Abstract
Mammals exist in a complicated symbiotic relationship with their gut microbiome, which is postulated to have broad impacts on host health and disease. As omics-based technologies have matured, the potential mechanisms by which the microbiome affects host physiology are being addressed. The gut microbiome, which provides environmental cues, can modify host cell responses to stimuli through alterations in the host epigenome and, ultimately, gene expression. Increasing evidence highlights microbial generation of bioactive compounds that impact the transcriptional machinery in host cells. Here, we review current understanding of the crosstalk between gut microbiota and the host epigenome, including DNA methylation, histone modification and non-coding RNAs. These studies are providing insights into how the host responds to microbial signalling and are predicted to provide information for the application of precision medicine.
Keywords: DNA methylation, epigenome, gut microbiome, histone modification, non-coding RNAs
There are as many as 1014 micro-organisms, including bacteria, archaea and viruses, living symbiotically with an individual human, in locations that include the skin, mouth, gut and other mucosal surfaces (1). In the gut, over 1,000 unique microbial species have already been identified, making the gastrointestinal tract the most heavily colonized organ (1, 2). Microbiota colonization begins after birth and is relatively stable throughout life, though it can be affected by diet, antibiotic use, infections, among other potential variables. The diversity and composition of gut microbiome have been studied for over a decade, but their precise roles in maintaining homeostasis and influencing host physiology are still poorly understood.
The gut microbiota resides on the intestinal mucosal surfaces and plays an integral role in digesting food, harvesting energy and regulating immune development (3). In particular, it can generate numerous bioactive compounds, including short chain fatty acids (SCFAs), choline metabolites and lipids (3), that are important to host physiology. Microbial metabolites are critical messengers in the crosstalk between microbiome and host cells. They induce not only local effects in the gut, but also changes in distant organs such as liver, heart and the central nervous system (4). Dysbiosis of the gut microbiome has been associated with numerous diseases including obesity, diabetes, metabolic syndrome and colorectal cancer (5–8).
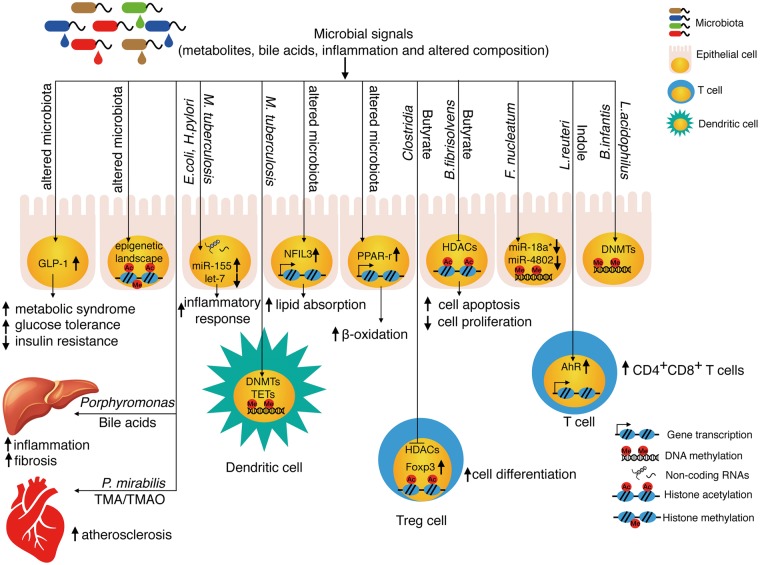
Microbiota, as one type of environment cue, can trigger host epigenetic modification. In particular, hosts have the ability to respond to environmental stimuli through the alterations of DNA methylation and histone modifications, which can have long-term effects on the host’s physiology (9). Given the important relationship between the gut microbiome and host physiology, our review will highlight the roles of various microbiota-derived metabolites in DNA methylation, histone modification and non-coding RNAs (Fig. 1).
Fig. 1.
Microbiota regulate the host epigenome through microbial signals including metabolites, bile acids, inflammation and altered composition. Dietary supplement, antibiotic treatment, infections, diurnal oscillations and other factors will affect the compositions of gut microbiota. The changed microbiome will generate different metabolites (such as butyrate and indoles), which will modify histone acetylation, nuclear receptors and transcription factors to reprogram the transcriptome in the host. Through affecting the methyl group donor SAM, microbiota also can impact the DNMTs and TETs. Certain non-coding RNAs can interact with bacteria to trigger host inflammation and autophagy. Microbial signals not only affect the transcriptome and chromatin modifications in local organs like intestine and innate lymphoid cells, but also regulate distant organ like liver and the cardiovascular system through metabolites. PPAR-γ, peroxisome proliferator activated receptor gamma.
Microbiota and DNA Methylation/Demethylation
DNA methylation is a crucial epigenetic control mechanism in mammals that can influence gene expression by regulating accessibility of transcription factors, histone modifiers and transcriptional machinery to chromatin (10, 11). This modification is itself highly regulated with multiple components adding and removing methyl groups. DNA methyltransferases (DNMTs) can add a methyl group from the donor S-adenosylmethionine (SAM) to the carbon-5 position of the cytosine (5mC), while the ten-eleven translocation enzyme (TET) dioxygenase family can actively reverse this process through oxidation of 5mC to 5-hydroxymethylcytosine (5hmC). The dynamics of 5mC and 5hmC synthesis are important for various biological processes and can be regulated by a number of small molecules (10).
Folate is a critical micronutrient and one of the water-soluble vitamins, which can be obtained from specific foods or dietary supplements. Folate supports one carbon metabolism and the generation of SAM, the primary methyl donor for DNMT. (12). Bifidobacterium and Lactobacillus, common probiotic bacteria, can produce vitamins including folate (13). In a pilot study, volunteers were administered three probiotic strains of Bifidobacterium and all three strains significantly increased the folic acid concentration in faeces (14). These results suggest that Bifidobacterium can generate folate in the gut, which can be utilized by the host and might affect DNA methylation patterns. In support of this notion, coculture of human foetal and adult intestinal epithelial cells with Lactobacillus acidophilus (L.acidophilus) and Bifidobacterium infantis (B.infantis) causes significant DNA methylation differences (Fig. 1) and triggers a bacteria-specific transcriptome (15).
Commensal gut microbes also generate signals that balance effective inflammatory responses against stress. TLR4, a member of the toll-like receptor family, recognizes lipopolysaccharide (LPS) and activates the innate immune system. Commensal microbes were reported to increase the methylation level in the TLR4 gene, which decreases transcriptional activity at this locus and leads to decreased responsiveness to LPS and the maintenance of bacterial insensitivity in the colon (16). Although the underlying mechanisms are not clear, it is possible that microbiota interact with the enzymes responsible for DNMTs to induce the host response. Bacteria also induce demethylation through the oxidation of 5mC to 5hmC, a process catalyzed by TET family proteins. Challenging human dendritic cells (DCs) with Mycobacterium tuberculosis (M.tuberculosis) induced DNA demethylation at thousands of loci, independent of cell division. Those demethylated regions were located at distal enhancer elements rather than at proximal promoter regions. Integrated analysis showed that these loci were enriched at genes expressing immune transcription factors, at sites of active histone marks, at areas of increased chromatin accessibility, thus leading to a strong correlation with gene expression (17). These data suggest that bacterial stimulation has the capacity to influence host 5mC and 5hmC localization leading to changes in epigenetic memory (Fig. 1).
Dietary methionine not only shapes the composition of the host microbiota (18), but also modulates bacterial metabolism (19) to generate substrates for SAM synthesis, which is required during DNA methylation and phosphatidylcholine biosynthesis. Escherichia coli (E.coli) lacking an external source of methionine reduced the efficiency of phosphatidylcholine synthesis in the host. Phosphatidylcholine can activate the nuclear receptor NR5A, which suppresses grl-21 expression to inhibit mitochondrial fission and host lipid accumulation. Thus, an interaction between microbiota and host not only modifies the DNA methylation dynamics, but also regulates nuclear receptor activity to control the gene transcriptions in response to environment signals (19).
In human studies, some groups found that Fusobacterium nucleatum (F.nucleatum) was enriched in colorectal cancer tissues and correlated with DNA methylation (20–22). In particular, F.nucleatum was over represented in a subtype of colorectal cancer with a specific mutational spectrum (20). F.nucleatum has been shown to target innate immune signalling and active autophagy (23), which suggests potential interplay with DNA methylation and tumourigenesis.
Microbiota and Histone Modification
Histones (H2A, H2B, H3 and H4) make up the core nucleosome which is encircled by DNA (11). Multiple modifications can be made to these histones including acetylation, methylation, phosphorylation, and ubiquitination, primarily on N-terminal histone tails. These covalent modifications are now known to be critical regulatory features of chromatin structure and function, which leads to changes in cell fate and tissue development (11). In many cases, single and/or multiple modifications occur at the same position on the histone tails and almost certainly function together to provide an exquisite level of genomic control. Epigenetic writers, readers and erasers can recognize specific combinations of histone modification to provide a dynamic system for modulating gene expression. In general, microbiota affect histone modification by (i) altering the activity of modification-related enzymes, and (ii) influencing the levels of substrates used by these enzymes.
The majority of histone modifications, such as histone acetylation, are reversible and responsive to metabolic changes. Histone acetylation is usually associated with active gene transcription, while deacetylation is associated with transcriptional repression (11). Deposition of acetyl groups is catalyzed by histone acetyltransferase (HAT), which can transfer an acetyl group from acetyl-coenzyme A to the ε amino group of lysine residues. Acetylation of histones also leads to exposure of target sites in nucleosomal DNA for transcription factors to initiate the assembly of transcription complexes. Deacetylation is catalyzed by histone deacetylase (HDAC), which removes acetyl groups from histone tails, thereby leading to a reduction in accessibility. There are several ways in which the microbiome can regulate this process. Microbial metabolism can generate a number of bioactive compounds, including the SCFAs, acetate, propionate, butyrate and other products. In healthy adults, the total concentration of acetate, propionate, butyrate in the colon is around 50–150 mM. Acetate is the most abundant SCFA—it is approximately three times more abundant than propionate and butyrate. In the systemic circulation, concentrations of butyrate and propionate are very low and acetate is around 0.1 mM (8). Butyrate is rapidly adsorbed from the colonic lumen and constitutes a preferred energy source for colonic epithelial cells. High concentrations of butyrate have the potential to inhibit the activity of HDACs, with a resulting impact on histone modifications and transcriptional regulation (24). Butyrate production depends on a both bacterial composition and dietary intake. For example, butyrate-producing bacteria represent considerable phylogenetic diversity: at the genus level this includes but is not limited to Roseburia, Odoribacter, Faecalibacterium, Eubacterium, Subdoligranulum, Peptoniphilus, Coprococcus, Fusobacterium, Porphyromonas, Clostridium, Anaerotruncus, Megasphaera and others (25). Dietary fibre is important for intestinal bacteria to produce butyrate, while high fat diets reduce the formation of butyrate. Using germfree mouse models colonized with wide type and mutant butyrate-producing bacteria Butyrivibrio fibrisolvens (B.fibrisolvens), Donohoe showed that dietary fibre could be fermented into butyrate in the lumen in a bacteria-dependent manner (26). Dietary supplementary of tributyrin to germfree mice, which also increases butyrate level without bacteria, can recapitulate the similar phenotype (26). Due to the complexity of this system (multiple species of bacteria, dietary components), it’s difficult to assign causality for the decreased butyrate level commonly observed in high fat/low fibre diet conditions.
However, some of the molecular mechanisms involved in microbiota-dependent modification of histones are beginning to be elucidated. As an HDAC inhibitor, butyrate exerts anti-inflammatory activity by suppressing NF-kB and STAT1 activation (27). In addition, butyrate-induced differentiation of colonic Treg cells (Fig. 1) through enhanced histone H3 acetylation in the promoter and conserved regions of the Foxp3 locus (28). Although butyrate-induced histone hyper acetylation leads to cell differentiation and inhibits proliferation of tumour cells, butyrate’s role in tumourigenesis is still controversial (29). Hu et al., (30) found that butyrate appeared to function as a tumour suppressor and could decrease colon cancer cell proliferation and stimulate apoptosis through inhibition of miR-92a transcription. These relationships suggest that the consumption of dietary fibre or colonization of the gut by a butyrate-producing bacterium might protect against colorectal cancer. Other studies have shown that butyrate accumulates in tumour cells and acted as an HDAC inhibitor to decrease cell proliferation and stimulate apoptosis due to the Warburg effect (26). In contrast, in genetically modified animals, butyrate supplementation promoted hyper proliferation of colon epithelial cells that appears to promote carcinogenesis (31). These seemingly contradictory results might result from the different genetic backgrounds of the mice, differences in microbial composition of animal facilities involved and different mechanisms involved in colon carcinogenesis (29).
Histone methylation/demethylation can be globally modulated by various cellular metabolites/cofactors including SAM, Fe2+/Fe3+ and α-KG (32). Gut microbiota, in combination with dietary substrates also produce metabolites that can affect histone methylation in multiple tissues of the host (33). In a germ-free mouse model, H3 methylation patterns were changed dependent on gut bacteria colonization status. The introduction of gut microbiota significantly increased levels of H3K27me3 and unmethylated H3K36 and decreased levels of H3K18me1 and unmethylated H3K23 in colon, liver and adipose tissues compared with germ-free mice. K27me2 and K36me1 on the variant H3.3 were also decreased in colon, liver and adipose tissues. Some histone methylation changes were tissue specific, for example, H3K27me1 and H3K36me2 were increased in adipose tissues, while decreased in colon and liver tissues (33). Although the detailed mechanism by which these changes at a distance occur is still unclear, it appears plausible that gut microbiota produces metabolites and/or cofactors that influence enzymes participating in histone modification (34). A major challenge for the future is identification of specific messenger molecules and definition of modes of action by which gut bacteria influence the epigenome in distant sites not in direct contact with the microbial flora.
Microbiota and Non-coding RNA
Non-coding RNAs, which do not encode proteins, also participate in important biological processes. Non-coding RNA comprises long non-coding RNAs, microRNAs and snoRNAs, as well as other small RNAs. Long non-coding RNAs have been reported to participate in the responses of intestinal epithelial cells to bacteria (35). Compared to germ-free mice, mice that have been colonized with specific bacteria displayed a significantly different profile of lncRNAs, most of which were transcribed from introns. Moreover, colonization with wild type E.coli or E.coli expressing bile salt hydrolase resulted in different effects on lncRNA expression. Although a small number of the observed lncRNAs overlapped with known inflammation-related lncRNAs, these changes in host epithelial cells were bacteria-species dependent (35).
miRNA is a small non-coding RNA molecule (containing about 22 nucleotides), which can regulate gene expression post-transcriptionally by binding to its targets’ 3’UTRs. MicroRNAs play important roles in regulating host immune functions, but how they respond to bacteria remains unclear. Infection with bacteria such as M.tuberculosis, Helicobacter and Salmonella enterica alters the cellular miRNA profiles (36, 37). Mycobacterium tuberculosis downregulated miR-let-7f in infected macrophages by secreting ESAT-6. MiR-let-7f targeted TNFAIP3, which is a negative regulator of the NF-κB pathway, to active host immune responses and decrease bacterial survival. A reduction in the miR-let-7f level by Mycobacterium tuberculosis decreased the production of cytokines including TNF and IL-1β and led to an ineffective immune response that predisposed the host to tuberculosis (38). F.nucleatum could regulate TLR4 and MYD88 innate immune signalling (Fig. 1) and activate autophagy by decreasing miR-18a* and miR- 4802 to mediate colorectal cancer chemoresistance (23). Helicobacter infection was reported to upregulate the miR-155 expression both in epithelial cells and macrophages (Fig. 1); this process was shown to be dependent on TLR4 and activation of the NF-κB pathway (39, 40). miR-155 (Fig. 1) likely participates in a negative-feedback loop to modulate inflammatory responses to Helicobacter infection. In addition to the regulation of miR-155, Helicobacter infection was also reported to down-regulate the let-7 miRNA family and thereby regulate the NF-κB inflammatory response (41).
It is interesting, if not surprising, that true crosstalk can occur between the host and the symbiotic bacterial population. For example, microRNAs from the host also can selectively regulate the function of microbiota. miRNAs from host faecal samples can enter bacteria, such as F.nucleatum and E.coli, thereby regulating bacterial gene transcription and bacterial growth (42). A conditional knockout of the miRNA-processing enzyme in intestinal epithelium exhibits uncontrolled gut microbiota and exacerbated colitis; faecal miRNA transplantation can rescue the phenotype. In support of this finding, coculture of the miRNAs with bacteria can induce significant changes in bacteria gene expression (42). These findings demonstrate that host secreted miRNA also feedback on to the gut bacteria to maintain the homeostasis of the intestine.
Microbial signals and host responses
Besides microbial derived SCFAs, microbiota can also generate other signals to modify the host epigenome (43–46). Like mammalian circadian rhythms, gut microbiota also undergoes diurnal oscillations, which are controlled by the diet (43). During different times of the day, the compositions and functions of gut bacteria change as does distance from the mucus. OTUs affected include: Lactobacillus reuteri (L.reuteri), Bacteroides acidifaciens, Mucispirillum schaedleri and Ruminococcus gnavus (R.gnavus). Diurnal fluctuations in the abundance of bacteria influence microbial derived metabolites, including serotonin, xylose, cytidine, proline and biotin that play key roles in biosynthetic pathways. These oscillating microbial signals could be expected to have an impact on the host epigenome and transcriptome. Actually, both the intestinal transcriptome and enhancer landscape demonstrated diurnal oscillations that were microbiota dependent. Antibiotic treatment eliminated the diurnal oscillations of transcriptomes and chromatin modifications in intestinal epithelial cells (43). Although the exact mechanism is still unknown, a contributory role of microbiota and their metabolites seems likely. A circadian transcription factors, nuclear factor interleukin-3-regulated protein (NFIL3) was recently reported (Fig. 1) to be regulated by microbiota (44). The microbiota triggered immune signals in innate lymphoid cells (ILCs) by flagellin or LPS. Subsequently, IL-22 was induced, leading to activation of STAT3, which could repress Rev-erbα transcription through binding to its promoter. Downregulating Rev-erbα induced epithelial NFIL3 expression to regulate lipid absorption and export in intestinal epithelium (44).
Double positive (CD4+CD8+) intraepithelial T lymphocytes (DP IELs) in the intestine promote tolerance to dietary antigens and have regulatory functions. There are no DP IELs in germ free mice and their numbers vary in different animal facilities, indicating that induction of DP IELs is microbiota dependent (45). Mechanistically, L.reuteri could metabolize tryptophan into indole-3-lactic that activates aryl-hydrocarbon receptor (AhR) (Fig. 1) to reprogram the CD4+ T cell into DP IELs by downregulation of transcription factor ThPOK. Unlike tributyrin, dietary supplement of tryptophan in the germ free mice can induce DP IELs only in a bacteria dependent manner (45).
Microbiome and metabolic diseases
There is a fine balance between the microbiota and the host to maintain a true symbiotic relationship. Gut microbiota can digest the host diet and secrete small molecules that in turn support host health. Once this equilibrium is disturbed, microbial activity can play a role in initiating and/or maintaining a series of metabolic diseases in host. Gut microbiota cannot only crosstalk locally with the intestine, but can also generate signals to communicate with distant organs including adipose, liver or cardiovascular system. These signals can consist of bacteria themselves (limited to local signalling), microbial derived metabolites, microbial induced inflammation and/or inflammatory cytokines and other factors [glucagon-like peptide-1 (GLP-1), 5-hydroxytryptamine] from the gut (47).
Obesity may be the most widely studied microbiota-associated metabolic disease. Over a decade ago, Bäckhed et al. (5), found that gut microbiota could regulate genes involved in lipogenesis and that there was significant decrease of bacterial community diversity, especially observed in the ratio of Bacteroidetes/Firmicutes in obese individuals (48). Although the nature of the relationship between the B/F ratio and obesity remains a topic of discussion, numerous additional studies have identified obesity-linked bacteria (49, 50). Fei et al., isolated the endotoxin-producing bacterium, Enterobacter cloacae B29, from a morbidly obese volunteer’s gut. Transplantation of this species to germ-free mice with HFD induced excessive fat accumulation and led to animals that developed both obesity and insulin resistance. In contrast, control mice colonized with B29 and fed a normal diet remained lean throughout the study. B29 bacteria also elevated serum endotoxin by chylomicrons induced by long-chain fatty acids in the HFD to enhance systemic inflammation (50). Circulating free fatty acids also inhibit GLP-1 secretion in intestinal enteroendocrine cells (Fig. 1), which may be causal for induction of metabolic syndrome. Increasing the GLP-1 level by changing the gut microbiota through an antibiotic cocktail treatment improved glucose intolerance, insulin resistance and increased other beneficial metabolites like succinic acid (51). These findings suggest that certain bacteria excel at extracting more energy from fat and regulating metabolites, thus promoting weight gain and metabolic dysregulation in the host. Diet, in turn, also can modify the bacteria and create a feed-forward loop that promotes obesity.
Besides obesity, gut microbiota has been linked to cardiometabolic diseases like atherosclerosis (52, 53). Through case control study, Jie et al., identified Enterobacteriaceae and Streptococcus species that were increased in atherosclerotic cardiovascular disease patients by whole metagenome sequencing. Atherosclerosis enriched bacteria like Streptococcus species, was positively correlated with blood pressure (53). KEGG pathways analysis using microbial genes indicated the enrichment of metabolism and transport of several molecules important for cardiovascular health, for example: enzymes involved in trimethylamine (TMA) synthesis, the precursor for the trimethylamine N-oxide (TMAO) (53). TMAO, which is associated with atherosclerosis and cardiovascular disease, is a gut microbiota dependent metabolite (Fig. 1). Proteus mirabilis (P.mirabilis) with cutC/D genes can generate TMA from dietary choline, which is subsequently converted into TMAO by hepatic FMO3 (52). Wang et al. (52), found that 3,3-dimethyl-1-butanol (DMB), a choline analogue, inhibited microbial derived TMA in vitro and in vivo. DMB also inhibited choline diet induced atherosclerotic lesion development in apoE−/− mice. Thus, targeting the microbial enzyme genes could be a useful treatment for atherosclerotic cardiovascular disease.
The gut microbiota has also been associated with non-alcoholic fatty liver disease (NAFLD), the most common liver disease worldwide (54). Gut microbiota can cause liver inflammation, which is associated with hepatic cell damage and fibrosis, leading to the development of NAFLD. Suppressing gut bacteria via antibiotics decreased secondary bile acid levels and attenuated hepatic inflammation and fibrosis (54). Dysbiosis of the gut microbiome has also been shown to damage the permeability of the intestinal barrier, thus allowing bacterial components such as LPS to permeate the gut lining, activate TLR signalling and stimulate liver inflammation (55). The mechanism of this process is becoming clearer, as it’s been shown that TLR4 cooperates with MyD88 to mediate signal transduction events, such as activation of NF-κB to enhance the expression of TNFα and IL6 to induce liver inflammation (56).
Conclusions and Perspective
Recent work highlights potential interactions between microbiome and host epigenome, which demonstrates a key role for the microbiota in regulating the host response against environment signals. Due to the frequent and direct contact of the microbiota and host cells in numerous organs, but especially the gut, microbiota-derived metabolites are thought to be a primary mechanism of regulating the host epigenome. Although SCFAs seem to be crucial in modulating the host epigenome through DNA methylation and histone modifications, many of the details of the mechanism remain unclear. For example, which bacteria or community can generate specific metabolites, and which nutritional components are critical primary substrates? Furthermore, it is currently unclear which signalling pathways bacterial metabolites impact to regulate host responses and what specific components of host transcriptional machinery are modulated to induce changes in host gene expression? Whether microbiota also effect changes in RNA modification or higher order chromatin structure is still to be determined. Further complicating this biological picture, a host’s life style, health status and exposures, such as antibiotics, also regulate the microbiota and alter their influence. Therefore, further studies deciphering specific microbiota-derived signals that affect the host epigenome will provide new insights in understanding the host–microbiota interactions and lead to new mechanistic insights into health and diseases.
Acknowledgements
The authors thank the members of the Wade laboratory for many useful discussions through the course of this work. We apologize for any omission of relevant publications due to space limitations.
Funding
This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (ES101965 to P.A.W.).
Conflict of Interest
None declared.
References
- 1. Sekirov I., Russell S.L., Antunes L.C., Finlay B.B. (2010) Gut microbiota in health and disease. Physiol. Rev. 90, 859–904 [DOI] [PubMed] [Google Scholar]
- 2. Rajilić-Stojanović M., de Vos W.M. (2014) The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol. Rev. 38, 996–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nicholson J.K., Holmes E., Kinross J., Burcelin R., Gibson G., Jia W., Pettersson S. (2012) Host-gut microbiota metabolic interactions. Science 336, 1262–1267 [DOI] [PubMed] [Google Scholar]
- 4. Galland L. (2014) The gut microbiome and the brain. J. Med. Food 17, 1261–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bäckhed F., Ding H., Wang T., Hooper L.V., Koh G.Y., Nagy A., Semenkovich C.F., Gordon J.I. (2004) The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. U S A 101, 15718–15723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cani P.D., Everard A., Duparc T. (2013) Gut microbiota, enteroendocrine functions and metabolism. Curr. Opin. Pharmacol. 13, 935–940 [DOI] [PubMed] [Google Scholar]
- 7. Le Chatelier E., Nielsen T., Qin J., Prifti E., Hildebrand F., Falony G., Almeida M., Arumugam M., Batto J-M., Kennedy S., Leonard P., Li J., Burgdorf K., Grarup N., Jørgensen T., Brandslund I., Nielsen H.B., Juncker A.S., Bertalan M., Levenez F., Pons N., Rasmussen S., Sunagawa S., Tap J., Tims S., Zoetendal E.G., Brunak S., Clément K., Doré J., Kleerebezem M., Kristiansen K., Renault P., Sicheritz-Ponten T., de Vos W.M., Zucker J-D., Raes J., Hansen T., Bork P., Wang J., Ehrlich S.D., Pedersen O., Guedon E., Delorme C., Layec S., Khaci G., van de Guchte M., Vandemeulebrouck G., Jamet A., Dervyn R., Sanchez N., Maguin E., Haimet F., Winogradski Y., Cultrone A., Leclerc M., Juste C., Blottière H., Pelletier E., LePaslier D., Artiguenave F., Bruls T., Weissenbach J., Turner K., Parkhill J., Antolin M., Manichanh C., Casellas F., Boruel N., Varela E., Torrejon A., Guarner F., Denariaz G., Derrien M., van Hylckama Vlieg J.E.T., Veiga P., Oozeer R., Knol J., Rescigno M., Brechot C., M’Rini C., Mérieux A., Yamada T. (2013) Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546 [DOI] [PubMed] [Google Scholar]
- 8. Louis P., Hold G.L., Flint H.J. (2014) The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 12, 661–672 [DOI] [PubMed] [Google Scholar]
- 9. Feil R., Fraga M.F. (2012) Epigenetics and the environment: emerging patterns and implications. Nat. Rev. Genet. 13, 97–109 [DOI] [PubMed] [Google Scholar]
- 10. Wu X., Zhang Y. (2017) TET-mediated active DNA demethylation: mechanism, function and beyond. Nat. Rev. Genet. 18: 517–534 [DOI] [PubMed] [Google Scholar]
- 11. Bernstein B.E., Meissner A., Lander E.S. (2007) The mammalian epigenome. Cell 128, 669–681 [DOI] [PubMed] [Google Scholar]
- 12. Crider K.S., Yang T.P., Berry R.J., Bailey L.B. (2012) Folate and DNA methylation: a review of molecular mechanisms and the evidence for folate’s role. Adv. Nutr. 3, 21–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rossi M., Amaretti A., Raimondi S. (2011) Folate production by probiotic bacteria. Nutrients 3, 118–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Strozzi G.P., Mogna L. (2008) Quantification of folic acid in human feces after administration of Bifidobacterium probiotic strains. J. Clin. Gastroenterol. 42(Suppl. 3), S179–S184 [DOI] [PubMed] [Google Scholar]
- 15. Cortese R., Lu L., Yu Y., Ruden D., Claud E.C. (2016) Epigenome-Microbiome crosstalk: a potential new paradigm influencing neonatal susceptibility to disease. Epigenetics 11, 205–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Takahashi K., Sugi Y., Nakano K., Tsuda M., Kurihara K., Hosono A., Kaminogawa S. (2011) Epigenetic control of the host gene by commensal bacteria in large intestinal epithelial cells. J. Biol. Chem. 286, 35755–35762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pacis A., Tailleux L., Morin A.M., Lambourne J., MacIsaac J.L., Yotova V., Dumaine A., Danckaert A., Luca F., Grenier J.C., Hansen K.D., Gicquel B., Yu M., Pai A., He C., Tung J., Pastinen T., Kobor M.S., Pique-Regi R., Gilad Y., Barreiro L.B. (2015) Bacterial infection remodels the DNA methylation landscape of human dendritic cells. Genome Res. 25, 1801–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Okubo H., Kushiyama A., Sakoda H., Nakatsu Y., Iizuka M., Taki N., Fujishiro M., Fukushima T., Kamata H., Nagamachi A., Inaba T., Nishimura F., Katagiri H., Asahara T., Yoshida Y., Chonan O., Encinas J., Asano T. (2016) Involvement of resistin-like molecule beta in the development of methionine-choline deficient diet-induced non-alcoholic steatohepatitis in mice. Sci. Rep. 6, 20157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lin C.J., Wang M.C. (2017) Microbial metabolites regulate host lipid metabolism through NR5A-Hedgehog signalling. Nat. Cell Biol. 19, 550–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tahara T., Yamamoto E., Suzuki H., Maruyama R., Chung W., Garriga J., Jelinek J., Yamano H.O., Sugai T., An B., Shureiqi I., Toyota M., Kondo Y., Estecio M.R., Issa J.P. (2014) Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 74, 1311–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tahara T., Hirata I., Nakano N., Tahara S., Horiguchi N., Kawamura T., Okubo M., Ishizuka T., Yamada H., Yoshida D., Ohmori T., Maeda K., Komura N., Ikuno H., Jodai Y., Kamano T., Nagasaka M., Nakagawa Y., Tuskamoto T., Urano M., Shibata T., Kuroda M., Ohmiya N. (2017) Potential link between Fusobacterium enrichment and DNA methylation accumulation in the inflammatory colonic mucosa in ulcerative colitis. Oncotarget 8, 61917–61926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ito M., Kanno S., Nosho K., Sukawa Y., Mitsuhashi K., Kurihara H., Igarashi H., Takahashi T., Tachibana M., Takahashi H., Yoshii S., Takenouchi T., Hasegawa T., Okita K., Hirata K., Maruyama R., Suzuki H., Imai K., Yamamoto H., Shinomura Y. (2015) Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int. J. Cancer 137, 1258–1268 [DOI] [PubMed] [Google Scholar]
- 23. Yu T., Guo F., Yu Y., Sun T., Ma D., Han J., Qian Y., Kryczek I., Sun D., Nagarsheth N., Chen Y., Chen H., Hong J., Zou W., Fang J. Y. (2017) Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 170, 548–563.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Donohoe D.R., Garge N., Zhang X., Sun W., O’Connell T.M., Bunger M.K., Bultman S.J. (2011) The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 13, 517–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Demehri F.R., Frykman P.K., Cheng Z., Ruan C., Wester T., Nordenskjold A., Kawaguchi A., Hui T.T., Granstrom A.L., Funari V., Teitelbaum D.H., Group H.C.R. (2016) Altered fecal short chain fatty acid composition in children with a history of Hirschsprung-associated enterocolitis. J. Pediatr. Surg. 51, 81–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Donohoe D.R., Holley D., Collins L.B., Montgomery S.A., Whitmore A.C., Hillhouse A., Curry K.P., Renner S.W., Greenwalt A., Ryan E.P., Godfrey V., Heise M.T., Threadgill D.S., Han A., Swenberg J.A., Threadgill D.W., Bultman S.J. (2014) A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov. 4, 1387–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kumar P., Gogulamudi V.R., Peryasamy R., Raghavaraju G., Subramania U., Pandey K.N. (2017) Inhibition of HDAC enhances STAT acetylation, blocks NF-KappaB, and suppresses the renal inflammation and fibrosis in Npr1 haplotype male mice. Am. J. Physiol. Renal Physiol. 313, F781–F795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Furusawa Y., Obata Y., Fukuda S., Endo T.A., Nakato G., Takahashi D., Nakanishi Y., Uetake C., Kato K., Kato T., Takahashi M., Fukuda N.N., Murakami S., Miyauchi E., Hino S., Atarashi K., Onawa S., Fujimura Y., Lockett T., Clarke J.M., Topping D.L., Tomita M., Hori S., Ohara O., Morita T., Koseki H., Kikuchi J., Honda K., Hase K., Ohno H. (2013) Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 [DOI] [PubMed] [Google Scholar]
- 29. Bultman S.J., Jobin C. (2014) Microbial-derived butyrate: an oncometabolite or tumor-suppressive metabolite? Cell Host Microbe 16, 143–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hu S., Liu L., Chang E.B., Wang J.Y., Raufman J.P. (2015) Butyrate inhibits pro-proliferative miR-92a by diminishing c-Myc-induced miR-17-92a cluster transcription in human colon cancer cells. Mol. Cancer 14, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Belcheva A., Irrazabal T., Robertson S.J., Streutker C., Maughan H., Rubino S., Moriyama E., Copeland J.K., Surendra A., Kumar S., Green B., Geddes K., Pezo R.C., Navarre W.W., Milosevic M., Wilson B.C., Girardin S.E., Wolever T.M.S., Edelmann W., Guttman D.S., Philpott D.J., Martin A. (2014) Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell 158, 288–299 [DOI] [PubMed] [Google Scholar]
- 32. Janke R., Dodson A.E., Rine J. (2015) Metabolism and epigenetics. Annu. Rev. Cell Dev. Biol. 31, 473–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Krautkramer K.A., Kreznar J.H., Romano K.A., Vivas E.I., Barrett-Wilt G.A., Rabaglia M.E., Keller M.P., Attie A.D., Rey F.E., Denu J.M. (2016) Diet-microbiota interactions mediate global epigenetic programming in multiple host tissues. Mol. Cell 64, 982–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Paul B., Barnes S., Demark-Wahnefried W., Morrow C., Salvador C., Skibola C., Tollefsbol T.O. (2015) Influences of diet and the gut microbiome on epigenetic modulation in cancer and other diseases. Clin. Epigenetics 7, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liang L., Ai L., Qian J., Fang J.Y., Xu J. (2015) Long noncoding RNA expression profiles in gut tissues constitute molecular signatures that reflect the types of microbes. Sci. Rep. 5, 11763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Maudet C., Mano M., Sunkavalli U., Sharan M., Giacca M., Forstner K.U., Eulalio A. (2014) Functional high-throughput screening identifies the miR-15 microRNA family as cellular restriction factors for Salmonella infection. Nat Commun. 5, 4718. [DOI] [PubMed] [Google Scholar]
- 37. Staedel C., Darfeuille F. (2013) MicroRNAs and bacterial infection. Cell. Microbiol. 15, 1496–1507 [DOI] [PubMed] [Google Scholar]
- 38. Kumar M., Sahu S.K., Kumar R., Subuddhi A., Maji R.K., Jana K., Gupta P., Raffetseder J., Lerm M., Ghosh Z., van Loo G., Beyaert R., Gupta U.D., Kundu M., Basu J. (2015) MicroRNA let-7 modulates the immune response to Mycobacterium tuberculosis infection via control of A20, an inhibitor of the NF-kappaB pathway. Cell Host Microbe 17, 345–356 [DOI] [PubMed] [Google Scholar]
- 39. Xiao B., Liu Z., Li B.S., Tang B., Li W., Guo G., Shi Y., Wang F., Wu Y., Tong W. D., Guo H., Mao X.H., Zou Q.M. (2009) Induction of microRNA-155 during Helicobacter pylori infection and its negative regulatory role in the inflammatory response. J. Infect. Dis. 200, 916–925 [DOI] [PubMed] [Google Scholar]
- 40. Koch M., Mollenkopf H.J., Klemm U., Meyer T.F. (2012) Induction of microRNA-155 is TLR- and type IV secretion system-dependent in macrophages and inhibits DNA-damage induced apoptosis. Proc. Natl. Acad. Sci. U S A 109, E1153–E1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Teng G., Wang W., Dai Y., Wang S., Chu Y., Li J., El-Rifai W. (2013) Let-7b is involved in the inflammation and immune responses associated with Helicobacter pylori infection by targeting Toll-like receptor 4. PLoS One 8, e56709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu S., da Cunha A.P., Rezende R.M., Cialic R., Wei Z., Bry L., Comstock L.E., Gandhi R., Weiner H.L. (2016) The Host Shapes the Gut Microbiota via Fecal MicroRNA. Cell Host Microbe 19, 32–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Thaiss C.A., Levy M., Korem T., Dohnalova L., Shapiro H., Jaitin D.A., David E., Winter D.R., Gury-BenAri M., Tatirovsky E., Tuganbaev T., Federici S., Zmora N., Zeevi D., Dori-Bachash M., Pevsner-Fischer M., Kartvelishvily E., Brandis A., Harmelin A., Shibolet O., Halpern Z., Honda K., Amit I., Segal E., Elinav E. (2016) Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell 167, 1495–1510.e12 [DOI] [PubMed] [Google Scholar]
- 44. Wang Y., Kuang Z., Yu X., Ruhn K.A., Kubo M., Hooper L.V. (2017) The intestinal microbiota regulates body composition through NFIL3 and the circadian clock. Science 357, 912–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cervantes-Barragan L., Chai J.N., Tianero M.D., Di Luccia B., Ahern P.P., Merriman J., Cortez V.S., Caparon M.G., Donia M.S., Gilfillan S., Cella M., Gordon J.I., Hsieh C.S., Colonna M. (2017) Lactobacillus reuteri induces gut intraepithelial CD4+CD8alphaalpha+ T cells. Science 357, 806–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Byndloss M.X., Olsan E.E., Rivera-Chávez F., Tiffany C.R., Cevallos S.A., Lokken K.L., Torres T.P., Byndloss A.J., Faber F., Gao Y., Litvak Y., Lopez C., Xu G., Napoli E., Giulivi C., Tsolis R.M., Revzin A., Lebrilla C.B., Bäumler A.J. (2017) Microbiota-activated PPAR-gamma signaling inhibits dysbiotic Enterobacteriaceae expansion. Science 357, 570–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schroeder B.O., Backhed F. (2016) Signals from the gut microbiota to distant organs in physiology and disease. Nat. Med. 22, 1079–1089 [DOI] [PubMed] [Google Scholar]
- 48. Ley R.E., Backhed F., Turnbaugh P., Lozupone C.A., Knight R.D., Gordon J.I. (2005) Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U S A 102, 11070–11075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Walters W.A., Xu Z., Knight R. (2014) Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett. 588, 4223–4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fei N., Zhao L. (2013) An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. ISME J. 7, 880–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hwang I., Park Y.J., Kim Y.R., Kim Y.N., Ka S., Lee H.Y., Seong J.K., Seok Y.J., Kim J.B. (2015) Alteration of gut microbiota by vancomycin and bacitracin improves insulin resistance via glucagon-like peptide 1 in diet-induced obesity. FASEB J. 29, 2397–2411 [DOI] [PubMed] [Google Scholar]
- 52. Wang Z., Roberts A.B., Buffa J.A., Levison B.S., Zhu W., Org E., Gu X., Huang Y., Zamanian-Daryoush M., Culley M.K., DiDonato A.J., Fu X., Hazen J.E., Krajcik D., DiDonato J.A., Lusis A.J., Hazen S.L. (2015) Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell 163, 1585–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jie Z., Xia H., Zhong S. L., Feng Q., Li S., Liang S., Zhong H., Liu Z., Gao Y., Zhao H., Zhang D., Su Z., Fang Z., Lan Z., Li J., Xiao L., Li J., Li R., Li X., Li F., Ren H., Huang Y., Peng Y., Li G., Wen B., Dong B., Chen J.Y., Geng Q.S., Zhang Z.W., Yang H., Wang J., Wang J., Zhang X., Madsen L., Brix S., Ning G., Xu X., Liu X., Hou Y., Jia H., He K., Kristiansen K. (2017) The gut microbiome in atherosclerotic cardiovascular disease. Nat. Commun. 8, 845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Janssen A.W.F., Houben T., Katiraei S., Dijk W., Boutens L., van der Bolt N., Wang Z., Brown J.M., Hazen S.L., Mandard S., Shiri-Sverdlov R., Kuipers F., Willems van Dijk K., Vervoort J., Stienstra R., Hooiveld G., Kersten S. (2017) Modulation of the gut microbiota impacts nonalcoholic fatty liver disease: a potential role for bile acids. J. Lipid Res. 58, 1399–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Frasinariu O.E., Ceccarelli S., Alisi A., Moraru E., Nobili V. (2013) Gut-liver axis and fibrosis in nonalcoholic fatty liver disease: an input for novel therapies. Dig. Liver Dis. 45, 543–551 [DOI] [PubMed] [Google Scholar]
- 56. Li L., Chen L., Hu L., Liu Y., Sun H.Y., Tang J., Hou Y. J., Chang Y.X., Tu Q.Q., Feng G.S., Shen F., Wu M.C., Wang H.Y. (2011) Nuclear factor high-mobility group box1 mediating the activation of Toll-like receptor 4 signaling in hepatocytes in the early stage of nonalcoholic fatty liver disease in mice. Hepatology 54, 1620–1630 [DOI] [PubMed] [Google Scholar]