodovsky A et al. Efficient induction of differentiation and growth inhibition in IDH1 mutant glioma cells by the DNMT inhibitor decitabine. Oncotarget. 2013;4(10):1729–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Feng J, Yan PF, Zhao HY, Zhang FC, Zhao WH, Feng M. Inhibitor of nicotinamide phosphoribosyltransferase sensitizes glioblastoma cells to temozolomide via activating ROS/JNK signaling pathway. Biomed Res Int. 2016;2016:1450843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Obata Y, Furusawa Y, Hase K. Epigenetic modifications of the immune system in health and disease. Immunol Cell Biol. 2015;93(3):226–232. [DOI] [PubMed] [Google Scholar]
- 95. Schumacher T, Bunse L, Pusch S et al. A vaccine targeting mutant IDH1 induces antitumour immunity. Nature. 2014;512(7514):324–327. [DOI] [PubMed] [Google Scholar]
- 96. Dimitrov L, Hong CS, Yang C, Zhuang Z, Heiss JD. New developments in the pathogenesis and therapeutic targeting of the IDH1 mutation in glioma. Int J Med Sci. 2015;12(3):201–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Tivnan A, Heilinger T, Lavelle EC, Prehn JH. Advances in immunotherapy for the treatment of glioblastoma. J Neurooncol. 2017;131(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Berghoff AS, Kiesel B, Widhalm G et al. Correlation of immune phenotype with IDH mutation in diffuse glioma. Neuro Oncol. 2017;19(11):1460–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Park J, Thomas S, Munster PN. Epigenetic modulation with histone deacetylase inhibitors in combination with immunotherapy. Epigenomics. 2015;7(4):641–652. [DOI] [PubMed] [Google Scholar]
- 100. Sahebjam S, McNamara MG, Mason WP. Emerging biomarkers in anaplastic oligodendroglioma: implications for clinical investigation and patient management. CNS Oncol. 2013;2(4):351–358. [DOI] [PMC free article] [PubMed] [Google Scholar]