Abstract
The de novo assembly of the red fox (Vulpes vulpes) genome has facilitated the development of genomic tools for the species. Efforts to identify the population history of red foxes in North America have previously been limited by a lack of information about the red fox Y-chromosome sequence. However, a megabase of red fox Y-chromosome sequence was recently identified over 2 scaffolds in the reference genome. Here, these scaffolds were scanned for repeated motifs, revealing 194 likely microsatellites. Twenty-three of these loci were selected for primer development and, after testing, produced a panel of 11 novel markers that were analyzed alongside 2 markers previously developed for the red fox from dog Y-chromosome sequence. The markers were genotyped in 76 male red foxes from 4 populations: 7 foxes from Newfoundland (eastern Canada), 12 from Maryland (eastern United States), and 9 from the island of Great Britain, as well as 48 foxes of known North American origin maintained on an experimental farm in Novosibirsk, Russia. The full marker panel revealed 22 haplotypes among these red foxes, whereas the 2 previously known markers alone would have identified only 10 haplotypes. The haplotypes from the 4 populations clustered primarily by continent, but unidirectional gene flow from Great Britain and farm populations may influence haplotype diversity in the Maryland population. The development of new markers has increased the resolution at which red fox Y-chromosome diversity can be analyzed and provides insight into the contribution of males to red fox population diversity and patterns of phylogeography.
Keywords: introgression, marker development, microsatellites, population history, Vulpes vulpes
The red fox (Vulpes vulpes) inhabits regions of Europe, Asia, Africa, North America, and, by introduction, Australia, making it the most geographically widespread wild carnivore species (Lariviere and Pasitschniak-Arts 1996; Schipper et al. 2008). The species has also successfully adapted to the urban environments of many major cities throughout Europe and North America (Harris and Rayner 1986; Harris and Trewhella 1988; Gloor et al. 2001). The North American red fox was successfully domesticated for fur farming (Ashbrook 1937; Westwood 1989), and foxes from this breeding stock (Statham et al. 2011) were even further selected for behavior in an experiment in Novosibirsk, Russia that produced foxes with tame, dog-like behavior (Trut et al. 2009). The status of the North American red fox has been subject to debate for many years, with early studies characterizing between 1 and 10 North American species distinct from the northern European V. vulpes (e.g., Merriam 1900); however, in the 20th century, the taxonomy shifted to identify the predominant North American fox population as the subspecies V. v. fulva, along with additional subspecies existing in relict populations (e.g., Churcher 1959). To further complicate the distinction between the populations, the North American red fox has at times been considered invasive outside of relict populations such as those in the Sacramento Valley and Intermountain West due to historical accounts of introduction of European foxes by colonists (Kamler and Ballard 2002; Frey 2013). Thus the red fox has a complex population history in North America.
Recent work has resolved some of the questions surrounding the origins of the red fox in North America. Studies of mitochondrial DNA (mtDNA) that corroborate the fossil record indicated that North America is dominated by 2 clades of red foxes that emerged while occupying distinct refugia during the Pleistocene: a Holarctic clade in Alaska and Western Canada that includes haplotypes shared with Eurasia, and a Nearctic clade specific to North America (Aubry et al. 2009). More recent nuclear genetic analysis further indicates that Afro-Eurasian and North American red foxes form 2 distinct lineages that are at least as divergent from one another as are some sister species, such as the American marten (Martes americana) and Pacific marten (Martes caurina) or the kit fox (Vulpes macrotis) and swift fox (Vulpes velox) (Statham et al. 2014). Today the North American red fox’s range covers most of the continent, including regions outside of its indigenous range (Lewis et al. 1999; Aubry et al. 2009; Statham et al. 2012; Frey 2013). Conservation efforts targeted at indigenous populations of foxes, such as the endangered Sierra Nevada red fox (V. vulpes necator) and the Sacramento Valley red fox (V. vulpes patwin), have drawn support from mtDNA and autosomal microsatellite studies that validate the unique genetic composition of these populations (Perrine et al. 2007; Sacks et al. 2010; Sacks et al. 2011). Today, nonnative red foxes from a variety of sources also occur in low-elevation, human-dominated landscapes in the West, Midwest, and eastern seaboard of the United States (Perrine et al. 2007; Aubry et al. 2009; Sacks et al. 2010; Statham et al. 2012; Frey 2013).
The nonnative genetic origins of North American red fox populations have been attributed to 2 potential sources: deliberate introduction of red foxes from Great Britain during the English colonial era, and the escape or release of farm-bred strains of V. vulpes during the past century (Lewis et al. 1999; Kamler and Ballard 2002; Frey 2013). Although mtDNA studies sampling broadly across the North American continent initially found no evidence supporting European introgression, a recent study based in the mid-Atlantic region of the United States found that 17% of the red foxes they sampled carried European mtDNA haplotypes (Kasprowicz et al. 2016). Mitochondrial DNA has also been used to confirm historical accounts that the stock of red fox fur farms in North America and in the experimental populations in Novosibirsk originated in North America, primarily from eastern Canada (Statham et al. 2011; Statham et al. 2012; Lounsberry et al. 2017). Fox farming, which was previously common throughout much of North America (Dearborn 1915; Laut 1921; Ashbrook 1937; Westwood 1989), is known to carry a risk of introduction into wild populations due to foxes escaping from farms and becoming feral (Lewis et al. 1999; Norén et al. 2005). Though no evidence for admixture from fur farms was found in wild Newfoundland populations (Lounsberry et al. 2017), analysis of red foxes from the mid-Atlantic region of the United States found that 13–48% carried mtDNA haplotypes associated with farms (Kasprowicz et al. 2016).
Examination of red fox patrilines would be expected to reveal different patterns of genetic diversity than those revealed through matrilines because male red foxes disperse more widely than females (Harris and Trewhella 1988). The development of tools for this analysis has been limited by a lack of known sequence from the red fox Y-chromosome, but the close phylogenetic relationship between dog and red fox (Wayne 1993) recently facilitated the development of 2 red fox Y-chromosome microsatellite markers (Statham et al. 2014) from 24 Kbp of dog Y-chromosome sequence (Natanaelsson et al. 2006). Analysis of these 2 markers in 84 globally distributed male red foxes revealed 16 haplotypes comprising 3 continent-specific clades, with all North American haplotypes clustering monophyletically (Statham et al. 2014). Genotyping of these markers in red foxes from the mid-Atlantic region supported the introgression of European red foxes into this region, although only 2% of foxes were found to carry European Y-haplotypes as opposed to the 17% carrying European mtDNA haplotypes (Kasprowicz et al. 2016). However, Y-chromosome haplotypes have not been characterized on fox farms, so rates of introgression from farms into the wild mid-Atlantic population could not be compared between male and female lineages.
The recent sequencing of the full genome of a male red fox (NCBI BioProject PRJNA378561; A.V. Kukekova et al. in review) presents an opportunity to expand the tools available for the assessment of Y-chromosome diversity in the red fox. The development of a wider panel of Y-chromosome-specific markers is expected to improve the resolution at which haplotypes can be identified, allow haplotype assignation even in cases where some markers fail to amplify, and reduce the likelihood that microsatellite back-mutations could result in homoplasy. To this end, 11 novel microsatellite markers were developed from microsatellites identified in red fox genomic scaffolds found to contain Y-chromosome sequence (NCBI BioProject PRJNA378561; A.V. Kukekova et al. in review). These markers were analyzed in conjunction with the 2 developed by Statham et al. (2014) to assess the potential for the expanded panel to elucidate the genetic origins of North American red foxes.
To test whether the markers were of sufficient resolution to identify specific populations, red foxes were sampled from 4 locations. The “Newfoundland” population was sampled from Newfoundland, Canada, which is located within the indigenous range of the North American red fox and has little to no introgression from farms (Lounsberry et al. 2017). Foxes from 2 potential sources of introgression were also sampled: the “Great Britain” samples were drawn from modern wild fox populations on the island of Great Britain, and the “Novosibirsk” samples were drawn from an experimental farm in Novosibirsk, Russia where foxes were bred for behavioral traits in a long-term experiment (Trut et al. 2009). The history of fox breeding and the genetic similarity between North American wild foxes and the farmed foxes in Novosibirsk indicate that the Novosibirsk population is derived from North American farm stock (Vahrameyev and Belyaev 1948; Statham et al. 2011). The “Maryland” samples were collected from sites in Maryland in the eastern United States, which is the region in which previous work has identified native, fur-farm, and introduced mitochondrial heritage (Kasprowicz et al. 2016). The newly developed markers could thus be tested by assigning haplotypes to the red foxes within each population and then comparing haplotypes across populations.
Methods
Sample Selection and DNA Extraction
Tissue samples included wild red foxes from the island of Great Britain (“Great Britain” samples, N = 9 males), including 6 previously genotyped at markers Y29 and Y30 by Statham et al. (2014). “Maryland” samples (N = 12 males) were wild red foxes collected by trappers in central Maryland and eastern West Virginia in the eastern United States. Males were identified at time of capture. “Newfoundland” samples (N = 8; 7 males and 1 female) were acquired from red fox trappers on the island of Newfoundland in eastern Canada, and sex was determined by Lounsberry et al. (2017). The “Novosibirsk” samples (N = 52; 48 males and 4 females) came from tame, aggressive, and conventional farm-bred populations of silver foxes (a coat color variant of the red fox) that have been bred for behavior at the Institute of Cytology and Genetics in Novosibirsk, Russia for decades (Trut et al. 2009) and are derived from eastern Canadian stock (Statham et al. 2011). Pedigrees were used to select males that would maximize the number of founding male lineages sampled from the Novosibirsk population. Additionally, samples from 4 other canid species that could serve as outgroups were analyzed (Supplementary Note) and included an Arctic Fox (Vulpes lagopus), a red wolf (Canis rufus), gray wolves (Canis lupus), and dogs (Canis lupus familiaris). DNA was extracted from the Maryland red foxes using a QuickGene DNA Tissue Kit (Fujifilm, Tokyo) and from the Great Britain, Newfoundland, and Novosibirsk foxes as well as the outgroup samples using a DNeasy Blood and Tissue kit (Qiagen, Valencia, CA).
Microsatellite Identification
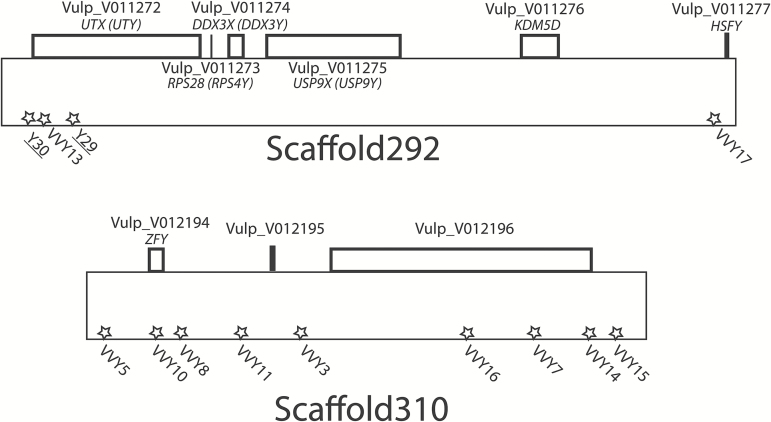
Scaffolds 292 and 310 (Figure 1) are the 2 scaffolds from the red fox genome identified as syntenic to the dog Y-chromosome (NCBI BioProject PRJNA37856; A.V. Kukekova et al. in review) based on similarity to the dog Y-chromosome sequence (Li et al. 2013). Together, they contain over 1 Mb of sequence. The scaffolds were soft-masked with RepeatMasker 4.0 (Smit et al. 2013) with the species parameter set to dog and scanned with an in-house Python script for 2–5 bp motifs of soft-masked sequence that were repeated at least 10 times. The motif, microsatellite sequence, and 250 bp of flanking sequence on either side of the 194 microsatellites detected were then examined, and candidate microsatellites were prioritized for screening based on the proportion of flanking sequence that was unmasked (i.e., likely to be non-repetitive).
Figure 1.
Relative positions of microsatellite markers on red fox Y-chromosome scaffolds. Stars indicate positions of microsatellite markers, and the names of the markers previously developed by Statham et al. (2014) are underlined. “Vulp_V” names correspond to the draft annotation (A.V. Kukekova et al. in review). For fox genes that were identified during annotation based on homology to human transcripts (A.V. Kukekova et al. in review), putative gene names are listed in italics; in cases where the predicted gene name corresponds to a paralog of a gene found on carnivore Y-chromosomes (Li et al. 2013), the most likely Y-chromosome gene is also identified in parentheses.
Primer Development
Primers were designed for 23 of the identified microsatellite loci using Primer3 (Untergasser et al. 2012). Primers were first tested in 2 male and 2 female Novosibirsk foxes with known sexes to confirm male specificity. PCRs were conducted at 25 µL with concentrations of 0.5 pmol of each primer and 12.5 μL of GoTAQ master mix (Promega, Madison, WI). The reactions were run in a thermal cycler under the following conditions: denaturation at 96 °C for 2 min followed by 30 cycles comprised by 20 s at 96 °C, 20 s at 58 °C, and 20 s at 72 °C, followed by a final extension step of 5 min at 72 °C. PCRs were analyzed on 1.8% agarose gels to score presence or absence of amplification. Primers were considered Y-chromosome-specific if they produced bands in only male foxes. Next, primers were tested in 2 Novosibirsk and 2 Newfoundland male foxes using the same PCR conditions as sex testing. The PCR products were then visualized on an 8% acrylamide gel. If the genotypes across all 4 foxes were monomorphic, the Novosibirsk foxes were then compared to 2 Great Britain males.
The 15 polymorphic microsatellites identified were then tagged with fluorescence by adding 18-bp M13 (sequence: TGTAAAACGACGGCCAGT) tails to the 5′ end of the forward primers (Schuelke 2000). PCR conditions were optimized for each primer set (Supplementary Note). Marker VVY10 was also tested with a VIC fluorescent tag directly attached to the forward primer due to interference from M13.
Inclusion of Markers From the Literature
Two markers for the red fox Y-chromosome were previously developed (Statham et al. 2014). One of the primers described, Y30_DogR, was developed for Y analysis in dogs (Natanaelsson et al. 2006) but was also found to be appropriate for analysis of the red fox (Statham et al. 2014). However, comparison of the sequence of this primer against the red fox genome revealed a single nucleotide transversion (G->C) at the seventh nucleotide of the reverse primer sequence (scaffold292: 33,966), and therefore an updated version of the primer specific to the red fox was used instead (Y30_FoxR: AGAGAGCTAAGGCATAGTTTG).
Testing Markers in a Panel
Thirteen Y-microsatellite markers (Table 1), 11 of which are novel, were multiplexed (Supplementary Table 1) and amplified (Supplementary Note) on a 96-well plate that included 76 male red foxes from the 4 populations, 5 female red fox negative controls, 1 male Arctic fox sample (Vulpes lagopus), and 8 male canids from 3 other species: 4 male dogs (Canis lupus familiaris), 3 male gray wolves (Canis lupus), and a male red wolf (Canis rufus) (Supplementary Note). The remaining 6 wells contained no-template negative controls placed throughout the plate. Fragment analysis was conducted on an ABI3730 Genetic Analyzer (PE Biosystems, Foster City, CA) using a LIZ500 size standard at the Keck Center at the University of Illinois at Urbana-Champaign. The fluorescent peaks were visualized and analyzed manually using Gene Mapper 3.5 (PE Biosystems). For each marker, the peak height in the negative controls and female red foxes was identified to determine the minimum threshold for genotype calls. Haplotypes were called based on the genotypes recorded.
Table 1.
Panel of 13 markers for the red fox Y-chromosome
| Marker | Position of microsatellite | Primer name | Primer sequence | Motif (repeats in reference) | Range of sizes observed, bp |
|---|---|---|---|---|---|
| VVY3 | scaffold310: 191,046–191,116 | Ba1F | ACCTGGGTATTTGGATCTTGGA | (aaaat)14 | 413–438 |
| Ba1R | ACTGACATTAACATTGTGTGCCA | ||||
| VVY5 | scaffold310: 16,171–16,221 | Da1F | AGCCTAACACTGAAGCATACCCA | (ac)25 | 218–232 |
| Db1R | TTTTCCTGTCGTTCCAAACCAA | ||||
| VVY7 | scaffold310: 399,903–399,951 | Fb1F | AGCAAGAATCTGCCACCAAATGA | (ttat)12 | 258–282 |
| Fb1R | CCCATACTCAGACAAGCTAATCTC | ||||
| VVY8 | scaffold310: 83,947–83,969 | Gb1F | GGCTTTGCATGTGTCTGCCATT | (ca)11 | 118–121 |
| Ga1R | GCAGGAATGTTTCAGTGTTTGGC | ||||
| VVY10 | scaffold310: 62,137–62,161 | Ib1F | ATCTTAAATGTCCTGTTACCACGT | (ag)12 | 298–314† |
| Ia1R | ATTTCTACCCAAGCCGCCTTCT | ||||
| VVY11 | scaffold310: 137,858–137,898 | J1F | AAACCAAACAACTAGGGAGCCC | (aaac)10 | 300–316 |
| J1R | TCCAGGGATTACTTACTGTTTTCC | ||||
| VVY13 | scaffold292: 37,829–37,849 | L1F | TGCCTTGATTCCTTTTTCCTTC | (ag)10 | 418–426 |
| L1R | TGGACAGAAAAGCCAGTGTAAC | ||||
| VVY14 | scaffold310: 448,786–448,808 | M1F | AGCAGACCATTGGCTATTTTCC | (ac)11 | 150–154 |
| M1R | GAAAGCTCCAAGCCATACTTCC | ||||
| VVY15 | scaffold310: 472,787–472,833 | N1F | AGTAAATGTCCGCTGGGGATAA | (ca)23 | 171–188 |
| N1R | TGTGAGCTAAACACCGCCTAAA | ||||
| VVY16 | scaffold310: 339,297–339,325 | O2F | TTGGGTGTTTTGTTTGAAAGTCT | (gt)14 | 213–243 |
| O2R | CTGCCAAAGAAGTGGTCCAAC | ||||
| VVY17 | scaffold292: 637,949–637,977 | P1F | GAAGACCCGGGCCTGAAGATC | (ag)14 | 241–255 |
| P1R | TCAACCATTTCTGCATGTTAGCCA | ||||
| Y29 | scaffold292: 63,770–63,808 | Y29-FoxF2* | AGTGCTTAGGCTCAGGATGC | (ag)19 | 189–195 |
| Y29-FoxR1* | TCCAGGTTTTATTTAGGGTCTT | ||||
| Y30 | scaffold292: 33,705–33,727 | Y30-FoxF2* | TCCTTTCCATTTTCAGAAAGC | (ga)11 | 399–423 |
| Y30-FoxR** | AGAGAGCTAAGGCATAGTTTG |
Number of repeats is based on the genome assembly of the reference individual. Sizes provided are the range of genotypes observed in red foxes in the present analysis. Primers marked with a single asterisk are those developed by Statham et al. (2014); the primer marked with a double asterisk is an adapted version of a dog-based primer used by Statham et al. (2014) that has been modified to match the sequence in the fox genome. The M13 tag adds 18 bp to the observed sizes of all markers except VVY10, marked with a †.
Diversity Analysis
Haplotypes were called manually based on all loci successfully genotyped for an individual. To identify similarity among haplotypes, genotypes were re-coded for Network 5.0.0.1 (Fluxus Technology Ltd., Clare, Suffolk, UK, http://www.fluxus-engineering.com), using a number of repeats estimated as: (g − 18 − f + 1)/m, where g is the length observed, f is the length of the flanking sequencing according to the reference genome, 18 is used to adjust the observed genotype length to account for the addition of M13, and m is the length of the motif, rounded to the nearest whole number. If 2 different genotypes rounded to the same number of repeats, the values used were pushed up or down to avoid conflict between distinct genotypes.
A haplotype network was drawn for the red foxes using star contraction (Forster et al. 1998), a median-joining tree (Bandelt et al. 1999) rooted in the Arctic Fox haplotype, and MP-contraction (Polzin and Vahdati Daneshmand 2003). Markers were weighted as (1/n)*10 rounded to the nearest whole number (Sacks et al. 2013), with n being the number of variants observed in V. vulpes. Haplotypes were also recalled using only the genotypes at Y29 and Y30 and were compared against the haplotype calls made with the full panel. Alleles were standardized with the genotypes reported for Y29 and Y30 by Statham et al. (2014) by comparing animals that were genotyped in both studies and adjusting genotype calls based on any differences in the size observed across technical replicates.
Population diversity was estimated using 2 metrics: the effective number of haplotypes and the haplotype richness. Estimating the effective number of haplotypes minimizes the effect of rare haplotypes by identifying the number of haplotypes at equal frequency that would be required to produce an equivalent level of diversity. It is estimated using the same formula as effective number of alleles (Ewens 1964):
where i is the number of haplotypes identified and pi is the frequency of the ith haplotype in the population. Haplotype richness is an estimator that facilitates cross-population comparisons of diversity by correcting the number of haplotypes observed in each population to that which would be expected if an identical number of individuals had been sampled from every population, correcting diversity to match all sample sizes to the population where the smallest number of individuals was sampled (Kalinowski 2004).
Results
Markers Developed
Scanning the Y-chromosome scaffolds identified 194 putative loci (Supplementary Table 2). Primers were designed and tested for 23 of these loci, and all primers amplified in the 2 males but not in the 2 females. After the monoallelic loci and those that failed during PCR with M13 extensions were eliminated, 11 novel microsatellite markers remained (Table 1; Figure 1). The 2 microsatellites developed by Statham et al. (2014), whose positions had also been flagged during the microsatellite scan, were included in the panel (Table 1; Figure 1).
At 2 loci, VVY5 and VVY10, some red foxes carried multiple, independent genotypes (Supplementary Table 3). Mapping the primers against the full draft red fox genome with the NCBI’s BLASTn program did not identify any predicted secondary hits, and the fact that these markers show in situ amplification only in males suggests that any secondary or tertiary amplifications are specific to the Y-chromosome. These results suggest potential segmental duplications of sequence in recent red fox Y-chromosome evolution.
Haplotype Identification and Diversity
Between 2 and 9 variants were identified at each locus among the 76 red fox males. For markers VVY5 and VVY10, the multiple peaks were broken up into size categories and coded as 3 distinct sites (Supplementary Note; Supplementary Table 3). The Arctic Fox was genotyped at all loci except VVY3, VVY10, and VVY11 (Supplementary Table 3).
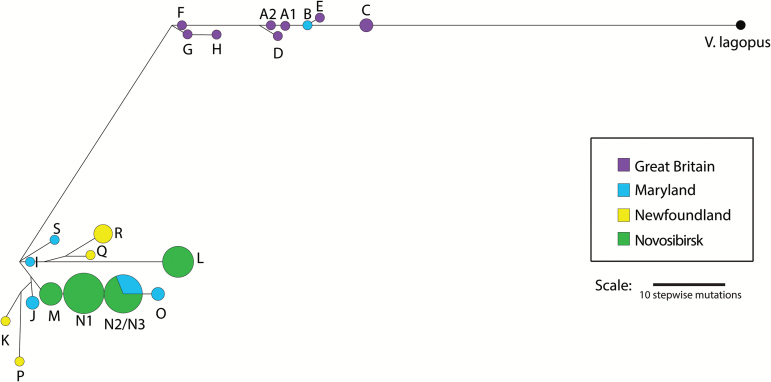
Among the red foxes, 22 haplotypes were identified (Supplementary Table 3). All haplotypes except one were population-specific (Table 2), the exception being haplotype N2, which was found in the Novosibirsk and Maryland populations. Analysis with Network revealed that the haplotypes clustered largely by continent (Figure 2), with the Great Britain samples separating out from all other populations except a single Maryland red fox.
Table 2.
Frequencies of each haplotype among the populations
| Haplotype | Great Britain | Maryland | Newfoundland | Novosibirsk | Total |
|---|---|---|---|---|---|
| A1 | 1 (11%) | 1 | |||
| A2 | 1 (11%) | 1 | |||
| B | 1 (8%) | 1 | |||
| C | 2 (22%) | 2 | |||
| D | 1 (11%) | 1 | |||
| E | 1 (11%) | 1 | |||
| F | 1 (11%) | 1 | |||
| G | 1 (11%) | 1 | |||
| H | 1 (11%) | 1 | |||
| I | 1 (8%) | 1 | |||
| J | 2 (17%) | 2 | |||
| K | 1 (14%) | 1 | |||
| L | 11 (23%) | 11 | |||
| M | 6 (13%) | 6 | |||
| N1 | 19 (40%) | 19 | |||
| N2 | 5 (42%) | 11 (23%) | 16 | ||
| N3 | 1 (1%) | 1 | |||
| O | 2 (17%) | 2 | |||
| P | 1 (14%) | 1 | |||
| Q | 1 (14%) | 1 | |||
| R | 4 (38%) | 4 | |||
| S | 1 (8%) | 1 | |||
| Total | 9 | 12 | 7 | 48 | 76 |
Percentages represent within-population frequencies and are approximate.
Figure 2.
Haplotype network for the 22 haplotypes identified in the red fox. The network is rooted in the Arctic Fox (V. lagopus). Nodes are scaled to be proportional to haplotype frequency. The distance between nodes is proportional to the number of mutational steps separating them. The “Great Britain” population is purple, “Maryland” is blue, “Newfoundland” is yellow, and “Novosibirsk” is green. Haplotypes N2 (found in 9 Novosibirsk foxes and 5 Maryland red foxes) and N3 (found in 1 Novosibirsk fox) were merged into a single node by Network.
Calling these haplotypes with only Y29 and Y30, which are the markers developed by Statham et al. (2014), yielded a total of 10 haplotypes (Supplementary Table 4). Comparing the haplotypes called with only Y29 and Y30 to the haplotype network constructed with the full panel revealed that the Y29/Y30 haplotypes did not always map to adjacent nodes in the network. Specifically, the haplotype defined as 172/397 using the Y29/Y30 nomenclature (Statham et al. 2014) is found in red foxes assigned haplotype C with the full 13-marker panel. Though C is the most diverged Great Britain haplotype, the same Y29/Y30 haplotype would also be assigned to the red foxes with haplotypes F, G, and H, which form their own cluster within the Great Britain samples. Similarly, Y29/Y30 haplotype 172/387 is found in L (a Novosibirsk haplotype) and P (a Newfoundland haplotype) that are not adjacent using the 13-marker panel. Thus the higher resolution of the new panel may reduce the influence of homoplasy in assigning haplotypes compared to Y29 and Y30 alone.
Both estimates of haplotype diversity calculated, namely the effective number of haplotypes and the haplotype richness, clearly reflected the higher resolution achieved with the full 13-marker panel compared to the original lower-resolution marker set (Supplementary Table 5). Of the wild populations, the Great Britain sample scored highest on both metrics, and all wild populations were found to be more diverse than the Novosibirsk population across both measures. This was not the case with the alternative, lower-resolution marker set, which produced very similar estimates of the effective numbers of haplotypes in the Novosibirsk and Maryland populations. In fact, using the higher-resolution panel increased the effective number of haplotypes from less than 2 haplotypes in all populations except Great Britain to more than 4 in all of the wild populations, with 6 effective haplotypes in the Great Britain population. Additionally, comparing the estimates of haplotype richness suggests that sampling even as few as 7 wild red foxes with the new marker set would be expected to reveal between 4 and 6 haplotypes, depending on the population, compared to 2–4 haplotypes with the lower-resolution set.
Discussion
Until now, population genetic studies of the red fox Y-chromosome have been limited to 2 markers, but use of the red fox Y-chromosome genomic sequence to identify 11 additional microsatellite markers has significantly increased the resolution at which diversity in red fox populations can be resolved. Among the 76 male red foxes analyzed here, the 2 previously developed markers, Y29 and Y30, would have identified 10 haplotypes, whereas the expanded panel identified 22 haplotypes. Within the 2 major clusters revealed in the haplotype network, one contained all of the Great Britain samples and the other contained only North American samples, supporting the idea, as presented by Statham et al. (2014), that the major clades are continent-specific.
The organization of the Maryland population with respect to the others offered additional support for a heterogeneous ancestry of red foxes in this region. One Maryland sample clustered with the red foxes from Great Britain; this finding was consistent with Kasprowicz et al. (2016), where analysis of Y-haplotypes supported historical accounts of English colonists releasing red foxes from Europe in what is now the eastern United States. The rate of discovery of European Y-haplotypes in Maryland red foxes was similar across the 2 studies, with 3 European Y-haplotypes discovered by Kasprowicz et al. (2016) using Y29 and Y30 in a sample of 135 foxes, and 1 Great Britain haplotype identified in the 12 red foxes from Maryland analyzed here. Additionally, a shared haplotype among the Maryland foxes and the Novosibirsk foxes suggested a high rate of introgression from feral foxes (farm-escaped foxes and their descendants): the common Novosibirsk haplotype N2 was the only one shared among any populations, and was found in 5 of the 12 wild Maryland foxes. For comparison, no haplotypes were shared between the Novosibirsk and Newfoundland populations, even though eastern Canada is thought to be the source of the Novosibirsk population (Statham et al. 2011). Feral mtDNA haplotypes have been found to be more common in human-altered landscapes (Statham et al. 2012; Kasprowicz et al. 2016; Sacks et al. 2016), with between 13% and 48% of foxes sampled in Maryland carrying feral mtDNA haplotypes (Kasprowicz et al. 2016). Future analyses of both Y-chromosome and mtDNA haplotypes in this heterogeneous population can compare the prevalences of European and feral fox lineages.
The development of robust Y-chromosome resources for the red fox thus provides a useful complement to the available mitochondrial tools. Microsatellites offer a cost-effective alternative to genome-wide SNP analyses that render them a preferred method for monitoring wildlife populations (DeYoung and Honeycutt 2005), and Y-microsatellites are especially needed given that little is known about nucleotide-level diversity on the fox Y-chromosome (A.V. Kukekova et al. in review). Nuclear microsatellites have been used in conservation to demonstrate the divergence of relict fox populations, such as the ones in the Greater Yellowstone Ecosystem (Swanson et al. 2005) and the Sacramento Valley (Sacks et al. 2011); adding Y-microsatellites to the tools available for these analyses could further empower studies to identify populations of interest. Because sex can influence behavior, diversity estimates in many species vary depending on whether the male or female lineage is analyzed. For example, unidirectional gene flow from polar bears into brown bear populations has created a system where an individual with a polar bear mtDNA haplotype may actually have less than 10% polar bear ancestry and carry a genome inherited almost entirely from brown bears (Cahill et al. 2015). Similarly, patterns of elephant mtDNA haplotypes within Africa were found to segregate differently than overall relatedness as determined by nuclear and sex chromosome markers (Ishida et al. 2011). In the red fox, sex is known to influence patterns of dispersal (Harris and Trewhella 1988), and sex-specific effects have been observed to influence patterns of genetic diversity (e.g., Sacks et al. 2016). Kasprowicz et al. (2016) also found differences in the frequencies of European mitochondrial and Y-chromosome haplotypes in mid-Atlantic red fox populations, suggesting that mitochondrial and Y-chromosome diversity must be analyzed together to capture a complete picture of modern red fox populations.
The genomic coming-of-age of the red fox provided an opportunity to develop 11 novel, robust Y-specific markers that help to elucidate Y-chromosome diversity at a higher resolution than was previously possible. Future analyses should seek to characterize the high-resolution haplotypes of additional populations, such as western North American red fox populations in the Rocky Mountains and Alaska, and in more foxes within each population, like in the large sample sizes of the analyses that used the Y29 and Y30 markers (Statham et al. 2014; Kasprowicz et al. 2016). Analysis of red foxes from a variety of populations will expand knowledge of red fox Y-chromosomal variation and the distribution of red fox diversity across North America. In particular, previous studies have reported higher frequencies of feral mitochondrial haplotypes in urbanized areas than in rural areas (Sacks et al. 2010; Kasprowicz et al. 2016; Lounsberry et al. 2017); analysis of male red fox lineages can identify whether sex-specific trends shape how wild North American populations respond to anthropogenic changes to the environment, therefore furthering our understanding of how humans have altered and continue to alter the genetic diversity of North American red fox populations. The increased resolution provided by the new panel of markers will allow for finer-scale ascertainment of population differentiation and within-population diversity in studies of males, which will in turn illuminate the complex population history of the red fox.
Supplementary Material
Supplementary data is available at Journal of Heredity online.
Funding
National Institutes of Health (grant number R01GM120782); the University of Illinois at Urbana-Champaign Campus Research Board; the United States Department of Agriculture Federal Hatch Project (grant number 538922); Project of the Institute of Cytology and Genetics of the Siberian Branch of the Russian Academy of Sciences (grant number 0324-2016-0002); the Department of Defense (DoD) National Defense Science & Engineering Graduate Fellowship (NDSEG) Program (H.M.R.); and the National Institutes of Health/National Institute on Alcohol Abuse and Alcoholism Intramural Program (to C.A.D. and C.S.B.).
Data Availability
The raw data have been made available in accordance with the policies of the Journal of Heredity (Baker, 2013). The genotype data used in the present analysis is available at Dryad: doi10.5061/dryad.7k749.
Supplementary Material
Acknowledgments
We thank N. Won, Q. Zhao, M. Wang, Z. Lounsberry, and M. Statham for their experimental work; A. Roca, and our reviewers for providing comments on an earlier version of the manuscript; and Anastasiya V. Kharlamova, Rimma G. Gulevich, Anastasiya V. Vladimirova, Irina V. Pivovarova, and Tatyana I. Semenova for their assistance in collecting fox samples at the experimental farm in Novosibirsk. We are grateful to the Illinois State Museum in Springfield, IL, the Rosamond Gifford Zoo in Syracuse, NY, and the Wolf Park in Battle Ground, IN for donating the tissue specimens used in the outgroup analysis. We would also like to thank the trappers in Maryland and Newfoundland who contributed samples, as well as Jessica and Jack Hekman.
References
- Ashbrook FG. 1937. The breeding of fur animals. Yearbook of Agriculture No. 1603. Washington (DC): U.S. Department of Agriculture; pp. 1379–1395. [Google Scholar]
- Aubry KB, Statham MJ, Sacks BN, Perrine JD, Wisely SM. 2009. Phylogeography of the North American red fox: vicariance in Pleistocene forest refugia. Mol Ecol. 18:2668–2686. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Röhl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 16:37–48. [DOI] [PubMed] [Google Scholar]
- Cahill JA, Stirling I, Kistler L, Salamzade R, Ersmark E, Fulton TL, Stiller M, Green RE, Shapiro B. 2015. Genomic evidence of geographically widespread effect of gene flow from polar bears into brown bears. Mol Ecol. 24:1205–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churcher CS. 1959. The specific status of the new world red fox. J Mammal. 40:513–520. [Google Scholar]
- Dearborn N. 1915. Silver fox farming in eastern North America, Vol. 301. Washington (DC): U.S. Department of Agriculture. [Google Scholar]
- DeYoung R. W, Honeycutt RL. 2005. The molecular toolbox: genetic techniques in wildlife ecology and management. J Wildl Manage. 69:1362–1384. [Google Scholar]
- Ewens WJ. 1964. The maintenance of alleles by mutation. Genetics. 50:891–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster P, Toth A, Bandelt H. 1998 Evolutionary network analysis of word lists: Visualising the relationships between alpine romance languages. J Quant Linguist. 5:174–187. [Google Scholar]
- Frey JK. 2013. Re-evaluation of the evidence for the importation of red foxes from Europe to colonial America: Origins of the southeastern red fox (Vulpes vulpes fulva). Biol Cons. 158:74–79. [Google Scholar]
- Gloor S, Bontadina F, Hegglin D, Deplazes P, Breitenmoser U. 2001. The rise of urban fox populations in Switzerland. Mamm Biol. 66:155–164. [Google Scholar]
- Harris S, Rayner JMV. 1986. Urban fox (Vulpes vulpes) population estimates and habitat requirements in several British cities. J Anim Ecol. 55:575–591. [Google Scholar]
- Harris S, Trewhella WJ. 1988. An analysis of some of the factors affecting dispersal in an urban fox (Vulpes vulpes) population. J Appl Ecol. 25:409–422. [Google Scholar]
- Ishida Y, Oleksyk TK, Georgiadis NJ, David VA, Zhao K, Stephens RM, Kolokotronis SO, Roca AL. 2011. Reconciling apparent conflicts between mitochondrial and nuclear phylogenies in African elephants. PLoS One. 6:e20642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowski ST. 2004. Counting alleles with rarefaction: Private alleles and hierarchical sampling designs. Conserv Genet. 5:539–543. [Google Scholar]
- Kamler JF, Ballard WB. 2002. A review of native and nonnative red foxes in North America. Wildl Soc Bull (1973–2006). 30:370–379. [Google Scholar]
- Kasprowicz AE, Statham MJ, Sacks BN. 2016. Fate of the other redcoat: remnants of colonial British foxes in the Eastern United States. J Mammal. 97:298–309. [Google Scholar]
- Lariviere S, Pasitschniak-Arts M. 1996. Vulpes vulpes. Mamm Species. 537: 1–11. [Google Scholar]
- Laut AC. 1921. The fur trade of America. New York: Macmillan. [Google Scholar]
- Lewis JC, Sallee KL, Golightly RT., Jr 1999. Introduction and range expansion of nonnative red foxes (Vulpes vulpes) in California. Midland Naturalist. 142:372–381. [Google Scholar]
- Li G, Davis BW, Raudsepp T, Wilkerson AJP, Mason VC, Ferguson-Smith M, O’Brien PC, Waters PD, Murphy WJ. 2013. Comparative analysis of mammalian Y chromosomes illuminates ancestral structure and lineage-specific evolution. Genome Res. 23:1486–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lounsberry ZT, Quinn CB, Statham MJ, Angulo CL, Kalani TJ, Tiller E, Sacks BN. 2017. Investigating genetic introgression from farmed red foxes into the wild population in Newfoundland, Canada. Conserv Genet. 18:383–392. [Google Scholar]
- Merriam CH. 1900. Preliminary revision of the North American red foxes. Proc Wash Acad Sci. 2:661–676. [Google Scholar]
- Natanaelsson C, Oskarsson MC, Angleby H, Lundeberg J, Kirkness E, Savolainen P. 2006. Dog Y chromosomal DNA sequence: identification, sequencing and SNP discovery. BMC Genet. 7:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norén K, Dalén L, Kvaløy K, Angerbjörn A. 2005. Detection of farm fox and hybrid genotypes among wild arctic foxes in Scandinavia. Conserv Genet. 6:885–894. [Google Scholar]
- Perrine JD, Pollinger JP, Sacks BN, Barrett RH, Wayne RK. 2007. Genetic evidence for the persistence of the critically endangered Sierra Nevada red fox in California. Conserv Genet. 8:1083–1095. [Google Scholar]
- Polzin T, Vahdati Daneshmand S. 2003. On Steiner trees and minimum spanning trees in hypergraphs. Oper Res Lett. 31:12–20. [Google Scholar]
- Sacks BN, Brazeal JL, Lewis JC. 2016. Landscape genetics of the nonnative red fox of California. Ecol Evol. 6:4775–4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacks BN, Brown SK, Stephens D, Pedersen NC, Wu J, Te Berry O. 2013. Y chromosome analysis of dingoes and Southeast Asian village dogs suggests a neolithic continental expansion from Southeast Asia followed by multiple Austronesian dispersals. Mol Biol Evol. 30:1103–1118. [DOI] [PubMed] [Google Scholar]
- Sacks BN, Moore M, Statham MJ, Wittmer HU. 2011. A restricted hybrid zone between native and introduced red fox (Vulpes vulpes) populations suggests reproductive barriers and competitive exclusion. Mol Ecol. 20:326–341. [DOI] [PubMed] [Google Scholar]
- Sacks BN, Statham MJ, Perrine JD, Wisely SM, Aubry KB. 2010. North American montane red foxes: expansion, fragmentation, and the origin of the Sacramento Valley red fox. Conserv Genet. 11:1523–1539. [Google Scholar]
- Schipper J, Chanson JS, Chiozza F, Cox NA, Hoffmann M, Katariya V, Lamoreux J, Rodrigues AS, Stuart SN, Temple HJ, et al. 2008. The status of the world’s land and marine mammals: diversity, threat and knowledge. Science. 322:225–230. [DOI] [PubMed] [Google Scholar]
- Schuelke M. 2000. An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol. 18:233–234. [DOI] [PubMed] [Google Scholar]
- Smit A, Hubley R, Green P. 2013. RepeatMasker Open-4.0.
- Statham MJ, Murdoch J, Janecka J, Aubry KB, Edwards CJ, Soulsbury CD, Berry O, Wang Z, Harrison D, Pearch M, et al. 2014. Range-wide multilocus phylogeography of the red fox reveals ancient continental divergence, minimal genomic exchange and distinct demographic histories. Mol Ecol. 23:4813–4830. [DOI] [PubMed] [Google Scholar]
- Statham MJ, Sacks BN, Aubry KB, Perrine JD, Wisely SM. 2012. The origin of recently established red fox populations in the United States: Translocations or natural range expansions? J. Mammal. 93:52–65. [Google Scholar]
- Statham MJ, Trut LN, Sacks BN, Kharlamova AV, Oskina IN, Gulevich RG, Johnson JL, Temnykh SV, Acland GM, Kukekova AV. 2011. On the origin of a domesticated species: Identifying the parent population of Russian silver foxes (Vulpes vulpes). Biol J Linn Soc Lond. 103:168–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson BJ, Fuhrmann RT, Crabtree RL. 2005. Elevational isolation of red fox populations in the Greater Yellowstone Ecosystem. Conserv Genet. 6:123–131. [Google Scholar]
- Trut L, Oskina I, Kharlamova A. 2009. Animal evolution during domestication: the domesticated fox as a model. Bioessays. 31:349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. 2012. Primer3–new capabilities and interfaces. Nucleic Acids Res. 40:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahrameyev K, Belyaev DK. 1948. Guide for fox breeding (in Russian). Moscow (Russia): International Book. [Google Scholar]
- Wayne RK. 1993. Molecular evolution of the dog family. Trends Genet. 9:218–224. [DOI] [PubMed] [Google Scholar]
- Westwood RE. 1989. Early fur farming in Utah. Utah Hist Q. 57, 320–339. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data have been made available in accordance with the policies of the Journal of Heredity (Baker, 2013). The genotype data used in the present analysis is available at Dryad: doi10.5061/dryad.7k749.