Abstract
Polysaccharide-based biopolymers have many material properties relevant to industrial and medical uses, including as drug delivery agents, wound-healing adhesives, and food additives and stabilizers. Traditionally, polysaccharides are obtained from natural sources. Microbial synthesis offers an attractive alternative for sustainable production of tailored biopolymers. Here, we review synthetic biology strategies for select “green” biopolymers: cellulose, alginate, chitin, chitosan, and hyaluronan. Microbial production pathways, opportunities for pathway yield improvements, and advances in microbial engineering of biopolymers in various hosts are discussed. Taken together, microbial engineering has expanded the repertoire of green biological chemistry by increasing the diversity of biobased materials.
Keywords: metabolic engineering, synthetic biology, polysaccharide, biomaterials, secretion
Context for polysaccharide-based biopolymers
Polysaccharide-based scaffolds have applications in medicine, as agricultural and food products, and as biomaterials with bioactive, biocompatible, and biodegradable properties (Table 1). Biopolymers, such as the chitin derivative chitosan, have been shown to accelerate wound healing (1) and offer opportunities for scalable manufacturing in the bioprinting industry (2). Traditionally, biopolymers are obtained from natural sources by extraction from the environment and require further downstream processing, including, in many cases, the use of harsh chemicals to obtain desired material properties. For example, chitin, which is traditionally sourced from shellfish, is a waste product resulting from the seafood industry that requires chemical protection and deprotection steps of various hydroxyl groups to impart desired functional properties (3).
Table 1.
Overview of microbial production of carbohydrate biopolymers: cellulose, alginate, chitin, chitosan, and hyaluronan
ManA is mannuronic acid.
| Primary chemical structure | Metabolic precursor | Polymerizing enzyme | Natural/Industrial source | Select examples: host(s), titer, scale | Industrial application | |
|---|---|---|---|---|---|---|
| Cellulose | β-(1,4)-Linked homopolymer of d-Glc | UDP-Glc | Cellulose synthase (BcsA) | Green plants; some algae; oomycetes | Acetobacter xylinum BRC5, 15.3 g/liter, 10-liter batch (26) | Electronics (diaphragms of acoustic transducers); food (nata de coco); medicine (scaffolds for tissue engineering, wound dressing, soft tissue replacement) |
| Alginate | β-(1,4)-Linked non-repeating heteropolymer of ManA and UDP-guluronic acid | GDP-ManA | Glycosyltransferase (Alg8) | Seaweed; some bacteria | Azotobacter vinelandii, 6.6 g/liter, 1.5-liter batch reactors (99) | Feed; food (gelling agents, stabilizers); medicine (tissue scaffolds, drug delivery; research |
| Chitin | β-(1,4)-Homopolymer of GlcNAc | UDP-GlcNAc | Chitin synthase (NodC) | Fungi; crustaceans; insects; beaks of cephalopods; some fish and amphibians | GlcN: E. coli, 17 g/liter, 1-liter fed batch (43) GlcNAc: E. coli, 110 g/liter, 1-liter two-phase fed batch (43) Mixture of chitin oligosaccharides penta-N-acetyl-chitopentaose and tetra-N-acetyl-chitopentaose: E. coli, 2.5 g/liter, 2-liter fed batch (64) | Agriculture (pesticides); manufacturing (bioprinting); medicine (wound healing, drug delivery) |
| Chitosan | β-(1,4)-Linked heteropolymer of GlcNAc and GlcN | |||||
| Hyaluronan | β-(1,4)-Linked repeating heteropolymer of disaccharide units of GlcUA and GlcNAc | UDP-GlcUA + UDP-GlcNAc | Hyaluronan synthase (HasA) | Vertebrates (e.g. rooster combs); some bacteria | C. glutamicum, 21.6 g/liter, 5-liter fed batch (77) | Cosmetics; medicine (visco-supplementation, tissue repair, drug delivery) |
Biobased production of chemicals from sugars and biomass is more sustainable than traditional non-renewable petrochemical routes (4). The cell factory approach, where a chemical is synthesized in vivo, utilizes simple and inexpensive starting materials like glucose. Metabolic pathways can be overexpressed and optimized in native organisms or reconstructed into heterologous hosts for improved yields. In the past 30 years, notable advances have been made in the microbial biosynthesis of building block chemicals, such as dicarboxylic acids (e.g. glucaric acid (5)), diamines (e.g. putrescine (6)), hydroxyacids (e.g. 3-hydroxybutyrate (7)), and diols (e.g. butanediol (8)). Whereas microbial production of bioplastics from hydroxyacid monomers is a keystone example of industrially relevant biopolymerization (9), this Minireview will not cover this area as other reviews sufficiently discuss this topic (10).
By harnessing nature's toolbox of diverse biochemistry, microbial production of building block monomers can be extended to in-cell functionalization and polymerization. One-step microbial production of biopolymers is a sustainable alternative to avoid the use of environmentally damaging chemicals and catalysts and offers a scalable process that does not depend on harvesting from fragile ocean ecosystems, as is the case for biopolymers chitin and alginate; competing for valuable land as is the case for cellulose; or interfering with ethics of animal-based products as is the case for hyaluronan. Synthesizing biopolymers through enzymatic or whole-cell biocatalysis allows for higher regio- and stereoselectivity for in-cell composition-tailoring of polymers, which can reduce downstream processing. Useful objectives for metabolic engineering are polymer chain length by molecular weight control, sequence of saccharide units for composition control, and yield improvements for increased economic feasibility. Although greener methods do exist for biopolymer extraction from natural sources, such as utilization of ionic liquids for extracting chitin from crustacean shells (11), the economic competitiveness versus synthetic biology strategies has not yet been demonstrated.
This Minireview highlights strategies for cellular biosynthesis of select industrially and medically relevant polysaccharides: cellulose, alginate, chitin, chitosan, and hyaluronan. These biopolymers are examples of polysaccharides that are synthesized by the synthase-dependent pathway where polymerization and translocation processes are performed by a single synthase protein complex (12). Native biosynthetic mechanisms, such as microbial exopolysaccharide (EPS)3 biosynthesis, serve as a template for biotechnological production of biomaterials. Typically, synthase-dependent pathways favor homopolymer formation, and the polymers are released into the extracellular environment as non-covalently associated EPS fibers. These fibers are secreted into the surrounding environment at high molecular weights and can be harvested from cell cultures in a cost-effective manner by filtration. Several studies have demonstrated the synthesis of natural or novel variants of biopolymers from engineered organisms (13). General production strategies for microbial biosynthesis of biopolymers, such as increasing the pool of metabolite precursor supply and carbon flux toward the end product (14), are discussed within. Because of the diversity of material design options, microbial production of biopolymers also offers an attractive opportunity toward the production of new, custom-made materials beyond those from natural sources, such as engineered biosynthesis of non-natural fluorinated polyhydroxyalkanoates for bioplastics (15).
Synthetic biology strategies for polysaccharide biosynthesis
Cellulose
Cellulose is the most abundant polymer on the planet and is one of the most widely used natural materials in products such as papers and textiles. The value of the global market size for cellulose fiber was USD 20.61 billion in 2015 and is projected to reach USD 48.37 billion by 2025 (16). Cellulose is a monomeric polymer of β(1→4) d-glucose units (Fig. 1A) forming chains that twist into higher crystalline structures of cellulose I (triclinic structures Iα and Iβ) and cellulose II. Cellulose crystals are formed by aggregation of nearby chains secreted from collections of cellulose synthase complexes arranged in the cell membrane, typically forming rosettes in plant cells and axially aligned lines in bacteria (17).
Figure 1.
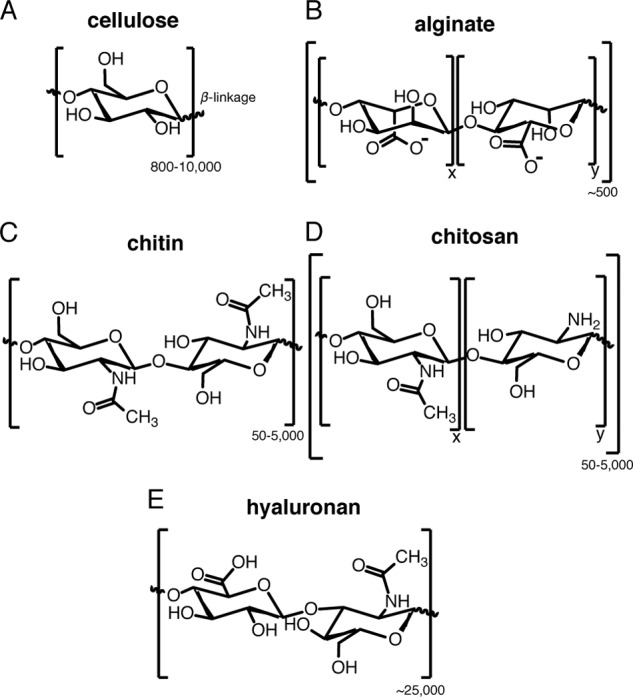
Chemical structures of biopolymers highlighted in this Minireview. A, cellulose; B, alginate; C, chitin, D, chitosan; E, hyaluronan. The number of units is indicated on the bottom right of each bracket. X and Y designate different monomeric units.
Bacterial cellulose (BC) production offers unique advantages over plant fiber processing by reducing the chemical and power input during purification and offering access to the cell surface through the media to modulate crystal formation during synthesis. Manual addition of hydrogen bonding molecules to the culture media can control Iα/Iβ ratios and molecular weights in resulting cellulose particles (18, 19). BC crystals interweave in random patterns according to bacterial movement and form a pellicle at the oxygen–media interface that grows to take the shape of the bioreactor. Pellicles (also called tea mushroom or Symbiotic Culture of Bacteria and Yeast (SCOBY)) occur naturally on the surface of Kombucha fermented tea cultures and have been used as materials for thousands of years (20). When grown in controlled conditions, regular cellulose films with complex structure can incorporate functional additives to achieve optical activity, conductivity, magnetism, and photo-catalytic degradation (21). The genetic tractability of bacteria makes cellulose material synthesis an attractive target for synthetic biology.
Bacterial cellulose synthase (BCS) requires cyclic di-GMP for activation and is internally regulated by accumulation of UDP, which is released from UDP-glucose during polymerization (Fig. 2) (22). Heterologous expression of Gluconacetobacter xylinus cellulose synthase genes from plasmids in Escherichia coli led to amorphous synthesis and a non-native cellulose II structure indicating the importance of membrane organization to crystallization (23). Bacteria that produce crystalline fibrils include a fourth gene bcsD that is likely to facilitate the formation of multienzyme structures in the membrane, as evidenced by knockouts that produce cellulose with reduced crystallinity (24). BCS operons are known to occasionally include a gene encoding endoglucanase, BcsZ, which is capable of specifically degrading amorphous improperly formed cellulose chains. Inclusion of BcsZ protein after fibrils are formed increases crystallinity in reconstituted in vitro systems (24). Induction by quorum-sensing molecule N-acyl homoserine lactone can be used to penetrate the pellicle layer and lead to effective control of engineered expression in a newly isolated strain of Komagataeibacter rhaeticus (25). Maximum titers of 15.3 g/liter at production rates of up to 3.1 g/liter/h have been achieved in bioreactor fermentations with Acetobacter (Table 1) (26).
Figure 2.
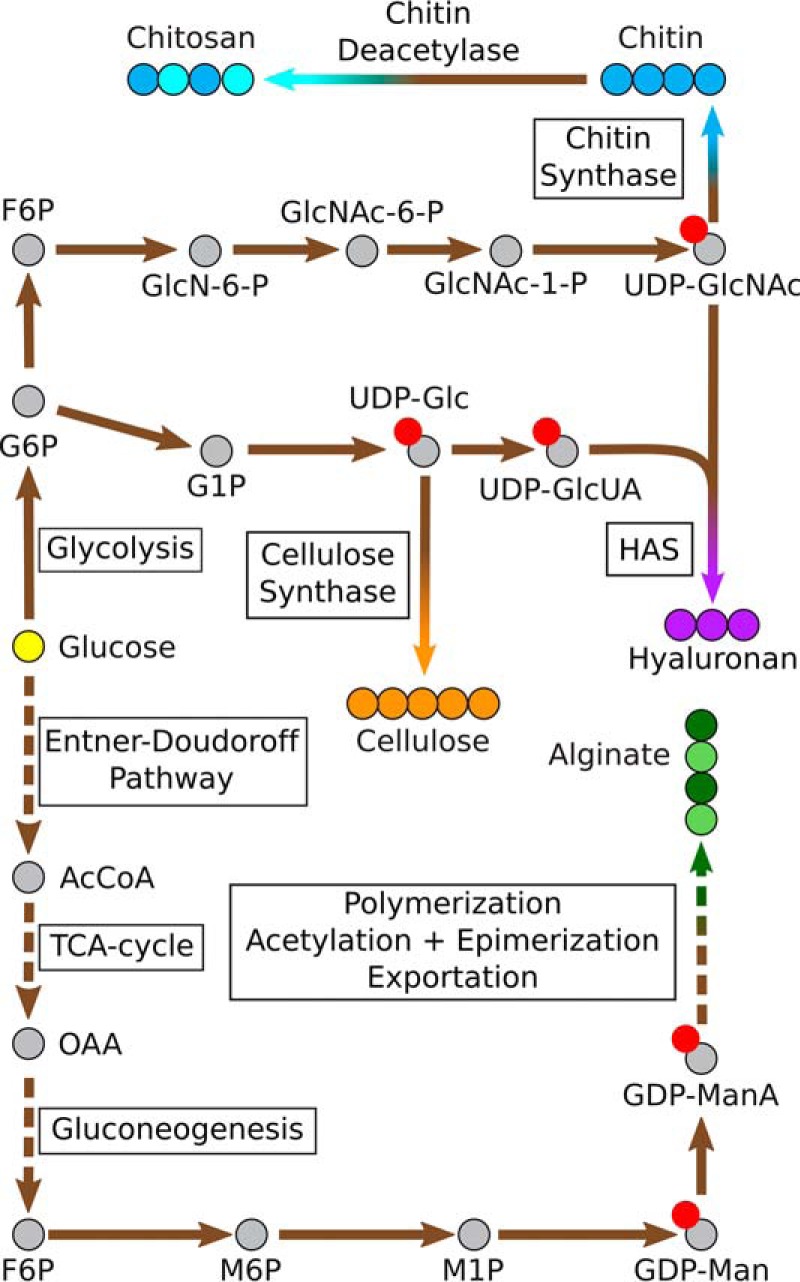
Overview of biosynthetic routes to biopolymers. The main steps in the microbial biosynthetic routes for cellulose, alginate, chitin, chitosan, and hyaluronan from glucose are briefly depicted. The solid arrow represents an enzymatic step, and the broken arrow represents a multistep pathway that includes a number of enzymatic steps. The yellow circle represents the starting material, glucose; gray circles represent intermediate metabolites; red circles represent a sugar nucleotide; and blue, orange, purple, and green circles represent product biopolymer molecules. Abbreviations used are as follows: AcCoA, acetyl coenzyme-A; BC, bacterial cellulose; F6P, fructose 6-phosphate; G1P, glucose 1-phosphate; G6P, glucose 6-phosphate; GDP-Man, GDP-mannose; GDP-ManA, GDP-mannuronic acid; GlcN-6-P, glucosamine 6-phosphate; GlcNAc-1-P, N-acetylglucosamine 1-phosphate; GlcNAc-6-P, N-acetylglucosamine 6-phosphate; M1P, mannose 1-phosphate; M6P, mannose 6-phosphate; OAA, oxaloacetate; TCA, tricarboxylic acid; UDP-Glc, UDP-glucose; UDP-GlcNAc, UDP-N-acetylglucosamine; UDP-GlcUA, UDP-glucuronic acid.
Alginate
Alginates are natural biopolymers that are abundant in marine brown algae (Phaeophyceae) (27) and in Pseudomonas and Azotobacter genera of bacteria (28). These EPS are the major structural component of algal cell walls, comprising up to 40–45% of the total algal dry matter (29), whereas in Pseudomonas and Azotobacter, alginates contribute to highly-structured biofilm-matrix and cyst-wall formations, respectively (30). Commercially, alginates have widespread use as stabilizers, viscosifiers, and gelling agents in food, cosmetics, beverage, paper, printing, and pharmaceutical industries (28). Globally, the demand for alginates was valued at USD 624 million in 2016, and the demand is projected to reach USD 923.8 million by 2025 with a consumption volume of 21,516 tons (31).
Structurally, alginates are a family of linear, non-repeating block copolymers consisting of variable ratios of β-d-mannuronic acid and its C5 epimer α-l-guluronic acid linked by β-(1,4)-glycosidic bonds (Fig. 1B) (32). Variation in the molar ratios of β-d-mannuronic acid to α-l-guluronic acid residues controls the molecular weight and material properties of alginates. Because of their unique water-retaining capability, biocompatibility, low toxicity, relatively low cost of production, and temperature-independent mild gelation (sol-gel transition) ability in the presence of multivalent cations (e.g. Ca2+), alginates are excellent biomaterials for use in biomedical applications, including wound healing, dental implants, drug delivery systems, tissue engineering, and regenerative medicine (33).
The alginate biosynthesis pathway has been extensively investigated in Pseudomonas aeruginosa (34) and Azotobacter vinelandii (35), but the complex polymerization, transport, and secretion system, as well as regulatory mechanisms controlling the pathway, are not fully understood. Metabolic engineering efforts for increasing alginate production are still in their infancy. Alginate production is tightly regulated in bacteria, and thus efforts have been made to characterize and engineer the regulatory system in Pseudomonas fluorescens (36) to identify the correlation between precursor availability and alginate production. Maleki et al. (37) showed increased alginate production (2.2 g/liter) from glycerol in an engineered P. fluorescens strain, in which deletion of glucose-6-phosphate dehydrogenase redirected more carbon flux through the Entner-Doudoroff pathway to produce alginate precursor fructose 6-phosphate (Fig. 2). A similar correlation between alginate production and precursor availability was observed in another recent study with P. fluorescens (38). Studies with A. vinelandii (39) utilized a different strategy, in which increased carbon fluxes were channeled toward alginate production by disrupting the competing polyhydroxybutyrate pathway. Implementing this strategy resulted in an alginate titer of up to 6.6 g/liter with a yield of 1.9 g/g sucrose. Single gene-deletion studies in A. vinelandii (39, 40) also increased alginate yield (0.66 g/g on sucrose) (Table 1) and lowered the degree of acetylation for altered molecular composition. More recently, alginate overproduction was reported during biofilm formation in a newly discovered strain Pseudomonas mandelii 6A1 (41) that was isolated from Antarctica (41). Biofilm formation by the strain was increased at lower temperatures due to increased alginate productivity, which in turn was correlated to the down-regulation of the regulatory protein MucA, which acts as a repressor in the alginate operon (41).
Synthetic biology strategies for aminopolysaccharide biosynthesis
Chitin and chitosan
The aminopolysaccharide monomer glucosamine (GlcN) and its derivative N-acetylglucosamine (GlcNAc) are attractive candidates for microbial biosynthesis because of the facility of the amine group for functionalization and the utility for subsequent polymerization into chitin and chitosan (Fig. 2). GlcN and GlcNAc are glucose moieties with C2 hydroxyl substitution by an amino group and acetylated amino group, respectively. Aminopolysaccharides and subsequent biopolymers have traditionally been obtained through strong acid hydrolysis of chitin from shellfish. In recent years, microbial production of GlcN and derivatives has been demonstrated in a variety of hosts including E. coli, Bacillus subtilis, and Saccharomyces cerevisiae (42).
In E. coli, expression of GlcNAc transferase and deletion of nagE, a GlcNAc transporter, increased GlcN titer to 17 g/liter (43). The GlcNAc synthesis module has been strengthened at the transcriptional level by increasing enzyme expression through testing a range of promoters for two key enzymes, glucosamine synthase and glucosamine acetyltransferase (44). Elimination of acidic by-products was accomplished by knocking out ldh and pta of the lactate and acetate synthetic pathways, respectively. By overexpressing glucosamine synthase (GlmS), inactivating catabolic genes, and utilizing a two-stage fed-batch fermentation, Deng et al. (43) achieved GlcN titers of up to 110 g/liter in E. coli in a fed-batch fermentation (Table 1).
In B. subtilis, expression of various combinations of synthetic small regulatory RNAs and the Hfq protein, designed to repress glycolysis by targeting pfk (encoding phosphofructokinase) and peptidoglycan synthesis by targeting glmM (encoding phosphoglucosamine mutase), improved GlcNAc titers to 31.65 g/liter in a 3-liter fed-batch bioreactor (45). With a dynamic metabolomics approach, Liu et al. (46) found that a futile cycle between N-acetylglucosamine-6-phosphate (GlcNAc-6-P) and GlcNAc is the primary challenge for pathway productivity, due to high energy demands of ATP phosphorylation–dephosphorylation. Deletion of the responsible glucokinase doubles GlcNAc productivity through a dual effect of increasing ATP and restoring healthy growth to the cell.
In S. cerevisiae, a synthetic suicide riboswitch that regulated growth in response to the precursor GlcN-6-P was applied to screen for overproducers of GlcNAc. The growth-coupled circuit allowed for screening of an effective glutamine-fructose-6-phosphate transaminase (GFA1) mutant and haloacid dehalogenase-like (HAD) phosphatase (47). The mutant contained changes in GFA1 expression, which is the first and rate-limiting step of chitin biosynthesis, along with overexpression of HAD phosphatase YqaB, specific for conversion of GlcNAc-6-P to GlcNAc. Subsequently, GlcNAc production was further improved by reducing glycolytic flux by the disruption of pfk-2, achieving titers of 1.2 g/liter when fed glucose and 1.8 g/liter when fed galactose, in shake flask fermentation (48). Under galactose feeding, deletion of pfk-2 allowed for enough reduction of glycolysis to activate gluconeogenesis thus allowing for galactose to be used as a sole carbon source.
Microbial production challenges in the biosynthesis of GlcN and GlcNAc include feedback inhibition effects where GlcN-6-P is a strong inhibitor of GlcN synthase and GlcN degradation, thus limiting the accumulation of GlcN inside the cell (Fig. 2) (49). Additionally, aminosugars can serve as alternative carbon and nitrogen sources, so it is difficult to achieve high titers in culture broth, unless a recovery strategy is incorporated in the fermentation (43). UDP-N-acetylglucosamine (UDP-GlcNAc) is normally maintained at high intracellular concentrations in growing bacterial cells to balance growth and production as the sugar donor for the synthesis of N-acetylated chitooligosaccharide, the precursor for the biosynthesis of peptidoglycan. Aminosugars containing free amino groups are unstable in aqueous solution at neutral pH where GlcN can undergo spontaneous rearrangement and dimerization to form fructosazine, d-arabinose, and pyrazine derivatives, among others (50). Thus, biopolymerization is advantageous to circumvent degradation issues. Opportunities exist for combining strategies for GlcN and GlcNAc overproduction for subsequent biopolymerization.
Chitin is the second most abundant biopolymer on the planet and is found in almost all fungi, many animals (invertebrates), several protists, and a few algae, playing an essential role in structure. Over 800 putative chitin synthases associated with 130 genomes have been identified (51). Chitin is a hexosamine biopolymer composed of as many as 5,000 β-(1,4)-glycosidically linked GlcNAc units cross-linked by hydrogen bonding (Fig. 1C). Chitosans are deacetylated chitin, as heteropolymers of GlcNAc and GlcN units (Fig. 1D). Chitosans are variable mixtures of molecules depending on the degree of polymerization and the degree of acetylation and are valuable functional biopolymers due to their physicochemical and biological compatibility. Chitosans have many agricultural, industrial, and biomedical applications, including use as an agricultural agent for plant defense and yield increase (52), drug delivery (53), wound healing (54), water filtration (55), and bio-printing (2). Chitosan trisaccharide is a valuable precursor for synthesis of epitopes such as type II blood group antigens (56). The global market size for chitin and chitosan was valued at USD 3.19 billion in 2015 and is projected to reach USD 17.84 billion by 2025 (57, 58). Microbial production of chitin and chitosan offers a green alternative to shellfish harvesting and allows for control of degree of polymerization and acetylation (59). van Montagu and co-workers (60) pioneered a cell factory approach for chitin oligosaccharide biosynthesis in E. coli by functional expression of the nod gene cluster from Rhizobium. NodC is a chitin oligomer synthase (Fig. 2) producing fully acetylated chitin oligomers of 2–5 saccharide residues (61), where the C-terminal domain of transmembrane NodC controls chain length (62). In vitro exposure of chitin oligosaccharides to enzymes NodB from Rhizobium sp. GRH2 and COD from Vibrio cholerae allowed for specific patterning of deacetylated chitosan oligomers (63). In 1997, Samain et al. (64) demonstrated the production of gram amounts of a perfectly defined, mono-deacetylated chito-pentose with a GlcN unit at the non-reducing end in E. coli (Table 1). Novel chitosan oligomers have been obtained, such as N-acetyl-lactosamine (65) and thio-chito-oligosaccharide analogs, where the oxygen glycosidic linkage is replaced with sulfur for improved stability against hydrolysis by chitinases (66).
Hyaluronan
Hyaluronic acid (HA), also known as hyaluronan, is known for its structural role in the extracellular matrix of vertebrate epithelial, neural, and connective tissues like cartilage. HA is a linear copolymer of disaccharide units of β(1→3)-GlcNAc and β(1→4)-glucuronic acid (Fig. 1E), produced in varying molecular weights, by vertebrates and prokaryotes. The repeating carboxylate groups from glucuronic acid moieties are highly hydrophilic and HA polymers have been incorporated for water retention and viscosity properties in cosmetics for more than 100 years. Purified high molecular weight preparations of HA elicit no detectable inflammatory response in mammalian cells, making HA a key functional material in designs for surgical biomaterials and cell scaffolds (67) such as HA-based hydrogels enabling the successful culture of rod photoreceptors in vitro (68). The global demand for HA was estimated to be worth USD 7.2 billion in 2016; thanks to the steady increase in demand, the global market size is projected to reach USD 15.4 billion by 2025 (69).
Traditionally, HA is purified from animal tissues such as rooster combs and umbilical cords. A lower cost bacterial fermentation method has been developed by leveraging the natural producer Streptococcus zooepidemicus. However, natural microbial HA pathways have typically evolved as a masking technique to hide invading cells within the human body, and S. zooepidemicus is itself a recognized human pathogen. Endotoxins and viral contaminants from animal and pathogen sources are a source of concern and have driven development of sustainable Generally Recognized as Safe (GRAS) alternatives. Synthesis of HA has been demonstrated in B. subtilis (70), Lactococcus lactis (71), and Pichia pastoris (72). In bacteria, HA is synthesized from UDP-glucuronic acid (UDP-GlcUA) and UDP-GlcNAc by a single enzyme complex hyaluronan synthase (HAS) (Fig. 2). UDP-GlcUA and UDP-GlcNAc occur naturally as part of cell wall synthesis, which directly competes with HA synthesis. The relative abundance of precursors and HAS has a definitive effect on chain length and average molecular weight of resulting HA polymers (73).
A key challenge for HA and most polysaccharide syntheses stems from the tradeoff between a high ATP and NAD+ requirements leading to high dissolved oxygen (DO) requirements for electron cycling versus low molecular diffusion in high-viscosity cultures as the concentration of high-molecular-weight polymer increases. DO has a critical effect on molecular weight, possibly by affecting the abundance of precursors (74). The desired outcome of high titer leads to poor mixing and an increasingly anaerobic environment. Countering this effect mechanically with increased aeration and mixing is energy-intensive and impractical at higher viscosities. In strains capable of anaerobic fermentation, this leads to elevated levels of fermentation products such as lactic acid, which limits HA production (S. zooepidemicus) (75). In strains that are sensitive to low DO, such as B. subtilis, the anaerobic environment leads to early cessation of production at 3 g/liter (76). This limitation in B. subtilis was overcome by controlled expression of hyaluronidase to reduce molecular weight and viscosity of the culture with the tradeoff that smaller chains are produced (6 × 103 Da versus 6 × 106 Da) (76). In anaerobic-tolerant Corynebacterium glutamicum, heterologous expression of the HA pathway with knockout of lactate dehydrogenase allowed for accumulation of 21 g/liter HA with a mid-range mass of 2 × 105–8 × 105 Da (77) (Table 1). In S. zooepidemicus, a recombinant suicide plasmid added to prevent natural expression of hyaluronidase, yielded 9 g/liter of a higher molecular weight product (78).
Summary and future outlook
Metabolic engineering and synthetic biology strategies have advanced the techniques for microbial production of biopolymers and promise sustainable and reliable alternatives to current production from natural sources. Synthetic biology strategies highlighted include the implementation of a riboswitch to balance glycolytic flux in the biosynthesis of chitin and HA precursor GlcNAc (47), expression balancing with promoter replacement of synthase and acetyltransferase genes in the biosynthesis of GlcNAc (45), redirection of carbon flux by deleting glycolytic genes in the biosynthesis of alginate (37), and preventing expression of degradative enzymes like hyaluronidase for HA biosynthesis (78).
Although efforts have successfully demonstrated microbial biopolymer production, there are still challenges to address: 1) understanding competition for endogenous cellular resources, such as precursor sugar nucleotide pools and energy requirements (e.g. ATP, NAD+); 2) transcriptional regulation, where synthesis is tightly regulated and controlled by complex regulatory machinery, which functions when cells need to construct structural components like EPS that relate to pathogenicity and defense mechanisms; 3) in vivo biopolymerization, where there is a need for better characterization of polymerization enzymes (79) and the steps in EPS biosynthesis, even though gene clusters have been known for several years (80); and finally, extracellular secretion, which poses process engineering challenges where titer is limited by viscosity leading to mass transfer issues (81). Host compatibility should also be considered, for example, the robustness of model organisms like E. coli to industrial conditions.
To address these challenges, the advent of new technologies and approaches is critical. Although dynamic regulation of carbon fluxes has been implemented in monomeric carbohydrate biosynthesis like glucaric acid (82), application of similar strategies for biopolymer synthesis has yet to be explored. Precursor supply by regenerating sugar nucleotides can balance cellular resources (42), and genome-level metabolic modeling of microbial cell factories is instrumental for optimizing the performance of heterologous biopolymer-producing pathways (83). For example, model-guided metabolic engineering followed by experimental validation of growth-coupled glycan-overproducing strains identified metabolic imbalances that rerouted flux toward glycan precursor synthesis (83). 13C metabolic flux analysis is another powerful tool for identifying pathway bottlenecks (84) in the optimization of microbial biopolymer synthesis (84). DNA sequencing (85) and synthesis technologies coupled to machine learning (86), along with the development of CRISPR–Cas9 gene-editing technologies, have allowed for increased engineering efficiency (87). Bioprospecting for new sequences and functions (88) can help characterize polymerization protein complexes and also help identify novel molecular targets for the potential of tailor-made mixed biopolymers of varying material properties made from functionalized block copolymers.
Copolymer formulations provide further opportunities for tailoring, where strategies of metabolic engineering and growth medium modifications can help control biopolymer compositions (89). Yadav et al. (90, 91) demonstrated bacterial production of a hybrid cellulose-chitin copolymer for biomedical applications where lysozyme susceptibility allowed for in vivo biodegradation. Structurally, the presence of GlcNAc in bacterial cellulose disrupts the highly ordered cellulose crystalline structure, thus transforming the cellulose type Iα structure to cellulose-chitin type II due to alterations in fibril–fibril interactions (92). Further functionalization of modified bacterial cellulose through deacetylation can generate materials with a reactive amine surface that allows for various applications, such as engineering novel biocomposites, tissue engineering scaffolding, biosensor small molecule detection, and drug delivery vehicles (90).
For microbial production of biopolymers to be considered a green technology, important criteria, including energy efficiency, material efficiency, land use, and costs metrics need to be assessed. Whereas building block chemicals such as lactic acid and isoprene have been assessed by green metrics (93), an opportunity exists for assessing other valuable biosynthesized materials. Microbially-produced lactic acid and the polymer polylactic acid have higher economic efficiency over chemically-produced similar materials due to increased energy efficiency and fermentation-driven stereoselection of d(−)- or l(+)-lactic acid (94). Moving beyond cellular control, more opportunities for green processing exist, including valorization with CO2 and utilization of feedstocks from biodegradation of waste products such as bioplastics (95). Other metrics such as life cycle assessment, the E-factor, and principles of green chemistry should be implemented to drive a circular and sustainable economy forward with reduced waste and conservation of resources. The environmental and health risk of biopolymers, for instance fluorine-containing bioplastics (15), need to be addressed. Biopolymer microbial synthesis could also be suitable in sustainable systems such as space stations or intergalactic habitats because of its renewability and replicability. There is an overlap between closed systems designed for space habitation and “green” technology on Earth (96).
Emerging technologies for in silico design and predictions of material properties will help advance the cell factory approach to biopolymer production such as in the creation of biomimetic scaffolds composed of 3D cell culture polysaccharide hydrogels (97). Opportunities also exist for combination of the cell factory approach with manufacturing such as in the controlled biofilm layering by EPS secreting microbes biofabricated by 3D printing, along with advances in 4D printed biomaterials with integrated “smart” diagnostics (98). Imagine a world where microbes are full cell factories, not just making single molecules but assembling entire functional materials.
Acknowledgments
We thank John W. K. Oliver, Kristina Haslinger (MIT ChemE Communication Lab Fellow), and Nicolas R. Ball-Jones for helpful discussions regarding the manuscript.
This work was supported by National Institutes of Health Grant P50 GM098792 from NIGMS and National Science Foundation Grant MCB-1517913. This is the fifth article in the Thematic Minireview series “Green biological chemistry.” The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- EPS
- exopolysaccharide
- BC
- bacterial cellulose
- BCS
- bacterial cellulose synthase
- DO
- dissolved oxygen
- GFA1
- glutamine-fructose-6-phosphate transaminase
- GlcN-6-P
- glucosamine 6-phosphate
- GlcNAc-6-P
- N-acetylglucosamine-6-phosphate
- HA
- hyaluronic acid
- HAD
- haloacid dehalogenase-like
- HAS
- hyaluronan synthase
- USD
- United States dollars
- CRISPR
- clustered regularly interspaced short palindromic repeat.
References
- 1. Patrulea V., Ostafe V., Borchard G., and Jordan O. (2015) Chitosan as a starting material for wound healing applications. Eur. J. Pharm. Biopharm. 97, 417–426 10.1016/j.ejpb.2015.08.004 [DOI] [PubMed] [Google Scholar]
- 2. Fernandez J. G., and Ingber D. E. (2014) Manufacturing of large-scale functional objects using biodegradable chitosan bioplastic. Macromol. Mater. Eng. 299, 932–993 10.1002/mame.201300426 [DOI] [Google Scholar]
- 3. Trombotto S., Ladavière C., Delolme F., and Domard A. (2008) Chemical preparation and structural characterization of a homogeneous series of chitin/chitosan oligomers. Biomacromolecules 9, 1731–1738 10.1021/bm800157x [DOI] [PubMed] [Google Scholar]
- 4. Morais A. R. C., Dworakowska S., Reis A., Gouveia L., Matos C. T., Bogdał D., and Bogel-Łukasik R. (2015) Chemical and biological-based isoprene production: Green metrics. Catal. Today 239, 38–43 10.1016/j.cattod.2014.05.033 [DOI] [Google Scholar]
- 5. Moon T. S., Yoon S.-H., Lanza A. M., Roy-Mayhew J. D., and Prather K. L. (2009) Production of glucaric acid from a synthetic pathway in recombinant Escherichia coli. Appl. Environ. Microbiol. 75, 589–595 10.1128/AEM.00973-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Qian Z.-G., Xia X.-X., and Lee S. Y. (2009) Metabolic engineering of Escherichia coli for the production of putrescine: a four carbon diamine. Biotechnol. Bioeng. 104, 651–662 [DOI] [PubMed] [Google Scholar]
- 7. Jung Y. K., Kim T. Y., Park S. J., and Lee S. Y. (2010) Metabolic engineering of Escherichia coli for the production of polylactic acid and its copolymers. Biotechnol. Bioeng. 105, 161–171 10.1002/bit.22548 [DOI] [PubMed] [Google Scholar]
- 8. Oliver J. W., Machado I. M., Yoneda H., and Atsumi S. (2013) Cyanobacterial conversion of carbon dioxide to 2,3-butanediol. Proc. Natl. Acad. Sci. U.S.A. 110, 1249–1254 10.1073/pnas.1213024110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Criddle C. S., Billington S. L., and Frank C. W. (2014) Renewable bioplastics and biocomposites from biogas methane and waste-derived feedstock: development of enabling technology, life cycle assessment, and analysis of costs. California Department of Resources Recycling and Recovery, State of California [Google Scholar]
- 10. Adkins J., Pugh S., McKenna R., and Nielsen D. R. (2012) Engineering microbial chemical factories to produce renewable “biomonomers.” Front. Microbiol. 3, 313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Qin Y., Lu X., Sun N., and Rogers R. D. (2010) Dissolution or extraction of crustacean shells using ionic liquids to obtain high molecular weight purified chitin and direct production of chitin films and fibers. Green Chem. 12, 968–971 10.1039/C003583A [DOI] [Google Scholar]
- 12. Zhao X., Chen Z., Gu G., and Guo Z. (2016) Recent advances in the research of bacterial glucuronosyltransferases. J. Carbohydr. Chem. 35, 201–223 10.1080/07328303.2016.1205597 [DOI] [Google Scholar]
- 13. Rehm B. H. (2015) Synthetic biology towards the synthesis of custom-made polysaccharides. Microb. Biotechnol. 8, 19–20 10.1111/1751-7915.12241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schmid J., Sieber V., and Rehm B. (2015) Bacterial exopolysaccharides: biosynthesis pathways and engineering strategies. Front. Microbiol. 6, 496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thuronyi B. W., Privalsky T. M., and Chang M. C. (2017) Engineered fluorine metabolism and fluoropolymer production in living cells. Angew. Chem. Int. Ed. Engl. 56, 13637–13640 10.1002/anie.201706696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cellulose Fiber Market Size & Share Industry Report, 2014–2025 (online) https://www.grandviewresearch.com/industry-analysis/cellulose-fibers-market (Accessed December 20, 2017) (Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.)
- 17. Brown R. M. Jr., Willison J. H., and Richardson C. L. (1976) Cellulose biosynthesis in Acetobacter xylinum: visualization of the site of synthesis and direct measurement of the in vivo process. Proc. Natl. Acad. Sci. U.S.A. 73, 4565–4569 10.1073/pnas.73.12.4565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dayal M. S., and Catchmark J. M. (2016) Mechanical and structural property analysis of bacterial cellulose composites. Carbohydr. Polym. 144, 447–453 10.1016/j.carbpol.2016.02.055 [DOI] [PubMed] [Google Scholar]
- 19. Lima F. M., Pinto F. C., Andrade-da-Costa B. L., Silva J. G., Campos O. Jr., and Aguiar J. L. (2017) Biocompatible bacterial cellulose membrane in dural defect repair of rat. J. Mater. Sci. Mater. Med. 28, 37 10.1007/s10856-016-5828-9 [DOI] [PubMed] [Google Scholar]
- 20. Ng A. (2017) ISWC '17, Proceedings of the 2017 ACM International Symposium on Wearable Computers, pp. 209–214, September 11–15, 2017, ACM, New York [Google Scholar]
- 21. Huang Y., Zhu C., Yang J., Nie Y., Chen C., and Sun D. (2014) Recent advances in bacterial cellulose. Cellulose 21, 1–30 10.1007/s10570-013-0088-z [DOI] [Google Scholar]
- 22. Omadjela O., Narahari A., Strumillo J., Mélida H., Mazur O., Bulone V., and Zimmer J. (2013) BcsA and BcsB form the catalytically active core of bacterial cellulose synthase sufficient for in vitro cellulose synthesis. Proc. Natl. Acad. Sci. U.S.A. 110, 17856–17861 10.1073/pnas.1314063110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Imai T., Sun S.-J., Horikawa Y., Wada M., and Sugiyama J. (2014) Functional reconstitution of cellulose synthase in Escherichia coli. Biomacromolecules 15, 4206–4213 10.1021/bm501217g [DOI] [PubMed] [Google Scholar]
- 24. Basu S., Omadjela O., Gaddes D., Tadigadapa S., Zimmer J., and Catchmark J. M. (2016) Cellulose microfibril formation by surface-tethered cellulose synthase enzymes. ACS Nano 10, 1896–1907 10.1021/acsnano.5b05648 [DOI] [PubMed] [Google Scholar]
- 25. Florea M., Hagemann H., Santosa G., Abbott J., Micklem C. N., Spencer-Milnes X., de Arroyo Garcia L., Paschou D., Lazenbatt C., Kong D., Chughtai H., Jensen K., Freemont P. S., Kitney R., Reeve B., and Ellis T. (2016) Engineering control of bacterial cellulose production using a genetic toolkit and a new cellulose-producing strain. Proc. Natl. Acad. Sci. U.S.A. 113, E3431–E3440 10.1073/pnas.1522985113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hwang J. W., Yang Y. K., Hwang J. K., Pyun Y. R., and Kim Y. S. (1999) Effects of pH and dissolved oxygen on cellulose production by Acetobacter xylinum BRC5 in agitated culture. J. Biosci. Bioeng. 88, 183–188 10.1016/S1389-1723(99)80199-6 [DOI] [PubMed] [Google Scholar]
- 27. Draget K. I., Smidsrød O., and Skjåk-Bræk G. (2005) Alginates from algae. in Biopolymers Online, Wiley-VCH Verlag GmbH & Co. KGaA; 10.1002/3527600035.bpol6008 [DOI] [Google Scholar]
- 28. Hay I. D., Ur Rehman Z., Moradali M. F., Wang Y., and Rehm B. H. (2013) Microbial alginate production, modification and its applications. Microb. Biotechnol. 6, 637–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Skjåk-Bræk G., and Draget K. I. (2012) Alginates: properties and applications. in Polymer Science: A Comprehensive Reference (Möller K. M., ed) ScienceDirect®, Elsevier B.V., Amsterdam: 10.1016/B978-0-444-53349-4.00261-2 [DOI] [Google Scholar]
- 30. Rehm B. H. (ed) (2009) in Alginates: Biology and Applications, pp. 55–71, Springer, Berlin [Google Scholar]
- 31. Alginate Market Size Worth $923.8 Million By 2025 | CAGR: 4.5% (online) https://www.grandviewresearch.com/press-release/global-alginate-market (Accessed December 20, 2017) (Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.)
- 32. Rehm B. H. (2010) Bacterial polymers: biosynthesis, modifications and applications. Nat. Rev. Microbiol. 8, 578–592 10.1038/nrmicro2354 [DOI] [PubMed] [Google Scholar]
- 33. Lee K. Y., and Mooney D. J. (2012) Alginate: properties and biomedical applications. Prog. Polym. Sci. 37, 106–126 10.1016/j.progpolymsci.2011.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chitnis C. E., and Ohman D. E. (1993) Genetic analysis of the alginate biosynthetic gene cluster of Pseudomonas aeruginosa shows evidence of an operonic structure. Mol. Microbiol. 8, 583–593 10.1111/j.1365-2958.1993.tb01602.x [DOI] [PubMed] [Google Scholar]
- 35. Remminghorst U., and Rehm B. H. (2006) In vitro alginate polymerization and the functional role of Alg8 in alginate production by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 72, 298–305 10.1128/AEM.72.1.298-305.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lien S. K., Sletta H., Ellingsen T. E., Valla S., Correa E., Goodacre R., Vernstad K., Borgos S. E., and Bruheim P. (2013) Investigating alginate production and carbon utilization in Pseudomonas fluorescens SBW25 using mass spectrometry-based metabolic profiling. Metabolomics 9, 403–417 10.1007/s11306-012-0454-0 [DOI] [Google Scholar]
- 37. Maleki S., Mærk M., Valla S., and Ertesvåg H. (2015) Mutational analyses of glucose dehydrogenase and glucose-6-phosphate dehydrogenase genes in Pseudomonas fluorescens reveal their effects on growth and alginate production. Appl. Environ. Microbiol. 81, 3349–3356 10.1128/AEM.03653-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ertesvåg H., Sletta H., Senneset M., Sun Y. Q., Klinkenberg G., Konradsen T. A., Ellingsen T. E., and Valla S. (2017) Identification of genes affecting alginate biosynthesis in Pseudomonas fluorescens by screening a transposon insertion library. BMC Genomics 18, 11 10.1186/s12864-016-3467-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Peña C., Miranda L., Segura D., Núñez C., Espín G., and Galindo E. (2002) Alginate production by Azotobacter vinelandii mutants altered in poly-β-hydroxybutyrate and alginate biosynthesis. J. Ind. Microbiol. Biotechnol. 29, 209–213 10.1038/sj.jim.7000310 [DOI] [PubMed] [Google Scholar]
- 40. Gaytán I., Peña C., Núñez C., Córdova M. S., Espín G., and Galindo E. (2012) Azotobacter vinelandii lacking the Na+-NQR activity: a potential source for producing alginates with improved properties and at high yield. World J. Microbiol. Biotechnol. 28, 2731–2740 10.1007/s11274-012-1084-4 [DOI] [PubMed] [Google Scholar]
- 41. Vásquez-Ponce F., Higuera-Llantén S., Pavlov M. S., Ramírez-Orellana R., Marshall S. H., and Olivares-Pacheco J. (2017) Alginate overproduction and biofilm formation by psychrotolerant Pseudomonas mandelii depend on temperature in Antarctic marine sediments. Electron. J. Biotechnol. 28, 27–34 10.1016/j.ejbt.2017.05.001 [DOI] [Google Scholar]
- 42. Hossain G. S., Shin H.-D., Li J., Wang M., Du G., Chen J., and Liu L. (2016) Metabolic engineering for amino-, oligo-, and polysugar production in microbes. Appl. Microbiol. Biotechnol. 100, 2523–2533 10.1007/s00253-015-7215-8 [DOI] [PubMed] [Google Scholar]
- 43. Deng M.-D., Severson D. K., Grund A. D., Wassink S. L., Burlingame R. P., Berry A., Running J. A., Kunesh C. A., Song L., Jerrell T. A., and Rosson R. A. (2005) Metabolic engineering of Escherichia coli for industrial production of glucosamine and N-acetylglucosamine. Metab. Eng. 7, 201–214 10.1016/j.ymben.2005.02.001 [DOI] [PubMed] [Google Scholar]
- 44. Liu Y., Zhu Y., Ma W., Shin H.-D., Li J., Liu L., Du G., and Chen J. (2014) Spatial modulation of key pathway enzymes by DNA-guided scaffold system and respiration chain engineering for improved N-acetylglucosamine production by Bacillus subtilis. Metab. Eng. 24, 61–69 10.1016/j.ymben.2014.04.004 [DOI] [PubMed] [Google Scholar]
- 45. Liu Y., Zhu Y., Li J., Shin H.-D., Chen R. R., Du G., Liu L., and Chen J. (2014) Modular pathway engineering of Bacillus subtilis for improved N-acetylglucosamine production. Metab. Eng. 23, 42–52 10.1016/j.ymben.2014.02.005 [DOI] [PubMed] [Google Scholar]
- 46. Liu Y., Link H., Liu L., Du G., Chen J., and Sauer U. (2016) A dynamic pathway analysis approach reveals a limiting futile cycle in N-acetylglucosamine overproducing Bacillus subtilis. Nat. Commun. 7, 11933 10.1038/ncomms11933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lee S.-W., and Oh M.-K. (2015) A synthetic suicide riboswitch for the high-throughput screening of metabolite production in Saccharomyces cerevisiae. Metab. Eng. 28, 143–150 10.1016/j.ymben.2015.01.004 [DOI] [PubMed] [Google Scholar]
- 48. Lee S.-W., and Oh M.-K. (2016) Improved production of N-acetylglucosamine in Saccharomyces cerevisiae by reducing glycolytic flux. Biotechnol. Bioeng. 113, 2524–2528 10.1002/bit.26014 [DOI] [PubMed] [Google Scholar]
- 49. Sitanggang A. B., Wu H.-S., Wang S. S., and Ho Y.-C. (2010) Effect of pellet size and stimulating factor on the glucosamine production using Aspergillus sp. BCRC 31742. Bioresour. Technol. 101, 3595–3601 10.1016/j.biortech.2009.12.084 [DOI] [PubMed] [Google Scholar]
- 50. Dutta U., and Dain J. A. (2005) Capillary electrophoretic analysis of advanced glycation end products formed from the reaction of reducing sugars with the amino group of glucosamine. Anal. Biochem. 343, 237–243 10.1016/j.ab.2005.05.026 [DOI] [PubMed] [Google Scholar]
- 51. Gonçalves I. R., Brouillet S., Soulié M.-C., Gribaldo S., Sirven C., Charron N., Boccara M., and Choquer M. (2016) Genome-wide analyses of chitin synthases identify horizontal gene transfers towards bacteria and allow a robust and unifying classification into fungi. BMC Evol. Biol. 16, 252 10.1186/s12862-016-0815-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang S.-L., Yen Y.-H., Tsiao W.-J., Chang W.-T., and Wang C.-L. (2002) Production of antimicrobial compounds by Monascus purpureus CCRC31499 using shrimp and crab shell powder as a carbon source. Enzyme Microb. Technol. 31, 337–344 10.1016/S0141-0229(02)00135-7 [DOI] [Google Scholar]
- 53. Kadgwaki S., Saskiawan I., Watanabe J., Yamamoto K., Bunn M., Ichihara Y., and Kumagai H. (1997) Transglycosylation activity of N-acetylhexosaminidase from Penicillium oxalicurnum and its application to synthesis of a drug carrier. J. Ferment. Bioeng. 4, 341–345 10.1016/S0922-338X(97)80139-0 [DOI] [Google Scholar]
- 54. Azuma K., Izumi R., Osaki T., Ifuku S., Morimoto M., Saimoto H., Minami S., and Okamoto Y. (2015) Chitin, chitosan, and its derivatives for wound healing: old and new materials. J. Funct. Biomater. 6, 104–142 10.3390/jfb6010104 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55. Bhatnagar A., and Sillanpää M. (2009) Applications of chitin- and chitosan-derivatives for the detoxification of water and wastewater–a short review. Adv. Colloid Interface Sci. 152, 26–38 10.1016/j.cis.2009.09.003 [DOI] [PubMed] [Google Scholar]
- 56. Cottaz S., and Samain E. (2005) Genetic engineering of Escherichia coli for the production of NI,NII-diacetylchitobiose (chitinbiose) and its utilization as a primer for the synthesis of complex carbohydrates. Metab. Eng. 7, 311–317 10.1016/j.ymben.2005.05.004 [DOI] [PubMed] [Google Scholar]
- 57. Chitin and Chitosan Derivatives: Technologies, Applications and Global Markets (online) https://www.reportbuyer.com/product/4806925/chitin-and-chitosan-derivatives-technologies-applications-and-global-markets.html (Accessed December 20, 2017) (Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.)
- 58. Chitosan Market Size To Reach USD 17.84 Billion By 2025 (online) https://www.grandviewresearch.com/press-release/chitosan-market-analysis (Accessed December 20, 2017) (Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.)
- 59. Naqvi S., and Moerschbacher B. M. (2017) The cell factory approach toward biotechnological production of high-value chitosan oligomers and their derivatives: an update. Crit. Rev. Biotechnol. 37, 11–25 10.3109/07388551.2015.1104289 [DOI] [PubMed] [Google Scholar]
- 60. Mergaert P., D'Haeze W., Geelen D., Promé D., Van Montagu M., Geremia R., Promé J. C., and Holsters M. (1995) Biosynthesis of Azorhizobium caulinodans Nod factors. Study of the activity of the NodABCS proteins by expression of the genes in Escherichia coli. J. Biol. Chem. 270, 29217–29223 10.1074/jbc.270.49.29217 [DOI] [PubMed] [Google Scholar]
- 61. Geremia R. A., Mergaert P., Geelen D., Van Montagu M., and Holsters M. (1994) The NodC protein of Azorhizobium caulinodans is an N-acetylglucosaminyltransferase. Proc. Natl. Acad. Sci. U.S.A. 91, 2669–2673 10.1073/pnas.91.7.2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kamst E., Pilling J., Raamsdonk L. M., Lugtenberg B. J., and Spaink H. P. (1997) Rhizobium nodulation protein NodC is an important determinant of chitin oligosaccharide chain length in Nod factor biosynthesis. J. Bacteriol. 179, 2103–2108 10.1128/jb.179.7.2103-2108.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hamer S. N., Cord-Landwehr S., Biarnés X., Planas A., Waegeman H., Moerschbacher B. M., and Kolkenbrock S. (2015) Enzymatic production of defined chitosan oligomers with a specific pattern of acetylation using a combination of chitin oligosaccharide deacetylases. Sci. Rep. 5, 8716 10.1038/srep08716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Samain E., Drouillard S., Heyraud A., Driguez H., and Geremia R. A. (1997) Gram-scale synthesis of recombinant chitooligosaccharides in Escherichia coli. Carbohydr. Res. 302, 35–42 10.1016/S0008-6215(97)00107-9 [DOI] [PubMed] [Google Scholar]
- 65. Bettler E., Samain E., Chazalet V., Bosso C., Heyraud A., Joziasse D. H., Wakarchuk W. W., Imberty A., and Geremia A. R. (1999) The living factory: in vivo production of N-acetyllactosamine containing carbohydrates in E. coli. Glycoconj. J. 16, 205–212 10.1023/A:1007024320183 [DOI] [PubMed] [Google Scholar]
- 66. Southwick A. M., Wang L.-X., Long S. R., and Lee Y. C. (2002) Activity of Sinorhizobium meliloti NodAB and NodH enzymes on thiochitooligosaccharides. J. Bacteriol. 184, 4039–4043 10.1128/JB.184.14.4039-4043.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kim I. L., Mauck R. L., and Burdick J. A. (2011) Hydrogel design for cartilage tissue engineering: a case study with hyaluronic acid. Biomaterials 32, 8771–8782 10.1016/j.biomaterials.2011.08.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mitrousis N., Tam R. Y., Baker A. E. G., van der Kooy D., and Shoichet M. S. (2016) Hyaluronic acid-based hydrogels enable rod photoreceptor survival and maturation in vitro through activation of the mTOR pathway. Adv. Funct. Mater. 26, 1975–1985 10.1002/adfm.201504024 [DOI] [Google Scholar]
- 69. Hyaluronic Acid Market Size Worth USD 15.4 Billion by 2025| CAGR: 8.8% (online) https://www.grandviewresearch.com/press-release/global-hyaluronic-acid-market (Accessed December 20, 2017) (Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.)
- 70. Jia Y., Zhu J., Chen X., Tang D., Su D., Yao W., and Gao X. (2013) Metabolic engineering of Bacillus subtilis for the efficient biosynthesis of uniform hyaluronic acid with controlled molecular weights. Bioresour. Technol. 132, 427–431 10.1016/j.biortech.2012.12.150 [DOI] [PubMed] [Google Scholar]
- 71. Kaur M., and Jayaraman G. (2016) Hyaluronan production and molecular weight is enhanced in pathway-engineered strains of lactate dehydrogenase-deficient Lactococcus lactis. Metab. Engin. Commun. 3, 15–23 10.1016/j.meteno.2016.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Jeong E., Shim W. Y., and Kim J. H. (2014) Metabolic engineering of Pichia pastoris for production of hyaluronic acid with high molecular weight. J. Biotechnol. 185, 28–36 10.1016/j.jbiotec.2014.05.018 [DOI] [PubMed] [Google Scholar]
- 73. de Oliveira J. D., Carvalho L. S., Gomes A. M., Queiroz L. R., Magalhães B. S., and Parachin N. S. (2016) Genetic basis for hyper production of hyaluronic acid in natural and engineered microorganisms. Microb. Cell Fact. 15, 119 10.1186/s12934-016-0517-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Marcellin E., Steen J. A., and Nielsen L. K. (2014) Insight into hyaluronic acid molecular weight control. Appl. Microbiol. Biotechnol. 98, 6947–6956 10.1007/s00253-014-5853-x [DOI] [PubMed] [Google Scholar]
- 75. Liu L., Liu Y., Li J., Du G., and Chen J. (2011) Microbial production of hyaluronic acid: current state, challenges, and perspectives. Microb. Cell Fact. 10, 99 10.1186/1475-2859-10-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Jin P., Kang Z., Yuan P., Du G., and Chen J. (2016) Production of specific-molecular-weight hyaluronan by metabolically engineered Bacillus subtilis 168. Metab. Eng. 35, 21–30 10.1016/j.ymben.2016.01.008 [DOI] [PubMed] [Google Scholar]
- 77. Cheng F., Luozhong S., Guo Z., Yu H., and Stephanopoulos G. (2017) Enhanced biosynthesis of hyaluronic acid using engineered Corynebacterium glutamicum via metabolic pathway regulation. Biotechnol. J. 12, 10.1002/biot.201700191 [DOI] [PubMed] [Google Scholar]
- 78. Pourzardosht N., and Rasaee M. J. (2017) Improved yield of high molecular weight hyaluronic acid production in a stable strain of Streptococcus zooepidemicus via the elimination of the hyaluronidase-encoding gene. Mol. Biotechnol. 59, 192–199 10.1007/s12033-017-0005-z [DOI] [PubMed] [Google Scholar]
- 79. Taguchi S. (2017) Designer enzyme for green materials innovation: lactate-polymerizing enzyme as a key catalyst. Front. Chem. Sci. Eng. 11, 139–142 10.1007/s11705-017-1636-0 [DOI] [Google Scholar]
- 80. Schmid J., Sieber V., and Rehm B. (2015) Bacterial exopolysaccharides: biosynthesis pathways and engineering strategies. Front. Microbiol. 6, 496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Seviour R. J., McNeil B., Fazenda M. L., and Harvey L. M. (2011) Operating bioreactors for microbial exopolysaccharide production. Crit. Rev. Biotechnol. 31, 170–185 10.3109/07388551.2010.505909 [DOI] [PubMed] [Google Scholar]
- 82. Gupta A., Reizman I. M., Reisch C. R., and Prather K. L. (2017) Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit. Nat. Biotechnol. 35, 273–279 10.1038/nbt.3796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wayman J. A., Glasscock C., Mansell T., DeLisa M. P., and Varner J. (2017) Improving designer glycan production in Escherichia coli through model-guided metabolic engineering. bioRxiv. 10.1101/160853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Stephanopoulos G., Aristidou A. A., and Nielsen J. (1998) in Metabolic Engineering: Principles and Methodologies (Aristidou A. A., and Nielsen J., eds) pg. iii, Academic Press, San Diego [Google Scholar]
- 85. Jeffries T. W., Grigoriev I. V., Grimwood J., Laplaza J. M., Aerts A., Salamov A., Schmutz J., Lindquist E., Dehal P., Shapiro H., Jin Y.-S., Passoth V., and Richardson P. M. (2007) Genome sequence of the lignocellulose-bioconverting and xylose-fermenting yeast Pichia stipitis. Nat. Biotechnol. 25, 319–326 10.1038/nbt1290 [DOI] [PubMed] [Google Scholar]
- 86. Patel K. G., Welch M., and Gustafsson C. (2016) in Metabolic Engineering for Bioprocess Commercialization (Van Dien, ed). Chapter 4, Springer, Cham, Switzerland [Google Scholar]
- 87. Jakočiūnas T., Jensen M. K., and Keasling J. D. (2016) CRISPR/Cas9 advances engineering of microbial cell factories. Metab. Eng. 34, 44–59 10.1016/j.ymben.2015.12.003 [DOI] [PubMed] [Google Scholar]
- 88. Lee S.-M., Jellison T., and Alper H. S. (2015) Xylan catabolism is improved by blending bioprospecting and metabolic pathway engineering in Saccharomyces cerevisiae. Biotechnol. J. 10, 575–583 10.1002/biot.201400622 [DOI] [PubMed] [Google Scholar]
- 89. Chien L.-J., Chen H.-T., Yang P.-F., and Lee C.-K. (2006) Enhancement of cellulose pellicle production by constitutively expressing vitreoscilla hemoglobin in Acetobacter xylinum. Biotechnol. Prog. 22, 1598–1603 10.1021/bp060157g [DOI] [PubMed] [Google Scholar]
- 90. Yadav V., Paniliatis B. J., Shi H., Lee K., Cebe P., and Kaplan D. L. (2010) Novel in vivo-degradable cellulose-chitin copolymer from metabolically engineered Gluconacetobacter xylinus. Appl. Environ. Microbiol. 76, 6257–6265 10.1128/AEM.00698-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Yadav V., Sun L., Panilaitis B., and Kaplan D. L. (2015) In vitro chondrogenesis with lysozyme susceptible bacterial cellulose as a scaffold. J. Tissue Eng Regen. Med. 9, E276–E288 10.1002/term.1644 [DOI] [PubMed] [Google Scholar]
- 92. Yamamoto H., Horii F., and Hirai A. (1996) In situ crystallization of bacterial cellulose II. Influences of different polymeric additives on the formation of celluloses Iα and Iβ at the early stage of incubation. Cellulose 3, 229–242 10.1007/BF02228804 [DOI] [Google Scholar]
- 93. Sheldon R. A., and Sanders J. P. M. (2015) Toward concise metrics for the production of chemicals from renewable biomass. Catal. Today 239, 3–6 10.1016/j.cattod.2014.03.032 [DOI] [Google Scholar]
- 94. Juodeikiene G., Vidmantiene D., Basinskiene L., Cernauskas D., Bartkiene E., and Cizeikiene D. (2015) Green metrics for sustainability of biobased lactic acid from starchy biomass vs chemical synthesis. Catal. Today 239, 11–16 10.1016/j.cattod.2014.05.039 [DOI] [Google Scholar]
- 95. Garcia-Gonzalez L., Mozumder M. S., Dubreuil M., Volcke E. I., and De Wever H. (2015) Sustainable autotrophic production of polyhydroxybutyrate (PHB) from CO2 using a two-stage cultivation system. Catal. Today 257, 237–245 10.1016/j.cattod.2014.05.025 [DOI] [Google Scholar]
- 96. Rothschild L. J. (2016) Synthetic biology meets bioprinting: enabling technologies for humans on Mars (and Earth). Biochem. Soc. Trans. 44, 1158–1164 10.1042/BST20160067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Tam R. Y., Fisher S. A., Baker A. E., and Shoichet M. S. (2016) Transparent porous polysaccharide cryogels provide biochemically defined, biomimetic matrices for tunable 3D cell culture. Chem. Mater. 28, 3762–3770 10.1021/acs.chemmater.6b00627 [DOI] [Google Scholar]
- 98. Morouço P., Lattanzi W., and Alves N. (2017) 4D bioprinting as a new era for tissue engineering and regenerative medicine. Front. Bioeng. Biotechnol. 5, 61 10.3389/fbioe.2017.00061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Trujillo-Roldán M. A., Moreno S., Segura D., Galindo E., and Espín G. (2003) Alginate production by an Azotobacter vinelandii mutant unable to produce alginate lyase. Appl. Microbiol. Biotechnol. 60, 733–737 10.1007/s00253-002-1173-7 [DOI] [PubMed] [Google Scholar]


