Abstract
Amino acid availability is sensed by GCN2 (general control nonderepressible 2) and mechanistic target of rapamycin complex 1 (mTORC1), but how these two sensors coordinate their respective signal transduction events remains mysterious. In this study we utilized mouse genetic models to investigate the role of GCN2 in hepatic mTORC1 regulation upon amino acid stress induced by a single injection of asparaginase. We found that deletion of Gcn2 prevented hepatic phosphorylation of eukaryotic initiation factor 2α to asparaginase and instead unleashed mTORC1 activity. This change in intracellular signaling occurred within minutes and resulted in increased 5′-terminal oligopyrimidine mRNA translation instead of activating transcription factor 4 synthesis. Asparaginase also promoted hepatic mRNA levels of several genes which function as mTORC1 inhibitors, and these genes were blunted or blocked in the absence of Gcn2, but their timing could not explain the early discordant effects in mTORC1 signaling. Preconditioning mice with a chemical endoplasmic reticulum stress agent before amino acid stress rescued normal mTORC1 repression in the liver of Gcn2−/− mice but not in livers with both Gcn2 and the endoplasmic reticulum stress kinase, Perk, deleted. Furthermore, treating wildtype and Gcn2−/− mice with ISRIB, an inhibitor of PERK signaling, also failed to alter hepatic mTORC1 responses to asparaginase, although administration of ISRIB alone had an inhibitory GCN2-independent effect on mTORC1 activity. Taken together, the data show that activating transcription factor 4 is not required, but eukaryotic initiation factor 2α phosphorylation is necessary to prevent mTORC1 activation during amino acid stress.
Keywords: eukaryotic initiation factor 2 (eIF2), mTOR complex (mTORC), liver, endoplasmic reticulum stress (ER stress), amino acid, translation regulation, ATF4, GCN2, PERK
Introduction
A variety of cytoplasmic and proteotoxic stresses activates the integrated stress response (ISR),2 a conserved survival strategy in eukaryotes. In mammals, the ISR is triggered by a family of kinases with a shared substrate; namely, the α subunit of eukaryotic initiation factor 2 (eIF2). Phosphorylation of eIF2 at serine 51 leads to global and gene-specific changes in translation that, depending on the timing and intensity of the stress, direct the cell/tissue/organism toward adaptation or cell death (1).
The translation factor eIF2 delivers initiator Met-tRNA and GTP to the 40S ribosomal subunit during the initiation step and is then released from the 80S monosome in the GDP-bound form. Participation of eIF2 in a new round of translation initiation requires GDP-GTP exchange by eIF2B, a guanine nucleotide exchange factor for eIF2. Phosphorylation of eIF2 reduces eIF2B activity (2), slowing reinitiation and resulting in preferential translation of mRNAs with specific inhibitory features. The best characterized member of this group of mRNAs is activating transcription factor 4 (ATF4), a basic leucine zipper DNA-binding protein. Newly synthesized ATF4 regulates transcription of genes containing cis-regulatory motifs called CAAT enhancer-binding protein ATF response element (3). The resulting transcriptional program alters cellular metabolism to alleviate cytoplasmic and proteotoxic stress.
Another important signal transduction response to cellular stress is the mechanistic target of rapamycin complex 1 (mTORC1) pathway. mTORC1 is a major positive regulator of anabolic processes under conditions of nutrient and especially amino acid sufficiency (4). Under nutrient-sufficient conditions, mTORC1 promotes cell growth and proliferation by phosphorylating multiple targets including ribosomal protein S6 kinase 1 (S6K1) and eIF4E-binding protein 1. A class of mRNAs specifically regulated by mTORC1 activity contains 5′-terminal oligopyrimidine (TOP) motifs (5). TOP mRNAs encode ribosomal proteins and other cellular machinery important for translational capacity. Previous work by this lab shows hepatic TOP mRNA translation is preferentially inhibited by amino acid deprivation in the liver of healthy rodents (6).
Inappropriate activation of mTORC1 during cytoplasmic or proteotoxic stress has deleterious effects for both cells (7) and organisms (8–12). Our lab was the first to identify the existence of coordinated cross-talk between the ISR and mTORC1 pathway in mammals following amino acid deprivation (10). Since then, multiple labs have explored the basis for this bidirectional relationship in animal tissues and in normal cells and cancer cells (8, 13–21). Despite these efforts, the core factor(s) coordinating ISR-mTORC1 communication during amino acid stress remain mysterious. Furthermore, it is unclear whether the ISR plays a role in regulating TOP mRNA translation during amino acid stress.
To better understand when and how activation of the ISR represses mTORC1 signaling during amino acid stress, we performed time-course experiments in mice, analyzing the hepatic response to amino acid depletion by asparaginase over a 24-h time period. These experiments showed that deletion of the eIF2 kinase GCN2 disallowed Atf4 translation and instead permitted rapid activation of mTORC1 signaling and increased TOP mRNA translation in response to asparaginase. We also observed that mTORC1 activation following asparaginase exposure in mice lacking Gcn2 preceded both translational control of Atf4 gene expression and transcriptional control of the mTORC1 modulators Sesn2 (sestrin 2), Redd1 (regulated in development and DNA damage response), 4ebp1, and Trib3 (tribbles 3), indicating that the acute mTORC1 repressor was not ATF4. Interestingly, mice lacking Perk in liver did not show enhanced mTORC1 activity following injection of tunicamycin (a pharmacological ER stress agent), signifying that communication between the ISR and mTORC1 is specific to amino acid stress. Furthermore, pretreatment of Gcn2−/− mice with tunicamycin before asparaginase disallowed hepatic mTORC1 hyperactivation, which was restored by the loss of both Perk and Gcn2 in liver. Finally, pretreatment with ISRIB, a potent inhibitor of PERK signaling, did not alter eIF2 phosphorylation or mTORC1 signaling in wildtype and Gcn2−/− mice administered asparaginase, but it did by itself reduce hepatic mTORC1 activity in both wildtype and Gcn2−/− mice. Taken together, we conclude that eIF2 phosphorylation, not ATF4 synthesis, is necessary for acute inhibition of hepatic mTORC1 and TOP mRNA translation to amino acid stress in mice. Understanding how healthy organisms respond and adapt to amino acid deficiency at the molecular level may help in the development of improved and targeted means to treat cancer and other chronic diseases.
Results
Loss of Gcn2 quickly unleashes hepatic mTORC1 activity during amino acid stress
To define the role of GCN2 in the regulation of hepatic mTORC1 in response to amino acid stress in an in vivo setting, we utilized asparaginase to activate hepatic GCN2 (Fig. 1A). The kinetics of amino acid changes in the blood induced by asparaginase injection were similar in wildtype and Gcn2−/− mice (Table S2 and Figs. S1 and S2). Notably, asparagine concentrations were consistently reduced over time, whereas glutamine concentrations were acutely reduced at 3 h after injection, which then recovered to serum levels in untreated mice by 12 h (Fig. S1). Circulating levels of most other amino acids and in particular the dietary essential amino acids steadily increased over 24 h (Fig. S2) (9).
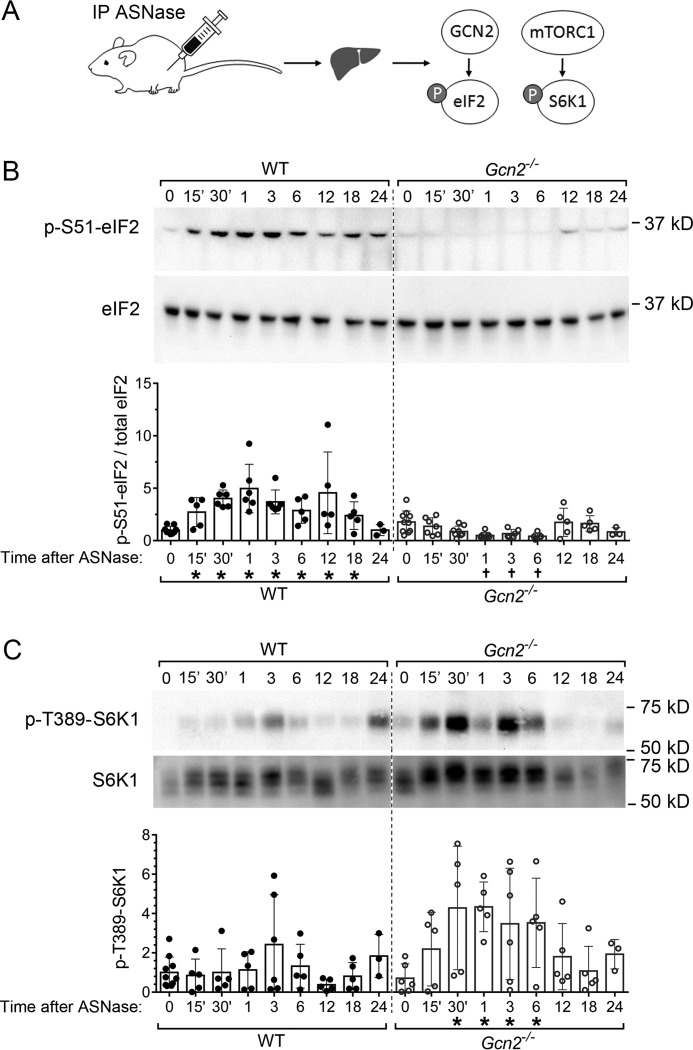
Figure 1.
Gcn2 status determines the hepatic eIF2 and mTORC1 response to asparaginase (ASNase). A, schematic diagram of experimental setup. The mice were injected intraperitoneally (IP) with ASNase and killed at regular intervals over 24 h. Liver samples were analyzed for phosphorylation of eIF2 (p-eIF2) and p-S6K1. B, upper panels show representative immunoblots displaying p-eIF2 at serine 51 and eIF2α total protein. Quantitative analysis of individual values for p-eIF2 levels normalized to total eIF2α at each time point is summarized in the bar graph in the lower panel. C, upper panels show representative immunoblots displaying p-S6K1 at threonine 389 and total levels of S6K1. Quantitative analysis of the Thr(P)-389 band intensities at each time point is summarized in the bar graph in the lower panel. Fig. S3 shows images of additional biological replicates. In B and C, circles represent individual liver samples, and bars represent average values across the treatment groups ± S.D. *, significantly higher than values at 0 which are PBS-treated, p < 0.05 by t test; †, significantly lower than values at 0 which are PBS-treated, p < 0.05 by t test.
Activation states of GCN2 and mTORC1 were tracked by assessing phosphorylation of their respective substrates, eIF2 and S6K1, respectively. Phosphorylation of eIF2 was increased within 15 min after injection and reached a maximum by 30 min (Fig. 1B), which corresponded with the nadir in circulating glutamine (Fig. S1). During this time frame, no significant change in mTORC1 activity was evident in the livers of wildtype mice (Fig. 1C and Fig. S3). In contrast, strong activation of mTORC1 was observed in the livers of Gcn2−/− mice as evidenced by the rapid increase in S6K1 phosphorylation at Thr-389 that lasted for at least 6 h (Fig. 1C and Fig. S3). Thus, Gcn2 deletion results in rapid and sustained activation of hepatic mTORC1 in response to amino acid stress.
Hepatic Gcn2 status dictates the response of translational machinery to amino acid stress
To assess the effect of amino acid depletion on gene-specific translation in liver, we conducted polysome profile analysis at multiple time points (Fig. 2A). Half an hour following asparaginase injection, we observed accumulation of 80S monosomes in wildtype livers, but not in Gcn2−/− livers (Fig. 2B), showing that GCN2 is required for repression of mRNA translation initiation in response to amino acid depletion. This effect was sustained for at least 3 h in wildtype mice, whereas in the livers of Gcn2−/− mice, little to no change in polysome profiles occurred in the period of 30 min to 3 h following asparaginase injection. Assessment of Atf4 mRNA distribution in polysome profile fractions revealed accumulation of Atf4 mRNA in the fractions heavily loaded with ribosomes as early as 1 h after asparaginase exposure (Fig. 2C, left panel). Importantly, Gcn2−/− livers did not exhibit this shift in Atf4 mRNA distribution (Fig. 2C, left panel). No apparent shift was found in the control mRNA encoding for actin (Fig. 2C, right panel). Fig. S4 shows the analysis of additional polysome profiles. These data indicate that 1) GCN2 kinase is required for translational induction of Atf4 mRNA following amino acid depletion by asparaginase, 2) gene-specific translation of Atf4 mRNA occurs very early following asparaginase injection, and 3) ATF4 synthesis following a single injection of asparaginase is a transient event, complete by 3 h after injection.
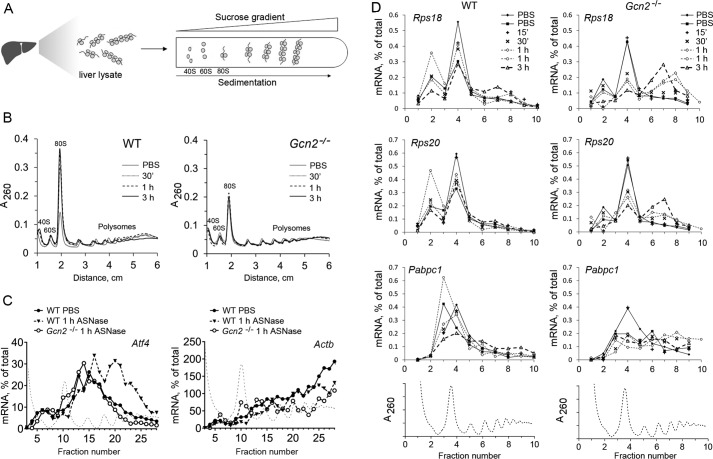
Figure 2.
Loss of Gcn2 prevents increased Atf4 mRNA translation efficiency in liver following ASNase exposure and instead permits enhanced TOP tract mRNA translation. A, schematic diagram of polysomal profiling method. Polysomes from liver lysates were separated based on their different migration rates through sucrose gradients. Polysomal RNA was isolated from fractions across the gradient, and translational efficiency was assessed by RT-qPCR analysis of RNA isolated from polysomal fractions. B, representative images of hepatic polysomal profiles from WT (left panel) and Gcn2−/− (right panel) mice treated with ASNase. An increase in the 80S peak height in WT reflects inhibition of mRNA translation initiation as early as 30 min after a single injection of asparaginase. This increase is not observed in Gcn2−/− following ASNase exposure. C, Atf4 mRNA levels are present in heavier polysomes at 1 h after ASNase in WT but not in Gcn2−/− mice. Black dotted lines of the polysomal profiles indicate the position of each fraction along the profile. The symbols represent values of relative Atf4 mRNA amount, expressed as a percentage of total mRNA value in all the fractions. D, assessment of select TOP mRNA (Rps18, Rps20, and Pabpc1) levels across sucrose gradient fractions. The polysomal profile is shown below to indicate relative position of each fraction. The symbols represent values of relative amounts of mRNAs in the collected fractions, expressed as percentages of total mRNA value in all the fractions.
We proceeded to test whether hyperactivation of mTORC1 in Gcn2−/− mice corresponded with maladaptive changes in gene-specific translation, namely the enhanced translation of TOP mRNAs (5). Quantitative analysis of the most responsive TOP mRNAs (encoding ribosomal proteins Rps18 and Rps20 and Pabpc1 (poly(A)-binding protein cytoplasmic 1)) in polysomal fractions revealed their accumulation in heavy polysomes 1–3 h following asparaginase injection (Fig. 2D) in samples with the highest mTORC1 activity. Thus, altered TOP mRNA translation is consequential to Gcn2 deletion during amino acid stress.
We also assessed polysomal distribution of mRNAs that are reportedly translationally enhanced in mouse livers upon perfusion with a methionine-deficient solution, namely chaperone Hspa5 (heat shock protein family A5), amino acid transporter Slc3a2 (solute carrier family 2 member 2), and transferrin receptor Trf2 (transferring receptor 2; Ref. 22). None of these mRNAs exhibited any changes in translational efficiency upon asparaginase exposure (Fig. S5). This observation suggests that hepatic response to amino acid stress by asparaginase is different from the one caused by methionine deficiency. In fact, recent studies demonstrate that hepatic responses to methionine restriction are mediated in a eIF2 phosphorylation-independent manner (23) and perhaps via its own unique pathway (24).
Hyperactivation of mTORC1 in Gcn2−/− livers precedes transcriptional control of mTORC1 repressors
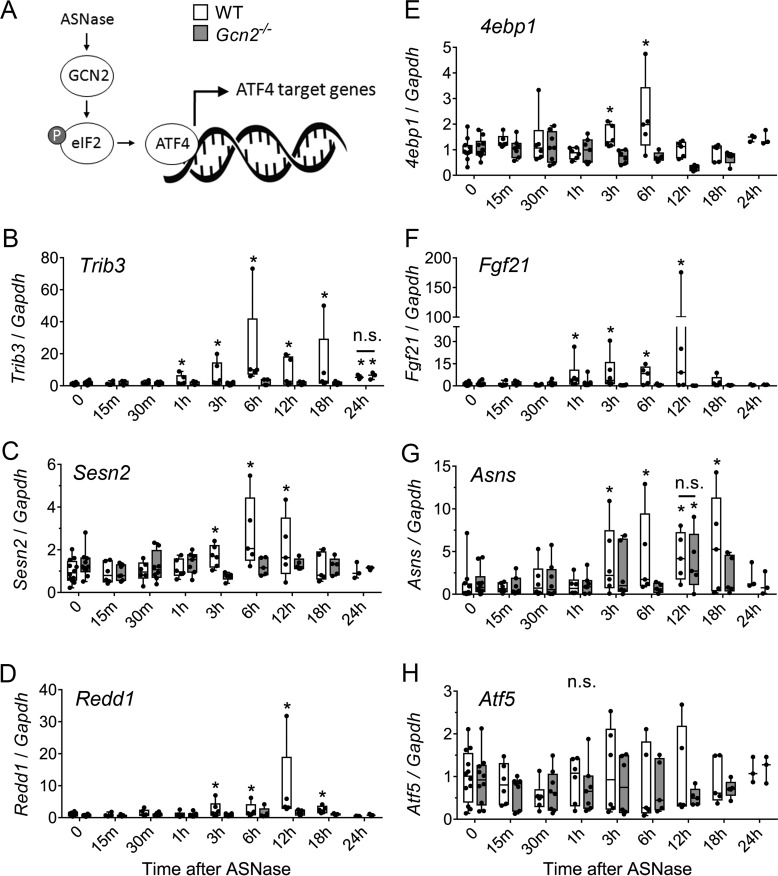
To assess when ATF4 target genes serve as mTORC1 regulators during amino acid stress, we examined mRNA levels of Sesn2, 4ebp1, Trib3, and Redd1, as well as a few classic targets of ATF4: Fgf21 (fibroblast growth factor 21), Asns (asparagine synthetase), and Atf5. No acute changes were observed in hepatic mRNA levels of any of the genes within the first hour following injection. A robust increase in Trib3 mRNA was observed 3–18 h following asparaginase exposure in wildtype (up to 80 times over untreated control) but not Gcn2−/− livers. In the same time frame, moderate elevation was seen in Redd1 mRNA levels (up to 30 times), and only mild elevations in Sesn2 and 4ebp1 mRNAs (four times the maximum) were observed in wildtype, but not in Gcn2−/− mice (Fig. 3). These data clearly show that control of mTORC1 activity by known ATF4 targets occurs 3 h or more following asparaginase exposure and thus cannot explain the immediate loss of control over mTORC1 activity in Gcn2−/− livers.
Figure 3.
Time-resolved changes in hepatic expression of select ATF4 target genes after ASNase exposure. A, total RNA was isolated from livers of mice after a single injection of ASNase and select ATF4 target genes were quantitatively assessed using RT-qPCR. B–H, Sesn2 (B), Redd1 (C), Asns (D), Trib3 (E), 4ebp1 (F), Fgf21 (G), and Atf5 (H). The boxes extend from the 25th to 75th percentiles as computed in GraphPad software using the following equation: r = P * (n + 1)/100, where P is the desired percentile, and n is the number of values in a data set. Whiskers extend to the maximum and minimum values of the data set. *, significantly different as compared with values at 0, which represents PBS injection, p < 0.05 by t test.
Exposure to chemical ER stress does not activate mTORC1 in livers lacking Perk
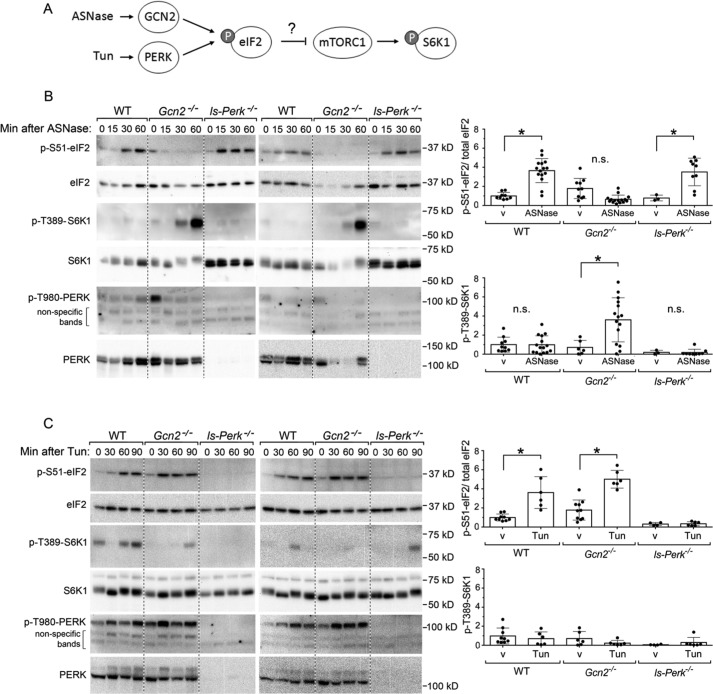
To check whether asparaginase may acutely induce ER stress, we analyzed PERK activation (Fig. 4A), which was assessed via its phosphorylation at Thr-980. No activation of PERK was observed in response to asparaginase exposure in either wildtype or Gcn2−/− livers (Fig. 4B), confirming that amino acid depletion by asparaginase increases eIF2 phosphorylation via GCN2 (25). Moreover, mice with genetic ablation of hepatic Perk via Cre-mediated recombination (liver-specific Perk−/−, ls-Perk−/−) displayed increased eIF2 phosphorylation and no hyperactivation of mTORC1 in response to asparaginase exposure, similar to wildtype animals (Fig. 4B).
Figure 4.
Loss of Perk does not unleash mTORC1 signaling upon ASNase exposure or upon ER stress by tunicamycin (Tun). A, schematic diagram of molecular cascades tested, namely the ISR and mTORC1 pathway after a single injection of either ASNase or Tun. B, p-S6K1 at threonine 389 is increased in livers of Gcn2−/− but not ls-Perk−/− mice. Two representative images are displayed next to mean values of band intensities for p-eIF2 and p-S6K1. v stands for PBS vehicle control. Mean values combine results at 15, 30, and 60 min after ASNase. C, no increase in p-S6K1 is noted in the livers of ls-Perk−/− mice injected with Tun. Two representative images are displayed next to mean values of band intensities for p-eIF2 and p-S6K1. v stands for 0.3% DMSO vehicle control. Mean values combine results at 30, 60, and 90 min after a single injection of Tun. Circles represent individual data points, and bars represent average values across the treatment groups ± S.D. *, significantly higher than values at the corresponding v groups, p < 0.05 by t test.
To better understand whether communication between the ISR and mTORC1 occurs more broadly in response to proteotoxic stress, we tested whether acute ER stress would cause similar activation of mTORC1 in livers bearing genetically disrupted Perk (Fig. 4A). To test this idea, we injected tunicamycin, a potent inhibitor of glycosylation and an ER stress–causing agent, to wildtype mice, Gcn2−/− mice and ls-Perk−/− mice. As expected, tunicamycin activated PERK, evidenced by its autophosphorylation at Thr-980 (Fig. 4C). The response was very similar in wildtype and Gcn2−/− livers, whereas in ls-Perk−/− mice, no phosphorylation of eIF2 was evident (26). Remarkably, no acute (over 90 min) activation of mTORC1 was observed in the livers of any mouse strains following tunicamycin injection. These data indicate that the type of cellular stress determines whether or not a functional ISR–mTORC1 communication axis exists.
Pretreatment of Gcn2−/− mice with tunicamycin rescues hepatic mTORC1 repression following asparaginase exposure
We then tested whether induction of eIF2 phosphorylation by alternative means in Gcn2−/− mice would help prevent activation of mTORC1 (i.e. ISR preconditioning). To do this, wildtype and Gcn2−/− mice were injected with tunicamycin 30 min prior to asparaginase injection to induce phosphorylation of eIF2 (Fig. 5A). Analysis of mTORC1 activity showed no hyperphosphorylation of S6K1 in the livers of Gcn2−/− mice preconditioned with tunicamycin (Fig. 5, B–D). However, genetic ablation of both Perk and Gcn2−/− in liver blocked phosphorylation of eIF2 to both tunicamycin and asparaginase and allowed for hyperactivation of mTORC1 once again (Fig. 5, B–D). These data suggest that ISR preconditioning by another kinase is sufficient to block mTORC1 activation in response to acute amino acid stress.
Figure 5.
Pretreatment with tunicamycin (Tun) rescues normal control of mTORC1 by ASNase in Gcn2−/− livers, whereas ISRIB does not. A, schematic diagram of the experiment. WT, Gcn2−/−, and ls-PERK−/− Gcn2−/− mice were injected with Tun 30 min prior to ASNase, after which livers were collected at the indicated time points. B, representative immunoblots display p-eIF2, p-S6K1, and p-PERK. C, mean values of band intensities for p-eIF2 levels normalized to total eIF2α at indicated time points. D, mean values of band intensities for p-S6K1 at threonine 389 at indicated time points. + indicates positive control, which is Gcn2−/− liver collected 30 min after ASNase; v represents vehicle control. Individual data points are shown in circles, and bars represent average values across treatment groups ± S.D. *, p < 0.05 by t test. E, schematic diagram of the experiment. WT and Gcn2−/− mice were administered ISRIB 30 min prior to ASNase, after which livers were collected at the indicated time points. F, representative immunoblots display p-eIF2 and p-S6K1 in WT and Gcn2−/− livers. G, mean values of band intensities for p-eIF2 levels normalized to total eIF2α protein. H, mean values of band intensities for p-S6K1 at threonine 389. I, mean values of p-S6K1 at threonine 389 in both genetic strains combined (WT and Gcn2−/−) following a single injection of ISRIB alone. Individual data points are shown in circles, and bars represent average values across treatment groups ± S.D. *, p < 0.05 by t test as compared with vehicle treated control.
ISRIB reduces mTORC1 activity by itself yet fails to prevent mTORC1 hyperactivation following asparaginase exposure in Gcn2−/− livers
Finally, to explore the relationship between eIF2B and mTORC1 in response to asparaginase, we utilized the small molecule inhibitor, ISRIB (Fig. 5E), which locks eIF2B in its active dimer form and renders cells insensitive to eIF2 phosphorylation by PERK. To our surprise, administration of ISRIB significantly reduced hepatic mTORC1 signaling independent of GCN2 status (Fig. 5, F–I, compare bar graphs of control versus ISRIB-treated animals). This noted decrease in mTORC1 activity upon ISRIB exposure is consistent with the published observation of ISRIB causing decreased translation of TOP mRNAs (27). Nevertheless, administration of ISRIB 30 min prior to asparaginase did not affect mTORC1 activity in wildtype mice, nor did it modify any measured outcome in wildtype and Gcn2−/− mice exposed to asparaginase (Fig. 5, F–I). Considering ISRIB functions as an inhibitor of PERK signaling, the results are consistent with asparaginase treatment of lsPerk−/− mice and lsPerk−/−-Gcn2−/− mice. Collectively, these data show that it is the phosphorylation of eIF2 and not ATF4 synthesis that directs mTORC1 regulation upon acute amino acid stress.
Discussion
This study utilized chemical activators of the amino acid response and the ER stress response to reveal new information regarding a vital role for the GCN2-eIF2 axis in acutely regulating hepatic mTORC1 in mice. The data show that hyperactivation of hepatic mTORC1 in Gcn2−/− mice exposed to asparaginase occurs within minutes, preceding the timing of enhanced translation of Atf4 mRNA and increased transcription of mTORC1 inhibitors in wildtype mice. The inappropriately high mTORC1 signaling in liver of Gcn2−/− mice corresponds with movement of TOP mRNAs into polysomes, identifying a physiologically maladaptive outcome resulting from enhanced mTORC1 signaling. Preconditioning the ISR in Gcn2−/− liver by activating PERK blocks mTORC1 activation following amino acid depletion, which is reversed in the liver of mice lacking both Gcn2 and Perk. Importantly, hepatic mTORC1 activity is not unleashed by ER stress in ls-Perk mice, signifying that communication between the ISR and mTORC1 is specific to sensing amino acid levels. Together these results suggest that eIF2 phosphorylation is necessary to impart negative control of the ISR over mTORC1 during amino acid stress.
Rapid and robust activation of hepatic mTORC1 in Gcn2−/− mice given asparaginase supports a model wherein the GCN2-eIF2 axis plays a suppressive, ATF4-independent role in regulating mTORC1 activity upon sensing amino acid stress. This suppressive role of GCN2 was first observed by us in mice fed a leucine-devoid diet to Gcn2−/− mice (10), then confirmed and further detailed using mice treated with asparaginase (8, 9, 19, 25). This work also strongly supports the findings of Averous et al. (20), who used a model of leucine deprivation in mouse embryonic fibroblasts (MEFs) to conclude that GCN2 and eIF2 phosphorylation, but not ATF4, are required for continuous suppression of mTORC1 during leucine or arginine deprivation. Deficiency in either of these two amino acids in Gcn2−/− MEFs strongly activated mTORC1 within 15 min. In our current and past in vivo models, we see hyperactivation of mTORC1 within the first 30–60 min of amino acid depletion, which is sustained for at least 6 h. Based on time-course results by ourselves and others, we conclude that ATF4 is not necessary to inhibit mTORC1 in response to amino acid deprivation. Our work also expands on the findings of Ye et al. (16), reporting that GCN2 induces the expression of Sestrin2 to inhibit mTORC1. No considerable induction of Sestrin2 mRNA levels was observed upon asparaginase exposure in our model of amino acid deprivation. Considering that mTORC1 repression can be rescued by ISR preconditioning via tunicamycin-induced PERK, we conclude that eIF2 phosphorylation is required for acute inhibition of mTORC1 upon amino acid stress, in agreement with (20).
In our previous study of chronic exposure to asparaginase (8 days), obese Gcn2−/− mice demonstrate PERK activation and eIF2 phosphorylation alongside highly activated mTORC1 (8). Thus, phosphorylation of eIF2 by PERK failed to prevent mTORC1 activation in obese mice. We reconcile these results by pointing out that the timing of PERK activation is very different in these two experimental models. In the current study, PERK is activated before exposure to amino acid stress, allowing it to function as a preconditioning agent. In the previous work, PERK is not activated in the excipient-treated mice, but only in asparaginase-treated mice, indicating that PERK becomes engaged after or in concert with GCN2. Overall, these data suggest that phosphorylation of eIF2 is necessary but not sufficient to coordinate mTORC1 signaling upon amino acid stress, and an intact GCN2-eIF2 axis is a vital necessity in this regard.
As part of the ISR to amino acid stress, ATF4-mediated induction of several mTORC1 inhibitors, namely Redd1 (28), Trib3 (29), Sesn2 (16), and 4ebp1 (30), is reported as playing a regulatory role. Our data show that of these four candidates, the most robust induction was observed in the expression of Trib3 gene, coding for a negative regulator of insulin signaling (31), whereas hepatic levels of Redd1, Sesn2, and 4ebp1 mRNAs showed only modest increases, if any. In all cases, these increases in mRNA levels were observed no earlier than 3 h following the injection of asparaginase, again confirming that acute inhibition of mTORC1 does not involve an ATF4-driven transcriptional program. Interestingly, our previous studies show that induction of Trib3 mRNA by asparaginase requires GCN2, but not ATF4, because Atf4−/− animals treated with asparaginase demonstrate 10-fold greater increases in Trib3 mRNA levels to asparaginase than wildtype animals, whereas no induction whatsoever is observed in Gcn2−/− animals (8, 32). This confirms that the Trib3 gene is controlled by other transcriptional factors in addition to ATF4 (18), but whether TRIB3 represents the major effector of GCN2 pathway that contributes to sustained inhibition of hepatic mTORC1 in WT, as well as in Atf4−/− animals, requires further investigation.
ATF4 is the central transcription factor of the ISR, but it is also utilized by the mTORC1 pathway to support anabolic processes (14, 15). For instance, stimulation of mTORC1 with insulin in MEFs leads to the enhanced synthesis of ATF4 without eIF2 phosphorylation. This increase in ATF4 can be blocked by either cycloheximide, an inhibitor of protein synthesis, or rapamycin (inhibitor of mTORC1), suggesting that both protein synthesis and mTORC1 signaling are required for the increased ATF4 protein levels (14). The proposed function of such signaling cascade is to increase cellular uptake of essential amino acids increase the synthesis of the non-essential ones (15), thereby supporting anabolic reactions during insulin stimulation of mTORC1. Our data demonstrate that hyperactive mTORC1 per se is not a sufficient signal to induce enhanced synthesis of ATF4 because we observed no translational induction of ATF4 in Gcn2−/− animals with high mTORC1 activity. The fact that the fundamentally distinct stimuli—insulin versus acute amino acid stress in the background of systemic Gcn2 disruption—produce one outcome in the form of hyperactive mTORC1 but distinct downstream response in the form of ATF4 production implies the existence of yet-to-be-identified factors, other than mTORC1, that influence cellular decision whether to up-regulate synthesis of ATF4.
Deregulation of mTORC1 is associated with many pathological conditions in the liver (11, 33, 34). Our study identifies GCN2 and its downstream effector eIF2 as essential components that coordinate hepatic mTORC1 during amino acid stress. These data complement and provide mechanistic insight as to why maladaptive activation of hepatic mTORC1 in Gcn2−/− mice associates with greater morbidity and metabolic toxicity (8) and have important implications for the clinical use of rapidly growing class of GCN2-activating drugs (35–44). Gcn2 is not an essential gene and so far is found to have 594 missense, 28 nonsense, and 33 frame-shifting mutations in the human population (45). Our study found that GCN2 governs important nutritional responses and, if dysfunctional, leads to altered whole body metabolism. It is tempting to speculate that as a result of this altered metabolism, subjects bearing deleterious mutations in the Gcn2 (EIF2AK4) gene have higher chances of developing metabolic conditions later in life such as pulmonary arterial hypertension (46). This work provides a foundation for personalized approaches in usage of other GCN2-activating drugs as well as dietary approaches.
Experimental procedures
Animals
All animals received humane care according to the criteria outlined in the Guide for the Care and use of Laboratory Animals prepared by the National Academy of Sciences (National Institutes of Health publication 86-23 revised 1985) and ARRIVE guidelines. Animal protocols were reviewed and approved by the Rutgers Institutional Animal Care and Use Committee. Adult (10–20 weeks old) male and female WT and Gcn2−/− mice on the C57Bl/6J genetic background (47) were used. Mice with albumin Cre-mediated deletion of Perk in liver (lsPerk−/−) were also used (48), and these mice were bred to Gcn2−/− mice to create a double knockdown of Perk and Gcn2 in liver. All mice were bred at the Bartlett animal facility on the Rutgers University Cook campus. All mice had free access to food (5001 laboratory rodent diet; LabDiet) and water and maintained on a 12-h light-dark cycle (7 a.m. to 7 p.m.) with same-sex littermates until experimental group assignment, wherein mice were housed in individual plastic cages with soft bedding.
Animals were administered a single injection of either bacterial asparaginase (3 IU/g body weight; Elspar, Merck), tunicamycin (1 mg/kg body weight; Sigma catalog no. T7765), or ISRIB (0.25 mg/kg; Sigma catalog no. SML0843) and euthanized by decapitation at indicated time points. The control groups were treated with PBS (vehicle for asparaginase) or 0.3% DMSO in PBS (vehicle for tunicamycin and ISRIB). All mice were killed between 12 and 4 p.m. (ZT5-9).
Serum amino acid profiling
Trunk blood was collected into microcentrifuge tubes. Serum was collected by centrifugation after blood clotting on ice. Proteins of the serum were precipitated by the addition of 180 μl of a solvent (0.1% formic acid in methanol) to 60 μl of serum. The resulting cloudy mixture was filtered using Captiva non-drip lipid filtration tubes (Agilent catalog no. A5400635). The recovered clear eluate in the amount of 20 μl was spiked with 5 μl of internal standard (equimolar mix of sarcosine and norvaline, 0.1 mm each) and analyzed using reversed-phase high pressure liquid chromatography (Agilent 1200 HPLC instrument) according to the manufacturer's protocol (Agilent publication catalog no. 5990-4547EN). Briefly, amino acid separation was carried out using a gradient mix of mobile phase A (10 mm Na2HPO4, 10 mm Na2B4O7, 5 mm NaN3, pH 8.2) and mobile phase B (acetonitrile/methanol/water = 45/45/10) on ZORBAX eclipse Plus C18 column at the speed of 0.42 ml/min. To allow for amino acid detection by fluorescence, derivatization was done automatically using autosampler according to the manufacturer's protocol mentioned above and consisted of two consequent steps: 1) conjugation to o-phthalaldehyde and 2) conjugation to fluorenylmethyloxycarbonyl chloride. Standard curves for each amino acid were retrieved from analyzing a set of equimolar standard mixtures. A total of 10 standard mixtures were used ranging from 2.25 to 900 μm of each amino acid. Analysis of chromatograms was performed using Agilent OpenLAB software.
Immunoblotting
Liver tissue was crushed using the Cellcrusher tissue pulverizer under liquid nitrogen conditions. The resulting tissue powder was lysed with radioimmune precipitation assay buffer (25 mm HEPES, pH 7.5, 10 mm DTT, 0.1% SDS, 1× protease inhibitor mixture (Sigma; P8340), 1 mm sodium orthovanadate, 0.5% deoxycholate, 50 mm β-glycerophosphate, 2 mm EDTA, 1 mm microcystin, 50 mm NaF, 3 mm benzamidine) using the Polytron bench top homogenizer followed by heating the lysates for 5 min in Laemmli buffer. Primary antibodies used were as follows: anti-total p70 S6K (Cell Signaling Technology, CST 9202), anti-phospho-(Thr-389)-S6K (Cell Signaling Technology, CST 9205), and anti-eIF4E-binding protein 1 (Bethyl Laboratories A300-501A), anti-phospho(S51)-eIF2 (Cell Signaling Technology, CST 3597), anti-eIF2α (Santa Cruz, sc-11386), anti-phospho(T980)-PERK (Santa Cruz, sc-3179S), and anti-PERK (Santa Cruz, sc-3192S). Secondary antibody was peroxidase-AffiniPure goat anti-rabbit TgC (H+L) from Jackson ImmunoResearch (111-035-003). The images were taken with Fluorchem M imager (ProteinSimple), and band densities were quantified using αView software.
RT-qPCR
RNA was isolated from 10 mg of frozen tissue powder using TRI reagent according to the manufacturer instructions. Quality of the isolated RNA was assessed by measuring A260/280 and A260/230 ratios; integrity was assessed by visualization of the rRNA on agarose gels. One microgram of the isolated RNA from each sample was used to generate cDNA using high-capacity cDNA reverse transcription kit (Thermo Fisher catalog no. 4368814). The resulting cDNA was used for quantitative analysis of gene expression using SYBR green system. Validated primers sequences are listed in the supporting information.
Polysomal profiling and RT-qPCR from sucrose gradients
Frozen mouse livers were crushed at liquid nitrogen temperature with tissue pulverizer. Approximately 50 mg of the frozen tissue powder were homogenized using blue plastic pestles in standard 1.5-ml microcentrifuge tubes using with lysis buffer (25 mm HEPES-KOH, pH 7.5, 100 mm KCl, 5 mm MgCl2, 0.5% deoxycholate, 1% Nonidet P-40, 1 mm DTT, 1× protease inhibitor (Sigma catalog no. P8340), SUPERase-In RNase inhibitor (Thermo Fisher catalog no. AM2696); tissue-to-lysis buffer ratio was 1:10. The lysates were incubated on ice for 5 min and centrifuged at 10,000 × g for 10 min to get rid of nuclei and cell debris. The cleared supernatants were loaded on 10–50% sucrose gradients and centrifuged at 100,000 × g for 3 h to resolve mRNA species loaded with different amounts of ribosomes. The resolved polysomes were collected in either 10 or 30 fractions from which RNA was isolated using TRI reagent for liquid samples according to the manufacturer's protocol. The RNA precipitates were washed twice with 75% ethanol and resuspended in 20 μl of nuclease-free water. Quality of the isolated RNA was assessed by measuring A260/280 and A260/230 ratios; integrity was assessed by visualization of the rRNA on agarose gels. RT-qPCR was performed as described above with the exception that the relative values of polysomal Atf4 mRNA were normalized to exogenous spike-in Luciferase instead of glyceraldehyde-3-phosphate dehydrogenase.
Statistics
In most graphical displays, liver samples from animal subjects are plotted individually alongside the mean values ± S.D. or plotted as box-whisker plots where boxes extend from the 25th to 75th percentiles and whiskers extend to the maximum and minimum values of the data set. Two group comparisons were analyzed by t test assuming two-tailed distribution with unequal variance and with an α level of p < 0.05 (GraphPad Prism, La Jolla, CA). A table listing the number of replicate mice in each strain and treatment group is included in Table S1. Additional details on experimental materials and procedures are available in the supporting information.
Author contributions
I. A. N. and T. G. A. conceptualization; I. A. N. data curation; I. A. N., E. T. M., C. C. S., and M. P. G. formal analysis; I. A. N. validation; I. A. N., E. T. M., C. C. S., and M. P. G. investigation; I. A. N. visualization; I. A. N., E. T. M., and C. C. S. methodology; I. A. N. writing-original draft; I. A. N., R. C. W., and T. G. A. writing-review and editing; R. C. W. and T. G. A. funding acquisition; T. G. A. resources; T. G. A. supervision; T. G. A. project administration.
Supplementary Material
This work was supported by National Institutes of Health Grants HD070487 (to T. G. A.) and DK109714 (to T. G. A. and R. C. W.). This work was also supported by Institutional Research and Academic Career Development Awards (IRACDA) New Jersey–New York for Science Partnerships in Research and Education National Institutes of Health postdoctoral training Grant K12GM093854 (to I. A. N.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Tables S1 and S2 and Figs. S1–S5.
- ISR
- integrated stress response
- ATF4
- activating transcription factor 4
- eIF2
- eukaryotic initiation factor 2
- mTORC1
- mechanistic target of rapamycin complex 1
- S6K1
- ribosomal protein S6 kinase 1
- ASNase
- asparaginase
- ER
- endoplasmic reticulum
- PERK
- protein kinase R-like ER resident kinase
- TOP
- 5′-terminal oligopyrimidine
- qPCR
- quantitative PCR
- MEF
- mouse embryonic fibroblast
- p
- phosphorylation.
References
- 1. Pakos-Zebrucka K., Koryga I., Mnich K., Ljujic M., Samali A., and Gorman A. M. (2016) The integrated stress response. EMBO Rep. 17, 1374–1395 10.15252/embr.201642195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kimball S. R., Fabian J. R., Pavitt G. D., Hinnebusch A. G., and Jefferson L. S. (1998) Regulation of guanine nucleotide exchange through phosphorylation of eukaryotic initiation factor eIF2a. J. Biol. Chem. 273, 12841–12845 10.1074/jbc.273.21.12841 [DOI] [PubMed] [Google Scholar]
- 3. Shan J., Zhang F., Sharkey J., Tang T. A., Örd T., and Kilberg M. S. (2016) The C/ebp-Atf response element (CARE) location reveals two distinct Atf4-dependent, elongation-mediated mechanisms for transcriptional induction of aminoacyl-tRNA synthetase genes in response to amino acid limitation. Nucleic Acids Res. 44, 9719–9732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sabatini D. M. (2017) Twenty-five years of mTOR: uncovering the link from nutrients to growth. Proc. Natl. Acad. Sci. U.S.A. 114, 11818–11825 10.1073/pnas.1716173114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Thoreen C. C., Chantranupong L., Keys H. R., Wang T., Gray N. S., and Sabatini D. M. (2012) A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113 10.1038/nature11083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Anthony T. G., Reiter A. K., Anthony J. C., Kimball S. R., and Jefferson L. S. (2001) Deficiency of dietary EAA preferentially inhibits mRNA translation of ribosomal proteins in liver of meal-fed rats. Am. J. Physiol. Endocrinol. Metab. 281, E430–E439 10.1152/ajpendo.2001.281.3.E430 [DOI] [PubMed] [Google Scholar]
- 7. Ng S., Wu Y.-T., Chen B., Zhou J., and Shen H.-M. (2011) Impaired autophagy due to constitutive mTOR activation sensitizes TSC2-null cells to cell death under stress. Autophagy 7, 1173–1186 10.4161/auto.7.10.16681 [DOI] [PubMed] [Google Scholar]
- 8. Nikonorova I. A., Al-Baghdadi R. J. T., Mirek E. T., Wang Y., Goudie M. P., Wetstein B. B., Dixon J. L., Hine C., Mitchell J. R., Adams C. M., Wek R. C., and Anthony T. G. (2017) Obesity challenges the hepatoprotective function of the integrated stress response to asparaginase exposure in mice. J. Biol. Chem. 292, 6786–6798 10.1074/jbc.M116.768408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wilson G. J., Bunpo P., Cundiff J. K., Wek R. C., and Anthony T. G. (2013) The eukaryotic initiation factor 2 kinase GCN2 protects against hepatotoxicity during asparaginase treatment. Am. J. Physiol. Endocrinol. Metab. 305, E1124–E1133 10.1152/ajpendo.00080.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Anthony T. G., McDaniel B. J., Byerley R. L., McGrath B. C., Cavener D. R., McNurlan M. A., and Wek R. C. (2004) Preservation of liver protein synthesis during dietary leucine deprivation occurs at the expense of skeletal muscle mass in mice deleted for eIF2 kinase GCN2. J. Biol. Chem. 279, 36553–36561 10.1074/jbc.M404559200 [DOI] [PubMed] [Google Scholar]
- 11. Menon S., Yecies J. L., Zhang H. H., Howell J. J., Nicholatos J., Harputlugil E., Bronson R. T., Kwiatkowski D. J., and Manning B. D. (2012) Chronic activation of mTOR complex 1 is sufficient to cause hepatocellular carcinoma in mice. Sci. Signal. 5, ra24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reith R. M., Way S., McKenna J. 3rd, Haines K., and Gambello M. J. (2011) Loss of the tuberous sclerosis complex protein tuberin causes Purkinje cell degeneration. Neurobiol. Dis. 43, 113–122 10.1016/j.nbd.2011.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gandin V., Masvidal L., Cargnello M., Gyenis L., McLaughlan S., Cai Y., Tenkerian C., Morita M., Balanathan P., Jean-Jean O., Stambolic V., Trost M., Furic L., Larose L., Koromilas A. E., et al. (2016) mTORC1 and CK2 coordinate ternary and eIF4F complex assembly. Nat. Commun. 7, 11127 10.1038/ncomms11127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ben-Sahra I., Hoxhaj G., Ricoult S. J. H., Asara J. M., and Manning B. D. (2016) mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science 351, 728–733 10.1126/science.aad0489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Adams C. M. (2007) Role of the transcription factor ATF4 in the anabolic actions of insulin and the anti-anabolic actions of glucocorticoids. J. Biol. Chem. 282, 16744–16753 10.1074/jbc.M610510200 [DOI] [PubMed] [Google Scholar]
- 16. Ye J., Palm W., Peng M., King B., Lindsten T., Li M. O., Koumenis C., and Thompson C. B. (2015) GCN2 sustains mTORC1 suppression upon amino acid deprivation by inducing Sestrin2. Genes Dev. 29, 2331–2336 10.1101/gad.269324.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Matsushima R., Harada N., Webster N. J., Tsutsumi Y. M., and Nakaya Y. (2006) Effect of TRB3 on insulin and nutrient-stimulated hepatic p70S6 kinase activity. J. Biol. Chem. 281, 29719–29729 10.1074/jbc.M511636200 [DOI] [PubMed] [Google Scholar]
- 18. Han J., Back S. H., Hur J., Lin Y.-H., Gildersleeve R., Shan J., Yuan C. L., Krokowski D., Wang S., Hatzoglou M., Kilberg M. S., Sartor M. A., and Kaufman R. J. (2013) ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell. Biol. 15, 481–490 10.1038/ncb2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Al-Baghdadi R. J. T., Nikonorova I. A., Mirek E. T., Wang Y., Park J., Belden W. J., Wek R. C., and Anthony T. G. (2017) Role of activating transcription factor 4 in the hepatic response to amino acid depletion by asparaginase. Sci. Rep. 7, 1272 10.1038/s41598-017-01041-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Averous J., Lambert-Langlais S., Mesclon F., Carraro V., Parry L., Jousse C., Bruhat A., Maurin A. C., Pierre P., Proud C. G., and Fafournoux P. (2016) GCN2 contributes to mTORC1 inhibition by leucine deprivation through an ATF4 independent mechanism. Sci. Rep. 6, 27698 10.1038/srep27698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xiao F., Huang Z., Li H., Yu J., Wang C., Chen S., Meng Q., Cheng Y., Gao X., Li J., Liu Y., and Guo F. (2011) Leucine deprivation increases hepatic insulin sensitivity via GCN2/mTOR/S6K1 and AMPK pathways. Diabetes 60, 746–756 10.2337/db10-1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dang Do A. N., Kimball S. R., Cavener D. R., and Jefferson L. S. (2009) eIF2α kinases GCN2 and PERK modulate transcription and translation of distinct sets of mRNAs in mouse liver. Physiol, Genomics 38, 328–341 10.1152/physiolgenomics.90396.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wanders D., Stone K. P., Forney L. A., Cortez C. C., Dille K. N., Simon J., Xu M., Hotard E. C., Nikonorova I. A., Pettit A. P., Anthony T. G., and Gettys T. W. (2016) Role of GCN2-independent signaling through a non-canonical PERK/NRF2 pathway in the physiological responses to dietary methionine restriction. Diabetes 65, 1499–1510 10.2337/db15-1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gu X., Orozco J. M., Saxton R. A., Condon K. J., Liu G. Y., Krawczyk P. A., Scaria S. M., Harper J. W., Gygi S. P., and Sabatini D. M. (2017) SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science 358, 813–818 10.1126/science.aao3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bunpo P., Dudley A., Cundiff J. K., Cavener D. R., Wek R. C., and Anthony T. G. (2009) GCN2 protein kinase is required to activate amino acid deprivation responses in mice treated with the anti-cancer agent l-asparaginase. J. Biol. Chem. 284, 32742–32749 10.1074/jbc.M109.047910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Harding H. P., Zhang Y., Bertolotti A., Zeng H., and Ron D. (2000) Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell 5, 897–904 10.1016/S1097-2765(00)80330-5 [DOI] [PubMed] [Google Scholar]
- 27. Sidrauski C., Mcgeachy A. M., Ingolia N. T., and Walter P. (2015) The small molecule ISRIB reverses the effects of eIF2 α phosphorylation on translation and stress granule assembly. Elife 4, e05033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kimball S. R., and Jefferson L. S. (2012) Induction of REDD1 gene expression in the liver in response to endoplasmic reticulum stress is mediated through a PERK, eIF2a phosphorylation, ATF4-dependent cascade. Biochem. Biophys. Res. Commun. 427, 485–489 10.1016/j.bbrc.2012.09.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ohoka N., Yoshii S., Hattori T., Onozaki K., and Hayashi H. (2005) TRB3, a novel ER stress-inducible gene, is induced via ATF4–CHOP pathway and is involved in cell death. EMBO J. 24, 1243–1255 10.1038/sj.emboj.7600596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mazor K. M., and Stipanuk M. H. (2016) GCN2- and eIF2α-phosphorylation-independent, but ATF4-dependent, induction of CARE-containing genes in methionine-deficient cells. Amino Acids 48, 2831–2842 10.1007/s00726-016-2318-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Du K., Herzig S., Kulkarni R. N., and Montminy M. (2003) TRB3: a tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science 300, 1574–1577 10.1126/science.1079817 [DOI] [PubMed] [Google Scholar]
- 32. Fusakio M. E., Willy J. A., Wang Y., Mirek E. T., Al Baghdadi R. J. T., Adams C. M., Al Baghdadi R. J., Adams C. M., Anthony T. G., and Wek R. C. (2016) Transcription factor ATF4 directs basal and select induced gene expression in the unfolded protein response and cholesterol metabolism in liver. Mol. Biol. Cell. 27, 1536–1551 10.1091/mbc.E16-01-0039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Laplante M., and Sabatini D. M. (2012) mTOR signaling in growth control and disease. Cell 149, 274–293 10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Umemura A., Park E. J., Taniguchi K., Lee J. H., Shalapour S., Valasek M. A., Aghajan M., Nakagawa H., Seki E., Hall M. N., and Karin M. (2014) Liver damage, inflammation, and enhanced tumorigenesis after persistent mTORC1 inhibition. Cell Metab. 20, 133–144 10.1016/j.cmet.2014.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kardos G. R., Wastyk H. C., and Robertson G. P. (2015) Disruption of proline synthesis in melanoma inhibits protein production mediated by the GCN2 pathway. Mol. Cancer Res. 13, 1408–1420 10.1158/1541-7786.MCR-15-0048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Keller T. L., Zocco D., Sundrud M. S., Hendrick M., Edenius M., Yum J., Kim Y. J., Lee H. K., Cortese J. F., Wirth D. F., Dignam J. D., Rao A., Yeo C. Y., Mazitschek R., and Whitman M. (2012) Halofuginone and other febrifugine derivatives inhibit prolyl-tRNA synthetase. Nat. Chem. Biol. 8, 311–317 10.1038/nchembio.790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sundrud M. S., Koralov S. B., Feuerer M., Calado D. P., Kozhaya A. E., Rhule-Smith A., Lefebvre R. E., Unutmaz D., Mazitschek R., Waldner H., Whitman M., Keller T., and Rao A. (2009) Halofuginone inhibits TH17 cell differentiation by activating the amino acid starvation response. Science 324, 1334–1338 10.1126/science.1172638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Peng W., Robertson L., Gallinetti J., Mejia P., Vose S., Charlip A., Chu T., and Mitchell J. R. (2012) Surgical stress resistance induced by single amino acid deprivation requires Gcn2 in mice. Sci. Transl. Med. 4, 118ra11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ni Y., Schwaneberg U., and Sun Z.-H. (2008) Arginine deiminase, a potential anti-tumor drug. Cancer Lett. 261, 1–11 10.1016/j.canlet.2007.11.038 [DOI] [PubMed] [Google Scholar]
- 40. Patil M. D., Bhaumik J., Babykutty S., Banerjee U. C., and Fukumura D. (2016) Arginine dependence of tumor cells: targeting a chink in cancer's armor. Oncogene 35, 4957–4972 10.1038/onc.2016.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Morrow K., Hernandez C. P., Raber P., Del Valle L., Wilk A. M., Majumdar S., Wyczechowska D., Reiss K., and Rodriguez P. C. (2013) Anti-leukemic mechanisms of pegylated arginase I in acute lymphoblastic T-cell leukemia. Leukemia 27, 569–577 10.1038/leu.2012.247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hernandez C. P., Morrow K., Lopez-Barcons L. A., Zabaleta J., Sierra R., Velasco C., Cole J., and Rodriguez P. C. (2010) Pegylated arginase I: a potential therapeutic approach in T-ALL. Blood 115, 5214–5221 10.1182/blood-2009-12-258822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yau T., Cheng P. N., Chan P., Chan W., Chen L., Yuen J., Pang R., Fan S. T., and Poon R. T. (2013) A phase 1 dose-escalating study of pegylated recombinant human arginase 1 (Peg-rhArg1) in patients with advanced hepatocellular carcinoma. Invest. New Drugs. 31, 99–107 10.1007/s10637-012-9807-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Suraweera A., Münch C., Hanssum A., and Bertolotti A. (2012) Failure of amino acid homeostasis causes cell death following proteasome inhibition. Mol Cell. 48, 242–253 10.1016/j.molcel.2012.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sherry S. T., Ward M. H., Kholodov M., Baker J., Phan L., Smigielski E. M., and Sirotkin K. (2001) dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 10.1093/nar/29.1.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hadinnapola C., Bleda M., Haimel M., Screaton N., Swift A., Dorfmüller P., Preston S. D., Southwood M., Hernandez-Sanchez J., Martin J., Treacy C., Yates K., Bogaard H., Church C., Coghlan G., et al. (2017) Phenotypic characterization of eIF2AK4 mutation carriers in a large cohort of patients diagnosed clinically with pulmonary arterial hypertension. Circulation 136, 2022–2033 10.1161/CIRCULATIONAHA.117.028351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang P., McGrath B. C., Reinert J., Olsen D. S., Lei L., Gill S., Wek S. A., Vattem K. M., Wek R. C., Kimball S. R., Jefferson L.S., and Cavener D. R. (2002) The GCN2 eIF2α kinase is required for adaptation to amino acid deprivation in mice. Mol. Cell. Biol. 22, 6681–6688 10.1128/MCB.22.19.6681-6688.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Teske B. F., Wek S. A., Bunpo P., Cundiff J. K., McClintick J. N., Anthony T. G., and Wek R. C. (2011) The eIF2 kinase PERK and the integrated stress response facilitate activation of ATF6 during endoplasmic reticulum stress. Mol. Biol. Cell 22, 4390–4405 10.1091/mbc.E11-06-0510 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.