Abstract
Circadian rhythms enable cells and organisms to coordinate their physiology with the cyclic environmental changes that come as a result of Earth's light/dark cycles. Cyanobacteria make use of a post-translational oscillator to maintain circadian rhythms, and this elegant system has become an important model for circadian timekeeping mechanisms. Composed of three proteins, the KaiABC system undergoes an oscillatory biochemical cycle that provides timing cues to achieve a 24-h molecular clock. Together with the input/output proteins SasA, CikA, and RpaA, these six gene products account for the timekeeping, entrainment, and output signaling functions in cyanobacterial circadian rhythms. This Minireview summarizes the current structural, functional and mechanistic insights into the cyanobacterial circadian clock.
Keywords: circadian rhythm, crystallography, nuclear magnetic resonance (NMR), bacterial protein kinase, ATPases associated with diverse cellular activities (AAA), bacterial signal transduction
Form and function of circadian clocks
Circadian rhythms are processes through which cells and organisms predict and adapt to the repetitive environmental changes incident to the 24-h solar day. These rhythms originate intracellularly and persist in the absence of external cues to orchestrate temporal organization of physiology (1). To achieve this, circadian clocks must perform three distinct but necessarily intertwined functions: timekeeping, entrainment, and output signaling.
Timekeeping is achieved through a cycle of slow biochemical processes that together set the period of oscillation close to 24 h. For this temporal information to be relevant, biological clocks must appropriately synchronize with their environment through entrainment. Temporal information runs from the timekeeping apparatus to the rest of the cell through output signaling mechanisms that impart circadian changes in physiology.
Timekeeping mechanisms that have been observed in circadian clocks are broadly categorized as transcription-translation feedback loops (TTFLs)2 or post-translational oscillators (PTOs) (2). TTFLs achieve oscillation via delayed negative feedback, where a gene product represses its own expression when its levels become high enough. This results in an oscillation in the expression level of the gene, with transitions between repression and derepression occurring at critical points in the oscillatory cycle. By contrast, PTOs utilize timekeeping mechanisms that are independent of transcription. Like TTFLs, PTOs involve a cycle of biochemical processes, but instead of being driven by changes in expression levels, their behavior is controlled by post-translational modifications, conformational changes, protein–protein interactions, and/or subcellular localization. It is important to note that although we make a distinction here between TTFLs and PTOs as general categories of negative feedback systems, many circadian clocks make use of both transcription/translation as well as post-translational timing steps to constitute a biochemical oscillator.
Post-translational steps in circadian clocks present unique biochemical challenges and opportunities because they link dynamic molecular processes to biological phenotypes through covalent and conformational changes in protein structure. PTOs can operate at constant protein concentrations and are therefore amenable to in vitro study. In the cyanobacterial Kai system, an entire PTO consisting of only three protein gene products (KaiA, KaiB, and KaiC or KaiABC) can reconstitute circadian biochemical oscillation in vitro (3). As a result, this system has become an important model for understanding how discrete biochemical steps are integrated to give rise to precise biological timing.
Circadian biology in cyanobacteria
Circadian rhythms are essentially ubiquitous in eukaryotes, but far fewer instances have been documented in prokaryotes (4). Cyanobacteria temporally segregate photosynthesis and nitrogen fixation, which require mutually exclusive oxidative environments (5). Evolutionary pressures such as this resulted in a cyanobacterial transcriptome in which at least one-third of transcripts undergo circadian variation in accumulation levels (6), and active transcription of nearly all-transcripts appears to be under circadian control (7). These changes in transcriptional output are linked to global changes in chromosome ultrastructure (8, 9), but they are controlled basally by a core biochemical oscillator (3, 10).
The KaiABC gene cluster was originally identified in genetic screens aimed at finding TTFLs involved in cyanobacterial circadian regulation, with deletion resulting in an arrhythmic phenotype (10). It was subsequently shown that the products of this gene cluster can recapitulate circadian biochemical oscillation in vitro (3) and that this circadian oscillation persists in vivo even when transcription and translation are inhibited (11). Its ability to be reconstituted in vitro demonstrates that KaiABC can act as a self-sustained PTO. In vivo, this PTO also functions within a TTFL framework, i.e. circadian variation of clock protein expression results in a more robust rhythm (4, 12–14). Together with three other effector proteins, the KaiABC oscillator comprises a self-contained minimal system that is capable of integrating timekeeping, entrainment, and output signaling processes into the cyanobacterial cell.
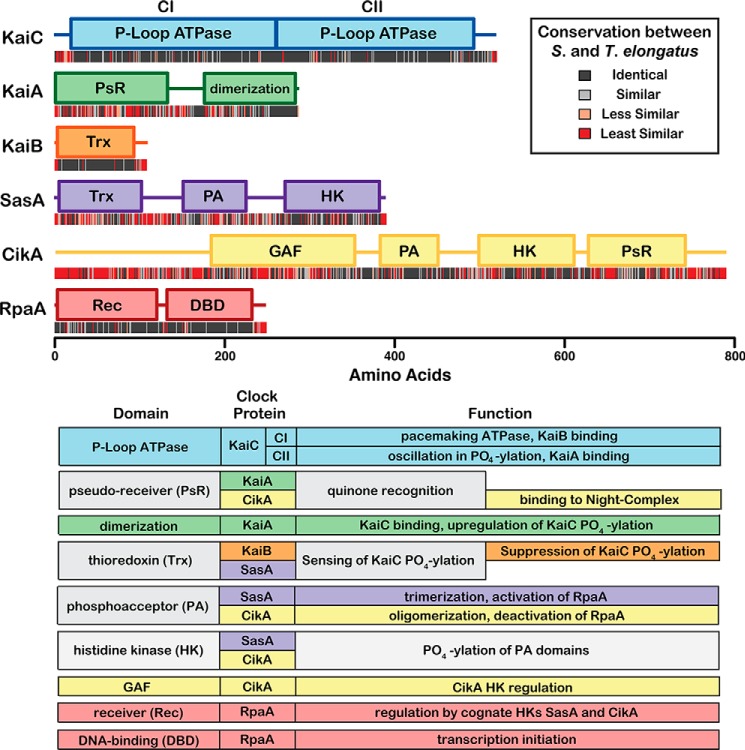
Domain structures of the six core clock proteins in cyanobacteria are shown in Fig. 1. Genetic studies on cyanobacterial rhythms have made extensive use of the mesophilic cyanobacterium Synechococcus elongatus, from which these genes were originally identified (10). Biophysical studies have often taken advantage of proteins produced from homologous gene sequences in the thermophilic variant Thermosynechococcus elongatus for their improved solubility and in vitro behavior (15). The six proteins shown in Fig. 1 are conserved between S. elongatus and T. elongatus and appear to comprise a core unit capable of accounting for all of the required functions of a circadian clock. Several additional genes have been identified that play roles in the circadian rhythms of cyanobacteria (16–21); however, this Minireview will focus on the six best-characterized gene products and the mechanisms through which they mediate timekeeping, entrainment, and output signaling functions in cyanobacterial circadian rhythms. For a broader review on circadian biology in cyanobacteria, see Refs. 4, 22.
Figure 1.
Domain structure, conservation, and function of core clock proteins. Domain structures of the six core clock proteins from S. elongatus are depicted as colored bars using boundaries and annotated functions from the Interpro Database (90). Sequences were aligned with homologues from the thermophilic variant T. elongatus using ClustalΩ (91). Identity scores at each position in the alignment are indicated below the domain maps as color-coded bars. Observed functions for each of the core clock protein domains are summarized below.
Structure and function of KaiC
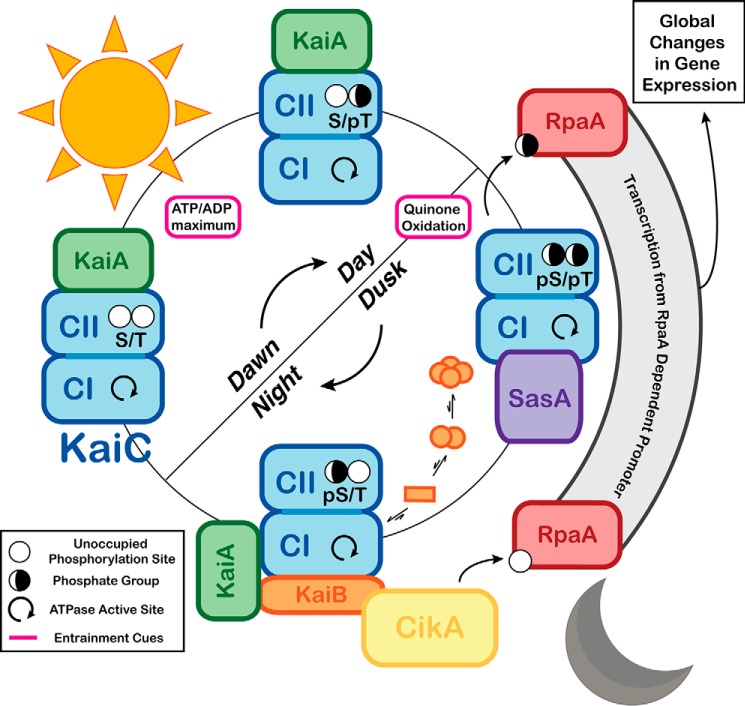
The core cyanobacterial clock is centered around the hexameric ATPase KaiC, which achieves timekeeping through circadian oscillations in phosphorylation that are intertwined with oscillations between the quaternary structures depicted in Fig. 2. The primary sequence of KaiC is a concatenation of two P-loop ATPase domains (10), termed CI and CII. In solution, KaiC binds ATP to form a hexamer through interactions at the CI subunit interfaces (23–25). Homologous ATPase active sites are present at the subunit interfaces of both CI and CII; however, these active sites catalyze distinct chemical transformations. CI transfers phosphate from ATP to water, and CII catalyzes transfer of phosphate between ATP and the side-chain hydroxyl groups of residues Ser-431 and Thr-432 (26, 27). Phosphorylation at these sites oscillates with a 24-h period in concert with interactions between KaiC and the other core clock proteins KaiA and KaiB (28, 29). These interactions are regulated by the phosphorylation state of KaiC and take place on non-homologous regions of the CI and CII domains, known as the respective B- and A-loops, where they in turn regulate the enzymatic activities on KaiC in a negative feedback loop.
Figure 2.
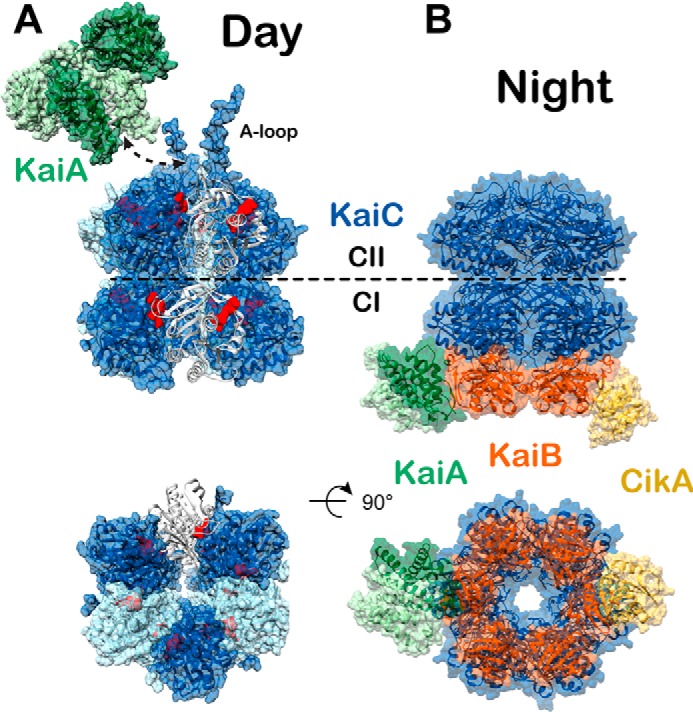
Day and night states of the cyanobacterial clock. High-resolution models of the day and nighttime states of the core oscillator are compared. Coloring scheme is the same as in Fig. 1. A, day complex. The CI and CII hexamers form stacked doughnuts (alternating subunits shown in light and dark blue for contrast, PDB code 3K0C). One subunit is shown in cartoon mode to highlight the nucleotide-binding sites (red) at the subunit interfaces. Phosphorylation of KaiC residues Thr-426 and Ser-432 takes place near the CII nucleotide-binding sites. The KaiA dimer (light and dark green for contrast) binds to C-terminal A-loops of KaiC (white, shown in complex with KaiA PDB code 5C5E). The dashed arrow represents the point of connection between the KaiA–CII loop structure and the CII A-loop extensions shown on the right. B, night complex. The intermediate-resolution cryo-EM model (PDB code 5N8Y) is combined with higher resolution models from studies on independent subcomplexes. The S431E phosphomimetic of the pS/T state of KaiC binds six molecules of KaiB (PDB code 5JWQ). This results in the recruitment and sequestration of KaiA near the KaiB–CI interface (PDB code 5JWR). Output signaling occurs through interactions between KaiB and the C-terminal PsR domain of CikA (PDB code 5JYV), as well as through interactions between KaiC and SasA at dusk, although no high-resolution structure exists for the latter.
During the day, KaiA is bound to the A-loops on CII where it promotes autophosphorylation (30–32). In the evening, KaiB binds to phosphorylated KaiC at the B-loops of the CI domain (33, 34), adding a third stacked toroid to the KaiC double-doughnut (Fig. 2) (35). At this point, KaiA is recruited to the KaiB–CI complex where it is inactivated and sequestered from CII throughout the night (33, 35). In the absence of KaiA, enzymatic equilibria at the CII-active sites favor the autodephosphorylation of Thr-432 and Ser-431 (31). Around dawn, CII phosphorylation is at a minimum, and affinity for KaiB is lost (30). The repressive complex is released, freeing KaiA to bind to the A-loops and begin the cycle anew.
Post-translational modification at residues Ser-431 and Thr-432 of KaiC occurs in a specific order, with Thr-432 acting as a substrate in the first reaction of both the phosphorylation and dephosphorylation arms of the cycle (29, 36). The order of the KaiC phosphorylation states is denoted in this Minireview using the shorthand: S/T → S/pT → pS/pT → pS/T. The S/T and S/pT states are dominant during the daytime phosphorylation arm of the oscillatory cycle, characterized by binding of KaiA at CII (26, 28). Accumulation of pS/pT initiates the nighttime state (28, 30, 35), characterized by the formation of the KaiABC complex, which persists through the pS/T state until phosphorylation reaches a minimum, and KaiA is recruited back to CII.
Sustained oscillation requires a power source, and KaiABC appears to achieve this through ATPase activity of the CI domain, whose enzymatic activity is correlated with a period both in vitro and in vivo (37, 38). CI ATPase activity oscillates as well, with the unphosphorylated forms of KaiC consuming ATP approximately twice as fast as the phosphorylated form (37). This enzymatic activity shows a relatively modest rate enhancement of ATP hydrolysis, with an apparent kcat/Km on the order of 10−4 m−1 s−1, about 6 orders of magnitude slower than the structurally similar F1-ATPase (38). High-resolution structures have been obtained for the isolated CI ring in the pre-ATP and post-ATP hydrolysis states (38) resulting in the three specific structural changes depicted in Fig. 3A. Together with studies showing that catalytically dead KaiC is unable to interact with KaiB (26), one emerging model is that CI ATPase functions in timing the recruitment of KaiB and resultant formation of the nighttime repressive complex, although no specific structural or biochemical information exists yet on how these two processes might be related.
Figure 3.
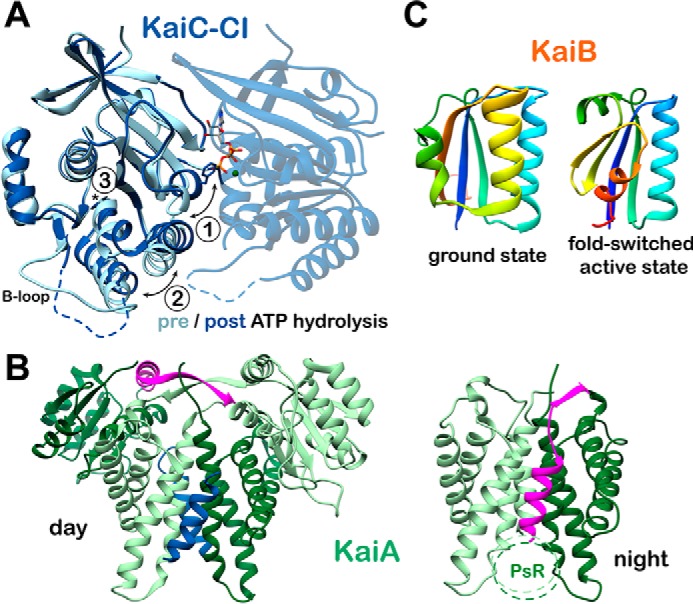
Conformational changes in the core oscillator. A, CI ATP hydrolysis. High-resolution crystal structures of the CI domain of KaiC in the ATP-bound (PDB code 4LTA) and ADP-bound (PDB code 4LT9, chain C) states are overlaid. Three major structural differences are noted upon ATP hydrolysis: 1) helix bearing Phe-199 is flipped into an alternative conformation; 2) α6-α7/α8 helices are repositioned away from the nucleotide-binding site; and 3) cis peptide bond between Asp-145 and Ser-146 in the ATP-bound state is found in the trans isomer in the ADP-bound state. B, KaiA inactivation. The daytime and nighttime states of KaiA are compared. During the day, the A-loops of KaiC (blue) are bound at the dimer interface, and the interdomain linker (pink) crosses over the complex. In the Night Complex, the linker shifts to occupy the A-loop binding site. C, KaiB fold-switching. KaiB undergoes a major structural reorganization between its free and KaiC-bound forms, involving changes in both secondary and tertiary structure for the C-terminal half of the protein. Both structures are represented with rainbow coloring, starting with dark blue at the N terminus. Note that the first half of the two proteins is identical (dark blue through light green), whereas the C-terminal halves are completely different. This interconversion is thought to happen spontaneously resulting in a conformational selection mechanism for formation of the KaiBC complex.
A direct structural connection between CII phosphorylation and the interactions of KaiC with other clock proteins has also proven difficult to pin down. High-resolution crystal structures exist of different phosphorylated and phosphomimetic states (39), but these structures are nearly identical. This is unexpected given the well-established functional differences between the phosphostates of KaiC. One possible explanation for this structural convergence is that crystallization of KaiC variants in these structures was performed in the presence of a non-hydrolyzable ATP analogue (24, 39), and structural changes associated with CII phosphorylation could be masked in the pre-ATP hydrolysis state. Alternatively, phosphorylation may influence dynamic structural equilibria that are not observable within a crystal lattice.
The latter idea is supported by lower-resolution studies on the overall shape and dynamic structural properties of KaiC. Time-resolved small-angle X-ray scattering data show an increase in the apparent radius of gyration in the pS/T phosphostate (40), providing evidence of a change in shape for KaiC in the night. Interactions between isolated CI and CII domains increase across the phosphorylation cycle with a minimum at the S/T state and a maximum in the pS/T state (41), and NMR line-shape analysis indicates a more rigid KaiC structure in these states as well. These structural changes, known collectively as “ring-ring stacking,” presumably affect the accessibility of the B-loops to regulate binding of thioredoxin domains.
One of the most surprising observations related to the enzymology of CII is that free radioactive ATP accumulates transiently in dephosphorylation reactions of 32P-labeled pS/pT KaiC (42), suggesting that dephosphorylation occurs by transfer of phosphate from side chains on KaiC to the β-phosphate of an ADP nucleophile. The surprising ability of KaiC to catalyze this energetically expensive reaction has garnered speculation of an evolutionary relationship with F1F0-ATP synthase (43), which also takes on a double doughnut-like structure (44). Because phosphorylation and dephosphorylation reactions on KaiC-CII appear to occur via the same chemistry going in opposite directions, the equilibrium can be regulated by substrate availability and ground state interactions without major restructuring of the active site. This model is consistent with the apparent mechanisms by which KaiA regulates CII phosphorylation, as described below.
KaiA: Primary regulator of KaiC phosphorylation
KaiA acts as the primary regulator of enzymatic activity on the CII domain of KaiC and therefore the phosphorylation cycle in general. Throughout the day, KaiA promotes phosphorylation of CII via two distinct mechanisms: 1) structural rearrangement of the CII catalytic core (32, 45), and 2) exchange of active-site ADP molecules for ATP (46). The enzymatic equilibrium of CII favors phosphatase activity when KaiC is in isolation (31), making KaiA occupancy the primary determinant of enzymatic equilibrium on CII.
Without KaiA bound, the A-loop residues project into the central cavity of KaiC (32). KaiA competes away interactions in this position, binding the loops and causing them to take on an α-helical conformation (47). When these loops are deleted, CII is in constitutive phosphorylation mode (32), indicating that the interaction of the A-loops with the cavity of KaiC abrogates CII phosphorylation and that KaiA disrupts those interactions. Additionally, KaiA has been shown to act as a nucleotide exchange factor, controlling the equilibrium of phosphorylation on CII at the level of substrate availability (46). KaiA increases the rate of ATP/ADP exchange on KaiC as well as the ratio of bound ATP at equilibrium (46). Hence, KaiA promotes CII phosphorylation by increasing the local concentration of ATP, as well as by direct perturbation of the active site.
Inactivation of KaiA during the night occurs through a conformational change that abrogates its A-loop binding activity (Fig. 3B). In the daytime state, the A-loops of KaiC form a helical bundle with the KaiA dimerization interface, flanked on either side by its N-terminal pseudo-receiver (PsR) domains. In this conformation, each β-strand from the interdomain linker crosses close to the other to form an intermolecular β-sheet, followed by a helical linker connecting the PsR domain. A high-resolution crystal structure of KaiA in the nighttime state was recently obtained by crystallizing its C-terminal dimerization domain and interdomain linker as part of a complex with KaiB and an engineered monomeric form of the KaiC-CI domain (35). KaiA docks onto an exposed β-sheet on KaiB in the KaiBC complex using the β-strand of its interdomain linker, which repositions the helical linker to dock into its own A-loop binding site. This intramolecular interaction between the α-helix of the interdomain linker and the dimerization interface precludes binding to an A-loop peptide from another KaiC hexamer, which demonstrates that binding to the KaiBC complex stabilizes an autoinhibited form of KaiA at night. The PsR domains of KaiA are not needed to bind KaiBC (48) and were omitted for this structure. However, this structure of an isolated CI–KaiB–KaiA complex agrees with a lower-resolution model obtained by cryo-electron microscopy using full-length wild-type KaiA, KaiB, and hexameric KaiC fully assembled in the nighttime state (49). Although the PsR domains were included here, they did not give sufficient density to be built into the model, suggesting that they are flexibly tethered to the complex.
KaiB: Attenuator of KaiC phosphorylation
The dephosphorylation arm of the KaiABC oscillatory cycle is initiated by the binding of KaiB at the CI domain of KaiC, followed by inactivation of KaiA. Historically, some discord has existed surrounding the location of KaiB binding on KaiC and how this leads to a shift of phosphorylation equilibrium that lasts through the night. Recently, a series of corroborative structural and biochemical studies (33–35, 49, 50) have put this controversy to rest. Our understanding of the role of KaiB in the clock has expanded immensely through the discovery of a major conformational rearrangement that occurs between the free and KaiC-bound states of KaiB. In the ground state, KaiB takes on the unique fold depicted in Fig. 3C and oligomerizes to form a dimer of dimers (51–53). When bound to the CI domain of KaiC, KaiB is in a monomeric thioredoxin fold (34), referred to as a “fold-switched” state because it comprises a major conformational shift, resulting in reassignment of secondary and tertiary structure in roughly 50 residues composing the C-terminal half of KaiB.
Binding of KaiB to KaiC occurs remarkably slowly, taking hours to go to completion in vitro (33, 53). This binding is quickened by mutations that favor the dimeric or thioredoxin-like monomeric states of KaiB (34, 53). Native mass spectrometry experiments have shown that binding occurs through a monomeric form of KaiB (54) and appears to be cooperative, as masses were obtained for KaiC bound to either one KaiB molecule or six, but none of the subsaturated stoichiometries in between (54). This is consistent with the observation of electrostatic complementarity between KaiB subunits in the recent structures of the KaiBC complex (35, 49).
The current biochemical data suggest that KaiB binds through a conformational selection mechanism, in which the ground state dimeric and tetrameric forms of KaiB act as a thermodynamic sink to decrease the concentration of the KaiC binding-competent KaiB monomer and limiting the rate of binding. Several different mutations have been identified that stabilize the KaiB monomer, each of which take on the fold-switched conformation (34, 35). In addition to increasing the rate of binding to KaiC in vitro, these mutations halt circadian oscillation in vitro and in vivo, resulting in a dominant arrhythmic phenotype that locks KaiC in the unphosphorylated state (34). In the fold-switched form, KaiB forms a ternary complex with KaiA and the CI domain of KaiC (34, 35), thus rendering KaiA inactive and mediating the dephosphorylation arm of the cycle. Together, these studies paint a unified picture of the repressive state of the cyanobacterial oscillator and highlight the clarifying role of structural biology, which together with genetic and biochemical studies have demonstrated how the three proteins of the cyanobacterial circadian PTO work in concert with one another to sustain biochemical oscillation.
Output signaling: SasA, CikA, and RpaA
Output signaling from the cyanobacterial PTO takes place through a two-component regulatory system (55). Two-component systems are pervasive in bacterial sensing and signal transduction (56). In the archetypal form, they consist of a sensor histidine kinase (HK) and a response regulator (RR) (57, 58). There is often overlap between cognate RRs and HKs; for example, an HK may activate multiple RRs, or an RR may be regulated by multiple HKs (59). Moreover, the relevant active domains of the components may reside within a single protein that has both kinase and substrate functions. Cross-talk between regulatory networks increases dramatically from prokaryotes to eukaryotes (60), and cyanobacteria appear to fall somewhere in the middle of this complexity. The circadian signaling network in cyanobacteria is not yet fully understood, and new components are still being identified (18, 19).
Despite the general complexity of regulatory networks in cyanobacteria, a two-component system consisting of two HKs and one RR stands out as being centrally involved in output signaling from the KaiABC oscillator (55, 61). RpaA is an RR that governs transcription from a locus encoding additional transcription factors that mediate the global changes in gene expression associated with circadian rhythms (62). This regulon includes the cotranscribed KaiBC genes, thus integrating the PTO into a TTFL (63).
RpaA is regulated by two cognate HKs, SasA and CikA (61). The sensor domain of SasA adopts a thioredoxin fold that is almost identical in structure to the fold-switched form of KaiB (34, 64) and binds to the pS/pT form of KaiC (34, 61, 65). Activation of SasA follows a canonical HK signaling mechanism. The sensor domains on SasA recognize and bind to phosphorylated KaiC, resulting in autophosphorylation of His-161 in the SasA kinase domain (61) and subsequent transfer of that phosphate to Asp-53 on RpaA, rendering the transcription factor competent for DNA binding (63). SasA has been shown by analytical ultracentrifugation to form a trimer in solution (65), and gel-filtration experiments support a stoichiometry of one SasA trimer per KaiC hexamer. HKs usually oligomerize as dimers, but in rare cases they have been found to function as higher order oligomers (66) or monomers (67). It is yet unknown whether this unusual oligomeric state for SasA is linked to its sensory function.
At the biochemical level, SasA competes with KaiB for the B-loops on the CI domain of phosphorylated KaiC (48). Binding of SasA is faster than that of KaiB, presumably due to the refolding requirement of KaiB for binding to KaiC (34, 65). This difference suggests an evolutionary selection for the metamorphic properties of KaiB in controlling the temporal window of SasA signaling, in addition to providing a timing cue for the oscillatory cycle, although more evidence is needed to support either of these models definitively. KaiA also plays a role in the competition between KaiB and SasA for the CI domain of KaiC. Although the presence of KaiA does not affect kobs for binding of KaiB to KaiC, it does make a difference in competitive KaiB binding when KaiC is pre-bound with SasA (48). This property implicates a role for KaiA in active eviction of SasA by KaiB, although no physical model for this currently exists.
The activating effect of SasA on RpaA is reversed by CikA, a multidomain protein that, in addition to its role in dephosphorylating RpaA, ties the clock to subcellular localization and entrainment functions (68–71). CikA is incorporated into the Night Complex through the interactions of its PsR domain with fold-switched KaiB at an interface that overlaps with the KaiA-binding site (Fig. 2). Consistent with a potential competitive mechanism, inclusion of the CikA PsR domain in an in vitro oscillation reaction results in a shortened period and decreased amplitude (34). Combined with the observation that CikA knockout results in a relatively severe gene expression phenotype (72), these data suggest that CikA has a more integral role in the timekeeping functions in vivo than suggested by its known roles in entrainment and output signaling. The integrated roles of CikA and the other core clock proteins are summarized in Fig. 4.
Figure 4.
Integrated timekeeping, entrainment, and output signaling functions of the cyanobacterial clock. Timekeeping, entrainment, and output signaling functions are highlighted within the oscillatory cycle of the cyanobacterial clock. Powered by ATPase activity on its CI domain, KaiC cycles through a series of phosphorylation states that is interdependent on its quaternary structure. KaiA is bound to the CII domain of KaiC during the day, stimulating phosphorylation. This process is sensitive to the ATP/ADP ratio, which peaks at midday, providing an entrainment cue. At dusk, levels of oxidized quinones rise in the cell, and the clock is entrained by this as well. Around this time, KaiC reaches the pS/pT state, and SasA binds to the CI domain to activate RpaA. CI-bound SasA is eventually competed away by KaiB. Binding of KaiB is slowed by its intrinsically unfavorable equilibrium that sequesters it in inactive states. Accumulation of KaiB in its KaiC-bound form recruits and inactivates KaiA, allowing CII dephosphorylation. The input/output protein CikA also interacts with the fold-switched form of KaiB, causing CikA to dephosphorylate RpaA, inactivating it.
Metabolic of entrainment of the clock
The cyanobacterial circadian clock achieves entrainment by linking the oscillatory cycle to photosynthesis through metabolic processes (73), rather than direct entrainment by light via sensory photoreceptors, as is commonly found in eukaryotic organisms (74). This metabolic entrainment of the cyanobacterial clock occurs in two ways: through sensitivity of the phosphorylation cycle to ATP/ADP ratios within the cell (75) and through the presence of nighttime-associated photosynthetic metabolites (71, 72, 76–78). The former entrainment cue occurs as a result of enzymatic sensitivity of the core oscillator to ATP/ADP ratios (72), causing it to align with the maximal photosynthetic period at midday when this ratio is highest (75, 79).
Cyanobacteria utilize quinone pools as intramembrane electron shuttles, reducing them during the process of photosynthesis (80). As a result, the concentration of oxidized quinones rises rapidly but briefly when photosynthesis shuts down with the onset of darkness before other pathways restore the redox steady state (76). Ectopic introduction of oxidized quinones can entrain the clock both in vivo and in vitro, and both KaiA and CikA play roles related to entrainment through binding of their PsR domains to oxidized quinones (71, 72, 76–78). The aggregation state and/or stability of the proteins is affected by quinone binding in vivo (71, 77). Still, the precise mechanism by which quinone binding influences the PTO is not currently well-understood.
Applications in synthetic biology
There is considerable interest in synthetic biology to control temporally regulated gene expression (81, 82). However, even the most robust synthetic oscillators are far less effective than the fully integrated oscillation of gene expression in cyanobacteria (83, 84). Because of its ability to function at constant protein concentrations and provide relatively stable temporal regulation at different temperatures, it could be enticing to reverse engineer synthetic oscillators from the cyanobacterial PTO. This has been achieved, on a rudimentary level, in a transplantation study where the core oscillator was introduced into E. coli, resulting in weak but measurable in vivo circadian rhythms in KaiC phosphorylation in a natively non-circadian organism (85). In this study, the clock was linked to transcriptional output signaling through a two-hybrid approach, although the researchers were unable to show circadian oscillation of gene expression originating from KaiABC. Thus, a great deal of opportunity still exists to integrate the cyanobacterial PTO more deeply into the physiology of the host organism and to achieve clock-driven oscillations in gene expression. Undeniably, these efforts benefit from the mechanistic advances in our understanding of the cyanobacterial PTO. Furthermore, a great deal of work has gone into computational modeling of the cyanobacterial PTO (36, 86–89). These studies can inform efforts to re-engineer the oscillatory period of the PTO, leading to future technologies in which the cyanobacterial PTO may be used as a starting point to introduce a flexible range of temporal routines of gene expression into target organisms.
Acknowledgments
We thank Roger Tseng, Joel Heisler, and Yong-Gang Chang for helpful conversations and comments on the manuscript.
This work was supported by National Institutes of Health Grants R35GM118290 (to S. S. G.), R01GM107521 (to A. L.), and R01GM121507 (to C. L. P.). This is the second article in the Thematic Minireview series “Green biological chemistry.” The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- TTFL
- transcription-translation feedback loop
- PTO
- post-translational oscillator
- PsR
- pseudo-receiver
- HK
- histidine kinase
- RR
- response regulator
- PDB
- Protein Data Bank.
References
- 1. Kuhlman S. J., Craig L. M., and Duffy J. F. (2017) Introduction to chronobiology. Cold Spring Harb. Perspect. Biol. 2017, a033613 10.1101/cshperspect.a033613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hurley J. M., Loros J. J., and Dunlap J. C. (2016) Circadian oscillators: around the transcription-translation feedback loop and on to output. Trends Biochem. Sci. 41, 834–846 10.1016/j.tibs.2016.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nakajima M., Imai K., Ito H., Nishiwaki T., Murayama Y., Iwasaki H., Oyama T., and Kondo T. (2005) Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 308, 414–415 10.1126/science.1108451 [DOI] [PubMed] [Google Scholar]
- 4. Johnson C. H., Zhao C., Xu Y., and Mori T. (2017) Timing the day: what makes bacterial clocks tick? Nat. Rev. Microbiol. 15, 232–242 10.1038/nrmicro.2016.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mitsui A., Kumazawa S., Takahashi A., Ikemoto H., Cao S., and Arai T. (1986) Strategy by which nitrogen-fixing unicellular cyanobacteria grow photoautotrophically. Nature 323, 720–722 10.1038/323720a0 [DOI] [Google Scholar]
- 6. Ito H., Mutsuda M., Murayama Y., Tomita J., Hosokawa N., Terauchi K., Sugita C., Sugita M., Kondo T., and Iwasaki H. (2009) Cyanobacterial daily life with Kai-based circadian and diurnal genome-wide transcriptional control in Synechococcus elongatus. Proc. Natl. Acad. Sci. U.S.A. 106, 14168–14173 10.1073/pnas.0902587106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu Y., Tsinoremas N. F., Johnson C. H., Lebedeva N. V., Golden S. S., Ishiura M., and Kondo T. (1995) Circadian orchestration of gene expression in cyanobacteria. Genes Dev. 9, 1469–1478 10.1101/gad.9.12.1469 [DOI] [PubMed] [Google Scholar]
- 8. Smith R. M., and Williams S. B. (2006) Circadian rhythms in gene transcription imparted by chromosome compaction in the cyanobacterium Synechococcus elongatus. Proc. Natl. Acad. Sci. U.S.A. 103, 8564–8569 10.1073/pnas.0508696103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vijayan V., Zuzow R., and O'Shea E. K. (2009) Oscillations in supercoiling drive circadian gene expression in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 106, 22564–22568 10.1073/pnas.0912673106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ishiura M., Kutsuna S., Aoki S., Iwasaki H., Andersson C. R., Tanabe A., Golden S. S., Johnson C. H., and Kondo T. (1998) Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science 281, 1519–1523 10.1126/science.281.5382.1519 [DOI] [PubMed] [Google Scholar]
- 11. Tomita J., Nakajima M., Kondo T., and Iwasaki H. (2005) No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science 307, 251–254 10.1126/science.1102540 [DOI] [PubMed] [Google Scholar]
- 12. Kitayama Y., Nishiwaki T., Terauchi K., and Kondo T. (2008) Dual KaiC-based oscillations constitute the circadian system of cyanobacteria. Genes Dev. 22, 1513–1521 10.1101/gad.1661808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Qin X., Byrne M., Mori T., Zou P., Williams D. R., McHaourab H., and Johnson C. H. (2010) Intermolecular associations determine the dynamics of the circadian KaiABC oscillator. Proc. Natl. Acad. Sci. U.S.A. 107, 14805–14810 10.1073/pnas.1002119107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hosokawa N., Kushige H., and Iwasaki H. (2013) Attenuation of the posttranslational oscillator via transcription-translation feedback enhances circadian-phase shifts in Synechococcus. Proc. Natl. Acad. Sci. U.S.A. 110, 14486–14491 10.1073/pnas.1302243110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Onai K., Morishita M., Itoh S., Okamoto K., and Ishiura M. (2004) Circadian rhythms in the thermophilic cyanobacterium Thermosynechococcus elongatus: compensation of period length over a wide temperature range. J. Bacteriol. 186, 4972–4977 10.1128/JB.186.15.4972-4977.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Taniguchi Y., Katayama M., Ito R., Takai N., Kondo T., and Oyama T. (2007) labA: a novel gene required for negative feedback regulation of the cyanobacterial circadian clock protein KaiC. Genes Dev. 21, 60–70 10.1101/gad.1488107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hanaoka M., Takai N., Hosokawa N., Fujiwara M., Akimoto Y., Kobori N., Iwasaki H., Kondo T., and Tanaka K. (2012) RpaB, another response regulator operating circadian clock-dependent transcriptional regulation in Synechococcus elongatus PCC 7942. J. Biol. Chem. 287, 26321–26327 10.1074/jbc.M111.338251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boyd J. S., Cheng R. R., Paddock M. L., Sancar C., Morcos F., and Golden S. S. (2016) A combined computational and genetic approach uncovers network interactions of the cyanobacterial circadian clock. J. Bacteriol. 198, 2439–2447 10.1128/JB.00235-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Espinosa J., Boyd J. S., Cantos R., Salinas P., Golden S. S., and Contreras A. (2015) Cross-talk and regulatory interactions between the essential response regulator RpaB and cyanobacterial circadian clock output. Proc. Natl. Acad. Sci. U.S.A. 112, 2198–2203 10.1073/pnas.1424632112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Osanai T., Shirai T., Iijima H., Kuwahara A., Suzuki I., Kondo A., and Hirai M. Y. (2015) Alteration of cyanobacterial sugar and amino acid metabolism by overexpression hik8, encoding a KaiC-associated histidine kinase. Environ. Microbiol. 17, 2430–2440 10.1111/1462-2920.12715 [DOI] [PubMed] [Google Scholar]
- 21. Ivleva N. B., Bramlett M. R., Lindahl P. A., and Golden S. S. (2005) LdpA: a component of the circadian clock senses redox state of the cell. EMBO J. 24, 1202–1210 10.1038/sj.emboj.7600606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cohen S. E., and Golden S. S. (2015) Circadian rhythms in cyanobacteria. Microbiol. Mol. Biol. Rev. 79, 373–385 10.1128/MMBR.00036-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mori T., Saveliev S. V., Xu Y., Stafford W. F., Cox M. M., Inman R. B., and Johnson C. H. (2002) Circadian clock protein KaiC forms ATP-dependent hexameric rings and binds DNA. Proc. Natl. Acad. Sci. U.S.A. 99, 17203–17208 10.1073/pnas.262578499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pattanayek R., Wang J., Mori T., Xu Y., Johnson C. H., and Egli M. (2004) Visualizing a circadian clock protein: crystal structure of KaiC and functional insights. Mol. Cell 15, 375–388 10.1016/j.molcel.2004.07.013 [DOI] [PubMed] [Google Scholar]
- 25. Hayashi F., Iwase R., Uzumaki T., and Ishiura M. (2006) Hexamerization by the N-terminal domain and intersubunit phosphorylation by the C-terminal domain of cyanobacterial circadian clock protein KaiC. Biochem. Biophys. Res. Commun. 348, 864–872 10.1016/j.bbrc.2006.07.143 [DOI] [PubMed] [Google Scholar]
- 26. Phong C., Markson J. S., Wilhoite C. M., and Rust M. J. (2013) Robust and tunable circadian rhythms from differentially sensitive catalytic domains. Proc. Natl. Acad. Sci. U.S.A. 110, 1124–1129 10.1073/pnas.1212113110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hayashi F., Itoh N., Uzumaki T., Iwase R., Tsuchiya Y., Yamakawa H., Morishita M., Onai K., Itoh S., and Ishiura M. (2004) Roles of two ATPase-motif-containing domains in cyanobacterial circadian clock protein KaiC. J. Biol. Chem. 279, 52331–52337 10.1074/jbc.M406604200 [DOI] [PubMed] [Google Scholar]
- 28. Kageyama H., Kondo T., and Iwasaki H. (2003) Circadian formation of clock protein complexes by KaiA, KaiB, KaiC, and SasA in cyanobacteria. J. Biol. Chem. 278, 2388–2395 10.1074/jbc.M208899200 [DOI] [PubMed] [Google Scholar]
- 29. Nishiwaki T., Satomi Y., Kitayama Y., Terauchi K., Kiyohara R., Takao T., and Kondo T. (2007) A sequential program of dual phosphorylation of KaiC as a basis for circadian rhythm in cyanobacteria. EMBO J. 26, 4029–4037 10.1038/sj.emboj.7601832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kageyama H., Nishiwaki T., Nakajima M., Iwasaki H., Oyama T., and Kondo T. (2006) Cyanobacterial circadian pacemaker: Kai protein complex dynamics in the KaiC phosphorylation cycle in vitro. Mol. Cell 23, 161–171 10.1016/j.molcel.2006.05.039 [DOI] [PubMed] [Google Scholar]
- 31. Iwasaki H., Nishiwaki T., Kitayama Y., Nakajima M., and Kondo T. (2002) KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 99, 15788–15793 10.1073/pnas.222467299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim Y. I., Dong G., Carruthers C. W. Jr., Golden S. S., and LiWang A. (2008) The day/night switch in KaiC, a central oscillator component of the circadian clock of cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 105, 12825–12830 10.1073/pnas.0800526105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chang Y. G., Tseng R., Kuo N. W., and LiWang A. (2012) Rhythmic ring-ring stacking drives the circadian oscillator clockwise. Proc. Natl. Acad. Sci. U.S.A. 109, 16847–16851 10.1073/pnas.1211508109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chang Y. G., Cohen S. E., Phong C., Myers W. K., Kim Y. I., Tseng R., Lin J., Zhang L., Boyd J. S., Lee Y., Kang S., Lee D., Li S., Britt R. D., Rust M. J., et al. (2015) Circadian rhythms. A protein fold switch joins the circadian oscillator to clock output in cyanobacteria. Science 349, 324–328 10.1126/science.1260031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tseng R., Goularte N. F., Chavan A., Luu J., Cohen S. E., Chang Y. G., Heisler J., Li S., Michael A. K., Tripathi S., Golden S. S., LiWang A., and Partch C. L. (2017) Structural basis of the day-night transition in a bacterial circadian clock. Science 355, 1174–1180 10.1126/science.aag2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rust M. J., Markson J. S., Lane W. S., Fisher D. S., and O'Shea E. K. (2007) Ordered phosphorylation governs oscillation of a three-protein circadian clock. Science 318, 809–812 10.1126/science.1148596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Terauchi K., Kitayama Y., Nishiwaki T., Miwa K., Murayama Y., Oyama T., and Kondo T. (2007) ATPase activity of KaiC determines the basic timing for circadian clock of cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 104, 16377–16381 10.1073/pnas.0706292104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Abe J., Hiyama T. B., Mukaiyama A., Son S., Mori T., Saito S., Osako M., Wolanin J., Yamashita E., Kondo T., and Akiyama S. (2015) Circadian rhythms. Atomic-scale origins of slowness in the cyanobacterial circadian clock. Science 349, 312–316 10.1126/science.1261040 [DOI] [PubMed] [Google Scholar]
- 39. Pattanayek R., Mori T., Xu Y., Pattanayek S., Johnson C. H., and Egli M. (2009) Structures of KaiC circadian clock mutant proteins: a new phosphorylation site at T426 and mechanisms of kinase, ATPase and phosphatase. PLoS ONE 4, e7529 10.1371/journal.pone.0007529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Murayama Y., Mukaiyama A., Imai K., Onoue Y., Tsunoda A., Nohara A., Ishida T., Maéda Y., Terauchi K., Kondo T., and Akiyama S. (2011) Tracking and visualizing the circadian ticking of the cyanobacterial clock protein KaiC in solution. EMBO J. 30, 68–78 10.1038/emboj.2010.298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chang Y. G., Kuo N. W., Tseng R., and LiWang A. (2011) Flexibility of the C-terminal, or CII, ring of KaiC governs the rhythm of the circadian clock of cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 108, 14431–14436 10.1073/pnas.1104221108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nishiwaki T., and Kondo T. (2012) Circadian autodephosphorylation of cyanobacterial clock protein KaiC occurs via formation of ATP as intermediate. J. Biol. Chem. 287, 18030–18035 10.1074/jbc.M112.350660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Egli M., Mori T., Pattanayek R., Xu Y., Qin X., and Johnson C. H. (2012) Dephosphorylation of the core clock protein KaiC in the cyanobacterial KaiABC circadian oscillator proceeds via an ATP synthase mechanism. Biochemistry 51, 1547–1558 10.1021/bi201525n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stock D., Leslie A. G., and Walker J. E. (1999) Molecular architecture of the rotary motor in ATP synthase. Science 286, 1700–1705 10.1126/science.286.5445.1700 [DOI] [PubMed] [Google Scholar]
- 45. Egli M., Pattanayek R., Sheehan J. H., Xu Y., Mori T., Smith J. A., and Johnson C. H. (2013) Loop-loop interactions regulate KaiA-stimulated KaiC phosphorylation in the cyanobacterial KaiABC circadian clock. Biochemistry 52, 1208–1220 10.1021/bi301691a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nishiwaki-Ohkawa T., Kitayama Y., Ochiai E., and Kondo T. (2014) Exchange of ADP with ATP in the CII ATPase domain promotes autophosphorylation of cyanobacterial clock protein KaiC. Proc. Natl. Acad. Sci. U.S.A. 111, 4455–4460 10.1073/pnas.1319353111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pattanayek R., and Egli M. (2015) Protein–protein interactions in the cyanobacterial circadian clock: structure of KaiA dimer in complex with C-terminal KaiC peptides at 2.8 A resolution. Biochemistry 54, 4575–4578 10.1021/acs.biochem.5b00694 [DOI] [PubMed] [Google Scholar]
- 48. Tseng R., Chang Y. G., Bravo I., Latham R., Chaudhary A., Kuo N. W., and Liwang A. (2014) Cooperative KaiA-KaiB-KaiC interactions affect KaiB/SasA competition in the circadian clock of cyanobacteria. J. Mol. Biol. 426, 389–402 10.1016/j.jmb.2013.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Snijder J., Schuller J. M., Wiegard A., Lössl P., Schmelling N., Axmann I. M., Plitzko J. M., Förster F., and Heck A. J. (2017) Structures of the cyanobacterial circadian oscillator frozen in a fully assembled state. Science 355, 1181–1184 10.1126/science.aag3218 [DOI] [PubMed] [Google Scholar]
- 50. Sugiyama M., Yagi H., Ishii K., Porcar L., Martel A., Oyama K., Noda M., Yunoki Y., Murakami R., Inoue R., Sato N., Oba Y., Terauchi K., Uchiyama S., and Kato K. (2016) Structural characterization of the circadian clock protein complex composed of KaiB and KaiC by inverse contrast-matching small-angle neutron scattering. Sci. Rep. 6, 35567 10.1038/srep35567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Iwase R., Imada K., Hayashi F., Uzumaki T., Morishita M., Onai K., Furukawa Y., Namba K., and Ishiura M. (2005) Functionally important substructures of circadian clock protein KaiB in a unique tetramer complex. J. Biol. Chem. 280, 43141–43149 10.1074/jbc.M503360200 [DOI] [PubMed] [Google Scholar]
- 52. Hitomi K., Oyama T., Han S., Arvai A. S., and Getzoff E. D. (2005) Tetrameric architecture of the circadian clock protein KaiB. A novel interface for intermolecular interactions and its impact on the circadian rhythm. J. Biol. Chem. 280, 19127–19135 10.1074/jbc.M411284200 [DOI] [PubMed] [Google Scholar]
- 53. Murakami R., Mutoh R., Iwase R., Furukawa Y., Imada K., Onai K., Morishita M., Yasui S., Ishii K., Valencia Swain J. O., Uzumaki T., Namba K., and Ishiura M. (2012) The roles of the dimeric and tetrameric structures of the clock protein KaiB in the generation of circadian oscillations in cyanobacteria. J. Biol. Chem. 287, 29506–29515 10.1074/jbc.M112.349092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Snijder J., Burnley R. J., Wiegard A., Melquiond A. S., Bonvin A. M., Axmann I. M., and Heck A. J. (2014) Insight into cyanobacterial circadian timing from structural details of the KaiB-KaiC interaction. Proc. Natl. Acad. Sci. U.S.A. 111, 1379–1384 10.1073/pnas.1314326111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Takai N., Nakajima M., Oyama T., Kito R., Sugita C., Sugita M., Kondo T., and Iwasaki H. (2006) A KaiC-associating SasA-RpaA two-component regulatory system as a major circadian timing mediator in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 103, 12109–12114 10.1073/pnas.0602955103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mitrophanov A. Y., and Groisman E. A. (2008) Signal integration in bacterial two-component regulatory systems. Genes Dev. 22, 2601–2611 10.1101/gad.1700308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gao R., and Stock A. M. (2009) Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 63, 133–154 10.1146/annurev.micro.091208.073214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zschiedrich C. P., Keidel V., and Szurmant H. (2016) Molecular mechanisms of two-component signal transduction. J. Mol. Biol. 428, 3752–3775 10.1016/j.jmb.2016.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Laub M. T., and Goulian M. (2007) Specificity in two-component signal transduction pathways. Annu. Rev. Genet. 41, 121–145 10.1146/annurev.genet.41.042007.170548 [DOI] [PubMed] [Google Scholar]
- 60. Ashby M. K., and Houmard J. (2006) Cyanobacterial two-component proteins: structure, diversity, distribution, and evolution. Microbiol. Mol. Biol. Rev. 70, 472–509 10.1128/MMBR.00046-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gutu A., and O'Shea E. K. (2013) Two antagonistic clock-regulated histidine kinases time the activation of circadian gene expression. Mol. Cell 50, 288–294 10.1016/j.molcel.2013.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Taniguchi Y., Takai N., Katayama M., Kondo T., and Oyama T. (2010) Three major output pathways from the KaiABC-based oscillator cooperate to generate robust circadian kaiBC expression in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 107, 3263–3268 10.1073/pnas.0909924107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Markson J. S., Piechura J. R., Puszynska A. M., and O'Shea E. K. (2013) Circadian control of global gene expression by the cyanobacterial master regulator RpaA. Cell 155, 1396–1408 10.1016/j.cell.2013.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Vakonakis I., Klewer D. A., Williams S. B., Golden S. S., and LiWang A. C. (2004) Structure of the N-terminal domain of the circadian clock-associated histidine kinase SasA. J. Mol. Biol. 342, 9–17 10.1016/j.jmb.2004.07.010 [DOI] [PubMed] [Google Scholar]
- 65. Valencia S. J., Bitou K., Ishii K., Murakami R., Morishita M., Onai K., Furukawa Y., Imada K., Namba K., and Ishiura M. (2012) Phase-dependent generation and transmission of time information by the KaiABC circadian clock oscillator through SasA-KaiC interaction in cyanobacteria. Genes Cells 17, 398–419 10.1111/j.1365-2443.2012.01597.x [DOI] [PubMed] [Google Scholar]
- 66. Wojnowska M., Yan J., Sivalingam G. N., Cryar A., Gor J., Thalassinos K., and Djordjevic S. (2013) Autophosphorylation activity of a soluble hexameric histidine kinase correlates with the shift in protein conformational equilibrium. Chem. Biol. 20, 1411–1420 10.1016/j.chembiol.2013.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rivera-Cancel G., Ko W. H., Tomchick D. R., Correa F., and Gardner K. H. (2014) Full-length structure of a monomeric histidine kinase reveals basis for sensory regulation. Proc. Natl. Acad. Sci. U.S.A. 111, 17839–17844 10.1073/pnas.1413983111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cohen S. E., Erb M. L., Selimkhanov J., Dong G., Hasty J., Pogliano J., and Golden S. S. (2014) Dynamic localization of the cyanobacterial circadian clock proteins. Curr. Biol. 24, 1836–1844 10.1016/j.cub.2014.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mackey S. R., Choi J. S., Kitayama Y., Iwasaki H., Dong G., and Golden S. S. (2008) Proteins found in a CikA interaction assay link the circadian clock, metabolism, and cell division in Synechococcus elongatus. J. Bacteriol. 190, 3738–3746 10.1128/JB.01721-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Schmitz O., Katayama M., Williams S. B., Kondo T., and Golden S. S. (2000) CikA, a bacteriophytochrome that resets the cyanobacterial circadian clock. Science 289, 765–768 10.1126/science.289.5480.765 [DOI] [PubMed] [Google Scholar]
- 71. Ivleva N. B., Gao T., LiWang A. C., and Golden S. S. (2006) Quinone sensing by the circadian input kinase of the cyanobacterial circadian clock. Proc. Natl. Acad. Sci. U.S.A. 103, 17468–17473 10.1073/pnas.0606639103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Williams S. B., Vakonakis I., Golden S. S., and LiWang A. C. (2002) Structure and function from the circadian clock protein KaiA of Synechococcus elongatus: a potential clock input mechanism. Proc. Natl. Acad. Sci. U.S.A. 99, 15357–15362 10.1073/pnas.232517099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Pattanayak G. K., Phong C., and Rust M. J. (2014) Rhythms in energy storage control the ability of the cyanobacterial circadian clock to reset. Curr. Biol. 24, 1934–1938 10.1016/j.cub.2014.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Golombek D. A., and Rosenstein R. E. (2010) Physiology of circadian entrainment. Physiol. Rev. 90, 1063–1102 10.1152/physrev.00009.2009 [DOI] [PubMed] [Google Scholar]
- 75. Rust M. J., Golden S. S., and O'Shea E. K. (2011) Light-driven changes in energy metabolism directly entrain the cyanobacterial circadian oscillator. Science 331, 220–223 10.1126/science.1197243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kim Y. I., Vinyard D. J., Ananyev G. M., Dismukes G. C., and Golden S. S. (2012) Oxidized quinones signal onset of darkness directly to the cyanobacterial circadian oscillator. Proc. Natl. Acad. Sci. U.S.A. 109, 17765–17769 10.1073/pnas.1216401109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wood T. L., Bridwell-Rabb J., Kim Y. I., Gao T., Chang Y. G., LiWang A., Barondeau D. P., and Golden S. S. (2010) The KaiA protein of the cyanobacterial circadian oscillator is modulated by a redox-active cofactor. Proc. Natl. Acad. Sci. U.S.A. 107, 5804–5809 10.1073/pnas.0910141107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gao T., Zhang X., Ivleva N. B., Golden S. S., and LiWang A. (2007) NMR structure of the pseudo-receiver domain of CikA. Protein Sci. 16, 465–475 10.1110/ps.062532007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Leypunskiy E., Lin J., Yoo H., Lee U., Dinner A. R., and Rust M. J. (2017) The cyanobacterial circadian clock follows midday in vivo and in vitro. Elife 6, e23539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bryant D. A. (1994) in The Molecular Biology of Cyanobacteria (Bryand D., ed) pp. 217–257, Kluwer/Academic Publishers, Dordrecht; Boston [Google Scholar]
- 81. Rosier B. J., and De Greef T. F. (2015) How to make an oscillator. Elife 2015;4:e12260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bartocci E., Bortolussi L., and Nenzi L. (2013) A temporal logic approach to modular design of synthetic biological circuits. Lect. N Bioinformat. 8130, 164–177 [Google Scholar]
- 83. Potvin-Trottier L., Lord N. D., Vinnicombe G., and Paulsson J. (2016) Synchronous long-term oscillations in a synthetic gene circuit. Nature 538, 514–517 10.1038/nature19841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Purcell O., Savery N. J., Grierson C. S., and di Bernardo M. (2010) A comparative analysis of synthetic genetic oscillators. J. R. Soc. Interface 7, 1503–1524 10.1098/rsif.2010.0183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Chen A. H., Lubkowicz D., Yeong V., Chang R. L., and Silver P. A. (2015) Transplantability of a circadian clock to a noncircadian organism. Sci. Adv. 1, e1500358 10.1126/sciadv.1500358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. van Zon J. S., Lubensky D. K., Altena P. R., and ten Wolde P. R. (2007) An allosteric model of circadian KaiC phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 104, 7420–7425 10.1073/pnas.0608665104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ma L., and Ranganathan R. (2012) Quantifying the rhythm of KaiB–C interaction for in vitro cyanobacterial circadian clock. PLoS ONE 7, e42581 10.1371/journal.pone.0042581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Lin J., Chew J., Chockanathan U., and Rust M. J. (2014) Mixtures of opposing phosphorylations within hexamers precisely time feedback in the cyanobacterial circadian clock. Proc. Natl. Acad. Sci. U.S.A. 111, E3937–E3945 10.1073/pnas.1408692111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Paijmans J., Lubensky D. K., and Ten Wolde P. R. (2017) A thermodynamically consistent model of the post-translational Kai circadian clock. PLoS Comput. Biol. 13, e1005415 10.1371/journal.pcbi.1005415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hunter S., Apweiler R., Attwood T. K., Bairoch A., Bateman A., Binns D., Bork P., Das U., Daugherty L., Duquenne L., Finn R. D., Gough J., Haft D., Hulo N., Kahn D., et al. (2009) InterPro: the integrative protein signature database. Nucleic Acids Res. 37, D211–D215 10.1093/nar/gkn785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 [DOI] [PMC free article] [PubMed] [Google Scholar]