Abstract
The dwindling wildlife species of our planet have become a cause célèbre for conservation groups, governments, and concerned citizens throughout the world. The application of powerful new genetic technologies to surviving populations of threatened mammals has revolutionized our ability to recognize hidden perils that afflict them. We have learned new lessons of survival, adaptation, and evolution from viewing the natural history of genomes in hundreds of detailed studies. A single case history of one species, the African cheetah, Acinonyx jubatus, is here reviewed to reveal a long-term story of conservation challenges and action informed by genetic discoveries and insights. A synthesis of 3 decades of data, interpretation, and controversy, capped by whole genome sequence analysis of cheetahs, provides a compelling tale of conservation relevance and action to protect this species and other threatened wildlife.
Keywords: Acinonyx jubatus; Cheetah genome, Indian Cheetah; population bottleneck
The cheetah remains emblematic of the threats facing wildlife, not only because of its unique adaptations but also its distinctive evolutionary history (Neff 1983; Marker and Eszterhas 2014). When breeding was attempted in zoos in the 1950–1980 period, cheetahs were unusual in that they bred poorly in captivity, rarely exceeding 15% success of attempted pairing. Even so, cub mortality was 30–40% higher than almost all zoo animals leading to a captive population that was hardly sustainable (Marker and O’Brien 1989; O’Brien et al. 1985). Cheetah males displayed a 10-fold reduction in sperm count plus an elevated incidence of malformed spermatozoa (~70–75% of sperm in any males had super large heads, tiny heads, coiled or bent tails, indicators of sterility in other Felidae species) (O’Brien et al. 1983, 1985; Wildt et al. 1993; Crosier et al. 2007). Part of the reason for the reproductive impairments was likely the relative paucity of overall genome variability in cheetahs sampled from zoos and in wild populations from southern and eastern Africa. Cheetahs displayed 90–99% less overall diversity than other cats and most other mammals based upon early surveys of nuclear allozymes, 2DE skin fibroblast proteins, and RFLP diversity in the major histocompatibility complex (MHC) (O’Brien et al. 1983, 1985; Yuhki et al. 1990; O’Brien and Johnson 2005). Perhaps the most remarkable indicator of the cheetah’s genetic impoverishment was the demonstration that cheetahs failed to reject surgically implanted skin allografts from unrelated cheetah donors, while their perfectly functional immune system adequately rejected xenograft skin patches from the domestic cat (O’Brien et al. 1985). The cheetah’s MHC, which mediates graft rejection in most species was so similar that their immune system failed to recognize “nonself,” as if the cheetahs tested were immunological clones or identical twins. It seemed as though the ancestors of modern cheetahs had offloaded most of their endemic genetic variability, leaving a species dramatically reduced in genetic diversity.
Reconstitution of allelic variation in rapidly mutating genetic markers (nuclear microsatellites and mitochondrial DNA which evolve 10–100 times faster than other chromosomal sequences) provided an innate chronometer that predicted the time elapsed since the population bottleneck which reduced the species’ genetic legacy. That time interval was first estimated at 10–12000 years ago in North America where the cheetah species had evolved (Menotti-Raymond and O’Brien 1993). Cheetah populations were widespread until this time, when a large mammal extinction event eliminated 75% of large mammals from North America, including mastodons, mammoths, giant ground sloth, short faced bears, saber toothed tiger, American lions, pumas, and cheetahs (Neff 1983; Gingerich 1984; Werdelin 1985; Martin and Wright 1967; Werdelin et al. 2009). Thankfully for the cheetah, many thousands of years earlier their forebears had migrated from North America across the Beringia straits to Asia (these exact geographic movements are controversial; O’Brien et al. 2016; Faurby et al. 2016) and dispersed southward to colonize Africa. That migration itself likely precipitated demographic and genetic reduction, but it nonetheless allowed the cheetah species to escape from the cataclysm in the North American lower Pleistocene, the most extreme species extinction in the 100 million year history of mammalian diversification.
When we first discovered the dramatic reproductive impairment of cheetah males coupled with their reduced fecundity and high mortality, we thought we had seen the worst consequences of this loss of diversity, but we were wrong. The homogenization of the cheetah genes, including those mediating immune defenses, showed itself again in a devastating outbreak of feline coronavirus (FeCV—a close virus relative of human SARS coronavirus) at a cheetah breeding facility in 1983 (O’Brien et al. 1985; Heeney et al. 1990; Pearks Wilkerson et al. 2004). FeCV causes feline infectious peritonitis in house cats, a progressive deadly pathology whereby the immune system produces virus-immunoglobulin deposits as a milky fluid in the peritoneum which strangulates kidneys, liver and internal organs. FeCV morbidity is usually less than 10% and mortality approximately 1% in domestic cat facilities or multi-cat households. The cheetah contagion was much worse. Within 6 months every cheetah at the breeding facility (45 individuals) was infected, all had symptoms (fever, diarrhea, twitches, seizers, and collapse) and within 3 years 60% of the cheetahs had died. This remains the worst case of FCoV infection ever reported in any species. The cheetahs genetic uniformity was clearly a determinant as an FeCV strain variant that adapted to evade the first victim’s immune system had inadvertently evolved a strategy to decimate all cheetahs.
The Cheetah Genome
Today, we enjoy unprecedented power to reconstruct the evolutionary history and predict the evolutionary potential of species through genome sequencing. In late 2015, Dobrynin et al. (2015) released the whole genome sequence assembly and annotation of 7 cheetahs including the reference genome of “Chewbacca,” the ambassador cheetah for the Cheetah Conservation Fund (see below). The initial genome analyses unraveled a plethora of fascinating insights around the cheetah’s past and also its remarkable specialization for dazzling speed. Multiple features of the cheetah’s genome were detailed and annotated in depth: 20343 protein-coding genes, the wide complexity of repetitive DNA families, noncoding RNA families, DNA variation within and outside of genes, copy number variation, and genes that showed evidence of recent selective pressures. The cheetah’s incredible specialization for running is likely influenced by selective retention of gene variants related to energetics and anabolism for producing muscle specialization. The genome analyses identified a group of 11 candidate genes that display evidence of selection involved in muscle contraction (5 genes), stress response (2 genes), and regulation of catabolic processes (4 genes), all now putative candidates for the cascade of sprinting adaptations we see in modern cheetahs.
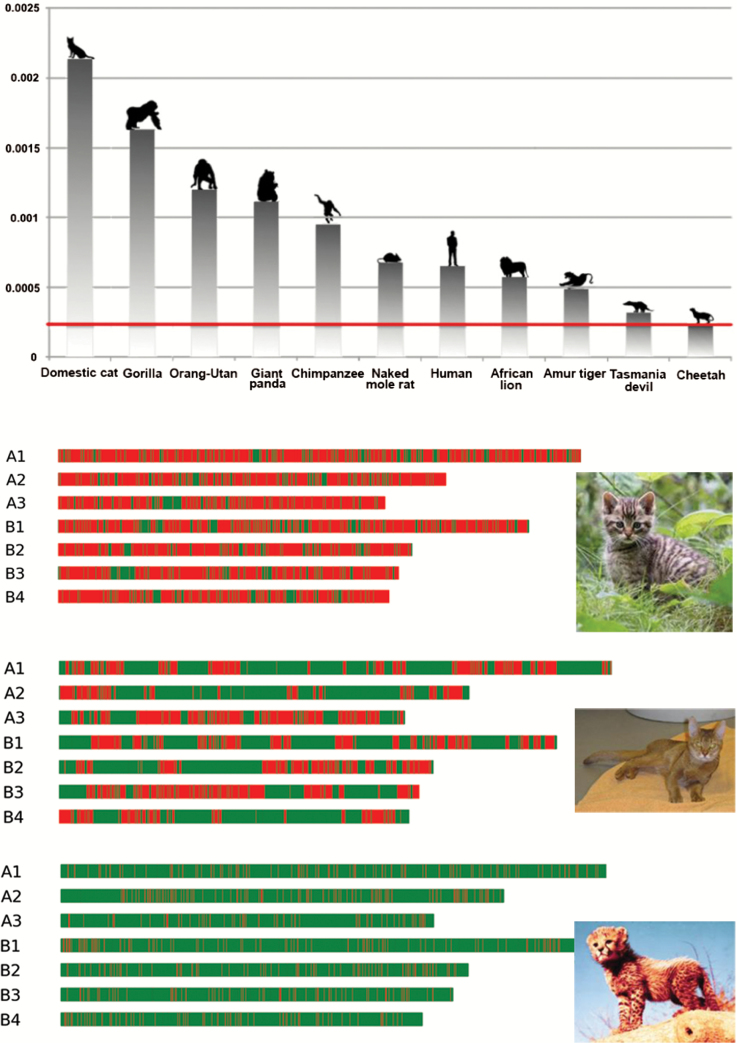
The cheetah’s genetic uniformity was confirmed by 7 different measures of genome-wide diversity. Cheetahs retain only 0.1–4% of overall genetic variation seen in most living species, much lower than other well-known examples of genetic impoverishment including Tasmanian devils, Virunga gorillas, Amur tigers, and even highly inbred domestic cats and dogs (Figure 1). Further mining of Chewbacca’s genome revealed a plausible explanation for the cheetahs’ ability to accept allogeneic skin grafts. Several genes that mediate graft rejection, the MHC (perhaps the most variable gene family in human and other mammal genomes) are nearly monomorphic across different cheetahs. In addition, all cheetahs have actually disarmed 4 MHC genes entirely (Dobrynin et al. 2015).
Figure 1.
(A) Estimates of diversity in the cheetah genome relative to other mammal genomes. The rate of single nucleotide variation (SNV) (x axis) for each individual was estimated using all variant positions, and repetitive regions were not filtered. (B) The genome of Boris, an outbred feral domestic cat living in St. Petersburg, Russia (top) is compared to Cinnamon, a highly inbred Abyssinian cat [Fca-6.2 reference for domestic cat genome sequence (Tamazian et al. 2014)] (middle) and Chewbacca, a captive cheetah (bottom) (Dobrynin et al. 2015). The first 7 chromosome homologues of the genomes of Boris, Cinnamon, and Chewbacca are displayed for direct comparison. Approximately 15000 regions of 100 Mb across the genome for each species were assessed for SNVs. Regions of high variability (>40 SNVs/100 kbp) are colored red (dark gray); highly homozygous regions (≤40 SNVs/100 kbp) are colored green (light gray). The cheetah genome is composed of 93% homozygous stretches. Reprinted from Dobrynin et al. (2015) with permission.
The estimate for the timing of the cheetahs’ historic bottlenecks were refined by coalescent analyses to suggest 2 historic population contractions: the earliest ~100000 years ago (coincident with the postulated migration from America to Africa) and the latest 11084–12589 years ago (the Pleistocene mammal extinction). One marked consequence of these bottlenecks and subsequent consanguineous matings is reproductive impairments including elevated incidence of malformed spermatozoa (O’Brien et al. 1983; Wildt et al. 1993; Crosier et al. 2007). The cheetah genome analyses revealed that AKAP4, a testis-expressed gene known to regulate sperm development in humans and mice, showed a remarkable signatures of selection due to 10 fixed substitutions in coding regions, 5 of which are function altering and likely compromise normal sperm development in the entire species (Dobrynin et al. 2015). This evolutionary gene alteration becomes a strong candidate to explain the spermatazoal abnormalities that were discovered in all cheetahs studied decades ago.
Moving Forward with Lessons Learned
The breadth and scope of the cheetah genome analysis and interpretation offers a rare insight into the silence of prehistory that molded modern species. The lessons for conservation from the cheetahs’ experience were chilling and clear. When a threatened population drops to very small numbers and survives, it can lose its endowment of genetic diversity, which otherwise provides an innate protection against rare recessive genetic abnormalities as well as a hedge against deadly infectious agents. With the example of the cheetah, the conservation community began to pay attention to genetic loss in small threatened populations.
The early studies of cheetahs made these points so persuasively that they were repeated in the popular media and many readers simply presumed that cheetahs were doomed. We do not agree that cheetahs are doomed by their genetic reduction, because the postulated bottleneck occurred at the latest some 10 millennia ago (O’Brien 2003; Dobrynin et al. 2015). Cheetah populations then grew to hundreds of thousands by the 19th century AD. Clearly, the physiological correlates of inbreeding that cheetahs experience were not rate-limiting to expansion in nature, or their numbers would never have risen so high. This misunderstanding by some led to a few deliberate assaults on the importance of genetic diversity compared to traditional ecological, demographic, or even stochastic threats to small endangered populations (Caro and Laurenson 1994; Caughley 1994; Lande 1988; Merola 1994). These arguments have been addressed, thoroughly in our view, by counter opinions and a more balanced interpretation of demographic and genetic risks, which nonetheless confirm the threat of inbreeding effects for a population’s survival (Frankham 1995; May 1995; O’Brien 1994, 1998; 2003; O’Brien and Johnson 2005; Frankham et al. 2010).
In 1990, one of us (L.M.) established the Cheetah Conservation Fund (CCF http://www.cheetah.org/), an international research and conservation organization based in the newly independent Republic of Namibia in Africa. CCF is dedicated to helping the cheetah species survive in Namibia and throughout its remaining range in Africa and in Iran, home to the last of the Asian population. Taking a holistic approach, the CCF worked with the fledgling democracy that had pledged to treasure all its wildlife, including the dwindling cheetah population, then numbering less than 15000 across Africa. Through education, communication, applied conservation strategies, science, and diligence, the CCF and sister organizations have changed attitudes in Namibia, so that cheetahs are viewed less often as “pests,” and have become a cherished symbol of Namibian natural resources. CCF has expanded its influence into other African countries and is now charged with opening new habitats for cheetahs in places where they presently and formerly existed. The work is ongoing and wide reaching, encompassing many conservation disciplines including molecular genetics and genomics. Today, cheetahs rescued from traps have been offered for reintroduction in Namibian habitats as well as in several other African countries as a source for species restoration across their former and recently truncated natural range.
New Opportunities: Replacing the Extinct Indian Cheetah
In early 2007, a courageous and determined effort by conservation-minded professionals of India was launched to consider the reintroduction of cheetahs into suitable habitat in India. The Asian cheetah was eliminated by sport hunting and exploitation in India before the 1940s, the only large mammal to go extinct in India in the past 1000 years. Spearheaded by noted Indian cheetah conservationists M. K. Ranjitsinh and Divyabhanusinh Chavda, a workshop to plan and facilitate the reintroduction of cheetah to suitable habitats of their former range in India nature reserves was convened in 2010 at Gajner, Rajasthan, a game reserve where the last Indian cheetah had survived. The workshop was attended by Indian conservation managers and scientists, by Nature Reserve superintendents and by cheetah researchers (Ranjitsinh and Jhala 2010).
A nattering issue that haunts all restoration initiatives remained. If Indian conservationists could identify regions/reserves with suitable habitat with adequate prey, from where should the translocated cheetah come? Conventional ecological wisdom says that one should strive to identify a stable population of animals that could be removed without harming the parent populations. Ideally the introduced animals should be genetically close to the original lost population so that any adaptations accumulated by the target (Indian cheetah) population over time would be retained. The obvious choice under these conditions would be the Iranian cheetah, the single living Asian cheetah population, a relict population of less than 50 animals clinging to survival in Iran since the overthrow of the Shah in 1960s (Farhadinia et al. 2016; Durant et al. 2017). However, the Iranian animals are not ideal candidates due to their endangered status, their precarious health, their present isolation into multiple small subpopulations and their political sensitive locale. So what to do?
It occurred to us that the well-known history of cheetahs’ historic bottleneck indicated that African populations were very closely related (<12000 years of separation), but the Asian cheetahs (modern or museum specimens from Iranian cheetahs) had not been examined. Were the Asian (Iranian and Indian) cheetahs really a continental subspecies of appreciable divergence from African cheetahs? Or alternatively, do they descend from the same recent Pleistocene population bottleneck as the African cheetahs? The answer to this question would bear importantly on choosing African versus Iranian cheetahs for Indian restoration.
To provide the best available science for this management decision, we and several colleagues obtained PCR generated DNA products from 21 African and Asian-Iranian cheetah specimens representing four mitochondrial genes (ATPase, ND5, 12s-RNA, 16S-RNA, control region) that were selected for their proven diagnostic values (Johnson et al. 2006).
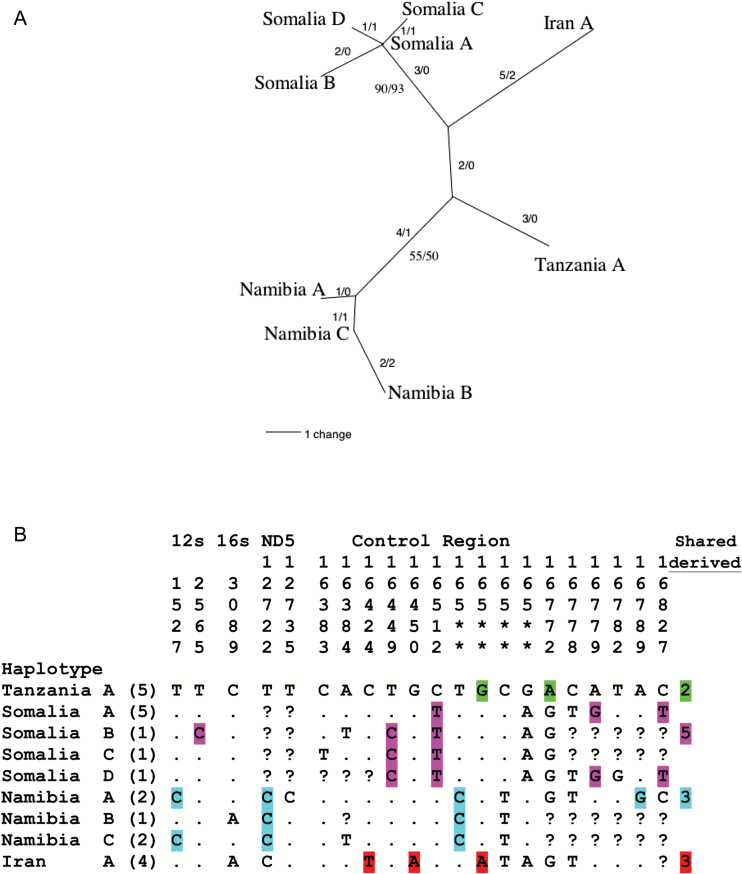
DNA sequences of these genes were analyzed phylogenetically and the results revealed a close relationship among for cheetah specimens from East Africa, South Africa, Somalia and Iran (Figure 2A). The number of mutational substitutions (i.e., genetic distance) between the 4 regions was similar, 3–5 steps between each African group or between the African populations and the Asian Iranian group. Diagnostic, shared derived (synapomorphic) substitutions for each population were few: 3 sites for Tanzania, 5 for Somalia, 3 for Namibia, and 3 for Iran (colored sites in Figure 2B). These substitutional distances allow for estimation of the time separations as between 4400 and 6100 years among any African population or between African and the Asian-Iranian specimens (Table 1). Such divergence times are quite recent, less than 1/20th of the comparable divergence times between Asian versus African lions or leopards or even human ethnic groups (Table 2).
Figure 2.
(A) Minimum parsimony spanning network of mtDNA haplotypes detected in 21 African and Asian cheetahs from the indicated geographic regions. Haplotypes were constructed from 1498 base pairs (bp) including fragments from mitochondrial gene segments: ATPase, ND5, 12s-RNA, 16S-RNA, and the control region. Sequences included 21 variable sites, and defined 9 haplotypes by 21 parsimony informative sites. Numbers on branches are the number of bp steps/number of homoplasies. Bootstrap values (>50) are shown in bold on the Somalia and Namibia lineages. (B) Variable sites from 4 mitochondrial DNA gene fragments defining cheetah mitochondrial DNA haplotypes. Nucleotide site numbers are based on the complete domestic cat mitochondrial DNA sequence (Lopez et al. 1996). A period (.) represents sites with the same base pair as the reference Tanzania haplotype. Question marks represent portions for which sequence was not obtained. Asterisks represent sites within the mtDNA control region that were too variable to confidently align with the domestic cat sequence. Synapomorphic sites for each region are colored. Number of cheetahs representing each haplotype is in parentheses.
Table 1.
Genetic distances in base pairs (bp) from mtDNA haplotypesa and estimated years of separation among geographically isolated populations of cheetahs
| Cheetah populations | No. bp steps | Time of separation (years) | Variance (years) |
|---|---|---|---|
| Tanzania vs. Namibia | 8–10 | 4383 | ±131 |
| Tanzania vs. Somalia | 9–10 | 4657 | ±274 |
| Namibia vs. Somalia | 9–11 | 4877 | ±145 |
| Iran vs. Somalia | 9–10 | 4657 | ±274 |
| Iran vs. Tanzania | 10 | 4931 | ±548 |
| Iran vs. Namibia | 12–13 | 6137 | ±439 |
aEstimates based on mtDNA divergence (1498 bp) in Figure 2, calibrated to the date of divergence of Acinonyx jubatus jubatus and Acinonyx jubatus raineyi at 4253–4514 years ago (Driscoll et al. 2002). mtDNA Sequences were deposited in GenBank (Johnson et al. 2006).
Table 2.
The time of separation for Asian and African cheetahs, with additional relevant examples of separation dates of other species and subspecies, as estimated by molecular phylogenetics
| Estimated | ||
|---|---|---|
| Genetic distance among: | Time interval | Citation |
| Clouded leopard species | 1.4 MYA | Buckley-Beason et al. (2006) |
| Wilting et al. (2007) | ||
| Orangutan species | 1.1–1.7 MYA | Janczewski et al. (1990) |
| Lu et al. (1996) | ||
| Asia vs. Africa humans | 700000 YBP | Malaspinas et al. (2016) |
| Mallick et al. (2016) | ||
| Pagani et al. (2016) | ||
| Asia vs. Africa lions | 100000 YBP | Antunes et al. (2008) |
| Asia vs. Africa leopard | 169000 YBP | Uphyrkina et al. (2001) |
| 5 Living tiger subspecies | 72000 YBP | Luo et al. (2004) |
| Asia vs. Africa cheetahs | 4500–6500 YBP | Table 1 |
MYA, million years ago; YBP, years before present.
If affirmed (see below) these results (albeit limited in specimen number and size of sequence assessed), would suggest that the Asia-Iranian cheetah is as close to modern African cheetah populations as the latter are to each other. Thus, we tentatively can conclude that the cheetahs in Iran did descend from the same historic bottleneck event which homogenized the African cheetah population ~10–12000 years ago. As such, in genetic terms, Iranian migrants (as a re-introduction source) would be no closer or “better adapted” than any African population to be considered for restoration to India. We presented these data and conclusions at the Gajner, Rajasthan restoration workshop in 2010. The workshop report noted that African cheetahs should be acceptable source for the Indian habitat restoration. The balance of the workshop business and proceedings concentrated on identifying suitable habitat and prey base among potential restoration site in Indian Nature reserves (Ranjitsinh and Jhala 2010). Then another shoe dropped.
Since the 2010 meeting, Charruau et al. (2011) published an independent detailed study of cheetah phylogeography examining 94 cheetah specimens including 12 Iranian cheetahs. This study included more individuals but shorter sequence and fewer informative sites (139 bp of control region for all, and 915 base pairs for some, with 12–14 parsimony informative sites) than our study. They also presented an analyses of 20 microsatellite loci for 92 cheetah specimens. Results of their phylogenetic analysis of mtDNA were in effect remarkably similar to ours. Their mtDNA analyses demonstrated monophyly of cheetahs from Asia, South Africa, and Somalia as well as differentiation of these populations using microsatellite genotypes. Southern and northeast African cheetah populations showed modest molecular genetic distances in comparison with Iranian cheetahs, similar to the distances among African population isolates. Their results affirm the distinction of Asian to African cheetah populations but also their shallow divergence, lending additional support to the interpretation that like African populations, Asiatic cheetahs derive from a post-bottleneck expansion. Charruau et al. (2011) also presented data on a museum specimen of extinct Indian cheetah showing it to align rather closely with the Iranian cheetah lineage. This result supports other evidence of a common Asian origin for Indian and Iranian cheetahs and discounts a previously unsettled postulate that the Iranian cheetahs are possibly descendants of African cheetahs that were re-located and released by Middle Eastern sheiks in historic times.
Charruau et al. (2011) also presented several coalescence calculations of the age of mtDNA and microsatellite diversity that puts the origin of cheetah genetic diversity at sometime between 4700 and 67000 ago, a rather wide confidence interval. The explanation seems that they employed multiple genetic models, which led to conflicting results. This seems due in part to the imprecision of microsatellite mutation rates estimates, which vary over 4 logs, and the short length mtDNA sequence (only 139 bp of control region). Nonetheless the new data from Charruau et al. (2011) are clearly consistent with our own previous findings and interpretations (Figure 2). The phylogenetic analyses of both mtDNA and microsatellite affirm the Asian monophyly and the relative similarity of Iranian, Indian and African cheetah specimens, consistent with a post bottleneck history for all living cheetahs.
For the case of the Indian Cheetah, the science was informative and definitive in leading to our strong recommendation to move forward with restoration using cheetahs from the Southern African cheetah population (South Africa Namibia and/or Botswana). Since 2010 there have been legal assaults on the restoration program for numerous reasons. In 2012, India’s Supreme Court suspended the proposed restoration based upon the ambiguous and imprecise dating calculations presented by Charruau et al. (2011). [A similar political stall occurred in the early 1990s with the Florida panther/puma recommendation for restoration. That ended happily with success for the 1996 puma restoration efforts (Johnson et al. 2010).]
Recently, a more precise computation of the date of the population bottleneck based upon cheetah whole genome sequence estimated a coalescence date as 11084–12589 years ago coincident with the late Pleistocene large mammal extinctions (Dobrynin et al. 2015). These new results combined with Figure 2 and the consistent phylogeography of Charruau et al. (2011) would affirm our conclusion that the Asian-Iranian and the extinct Indian cheetahs descend from the global population bottleneck that afflicted the African cheetah populations.
We would urge Indian conservationists to join forces to assist in using the new definitive genetic data, combined with habitat assessment (Ranjitsinh and Jhala 2010; Charruau et al. 2011; Dobrynin et al. 2015), to implement the proposal to restore cheetah to the Indian nature reserves in the near future. It is now time to reverse the ill-conceived attempt to suspend cheetah restoration into Indian habitat, which was based on flawed interpretations of the data available at the time.
Conclusions
New data from conservation genetics are weighing in on a wide range of management plans. In some cases it can be very important, but in others less so. We cannot always predict which endangered species will benefit but if history is a lesson some, perhaps many, will benefit from a robust genetic assessment and informed interpretation. Conservationists of all disciplines claim to want the same thing—species preservation and stabilization of natural ecosystems. Should new generations of wildlife managers look at all the data from different fields including genetics, they are better equipped to make informed management decisions, often having to make the best of some very bad situations. When that happens, species and wild areas will benefit, as genetic and genomic tools become better appreciated for their informed interpretations as well as their limitations.
In the case of the cheetah, the data are clear on 3 principal points: 1) All modern cheetahs descend from a late Pleistocene bottleneck that reduced genomic diversity by 1/10th to 1/100th in the surviving species; 2) The genetic loss was not rate-limiting in nature, meaning that ecological stabilization and habitat protection are key to cheetah conservation; and 3) African cheetahs rescued from the wild are suitable in genetic terms as founders of restored Asian populations since both African and Asian cheetahs descend from the recent population bottleneck of <12000 years ago.
The way forward we would recommend is to proceed with re-introduction of suitable African cheetahs, themselves descendent of a successful natural history expansion, to select Indian wildlife reserves (Ranjitsinh and Jhala 2010).
Funding
S.J.O. and P.D. were supported by the Russian Science Foundation grant (project no. 17-14-01138). S.J.O. was the principal investigator.
Acknowledgments
All tissue samples were collected in full compliance with specific Federal Fish and Wildlife permits [Conservation on International Trade in Endangered Species of Wild Fauna and Flora (CITES); Endangered and Threatened Species] issued to the National Cancer Institute, National Institutes of Health (Principal officer S.J.O.) by the U.S. Fish and Wildlife Service of the Department of the Interior.
References
- Antunes A, Troyer JL, Roelke ME, Pecon-Slattery J, Packer C, Winterbach C, Winterbach H, Hemson G, Frank L, Stander P et al. . 2008. The evolutionary dynamics of the lion Panthera leo revealed by host and viral population genomics. PLoS Genet. 4:e1000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley-Beason VA, Johnson WE, Nash WG, Stanyon R, Menninger JC, Driscoll CA, Howard J, Bush M, Page JE, Roelke ME et al. . 2006. Molecular evidence for species-level distinctions in clouded leopards. Curr Biol. 16:2371–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caro TM, Laurenson MK. 1994. Ecological and genetic factors in conservation: a cautionary tale. Science. 263:485–486. [DOI] [PubMed] [Google Scholar]
- Caughley G. 1994. Directions in conservation biology. J Anim Ecol. 63:215–244. [Google Scholar]
- Charruau P, Fernandes C, Orozco-Terwengel P, Peters J, Hunter L, Ziaie H, Jourabchian A, Jowkar H, Schaller G, Ostrowski S et al. . 2011. Phylogeography, genetic structure and population divergence time of cheetahs in Africa and Asia: evidence for long-term geographic isolates. Mol Ecol. 20:706–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosier AE, Marker L, Howard J, Pukazhenthi BS, Henghali JN, Wildt DE. 2007. Ejaculate traits in the Namibian cheetah (Acinonyx jubatus): influence of age, season and captivity. Reprod Fertil Dev. 19:370–382. [DOI] [PubMed] [Google Scholar]
- Dobrynin P, Liu S, Tamazian G, Xiong Z, Yurchenko AA, Krasheninnikova K, Kliver S, Schmidt-Küntzel A, Koepfli KP, Johnson W et al. . 2015. Genomic legacy of the African cheetah, Acinonyx jubatus. Genome Biol. 16:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durant SM, Mitchell N, Groom R, Pettorelli N, Ipavec A, Jacobson AP, Woodroffe R, Böhm M, Hunter LT, Becker MS et al. . 2017. The global decline of cheetah Acinonyx jubatus and what it means for conservation. Proc Natl Acad Sci U S A. 114:528–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhadinia MS, Akbari H, Eslami M, Adibi MA. 2016. A review of ecology and conservation status of Asiatic cheetah in Iran. Cat News Special Issue Iran. 10:18–26. [Google Scholar]
- Faurby S, Werdelin L, Svenning JC. 2016. The difference between trivial and scientific names: there were never any true cheetahs in North America. Genome Biol. 17:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankham R. 1995. Conservation genetics. Annu Rev Genet. 29:305–327. [DOI] [PubMed] [Google Scholar]
- Frankham R, Ballou JD, Briscoe DA. 2010. Introduction to conservation genetics. 2nd ed Cambridge University Press. [Google Scholar]
- Gingerich PG. 1984. Pleistocene extinctions in the context of origination-extinction equilibria in cenozoic mammals. In: Martin PS and Klein RG, editors. Quaternary extinctions. Tucson (AZ): University of Arizona Press. [Google Scholar]
- Heeney JL, Evermann JF, McKeirnan AJ, Marker-Kraus L, Roelke ME, Bush M, Wildt DE, Meltzer DG, Colly L, Lukas J. 1990. Prevalence and implications of feline coronavirus infections of captive and free-ranging cheetahs (Acinonyx jubatus). J Virol. 64:1964–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janczewski DN, Goldman D, O’Brien SJ. 1990. Molecular genetic divergence of orang utan (Pongo pygmaeus) subspecies based on isozyme and two-dimensional gel electrophoresis. J Hered. 81:375–387. [PubMed] [Google Scholar]
- Johnson WE, Eizirik E, Pecon-Slattery J, Murphy WJ, Antunes A, Teeling E, O’Brien SJ. 2006. The late Miocene radiation of modern Felidae: a genetic assessment. Science. 311:73–77. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Onorato DP, Roelke ME, Land ED, Cunningham M, Belden RC, McBride R, Jansen D, Lotz M, Shindle D et al. . 2010. Genetic restoration of the Florida panther. Science. 329:1641–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R. 1988. Genetics and demography in biological conservation. Science. 241:1455–1460. [DOI] [PubMed] [Google Scholar]
- Lu Zhi, Karish WB, Janczewski DN, Frazier-Taylor H, Sajuthi D, Gombek F, Andau M, Martenson JS, O’Brien SJ. 1996. Genomic differentiation among natural populations of orang-utan (Pongo pygmaeus). Curr Biol. 6:1326–1336. [DOI] [PubMed] [Google Scholar]
- Luo SJ, Kim JH, Johnson WE, van der Walt J, Martenson J, Yuhki N, Miquelle DG, Uphyrkina O, Goodrich JM, Quigley HB et al. . 2004. Phylogeography and genetic ancestry of tigers (Panthera tigris). PLoS Biol. 2:e442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaspinas AS, Westaway MC, Muller C, Sousa VC, Lao O, Alves I, Bergström A, Athanasiadis G, Cheng JY, Crawford JE et al. . 2016. A genomic history of Aboriginal Australia. Nature. 538:207–214. [DOI] [PubMed] [Google Scholar]
- Mallick S, Li H, Lipson M, Mathieson I, Gymrek M, Racimo F, Zhao M, Chennagiri N, Nordenfelt S, Tandon A et al. . 2016. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature. 538:201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marker L, Eszterhas S. 2014. Future for cheetahs. Otjiwarongo (Namibia): Cheetah Conservation Fund. [Google Scholar]
- Marker L, O’Brien SJ. 1989. Captive breeding of the cheetah (Acinonyx jubatus) in North American zoos (1871–1986). Zoo Biol. 8:3–16. [Google Scholar]
- Martin PS, Wright HE. 1967. Pleistocene extinctions: the search for a cause. New Haven (CT): Yale University Press. [Google Scholar]
- May R. 1995. The cheetah controversy. Nature. 374:309–310. [DOI] [PubMed] [Google Scholar]
- Menotti-Raymond M, O’Brien SJ. 1993. Dating the genetic bottleneck of the African cheetah. Proc Natl Acad Sci U S A. 90:3172–3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merola M. 1994. A reassessment of homozygosity and the case for inbreeding depression in the cheetah Acinonyx jubatus: implications for conservation. Conserv Biol. 8:961–971. [Google Scholar]
- Neff N. 1983. The big cats: the paintings of Guy Coheleach. New York: Harry N. Abrams Publ. p. 57. [Google Scholar]
- O’Brien SJ. 1994. The cheetah’s conservation controversy. Conserv Biol. 8:1153–1155. [Google Scholar]
- O’Brien SJ. 1998. Intersection of population genetics and species conservation: The cheetah’s dilemma. In: Hecht MK, MacIntrye RJ, Clegg MT, editors. Evolutionary biology. Vol. 30 New York: Plenum Press. [Google Scholar]
- O’Brien SJ. 2003. Tears of the cheetah and other tales from the genetic frontier. New York: St. Martin’s Press. [Google Scholar]
- O’Brien SJ, Johnson WE. 2005. Big cat genomics. Annu Rev Genomics Hum Genet. 6:407–429. [DOI] [PubMed] [Google Scholar]
- O’Brien SJ, Koepfli KP, Eizirik E, Johnson W, Driscoll C, Antunes A, Schmidt-Kuntzel A, Marker L, Dobrynin P. 2016. Response to comment by Faurby, Werdelin and Svenning. Genome Biol. 17:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien SJ, Roelke ME, Marker L, Newman A, Winkler CA, Meltzer D, Colly L, Evermann JF, Bush M, Wildt DE. 1985. Genetic basis for species vulnerability in the cheetah. Science. 227:1428–1434. [DOI] [PubMed] [Google Scholar]
- O’Brien SJ, Wildt DE, Goldman D, Merril CR, Bush M. 1983. The cheetah is depauperate in genetic variation. Science. 221:459–462. [DOI] [PubMed] [Google Scholar]
- Pagani L, Lawson DJ, Jagoda E, Mörseburg A, Eriksson A, Mitt M, Clemente F, Hudjashov G, DeGiorgio M, Saag L et al. . 2016. Genomic analyses inform on migration events during the peopling of Eurasia. Nature. 538:238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearks Wilkerson AJ, Teeling EC, Troyer JL, Bar-Gal GK, Roelke M, Marker L, Pecon-Slattery J, O’Brien SJ. 2004. Coronavirus outbreak in cheetahs: lessons for SARS. Curr Biol. 14:R227–R228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjitsinh MK, Jhala YV. 2010. Assessing the potential for reintroducing the cheetah in India. Wildlife Trust of India, Noida, & the Wildlife Institute of India, Dehradun. TR2010/001. [Google Scholar]
- Tamazian G, Simonov S, Dobrynin P, Makunin A, Logachev A, Komissarov A, Shevchenko A, Brukhin V, Cherkasov N, Svitin A et al. . 2014. Annotated features of the domestic cat (Felis catus) genome. GigaScience. 3:13 Available from: http://www.gigasciencejournal.com/content/3/1/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uphyrkina O, Johnson WE, Quigley H, Miquelle D, Marker L, Bush M, O’Brien SJ. 2001. Phylogenetics, genome diversity and origin of modern leopard, Panthera pardus. Mol Ecol. 10:2617–2633. [DOI] [PubMed] [Google Scholar]
- Werdelin L. 1985. Small Pleistocene felines of North America. J Vert Paleontol. 5:194–210. [Google Scholar]
- Werdelin L, O’Brien SJ, Johnson WE, Yamaguchi N. 2009. Phylogeny and evolution of cats (Felidae). In: Macdonald D and Loverage A, editors. Biology and conservation of wild felids. Oxford: Oxford University Press; p. 59–82. [Google Scholar]
- Wildt DE, Brown JL, Bush M, Barone MA, Cooper KA, Grisham J, Howard JG. 1993. Reproductive status of cheetahs (Acinonyx jubatus) in North American zoos: the benefits of physiological surveys for strategic planning. Zoo Biol. 12:45–80. [Google Scholar]
- Wilting A, Buckley-Beason VA, Feldhaar H, Gadau J, O’Brien SJ, Linsenmair KE. 2007. Clouded leopard phylogeny revisited: support for species recognition and population division between Borneo and Sumatra. Front Zool. 4:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuhki N, O’Brien SJ. 1990. DNA variation of the mammalian major histocompatibility complex reflects genomic diversity and population history. Proc Natl Acad Sci U S A. 87:836–840. [DOI] [PMC free article] [PubMed] [Google Scholar]