Abstract
The molecular mechanisms regulating hemicelluloses and pectin biosynthesis are poorly understood. An important question in this regard is how glycosyltransferases are oriented in the Golgi cisternae, and how nucleotide sugars are made available for the synthesis of the polymers. Here we show that the branching enzyme xyloglucan α,1–2 fucosyltransferase (XG-FucTase) from growing pea (Pisum sativum) epicotyls was latent and protected against proteolytic inactivation on intact, right-side-in pea stem Golgi vesicles. Moreover, much of the XG-FucTase activity was membrane associated. These data indicate that XG-FucTase is a membrane-bound luminal enzyme. GDP-Fuc uptake studies demonstrated that GDP-Fuc was taken up into Golgi vesicles in a protein-mediated process, and that this uptake was not competed by UDP-Glc, suggesting that a specific GDP-Fuc transporter is involved in xyloglucan biosynthesis. Once in the lumen, Fuc was transferred onto endogenous acceptors, including xyloglucan. GDPase activity was detected in the lumen of the vesicles, suggesting than the GDP produced upon transfer of Fuc was hydrolyzed to GMP and inorganic phosphate. We suggest than the GDP-Fuc transporter and GDPase may be regulators of xyloglucan fucosylation in the Golgi apparatus from pea epicotyls.
Plant cells are surrounded by an extracellular matrix known as the cell wall, which plays an important role in development, defense against pathogen attack, and mechanical resistance (Brett and Waldron, 1996). This matrix is composed mainly of polysaccharides, the most abundant of which are cellulose, hemicelluloses, and pectin. The synthesis of these polymers takes place in different subcellular compartments. Whereas cellulose is made at the plasma membrane, hemicelluloses and pectin are synthesized in the Golgi apparatus (Carpita and Gibeaut, 1993).
Mutants deficient in the synthesis of cellulose and hemicelluloses are contributing to our efforts to understand how and when these polysaccharides are synthesized (Reiter et al., 1993; Reiter et al., 1997; Turner and Somerville, 1997; Arioli et al., 1998). Additionally, key genes involved in the synthesis of cellulose and hemicellulose have been recently cloned (Pear et al., 1996; Arioli et al., 1998; Perrin et al., 1999; Taylor et al., 1999). However, little is known about the molecular and cellular mechanisms involved in the synthesis of these polysaccharides. We have focused our attention on the topological constraints associated with the synthesis of these polymers. This interest comes from the observation that nucleotide sugars utilized for polysaccharide biosynthesis are made in the cytoplasm (Coates et al., 1980; Bonin et al., 1997), while the products of this biosynthesis are found in the extracellular space (cellulose) and in the lumen of the Golgi apparatus (hemicelluloses and pectin) (Zhang and Staehelin, 1992).
A model to explain this topological constraint in the plasma membrane has been proposed for cellulose biosynthesis (Delmer and Amor, 1995). According to this model, cellulose synthase, a processive glycosyltransferase located in the plasma membrane, takes UDP-Glc and transfers Glc into cellulose on the cytosolic face of the membrane. The elongating polymer then crosses the membrane and is finally deposited on the outer face of the plasma membrane, becoming part of the cell wall. Thus, the question is whether glycosyltransferases involved in hemicellulose and pectin biosynthesis in the Golgi apparatus transfer the sugars into polysaccharides in a manner similar to cellulose synthase. Alternatively, the glycosyltransferases involved in hemicellulose and pectin biosynthesis may utilize a different mechanism to deal with the topological constraints of the subcellular location of substrate and end product.
One of the most abundant hemicelluloses in the primary cell wall of dicots is xyloglucan, a branched polymer that contains a β-1,4-Glc backbone substituted with α-1,6-Xyl or α–1,6-Xyl-β–1,2-Gal-α–1,2-Fuc side branches. The xyloglucan backbone resembles the structure of cellulose, and it is possible that a processive glycosyltransferase with similar characteristics to cellulose synthase is involved in this process. However, glucan synthase I, an enzyme that has been postulated to participate in the synthesis of the xyloglucan backbone (White et al., 1993), is detected in the lumen of the Golgi cisternae (Muñoz et al., 1996). In addition, UDP-Glc, the substrate for this enzyme, is transported into the lumen of the Golgi apparatus (Muñoz et al., 1996; Neckelmann and Orellana, 1998). Therefore, we have proposed that the synthesis of the Glc backbone of xyloglucan occurs in the lumen of the Golgi apparatus (Neckelmann and Orellana, 1998) by a mechanism topologically different from the one proposed for the synthesis of cellulose (Chrispeels et al., 1999). According to this model, the transport of UDP-Glc into the lumen of the Golgi cisternae via a UDP-Glc transporter is a necessary and important step. Moreover, upon transfer of Glc into the elongating polymer, UDP, the nucleotide moiety of UDP-Glc, is produced in the lumen and quickly hydrolyzed by a UDPase located in the lumen of the Golgi cisternae to UMP and inorganic phosphate (Pi) (Orellana et al., 1997). It is thought that this reaction drives the polymerization of Glc, favoring the synthesis of the polysaccharide. Therefore, UDPase may also play an important role in the mechanism of polysaccharide biosynthesis.
The model described above has been proposed based on studies of the synthesis of a β,1–4 Glc-polymer made by a processive enzyme, and the utilization of UDP-Glc. However, there are a number of glycosyltransferases involved in branching reactions that use uridine- and guanosine-derived nucleotide sugars, raising further questions about the model. Does this model apply to glycosyltransferases involved in branching reactions? Are nucleotide sugar transporters required for the uptake of all nucleotide sugars? How specific are these putative nucleotide sugar transporters? Are there different nucleoside diphosphatases in the Golgi? To answer some of these questions, we analyzed the orientation of the branching enzyme xyloglucan α,1–2 fucosyltransferase (XG-FucTase) in Golgi vesicles from pea (Pisum sativum) stems. This enzyme uses GDP-Fuc as a substrate and is responsible for the addition of the terminal Fuc onto xyloglucan (Camirand et al., 1987; Farkas and Maclachlan, 1988; Hanna et al., 1991). Our results indicate that XG-FucTase has its active site in the lumen of the Golgi cisternae. We also found that GDP-Fuc, the substrate for this enzyme, is taken up into Golgi vesicles in a protein-mediated process that is distinct from the UDP-Glc transporter, suggesting that a different transporter would be responsible for the transport of GDP-Fuc into the Golgi cisternae. In addition, a GDPase activity indistinguishable from the Golgi UDPase by electrophoresis in native gels is located in the lumen of the Golgi cisternae, suggesting that the enzyme responsible for the metabolism of the GDP released during the transfer of Fuc to its acceptor could be the same enzyme responsible for the hydrolysis of UDP.
MATERIALS AND METHODS
GDP-β-l-[3H]Fuc (5.2 Ci mmol−1) was purchased from DuPont-New England Nuclear (Boston), and UDP-[3H]Glc (4.5 Ci mmol−1) was purchased from Amersham (Buckinghamshire, UK). Non-radioactive GDP-Fuc, UDP, GDP, Triton X-100, glass fiber filters, and Trizma were purchased from Sigma-Aldrich (St. Louis). Xyloglucan from tamarind and cellulase from Trichoderma sp. were obtained from Megazyme International (Boronia, Australia). High-purity Suc was obtained from ICN (Costa Mesa, CA). Proteinase K was from Merck (Darmstadt, Germany).
Plant Material
Pea (Pisum sativum var Alaska) seeds were obtained from Instituto de Investigaciones Agropecuarias-Carillanca. Seeds were grown in moist vermiculite for 7 to 8 d in the dark at 25°C. Stem segments (1 cm) were excised from the elongating region of the epicotyls and kept on ice until homogenization.
Isolation of a Golgi Vesicle Fraction and a Subcellular Fractionation
Pea stems were homogenized using razor blades, and the vesicles were obtained by step Suc gradients following the same procedure described by Muñoz et al., (1996). Subcellular fractionation was performed by separating the organelles on linear Suc gradients (20–50%, w/w), as described by Orellana et al. (1997). Marker enzymes for Golgi, mitochondria, and ER were measured as described by Briskin et al. (1987). Proteins were measured by the bicinchoninic acid method.
XG-FucTase Activity
XG-FucTase activity was measured at 25°C for 30 or 60 min in a final volume of 100 μL, as described by Farkas and Maclachlan (1988). The incubation was in the presence of 1 μm GDP-[3H]Fuc (1.5 × 105 cpm nmol−1), 100 μg tamarind xyloglucan, 10 mm 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)/KOH, pH 7.0, 10 mm MnCl2, and 0.05% (v/v) Triton X-100. The reaction was terminated by adding 250 μL of 95% (v/v) ethanol. The samples were kept on ice for 30 min, and then filtered through 2.4-μm glass fiber filters using a filtration system (model FH225V, Hoefer, San Francisco). The filter were washed three times with 1 mL of 70% (v/v) ethanol containing 1 mm EDTA, dried, and the radioactivity was estimated by liquid scintillation counting. To estimate the latency of XG-FucTase, the activity was measured on intact vesicles using the same procedure except that 0.05% (v/v) Triton X-100 was not included in the incubation medium. The enzymatic activity was expressed in nanokatals, as described by Perrin et al. (1999).
Uptake of GDP-[3H]Fuc and UDP-[3H]Glc into Golgi Vesicles
Golgi vesicles (100 μg of protein) were incubated with 1 μm GDP-[3H]Fuc or 1 μm UDP-[3H]Glc (1.0 × 106 cpm nmol−1) in a medium containing 0.25 m Suc, 1 mm MgCl2, and 10 mm Tris-HCl, pH 7.5. The incubation was finished by diluting with 10 volumes of a cold solution containing 0.25 m Suc, 1 mm MgCl2, and 10 mm Tris-HCl, pH 7.5, and immediately filtered through 0.7-μm glass fiber filters using a filtration system (model FH225V, Hoefer). The filters were washed with an additional 10 volumes of the same solution, dried, and the radioactivity determined by liquid scintillation counting.
Proteolysis of the Golgi Vesicles
Golgi vesicles were incubated with proteinase K in a solution containing 0.25 m Suc, 1 mm MgCl2, and 10 mm Tris-HCl, pH 7.5, for 30 min at 30°C in a volume of 250 μL. The incubation was in the presence or absence of 0.05% (v/v) Triton X-100. The reaction was stopped by adding 2.5 μL of 50 mm phenylmethylsulfonyl fluoride (PMSF). The samples were kept on ice until the enzymatic reactions were performed.
Native PAGE and GDPase Activity
Native PAGE and the detection of UDPase and GDPase activities using 3 mm UDP or GDP were as described by Orellana et al. (1997).
Triton X-114 Partitioning of Golgi Proteins
The separation of proteins based on their ability to partition in Triton X-114 (Bordier, 1981) was performed as described in Orellana et al. (1997). Vesicles were solubilized in three volumes of a solution containing 1% (v/v) Triton X-114, incubated for 3 min at 30°C, and centrifuged at 1,000g for 3 min. The aqueous and detergent phases were separated, and aliquots from both phases were utilized to measure GDPase and UDPase activities.
Separation of the Membrane and Soluble Fraction from Golgi Vesicles
Freeze-thawing was use to lyse open the Golgi vesicles. Golgi vesicles were resuspended in 0.25 m Suc, 1 mm MgCl2, and 10 mm Tris-HCl, pH 7.5, and stored overnight in liquid nitrogen. After thawing, 250 μL of Golgi vesicles (200 μg of protein) were centrifuged at 100,000g for 50 min at 4°C to separate the soluble fraction (supernatant) and the membrane fraction (pellet). The supernatant was removed and the pellet was resuspended in 250 μL of 0.25 m Suc, 1 mm MgCl2, and 10 mm Tris-HCl, pH 7.5. XG-FucTase activity was measured as described above. UDPase activity was measured as described in Orellana et al. (1997).
Sensitivity of Fucosylated Polymers to Cellulase
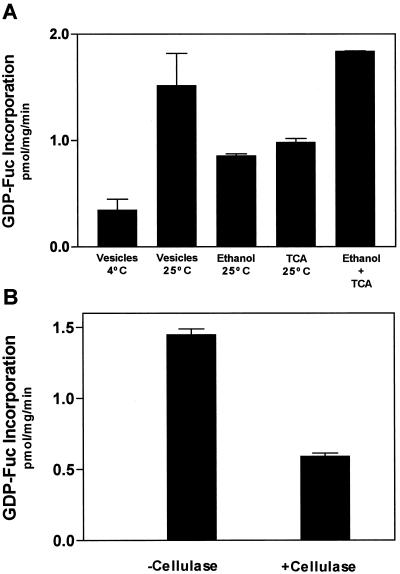
Golgi vesicles (250 μg of protein) were incubated with 1 μm of GDP-[3H]Fuc for 6 min at 25°C. The reaction was terminated by boiling for 3 min, and then adjusted to pH 5.0 with sodium acetate. The sample was then incubated at 40°C for 18 h in the presence and absence of 15 units of cellulase from Trichoderma sp. The incubation was terminated by adding 70% (v/v) ethanol. The samples were boiled for 1 min, kept at −20°C for 15 min, and then filtered on 0.7-μm glass fiber filters. The filters were dried and the radioactivity associated with the filters was detected by liquid scintillation counting.
All of the experiments were done at least twice, performing duplicate and triplicate assays.
RESULTS
XG-FucTase Is a Latent Enzyme
XG-FucTase, an enzyme located in the Golgi apparatus, functions by adding the terminal fucosyl residue on the side chain of xyloglucan (Camirand et al., 1987). XG-FucTase uses GDP-Fuc as a substrate. GDP-Fuc is a nucleotide sugar made from GDP-Man in the cytoplasm of plant cells (Bonin et al., 1997). Since the growing xyloglucan is located in the lumen of the Golgi cisternae (Zhang and Staehelin, 1992), the elongating polysaccharide and GDP-Fuc would be located in different cell compartments. Thus, questions arise regarding the orientation of XG-FucTase in the Golgi cisternae, and whether GDP-Fuc is incorporated into the lumen of the Golgi cisternae. We took two different approaches to addressing the question of the orientation of XG-FucTase.
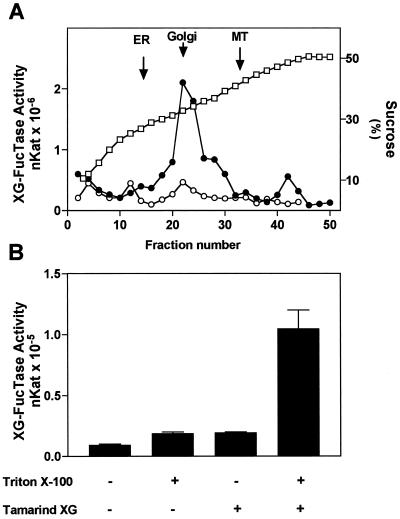
First, we analyzed the latency of the enzyme using tamarind xyloglucan, a polymer that lacks the terminal Fuc and is utilized as a substrate. Tamarind XG has a large Mr and we presume that it cannot penetrate the membrane of Golgi vesicles. Figure 1A shows the distribution in an isopycnic gradient of XG-FucTase from pea stems when the enzymatic activity was measured in the presence of tamarind xyloglucan in intact and permeabilized membranes. XG-FucTase activity was detected in the gradient only when the membranes were permeabilized with 0.05% Triton X-100 and it co-migrated with a Golgi marker. In the absence of Triton X-100 the activity was almost undetectable, indicating that detergent permits XG-FucTase to come into contact with the substrate.
Figure 1.
Subcellular distribution of XG-FucTase activity in pea stems. A, Organelles were separated on a linear Suc gradient (20%–50%) fractionated from the top of the tube. XG-FucTase was measured in the absence (○) and presence (●) of 0.05% (v/v) Triton X-100. Marker enzyme activity for ER (NADPH cytochrome c reductase antymicin A insensitive), Golgi (latent UDPase), and mitochondria (MT) (cytochrome c oxidase) were measured. The peak of the marker enzyme activity is indicated with arrows. The Suc concentration in each fraction is shown (□). A representative experiment is shown, and different experiments gave similar results. B, XG-FucTase was measured for 30 min on intact or permeabilized Golgi vesicles (100 μg of protein) in the presence (+) or absence (−) of 100 μg of tamarind xyloglucan (XG). Vesicles were permeabilized using 0.05% (v/v) of Triton X-100.
Second, using a preparation enriched in Golgi vesicles from pea stems, we evaluated in more detail the effect of adding tamarind xyloglucan and detergent into the XG-FucTase activity assay. Figure 1B shows that the addition of 0.05% Triton X-100 in the absence of tamarind xyloglucan resulted in a slight increase in activity compared with controls containing only Golgi vesicles and GDP-Fuc. This result could be explained by an increase in fucosylation of endogenous xyloglucan (or other potential endogenous acceptors) due to free access of GDP-Fuc into the lumen of Golgi vesicles. The XG-FucTase activity in the presence of tamarind xyloglucan and in the absence of 0.05% Triton X-100 was also slightly higher than the control, a result that can be explained by a small percentage of leaky vesicles that allowed the entrance of tamarind xyloglucan into the vesicles. The addition of both 0.05% Triton X-100 and tamarind xyloglucan into the assay resulted in an increase in activity close to 10-fold compared with control. We interpreted this higher activity as an increase in the tamarind xyloglucan available for fucosylation upon permeabilization of the Golgi membrane. This result also shows that upon addition of tamarind xyloglucan, most of the fucosylation is due to the addition of Fuc onto the exogenous acceptor. In fact, under our assay conditions, XG-FucTase activity increased linearly with increasing concentrations of tamarind xyloglucan added into the assay (data not shown).
The Active Site of XG-FucTase Faces the Lumen of the Golgi
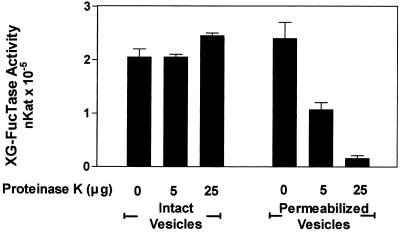
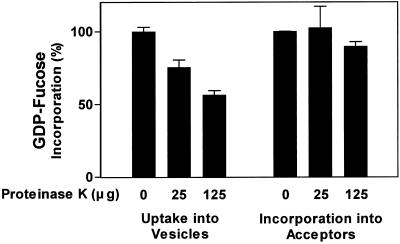
The results described above suggested that the active site of the XG-FucTase faces the lumen of Golgi cisternae. To confirm the orientation of XG-FucTase, sensitivity of the enzymatic activity to proteolytic inactivation on intact or permeabilized Golgi vesicles was tested. Intact and permeabilized Golgi vesicles were treated with increasing concentrations of proteinase K. After protease treatment, the vesicles were permeabilized to measure the total remaining XG-FucTase activity. GDP-[3H]Fuc and tamarind xyloglucan were used as exogenous acceptors. The results indicated that XG-FucTase activity was not affected by proteolysis when carried out on intact Golgi vesicles (Fig. 2). In contrast, XG-FucTase activity decreased in a proteinase K concentration-dependent manner when proteolysis was performed on permeabilized Golgi vesicles (Fig. 2), suggesting that the Golgi membrane protects XG-FucTase against proteolytic inactivation. Additionally, these results indicate that the enzyme was proteolytically inactivated only when the protease had access to the lumen of the Golgi vesicles. These results, in conjunction with the latency of the enzyme, strongly suggest that the active site of XG-FucTase is oriented toward the lumen of the Golgi cisternae.
Figure 2.
XG-FucTase is sensitive to proteinase K only in permeabilized Golgi vesicles. Golgi vesicles (62.5 μg of protein) were incubated in 125 μL for 30 min with 0, 5, and 25 μg of proteinase K (total amount) in the absence and presence of 0.05% (v/v) Triton X-100. The reaction was terminated by the addition of 1 mm PMSF. Then, the total XG-FucTase activity remaining in each sample was measured for 30 min in the presence of tamarind xyloglucan, GDP-[3H]Fuc, and 0.05% (v/v) Triton X-100, as described in “Materials and Methods.”
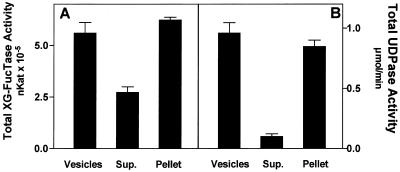
To further characterize XG-FucTase activity, we sought to determine if the enzyme is a soluble or a membrane-bound protein. Golgi vesicles were permeabilized by freezing in liquid nitrogen and thawing. This procedure decreased the latency of XG-FucTase. In addition, it decreased the latency of UDPase, a luminal, membrane-bound protein (Orellana et al., 1997), from 90% to 10% to 20%, indicating that the vesicles were indeed permeabilized (data not shown). The membranes were then separated from the soluble fraction by ultracentrifugation, and the XG-FucTase activity was measured, adding GDP-[3H]Fuc and tamarind xyloglucan in both fractions. The results showed that much of the XG-FucTase activity remained in the membrane fraction, although some activity was also detected in the soluble fraction (Fig. 3). However, the recovery of total XG-FucTase activity was higher than expected, possibly indicating that enzyme activation took place when the soluble fraction was separated from the membrane fraction. This activation was not due to the permeabilization procedure, because there was no significant increase in the activity before and after freezing and thawing (data not shown). This suggests that activation occurred due to a separation of the soluble and membrane fractions. The reason for this enzymatic activation remains unknown.
Figure 3.
Distribution of XG-FucTase and UDPase in a membrane and soluble fraction. Golgi vesicles kept overnight in liquid nitrogen were thawed, centrifuged at 100,000g for 1 h, and the supernatant and pellet separated. XG-FucTase (A) and UDPase (B) activities were measured in the vesicles, supernatant (Sup.), and pellet. The total activity in the supernatant and pellet are compared with the total activity present in the vesicles before centrifugation. The data are the average activity for triplicate samples from one experiment out of two independent experiments with similar results.
The distribution of UDPase, an enzyme previously shown to be a membrane-bound Golgi protein (Orellana et al., 1997), was measured as a control in the same experiment. As expected, UDPase remained almost completely in the membrane fraction, with only 10% of the activity detected in the soluble fraction. Comparing the results obtained for XG-FucTase and UDPase suggests that much of the XG-FucTase activity is membrane associated. The activity that is present in the soluble fraction may correspond to an active fragment that was cleaved off the membrane, a phenomenon that has been described for glycosyltransferases located in the Golgi apparatus from mammalian cells (Jaskiewicz et al., 1996). Alternative approaches to determine whether XG-FucTase is an integral membrane protein, such as partitioning in Triton X-114, resulted in a loss of the enzymatic activity. The results presented above, based only on the enzymatic activity, do not allow us to completely rule out that a soluble isoform of the enzyme may exist, however, it seems unlikely since there is no evidence that a Golgi-located glycosyltransferase exists in a soluble form. Enzymes involved in pectin biosynthesis have recently been characterized, and were found to behave as membrane-bound proteins (Doong and Mohnen, 1998; Goubet and Mohnen, 1999). These results support the idea that enzymes involved in the biosynthesis of both hemicelluloses and pectin are associated with the Golgi membrane.
GDP-Fuc Is Taken up into Golgi Vesicles and Fuc Is Transferred to Acceptors
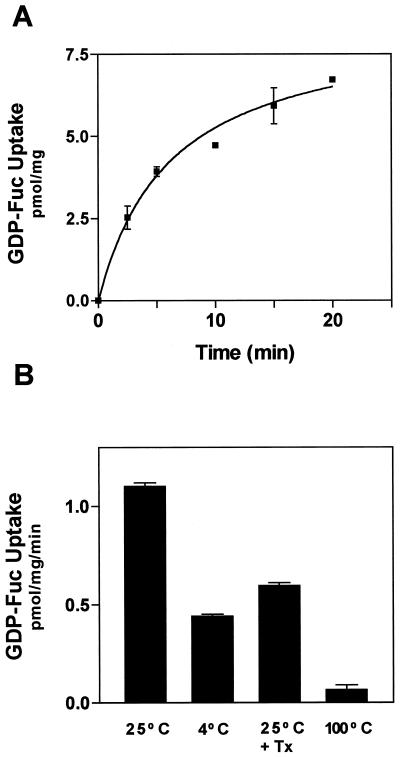
The luminal orientation of the active site of XG-FucTase poses a subcellular location dilemma. How does the cytosolic substrate GDP-Fuc become available to the luminal active site of the XG-FucTase? GDP-Fuc is synthesized from GDP-Man in the cytoplasm of the cell (Bonin et al., 1997), so it would have to cross the Golgi membrane to reach the active site of the enzyme. To determine whether GDP-Fuc is incorporated into Golgi vesicles, we incubated these vesicles with radiolabeled GDP-[3H]Fuc. The vesicles were separated from the incubation medium by filtration, and the radioactivity from GDP-[3H]Fuc taken up into the vesicles was determined. GDP-[3H]Fuc was incorporated into the vesicles at 25°C in a time-dependent manner (Fig. 4A). The uptake was sensitive to temperature, since it decreased at 4°C compared with 25°C (Fig. 4B). Treatment of the vesicles with low concentrations of detergent that did not disrupt the vesicles, just made them permeable, decreased the amount of radioactivity associated with the vesicles (Fig. 4B).
Figure 4.
Uptake of GDP-[3H]Fuc into Golgi vesicles. A, GDP-[3H]Fuc (1 μm, Sp. Act. 1 μCi/nmol) was incubated with Golgi vesicles (100 μg of protein) in a final volume of 0.1 mL for the times indicated. The reaction was terminated by 10-fold dilution of the reaction and filtering the incubation medium through 0.7-μm glass fiber filters. The filters were washed with cold buffer, dried, and the radioactivity associated with them estimated in a liquid scintillation counter. B, Effect of temperature and permeabilizing agents on the uptake of GDP-[3H]Fuc into Golgi vesicles was determined. The assay was for 5 min under the same conditions described in A.
In different experiments the decrease caused by permeabilization of the vesicles ranged from 30% to 50% of the control values using intact vesicles. This result suggest that [3H]Fuc was transferred from GDP-[3H]Fuc onto endogenous acceptors that remained associated with the permeabilized vesicles. The amount of radioactivity that was released from the permeabilized vesicles could be accounted for by either GDP-[3H]Fuc that was soluble in the lumen or by [3H]fucosylated acceptors that were soluble in the Golgi lumen and upon permeabilization were released from the vesicles.
To analyze how much [3H]Fuc was transferred onto endogenous acceptors, we analyzed the amount of [3H]Fuc incorporated onto polysaccharide and proteins by measuring the amount of radioactive material insoluble in 70% (v/v) ethanol or 10% trichloroacetic acid (TCA). The amount of radioactivity present in the 70% (v/v) ethanol and 10% TCA-insoluble fractions were similar. The sum of both insoluble fractions accounted for all of the GDP-[3H]Fuc incorporated into Golgi vesicles (Fig. 5A). This result indicates that most of the [3H]Fuc from GDP-[3H]Fuc was transferred onto endogenous acceptors and, therefore, the amount of free GDP-[3H]Fuc remaining in the vesicles was very low. Treatment of 70% (v/v) ethanol-insoluble material with cellulase, an enzyme that degrades the backbone of xyloglucan, reduced the incorporation of [3H]Fuc by 60%. This result suggest that at least 60% of the [3H]Fuc was incorporated into xyloglucan (Fig. 5B).
Figure 5.
Radioactivity from GDP-[3H]Fuc is incorporated into endogenous acceptors. A, Golgi vesicles (100 μg of protein) were incubated for 5 min at 25°C or 4°C with 1 μm GDP-[3H]Fuc (Sp. Act. 1 μCi/nmol). The reaction was terminated by different means. To analyze the uptake of GDP-Fuc into vesicles the reaction was diluted and filtered through glass fiber filters as described Figure 4. To measure the incorporation of radioactivity into endogenous acceptors, the reaction was terminated by adding 70% (v/v) ethanol or 10% (v/v) TCA. The insoluble material was collected in 1.5-μm glass fiber filters and the radioactivity measured in a liquid scintillation counter. Ethanol + TCA shows the addition of the material insoluble in 70% (v/v) ethanol and 10% (v/v) TCA. B, To estimate the incorporation of Fuc into xyloglucan, Golgi vesicles (250 μg of protein) were incubated with 1 μm GDP-[3H]Fuc, the reaction was stopped by boiling, and the pH was adjusted to pH 5.0. The sample was incubated for 18 h in the presence and absence of 15 units of cellulase from Trichoderma sp. After incubation, the samples were brought to 70% (v/v) ethanol, kept at −20°C for 15 min, and filtered. The filters were dried and the radioactivity associated to the filters was estimated in a liquid scintillation counter. The incorporation is expressed in equivalents of GDP-[3H]Fuc present in the incubation medium. The data are the average from triplicate samples, and the experiment was repeated twice.
GDP-Fuc Transport into Vesicles Is Protein Mediated
How is GDP-Fuc transported into Golgi vesicles? Evidence from mammalian cells indicates that GDP-Fuc is transported into the Golgi lumen by a transporter protein located in the membrane (Capasso and Hirschberg, 1984a). To determine whether the uptake of GDP-Fuc into pea stem Golgi vesicles is also mediated by a putative membrane protein, we analyzed the protease sensitivity of GDP-Fuc uptake into Golgi vesicles. Figure 6 shows that the uptake of GDP-Fuc into Golgi vesicles decreased as the concentration of proteinase K increased, suggesting that a membrane protein was required. The amount of proteinase K required to inactivate the uptake of GDP-Fuc was 5 times higher than the amount utilized to inactivate XG-FucTase (Fig. 2). There are several reasons why more proteinase K was required in this experiment. One reason could be the lower accessibility of the protease to a protein buried in the Golgi membrane. To make sure that the lower uptake into the vesicles was not caused by a proteolytic inactivation of the fucosyltransferases located in the Golgi lumen, we bypassed the transport step of the proteinase K-treated Golgi vesicles by permeabilizing the vesicles with 0.05% (v/v) Triton X-100 once the proteolysis was finished. The incorporation of [3H]Fuc onto endogenous acceptors that remained associated with the permeabilized vesicles was measured. Treatment with proteinase K did not affect the transfer of [3H]Fuc into endogenous acceptors, suggesting that the fucosyltransferases were not affected by this enzyme (data not shown). These results support the idea that the lower uptake of GDP-[3H]Fuc into Golgi vesicles was produced by the proteolytic inactivation of a protein or proteins involved in the transport of GDP-Fuc into the lumen of the Golgi vesicles.
Figure 6.
Proteinase K sensitivity of the uptake of GDP-[3H]Fuc into Golgi vesicles. Golgi vesicles were incubated for 30 min with 25 and 125 μg of proteinase K (total amount in the assay). The reaction was terminated by the addition of 1 mm PMSF. The uptake of GDP-[3H]Fuc into vesicles (100 μg) was determined by incubating the vesicles with 1 μm GDP-[3H]Fuc for 6 min at 25°C. The incubation was finished by diluting the medium 10-fold and filtering through 0.7-μm glass fiber filters, and the radioactivity in the filters was estimated by liquid scintillation counting. To analyze the transfer of radioactivity into endogenous acceptors, the vesicles treated with proteinase K were then permeabilized with 0.05% (v/v) Triton X-100 to bypass the membrane barrier. These Golgi vesicles were incubated with 1 μm GDP-[3H]Fuc for 6 min at 25°C. The reaction was terminated as described above, and the radioactivity transferred to endogenous acceptor was estimated by liquid scintillation counting. The incorporations are expressed as a percentage of the control not treated with proteinase K (vesicles, 1.09 ± 0.01 pmol mg−1 min−1; endogenous acceptors, 0.82 ± 0.01 pmol mg−1 min−1).
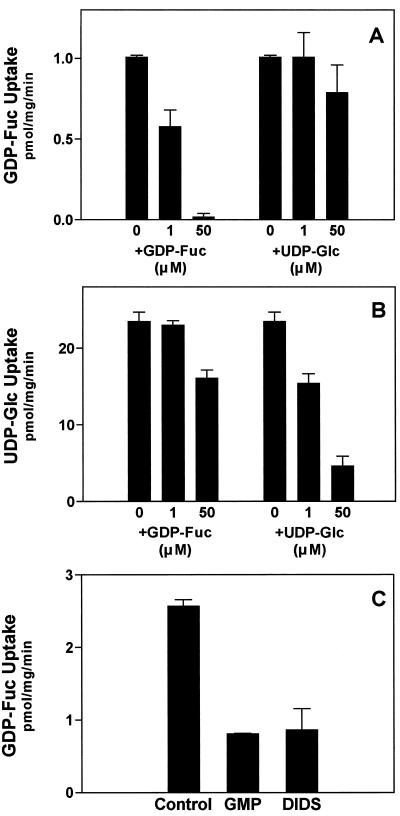
The results above suggest that GDP-Fuc is incorporated into the lumen of Golgi vesicles by a membrane-localized protein, possibly a nucleotide sugar transporter. Recently, we described a UDP-Glc transporter located in the Golgi membrane (Muñoz et al., 1996; Neckelmann and Orellana, 1998). To determine if this previously described UDP-Glc transporter also functions in the uptake of GDP-Fuc, we performed competition assays using GDP-Fuc and UDP-Glc. As expected, Figure 7A shows that the uptake of GDP-Fuc was competed for by the same substrate. Also, UDP-Glc added at increasing concentrations did not compete with the uptake of GDP-Fuc, and a slight effect may be seen at a concentration 50 times higher than GDP-Fuc. On the other hand, the uptake of UDP-Glc was competed for by UDP-Glc, and that GDP-Fuc had some inhibitory effect at a concentration 50 times higher than that of UDP-Glc (Fig. 7B). These results suggest that GDP-Fuc and UDP-Glc are transported by different proteins. Therefore, the mechanisms of uptake for GDP-Fuc and UDP-Glc into the lumen of the Golgi cisternae are independent. Moreover, the above results suggest that the Golgi apparatus from plant cells contains a GDP-Fuc transporter, which is different from the UDP-Glc transporter. To further characterize the uptake of GDP-Fuc, we tested the effect of GMP and 4,4′-diisothiocyanato-stilbene-2–2′-disulfonic acid (DIDS), two well-known inhibitors of the GDP-Fuc transporter present in the Golgi cisternae of mammals (Capasso and Hirschberg, 1984b, 1984c), and our results showed that both compounds inhibited the uptake of GDP-Fuc into Golgi vesicles (Fig. 7C).
Figure 7.
Uptake of GDP-Fuc into Golgi vesicles in the presence of UDP-Glc, GMP, and DIDS. A, Golgi vesicles (100 μg of protein) were incubated with 1 μm GDP-[3H]Fuc (specific activity, 1 μCi/nmol) for 5 min at 25°C in the presence of increasing concentrations of cold GDP-Fuc or UDP-Glc. The reaction was terminated and the uptake determined as described in the legend to Figure 4. B, Uptake of 1 μm UDP-[3H]Glc (specific activity, 1 μCi/nmol) into Golgi vesicles in the presence of cold GDP-Fuc or UDP-Glc was performed as described in A. C, Effect of 50 μm GMP and 20 μm DIDS on the uptake of GDP-Fuc was determined by adding them separately in the incubation medium.
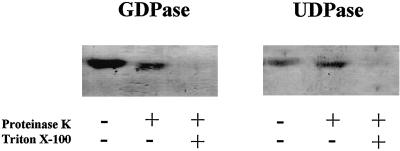
Fuc is mostly transferred into endogenous acceptors upon uptake of GDP-Fuc into Golgi vesicles; therefore, GDP should be produced in the lumen of the Golgi cisternae. Results from our laboratory indicate that a luminal membrane-bound UDPase hydrolyzes UDP produced during UDP-Glc metabolism in the Golgi cisternae (Orellana et al., 1997; Neckelmann and Orellana, 1998). Therefore, we assayed for the presence of a luminal Golgi GDPase. Proteins from pea Golgi vesicles were separated in a non-denaturing native polyacrylamide gel, and the GDPase activity was measured in-gel. A protein containing GDPase activity and having the same electrophoretic mobility as Golgi UDPase (Fig. 8) was detected. The topology of this enzyme was analyzed by measuring the sensitivity to proteolytic inactivation of the GDPase on intact and permeabilized Golgi vesicles. The results showed that proteinase K treatment of intact vesicles did not affect enzymatic activity; however, when the membrane was permeabilized, allowing the entrance of proteinase K into the lumen, the activity was lost, suggesting that GDPase was proteolytically inactivated (Fig. 8).
Figure 8.
Topology of GDPase in Golgi vesicles. Intact (−) and 0.05% (v/v) Triton X-100 permeabilized (+) Golgi vesicles (50 μg of protein) were incubated in the presence and absence of 2 μg of proteinase K. The reactions were terminated by adding 1 mm PMSF (final concentration) and the proteins were separated on acrylamide gels under non-denaturing electrophoresis conditions. After the run, the gel was incubated with 2 mm GDP (GDPase) or 2 mm UDP (UDPase) and the activity was determined in situ.
A small change in mobility was observed when intact vesicles were treated with proteinase K. This change can be explained by the cleavage of a protein fragment, most likely facing the outside of the vesicles, that did not affect the enzymatic activity. In these experiments GDPase behaved exactly as UDPase, a known luminal protein, suggesting that GDPase is located in the lumen of the organelle. We also analyzed whether GDPase is an integral membrane protein. Experiments using Triton X-114 partitioning of the proteins from Golgi vesicles, followed by in-gel detection of the GDPase and UDPase activities, showed that GDPase partitioned in the detergent fraction as an integral membrane-bound protein (no activity was detected in the soluble fraction) (Fig. 9). The distribution pattern of GDPase upon Triton X-114 partitioning was the same for UDPase, a known integral membrane-bound protein (Orellana et al., 1997). Since the GDPase and UDPase activities behaved almost identically, we suggest that the same enzyme may be responsible for both activities. Further studies including the cloning and expression of this enzyme(s) should confirm or discard this hypothesis.
Figure 9.
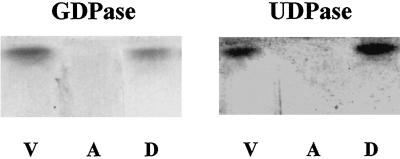
Triton X-114 partitioning of GDPase. Golgi vesicles (300 μg of protein) were incubated with Triton X-114, as described Orellana et al. (1997). Then the aqueous (A) and detergent (D) phases were separated and an aliquot of each fraction was separated in non-denaturing polyacrylamide gels. The GDPase and UDPase activities of each fraction were measured in gel and compared with the activity present in the vesicles (V) prior to the partitioning.
DISCUSSION
Topology of XG-FucTase
Hemicellulose and pectin are synthesized in the Golgi apparatus by glycosyltransferases that use nucleotide sugars as substrates. The mechanism of polysaccharide biosynthesis in this organelle, however, is still poorly understood. The growing polysaccharides are found in the lumen of the Golgi cisternae, whereas the nucleotide sugars are synthesized in the cytoplasm (Coates et al., 1980; Zhang and Staehelin, 1992; Bonin et al., 1997). This differential localization of the substrates and products raises a number of questions regarding the orientation of the enzymes and how the nucleotide sugars have access to the active site of the enzymes. A similar situation in terms of different localization of the substrate and product is also observed in cellulose biosynthesis.
To explain the topology of this process, a model has been proposed in which the substrate is used in the cytoplasm and the polymer elongates outside the plasma membrane (Delmer and Amor, 1995). The question that arises is whether the cell, in different subcellular localization, uses the same mechanisms to deal with topological constraints involved in the synthesis of cell wall polysaccharides. Studies of the orientation of glucan synthase I, a Golgi-located enzyme, suggested a luminal orientation of this activity (Muñoz et al., 1996). The polymers synthesized under conditions that favor glucan synthase I activity are mainly β,1–4 glucans, polymers that resemble the backbone of xyloglucan and are thought to be assembled by processive glycosyltransferases.
In this work we have specifically analyzed the orientation of XG-FucTase, a branching and non-processive glycosyltransferase involved in the addition of terminal Fuc onto xyloglucan (Camirand et al., 1987). A similar enzyme has been recently cloned from Arabidopsis and shown to be active in mammalian cells upon heterologous expression (Perrin et al., 1999). Despite the cloning and expression of the enzyme, there is no direct evidence regarding its actual topology in the Golgi apparatus. Our results indicate that the active site of the pea XG-FucTase faces the lumen of the Golgi cisternae, and that it is most likely a membrane-bound protein. The information obtained from the primary sequence of the Arabidopsis XG-FucTase suggests that this enzyme would have a structure and topology similar to Golgi glycosyltransferases from mammals (Perrin et al., 1999).
Our results on the topology of the pea XG-FucTase agree with a model for a type II membrane protein with the active site facing the lumen of the organelle. Despite the addition of protease inhibitors, we detected some activity that was not associated with membrane fractions. This soluble activity may correspond to the release of an active fragment due to proteolysis. This phenomenon has been described for type II membrane-bound Golgi proteins (Yanagisawa et al., 1990). To our knowledge, there is no example of a Golgi-localized glycosyltransferase that corresponds to a soluble protein in the lumen of the Golgi cisternae. Thus, we believe that pea XG-FucTase is a membrane-bound Golgi protein with its active site facing the lumen of the Golgi cisternae. Whether the pea XG-FucTase corresponds to the ortholog or an isoform of the Arabidopsis XG-FucTase remains to be determined.
Transport of GDP-Fuc into the Lumen of Golgi Cisternae
If the active site of XG-FucTase is located in the Golgi lumen, the substrate GDP-Fuc needs to be available in that compartment. GDP-Fuc is synthesized mainly from GDP-Man through a series of reactions involving dehydratase, epimerase, and reductase activities. This pathway has been well characterized in the Arabidopsis mutant mur-1, in which the dehydratase step is altered (Bonin et al., 1997). In addition, this dehydratase is located in the cytosolic fraction, and it is likely that the epimerase and reductase are also present in the same fraction. Our results indicate that GDP-Fuc is incorporated into Golgi vesicles and Fuc is then transferred onto xyloglucan; therefore, GDP-Fuc must be transported through the Golgi membrane. The results presented in the present study also suggest that this transport is protein mediated, thus opening up the possibility that a GDP-Fuc transporter could regulate the availability of GDP-Fuc during xyloglucan biosynthesis.
A UDP-Glc transporter thought to be involved in the biosynthesis of Glc-containing polysaccharides was recently identified in Golgi vesicles from pea stems (Muñoz et al., 1996); however, the results presented here indicate that it is unlikely that the same transporter is responsible for the transport of both GDP-Fuc and UDP-Glc. Thus, two different nucleotide transporters would be involved in the uptake of both nucleotide sugars. A number of different Golgi transporters for nucleotide sugars have been characterized and recently cloned in mammals, yeast, and Leishmannia (Hirschberg at al., 1998). These nucleotide sugar transporters seem to be quite specific. Our findings therefore support the idea that in plants every distinct nucleotide sugar utilized for polysaccharide biosynthesis in the Golgi apparatus is transported by a specific nucleotide sugar transporter. We have searched the Arabidopsis database looking for homologous sequences of genes encoding for nucleotide sugar transporters already cloned. So far, we have been able to identify some sequences with high homology, suggesting that, indeed, different nucleotide sugar transporters would be located in the Golgi apparatus. The characterization and search for knockouts of these genes should give us an idea of the role of these transporters in polysaccharide biosynthesis.
Golgi GDPase
A GDP-Fuc transporter most likely involved in the translocation of GDP-Fuc for fucosylation of glycoproteins and lipids was described in a mammalian Golgi apparatus (Sommers and Hirschberg, 1982). This GDP-Fuc transporter, like every nucleotide sugar transporter described so far, works as an antiporter between the nucleotide sugar (GDP-Fuc) and the nucleoside monophosphate (GMP) (Capasso and Hirschberg, 1984c). A similar antiporter mechanism could be responsible for the entrance of GDP-Fuc in plants. The GMP required as a substrate for the antiporter would be produced by the GDPase-catalyzed hydrolysis of the GDP released during the XG-FucTase reaction. Based on different results obtained during characterization of the GDPase activity, we believe that the same enzyme present in the lumen of the Golgi cisternae may be responsible for both the GDPase and the UDPase activities. We are currently working on the purification of the Golgi UDPase from pea stems, and fractions from the last steps of purification show activity toward UDP and GDP but not ADP, supporting the idea that one enzyme is responsible for the hydrolysis of nucleoside diphosphate released from uridine- and guanosine-containing nucleotide sugars (Norambuena and Orellana, manuscript in preparation). The cloning and active expression of this enzyme will allow us to determine whether the Golgi apparatus contains a single enzyme that utilizes both UDP and GDP or one that utilizes two distinct enzymes. A human Golgi luminal UDPase has recently been cloned and expressed in Cos-7 cells (Wang and Guidotti, 1998), and shown to exhibit activity toward UDP and GDP. These results support the possibility that, indeed, the plant Golgi UDPase may be responsible for both activities. In addition, a yeast GDPase has been purified to homogeneity and shown to exhibit both GDPase and UDPase activity (Yanagisawa et al., 1990).
Mechanism for Polysaccharide Biosynthesis in the Golgi Apparatus
We have recently proposed a model to explain the metabolism of UDP-Glc during the biosynthesis of polysaccharides in the Golgi apparatus (Neckelmann and Orellana, 1998). This model takes into account the role of the UDP-Glc transporter, the glucosyltransferase(s), and the UDPase. The results presented in this paper indicate that during the process of xyloglucan fucosylation in the Golgi apparatus, a GDP-Fuc transporter and GDPase are also required. This suggests that the role of nucleotide sugar transporters and NDPase could be part of a general mechanism associated with the synthesis of sugar-containing cell wall polymers in the Golgi apparatus from plant cells.
Distinct and specific nucleotide sugar transporters may be involved in the uptake of nucleotide sugars required for the synthesis of different polysaccharides. In that case, NDPase would be responsible for the hydrolysis of uridine- or guanosine-diphosphate released upon transfer of the sugar by glycosyltransferases. Thus, the activity of nucleotide sugar transporters and NDPase may pose additional steps for the regulation of polysaccharide biosynthesis in the Golgi apparatus. Therefore, these findings raise the possibility that some of the Arabidopsis primary cell wall mutants deficient in Fuc, Ara, and Rha (mur mutants) (Reiter et al., 1993; Reiter et al., 1997) could be due to an impairment in the transport of the nucleotide sugars into the lumen of the Golgi cisternae. Efforts to obtain the genes for nucleotide sugar transporters and NDPase, and to test their function in vivo by reverse-genetic approaches, should shed more light on the impact that their function has on cell wall polysaccharide biosynthesis.
ACKNOWLEDGMENTS
Thanks to Dr. Debra Mohnen, Dr. Lee Meisel, and Dr. Herman Silva for helpful discussion and reading the manuscript. Thanks to Dr. Mario Mera for providing the seeds.
Footnotes
This work was supported by Fondecyt grant no. 1970494, Chile.
LITERATURE CITED
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Hofte H, Plazinski J, Birch R, Cork A, Glover J, Redmond J, Williamson RE. Molecular analysis of cellulose biosynthesis in Arabidopsis. Science. 1998;279:717–720. doi: 10.1126/science.279.5351.717. [DOI] [PubMed] [Google Scholar]
- Bonin CP, Potter I, Vanzin GF, Reiter W-D. The MUR1 gene of Arabidopsis thaliana encodes an isoform of GDP-d-mannose-4,6-dehydratase, catalyzing the first step in the de novo synthesis of GDP-l-fucose. Proc Natl Acad Sci USA. 1997;94:2085–2090. doi: 10.1073/pnas.94.5.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- Brett CT, Waldron KW. Cell wall formation. In: Brett C, Waldron K, editors. Physiology and Biochemistry of Plant Cell Walls. Ed 2. London: Chapman & Hall; 1996. pp. 75–111. [Google Scholar]
- Briskin DP, Leonard RT, Hodges TK. Isolation of the plasma membrane: membrane markers and general principles. Methods Enzymol. 1987;148:542–558. [Google Scholar]
- Camirand A, Brumell D, Maclachlan G. Fucosylation of xyloglucan: localization of the transferase in dictyosomes of pea stem cells. Plant Physiol. 1987;84:753–756. doi: 10.1104/pp.84.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capasso JM, Hirschberg CB. Mechanism of glycosylation and sulfation in the Golgi apparatus: evidence for nucleotide sugar/nucleoside monophosphate and nucleotide sulfate/ nucleoside monophosphate antiports in the Golgi apparatus membrane. Proc Natl Acad Sci USA. 1984a;81:7051–7055. doi: 10.1073/pnas.81.22.7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capasso JM, Hirschberg CB. Effect of nucleotides on translocation of sugar nucleotides and adenosine 3′-phosphate 5′-phosphosulfate into Golgi apparatus vesicles. Biochim Biophys Acta. 1984b;777:133–139. doi: 10.1016/0005-2736(84)90505-4. [DOI] [PubMed] [Google Scholar]
- Capasso JM, Hirschberg CB. Effect of atractylosides, palmitoyl coenzyme A, and anion transport inhibitors on translocation of nucleotide sugars and nucleotide sulfate into Golgi vesicles. J Biol Chem. 1984c;259:4263–4266. [PubMed] [Google Scholar]
- Carpita N, Gibeaut DM. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the wall during growth. Plant J. 1993;3:1–30. doi: 10.1111/j.1365-313x.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- Chrispeels M, Holuigue L, Latorre R, Luan S, Orellana A, Peña-Cortés H, Raikhel N, Ronald P, Trewavas A. Signal transduction networks and the biology of plant cells. Biol Res. 1999;32:35–60. [PubMed] [Google Scholar]
- Coates SW, Gurney T, Jr, Sommers LW, Yeh M, Hirschberg CB. Subcellular localization of sugar nucleotide synthetases. J Biol Chem. 1980;255:9225–9229. [PubMed] [Google Scholar]
- Delmer DP, Amor Y. Cellulose biosynthesis. Plant Cell. 1995;7:987–1000. doi: 10.1105/tpc.7.7.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doong RL, Mohnen D. Solubilization and characterization of a galacturonosyltransferase that synthesizes the pectic polysaccharide homogalacturonan. Plant J. 1998;13:115–123. [Google Scholar]
- Farkas V, Maclachlan G. Fucosylation of exogenous xyloglucans by pea microsomal membranes. Arch Biochem Biophys. 1988;264:48–53. doi: 10.1016/0003-9861(88)90568-1. [DOI] [PubMed] [Google Scholar]
- Goubet F, Mohnen D. Solubilization and partial characterization of homogalacturonan-methyltransferase from microsomal membranes of suspension-cultured tobacco cells. Plant Physiol. 1999;121:281–290. doi: 10.1104/pp.121.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna R, Brummel DA, Camirand A, Hensel A, Russell EF, Maclachlan G. Solubilization and properties of GDP-fucose: xyloglucan 1,2-α-l-fucosyltransferase from pea epicotyl membranes. Arch Biochem Biophys. 1991;290:7–13. doi: 10.1016/0003-9861(91)90584-6. [DOI] [PubMed] [Google Scholar]
- Hirschberg CB, Robbins PW, Abeijon C. Transporters of nucleotide sugars, ATP, and nucleotide sulfate in the endoplasmic reticulum and Golgi apparatus. Annu Rev Biochem. 1998;67:49–69. doi: 10.1146/annurev.biochem.67.1.49. [DOI] [PubMed] [Google Scholar]
- Jaskiewicz E, Zhu G, Bassi R, Darling DS, Young WW., Jr Beta1,4-N-acetylgalactosaminyltransferase (GM2 synthase) is released from Golgi membranes as a neuraminidase-sensitive, disulfide-bonded dimer by a cathepsin D-like protease. J Biol Chem. 1996;271:26395–26403. [PubMed] [Google Scholar]
- Muñoz P, Norambuena L, Orellana A. Evidence for a UDP-glucose transporter in Golgi apparatus-derived vesicles from pea and its possible role in polysaccharide biosynthesis. Plant Physiol. 1996;112:1585–1594. doi: 10.1104/pp.112.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neckelmann G, Orellana A. Metabolism of uridine 5′-diphosphate-glucose in Golgi vesicles from pea stems. Plant Physiol. 1998;117:1007–1014. doi: 10.1104/pp.117.3.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orellana A, Neckelmann G, Norambuena L. Topography and function of Golgi uridine-5′-diphosphatase from pea stems. Plant Physiol. 1997;114:99–107. doi: 10.1104/pp.114.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM. Higher plants contain homologs of the bacterial celAgenes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci USA. 1996;93:12637–12642. doi: 10.1073/pnas.93.22.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin RM, DeRocher AE, Bar-Peled M, Zeng W, Norambuena L, Orellana A, Raikhel NV, Keegstra K. Xyloglucan fucosyltransferase, an enzyme involved in plant cell wall biosynthesis. Science. 1999;284:1976–1979. doi: 10.1126/science.284.5422.1976. [DOI] [PubMed] [Google Scholar]
- Reiter W-D, Chapple CCS, Somerville CR. Altered growth and cell walls in a fucose-deficient mutant of Arabidopsis. Science. 1993;261:1032–1035. doi: 10.1126/science.261.5124.1032. [DOI] [PubMed] [Google Scholar]
- Reiter WD, Chapple CCS, Somerville CR. Mutants of Arabidopsis thalianawith altered cell wall polysaccharide composition. Plant J. 1997;12:335–345. doi: 10.1046/j.1365-313x.1997.12020335.x. [DOI] [PubMed] [Google Scholar]
- Sommers LW, Hirschberg CB. Transport of sugar nucleotides into rat liver Golgi. J Biol Chem. 1982;257:10811–10817. [PubMed] [Google Scholar]
- Taylor NG, Scheible WR, Cutler S, Somerville CR, Turner SR. The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell. 1999;11:769–780. doi: 10.1105/tpc.11.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SR, Somerville CR. Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell. 1997;9:689–701. doi: 10.1105/tpc.9.5.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T-F, Guidotti G. Golgi localization and functional expression of human uridine diphosphatase. J Biol Chem. 1998;273:11392–11399. doi: 10.1074/jbc.273.18.11392. [DOI] [PubMed] [Google Scholar]
- White AR, Xin Y, Pezeshk V. Xyloglucan glucosyltransferase in Golgi membranes from Pisum sativum(pea) Biochem J. 1993;294:231–238. doi: 10.1042/bj2940231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa K, Resnick D, Abeijon C, Robbins PW, Hirschberg CB. A guanosine diphosphatase enriched in Golgi vesicles of Saccharomyces cerevisiae. J Biol Chem. 1990;265:19351–19355. [PubMed] [Google Scholar]
- Zhang GF, Staehelin LA. Functional compartmentation of the Golgi apparatus of plant cells. Plant Physiol. 1992;99:1070–1083. doi: 10.1104/pp.99.3.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]