Abstract
Heart failure is an aging-associated disease that is the leading cause of death worldwide. Sirtuin family members have been largely studied in the context of aging and aging-associated diseases. Sirtuin 2 (SIRT2) is a cytoplasmic protein in the family of sirtuins that are NAD+-dependent class III histone deacetylases. In this work, we studied the role of SIRT2 in regulating nuclear factor of activated T-cells (NFAT) transcription factor and the development of cardiac hypertrophy. Confocal microscopy analysis indicated that SIRT2 is localized in the cytoplasm of cardiomyocytes and SIRT2 levels are reduced during pathological hypertrophy of the heart. SIRT2-deficient mice develop spontaneous pathological cardiac hypertrophy, remodeling, fibrosis, and dysfunction in an age-dependent manner. Moreover, young SIRT2-deficient mice develop exacerbated agonist-induced hypertrophy. In contrast, SIRT2 overexpression attenuated agonist-induced cardiac hypertrophy in cardiomyocytes in a cell-autonomous manner. Mechanistically, SIRT2 binds to and deacetylates NFATc2 transcription factor. SIRT2 deficiency stabilizes NFATc2 and enhances nuclear localization of NFATc2, resulting in increased transcription activity. Our results suggest that inhibition of NFAT rescues the cardiac dysfunction in SIRT2-deficient mice. Thus, our study establishes SIRT2 as a novel endogenous negative regulator of NFAT transcription factor.
Keywords: heart failure, cardiac hypertrophy, sirtuin, NFAT transcription factor, cardiomyocyte, SIRT2, lysine acetylation
Introduction
The term heart failure denotes loss of function of myocardium, which is the culmination of complex remodeling that occurs in the heart, initiated as an adaptive response to pressure overload or cardiac stressors in humans. In the failing myocardium, several functional abnormalities, including disruption of calcium homeostasis, metabolic malfunction, energy deficits, and contractile dysfunction have been recognized (1, 2). Notably, pathological cardiac remodeling involves activation of cell surface receptors, signaling mediators, cellular protein synthesis, and transcriptional machinery (3, 4). Although there has been an advancement in the understanding of risk factors for heart failure, the basic molecular events responsible for initiation of heart failure are poorly recognized.
Heart failure is reversible by calorie restriction, a feeding regimen that limits calorie intake, or by physical exercise in animal models (5–7). Evidence suggests that health benefits of calorie restriction and exercise are mediated through the activation of NAD+-dependent class III histone deacetylases, called sirtuins. So far, seven different sirtuin isoforms (SIRT1 to 7), have been identified in mammals (8). Sirtuin 2 (SIRT2) is highly expressed in the brain and the heart of humans (9). SIRT2 regulates nuclear envelope dynamics, cell metabolism, and autophagy (10). SIRT2 levels increase during calorie restriction and nutrient deprivation (11). On the other hand, SIRT2 levels are reduced in the visceral adipose tissue of human obese subjects, human liver tissues of iron overload, hepatocellular carcinoma samples, and cardiomyocytes from an animal model of type 1 diabetic mellitus (12–15). SIRT2-deficient mice show an increased incidence of mammary tumors and hepatocellular carcinoma (14). Similarly, SIRT2 deficiency increases susceptibility to colitis and iron deficiency–induced hepatocyte death (13, 16). Recently it has been demonstrated that SIRT2 deficiency promotes cardiac hypertrophy through impaired activation of AMP-activated protein kinase (AMPK)4 signaling in the heart (17). AMPK is the key regulator of cardiac energy homeostasis, and it regulates protein synthesis in heart during hypertrophy (18, 19). However, it is not clear how AMPK regulates the other events, such as cardiac fetal gene expression, involved in the hypertrophy of the heart.
In the hypertrophic myocardium, activation of nuclear factor of activated T-cells (NFAT) as a transcription factor plays a key role in the expression of cardiac fetal genes. Roughly 13% of the genes expressed in the human heart have the binding site for NFAT in their promoter region, whereas in advanced stages of heart failure, ∼20–40% of these genes have modified expression (20). NFAT has five different isoforms, NFATc1–4 and NFAT5. NFATc1–4 are regulated by calcium signaling, whereas NFAT5 is regulated by the hyperosmotic environment of the cell (21). The protein levels and activity of NFAT is up-regulated in age-related diseases and heart failure (22). Of note, NFAT transcription factors are involved in pathological, but not physiological, forms of cardiac hypertrophy (23).
In the present work, we studied the role of SIRT2 in the development of heart failure. Our results suggest that SIRT2 deficiency induces spontaneous heart failure in mice. SIRT2 deficiency also exacerbates agonist-induced cardiac hypertrophy. On the other hand, overexpression of SIRT2 in cardiomyocytes antagonizes the agonist-induced cardiac hypertrophy through repression of NFAT.
Results
SIRT2 is a cytoplasmic protein in cardiomyocytes
In dividing cells, SIRT2 is predominantly a cytoplasmic protein, but shuttles between the nucleus and cytoplasm during G2/M transition (24). Because cardiomyocytes are postmitotic, we tested the localization of SIRT2 in cardiomyocytes by confocal microscopy. Our results suggest that SIRT2 is mostly cytoplasmic under basal conditions (Fig. 1A). Next, we tested whether SIRT2 localization is influenced by cardiac stressors, as previous reports suggested SIRT1, another sirtuin isoform, localization to be regulated by stress (25). Cardiomyocytes were treated with phenylephrine (PE), α1-adrenergic receptor agonist, and its localization was tested. We did not see any major changes in the localization of SIRT2 when cells were treated with PE (Fig. 1A). These findings indicate that SIRT2 is a major cytoplasmic protein in cardiomyocytes under basal as well as stressed conditions.
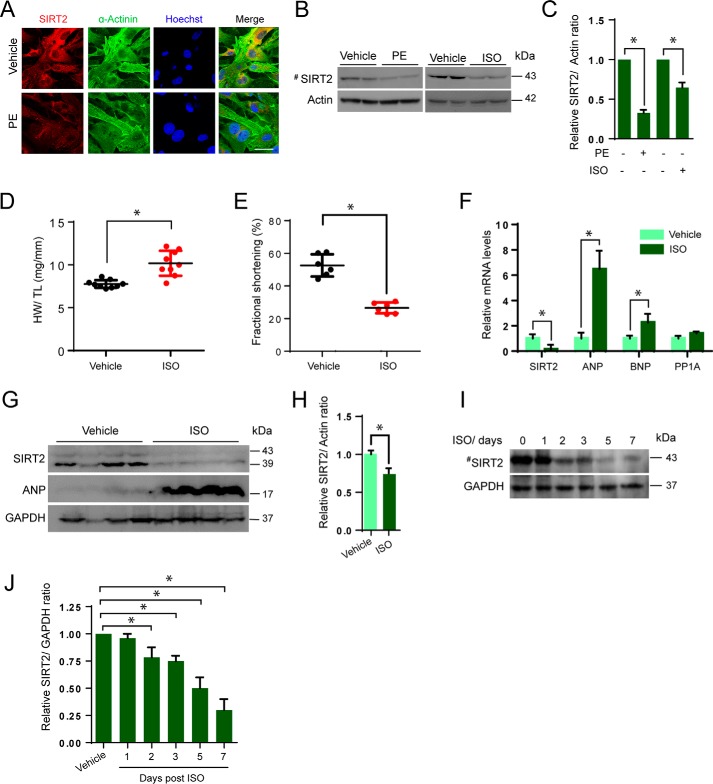
Figure 1.
Pathological hypertrophy reduces SIRT2 levels in heart. A, representative confocal images of vehicle or PE (20 μm for 48 h) treated neonatal primary cardiomyocytes stained with α-actinin and SIRT2. Scale bar = 20 μm. B, Western blot analysis of vehicle- or PE- or ISO- treated (20 μm for 48 h) primary cardiomyocytes probed for SIRT2. C, quantification of SIRT2 from B. Error bars, mean ± S.D.; *, p < 0.05. D, scatter plot representing HW/TL ratio of vehicle- or ISO-treated (10 mg/kg/day for 7 days) mice. n = 9 mice; error bars, mean ± S.D.; *, p < 0.05. E, scatter plot representing fractional shortening of vehicle- or ISO-treated (10 mg/kg/day for 7 days) mice. n = 6 mice; error bars, mean ± S.D.; *, p < 0.05. F, qPCR analysis of SIRT2, ANP, BNP, and PPIA in vehicle- or ISO-treated mice hearts. Cyclophilin A (PPIA) was used as negative control. n = 4 mice; error bars, mean ± S.D.; *, p < 0.05. G, Western blot analysis of vehicle- and/or ISO-treated mice heart samples for SIRT2 and ANP. n = 4 mice. H, quantification of SIRT2 from G. I, Western blot analysis of vehicle- or ISO-treated mice for SIRT2, n = 4 mice. J, quantification of SIRT2 from I.
SIRT2 levels are reduced during pathological hypertrophy of the heart
To understand the role of SIRT2 in the development of cardiac hypertrophy, we first tested the levels of SIRT2 in cardiomyocytes treated with PE or isoproterenol (ISO). We found reduced levels of SIRT2 in PE-treated cells by confocal microscopy (Fig. 1A). Similarly, we found markedly low levels of SIRT2 in PE- or ISO-treated cells by Western blotting (Fig. 1, B and C). In the next set of experiments, we tested whether the cardiac hypertrophy is associated with reduced levels of SIRT2. We induced cardiac hypertrophy by chronically infusing the hypertrophic agonist, ISO by implanting osmotic minipumps into the peritoneal cavity of mice (26). Our results suggest that chronic ISO infusion significantly increased the heart weight to tibia length (HW/TL) ratio, and contractile dysfunction (Fig. 1, D and E). Further quantitative PCR (qPCR) analysis suggested that levels of fetal genes, ANP, and BNP were significantly increased in ISO-infused mice, indicating the development of cardiac hypertrophy (Fig. 1F). Next, we analyzed the mRNA and protein levels of SIRT2 in the hypertrophied hearts. We found significantly lower levels of SIRT2 mRNA in ISO-infused heart tissues by real-time qPCR analysis (Fig. 1F). Similarly, we found markedly low levels of SIRT2 protein in ISO-infused heart samples by Western blotting (Fig. 1, G and H). In order to measure the SIRT2 levels during the initial stages of the development of cardiac hypertrophy, we analyzed the SIRT2 levels at different time points post–ISO infusion by Western blotting. Western blotting analysis suggested that SIRT2 levels reduced significantly post 2 days ISO infusion and decreased further until the day tested (Fig. 1, I and J). These data indicate that SIRT2 levels decrease during cardiac hypertrophy and its deficiency might be associated with the development of cardiac hypertrophy.
SIRT2 deficiency induces spontaneous cardiac hypertrophy and failure
To test whether SIRT2 deficiency causes heart failure, we analyzed the cardiac structure and function of C57BL/6 SIRT2-deficient (SIRT2-KO) mice at 2 and 9 months of age. Western blotting analysis confirmed deficiency of SIRT2 in SIRT2-KO mice hearts (Fig. 2A). At 2 months of age, SIRT2-deficient mice showed no obvious cardiac phenotype (Fig. 2, B–E). Interestingly, SIRT2-deficient mice developed cardiac hypertrophy with significantly increased HW/TL ratio, wall thickness, and left ventricular internal diameter at 9 months of age (Fig. 2, B–D). Further, echocardiographic analysis of 9-month-old SIRT2-deficient mice indicated significantly reduced fractional shortening, an indicator of cardiac contractile dysfunction (Fig. 2E). SIRT2-deficient mice hearts exhibited significant down-regulation in the expression of α-MHC and up-regulation of BNP, as measured by real-time qPCR in 9-month-old SIRT2-deficient mice heart (Fig. 2F). Histological analysis indicated a significant increase in cardiomyocyte cross-sectional area in SIRT2-deficient mice hearts at 9 months of age (Fig. 2, G and H). In addition, we found prominent interstitial and replacement fibrosis in the heart tissues of SIRT2-deficient mice (Fig. 2, G–I). The expression of fibrosis-associated genes, α-SMA, fibronectin-1, and collagen-1 was also up-regulated in SIRT2-deficient mice heart samples at 9 months of age (Fig. 2J). Collectively, these findings suggest that SIRT2 deficiency leads to spontaneous pathological cardiac hypertrophy, remodeling, fibrosis, and subsequent heart failure.
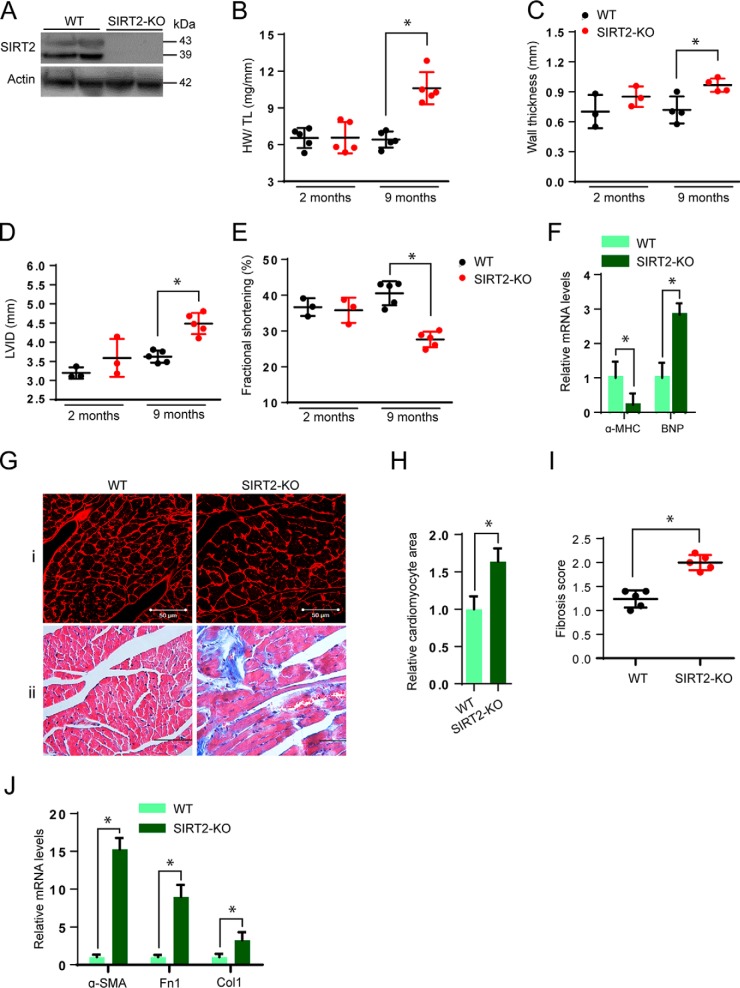
Figure 2.
SIRT2 deficiency induces cardiac hypertrophy. A, Western blot analysis of WT and SIRT2-KO mice hearts for SIRT2. B, scatter plot representing HW/TL ratio of WT and SIRT2-KO mice at 2 months and 9 months of age. Error bars, mean ± S.D.; n = 5 mice; *p < 0.05. C–E, scatter plots depicting left ventricular wall thickness (C), left ventricular internal diameter (D), and fractional shortening (E) of WT and SIRT2-KO mice at 2 months and 9 months of age. n = 3–5 mice; error bars, mean ± S.D.; *, p < 0.05. F, qPCR analysis of α-MHC and BNP in WT and SIRT2-KO mice hearts at 9 months of age. n = 4 mice; error bars, mean ± S.D.; *, p < 0.05. G(i), representative confocal images showing WGA staining in sections of WT and SIRT2-KO mice hearts at 9 months of age. Scale bar = 50 μm. G(ii), heart sections of 9-month-old WT and SIRT2-KO mice stained with Masson's trichrome stain showing cardiac fibrosis. n = 3–5 mice. H, graph showing relative cardiomyocyte cross-sectional area measured from G(i). I, scatter plot showing fibrosis scored in a blinded fashion from G(ii). n = 5 mice; error bars, mean ± S.D.; *, p < 0.05. J, qPCR analysis of alpha smooth muscle actin (α-SMA), fibronectin 1 (Fn1) and collagen I (Col1) in WT and SIRT2-KO mice hearts at 9 months of age. n = 3–4 mice; error bars, mean ± S.D.; *, p < 0.05.
SIRT2 deficiency exacerbates isoproterenol-induced hypertrophy
Because SIRT2-deficient mice do not show any obvious cardiac phenotype at 2 months of age, we intended to test whether SIRT2 deficiency exacerbates agonist-induced cardiac hypertrophy. Chronic ISO infusion by implanting osmotic minipumps into the peritoneal cavity of mice resulted in nearly 40% cardiac hypertrophy in SIRT2-KO mice, whereas wildtype mice produced almost 20% cardiac hypertrophy, as assessed by HW/TL ratio and wall thickness (Fig. 3, A and B). In SIRT2-KO mice, fractional shortening was significantly low after treatment with ISO, when compared with controls, suggesting that SIRT2 deficiency markedly impairs cardiac functions following chronic hypertrophic agonist infusion (Fig. 3C). Similarly, fibrosis was found to be significantly high in SIRT2-KO mice hearts as compared with control mice after ISO treatment (Fig. 3, D and E). These results suggest that SIRT2 deficiency exacerbates agonist-induced cardiac hypertrophic response in mice. These findings were consistent with the previous work, where SIRT2 deficiency exacerbated Ang II–induced cardiac hypertrophy (17).
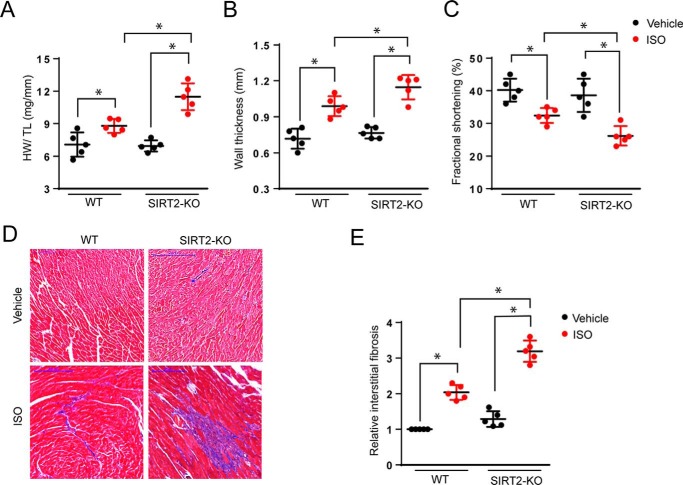
Figure 3.
SIRT2 deficiency augments agonist-induced cardiac hypertrophy. A, scatter plot showing HW/TL ratio of 2-month-old WT and SIRT2-KO mice treated with vehicle or isoproterenol (5 mg/kg/day) for 7 days. n = 5 mice; error bars, mean ± S.D.; *, p < 0.05. B, scatter plot depicting left ventricular wall thickness of 2-month-old WT and SIRT2-KO mice treated with vehicle or ISO. n = 5 mice; error bars, mean ± S.D.; *, p < 0.05. C, scatter plot depicting fractional shortening of 2-month-old WT and SIRT2-KO mice treated with vehicle or ISO. n = 5 mice; error bars, mean ± S.D.; *, p < 0.05. D, heart sections of 2-month-old WT and SIRT2-KO mice treated with vehicle or ISO stained with Masson's trichrome stain to detect fibrosis. n = 5 mice. E, graph showing relative fibrosis scored in a blinded fashion from D. n = 5 mice; error bars, mean ± S.D.; *, p < 0.05.
SIRT2 depletion causes hypertrophy of cardiomyocytes in a cell-autonomous manner
Because SIRT2-deficient mice lack SIRT2 in all the tissues, the role of SIRT2 on cardiac functions may or may not be directly related to its role in cardiomyocytes. To test whether SIRT2 is involved in cardiomyocyte hypertrophy, we performed experiments in cultured murine cardiomyocytes. SIRT2 levels were reduced in cardiomyocytes by transfecting specific siRNA pools and their susceptibility to cardiac hypertrophy was tested. SIRT2 depletion in cardiomyocytes was confirmed by Western blotting analysis (Fig. 4A). Interestingly, depletion of SIRT2 in cardiomyocytes significantly increased protein synthesis as measured by [3H]leucine incorporation (Fig. 4B). Similarly, Western blotting analysis suggested that SIRT2-depleted cardiomyocytes exhibited increased expression of ANP at basal as well as PE- or ISO-treated conditions (Fig. 4C). Moreover, SIRT2-depleted cardiomyocytes showed perinuclear localization of ANP and sarcomere reorganization, which are hallmarks of cardiomyocyte hypertrophy (26, 27) (Fig. 4D). These findings indicate that SIRT2 depletion causes cardiomyocyte hypertrophy in a cell-autonomous manner.
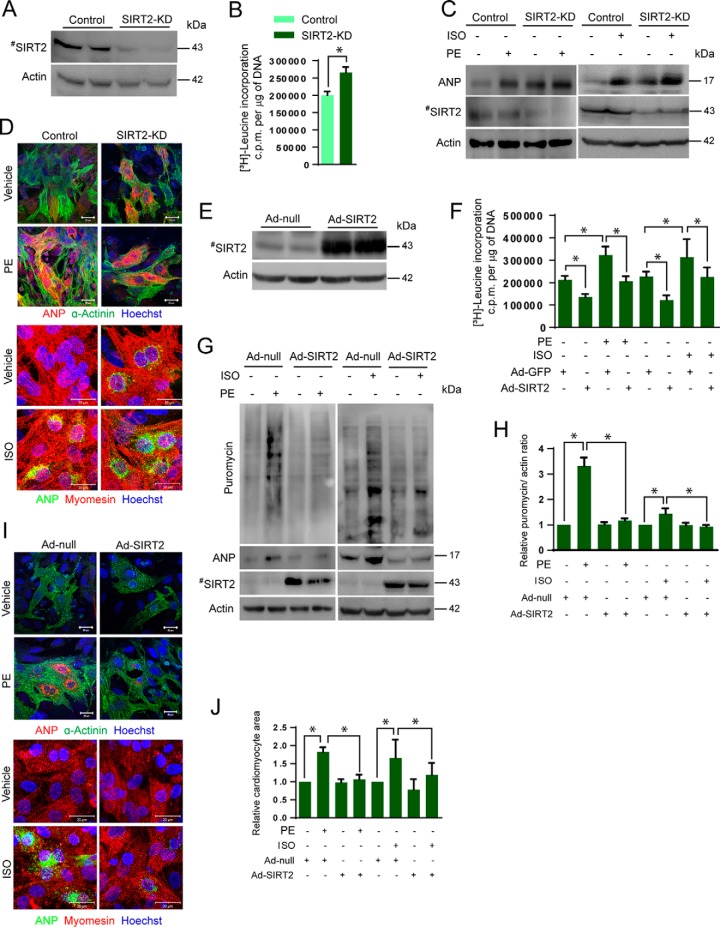
Figure 4.
SIRT2 depletion induces hypertrophy in cardiomyocytes. A, Western blot analysis confirming depletion of SIRT2 in cardiomyocytes transfected with SIRT2-specific siRNA. B, [3H]leucine incorporation into cellular proteins of neonatal rat cardiomyocytes transfected with control or SIRT2 siRNA. n = 5 mice; error bars, mean ± S.D.; *, p < 0.05. c.p.m., counts per minute. C, Western blot analysis of control and SIRT2-knockdown (SIRT2-KD) cardiomyocytes treated with vehicle or PE (20 μm) or ISO (20 μm) for 48 h and probed for ANP and SIRT2. D, representative confocal images of control and SIRT2-KD cardiomyocytes treated with vehicle or PE (20 μm) or ISO (20 μm) for 48 h and further stained with ANP and α-actinin or myomesin. Scale bar = 20 μm. E, Western blot analysis showing overexpression of SIRT2 in cardiomyocytes. F, [3H]leucine incorporation into total cellular protein of neonatal rat cardiomyocytes infected with SIRT2 (Ad-SIRT2) or control adenovirus (Ad-GFP) and then treated with vehicle or PE (20 μm) or ISO (20 μm) for 48 h. n = 5; error bars, mean ± S.D.; *, p < 0.05. c.p.m., counts per minute. G, Western blot analysis of Ad-null or Ad-SIRT2 expressing cardiomyocytes treated with vehicle or PE (20 μm) or ISO (20 μm) for 48 h and harvested after puromycin treatment (1 μm), and probed for puromycin, ANP, and SIRT2. H, quantification of puromycin incorporation from G. I, representative confocal images of control and SIRT2 overexpressing cardiomyocytes treated with vehicle or PE (20 μm) or ISO (20 μm) for 48 h and stained with ANP and α-actinin or myomesin. Scale bar = 20 μm. J, graph showing quantification of relative cardiomyocyte size measured from I.
SIRT2 overexpression blocks agonist-induced cardiomyocyte hypertrophy
To test whether SIRT2 activation blocks the cardiac hypertrophy, we analyzed the effect of SIRT2 overexpression in PE-induced cardiomyocyte hypertrophy. We infected cardiomyocytes with adenovirus expressing SIRT2 and studied the hypertrophic response to PE or ISO treatment. SIRT2 overexpression in cardiomyocytes was confirmed by Western blotting (Fig. 4E). Cardiomyocytes were treated with PE or ISO for 48 h, and their hypertrophic response was assessed. We found that SIRT2 overexpression attenuates PE as well as ISO-stimulated protein synthesis in cardiomyocytes as measured by [3H]leucine incorporation assay (Fig. 4F). To further validate the results of leucine incorporation assay, we used an alternative nonradioactive technique, which measures global protein synthesis by monitoring the labeling of nascent peptide to puromycin (28). Primary cardiomyocytes were infected with adenovirus-expressing SIRT2 and the basal and PE- or ISO-induced puromycin incorporation was measured by Western blotting using specific antibody against puromycin. Western blot analysis suggested that SIRT2 overexpression attenuates PE- as well as ISO-stimulated protein synthesis in cardiomyocytes (Fig. 4, G and H). Further, we noted a marked reduction in PE- or ISO-induced expression and perinuclear localization of ANP, cardiomyocyte size, and sarcomere reorganization in SIRT2 overexpressing cardiomyocytes (Fig. 4, G, I, and J). Collectively, these results indicate that SIRT2 overexpression is sufficient to block agonist-induced cardiomyocyte hypertrophy.
SIRT2 deficiency hyperactivates NFAT signaling in heart
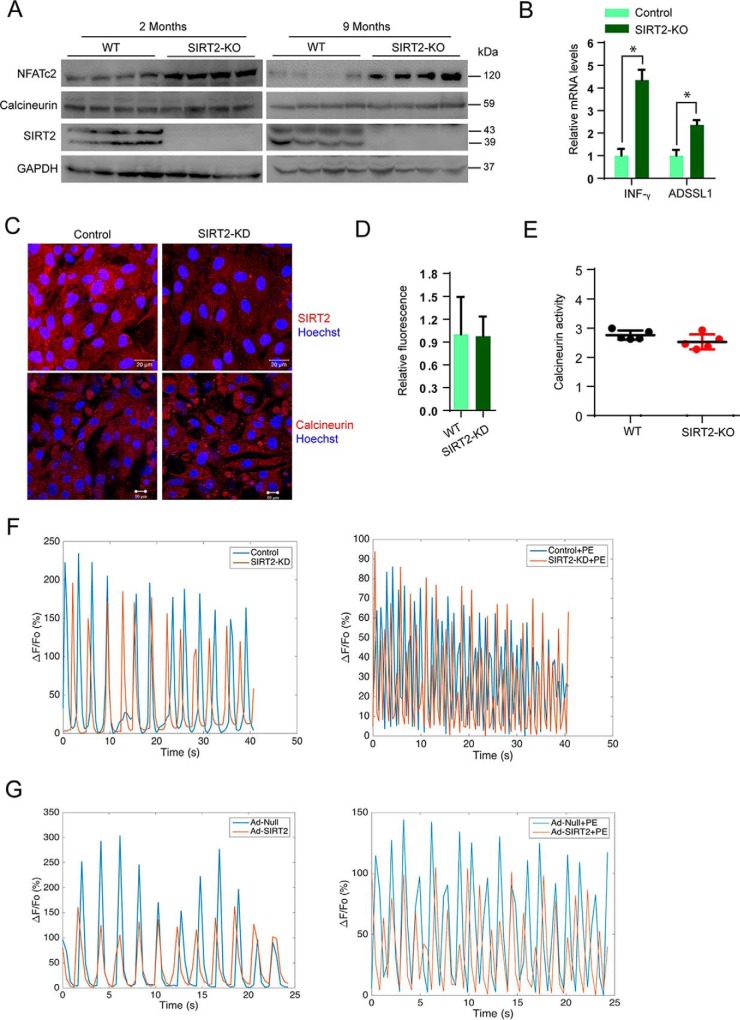
Previous studies indicate that calcineurin-NFAT signaling regulates the cardiac hypertrophic response. NFAT transcriptionally regulates the expression of fetal genes such as ANP, Myh7, and CARP in cardiomyocytes (29). Because our findings suggest that SIRT2 deficiency spontaneously activates fetal gene program in the heart, we hypothesized that SIRT2 might directly control NFAT signaling in hearts. We performed Western blotting analysis to detect the expression of NFATc2, a major NFAT isoform in heart tissue of 2- and 9-month-old SIRT2-deficient mice. Results suggested that SIRT2-deficient hearts exhibit increased levels of NFATc2 at both age groups (Fig. 5A). To test the endogenous NFAT activity, we quantified the expression of IFN-γ and ADSSL1, which are well-characterized targets of NFAT in the heart (30, 31). Our qPCR results suggested that SIRT2-deficient hearts express significantly higher levels of NFAT target genes, suggesting that SIRT2 deficiency hyperactivated NFAT in the heart (Fig. 5B).
Figure 5.
SIRT2 deficiency increases NFATc2 levels in mice hearts. A, Western blot analysis of 2- and 9-month-old WT and SIRT2-KO mice hearts samples for NFATc2, calcineurin, and SIRT2. B, qPCR analysis of IFN-γ and ADSSL1 in 9-month-old WT and SIRT2-KO mice hearts. n = 3–4 mice; error bars, mean ± S.D.; *, p < 0.05. C, representative confocal images showing control and SIRT2-KD cardiomyocytes stained for calcineurin and SIRT2. Scale bar = 20 μm for upper panel, 10 μm for lower panel. D, graph showing quantification of calcineurin from C. Error bars, mean ± S.D.; *, p < 0.05. E, scatter plot showing calcineurin activity assay in 9-month-old WT and SIRT2-KO mice hearts. n = 5 mice; error bars, mean ± S.D.; *, p < 0.05. F, graph showing the calcium transients in control and SIRT2-KD cardiomyocytes treated with either vehicle or PE. G, graph showing the calcium transients in Ad-null and Ad-SIRT2 overexpressing cardiomyocytes treated with either vehicle or PE.
In hypertrophic hearts, dephosphorylation of NFAT transcription factor by calcineurin, a Ca2+-dependent phosphatase, enhances nuclear localization and transcriptional activity of NFAT (23, 29). Because our results show increased nuclear localization of NFAT in SIRT2-deficient cardiomyocytes, we expected that SIRT2 might modulate the levels and the activity of calcineurin. Interestingly, we do not find marked changes in the protein levels of calcineurin in SIRT2-deficient mice hearts at 2 months and 9 months of age (Fig. 5A). Further, confocal microscopy analysis suggested similar results (Fig. 5, C and D). Moreover, we do not see any marked changes in the activity of calcineurin in SIRT2-deficient mice hearts (Fig. 5E). These findings suggest that the increased activity of NFATc2 observed in SIRT2-deficient conditions is not associated with hyperactive calcineurin.
It is well-accepted that increased calcium concentration activates calcineurin signaling in the heart (32). Therefore, we measured the calcium transients in control and SIRT2-depleted cardiomyocytes by live confocal microscopy. Under basal conditions, we do not find marked differences in calcium transients in wildtype and SIRT2-depleted cardiomyocytes. Similarly, treatment of PE in SIRT2-depleted cardiomyocytes does not change calcium transients (Fig. 5F). Furthermore, cardiomyocytes overexpressing SIRT2 do not show any change in calcium transients at basal and PE-induced conditions (Fig. 5G). These findings suggest that increased nuclear localization of NFAT is not related to calcium transients in SIRT2-deficient cardiomyocytes.
SIRT2 regulates the transcriptional activity of NFAT transcription factor
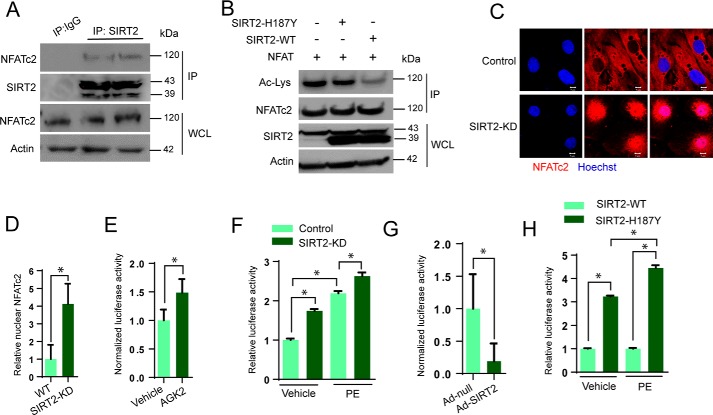
NFAT is known to be an acetylated protein, and acetylation of NFATc1 by acetyl transferases p300 and PCAF promotes its stability, and thereby enhances the transcriptional activity of NFATc1 during osteoclast differentiation (33). Because SIRT2 is a cytoplasmic deacetylase, and our results suggest increased levels of NFATc2 in SIRT2-deficient mice, we suspected that SIRT2 might regulate the acetylation of NFATc2. To test whether SIRT2 interacts with NFATc2, we immunoprecipitated NFATc2 from heart samples and tested its binding with SIRT2 by Western blotting. Our results suggest that SIRT2 interacts with NFATc2 (Fig. 6A). To test whether SIRT2 deacetylates NFATc2, we performed an in vitro deacetylation assay, where NFATc2 was first acetylated by p300 acetyltransferase. Subsequently, acetylated NFATc2 was incubated with either WT or SIRT2-H187Y, a catalytic mutant of SIRT2. Western blotting analysis indicated WT, but not catalytic mutant-SIRT2 markedly reduced the acetylation status of NFATc2 (Fig. 6B). These findings indicate that SIRT2 is an NFATc2 deacetylase.
Figure 6.
SIRT2 deacetylates NFATc2 and inhibits its activity. A, Western blot analysis of SIRT2 interaction with NFATc2. SIRT2 was immunoprecipitated from heart lysates and probed for its interaction with NFATc2. B, in vitro deacetylation assay where recombinant acetylated-NFATc2 was incubated with wildtype or SIRT2-H187Y in the presence of NAD+. NFATc2 acetylation was analyzed by Western blotting. C, representative confocal images showing control and SIRT2-KD cardiomyocytes stained for NFATc2. Scale bar = 5 μm. D, graph showing quantification of NFATc2 from Fig. 4C. Error bars, mean ± S.D.; *, p < 0.05. E, luciferase activity in vehicle- or AGK2-treated cells transfected with NFAT-Luc plasmid. n = 3–5; error bars, mean ± S.D.; *, p < 0.05. F, luciferase activity in control and SIRT2-KD cells treated with vehicle or PE (20 μm) for 48 h. n = 3; error bars, mean ± S.D.; *, p < 0.05. G, luciferase activity in control and SIRT2 overexpressing cardiomyocytes transfected with NFAT-Luc plasmid. Error bars, mean ± S.D.; n = 4; *, p < 0.05. H, luciferase activity in wildtype SIRT2 and SIRT2-H187Y overexpressing cells treated with vehicle or PE (20 μm) for 48 h. n = 3; error bars, mean ± S.D.; *, p < 0.05.
Because the nuclear localization of NFAT is required for its transcriptional activation, we tested the endogenous localization of NFATc2 in SIRT2-depleted murine cardiomyocytes by confocal microscopy. We found increased nuclear localization of NFATc2 in SIRT2-depleted cardiomyocytes (Fig. 6, C and D). We further validated the effect of SIRT2 on the transcription activity of NFAT by an NFAT-responsive promoter/reporter (NFAT-Luc) assay. Our result suggested that SIRT2 inhibition by AGK2, a pharmacological inhibitor of SIRT2, significantly increased NFAT-Luc activity (Fig. 6E). Moreover, SIRT2 depletion significantly increased the basal and PE-induced transcription activity of NFAT (Fig. 6F). On the other hand, adenovirus-mediated overexpression of SIRT2 attenuated the activity of NFAT-Luc, suggesting that SIRT2 blocks the transcription activity of NFAT transcription factor in cardiomyocytes (Fig. 6G). To further validate whether SIRT2-mediated repression of NFAT requires its deacetylase activity, we performed reporter assay with wildtype and catalytically inactive mutant of SIRT2. Cells were transiently overexpressed with either SIRT2-WT or SIRT2-H187Y mutant, and NFAT transcriptional activity was measured. Our result suggested that wildtype, but not SIRT2-H187Y, attenuated PE-induced increased transcriptional activity of NFAT (Fig. 6H). Collectively, our findings indicate that SIRT2 binds to and deacetylases NFATc2 transcription factor, and its deficiency might increase protein levels and nuclear translocation of NFATc2, resulting in an augmented transcriptional activity of NFATc2.
Inhibition of NFAT signaling rescues hypertrophy in SIRT2-deficient cardiomyocytes
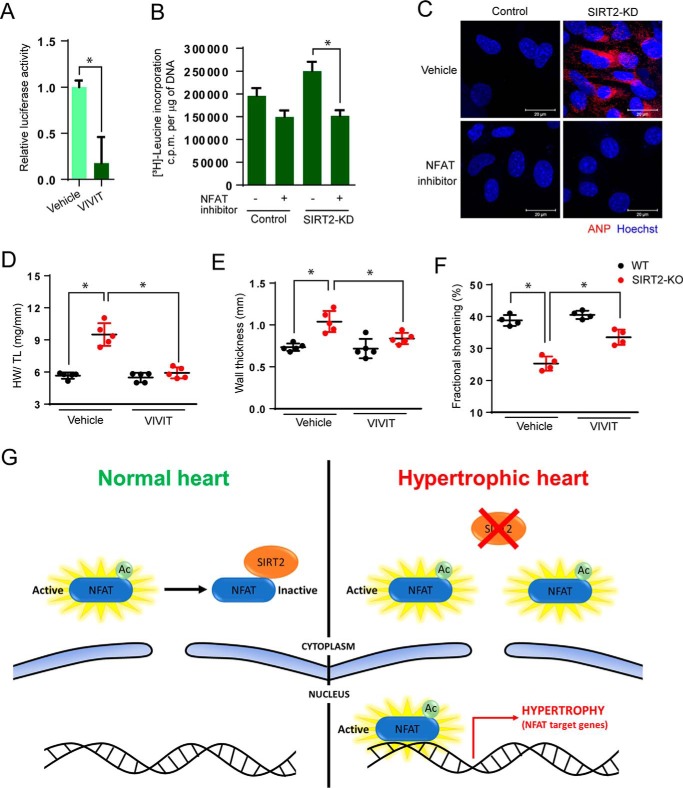
To confirm the role of NFAT in the induction of hypertrophy in SIRT2-deficient cardiomyocytes, we tested the effects of VIVIT, a cell-permeable peptide inhibitor of NFAT (34). SIRT2-depleted cardiomyocytes were treated with VIVIT and tested for the markers of cardiac hypertrophy. Our results suggest that treatment with VIVIT markedly reduced the endogenous transcriptional activity of NFAT, as measured by luciferase reporter assay (Fig. 7A). Interestingly, treatment with VIVIT significantly attenuated the protein synthesis in SIRT2-depleted cardiomyocytes (Fig. 7B), suggesting that inhibition of NFAT reduces cardiac hypertrophy in SIRT2-depleted cardiomyocytes. Similarly, treatment with VIVIT markedly reduced the perinuclear localization of ANP in SIRT2-depleted cardiomyocytes (Fig. 7C), suggesting that inhibition of NFAT rescues the expression of fetal genes in SIRT2-depleted cardiomyocytes. To validate our findings in vivo, we performed rescue experiments with VIVIT in SIRT2-KO mice (34). Treatment with VIVIT significantly reduced HW/TL ratio and wall thickness, while increasing fractional shortening in SIRT2-deficient mice (Fig. 7, D–F), indicating that NFAT inhibition rescue the cardiac hypertrophy and contractile dysfunction resulting from SIRT2 deficiency.
Figure 7.
NFAT inhibition rescues hypertrophy in SIRT2-depleted cardiomyocytes. A, luciferase reporter assay for cells treated with vehicle or NFAT inhibitor (VIVIT, 1 μm). B, [3H]leucine incorporation into total cellular protein of control and SIRT2-KD cardiomyocytes treated with vehicle or NFAT inhibitor (VIVIT, 1 μm). n = 5; error bars, mean ± S.D.; *, p < 0.05. c.p.m., counts per minute. C, representative confocal images showing ANP expression in vehicle or NFAT inhibitor (VIVIT, 1 μm) treated control and SIRT2-KD cardiomyocytes. D, scatter plot representing HW/TL ratio of 9-month-old WT and SIRT2-KO mice treated with either vehicle or NFAT inhibitor VIVIT for 14 days. n = 5 mice; error bars, mean ± S.D.; *, p < 0.05. E, scatter plot depicting left ventricular wall thickness of 9-month-old WT and SIRT2-KO mice treated with either vehicle or NFAT inhibitor VIVIT for 14 days. n = 5 mice; error bars, mean ± S.D.; *, p < 0.05. F, scatter plot depicting fractional shortening of 9-month-old WT and SIRT2-KO mice treated with either vehicle or NFAT inhibitor VIVIT (10 mg/kg) for 14 days. n = 5 mice; error bars, mean ± S.D.; *, p < 0.05. G, schematic representation of the proposed mechanism. Under physiological conditions, SIRT2 deacetylases NFATc2 and excludes it from the nucleus, leading to its degradation, and represses its transcriptional activity. During cardiac stress SIRT2 levels decrease in heart. resulting in enhanced acetylation of NFATc2 and transcriptional hyperactivation. Therefore, NFAT remains active inside the nucleus and induces cardiac hypertrophy program.
Discussion
We found that SIRT2 deacetylates and regulates the activity of NFATc2, a key effector of pathological cardiac remodeling. In previous reports, it has been shown that NFAT activity is up-regulated in age-related diseases and heart failure (22). Moreover, the protein level of NFATc2 was increased in failing and aging hearts (35). Increased calcineurin/NFAT signaling in the diseased human and mouse myocardium reactivates the transcription factor Hand2 and fetal gene program, intricately linked to pathological heart disease (36). Previous reports have shown that SIRT1, a founding member of sirtuins, suppresses NFAT activity during inflammation in atherosclerosis (37, 38). Similarly, SIRT3, a mitochondrial deacetylase deficiency in cardiomyocytes is known to up-regulate the activity of NFAT (26). In our study, we found that SIRT2 is required to repress NFAT activity in the heart. Our finding is unique among sirtuins, as our report suggests that SIRT2 binds to and deacetylates NFAT to regulate its transcription activity, which has not been demonstrated previously.
In our current work, we have shown that SIRT2 deficiency promotes cardiac hypertrophy through activation of NFAT transcription factors. Binding of PE and ISO to α-adrenergic receptor (AR) and β-adrenergic receptor, respectively, results in activation of G protein–coupled receptor via specific Gα subunit and downstream signaling. Second messengers such as cyclic AMP are produced by α-AR, whereas inositol 1,4,5-trisphosphate (InsP3) and diacylglycerol (DAG) are produced by stimulated β-AR (39). Both PE and ISO increase calcium concentration and hence hypertrophic response. Stimulation of both α-AR and β-AR results in an increase in the organization of individual contractile myosin light chain into sarcomere and activate the immediate early gene program (40). Interestingly, NFAT transcription factor activation plays a key role in the cardiac hypertrophy induced by PE and ISO. PE induces NFAT translocation to the nucleus, leading to hypertrophy (41, 42). Similarly, ISO activates NFAT and induces myocardial hypertrophy by increasing the expression of NFAT target fetal genes (43). Our data suggest that SIRT2 activation might antagonize NFAT-dependent cardiac hypertrophy independent of upstream cardiac stress stimuli.
Sirtuins have been known to play diverse roles in heart failure. SIRT3 and SIRT6 are known to be protective against hypertrophy and heart failure (26, 27). SIRT7 is protective against inflammatory cardiomyopathy (44). A recent report shows the protective nature of SIRT5 in the heart (45). In contrast, SIRT1 and SIRT4 are known to induce hypertrophy (46, 47). On similar lines, previous studies have shown that SIRT1 is involved in the progression of cardiac hypertrophy through the suppression of ERR transcriptional pathway by SIRT1, thus inducing mitochondrial dysfunction and heart failure (46). Our previous work has also shown that SIRT1 induces cardiac hypertrophy (47). Therefore, SIRT1 and SIRT2, although both are localized in cardiomyocyte cytoplasm, may have a contrasting role in the regulation of cardiac hypertrophy. Because cardiomyocytes are post-mitotic cells, SIRT2 is localized in the cytoplasm, even upon induction with hypertrophic agonists. This is in concurrence with the previous reports that SIRT2 is a cytoplasmic protein and shuttles to the nucleus during G2/M transition (24). In the current work, we used whole-body SIRT2-deficient mice, which lack SIRT2 in all the cell types in heart, including cardiac fibroblasts. Because SIRT2 is expressed in fibroblasts as well, the phenotype found in SIRT2-KO mice might be because of the cumulative role of SIRT2 in cardiomyocytes and fibroblasts.
SIRT2 levels have been seen to be down-regulated in aging-related disorders like diabetes, metabolic regulation, and age-related immune and inflammatory responses (48, 49). SIRT2 has also been shown to act as a tumor suppressor (50). Ectopic expression of SIRT2 led to a reduction in the colony formation ability in glioma cell lines (51). SIRT2 deficiency is known to promote genomic instability, which is an early event in carcinogenesis (14, 52–54). Consistently, genomic data analysis has shown SIRT2 expression to be down-regulated in human hepatocellular carcinoma and breast cancer (14). Similarly, microarray analysis has shown reduced SIRT2 expression at the mRNA level in glioblastoma, prostate cancer, oligodendroglioma, and clear cell renal carcinoma (55). In our study too, we found SIRT2 to be down-regulated in the later stages of hypertrophic heart samples. These findings suggest that SIRT2 can be used as a biomarker to hint at the onset of hypertrophy and heart failure.
Recently, SIRT2 has been shown to protect heart from Ang II–induced hypertrophic stimuli by promoting the activity of AMPK by deacetylating its upstream kinase LKB1 (17). Similarly, our work demonstrates that SIRT2 activation protects the heart from aging-associated and isoproterenol-induced pathological cardiac hypertrophy by repressing NFAT transcription factor. It is well-documented that induction of cardiac hypertrophy involves activation of two key events, (1) fetal gene transcription and (2) cellular protein synthesis. Inhibiting either of these events blunts the pathological cardiac growth. However, AMPK is defined as a key regulator of cardiac energy homeostasis; it regulates cardiac protein synthesis associated with cardiomyocyte hypertrophy (18, 19). On the other hand, NFAT transcription factors are master regulators of cardiac fetal gene expression (56). Our findings indicate that SIRT2 regulates the cardiac hypertrophy by inhibiting both fetal gene transcription and cellular protein synthesis. It is possible that SIRT2 might regulate these processes through two different arms, i.e. AMPK and NFAT transcription factors. Because, NFAT inhibition rescues the cardiac hypertrophy in SIRT2-deficient mice, we cannot rule out this possibility. Overall, our work identified SIRT2 as a novel regulator of NFAT transcription factors and the development of pathological cardiac hypertrophy. We believe that SIRT2 activators may protect the heart against adverse remodeling and failure.
Experimental procedures
Animals, cell culture, reagents, and methods
All animal experiments carried out were with the approval of the Institutional Animal Ethics Committee of Indian Institute of Science, Bengaluru, India, constituted as per Article 13 of the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA), Government of India. SIRT2 knockout mice were purchased from The Jackson Laboratory. All animals were housed in the clean air facility in Indian Institute of Science in individually ventilated cages. Mice were sacrificed using CO2 and heart tissues were harvested. Primary cardiomyocytes were isolated from neonatal rats and mice using our previous publication (57). Adenovirus for SIRT2 overexpression was purchased from Vector BioLabs. Phenylephrine was used at a concentration of 20 μm. [3H]leucine incorporation assay was described in our earlier work (58). Plasmids used in the study were procured from Addgene. Wheat germ agglutinin (WGA) was purchased from Molecular Probes. NFAT inhibitor (VIVIT-N7032) was purchased from Sigma. Puromycin was purchased from Amresco. All chemicals were purchased from Sigma unless otherwise specified.
Western blot analysis
Protocol for Western blot analysis has been previously reported (27, 47). Protein concentration was determined by Bradford assay. The following antibodies were used: α-actinin (Sigma-Aldrich), SIRT2 (EMD Millipore, Sigma-Aldrich, Cell Signaling Technology#), ANP (Abcam, Sunny Corp*), NFATc2 (Thermo Scientific), calcineurin (Thermo Scientific), NFAT (Thermo Scientific), acetyl lysine (EMD Millipore, Cell Signaling Technology), puromycin (Developmental Studies Hybridoma Bank), α-SMA (EMD Millipore), Fn1 (Santa Cruz Biotechnology), Col1a (Sigma-Aldrich), Col3a (Santa Cruz Biotechnology). GAPDH (Santa Cruz Biotechnology) and actin (Thermo Scientific, Sunny Corp). Number sign (#)–marked antibody gives single band for SIRT2 as shown in figures. Asterisk (*)-marked antibody give double bands for ANP as shown in figures. ECL reagents were used to develop the blots in a ChemiDoc Touch chemiluminescence detection system (Bio-Rad).
Confocal microscopy and live imaging
We have described the protocol for immunofluorescence in our previous publications (26, 59). Cells were fixed with formaldehyde (4%) and were permeabilized using 0.2% Triton X-100. Cells were incubated with the primary antibodies, followed by incubation with secondary antibodies conjugated with Alexa Fluor 488 and/or 546. Hoechst was used to stain the nuclei. Carl Zeiss LSM 710 confocal microscope was used to acquire images. The detailed protocol for measurement of cytosolic Ca2+ transients has been described in our previous work (57). Imaging was done in cell culture dishes placed at 37 °C and 5% CO2 in a special chamber incubator mounted on the Zeiss LSM 880 microscope. The fluorescence emission was measured with 488 nm excitation laser for Fluo-4 for control or PE-treated cells with high-speed imaging.
Luciferase reporter assay
The protocol for NFAT luciferase reporter assay was described in our previous publication (60). We used Renilla luciferase for normalizing the luciferase signal. Luciferase activity was measured using a luminometer (Pharmingen Moonlight 3010, BD Biosciences).
SUnSET assay
Surface sensing of translation (SUnSET), a nonradioactive method, was performed to measure in vitro protein synthesis (28). For puromycin incorporation into nascent peptides, cardiomyocytes were treated with 1 μm puromycin for 30 min. Cells were harvested in lysis buffer. Total protein in the lysate was quantified by Bradford assay, and samples were boiled in Laemmli Sample Buffer (Bio-Rad) supplemented with 5% β-mercaptoethanol for 5 min at 96 °C. Puromycin signal was captured after Western blotting using anti-puromycin antibody (Developmental Studies Hybridoma Bank, University of Iowa) using a ChemiDoc Touch (Bio-Rad).
Histology
Age-matched wildtype and SIRT2-KO mice hearts were collected and fixed in 10% neutral buffered formalin. An automated tissue processer (Leica) was used for tissue processing. Fibrosis was observed and measured by staining paraffin-embedded 4 μm-thick sections with Masson's trichrome stain (Sigma). Fibrosis was scored in a blinded fashion as described in our previous publications (27, 47). Myocyte cell size was measured in sections stained with WGA (5 μg/ml).
Echocardiography of mice
Isoflurane (∼1%) inhalation was used for anesthetizing the mice. A topical depilatory agent was used for removing chest hair of the mice. Limb leads were attached for electronic grating and VisualSonics Vevo 1100 was used for the imaging of the animals in the left lateral decubitus position. Warming lamps along with a heated imaging platform was used for maintaining the body temperature of the mice. Wall thickness, left ventricular chamber size, and fractional shortening were measured (27, 47).
Real-time qPCR analysis
Our previous publications describe the detailed protocol for real-time qPCR (26, 59). The primer sequences used are SIRT2 (forward), GCAGTGTCAGAGCGTGGTAA, SIRT2 (reverse), CTAGTGGTGCCTTGCTGATG; ANP (forward), CCTGTGTACAGTGCGGTGTC, ANP (reverse), CCTCATCTTCTACCGGCATC; BNP (forward), AAGGGAGAATACGGCATCATTG, BNP (reverse), ACAGCACCTTCAGGAGATCCA; α-MHC (forward), TAAAATTGAGGACGAGCAGGC, α-MHC (reverse), TCCAGCTCCTCGATGCGT; α-SMA (forward), CTGACAGAGGCACCACTGAA, α-SMA (reverse), CATCTCCAGAGTCCAGCACA; fibronectin-1 (forward), GCGGTTGTCTGACGCTGGCT, fibronectin-1 (reverse), TGGGTTCAGCAGCCCCAGGT; collagen-1 (forward), TGCTGCTTGCAGTAACGTCG, collagen-1 (reverse), TCAACACCATCTCTGCCTCG; IFN-γ (forward), AACTGGCAAAAGGATGGTGAC, IFN-γ (reverse), TTGCTGATGGCCTTGATTGTC; ADSSL1 (forward), GGCTCACCTTGTGTTCGACT, ADSSL1 (reverse), TTCCCCTCTTGTGCTTGACG; actin (forward), TTCTACAATGAGCTGCGTGTG, actin (reverse), GGGGTGTTGAAGGTCTCAAA; GAPDH (forward). TATGTCGTGGAGTCTACTGGT, GAPDH (reverse), GAGTTGTCATATTTCTCGTGG.
Quantification and statistical analysis
GraphPad Prism version 6.04 software was used for data analysis and for graph preparation. Data analysis was performed by t test, one-way analysis of variance (ANOVA), and two-way ANOVA. ZEN and ImageJ software were used for analyzing confocal images. Densitometric analysis was performed by ImageJ.
Author contributions
M. S., S. Maity, S. Mishra, A. J., P. A. D., and S. K. data curation; M. S., S. Maity, S. Mishra, A. J., A. K. T., V. R., M. S. K., P. A. D., D. K., S. K., S. R., M. I., and A. S. P. formal analysis; M. S., V. R., and M. S. K. investigation; M. S., S. Maity, A. J., A. K. T., M. S. K., and A. S. P. methodology; S. Mishra writing-original draft; A. K. T., V. R., D. K., S. R., M. I., A. S. P., and N. R. S. resources; N. R. S. conceptualization; N. R. S. supervision; N. R. S. funding acquisition; N. R. S. writing-review and editing.
This work was supported by research funding from the Department of Science and Technology Extra Mural Research Funding EMR/2014/000065, the Department of Biotechnology Extramural Research Grants BRB/10/1294/2014 and MED/30/1454/2014, and the Council for Scientific and Industrial Research extramural research support 37(1646)/15/EMR-II (to N. R. S.). This work was supported by the SERB-National Post-Doctoral Fellowship (N-PDF) (to M. S. K. and S. M.) and by the Ramalingaswami Re-entry Fellowship and the Innovative Young Biotechnologist Award (IYBA) from the Department of Biotechnology, Government of India (to N. R. S.). The authors declare that they have no conflicts of interest with the contents of this article.
- AMPK
- AMP-activated protein kinase
- NFAT
- nuclear factor of activated T-cells
- PE
- phenylephrine
- ISO
- isoproterenol
- HW/TL
- heart weight to tibia length
- NFAT-Luc assay
- NFAT-responsive promoter/reporter assay
- AR
- adrenergic receptor
- WGA
- wheat germ agglutinin
- qPCR
- quantitative PCR.
References
- 1. Liew C. C., and Dzau V. J. (2004) Molecular genetics and genomics of heart failure. Nat. Rev. Genet. 5, 811–825 10.1038/nrg1470 [DOI] [PubMed] [Google Scholar]
- 2. Heineke J., and Molkentin J. D. (2006) Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat. Rev. Mol. Cell Biol. 7, 589–600 10.1038/nrm1983 [DOI] [PubMed] [Google Scholar]
- 3. Harvey P. A., and Leinwand L. A. (2011) Cellular mechanisms of cardiomyopathy. J. Cell Biol. 194, 355–365 10.1083/jcb.201101100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mittmann C., Eschenhagen T., and Scholz H. (1998) Cellular and molecular aspects of contractile dysfunction in heart failure. Cardiovasc. Res. 39, 267–275 10.1016/S0008-6363(98)00139-4 [DOI] [PubMed] [Google Scholar]
- 5. Han X. F., and Ren J. (2010) Caloric restriction and heart function: Is there a sensible link? Acta Pharmacol. Sin. 31, 1111–1117 10.1038/aps.2010.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dolinsky V. W., and Dyck J. R. B. (2011) Calorie restriction and resveratrol in cardiovascular health and disease. Biochim. Biophys. Acta 1812, 1477–1489 10.1016/j.bbadis.2011.06.010 [DOI] [PubMed] [Google Scholar]
- 7. Piña I. L., and Fitzpatrick J. T. (1996) Exercise and heart failure—a review. Chest 110, 1317–1327 10.1378/chest.110.5.1317 [DOI] [PubMed] [Google Scholar]
- 8. Guarente L. (2007) Sirtuins in aging and disease. Cold Spring Harb. Symp. Quant. Biol. 72, 483–488 10.1101/sqb.2007.72.024 [DOI] [PubMed] [Google Scholar]
- 9. Outeiro T. F., Kontopoulos E., Altmann S. M., Kufareva I., Strathearn K. E., Amore A. M., Volk C. B., Maxwell M. M., Rochet J. C., McLean P. J., Young A. B., Abagyan R., Feany M. B., Hyman B. T., and Kazantsev A. G. (2007) Sirtuin 2 inhibitors rescue α-synuclein-mediated toxicity in models of Parkinson's disease. Science 317, 516–519 10.1126/science.1143780 [DOI] [PubMed] [Google Scholar]
- 10. de Oliveira R. M., Sarkander J., Kazantsev A. G., and Outeiro T. F. (2012) SIRT2 as a therapeutic target for age-related disorders. Front. Pharmacol. 3, 82 10.3389/fphar.2012.00082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yu W., Zhou H. F., Lin R. B., Fu Y. C., and Wang W. (2014) Short-term calorie restriction activates SIRT1–4 and -7 in cardiomyocytes in vivo and in vitro. Mol. Med. Rep. 9, 1218–1224 10.3892/mmr.2014.1944 [DOI] [PubMed] [Google Scholar]
- 12. Krishnan J., Danzer C., Simka T., Ukropec J., Walter K. M., Kumpf S., Mirtschink P., Ukropcova B., Gasperikova D., Pedrazzini T., and Krek W. (2012) Dietary obesity-associated Hif1α activation in adipocytes restricts fatty acid oxidation and energy expenditure via suppression of the Sirt2-NAD+ system. Genes Dev. 26, 259–270 10.1101/gad.180406.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang X., Park S. H., Chang H. C., Shapiro J. S., Vassilopoulos A., Sawicki K. T., Chen C., Shang M., Burridge P. W., Epting C. L., Wilsbacher L. D., Jenkitkasemwong S., Knutson M., Gius D., and Ardehali H. (2017) Sirtuin 2 regulates cellular iron homeostasis via deacetylation of transcription factor NRF2. J. Clin. Invest. 127, 1505–1516 10.1172/JCI88574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim H. S., Vassilopoulos A., Wang R. H., Lahusen T., Xiao Z., Xu X., Li C., Veenstra T. D., Li B., Yu H., Ji J., Wang X. W., Park S. H., Cha Y. I., Gius D., and Deng C. X. (2011) SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell 20, 487–499 10.1016/j.ccr.2011.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yuan Q., Zhan L., Zhou Q. Y., Zhang L. L., Chen X. M., Hu X. M., and Yuan X. C. (2015) SIRT2 regulates microtubule stabilization in diabetic cardiomyopathy. Eur. J. Pharmacol. 764, 554–561 10.1016/j.ejphar.2015.07.045 [DOI] [PubMed] [Google Scholar]
- 16. Dieleman L. A., Ridwan B. U., Tennyson G. S., Beagley K. W., Bucy R. P., and Elson C. O. (1994) Dextran sulfate sodium-induced colitis occurs in severe combined immunodeficient mice. Gastroenterology 107, 1643–1652 10.1016/0016-5085(94)90803-6 [DOI] [PubMed] [Google Scholar]
- 17. Tang X., Chen X. F., Wang N. Y., Wang X. M., Liang S. T., Zheng W., Lu Y. B., Zhao X., Hao D. L., Zhang Z. Q., Zou M. H., Liu D. P., and Chen H. Z. (2017) SIRT2 acts as a cardioprotective deacetylase in pathological cardiac hypertrophy. Circulation 136, 2051–2067 10.1161/CIRCULATIONAHA.117.028728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Noga A. A., Soltys C. L., Barr A. J., Kovacic S., Lopaschuk G. D., and Dyck J. R. (2007) Expression of an active LKB1 complex in cardiac myocytes results in decreased protein synthesis associated with phenylephrine-induced hypertrophy. Am. J. Physiol. Heart Circ. Physiol. 292, H1460–H1469 10.1152/ajpheart.01133.2006 [DOI] [PubMed] [Google Scholar]
- 19. Chan A. Y., Soltys C. L., Young M. E., Proud C. G., and Dyck J. R. (2004) Activation of AMP-activated protein kinase inhibits protein synthesis associated with hypertrophy in the cardiac myocyte. J. Biol. Chem. 279, 32771–32779 10.1074/jbc.M403528200 [DOI] [PubMed] [Google Scholar]
- 20. Putt M. E., Hannenhalli S., Lu Y., Haines P., Chandrupatla H. R., Morrisey E. E., Margulies K. B., and Cappola T. P. (2009) Evidence for coregulation of myocardial gene expression by MEF2 and NFAT in human heart failure. Circ. Cardiovasc. Genet. 2, 212–219 10.1161/CIRCGENETICS.108.816686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Woo S. K., Lee S. D., and Kwon H. M. (2002) TonEBP transcriptional activator in the cellular response to increased osmolality. Pflugers Arch. 444, 579–585 10.1007/s00424-002-0849-2 [DOI] [PubMed] [Google Scholar]
- 22. Molkentin J. D. (2004) Calcineurin-NFAT signaling regulates the cardiac hypertrophic response in coordination with the MAPKs. Cardiovasc. Res. 63, 467–475 10.1016/j.cardiores.2004.01.021 [DOI] [PubMed] [Google Scholar]
- 23. Wilkins B. J., Dai Y. S., Bueno O. F., Parsons S. A., Xu J., Plank D. M., Jones F., Kimball T. R., and Molkentin J. D. (2004) Calcineurin/NFAT coupling participates in pathological, but not physiological, cardiac hypertrophy. Circ. Res. 94, 110–118 10.1161/01.RES.0000109415.17511.18 [DOI] [PubMed] [Google Scholar]
- 24. North B. J., and Verdin E. (2007) Interphase nucleo-cytoplasmic shuttling and localization of SIRT2 during mitosis. PLoS One 2, e784 10.1371/journal.pone.0000784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tanno M., Sakamoto J., Miura T., Shimamoto K., and Horio Y. (2007) Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J. Biol. Chem. 282, 6823–6832 10.1074/jbc.M609554200 [DOI] [PubMed] [Google Scholar]
- 26. Sundaresan N. R., Gupta M., Kim G., Rajamohan S. B., Isbatan A., and Gupta M. P. (2009) Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J. Clin. Invest. 119, 2758–2771 10.1172/JCI39162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sundaresan N. R., Vasudevan P., Zhong L., Kim G., Samant S., Parekh V., Pillai V. B., Ravindra P. V., Gupta M., Jeevanandam V., Cunningham J. M., Deng C. X., Lombard D. B., Mostoslavsky R., and Gupta M. P. (2012) The sirtuin SIRT6 blocks IGF-Akt signaling and development of cardiac hypertrophy by targeting c-Jun. Nat. Med. 18, 1643–1650 10.1038/nm.2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schmidt E. K., Clavarino G., Ceppi M., and Pierre P. (2009) SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Meth. 6, 275–277 10.1038/nmeth.1314 [DOI] [PubMed] [Google Scholar]
- 29. Schulz R. A., and Yutzey K. E. (2004) Calcineurin signaling and NFAT activation in cardiovascular and skeletal muscle development. Dev. Biol. 266, 1–16 10.1016/j.ydbio.2003.10.008 [DOI] [PubMed] [Google Scholar]
- 30. Wen H. Y., Xia Y., Young M. E., Taegtmeyer H., and Kellems R. E. (2002) The adenylosuccinate synthetase-1 gene is activated in the hypertrophied heart. J. Cell. Mol. Med. 6, 235–243 10.1111/j.1582-4934.2002.tb00190.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sica A., Dorman L., Viggiano V., Cippitelli M., Ghosh P., Rice N., and Young H. A. (1997) Interaction of NF-κB and NFAT with the interferon-γ promoter. J. Biol. Chem. 272, 30412–30420 10.1074/jbc.272.48.30412 [DOI] [PubMed] [Google Scholar]
- 32. Perrino B. A., Ng L. Y., and Soderling T. R. (1995) Calcium regulation of calcineurin phosphatase activity by its B subunit and calmodulin. Role of the autoinhibitory domain. J. Biol. Chem. 270, 340–346 10.1074/jbc.270.1.340 [DOI] [PubMed] [Google Scholar]
- 33. Kim J. H., Kim K., Youn B. U., Jin H. M., Kim J. Y., Moon J. B., Ko A., Seo S. B., Lee K. Y., and Kim N. (2011) RANKL induces NFATc1 acetylation and stability via histone acetyltransferases during osteoclast differentiation. Biochem. J. 436, 253–262 10.1042/BJ20110062 [DOI] [PubMed] [Google Scholar]
- 34. Fang J., Li T. Y., Zhu X. H., Deng K. Q., Ji Y. X., Fang C., Zhang X. J., Guo J. H., Zhang P., Li H. L., and Wei X. (2017) Control of pathological cardiac hypertrophy by transcriptional corepressor IRF2BP2 (interferon regulatory factor-2 binding protein 2). Hypertension 70, 515–523 10.1161/HYPERTENSIONAHA.116.08728 [DOI] [PubMed] [Google Scholar]
- 35. Bourajjaj M., Armand A. S., da Costa Martins P. A., Weijts B., van der Nagel R., Heeneman S., Wehrens X. H., and De Windt L. J. (2008) NFATc2 is a necessary mediator of calcineurin-dependent cardiac hypertrophy and heart failure. J. Biol. Chem. 283, 22295–22303 10.1074/jbc.M801296200 [DOI] [PubMed] [Google Scholar]
- 36. Dirkx E., Gladka M. M., Philippen L. E., Armand A. S., Kinet V., Leptidis S., El Azzouzi H., Salic K., Bourajjaj M., da Silva G. J. J., Olieslagers S., van der Nagel R., de Weger R., Bitsch N., Kisters N., et al. (2013) Nfat and miR-25 cooperate to reactivate the transcription factor Hand2 in heart failure. Nat. Cell Biol. 15, 1282–1293 10.1038/ncb2866 [DOI] [PubMed] [Google Scholar]
- 37. Kitada M., Ogura Y., and Koya D. (2016) The protective role of Sirt1 in vascular tissue: Its relationship to vascular aging and atherosclerosis. Aging 8, 2290–2307 10.18632/aging.101068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stein S., and Matter C. M. (2011) Protective roles of SIRT1 in atherosclerosis. Cell Cycle 10, 640–647 10.4161/cc.10.4.14863 [DOI] [PubMed] [Google Scholar]
- 39. Rockman H. A., Koch W. J., and Lefkowitz R. J. (2002) Seven-transmembrane-spanning receptors and heart function. Nature 415, 206–212 10.1038/415206a [DOI] [PubMed] [Google Scholar]
- 40. Iwaki K., Sukhatme V. P., Shubeita H. E., and Chien K. R. (1990) α- and β-adrenergic stimulation induces distinct patterns of immediate early gene expression in neonatal rat myocardial cells. fos/jun expression is associated with sarcomere assembly; Egr-1 induction is primarily an α1-mediated response. J. Biol. Chem. 265, 13809–13817 [PubMed] [Google Scholar]
- 41. Pu W. T., Ma Q., and Izumo S. (2003) NFAT transcription factors are critical survival factors that inhibit cardiomyocyte apoptosis during phenylephrine stimulation in vitro. Circ. Res. 92, 725–731 10.1161/01.RES.0000069211.82346.46 [DOI] [PubMed] [Google Scholar]
- 42. van Rooij E., Doevendans P. A., de Theije C. C., Babiker F. A., Molkentin J. D., and de Windt L. J. (2002) Requirement of nuclear factor of activated T-cells in calcineurin-mediated cardiomyocyte hypertrophy. J. Biol. Chem. 277, 48617–48626 10.1074/jbc.M206532200 [DOI] [PubMed] [Google Scholar]
- 43. Hojayev B., Rothermel B. A., Gillette T. G., and Hill J. A. (2012) FHL2 binds calcineurin and represses pathological cardiac growth. Mol. Cell. Biol. 32, 4025–4034 10.1128/MCB.05948-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vakhrusheva O., Smolka C., Gajawada P., Kostin S., Boettger T., Kubin T., Braun T., and Bober E. (2008) Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ. Res. 102, 703–710 10.1161/CIRCRESAHA.107.164558 [DOI] [PubMed] [Google Scholar]
- 45. Sadhukhan S., Liu X., Ryu D., Nelson O. D., Stupinski J. A., Li Z., Chen W., Zhang S., Weiss R. S., Locasale J. W., Auwerx J., and Lin H. (2016) Metabolomics-assisted proteomics identifies succinylation and SIRT5 as important regulators of cardiac function. Proc. Natl. Acad. Sci. U.S.A. 113, 4320–4325 10.1073/pnas.1519858113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Oka S., Alcendor R., Zhai P., Park J. Y., Shao D., Cho J., Yamamoto T., Tian B., and Sadoshima J. (2011) PPARα-Sirt1 complex mediates cardiac hypertrophy and failure through suppression of the ERR transcriptional pathway. Cell Metab. 14, 598–611 10.1016/j.cmet.2011.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sundaresan N. R., Pillai V. B., Wolfgeher D., Samant S., Vasudevan P., Parekh V., Raghuraman H., Cunningham J. M., Gupta M., and Gupta M. P. (2011) The deacetylase SIRT1 promotes membrane localization and activation of Akt and PDK1 during tumorigenesis and cardiac hypertrophy. Sci. Signal. 4, ra46 10.1126/scisignal.2001465 [DOI] [PubMed] [Google Scholar]
- 48. Jiang W., Wang S., Xiao M., Lin Y., Zhou L., Lei Q., Xiong Y., Guan K. L., and Zhao S. (2011) Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol. Cell 43, 33–44 10.1016/j.molcel.2011.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rothgiesser K. M., Erener S., Waibel S., Lüscher B., and Hottiger M. O. (2010) SIRT2 regulates NF-κB-dependent gene expression through deacetylation of p65 Lys310. J. Cell Sci. 123, 4251–4258 10.1242/jcs.073783 [DOI] [PubMed] [Google Scholar]
- 50. Park S. H., Zhu Y. M., Ozden O., Kim H. S., Jiang H. Y., Deng C. X., Gius D., and Vassilopoulos A. (2012) SIRT2 is a tumor suppressor that connects aging, acetylome, cell cycle signaling, and carcinogenesis. Transl. Cancer Res. 1, 15–21 [PMC free article] [PubMed] [Google Scholar]
- 51. Hiratsuka M., Inoue T., Toda T., Kimura N., Shirayoshi Y., Kamitani H., Watanabe T., Ohama E., Tahimic C. G. T., Kurimasa A., and Oshimura M. (2003) Proteomics-based identification of differentially expressed genes in human gliomas: Down-regulation of SIRT2 gene. Biochem. Biophys. Res. Commun. 309, 558–566 10.1016/j.bbrc.2003.08.029 [DOI] [PubMed] [Google Scholar]
- 52. Guarente L. (2000) Sir2 links chromatin silencing, metabolism, and aging. Gene Dev. 14, 1021–1026 [PubMed] [Google Scholar]
- 53. Kyrylenko S., Kyrylenko O., Suuronen T., and Salminen A. (2003) Differential regulation of the Sir2 histone deacetylase gene family by inhibitors of class I and II histone deacetylases. Cell Mol. Life Sci. 60, 1990–1997 10.1007/s00018-003-3090-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lombard D. B., Chua K. F., Mostoslavsky R., Franco S., Gostissa M., and Alt F. W. (2005) DNA repair, genome stability, and aging. Cell 120, 497–512 10.1016/j.cell.2005.01.028 [DOI] [PubMed] [Google Scholar]
- 55. Rhodes D. R., Yu J. J., Shanker K., Deshpande N., Varambally R., Ghosh D., Barrette T., Pandey A., and Chinnaiyan A. M. (2004) ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia 6, 1–6 10.1016/S1476-5586(04)80047-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dirkx E., da Costa Martins P. A., and De Windt L. J. (2013) Regulation of fetal gene expression in heart failure. Biochim. Biophys. Acta 1832, 2414–2424 10.1016/j.bbadis.2013.07.023 [DOI] [PubMed] [Google Scholar]
- 57. Jain A., Ravi V., Muhamed J., Chatterjee K., and Sundaresan N. R. (2017) A simplified protocol for culture of murine neonatal cardiomyocytes on nanoscale keratin coated surfaces. Int. J. Cardiol. 232, 160–170 10.1016/j.ijcard.2017.01.036 [DOI] [PubMed] [Google Scholar]
- 58. Pillai V. B., Sundaresan N. R., Samant S. A., Wolfgeher D., Trivedi C. M., and Gupta M. P. (2011) Acetylation of a conserved lysine residue in the ATP binding pocket of p38 augments its kinase activity during hypertrophy of cardiomyocytes. Mol. Cell. Biol. 31, 2349–2363 10.1128/MCB.01205-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sundaresan N. R., Samant S. A., Pillai V. B., Rajamohan S. B., and Gupta M. P. (2008) SIRT3 is a stress-responsive deacetylase in cardiomyocytes that protects cells from stress-mediated cell death by deacetylation of Ku70. Mol. Cell. Biol. 28, 6384–6401 10.1128/MCB.00426-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pillai V. B., Samant S., Sundaresan N. R., Raghuraman H., Kim G., Bonner M. Y., Arbiser J. L., Walker D. I., Jones D. P., Gius D., and Gupta M. P. (2015) Honokiol blocks and reverses cardiac hypertrophy in mice by activating mitochondrial Sirt3. Nat. Commun. 6, 6656 10.1038/ncomms7656 [DOI] [PMC free article] [PubMed] [Google Scholar]