Abstract
Transient receptor potential vanilloid 4 (TRPV4) is a Ca2+-permeable channel of the transient receptor potential (TRP) superfamily activated by diverse stimuli, including warm temperature, mechanical forces, and lipid mediators such as arachidonic acid (AA) and its metabolites. This activation is tightly regulated by protein phosphorylation carried out by various serine/threonine or tyrosine kinases. It remains poorly understood how phosphorylation differentially regulates TRPV4 activation in response to different stimuli. We investigated how TRPV4 activation by AA, an important signaling process in the dilation of coronary arterioles, is affected by protein kinase A (PKA)–mediated phosphorylation at Ser-824. Wildtype and mutant TRPV4 channels were expressed in human coronary artery endothelial cells (HCAECs). AA-induced TRPV4 activation was blunted in the S824A mutant but was enhanced in the phosphomimetic S824E mutant, whereas the channel activation by the synthetic agonist GSK1016790A was not affected. The low level of basal phosphorylation at Ser-824 was robustly increased by the redox signaling molecule hydrogen peroxide (H2O2). The H2O2-induced phosphorylation was accompanied by an enhanced channel activation by AA, and this enhanced response was largely abolished by PKA inhibition or S824A mutation. We further identified a potential structural context dependence of Ser-824 phosphorylation–mediated TRPV4 regulation involving an interplay between AA binding and the possible phosphorylation-induced rearrangements of the C-terminal helix bearing Ser-824. These results provide insight into how phosphorylation specifically regulates TRPV4 activation. Redox-mediated TRPV4 phosphorylation may contribute to pathologies associated with enhanced TRPV4 activity in endothelial and other systems.
Keywords: arachidonic acid (AA) (ARA), endothelial cell, hydrogen peroxide, protein kinase A (PKA), protein phosphorylation, signal transduction, transient receptor potential channels (TRP channels)
Introduction
TRPV4, the fourth member of transient receptor potential (TRP)2 vanilloid subfamily, is a Ca2+-permeable cation channel involved in multiple physiological processes and diseases (1–4). Since it was first identified in 2000, TRPV4 has been found in various cell types/tissues, such as epithelial cells of kidney and airways, sensory neurons, keratinocytes, smooth muscle, and the vascular endothelium (1–3). In accordance with this broad distribution, TRPV4 has been implicated in nociception (5), systemic osmotic regulation (6, 7), epithelial ciliary activity (8), and shear-induced vasodilation and blood flow regulation (3, 9–13). Mutations in TRPV4, many being gain-of-function variants, cause a panel of neuromuscular dystrophies and skeletal dysplasia (14, 15). Emerging evidence indicates that nongenetic gain-of-function of TRPV4 also contributes to pathological processes such as neurogenic inflammation and hyperalgesia (5, 16–18), adipose cell inflammation (19), pulmonary and cardiac tissue fibrosis (20, 21), pathologies associated with diet-induced obesity (22), and angiotensin II–induced endothelial dysfunction (23).
The TRP channel, such as TRPV4, is a tetramer and has an overall architecture similar to that of voltage-gated potassium (KV) channels (24), with each monomer possessing six transmembrane (TM) domains, a pore loop between TM5 and TM6, and intracellular N- and C-terminal tails (25). TRPV4 can be activated by diverse physical and chemical stimuli including hypotonic cell swelling (26–28), moderate heat (29, 30), mechanical stimuli (e.g. membrane stretch and shear stress) (6, 7, 12, 31–33), low pH (6), and lipid mediators such as arachidonic acid (AA) and its metabolites (34–37). The mechanisms by which TRPV4 is activated by such diverse stimuli are currently poorly understood, yet evidence indicates at least two independent pathways of activation. The synthetic agonist 4α-PDD activates TRPV4 by directly binding to its TM3 and TM4 segments in a ligand-like manner (38). Although direct gating by mechanical force has been observed in various settings (32, 33), TRPV4 activation in response to hypotonic cell swelling, high viscous load, and shear stress appears to depend on phospholipase A2 (PLA2) activity and the associated release of AA and its metabolites (8, 10, 39). Mutating a tyrosine residue (Y555A) in the TM3 strongly impairs activation of TRPV4 by 4α-PDD but has no effect on activation by cell swelling or AA, a finding consistent with an independent activation mechanism by AA or its metabolites (39).
In addition to stimulus-dependent activation described above, TRPV4 can also be regulated by protein phosphorylation via tyrosine and serine/threonine kinases (40–42), which seems to be a conserved mechanism contributing to the sensitization and enhanced activity of TRPV4 and TRPV1 channels in conditions such as inflammation and hyperalgesia (17, 18, 43). It is well established that phosphorylation by protein kinase C (PKC) and protein kinase A (PKA) can phosphorylate and sensitize TRPV1 channel in the development of heat hyperalgesia (43). Phosphorylation of TRPV4 by these protein kinases, although at different sites (Ser-162, Thr-175, and Ser-189 residues by PKC and Ser-824 residue by PKA), enhances TRPV4 activation induced by hypotonic swelling (40). A subsequent study confirmed that PKC and possibly PKA phosphorylate TRPV4 at Ser-824 resulting in its sensitization to the hypotonic solution (42). However, the mechanisms by which phosphorylation regulates TRPV4 activation in response to different stimuli, as well as the exact pathways responsible for TRPV4 phosphorylation under physiological or pathophysiological conditions, remain to be defined.
Using native human coronary artery endothelial cells (HCAECs) and TRPV4-overexpressing HCAECs, we recently reported that AA activates TRPV4, and this activation requires PKA-mediated protein phosphorylation (36), similarly as that described for arachidonate-regulated Ca2+ (ARC) channel (44). Furthermore, TRPV4 mediates shear stress–induced and endothelium-dependent vasodilation in human coronary arterioles (HCAs) isolated from subjects with coronary artery disease, a condition associated with enhanced production of reactive oxygen species (ROS) such as hydrogen peroxide (H2O2) (45). Of note, H2O2, an endogenous redox signaling molecule involved in various pathophysiological conditions (46, 47) and a regulator of protein kinases (48), enhances Src-dependent tyrosine phosphorylation of TRPV4 (41). In this study, we explored the role and mechanism of PKA-mediated Ser-824 phosphorylation in TRPV4 activation by AA. The present study shows that AA-induced TRPV4 activation is strongly and specifically regulated by Ser-824 phosphorylation of the channel. We demonstrate that H2O2 stimulates PKA-dependent phosphorylation of TRPV4 at Ser-824 and consequently enhances AA-induced channel activation in both cultured and native HCAECs. To further identify the potential structural context by which Ser-824 phosphorylation confers specific regulation of TRPV4 activation by AA, we have developed a homology model of the human TRPV4, including the domain harboring the previously postulated arachidonate recognition sequence (ARS) (1), and the possible arrangements for the C-terminal domain bearing Ser-824 residue. We have used an automated docking approach to infer a likely conformation of AA in the putative binding region and hypothesized on a potential mechanism of TRPV4 activation by AA binding, the restraint on activation by the C-terminal domain, and facilitation of gating by Ser-824 phosphorylation that might dislodge the C-terminal domain from its inhibitory location.
Results
Ser-824 phosphorylation regulates TRPV4 activation by AA in HCAECs
TRPV4 is expressed in endothelial cells of different species and vascular beds including HCAECs (12, 13, 45). Using native HCAECs and cells overexpressing human TRPV4 with C-terminal green fluorescent protein (TRPV4-GFP), we previously identified an essential role of the endogenous lipid mediator AA in TRPV4 activation (36). The TRPV4 overexpression in these primary endothelial cells offers a favorable signal-to-noise ratio along with well-preserved signaling pathways as compared with commonly used HEK 293 cells. For example, AA, as well as the synthetic TRPV4 agonist GSK1016790A, increases intracellular Ca2+ concentration [Ca2+]i in native HCAECs that is sensitive to different TRPV4 blockers such as RN-1734, HC067047, and ruthenium red (36). Transfection of these endothelial cells with exogenous TRPV4 greatly increases (by ≥ 5- to 10-fold) Ca2+ responses to AA and GSK1016790A, which are also inhibited by TRPV4 blockers.
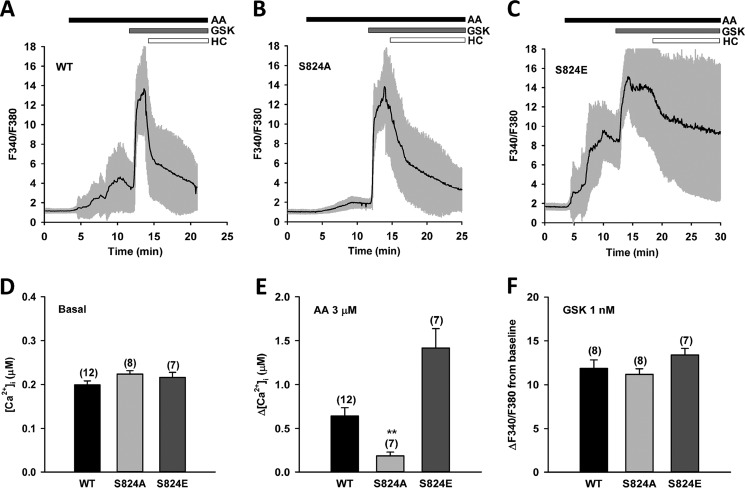
Furthermore, our previous work revealed that AA-induced TRPV4 activation requires PKA-mediated protein phosphorylation (36). To determine whether TRPV4 activation by AA involves phosphorylation at the specific amino acid residue Ser-824, a previously identified PKA phosphorylation site (40, 42), we analyzed TRPV4-dependent Ca2+ influx, using ratiometric fura-2 calcium imaging in HCAECs stably expressing the following TRPV4 constructs: wildtype (WT), nonphosphorylatable alanine mutant (S824A) and phosphomimetic glutamic acid mutant (S824E). Compared with HCAECs overexpressing WT TRPV4, AA (3 μm)-induced increase in [Ca2+]i was largely blunted but not eliminated in the nonphosphorylatable S824A mutant. Conversely, a significantly enhanced response to AA was observed in the phosphomimetic S824E mutant (Fig. 1, A–C and E). These results suggest that Ser-824 phosphorylation tightly regulates TRPV4 activation by AA. In contrast, subsequent addition of the synthetic agonist GSK1016790A (1 nm) further increased [Ca2+]i to similar levels in WT, S824A, and S824E mutants (Fig. 1, A–C and F), indicating that Ser-824 phosphorylation specifically regulates TRPV4 activation in response to AA. With a lower-affinity Ca2+ indicator fura-4F, we found that addition of GSK1016790A (1 nm) alone also induced similar Ca2+ responses in WT, S824A, and S824E mutants (Fig. S7).
Figure 1.
Regulation of AA-induced TRPV4 activation by Ser-824 phosphorylation in HCAECs. A–C, representative traces of fura-2 calcium assay of HCAECs transfected with TRPV4 (C-terminal GFP fusion protein) wildtype (WT), S824A, or S824E mutants in a lentiviral vector. The black line denotes mean F340/F380 ratio and the gray area 1× S.D. Cells were treated sequentially with AA (3 μm), the synthetic TRPV4 activator GSK1016790A (GSK) (1 nm), and the TRPV4 blocker HC067047 (HC) (1 μm). D–F, summarized data for AA- and GSK-induced Ca2+ increases (Δ[Ca2+]i and ΔF340/F380 from baseline, respectively) and basal intracellular Ca2+ concentrations ([Ca2+]i) in WT, S824A, and S824E mutants. All data represent mean ± S.E., with the number of independent experiments indicated in brackets above error bars. **, p < 0.01 compared with WT.
The basal [Ca2+]i was higher in HCAECs overexpressing WT TRPV4 (Fig. 1D) ([Ca2+]i, 199 ± 9 versus 89 ± 4 nm in nontransfected cells, respectively; n = 12 independent experiments; p < 0.05), reflecting the spontaneous activity of TRPV4 reported previously in these and other cells (36, 41). Although the basal Ca2+ concentrations in cells expressing TRPV4 WT and S824A mutant were comparable (Fig. 1D), they were substantially elevated in cells expressing the S824E mutant (Fig. S1, A–C). To minimize potential variations originating from factors other than TRPV4 channel activity, we excluded cells with very high basal [Ca2+]i (F340/F380 > 2.0) from our analysis (Fig. 1). A similar approach has been reported previously (42).
TRPV4 S824A and S824E mutants localize on the cell surface
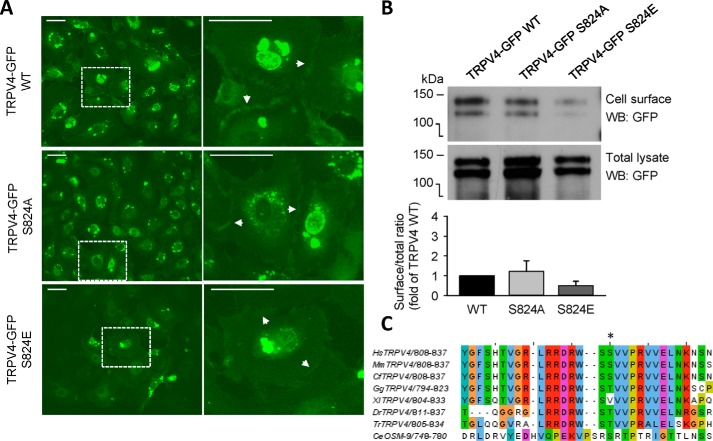
TRPV4 function requires the expression of channel proteins on the plasma membrane. To investigate whether mutation of Ser-824 affects protein expression and trafficking of the channels to the cell surface, we performed fluorescence imaging of HCAECs expressing TRPV4-GFP (WT, S824A, and S824E) fusion proteins (Fig. 2A). These fluorescence images revealed plasma membrane protein expression of WT and both Ser-824 mutants. Furthermore, cell surface biotinylation labeling followed by Western blot analysis confirmed the plasma membrane expression of WT and mutant TRPV4-GFP fusion proteins (Fig. 2B). Moreover, we observed a trend toward reduced membrane and total TRPV4 expression in the S824E mutant as compared with WT and S824A mutant, which may be because of high [Ca2+]i-associated cell toxicity in culture. Using the same cell surface biotinylation method, we found that similar levels of PECAM-1, an endothelial cell marker protein, were expressed at the cell surface of HCAECs expressing TRPV4 WT, S824A, and S824E mutants. As an additional control, we did not detect β-actin, a cytosolic protein, in the cell surface protein fraction (data not shown).
Figure 2.
TRPV4 containing S824A or S824E substitutions localizes at the cell surface of HCAECs. A, fluorescence imaging of HCAECs overexpressing TRPV4-GFP WT (top), S824A (middle), or S824E (bottom) mutants. Images on the right correspond to a zoomed view of the dashed box from the left. White arrows indicate plasma membrane localization of TRPV4 proteins. Scale bar, 50 μm. Data in A are representative of at least three independent experiments. B, Western blots with GFP antibodies of cell surface and total TRPV4-GFP WT, S824A, or S824E mutant proteins expressed in HCAECs. Cell surface proteins were labeled using a cell surface biotinylation method and captured with NeutrAvidin agarose beads. Total cellular lysates were analyzed in parallel. Lower, ratios of cell surface to total levels of TRPV4 protein for S824A and S824E mutants as compared with WT TRPV4 (mean ± S.E., n = 2). C, alignment of amino acid sequences surrounding Ser-824 from human (Hs, Homo sapiens), mouse (Mm, Mus musculus), dog (Cf, Canis familiaris), chicken (Gg, Gallus gallus), frog (Xl, Xenopus laevis), fish such as zebrafish (Dr, Danio rerio), and pufferfish (Tr, Takifugu rubripes) TRPV4 proteins, and worm (Ce, Caenorhabditis elegans) OSM-9 (TRPV4 homolog). The Ser-824 residue of human TRPV4 is indicated with an asterisk.
Activation of PKC and PKA induces TRPV4 phosphorylation at Ser-824
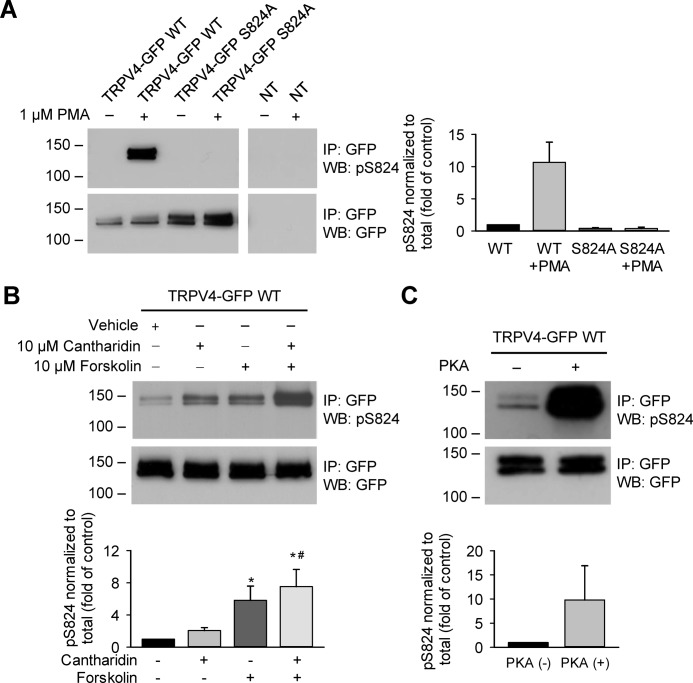
To directly examine TRPV4 phosphorylation at Ser-824, HCAECs were grown under standard conditions (without prior serum starvation) and exposed to PKC selective activators, which have been shown previously to induce Ser-824 phosphorylation in HEK 293 cells (42). Protein lysates of TRPV4-GFP–expressing HCAECs were immunoprecipitated with an antibody against GFP and then blotted for phosphorylated Ser-824 (pSer-824) using a motif-specific antibody. We consistently detected basal levels of Ser-824 phosphorylation in HCAECs, as indicated by more prolonged exposure of blots (data not shown). The phosphorylation at Ser-824 in TRPV4-GFP WT acutely increased following 1 μm phorbol 12-myristate 13-acetate (PMA, a PKC activator) treatment (Fig. 3A). In HCAECs expressing the phosphorylation-deficient TRPV4 variant (S824A), no phospho-specific signal could be detected after PMA stimulation. These results confirmed Ser-824 as a target residue for PKC, as well as the specificity of the anti–pSer-824 antibody.
Figure 3.
TRPV4 phosphorylation at Ser-824 in HCAECs in response to activation of PKC and PKA. A, HCAECs were transfected with plasmids encoding TRPV4-GFP wildtype (WT), S824A, or K407T mutants in a lentiviral vector. NT indicates nontransfected. Cells were exposed to the PKC activator PMA (1 μm) for 30 min. TRPV4 Ser-824 phosphorylation was analyzed by Western blotting with a phosphoserine motif antibody against the motif RXRXXS*/T* (pSer-824 antibodies), and the same blot was reprobed with GFP antibodies to detect total cellular TRPV4 proteins. B, similar experiments were conducted on TRPV4-transfected HCAECs exposed to the adenylate cyclase activator forskolin (10 μm) for 30 min, with or without co-incubation with the protein phosphatase (PP) 1/2A inhibitor cantharidin (10 μm). C, PKA phosphorylated TRPV4 at Ser-824 in vitro. The protein lysates of TRPV4-GFP–transfected HCAECs were immunoprecipitated and treated with PKA catalytic subunit (+) (2500 units for 50 μg input protein lysate) for 30 min at 30 °C. A–C, immunoblots are representative of three (A), five (B), or two (C) independent experiments. Quantification of -fold change in Ser-824 phosphorylation after different schemes of treatment is presented below or beside the representative immunoblots shown in panels A, B, and C, respectively. Data represent mean ± S.E. of pSer-824 band densities normalized to total TRPV4 expression levels and then converted to relative phosphorylation levels as compared with WT vehicle control. *, p < 0.05 compared with vehicle; #, p < 0.05 compared with cantharidin.
We also analyzed TRPV4 phosphorylation at Ser-824 in response to forskolin (10 μm, adenylate cyclase activator) treatment (Fig. 3B). We found that forskolin increased phosphorylation of Ser-824 in HCAECs, albeit to a smaller extent tha PMA, a finding similar to that obtained in HEK 293 and Madin-Darby canine kidney (MDCK) cells (42). Furthermore, forskolin-induced Ser-824 phosphorylation was more robustly detected in the presence of the protein phosphatase (PP) 1/2A inhibitor cantharidin (10 μm). Cantharidin alone induced a time-dependent increase in Ser-824 phosphorylation in HCAECs (data not shown). These results indicate that PP1/2A negatively regulates Ser-824 phosphorylation stimulated by the PKA pathway. The effect of cantharidin on PKA-mediated Ser-824 phosphorylation did not require prolonged incubation of cantharidin (e.g. 30 min in Fig. 3B) but only needed a brief pretreatment (e.g. 2 min before cell lysis in Figs. 5 and 6). The mechanisms responsible for this acute effect remain to be determined. To examine whether PKA can directly phosphorylate TRPV4 at Ser-824, TRPV4-GFP WT proteins were immunoprecipitated with anti-GFP antibodies and incubated with PKA catalytic subunits. TRPV4 phosphorylation at Ser-824 was markedly enhanced after exposure to PKA in vitro (Fig. 3C). Altogether, the presented results demonstrate that PKC and PKA can phosphorylate Ser-824 of TRPV4 in HCAECs.
Figure 5.
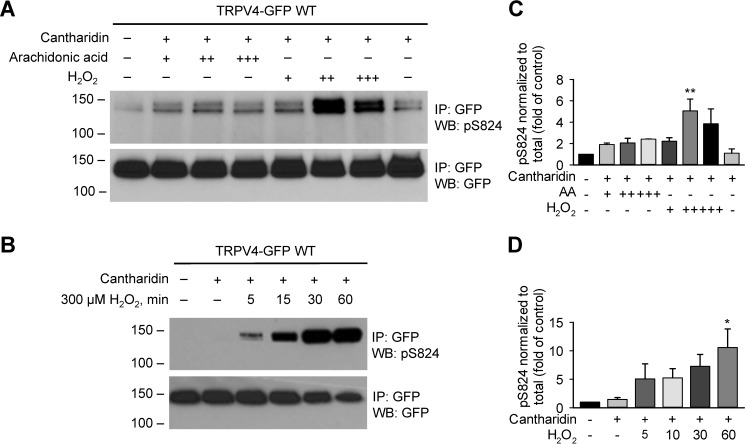
H2O2 and AA to a less extent stimulate Ser-824 phosphorylation of TRPV4 in HCAECs. A, TRPV4-GFP–transfected HACECs were treated with AA at 1, 3, 10 μm (+, ++, +++, respectively), or H2O2 at 100, 300, 1000 μm (+, ++, +++, respectively) for 30 min, along with 10 μm cantharidin (+). B, time course of TRPV4 Ser-824 phosphorylation induced by H2O2. TRPV4-transfected HACECs were exposed to 300 μm H2O2 for the indicated amount of time (5–60 min), with the addition of 10 μm cantharidin (+) during the final 2 min. Immunoblots in A and B are representative of three independent experiments. C and D, quantification of -fold change in Ser-824 phosphorylation after different schemes of treatment with AA or H2O2 as shown in panels A and B, respectively. Data represent mean ± S.E. of pSer-824 band densities that is normalized to total TRPV4 expression levels and then converted to relative phosphorylation levels as compared with WT vehicle control. *, p < 0.05 and **, p < 0.01 compared with vehicle control and cantharidin.
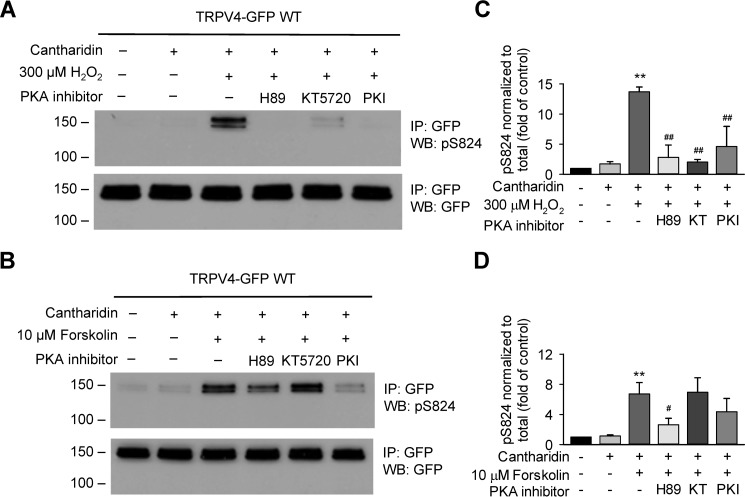
Figure 6.
PKA is involved in H2O2-stimulated Ser-824 phosphorylation of TRPV4 in HCAECs. A, H2O2-stimulated Ser-824 phosphorylation of TRPV4 was almost eliminated in TRPV4-GFP–overexpressing HCAECs pretreated for 30 min with three chemically distinct PKA inhibitors, H89 (10 μm), KT5720 (10 μm), and PKI 14–22 amide, myristoylated (PKI, 10 μm). Cells were then stimulated with 300 μm H2O2 for 15 min, with the addition of 10 μm cantharidin (+) during the final 2 min. B, experiments performed in parallel with those in panel A indicate that forskolin-stimulated Ser-824 phosphorylation of TRPV4 was reduced by pretreatment of cells for 30 min with PKA inhibitor PKI (10 μm) and to a lesser extent by H89 (10 μm). KT5720 (10 μm for 30 min) had no inhibition on Ser-824 phosphorylation induced by forskolin. Immunoblots in A and B are representative of four (C) and six (D) independent experiments. C and D, quantification of -fold change in Ser-824 phosphorylation after treatment with H2O2 in the absence or presence of PKA inhibitors. Data represent mean ± S.E. of pSer-824 band densities that is normalized to total TRPV4 expression levels and then converted to relative phosphorylation levels as compared with WT vehicle control. **, p ≤ 0.01 compared with vehicle control and cantharidin (C and D); #, p < 0.05 compared with forskolin + cantharidin (D), ##, p < 0.01 compared with H2O2 + cantharidin (C).
H2O2–PKA pathway regulates AA-induced TRPV4 activation in HCAECs
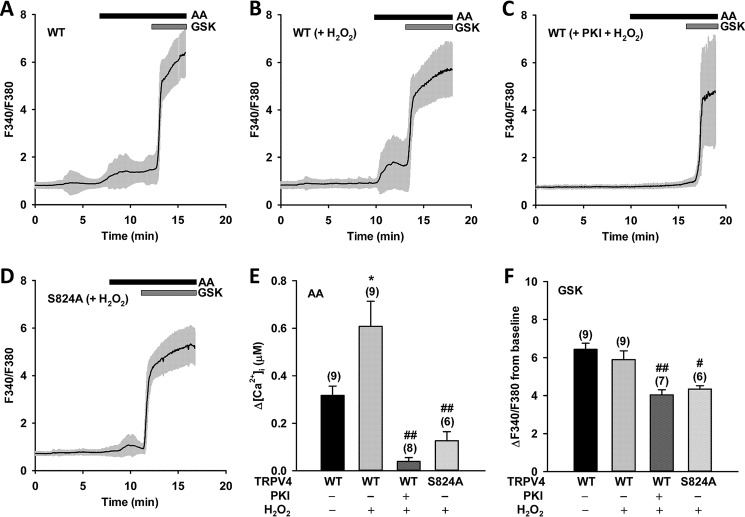
As discussed above, indirect evidence suggests that H2O2 may be an endogenous factor that regulates TRPV4 activation and phosphorylation at Ser-824. Thus, we examined Ca2+ responses to AA and GSK1016790A in WT TRPV4-expressing HCAECs with or without H2O2 pretreatment. Compared with untreated controls, pretreatment of HCAECs with H2O2 at a pathophysiologically relevant concentration (100 μm) (48) significantly enhanced [Ca2+]i response to AA (3 μm) (Fig. 4, A, B, and E) but not to GSK1016790A (1 nm) (Fig. 4, A, B, and F). These results indicate that H2O2 can specifically potentiate TRPV4 activation by AA. The AA-induced [Ca2+]i responses were almost entirely abolished when cells were pretreated with H2O2 plus PKI (10 μm) (Fig. 4, C and E), a peptide inhibitor of PKA, demonstrating that H2O2-potentiated and AA-induced TRPV4 activation involves PKA activity. In addition, the amplitudes of Ca2+ transients in response to AA (in the presence of H2O2) were significantly lower in HCAECs expressing TRPV4 S824A than in cells expressing TRPV4 WT (Fig. 4, D and E), indicating that Ser-824 is a major phosphorylation site involved in H2O2 regulation of AA-induced activation.
Figure 4.
H2O2-mediated Ser-824 phosphorylation regulates AA-induced TRPV4 activation in HCAECs. A–D, representative traces of fura-2 calcium assay of wildtype (WT) and S824A mutant TRPV4-GFP–overexpressing HCAECs pretreated with vehicle or the PKA inhibitor PKI (10 μm) for 30 min, followed by H2O2 (100 μm) for 10 min. Cells were then stimulated sequentially with AA (3 μm), and the synthetic TRPV4 activator GSK1016790A (GSK) (1 nm). The black line denotes mean F340/F380 ratio and the gray area 1× S.D. E and F, summarized data for AA- and GSK-induced Ca2+ increases (Δ[Ca2+]i and ΔF340/F380 from baseline, respectively) in HCAECs transfected with WT or S824A mutant TRPV4-GFP and treated as indicated in panels A–D. All data represent mean ± S.E., with the number of independent experiments indicated in brackets above error bars. *, p < 0.05 compared with WT; #, p < 0.05; and ##, p < 0.01 compared with WT (+H2O2).
HCAECs with TRPV4 S824A mutation or PKI pretreatment (both in the presence of H2O2) exhibited marked reduction or almost complete loss of Ca2+ responses to AA, to a level that was even lower than that of WT TRPV4 controls without H2O2 pretreatment (Fig. 4E and Fig. S1D). Similarly, AA-induced responses in cells expressing TRPV4 S824A were markedly reduced as compared with WT TRPV4 (Fig. 1E). These results indicate that a significant portion of TRPV4 channels is phosphorylated at Ser-824 in HCAECs under resting conditions. Together, these results suggest that the H2O2–PKA–TRPV4 pSer-824 signaling axis can provide strong and bidirectional control of AA-induced TRPV4 activation.
H2O2 and AA stimulate TRPV4 phosphorylation at Ser-824
To directly examine whether H2O2 induces phosphorylation at Ser-824, HCAECs overexpressing WT TRPV4 were stimulated with H2O2 (100, 300, and 1000 μm) for 30 min. TRPV4 phosphorylation was detected by immunoprecipitation followed by Western blotting with antibodies against pSer-824 as described above. H2O2 induced robust Ser-824 phosphorylation, and the maximal stimulation was observed with around 300 μm H2O2 (Fig. 5, A and C). Further time course study indicated that H2O2 (300 μm) enhanced Ser-824 phosphorylation in a time-dependent manner; the phosphorylation of Ser-824 was detected as early as 5 min after addition of H2O2, which continued to rise for up to 30 min and remained elevated at 60 min (Fig. 5, B and D). We also examined Ser-824 phosphorylation in HCAECs in response to AA (1–10 μm) in the presence of cantharidin (10 μm). Interestingly, AA treatment also led to enhanced phosphorylation at Ser-824 compared with untreated controls (Fig. 5, A and C). The stimulation of pSer-824 was concentration dependent at 1–3 μm AA, and no further enhancement was observed with a higher concentration (10 μm).
H2O2-stimulated TRPV4 Ser-824 phosphorylation involves PKA
Evidence from other studies suggests that H2O2 can activate PKA through the formation of an intermolecular disulfide bond between regulatory subunits via cysteine oxidation (48). Given the robust stimulation of Ser-824 phosphorylation by H2O2, we next examined whether PKA is indeed involved in H2O2-induced phosphorylation of TRPV4 at Ser-824. HCAECs were separately pretreated with three chemically distinct PKA inhibitors, PKI, H89, and KT5720, at a concentration of 10 μm each for 30 min. Western blot analysis revealed that H2O2 (100 μm)-induced phosphorylation of TRPV4 at Ser-824 was almost eliminated by all three PKA inhibitors, strongly supporting the involvement of PKA (Fig. 6, A and C). Additional proof was provided by parallel experiments wherein H2O2 was replaced by forskolin (10 μm). We found that forskolin-stimulated Ser-824 phosphorylation of TRPV4 was reduced by pretreatment of cells for 30 min with the PKA inhibitor H89 (10 μm). The inhibition of forskolin-induced phosphorylation by PKI (10 μm) was variable between experiments and thus did not reach statistical significance. The factors underlying this variability remain to be determined in future studies. However, these results confirm that at least one of the three PKA inhibitors used could effectively inhibit PKA activation by the classical cAMP signaling (Fig. 6, B and D).
H2O2 regulates AA-induced TRPV4 activation in native endothelial cells
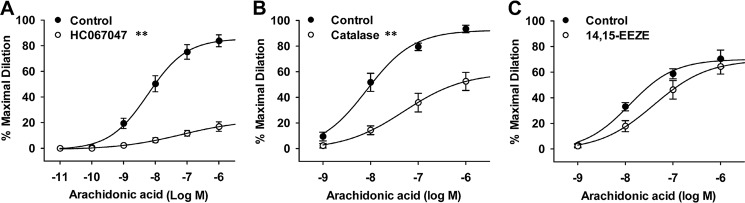
To determine whether H2O2 regulates AA-induced TRPV4 activation in native endothelial cells that endogenously express TRPV4 and its associated signaling pathways, we examined TRPV4-mediated vasodilatory responses in freshly isolated HCAs. Previous studies have shown that AA induces potent dilation of HCAs through the activation of endothelial TRPV4 and subsequent release of endothelial relaxing factors or hyperpolarization of vascular cells. This TRPV4-mediated response is more prominent in HCAs of subjects with coronary artery disease (45), a condition associated with increased production of reactive oxygen species such as H2O2, thereby providing an ex vivo bioassay system for examining the modulation of TRPV4 function by H2O2 under pathophysiologically relevant conditions. We first verified the mediating role of TRPV4 in vasodilatory responses to AA. In HCAs cannulated and pressurized under physiological conditions, AA (10−11 to 10−6 m) caused a potent and concentration-dependent dilation, and this dilation was largely blocked by the TRPV4 antagonist HC067047 (Fig. 7A). AA-induced dilation was significantly reduced when HCAs were incubated intraluminally with catalase, an H2O2-metabolizing enzyme that scavenges endogenously generated H2O2 (Fig. 7B). AA-induced vasodilation was only marginally attenuated by pretreatment of HCAs with 14,15-EEZE (49), a selective epoxyeicosatrienoic acid (EET) antagonist (Fig. 7C). Using a fura-2 assay similar as cultured HCAECs, we confirmed AA-induced and TRPV4-mediated Ca2+ response in endothelial cells in situ of cannulated arteries (Fig. S2). These results indicate that H2O2 regulates AA-induced TRPV4 activation in native endothelial cells as well. In addition, the P450 metabolites of AA such as EETs do not significantly contribute to endothelial TRPV4 activation by AA in HCAs.
Figure 7.
Role of TRPV4, H2O2, and cytochrome P450 metabolite EETs in AA-induced TRPV4 activation in native endothelial cells. TRPV4 activation was indicated by AA-induced and endothelial TRPV4-mediated dilation of coronary arterioles from human subjects with coronary artery disease. A, AA dilated coronary arterioles in a concentration-dependent manner. The dilation was markedly inhibited by the TRPV4 antagonist HC067047 (1 μm). B, AA-induced dilation was inhibited by the H2O2-metabolizing enzyme catalase (500 units/ml). C, AA-induced dilation was only slightly attenuated by 14,15-EEZE (10 μm), a specific EET antagonist. Data in A–C, n = 5 to 7 patients/each group. **, p < 0.01 versus control.
Potential structural basis of AA-induced TRPV4 activation and its regulation by Ser-824 phosphorylation
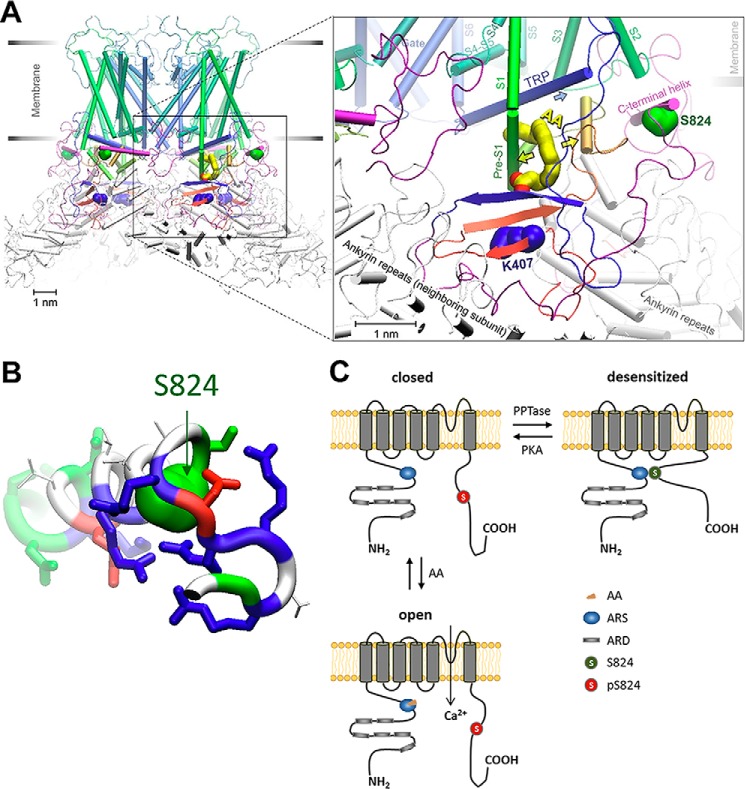
To gain insights into the specific interplay of AA binding and Ser-824 phosphorylation in TRPV4 activation, we performed homology modeling of human TRPV4, including the putative AA-binding ARS region (1) and C-terminal domain that harbors Ser-824. The ARS region contains Lys-407, and mutation in this residue has been associated with hereditary disease (50). We have employed seven leading structure prediction servers (see “Experimental Procedures” for details). The model that has captured the predominant trend and was in good agreement with the available cryo-EM structures of homologous channels has been delivered by SWISS-MODEL (51). Among all the models, there was a clear consensus for the region starting from N-terminal ankyrin repeat domain (ARD) and up to characteristic TRP helix. The ARS region that contains Lys-407 was modeled as a part of a tripartite antiparallel beta-strand domain. Two beta-strands of that domain belonged to N-terminal region between the ARD and pre-S1 helix, whereas the third strand arrived from the C terminus (Fig. 8A and Fig. S3). In contrast, the majority of C-terminal region after the TRP helix was poorly modeled by all the servers, which was often unresolved in the template structures available in the Protein Data Bank (PDB). From several templates, it was clear that the region contains a beta-strand (residues Trp-776 to Val-783, participating in the above-mentioned beta-strand domain), although the low-conservancy regions before and after that beta-strand were not consistently structured in predictions.
Figure 8.
Hypothetical mechanism of AA-induced TRPV4 activation and its modulation by Ser-824 phosphorylation. A, homology model of human TRPV4. The location and conformation of AA (based on automated docking simulations) in the hydrophobic crevice in proximity to the region of arachidonate recognition sequence (ARS) suggests that the AA binding might widen the crevice (yellow arrows) and facilitate an outward motion of TRP helix (blue arrow), leading to the channel activation. The channel is presented in a desensitized conformation where C-terminal helix (colored purple) harboring Ser-824 (green) hinders the expansion of the crevice. Ser-824 phosphorylation could prevent the docking and sensitize TRPV4 for activation by AA. B, analysis of the structural features of the Ser-824–harboring helix reveals the abundance of basic residues (blue sticks), which are hypothesized to mediate the docking of the helix in the crevice under S2-S3 loop through several salt bridges to the negatively charged residues at the binding cavity. Acidic (red sticks) and polar (green sticks) residues on Ser-824 helix are modeled to provide a few additional polar contacts to the rest of the protein, whereas the nonpolar patch (colored white) at the top of Ser-824 helix is expected to form a hydrophobic contact with S2-S3 loop (see details on Fig. S5). C, linear subunit model of TRPV4 activation by AA and the prerequisite sensitization by PKA-catalyzed Ser-824 phosphorylation. Positions of the putative ARS and Ser-824 residue mapped onto the predicted membrane topology of a human TRPV4 monomeric subunit.
To obtain better predictions for the unresolved C-terminal region from Arg-779 to Leu-871, we have performed a separate set of structural modeling (essentially a de novo approach; see “Experimental Procedures”) using structure prediction servers IntFOLD-TS (52), I-TASSER (53), Phyre2 (54), RaptorX (55), Robetta (56), and SWISS-MODEL (51). Following the beta-strand at the beginning of the region, the predominant trend was the largely disordered C-terminal region with only one persistent secondary structure element, a short (four to six turns) alpha-helix in the middle, typically residues Trp-822 to Ser-836 (Fig. 8A). This alpha-helical domain contained residue Ser-824, and we hypothesized that the alpha-helix normally resides somewhere in a reasonable vicinity of a putative AA-binding region (somewhere between Lys-407 and TRP helix, see below), whereas phosphorylation would dislodge it from that location. Given that there are six basic residues in the sequence vicinity of the Ser-824–harboring alpha-helix (Fig. 8B), we assumed that it binds to a region with predominant acidic residues. In the vicinity of the putative AA-binding region, there are two suitable locations for the C-terminal alpha-helix, one on the external side of the channel, facing the interface between the lipids and the membrane, and another inside the pore vestibule. Comparison of all the available structural models for the C-terminal domain has revealed the preferred candidates for each location. We have introduced the domain structures into homology models of TRPV4 channel and optimized the structure using 5000-step energy minimization. In both external (Fig. 8A) and internal (Fig. S4) locations, the Ser-824–bearing alpha-helix is stabilized in the position by five or six salt bridges. The side chain of Ser-824 in the interior location faces to a hydrophobic site flanked by the negatively charged groups, whereas in the external it is in direct contact with an acidic side chain Glu-430 (Fig. S5), suggesting that phosphorylation of Ser-824 that introduces large hydrophilic and negatively charged phosphate group might prevent docking of the Ser-824 helix and dislodge the C terminus that is connected by a flexible disordered linker to the rest of the channel.
We should note that the external location of the Ser-824 helix is more accessible for kinases, as well as other potential interaction partners of the C terminus, which advances this location as the preferred one. However, the C-terminal domain was not completely resolved in any of the available structures in the PDB, and therefore is likely to be dynamic, with a low binding energy. In this setting, there might be a dynamic equilibrium between the bound and detached C-terminal domain. The phosphorylation might occur in a detached conformation, suggesting that there might be no need for high accessibility of the docked conformation for phosphorylation.
To assess the possible binding site and conformation of AA, we have performed a set of automated docking calculations on the two homology models of TRPV4 with an external and internal placement of the C terminus. Considering that TRP helix is a putative integration domain for various stimuli activating TRPV4, and that Lys-407 residue (and the ARS segment in general) is likely implicated in the binding of TRPV4-activating AA, the search region was set to comprise both the TRP helix and Lys-407 (details provided in “Experimental Procedures” and in Fig. S3A). The predicted binding region in both models was very similar (Fig. S3, B and C). The exact conformation varied among the obtained predictions; however, the consensus placement for the aliphatic tail of AA was the hydrophobic crevice that spans from the TRP helix to the beta-sheet bearing Lys-407. The most favorable binding conformations had the carboxyl group of AA oriented toward Lys-407 (Fig. 8A). Although there was no direct contact between the two, AA carboxyl is within reach of a flexible Lys-407 side chain to form a salt bridge, which would stabilize the bound AA.
Discussion
Regulation of AA-induced TRPV4 activation by Ser-824 phosphorylation
Although the importance of protein phosphorylation in regulating TRPV4 function is well documented (40–42), the mechanisms by which phosphorylation regulates TRPV4 activation in response to different stimuli remain less well understood. Our previous studies revealed that TRPV4 in endothelial cells serves as a critical signaling component in mediating AA-induced dilation of HCAs (36). Furthermore, we found that AA acts as an endogenous lipid activator of TRPV4 channels in HCAECs and this activation requires PKA-mediated protein phosphorylation. In the present study, we identified the PKA phosphorylation site in TRPV4 (i.e. Ser-824) that is essential for channel activation by AA in coronary endothelial cells. Ser-824, an evolutionarily conserved residue between mammalian and some lower vertebrate (e.g. chicken and fish) TRPV4 proteins (Fig. 2C), could not be aligned to any serine or threonine in other TRP channels of the vanilloid subfamily, suggesting that phosphorylation at this site may be TRPV4-specific. Substitution of serine 824 with nonphosphorylatable alanine (S824A) or phosphomimetic glutamic acid (S824E) in TRPV4 markedly altered the function of this Ca2+-permeable channel in a stimulus-dependent manner. In comparison to WT TRPV4, S824A mutant displayed strongly reduced Ca2+ transients in response to AA, whereas an enhanced response to AA was observed in the S824E mutant. Importantly, we confirmed that both mutants did not affect GSK1016790A-induced TRPV4 activation or TRPV4 protein expression on the plasma membrane, using immunoblotting and fluorescence imaging. Thus, replacement of Ser-824 with alanine or glutamic acid changes AA-induced TRPV4 channel activation, without affecting its overall channel function or secretory trafficking.
Our data confirmed that both PKC and PKA induce TRPV4 phosphorylation at Ser-824 (40, 42). For this analysis, a commercially available anti–pSer-824 antibody was used, which can specifically recognize phosphorylated serine or threonine in the RXRXXS/T sequence motif. The phosphorylation at Ser-824 was strongly enhanced following PMA treatment. In HCAECs expressing the phosphorylation-deficient TRPV4 variant (S824A), no phospho-specific signal could be detected after PMA stimulation. These results confirmed Ser-824 as a target site for PKC, as well as the specificity of the anti–pSer-824 antibody used to detect phosphorylation of this residue. Similarly, activation of PKA by forskolin, the adenylate cyclase activator, also enhanced the phosphorylation of TRPV4 at Ser-824 to a smaller extent. Interestingly, forskolin-induced Ser-824 phosphorylation was enhanced by the presence of the protein phosphatase 1/2A inhibitor cantharidin, indicating that PP1/2A negatively regulates Ser-824 phosphorylation stimulated by the PKA pathway. The PKA inhibitors, PKI and H89, could reduce the effect of forskolin, confirming that PKA is involved in the phosphorylation of Ser-824. However, we cannot exclude the existence of other sites in TRPV4 that could be targeted by PKC and PKA and the possibility that Ser-824 could be phosphorylated by different kinase pathways.
The role of protein phosphorylation in TRPV4 function has been examined in previous studies using HEK 293 or other cell lines expressing human or murine TRPV4 untagged, with C-terminal FLAG or V5 tag, or with C-terminal IRES-GFP (17, 40–42). Equivalent results were obtained with different constructs of TRPV4, and one study verified that the addition of the C-terminal FLAG tag did not alter TRPV4 function (41). In this study, our results indicate that placing GFP at the C terminus of TRPV4 did not affect AA- and GSK1016790A-induced activation and PMA-induced phosphorylation of TRPV4 as compared with the small peptide FLAG tag (Fig. S6).
H2O2–PKA pathway in TRPV4 phosphorylation and activation
The present study provides the first evidence for a regulatory role of H2O2 in Ser-824 phosphorylation of TRPV4 channels. H2O2 at pathophysiologically relevant concentrations (100–300 μm) induced robust Ser-824 phosphorylation that could be detected as early as 5 min after addition of H2O2. The effect of H2O2 was inhibited by three chemically distinct PKA inhibitors, PKI, H89, and KT5720, strongly supporting the involvement of PKA in H2O2-induced phosphorylation of TRPV4 at Ser-824. Furthermore, compared with untreated WT TRPV4-overexpressing HCAECs, pretreatment of cells with H2O2 led to significant enhancement of Ca2+ response to AA, indicating H2O2 can potentiate TRPV4 activation by AA. This effect was almost entirely abolished by the PKA inhibitor, PKI, demonstrating the involvement of PKA activity. Together with the findings discussed above, this strongly suggests that the H2O2–PKA signaling axis may represent a powerful mechanism regulating AA-induced TRPV4 activation. In addition, observation of reduced Ca2+ transients in HCAECs expressing the S824A mutant channel as compared with WT TRPV4 further proves that Ser-824 is a major phosphorylation site involved in H2O2 regulation of AA-induced activation.
Pathophysiological significance of H2O2–PKA–TRPV4 coupling in vascular and other systems
TRPV4 mediates shear stress–induced and endothelium-dependent dilation of HCAs, and this functional role is more pronounced in subjects with coronary artery disease (45). Although precise mechanisms by which shear stress preferentially activates endothelial TRPV4 in disease remain unclear, evidence indicates that phospholipase A2 activation and the associated release of AA or its metabolites may be involved in HCAs3 and other vascular beds (10). Consistent with this proposed role of AA in TRPV4 activation, AA induced potent vasodilation and endothelial [Ca2+]i increase in HCAs, and these responses were blocked by the selective TRPV4 antagonists HC067047 (Fig. 7 and Fig. S2) and RN-1747 (36). The TRPV4-mediated vascular response to AA was partially inhibited by catalase, suggesting that H2O2 regulates AA-induced TRPV4 activation in native endothelial cells. As discussed above, a gain-of-function of TRPV4 contributes to several pathological processes such as neurogenic inflammation and hyperalgesia (5, 16–18), adipose cell inflammation (19), and pulmonary and cardiac tissue fibrosis (20, 21). In the vascular system, prolonged or excessive activation of endothelial TRPV4 causes disruption of endothelial barrier, tissue edema, and endothelial dysfunction in inflammation and other pathological conditions (57, 58). It remains of interest to determine whether H2O2-induced enhancement of TRPV4 activation by AA contributes to excessive TRPV4 activity under the above pathophysiological conditions.
There is increasing evidence that H2O2 regulates various protein kinases including PKA (48). Although not directly tested in HCAs because of the interference of PKA inhibitors on smooth muscle function, H2O2 regulation of TRPV4-mediated vascular response may involve PKA signaling, similar to that observed in cultured HCAECs. A previous study showed that H2O2 strongly up-regulates Src-dependent tyrosine phosphorylation of TRPV4, including the Tyr-110 residue (41). This Src-mediated phosphorylation has been suggested to contribute to the sensitization of TRPV4 in hyperalgesia (17, 41). It has yet to be determined whether serine/threonine and tyrosine kinase–mediated phosphorylation of TRPV4 may coordinate to regulate TRPV4 function in vascular and other cell types.
Hypothesized mechanisms involved in TRPV4 activation by AA and its regulation by Ser-824 phosphorylation
The developed homology model of TRPV4 channel has high confidence of prediction for the region that includes most of the N-terminal cytoplasmic domain (starting from ARD) and the whole transmembrane domains (including the “force hub” TRP helix (59, 60)). The structure is less certain for the C-terminal domain, but likely contains an alpha-helix with Ser-824, flanked by the flexible linkers. Based on structural analysis of the homology models, we propose two possible binding positions near the TRP and pre-S1 helices. This model of C-terminal arrangement, together with the predicted binding location for AA in the hydrophobic groove between the TRP helix and ARS region, allows us to suggest a possible mechanism for TRPV4 activation and its regulation by Ser-824 phosphorylation (Fig. 8). We hypothesize that binding of AA favors expansion of the hydrophobic crevice under TRP helix, thus decreasing the strength of van der Waals interactions with the short alpha-helices that contact TRP helix. It was shown previously that weakening the “latch” that holds TRP helix in place favors the opening of the channel gate that is located on S6 helix (directly connected to TRP helix) (59, 60). We suggest that similar effect might be caused by AA-induced widening of the hydrophobic crevice, allowing the outward motion of TRP helix and activation of the channel. This widening of the crevice might be contained if the C-terminal domain is lodged to one or another side of the crevice, as suggested by our structural models. However, phosphorylation of Ser-824 residue that is located on the alpha-helix in the middle of the C terminus might prevent the domain docking (a disinhibition mechanism), thus enabling the expansion of the hydrophobic crevice and sensitizing TRPV4 to activation by AA. Although this activation mechanism is hypothetical, the atomistic structural models allow specific predictions that can be tested experimentally in future studies.
The alpha-helix with Ser-824 also resides in the previously reported calmodulin (CaM)-binding domain (CBD), which is thought to be involved in Ca2+-dependent regulation of TRPV4 (61–63). Interestingly, TRPV4 mutants with deleted CBD (63) or charge-reversal (5R/E) within CBD (61) show increased channel activity in two different expression systems. This gain-of-function phenotype somewhat mimics what we observed in the S824E mutant and WT TRPV4 with Ser-824 phosphorylation, suggesting that a potentially similar disinhibition mechanism through dissociation of C terminus upon CaM binding or Ser-824 phosphorylation may be involved. However, disparate results have been obtained in the previous studies regarding the potential sites that the CBD might interreact with (62, 63). It remains largely unclear how Ser-824 phosphorylation and CaM binding in the C-terminal region of TRPV4 might be simultaneously regulated under physiological conditions. A previous study has reported that CaM binding at the C terminus (residues 696 to 729) negatively modulates the activity of TRPV5 and this effect can be reversed by PKA-dependent phosphorylation of residue Thr-709 (64).
Further structure-function studies are required to elucidate the precise mechanisms by which the lipid mediator AA and its metabolites interact with TRPV4 to cause channel opening. Although the synthetic agonist 4α-PDD activates TRPV4 by directly binding to its TM3 and TM4 segments in a ligand-like manner (38), earlier studies indicate that AA-induced TRPV4 activation involves metabolism of AA by cytochrome P450 into 5,6-epoxyeicosatrienoic acid (5,6-EET) (34, 35, 39). A recent study has further proposed a potential 5,6-EET-binding site in TRPV4, which is formed by residues from the TM2-TM3 linker, TM4, and TM4-TM5 linker (65). However, other studies indicate that TRPV4 can also be activated by other EET regioisomers such as 11,12-EET (66, 67), AA (36, 68), and endocannabinoids such as 2-arachidonoylglycerol (37). The reason for this discrepancy is presently unknown but may relate to the potential variable sensitivity of TRPV4 to AA and its metabolites in different experimental systems. As shown in this and a previous study (36), TRPV4 activation by AA in native human endothelial cells was only marginally affected by EET antagonism (Fig. 7C), a finding consistent with a more direct role of AA rather than its P450 metabolites in TRPV4 activation. Furthermore, AA-induced TRPV4 activation requires only nanomolar concentrations of AA in native endothelial cells (Fig. 7A) whereas much higher concentrations (1–3 μm) are needed to activate TRPV4 in cultured HCAECs, indicating an exquisite sensitivity of this ex vivo system to AA as compared with cultured endothelial or other cells.
In summary, the activation of TRPV4 by AA in endothelial cells is tightly regulated by PKA-dependent phosphorylation, in which Ser-824 is a major phosphorylation site. We present evidence supporting the notion that the H2O2–PKA–TRPV4 Ser-824 phosphorylation signaling axis may represent an important mechanism regulating AA-induced TRPV4 activation under pathophysiological conditions. Finally, we provide a structural model supporting the proposal that AA may activate TRPV4 through direct binding to the hydrophobic crevice between TRP helix and cytosolic ARS region and such binding or channel gating is enhanced by phosphorylation on the Ser-824 residue via a disinhibition mechanism. Together, these results give insight into the mechanisms of TRPV4 activation and its regulation by protein phosphorylation. A better understanding of these mechanisms may reveal novel therapeutic targets to specifically reduce augmented signaling under pathophysiological conditions while preserving normal physiological functions.
Experimental procedures
Cell culture
HCAECs were obtained from Lonza (Walkersville, MD) and maintained in complete EGM-2MV growth medium (Lonza) according to the manufacturer's instructions. In brief, HCAECs were grown in a humidified incubator at 37 °C and 5% CO2 and split at 1:3 to 1:4 ratios once the cells reached 90–95% confluence. Cells between passages 6 and 7 were used for experiments.
Plasmids, mutagenesis, and lentiviral production
The full-length human TRPV4 (NM_021625) cDNA clone was obtained from OriGene (Rockville, MD) and shuttled into a mammalian expression vector resulting in a TRPV4 fusion protein with its COOH terminus tagged with turbo GFP. The TRPV4-GFP construct was then cloned into the pWPTS lentiviral vector as described previously (36). Point mutations (e.g. S824A and S824E) were introduced into wildtype TRPV4 using site-directed mutagenesis service (Mutagenex, Suwanee, GA). All constructs were verified by DNA sequencing. Recombinant lentiviruses were produced from HEK 293 cells as described in our previous work (69).
Transient and stable transgene expression
For stable TRPV4 overexpression experiments, HCAECs at passage 6 were grown to 50–60% confluence before being transduced with recombinant lentiviruses at an m.o.i. (multiplicity of infection) of 50–100. Sixteen h after transduction, the concentration of free calcium in the culture medium was reduced to ∼0.6 mm by the addition of 1.2 mm EDTA, and the medium pH was readjusted. This low-calcium medium has been used previously to minimize potential calcium overload from TRPV4 overexpression in HCAECs (36). Cells were used for calcium imaging 3 to 4 days after transduction (without cell splitting) and for immunoblotting experiments 4 to 5 days after transduction (with one splitting at a ratio of 1:3).
To determine whether GFP affects TRPV4 function in TRPV4-GFP fusion constructs, we performed control experiments using HEK 293 cells transiently transfected with TRPV4-FLAG-IRES–turbo GFP pCMV6 (OriGene), or TRPV4-mGFP pCMV6 (OriGene). HEK 293 cells were grown in DMEM containing 10% FBS. Cells were transfected using Lipofectamine 2000 reagent (Invitrogen) and incubated in DMEM with reduced Ca2+ (0.6 mm) for an additional 16–20 h prior to calcium imaging, or for an additional 48 h prior to immunoblotting experiments.
Calcium imaging
HCAECs were plated onto 35-mm glass-bottom Petri dishes and grown to 60–70% confluence. The cells were incubated with fura-2 AM (5 μm) (Molecular Probes) in the presence of Pluronic F-127 (0.01%) at 37 °C for 30 min in a modified Hanks' balanced salt solution (HBSS) that contained (in mm): 123 NaCl, 5.4 KCl, 1.6 CaCl2, 0.5 MgCl2, 0.4 MgSO4, 4.2 NaHCO3, 0.3 NaH2PO4, 0.4 KH2PO4, 5.5 glucose, and 20 HEPES (pH 7.4 with NaOH). A fura-2 assay was used to monitor cytosolic Ca2+ signals as described previously (12, 36). MetaFluor software (Molecular Devices) was used to record and analyze the emitted fura-2 fluorescence at 510 nm in cells that are alternately exposed to 340 and 380 nm excitation wavelengths. Fluorescence images were acquired every 3 s for 20 to 30 min. Background fluorescence was subtracted before calculation of the ratio of the fluorescence intensity at 340 nm versus 380 nm excitation (F340/F380). Experiments were performed at 27 °C. The intracellular calcium concentration ([Ca2+]i) was calculated according to Equation 1,
| (1) |
where Kd is the Ca2+ dissociation constant of fura-2 (224 nm), R is the F340/F380 ratio at any time, Rmin and Rmax are minimal and maximal F340/F380 ratio under Ca2+-depleted (1 mm EGTA and 10 μm ATP in calcium-free HBSS) and Ca2+-saturated (5 mm CaCl2 and 2 μm ionomycin in normal HBSS) conditions, respectively, and Sf,2/Sb,2 represents the ratio of signal intensity at 380 nm excitation (F380) under Ca2+-depleted and Ca2+-saturated conditions, respectively (70). With a Kd value of around 224 nm, Equation 1 can accurately quantify [Ca2+]i in the range of 20 nm to 2 μm (or 0.1× Kd to 10× Kd). For GSK1016790A-induced Ca2+ responses in TRPV4-transfected HCAEC (Figs. 1F and 4F), which can be obtained at a level close to or above 1–2 μm, conversion of F340/F380 ratio to [Ca2+]i will introduce large errors, and we, therefore, presented the averaged changes in F340/F380 ratios. To ascertain that the fura-2 assay does not blunt the potential difference in GSK1016790A-induced responses between WT, S824A, and S824E constructs, additional experiments were performed with fura-4F, which has a lower affinity for Ca2+ (Kd around 0.77 μm) and thus is less likely to be saturated at 1–2 μm [Ca2+]i (Fig. S7).
Immunoprecipitation and immunoblotting
Following treatment as specified in the figure legends, endothelial cells expressing the TRPV4-GFP transgene were rinsed with ice-cold phosphate-buffered saline (PBS, pH 7.4), and solubilized in a lysis buffer (25 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1.0% Nonidet P-40, 0.5% sodium deoxycholate, 1 mm EDTA, and 5% glycerol) supplemented with 10 μm cantharidin, and a mixture of 1× protease inhibitors (Roche) and 1× phosphatase inhibitors (Pierce). Cell lysates were centrifuged at 12,000 × g for 10 min, and protein samples (150 μg) were mixed with 0.5 μg of a rabbit polyclonal anti-GFP antibody (Evrogen, AB513) for 1 h and then with 5 μl of protein A/G magnetic beads (Pierce, cat. no. 88802) for an additional 3 h at 4 °C to precipitate TRPV4-GFP immunocomplexes. The magnetic beads were then washed with the lysis buffer four times and water one time, and proteins were eluted with 60 μl of 1× LDS sample buffer (25 mm Tris-HCl, pH 8.0, 1% LDS, 10% glycerol, and 0.0045% bromophenol blue) supplemented with 50 mm DTT for 15 min at room temperature.
Protein eluates (20 μl, equivalent to 50 μg of total input protein) were separated by SDS-PAGE on 10% TGX precast gels (Bio-Rad) and transferred to PVDF membranes. Membranes were blocked with 5% BSA at room temperature for 1 h and then incubated with diluted rabbit monoclonal anti-Phospho-Akt Substrate antibody (1:1,000 in TBST with 5% BSA) (cat. no. 10001, clone 23C8D2, Cell Signaling Technology) at 4 °C for overnight. This anti–phospho-serine/threonine antibody, which is raised against the motif RXRXXS*/T*, was used to identify phosphorylated serine 824 of TRPV4. A similar antibody (cat. no. 9611 from Cell Signaling Technology against the motif [K/R]X[K/R]XXS*/T*) was used in a previous study (42). Blots were then washed with TBST prior to the addition of an HRP-conjugated mouse anti-rabbit IgG (conformation-specific) antibody (1:2000 in TBST with 5% NFDM) (cat. no. 5127S, clone L27A9, Cell Signaling Technology) at room temperature for 1 h. Membranes were repeatedly washed and developed using the ECL Plus reagent (Amersham Biosciences). The same membrane was stripped in Restore Western Blot Stripping Buffer (Pierce) and reprobed with a rabbit anti-GFP antibody (1:15,000 in TBST with 5% NFDM) (Evrogen, AB513), followed by a mouse anti-rabbit antibody as above (1:2000 in TBST with 5% NFDM) to obtain the total TRPV4 signal.
To assess protein phosphorylation of TRPV4 by PKA in vitro, protein lysates of HCAECs expressing TRPV4-GFP (150–200 μg total protein) were immunoprecipitated with anti-GFP antibody and protein A/G magnetic beads as described above. After washing with the lysis buffer three to four times and once with water, the beads were incubated in a kinase reaction buffer (50 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 0.1 mm EDTA, 2 mm DTT, 0.01% Brij 35), supplemented with 2500 units of a recombinant PKA, catalytic subunit (New England Biolabs) and 200 μm ATP, for 30 min at 30 °C prior to SDS-PAGE.
Biotinylation of cell surface proteins
A cell surface protein isolation kit from Pierce was used to isolate plasma membrane proteins from HCAECs, according to the manufacturer's instructions with slight modification (23). Briefly, cells were incubated in PBS containing 0.5 mg/ml sulfosuccinimidyl 2-(biotiamido)ethyl-1.3-dithiopropionate (Sulfo-NHS-SS-Biotin) for 30 min at 4 °C with gentle rocking. The biotinylation reaction was stopped by a quenching solution, and cells were washed with ice-cold Tris-buffered saline (TBS), followed by total protein preparation using a lysis buffer described above. To isolate biotinylated proteins, protein samples (150–200 μg input protein) were mixed with 50 μl of NeutrAvidin agarose beads (supplied as 50% slurry) and rotated for 3 h at 4 °C. The beads were collected by centrifugation at 2500 × g for 2 min and washed four to five times with a wash buffer. Proteins were then eluted with 100 μl of 1× Laemmli sample buffer (Bio-Rad) supplemented with 50 mm DTT by heating the samples for 5 min at 95 °C. The eluted proteins were analyzed by Western blotting as described earlier.
Vascular reactivity
Vasomotor response was assessed in coronary arterioles (100–200 μm) isolated from human atrial surgical specimens and mounted on pressure myograph as described previously in detail (36). All protocols were approved by the local institutional review boards on the use of human subjects in research. Arterioles were pressurized under 60 mm Hg and precontracted with endothelin-1, and relaxation responses to cumulative concentrations of AA (10−10 to 10−6 m) were determined in the presence or absence of HC-067046 (1 μm, intraluminal and in the bath), a selective antagonist of TRPV4. To examine the modulation of endogenous H2O2 on TRPV4-mediated dilation, vasodilatory responses to AA were examined before and after a 30-min incubation with catalase (500 units/ml, intraluminal), an H2O2-metabolizing enzyme. To determine the role of cytochrome P450 metabolites EETs, arterioles were incubated for 30 min with 14,15-EEZE (10 μm), a selective EET antagonist (49). At the end of each experiment, papaverine (10−4 m), an endothelium-independent vasodilator, was added to determine the maximal dilation for normalization of dilator responses. Vasodilator responses are expressed as a percentage of maximal relaxation relative to endothelin-1 constriction, with 100% representing full relaxation to basal tension or the maximal diameter.
Human TRPV4 homology model and molecular docking
Homology models of the human TRPV4 channel were developed using seven structure prediction servers: I-TASSER (53), Phyre2 (54), M4T (71), IntFOLD-TS (52), RaptorX (55), SWISS-MODEL (51), and Robetta (56). The complete protein sequence (UniProt ID: Q9HBA0) was submitted for processing. Comparison of the predictions (47 in total) has revealed a high consistency in the ankyrin repeats and transmembrane domains, whereas the ∼100-residue N-terminal domain and C-terminal region after TRP helix (characterized by the lower evolutionary conservancy and mostly unresolved in the experimental structures) showed significant variability in the models. The structure delivered by SWISS-MODEL server, which was based on the cryo-EM structure of TRPV2 channel (PDB ID: 5HI9) as the template, was most consistent with both the predictions of different servers and the available structures in the PDB. Taking this model as the basis for the N-terminal and the transmembrane parts, we have performed a separate round of modeling for an isolated C-terminal sequence (residues Arg-779 to Leu-871) to avoid bias by the preceding part with high homology. The domain was submitted to structure prediction servers IntFOLD-TS (52), I-TASSER (53), Phyre2 (54), RaptorX (55), Robetta (56), and SWISS-MODEL (51). The produced models (80 in total) were analyzed for compatibility with the rest of the channel structure, in particular, the ability to dock in the putative internal and external locations (see “Results”), the ability to be linked to the end of the beta-strand after the TRP helix without structural strain, and the absence of steric conflicts with the other subunits in tetrameric arrangement. Based on the inspection we have selected the best fitting structural model of the C-terminal region for each of the two target locations, and integrated them with the homology model using visual molecular dynamics (VMD) software (72). The complete homology models (Fig. 8A and Figs. S3 and S4) were subjected to 5000 steps of energy minimization to remove minor sterical conflicts. The minimization was performed in NAMD (73) using conjugate gradient algorithm and CHARMM36 force field (74).
Based on empirical evidence (1, 59, 60) we have suggested that the binding spot for AA might be located somewhere in the region between the TRP helix and ARS domain. To explore the possible binding conformations, we have used automated docking of AA molecule to the homology models of TRPV4 in AutoDock Vina software (75). The search box for the center of AA was set to be 30 Å × 30 Å × 30 Å, which covered the whole target region including TRP helix and the beta-sheet containing Lys-407. With that box, the volume that the carboxyl group or tail of AA could reach was nearly 50 Å × 50 Å × 50 Å (see Fig. S3). The protein structure was held rigid whereas AA molecule was flexible with rotations allowed around all the appropriate chemical bonds. The docking was performed with high exhaustiveness parameter (160), producing 20 most likely binding modes per run. Considering the stochastic nature of this docking approach, the whole procedure was repeated 10 times for every homology model, bringing a family of 200 predicted docking conformations for each of them (Fig. 8A and Figs. S3 and S4). The results of the homology modeling and automated docking were visualized using VMD (72).
Reagents
GSK1016790A was kindly provided by GlaxoSmithKline Pharmaceuticals, and 14,15-epoxyeicosa-5(Z)-enoic acid (14,15-EEZE) was provided by John R. Falck. AA sodium salt and H2O2 were obtained from Sigma-Aldrich. HC067047, cantharidin, forskolin, PMA, H89 dihydrochloride, and PKI 14–22 amide were obtained from Tocris. KT5720 was obtained from Alomone Labs (Jerusalem, Israel). Stock solutions of AA sodium salt and 14,15-EEZE were prepared in double-distilled ethanol. H2O2 (30% solution) was diluted in distilled water immediately before use. All other chemicals were dissolved in DMSO as stock solutions (1000× or higher). Vehicle control studies indicated that the addition of DMSO (0.01–0.1%) or ethanol (0.01–0.1%) did not affect the cellular responses examined in this study.
Data and statistical analysis
All Ca2+ responses are presented as mean ± S.E., with the number of independent experiments (n) included in the brackets above error bars. In each experiment, ≥ 20 randomly chosen cells were recorded and averaged as one measurement. All other molecular experiments were repeated at least three times unless otherwise stated. Statistical comparisons of vasodilatory responses were performed by two-way (factor) repeated measures ANOVA (factor 1, doses; factor 2, different treatments such as a vehicle or an inhibitor), followed by the Holm-Sidak test for pairwise comparisons or comparisons versus control. For other nonvascular studies, statistical comparisons were made by Student's t test or one-way ANOVA, where appropriate, using the statistical analysis programs provided in SigmaPlot (version 12). Significance in figures was depicted as * or #, p < 0.05; ** or ##, p < 0.01.
Author contributions
S. C., A. A., and D. X. Z. designed the study and wrote the paper. S. C. and N. S. Z. conducted and analyzed the experiments shown in Figures 1–7 and Figures S1 and S2, A. K. and Z. W. performed and analyzed the experiments shown in Figures S6 and S7, and A. A. performed the computational modeling of human TRPV4 shown in Figure 8 and Figures S3–S5. Y. N. and J. F. performed cloning and generated lentiviral vectors encoding WT and mutant human TRPV4 channels. D. A. W. provided intellectual input or technical advice. D. X. Z. conceived the idea for the project, generated research funds, and led and coordinated the study. All authors reviewed the results, commented on the manuscript, and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank the Division of Cardiothoracic Surgery at the Medical College of Wisconsin and the Veterans Affairs Medical Center, the Cardiothoracic Surgery Group of Milwaukee, the Cardiovascular Surgery Associates of Milwaukee, the Midwest Heart Surgery Institute, the Wisconsin Heart Group, Froedtert Memorial Lutheran Hospital, and the Aurora St. Luke's Medical Center in Milwaukee for providing surgical specimens. This research was completed in part with computational resources and technical support provided by the Research Computing Center at the Medical College of Wisconsin. We thank the Children's Hospital Foundation for a generous gift from John B. and Judith A. Gardetto.
This work was supported by National Institute of Health Grant HL096647 (to D. X. Z.) and by the Biology Department, University of Maryland, College Park (to A. A.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S7 and supporting references.
N. S. Zinkevich and D. X. Zhang, unpublished observations.
- TRP
- transient receptor potential
- AA
- arachidonic acid
- ARD
- ankyrin repeat domain
- ARS
- arachidonate recognition sequence
- CaM
- calmodulin
- CBD
- CaM-binding domain
- EET
- epoxyeicosatrienoic acid
- HCA
- human coronary arteriole
- HCAEC
- human coronary artery endothelial cell
- H2O2
- hydrogen peroxide
- PKI
- PKA inhibitor
- PMA
- phorbol 12-myristate 13-acetate
- TRPV4
- transient receptor potential vanilloid 4.
References
- 1. Nilius B., Vriens J., Prenen J., Droogmans G., and Voets T. (2004) TRPV4 calcium entry channel: A paradigm for gating diversity. Am. J. Physiol. Cell Physiol. 286, C195–C205 10.1152/ajpcell.00365.2003 [DOI] [PubMed] [Google Scholar]
- 2. Garcia-Elias A., Mrkonjić S., Jung C., Pardo-Pastor C., Vicente R., and Valverde M. A. (2014) The TRPV4 channel. Handb. Exp. Pharmacol. 222, 293–319 10.1007/978-3-642-54215-2_12 [DOI] [PubMed] [Google Scholar]
- 3. Randhawa P. K., and Jaggi A. S. (2015) TRPV4 channels: Physiological and pathological role in cardiovascular system. Basic Res. Cardiol. 110, 54 10.1007/s00395-015-0512-7 [DOI] [PubMed] [Google Scholar]
- 4. White J. P., Cibelli M., Urban L., Nilius B., McGeown J. G., and Nagy I. (2016) TRPV4: Molecular conductor of a diverse orchestra. Physiol. Rev. 96, 911–973 10.1152/physrev.00016.2015 [DOI] [PubMed] [Google Scholar]
- 5. Alessandri-Haber N., Dina O. A., Joseph E. K., Reichling D., and Levine J. D. (2006) A transient receptor potential vanilloid 4–dependent mechanism of hyperalgesia is engaged by concerted action of inflammatory mediators. J. Neurosci. 26, 3864–3874 10.1523/JNEUROSCI.5385-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Suzuki M., Mizuno A., Kodaira K., and Imai M. (2003) Impaired pressure sensation in mice lacking TRPV4. J. Biol. Chem. 278, 22664–22668 10.1074/jbc.M302561200 [DOI] [PubMed] [Google Scholar]
- 7. Liedtke W., and Friedman J. M. (2003) Abnormal osmotic regulation in trpv4−/− mice. Proc. Natl. Acad. Sci. U.S.A. 100, 13698–13703 10.1073/pnas.1735416100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Andrade Y. N., Fernandes J., Vázquez E., Fernández-Fernández J. M., Arniges M., Sánchez T. M., Villalón M., and Valverde M. A. (2005) TRPV4 channel is involved in the coupling of fluid viscosity changes to epithelial ciliary activity. J. Cell Biol. 168, 869–874 10.1083/jcb.200409070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Köhler R., Heyken W. T., Heinau P., Schubert R., Si H., Kacik M., Busch C., Grgic I., Maier T., and Hoyer J. (2006) Evidence for a functional role of endothelial transient receptor potential V4 in shear stress-induced vasodilatation. Arterioscler. Thromb. Vasc. Biol. 26, 1495–1502 10.1161/01.ATV.0000225698.36212.6a [DOI] [PubMed] [Google Scholar]
- 10. Hartmannsgruber V., Heyken W. T., Kacik M., Kaistha A., Grgic I., Harteneck C., Liedtke W., Hoyer J., and Köhler R. (2007) Arterial response to shear stress critically depends on endothelial TRPV4 expression. PLoS One 2, e827 10.1371/journal.pone.0000827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Loot A. E., Popp R., Fisslthaler B., Vriens J., Nilius B., and Fleming I. (2008) Role of cytochrome P450-dependent transient receptor potential V4 activation in flow-induced vasodilatation. Cardiovasc. Res. 80, 445–452 10.1093/cvr/cvn207 [DOI] [PubMed] [Google Scholar]
- 12. Mendoza S. A., Fang J., Gutterman D. D., Wilcox D. A., Bubolz A. H., Li R., Suzuki M., and Zhang D. X. (2010) TRPV4-mediated endothelial Ca2+ influx and vasodilation in response to shear stress. Am. J. Physiol. Heart Circ. Physiol. 298, H466–H476 10.1152/ajpheart.00854.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang D. X., and Gutterman D. D. (2011) Transient receptor potential channel activation and endothelium-dependent dilation in the systemic circulation. J. Cardiovasc. Pharmacol. 57, 133–139 10.1097/FJC.0b013e3181fd35d1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Loukin S., Su Z., and Kung C. (2011) Increased basal activity is a key determinant in the severity of human skeletal dysplasia caused by TRPV4 mutations. PLoS One 6, e19533 10.1371/journal.pone.0019533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nilius B., and Voets T. (2013) The puzzle of TRPV4 channelopathies. EMBO Rep. 14, 152–163 10.1038/embor.2012.219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grant A. D., Cottrell G. S., Amadesi S., Trevisani M., Nicoletti P., Materazzi S., Altier C., Cenac N., Zamponi G. W., Bautista-Cruz F., Lopez C. B., Joseph E. K., Levine J. D., Liedtke W., Vanner S., Vergnolle N., Geppetti P., and Bunnett N. W. (2007) Protease-activated receptor 2 sensitizes the transient receptor potential vanilloid 4 ion channel to cause mechanical hyperalgesia in mice. J. Physiol. 578, 715–733 10.1113/jphysiol.2006.121111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Poole D. P., Amadesi S., Veldhuis N. A., Abogadie F. C., Lieu T., Darby W., Liedtke W., Lew M. J., McIntyre P., and Bunnett N. W. (2013) Protease-activated receptor 2 (PAR2) protein and transient receptor potential vanilloid 4 (TRPV4) protein coupling is required for sustained inflammatory signaling. J. Biol. Chem. 288, 5790–5802 10.1074/jbc.M112.438184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhao P., Lieu T., Barlow N., Sostegni S., Haerteis S., Korbmacher C., Liedtke W., Jimenez-Vargas N. N., Vanner S. J., and Bunnett N. W. (2015) Neutrophil elastase activates protease-activated receptor-2 (PAR2) and transient receptor potential vanilloid 4 (TRPV4) to cause inflammation and pain. J. Biol. Chem. 290, 13875–13887 10.1074/jbc.M115.642736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ye L., Kleiner S., Wu J., Sah R., Gupta R. K., Banks A. S., Cohen P., Khandekar M. J., Boström P., Mepani R. J., Laznik D., Kamenecka T. M., Song X., Liedtke W., Mootha V. K., Puigserver P., Griffin P. R., Clapham D. E., and Spiegelman B. M. (2012) TRPV4 is a regulator of adipose oxidative metabolism, inflammation, and energy homeostasis. Cell 151, 96–110 10.1016/j.cell.2012.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Adapala R. K., Thoppil R. J., Luther D. J., Paruchuri S., Meszaros J. G., Chilian W. M., and Thodeti C. K. (2013) TRPV4 channels mediate cardiac fibroblast differentiation by integrating mechanical and soluble signals. J. Mol. Cell Cardiol. 54, 45–52 10.1016/j.yjmcc.2012.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rahaman S. O., Grove L. M., Paruchuri S., Southern B. D., Abraham S., Niese K. A., Scheraga R. G., Ghosh S., Thodeti C. K., Zhang D. X., Moran M. M., Schilling W. P., Tschumperlin D. J., and Olman M. A. (2014) TRPV4 mediates myofibroblast differentiation and pulmonary fibrosis in mice. J. Clin. Invest. 124, 5225–5238 10.1172/JCI75331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kusudo T., Wang Z., Mizuno A., Suzuki M., and Yamashita H. (2012) TRPV4 deficiency increases skeletal muscle metabolic capacity and resistance against diet-induced obesity. J. Appl. Physiol. 112, 1223–1232 10.1152/japplphysiol.01070.2011 [DOI] [PubMed] [Google Scholar]
- 23. Nishijima Y., Zheng X., Lund H., Suzuki M., Mattson D. L., and Zhang D. X. (2014) Characterization of blood pressure and endothelial function in TRPV4-deficient mice with l-NAME- and angiotensin II-induced hypertension. Physiol. Rep. 2, e00199 10.1002/phy2.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Long S. B., Tao X., Campbell E. B., and MacKinnon R. (2007) Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature 450, 376–382 10.1038/nature06265 [DOI] [PubMed] [Google Scholar]
- 25. Ramsey I. S., Delling M., and Clapham D. E. (2006) An introduction to TRP channels. Annu. Rev. Physiol. 68, 619–647 10.1146/annurev.physiol.68.040204.100431 [DOI] [PubMed] [Google Scholar]
- 26. Wissenbach U., Bödding M., Freichel M., and Flockerzi V. (2000) Trp12, a novel Trp related protein from kidney. FEBS Lett. 485, 127–134 10.1016/S0014-5793(00)02212-2 [DOI] [PubMed] [Google Scholar]
- 27. Strotmann R., Harteneck C., Nunnenmacher K., Schultz G., and Plant T. D. (2000) OTRPC4, a nonselective cation channel that confers sensitivity to extracellular osmolarity. Nat. Cell Biol. 2, 695–702 10.1038/35036318 [DOI] [PubMed] [Google Scholar]
- 28. Liedtke W., Choe Y., Martí-Renom M. A., Bell A. M., Denis C. S., Sali A., Hudspeth A. J., Friedman J. M., and Heller S. (2000) Vanilloid receptor-related osmotically activated channel (VR-OAC), a candidate vertebrate osmoreceptor. Cell 103, 525–535 10.1016/S0092-8674(00)00143-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Güler A. D., Lee H., Iida T., Shimizu I., Tominaga M., and Caterina M. (2002) Heat-evoked activation of the ion channel, TRPV4. J. Neurosci. 22, 6408–6414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Watanabe H., Vriens J., Suh S. H., Benham C. D., Droogmans G., and Nilius B. (2002) Heat-evoked activation of TRPV4 channels in a HEK293 cell expression system and in native mouse aorta endothelial cells. J. Biol. Chem. 277, 47044–47051 10.1074/jbc.M208277200 [DOI] [PubMed] [Google Scholar]
- 31. Gao X., Wu L., and O'Neil R. G. (2003) Temperature-modulated diversity of TRPV4 channel gating: Activation by physical stresses and phorbol ester derivatives through protein kinase C-dependent and -independent pathways. J. Biol. Chem. 278, 27129–27137 10.1074/jbc.M302517200 [DOI] [PubMed] [Google Scholar]
- 32. Loukin S., Zhou X., Su Z., Saimi Y., and Kung C. (2010) Wild-type and brachyolmia-causing mutant TRPV4 channels respond directly to stretch force. J. Biol. Chem. 285, 27176–27181 10.1074/jbc.M110.143370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Servin-Vences M. R., Moroni M., Lewin G. R., and Poole K. (2017) Direct measurement of TRPV4 and PIEZO1 activity reveals multiple mechanotransduction pathways in chondrocytes. Elife 6, e21074 10.7554/eLife.21074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Watanabe H., Vriens J., Prenen J., Droogmans G., Voets T., and Nilius B. (2003) Anandamide and arachidonic acid use epoxyeicosatrienoic acids to activate TRPV4 channels. Nature 424, 434–438 10.1038/nature01807 [DOI] [PubMed] [Google Scholar]
- 35. Vriens J., Owsianik G., Fisslthaler B., Suzuki M., Janssens A., Voets T., Morisseau C., Hammock B. D., Fleming I., Busse R., and Nilius B. (2005) Modulation of the Ca2 permeable cation channel TRPV4 by cytochrome P450 epoxygenases in vascular endothelium. Circ. Res. 97, 908–915 10.1161/01.RES.0000187474.47805.30 [DOI] [PubMed] [Google Scholar]
- 36. Zheng X., Zinkevich N. S., Gebremedhin D., Gauthier K. M., Nishijima Y., Fang J., Wilcox D. A., Campbell W. B., Gutterman D. D., and Zhang D. X. (2013) Arachidonic acid-induced dilation in human coronary arterioles: Convergence of signaling mechanisms on endothelial TRPV4-mediated Ca2+ entry. J. Am. Heart Assoc. 2, e000080 10.1161/JAHA.113.000080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ho W. S., Zheng X., and Zhang D. X. (2015) Role of endothelial TRPV4 channels in vascular actions of the endocannabinoid, 2-arachidonoylglycerol. Br. J. Pharmacol. 172, 5251–5264 10.1111/bph.13312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vriens J., Owsianik G., Janssens A., Voets T., and Nilius B. (2007) Determinants of 4 alpha-phorbol sensitivity in transmembrane domains 3 and 4 of the cation channel TRPV4. J. Biol. Chem. 282, 12796–12803 10.1074/jbc.M610485200 [DOI] [PubMed] [Google Scholar]
- 39. Vriens J., Watanabe H., Janssens A., Droogmans G., Voets T., and Nilius B. (2004) Cell swelling, heat, and chemical agonists use distinct pathways for the activation of the cation channel TRPV4. Proc. Natl. Acad. Sci. U.S.A. 101, 396–401 10.1073/pnas.0303329101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fan H. C., Zhang X., and McNaughton P. A. (2009) Activation of the TRPV4 ion channel is enhanced by phosphorylation. J. Biol. Chem. 284, 27884–27891 10.1074/jbc.M109.028803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wegierski T., Lewandrowski U., Müller B., Sickmann A., and Walz G. (2009) Tyrosine phosphorylation modulates the activity of TRPV4 in response to defined stimuli. J. Biol. Chem. 284, 2923–2933 10.1074/jbc.M805357200 [DOI] [PubMed] [Google Scholar]
- 42. Peng H., Lewandrowski U., Müller B., Sickmann A., Walz G., and Wegierski T. (2010) Identification of a protein kinase C-dependent phosphorylation site involved in sensitization of TRPV4 channel. Biochem. Biophys. Res. Commun. 391, 1721–1725 10.1016/j.bbrc.2009.12.140 [DOI] [PubMed] [Google Scholar]
- 43. Zhang X., and McNaughton P. A. (2006) Why pain gets worse: The mechanism of heat hyperalgesia. J. Gen. Physiol. 128, 491–493 10.1085/jgp.200609676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shuttleworth T. J. (2009) Arachidonic acid, ARC channels, and Orai proteins. Cell Calcium 45, 602–610 10.1016/j.ceca.2009.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bubolz A. H., Mendoza S. A., Zheng X., Zinkevich N. S., Li R., Gutterman D. D., and Zhang D. X. (2012) Activation of endothelial TRPV4 channels mediates flow-induced dilation in human coronary arterioles: Role of Ca2+ entry and mitochondrial ROS signaling. Am. J. Physiol. Heart Circ. Physiol. 302, H634–H642 10.1152/ajpheart.00717.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Holmström K. M., and Finkel T. (2014) Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol. 15, 411–421 10.1038/nrm3801 [DOI] [PubMed] [Google Scholar]
- 47. Sies H. (2014) Role of metabolic H2O2 generation: Redox signaling and oxidative stress. J. Biol. Chem. 289, 8735–8741 10.1074/jbc.R113.544635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Burgoyne J. R., Oka S., Ale-Agha N., and Eaton P. (2013) Hydrogen peroxide sensing and signaling by protein kinases in the cardiovascular system. Antioxid. Redox Signal. 18, 1042–1052 10.1089/ars.2012.4817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gauthier K. M., Deeter C., Krishna U. M., Reddy Y. K., Bondlela M., Falck J. R., and Campbell W. B. (2002) 14,15-Epoxyeicosa-5(Z)-enoic acid: A selective epoxyeicosatrienoic acid antagonist that inhibits endothelium-dependent hyperpolarization and relaxation in coronary arteries. Circ. Res. 90, 1028–1036 10.1161/01.RES.0000018162.87285.F8 [DOI] [PubMed] [Google Scholar]
- 50. Andreucci E., Aftimos S., Alcausin M., Haan E., Hunter W., Kannu P., Kerr B., McGillivray G., McKinlay Gardner R. J., Patricelli M. G., Sillence D., Thompson E., Zacharin M., Zankl A., Lamandé S. R., and Savarirayan R. (2011) TRPV4 related skeletal dysplasias: A phenotypic spectrum highlighted by clinical, radiographic, and molecular studies in 21 new families. Orphanet J. Rare Dis. 6, 37 10.1186/1750-1172-6-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L., and Schwede T. (2014) SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 10.1093/nar/gku340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. McGuffin L. J., Atkins J. D., Salehe B. R., Shuid A. N., and Roche D. B. (2015) IntFOLD: An integrated server for modelling protein structures and functions from amino acid sequences. Nucleic Acids Res. 43, W169–W173 10.1093/nar/gkv236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yang J., Yan R., Roy A., Xu D., Poisson J., and Zhang Y. (2015) The I-TASSER Suite: Protein structure and function prediction. Nat. Meth. 12, 7–8 10.1038/nmeth.3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kelley L. A., Mezulis S., Yates C. M., Wass M. N., and Sternberg M. J. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Källberg M., Margaryan G., Wang S., Ma J., and Xu J. (2014) RaptorX server: A resource for template-based protein structure modeling. Methods Mol. Biol. 1137, 17–27 10.1007/978-1-4939-0366-5_2 [DOI] [PubMed] [Google Scholar]
- 56. Kim D. E., Chivian D., and Baker D. (2004) Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 32, W526–W531 10.1093/nar/gkh468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Alvarez D. F., King J. A., Weber D., Addison E., Liedtke W., and Townsley M. I. (2006) Transient receptor potential vanilloid 4-mediated disruption of the alveolar septal barrier: A novel mechanism of acute lung injury. Circ. Res. 99, 988–995 10.1161/01.RES.0000247065.11756.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dalsgaard T., Sonkusare S. K., Teuscher C., Poynter M. E., and Nelson M. T. (2016) Pharmacological inhibitors of TRPV4 channels reduce cytokine production, restore endothelial function and increase survival in septic mice. Sci. Rep. 6, 33841 10.1038/srep33841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Teng J., Loukin S. H., Anishkin A., and Kung C. (2015) L596-W733 bond between the start of the S4-S5 linker and the TRP box stabilizes the closed state of TRPV4 channel. Proc. Natl. Acad. Sci. U.S.A. 112, 3386–3391 10.1073/pnas.1502366112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Teng J., Loukin S. H., Anishkin A., and Kung C. (2016) A competing hydrophobic tug on L596 to the membrane core unlatches S4-S5 linker elbow from TRP helix and allows TRPV4 channel to open. Proc. Natl. Acad. Sci. U.S.A. 113, 11847–11852 10.1073/pnas.1613523113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Strotmann R., Schultz G., and Plant T. D. (2003) Ca2+-dependent potentiation of the nonselective cation channel TRPV4 is mediated by a C-terminal calmodulin binding site. J. Biol. Chem. 278, 26541–26549 10.1074/jbc.M302590200 [DOI] [PubMed] [Google Scholar]
- 62. Strotmann R., Semtner M., Kepura F., Plant T. D., and Schöneberg T. (2010) Interdomain interactions control Ca2+-dependent potentiation in the cation channel TRPV4. PLoS One 5, e10580 10.1371/journal.pone.0010580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Loukin S. H., Teng J., and Kung C. (2015) A channelopathy mechanism revealed by direct calmodulin activation of TrpV4. Proc. Natl. Acad. Sci. U.S.A. 112, 9400–9405 10.1073/pnas.1510602112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. de Groot T., Kovalevskaya N. V., Verkaart S., Schilderink N., Felici M., van der Hagen E. A., Bindels R. J., Vuister G. W., and Hoenderop J. G. (2011) Molecular mechanisms of calmodulin action on TRPV5 and modulation by parathyroid hormone. Mol. Cell Biol. 31, 2845–2853 10.1128/MCB.01319-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Berna-Erro A., Izquierdo-Serra M., Sepúlveda R. V., Rubio-Moscardo F., Doñate-Macián P., Serra S. A., Carrillo-Garcia J., Perálvarez-Marín A., González-Nilo F., Fernández-Fernández J. M., and Valverde M. A. (2017) Structural determinants of 5′,6′-epoxyeicosatrienoic acid binding to and activation of TRPV4 channel. Sci. Rep. 7, 10522 10.1038/s41598-017-11274-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Earley S., Heppner T. J., Nelson M. T., and Brayden J. E. (2005) TRPV4 forms a novel Ca2+ signaling complex with ryanodine receptors and BKCa channels. Circ. Res. 97, 1270–1279 10.1161/01.RES.0000194321.60300.d6 [DOI] [PubMed] [Google Scholar]
- 67. Sonkusare S. K., Bonev A. D., Ledoux J., Liedtke W., Kotlikoff M. I., Heppner T. J., Hill-Eubanks D. C., and Nelson M. T. (2012) Elementary Ca2+ signals through endothelial TRPV4 channels regulate vascular function. Science 336, 597–601 10.1126/science.1216283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zuccolo E., Dragoni S., Poletto V., Catarsi P., Guido D., Rappa A., Reforgiato M., Lodola F., Lim D., Rosti V., Guerra G., and Moccia F. (2016) Arachidonic acid-evoked Ca2+ signals promote nitric oxide release and proliferation in human endothelial colony forming cells. Vascul. Pharmacol. 87, 159–171 10.1016/j.vph.2016.09.005 [DOI] [PubMed] [Google Scholar]
- 69. Fang J., Hodivala-Dilke K., Johnson B. D., Du L. M., Hynes R. O., White G. C. 2nd, and Wilcox D. A. (2005) Therapeutic expression of the platelet-specific integrin, αIIbβ3, in a murine model for Glanzmann thrombasthenia. Blood 106, 2671–2679 10.1182/blood-2004-12-4619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Grynkiewicz G., Poenie M., and Tsien R. Y. (1985) A new generation of Ca2+ indicators with greatly improved fluorescence properties. J. Biol. Chem. 260, 3440–3450 [PubMed] [Google Scholar]
- 71. Rykunov D., Steinberger E., Madrid-Aliste C. J., and Fiser A. (2009) Improved scoring function for comparative modeling using the M4T method. J. Struct. Funct. Genomics 10, 95–99 10.1007/s10969-008-9044-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Humphrey W., Dalke A., and Schulten K. (1996) VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33–38 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- 73. Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., and Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 10.1002/jcc.20289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Best R. B., Zhu X., Shim J., Lopes P. E., Mittal J., Feig M., and Mackerell A. D. Jr. (2012) Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 10.1021/ct300400x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Trott O., and Olson A. J. (2010) AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.