Abstract
Cyanobacteria are photosynthetic prokaryotes showing great promise as biocatalysts for the direct conversion of CO2 into fuels, chemicals, and other value-added products. Introduction of just a few heterologous genes can endow cyanobacteria with the ability to transform specific central metabolites into many end products. Recent engineering efforts have centered around harnessing the potential of these microbial biofactories for sustainable production of chemicals conventionally produced from fossil fuels. Here, we present an overview of the unique chemistry that cyanobacteria have been co-opted to perform. We highlight key lessons learned from these engineering efforts and discuss advantages and disadvantages of various approaches.
Keywords: biofuel, cyanobacteria, metabolic engineering, natural product, photosynthesis
Introduction
In 1998, Anastas and Warner published a list of 12 principles of green chemistry as guidelines to help researchers develop more sustainable chemical processes (1). Cyanobacteria provide a powerful platform for the development of green catalysts that utilize renewable feedstock in the form of atmospheric carbon dioxide (CO2) and convert it into fuels, commodity chemicals, and value-added products using (sun)light as the energy source. Cyanobacterial catalysts are expected to meet several of the green chemistry principles given the benign nature of the processes. The resultant carbon capture and utilization technologies have the potential to play an important role in mitigating the harmful effects of elevated CO2 levels if the technology advances to an industrial scale. Despite the potential, a number of technological challenges need to be overcome before cyanobacteria-based processes become commercially viable. In this Minireview, we present an overview of metabolic engineering of cyanobacteria and discuss some of the chemistry that these photosynthetic microbes have been engineered to perform. The studies reviewed herein are proof of concept for photosynthetic chemical production platforms, but industrial production systems have yet to be realized.
Cyanobacteria are ancient photosynthetic prokaryotes that are the progenitors of the higher plant chloroplast. They inhabit virtually any environment that contains water and can grow under diverse conditions (2). These organisms are the originators of photosynthesis and are responsible for generating the planet's original oxygen supply (3). Currently, cyanobacteria account for as much as for 25% of the planet's primary productivity and about 2/3 of the primary productivity in the open ocean (4, 5). Cyanobacteria use photosynthesis and the Calvin-Benson cycle (CBC)3 to generate biomass using only CO2 and sunlight as carbon and energy sources (Fig. 1). Manipulating the metabolism of these photosynthetic prokaryotes provides the opportunity for direct conversion of CO2 into commodity chemicals. This strategy may be advantageous over heterotrophic bioproduction platforms that require plant-derived fermentable sugars and compete with food production. Eukaryotic green algae have also been pursued for the production of lipid biofuels or biohydrogen (6, 7). Cyanobacteria offer distinct advantages over both plants and green algae. Cyanobacteria are more efficient at solar energy capture than plants, converting as much as 9% of the solar energy into biomass compared with only 0.5–3% for higher plants (8, 9). Additionally, cyanobacteria acquire their carbon through a bicarbonate intermediate, which presents a unique opportunity to supply carbon enrichment by the addition of bicarbonate derived from atmospheric CO2 or factory emissions (10). Cyanobacteria also grow faster than higher plants and maximize atom economy by not producing wasteful biomass such as roots and stems. This directs a higher amount of fixed carbon to desired products. Cyanobacteria are also readily genetically tractable, which provides ease of genetic manipulations to alter their metabolism. In contrast, the genetic complexity of eukaryotic algae has made metabolic engineering more challenging (11). Importantly, cyanobacteria can be cultivated in bioreactors in arid or otherwise unfarmable land, which minimizes the competition with food crops (12). However, these organisms, like plants or eukaryotic green algae, still require significant nitrogen and phosphorus inputs, which are limited and expensive resources that must be conserved (12, 13). Culturing cyanobacteria in waste or saltwater and/or using nitrogen-fixing strains could present a partial solution to this problem (14).
Figure 1.
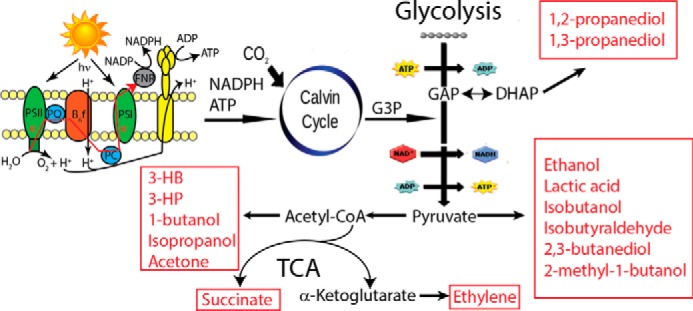
Overview of photosynthetic metabolism and production of green chemicals in cyanobacteria. Energy and reducing equivalents are generated by photosynthetic and respiratory complexes in the thylakoid membrane (cartoon top left). ATP, NADPH, and CO2 feed into the Calvin-Benson cycle and glycolysis. Target chemicals produced in cyanobacteria either through native metabolism or engineering are shown in the red boxes. G3P, 3-phosphoglycerate; PSI, photosystem I; PSII, photosystem II.
In 1999, Deng and Coleman (15) reported the first metabolic engineering of a cyanobacterium to produce ethanol. Subsequent studies over the last 2 decades have demonstrated heterologous expression of pathways for the production of compounds such as alcohols, diols, fatty acids, and organic acids (Table 1 and references cited therein). These works cover fuel as well as non-fuel commodity chemicals. In addition, cyanobacteria synthesize thousands of bioactive molecules (16, 17), which are synthesized from pathways that, for the most part, have not been targeted for production. Although the list of engineered chemicals is long, the majority is derived from a small number of central metabolites. Below, we discuss some of the production pathways that have been engineered in cyanobacteria and thereby hope to illustrate the variety of engineering strategies and an underlying set of metabolic “rules” for working with photosynthetic microbes.
Table 1.
List of chemicals produced by metabolic engineering of cyanobacteria
| Chemical | Highest titer | Refs. |
|---|---|---|
| Εthanol | 54 nmol OD730−1 liter−1 day−1 | 15 |
| Ιsobutanol | 450 mg liter−1 | 37, 43 |
| Isopropanol | 288 mg liter−1 | 25, 47 |
| 1,2-Propanediol | 150 mg liter−1 | 22 |
| 1,3-Propanediol | 1.22 g liter−1 | 25, 39 |
| 1-Butanol | 29.9 mg liter−1 | 21 |
| Free fatty acids | 130 mg liter−1 | 30, 49, 71, 76, 77 |
| Ιsoprene and isoprenoids | 1.26 g liter−1 | 63,78–80, 82–86 |
| Βiohydrogen | 54 mol/1017 cells | 57 |
| l-Lactate | 0.0178 mmol gDW−1 h−1 | 23, 42 |
| Succinate | 430 mg liter−1 | 56 |
| Ιsobutyraldehyde | 6230 μg liter−1 h−1 | 37 |
| Αcetone | 22.48 mg liter−1 | 50, 53 |
| Εthylene | 5650 μl liter−1 h−1 | 55 |
| Sugars and sugar alcohols | 35.5 mg liter−1 h−1 | 31, 59, 64 |
| Glycerol | 7733 μg liter−1 h−1 | 25, 45 |
| 3-Hydroxypropionic acid | 837.18 mg liter−1 | 52, 54 |
| 3-Hydroxybutyrate | 533.4 mg liter−1 | 48 |
| Αlka/enes | 1200 μg gDW−1 | 51, 67, 74 |
| Νatural products | 5 mg liter−1 | 79, 99, 100 |
| Phenylpropanoids | 7.2 mg liter−1 | 98 |
General comments on metabolic engineering of cyanobacteria
Metabolic engineering of cyanobacteria presents several unique challenges posed by their photoautotrophic lifestyle. Among these are the following: (i) carefully partitioning the flux of CO2-derived carbon between biomass and chemical production (18, 19); (ii) the high level of O2 produced in photosynthesis will inhibit O2-sensitive enzymes and reactions (20); (iii) photosynthesis produces NADPH rather than NADH, which can make NADH-dependent reactions rate-limiting (21–24); and (iv) there can be radically different metabolic behavior of the production host in the dark versus light conditions (25, 26). Furthermore, some core metabolic pathways in cyanobacteria behave differently than in heterotrophic organisms or are missing some enzymatic steps. For instance, cyanobacteria do not have a traditional TCA cycle and are lacking α-ketoglutarate dehydrogenase. As a consequence, in cyanobacteria the TCA cycle functions as a bifurcated pathway for production of biomass precursors rather than a complete cycle (27, 28). Redox balance in cyanobacteria is also a key consideration, as photosynthesis can generate an overabundance of reducing equivalents in the absence of sufficient catabolic processes, leading to stunted growth (29). Finally, it is worth considering that some cyanobacterial strains may be better equipped to produce certain types of metabolites due to differences in intracellular metabolite pools or cell physiology (23, 29–31).
Metabolic flux analysis of cyanobacteria
A number of studies report intracellular reaction rate analysis of model strains of cyanobacteria either with constraint-based modeling such as flux balance analysis (32) or isotopic 13C metabolic flux analysis (Fig. 2) (33–35). Notably, glycolysis, pentose phosphate pathway, and the TCA cycle are far less active in cyanobacteria during photoautotrophic growth compared with those in model heterotrophs (28). In non-stationary isotopic 13C-labeling experiments, the intermediates of the CBC and gluconeogenesis pathway show rapid accumulation of 13C with no detectable label accumulation in the TCA cycle intermediates, suggesting a slow turnover of these metabolites. However, some of the recent studies demonstrate plasticity in cyanobacterial metabolism resulting in a significantly higher flux through TCA cycle in engineered cyanobacteria (36). This flexibility in metabolism may be the key to success of the ongoing metabolic engineering efforts.
Figure 2.
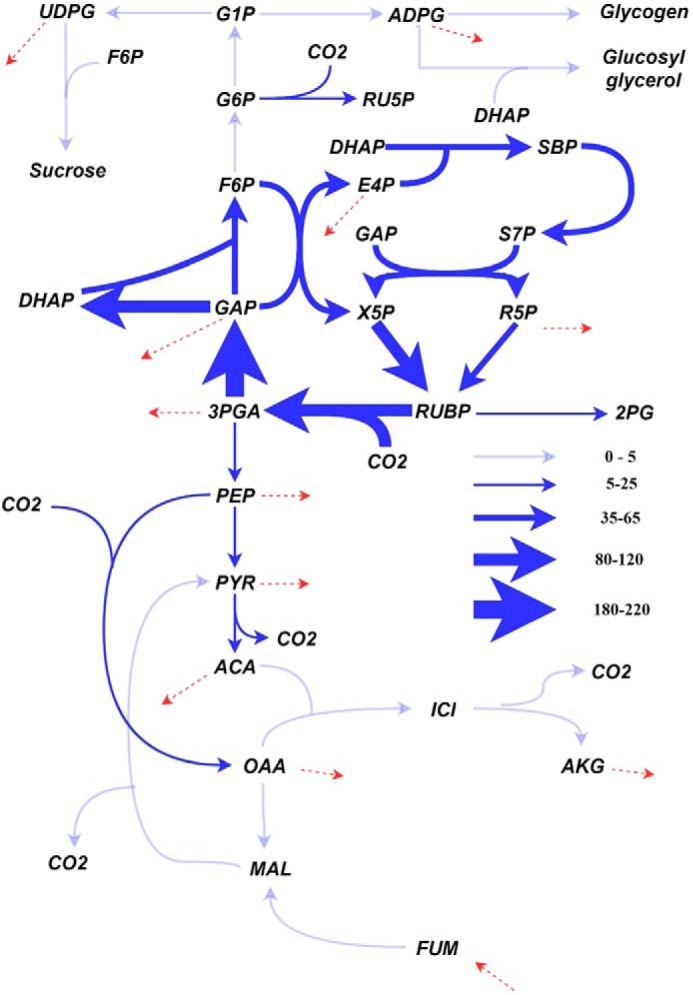
Generalized flux map for cyanobacterial photoautotrophic metabolism. The arrow thicknesses are proportional to the flux through the reactions. The flux values shown here are normalized to a CO2 uptake rate of 100 mmol/gDW/h and are averages of two studies involving 13C metabolic flux analysis performed on Synechocystis sp. PCC 6803 (33) and Synechococcus sp. PCC 7002 (35). The dotted arrows indicate drawdown of carbon for biomass synthesis. 2PG, 2-phosphoglycerate; 3PGA, 3-phosphoglycerate; ACA, acetyl-CoA; ADPG, ADP-glucose; AKG, α-ketoglutarate; E4P, erythrose-4-phosphate; F6P, fructose-6-phosphate; FUM, fumarate; G1P, glucose-1-phosphate; G6P, glucose-6-phosphate; ICI, isocitrate; MAL, malate; OAA, oxaloacetate; PEP, phosphoenolpyruvate; PYR, pyruvate; R5P, ribose-5-phosphate; RU5P, ribulose-5-phosphate; RUBP, ribulose-1,5 bisphosphate; S7P, sedoheptulose-7-phosphate; SBP, sedoheptulose-1,7-bisphosphate; UDPG, UDP-glucose; X5P, xylulose-5-phosphate.
Bioproduction strategies in cyanobacteria from CO2
Pyruvate- and DHAP-derived products
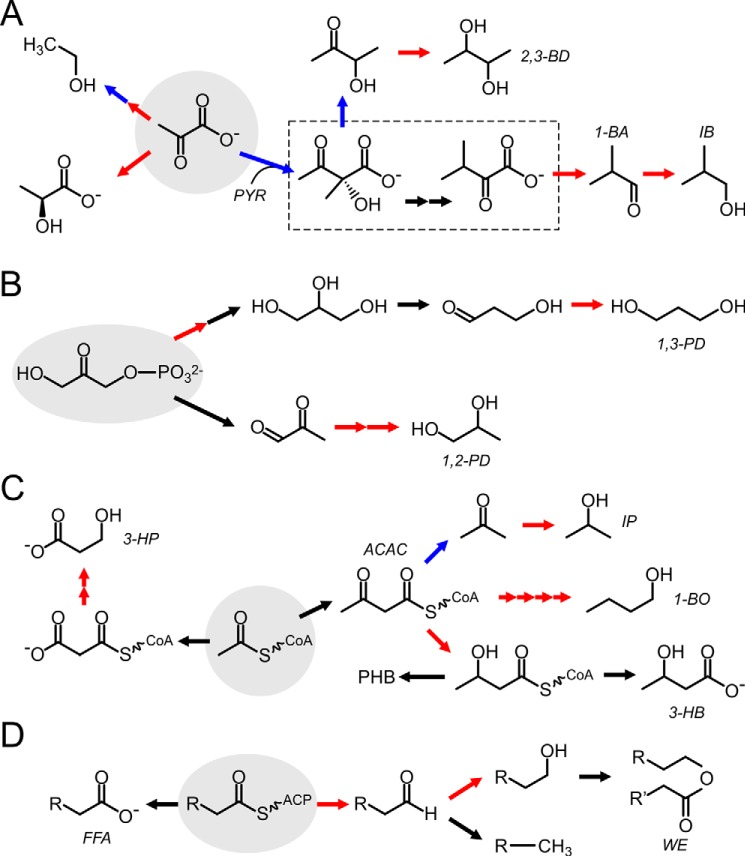
Pyruvate and DHAP are positioned in close proximity to the critical carbon-fixation reactions and are generated from glyceraldehyde 3-phosphate (GAP) via one or five chemical steps, respectively. In general, titers for chemicals produced from pyruvate or DHAP are among the highest reported for alcohols in cyanobacteria (15, 37–40). Pyruvate-derived production pathways are shown in Fig. 3A. In one of the first cyanobacterial metabolic engineering projects, ethanol was produced by expressing pyruvate decarboxylase and aldehyde dehydrogenase (15). More recent projects have applied systems-level analysis to study the whole-cell metabolic effects of ethanol production (41). l-Lactic acid has been produced by integrating an NADH-dependent dehydrogenase and transporters (23, 42). Yields were further improved by introducing a trans-hydrogenase to convert NADPH to NADH (23). The presence of the trans-hydrogenase caused a growth defect in cells unless the NADH-consuming dehydrogenase was also present to balance redox levels (23). Bioproduction of 1-butyraldehyde (37), isobutanol (37, 43), 2-methyl-1-butanol (40), and 2,3-butanediol (2,3-BD) (38, 44) from pyruvate was engineered by wholly or partly co-opting the parallel branched-chain amino acid (BCAA) pathways for valine and isoleucine. The first step in biosynthesis is an irreversible decarboxylative condensation that provides a strong thermodynamic driving force for the production pathways (21, 37). The alcohols and aldehydes are generated from BCAA intermediates by further decarboxylation and reduction. The production of 2,3-BD follows from decarboxylation of 2-acetolactate to acetoin and reduction to 2,3-BD (38). A common feature of each of these engineered pathways is the presence of at least one decarboxylation step and reduction using NADPH as a cofactor. The high yield observed for these pyruvate-derived products may be attributable to the following: (i) the high concentration of substrate (pyruvate) available during active photosynthesis; (ii) the presence of an early irreversible decarboxylation step in some of the pathways; and (iii) utilization of NADPH as a redox cofactor, which is abundantly available under photoautotrophic growth.
Figure 3.
Representative engineered and native production pathways for chemicals in cyanobacteria. Panels show bioproduction pathways derived from pyruvate (A), dihydroxyacetone phosphate (B), acetyl-CoA (C), and fatty acyl-ACPs (D). Red arrows indicate NAD(P)H-dependent oxidation or reduction steps. Blue arrows indicate decarboxylation steps. Black arrows are other types of enzymatic steps. A, branched-chain amino acid pathway is boxed. Starting metabolites are in gray circles. ACAC, acetoacetate; 1-BA, 1-butyraldehyde; 1-BO, 1-butanol; IB, isobutanol; IP, isopropanol; PYR, pyruvate; WE, wax ester.
1,2-Propanediol and 1,3-propanediol have been produced from DHAP (Fig. 3B) (22, 25). 1,3-PD is produced from DHAP via glycerol, which is generated as a side product in considerable quantities (25). Increasing expression of the bottleneck enzymes that convert glycerol to 1,3-PD resulted in a 4-fold increase in 1,3-PD and a concomitant decrease in glycerol (39). Glycerol production has been engineered as a useful 3-carbon precursor for a variety of chemicals (45). 1,2-PD was produced via methylglyoxal and acetol in two reductive steps (22). Swapping of NADH-dependent enzymes for those that utilize NADPH increased product titers of 1,2-PD nearly 10-fold (22), providing another example of the importance of matching cofactors to the production host.
Acetyl-CoA–derived products
Overall, product titers from acetyl-CoA–derived metabolites are lower than those derived from pyruvate, likely due to the low carbon flux to the TCA cycle and acetyl-CoA during light periods (Fig. 2) (27). It has been suggested that under light conditions cyanobacteria primarily utilize these pathways to generate carbon precursors for cellular components, as photosynthesis can generate sufficient reducing equivalents and energy (27). Thus, a key limitation is driving sufficient carbon to acetyl-CoA. One method is to activate glycolysis by inducing dark fermentation of the stored glycogen reserves generated during light periods (46). Alternatively, acetyl-CoA production can be triggered by nitrogen starvation of the cells (29, 47), although this can result in lower overall product yields due to decreased cell growth. Meaningful production levels of some chemicals were achieved only when cells were exposed to dark periods and nutrient starvation to increase the available acetyl-CoA pool (20, 29, 46–48). Without intervention, dark fermentation can result in the wasteful excretion of acetate and other compounds by cyanobacteria (26). In some cases, deletion of enzymes catalyzing the conversion of acetyl-CoA to acetate or storage polyhydroxybutyrates (PHB) has led to increased production of target chemicals (29, 49–52). Alternatively, Anfelt et al. (29) introduced a shunt by expressing phosphoketolase and phosphate acetyltransferase to directly convert CBC intermediates to acetyl-CoA.
A common theme in the bioproduction of acetyl-CoA is to convert it to acetoacetyl-CoA, which is then converted to 4-carbon products by reduction or 3-carbon products by decarboxylation and reduction (Fig. 3C). Acetone is produced from acetoacetate by decarboxylation (50, 53). Isopropanol can then be produced from acetone by reduction. The highest yields of isopropanol required culturing under light conditions followed by dark incubation to ferment the carbon stores generated during the light period into product (25, 47). 1-Butanol has been produced by introduction of an NADH-dependent fermentative Clostridium pathway under anoxic or under nitrogen-starved conditions (20, 29). Lan and Liao (21) improved 1-butanol production by introducing an ATP-dependent irreversible step to drive formation of acetoacetyl-CoA and by using NADPH-dependent enzymes in the subsequent steps. This strategy of introducing an irreversible step was also utilized by Chwa et al. (53) to increase acetone production from acetoacetyl-CoA. 3-Hydroxybutyrate (3-HB) production was engineered by increasing flux to 3-HB via acetoacetyl-CoA, expressing thioesterase, and by deleting the enzyme that catalyzes the polymerization of 3-HBs to the storage molecule PHB (48). This led to excretion of 3-HB from the cells. The product titers improved when the culture was subjected to phosphate starvation (48). 3-Hydroxypropionic acid (3-HP) has been produced from malonyl-CoA via two successive reductions with malonate semialdehyde as an intermediate (54). The authors also introduced a parallel pathway to 3-HP via β-alanine to avoid the feedback-regulated synthesis of malonyl-CoA from acetyl-CoA (54). 3-HP titers have been improved by increasing flux to malonyl-CoA production and eliminating competing pathways (52).
TCA cycle–derived products
The TCA cycle in cyanobacteria is under-utilized compared with that in heterotrophic organisms but is activated in response to certain growth conditions (27). Succinate and ethylene have been produced by intervening at the level of the TCA cycle (55, 56). Succinate can be produced during dark fermentation in some cyanobacterial strains (26, 57). Increased succinate production via the oxidative TCA cycle branch was engineered by introducing α-ketoglutarate decarboxylase and succinic semialdehyde dehydrogenase (56). Product titers were improved nearly 4-fold by increasing carbon flux by overexpressing phosphoenolpyruvate carboxylase and citrate synthase (56). Succinate was secreted from the cells into the medium. Ethylene is the most widely produced chemical feedstock on the planet. Inclusion of a single gene, ethylene-forming enzyme, converts α-ketoglutarate into ethylene, which is then collected directly from the culture headspace (55). A challenge of working in the TCA cycle is that α-ketoglutarate and other intermediates serve as carbon skeletons for amino acids and other cellular components. Thus, the engineered catabolic processes must be balanced by regeneration of the substrate to sustain biomass production (27, 56).
Glycogen and sugar products
During light periods, cyanobacteria store excess carbon and energy in the form of intracellular glycogen granules (58). Typically, between 5 and 15% of the fixed carbon is stored via this pathway with the glycogen content reaching 50% under certain growth conditions (31, 59). The diurnal lifestyle then mandates that the stored chemical energy is utilized during dark periods similar to that during heterotrophic growth. Additionally, freshwater cyanobacteria naturally accumulate intracellular sucrose as an osmoregulator when exposed to salt stress (60). This property has been utilized to induce sugar excretion by integrating transporters and exposing cells to high-salt conditions (31, 42, 61). This results in considerable accumulation of sugars in the growth medium. It may be possible to use a cyanobacterial co-culture to produce sugar feedstock for fermentative bioproduction of commodity chemicals by heterotrophs (61).
Interestingly, glycogen knockout mutants grow well under laboratory conditions even under diurnal growth conditions. Thus, a glycogen knockout strategy has been utilized to divert a greater fraction of the fixed carbon to products of interest. To exemplify this approach, the sugar alcohol mannitol has been produced from fructose 6-phosphate by introduction of mannitol dehydrogenase and phosphatase and by knocking out glycogen synthesis (62). Deleting glycogen synthesis is an attractive strategy as it can force cells to utilize other carbon sinks–such as engineered production pathways–to balance carbon fixation and consumption processes (43, 62, 63). Indeed, carbon and redox equivalent production by the CBC and photosynthesis may actually exceed catabolic processes and thereby act as a bottleneck to production. Integration of carbon-consuming production pathways has, in some cases, been shown to increase cell growth rate and photosynthetic activity, possibly by alleviating feedback photosynthetic inhibition (61, 64). Although promising, this strategy requires careful carbon partitioning to maintain cell growth and to minimize accumulation of undesirable fermentative metabolites (63). It remains to be seen whether a similar knockout of other carbon storage pathways or non-essential genes (58) would lead to greater flux toward the desired products.
Fatty acyl-ACP–derived products
Fatty acyl-ACP molecules are produced via the fatty-acid synthase (FAS) pathway in cyanobacteria using acetyl-CoA and malonyl-CoA as building blocks (65). Unlike eukaryotic microalgae, cyanobacteria do not synthesize triacylglycerols as a carbon storage (6). However, cyanobacteria naturally produce alkanes and alkenes (C15–C19) from acyl-ACPs (66–68). Production of several fatty acyl-ACP–derived products has been engineered in cyanobacteria, including free fatty acids (FFAs) (30, 49, 69–71), fatty alcohols and aldehydes (72, 73), hydrocarbons (51, 74), and wax esters and triacylglycerols (Fig. 3D) (73). The FAS pathway is tightly feedback-regulated, and overcoming this bottleneck is one of the challenges for increasing production yields (75). Engineering generally relies on first releasing the fatty acyl-ACP from the carrier protein either by hydrolysis to FFAs or reduction to aldehydes. FFAs are produced by introducing acyl-ACP thioesterase(s) to cleave fatty acids from ACP and deleting enzymes that recycle FFAs back to acyl-ACPs (49, 69, 76). This strategy relieves feedback inhibition of FAS and results in excretion of FFAs into the growth medium (49, 76). FFA excretion has also been accomplished by expressing lipases to cleave FFAs from membrane-bound diacylglycerol lipids (77), although this method does result in cell lysis. Alternatively, acyl-ACP reductase reduces ACP-linked fatty acids to the corresponding aldehydes (67, 73), a versatile substrate that can be converted to hydrocarbons by aldehyde-deformylating oxygenase (51), fatty alcohols by reduction (72, 73), or recycled to FFAs by oxidation (73). Alkane overproduction has been explored by overexpressing acyl-ACP reductase, aldehyde deformylating oxygenase, and increasing availability of acyl-ACP substrate by overexpressing acyl-ACP synthase and FAS complex enzymes (51). Changing culture conditions by increasing light or nutrient starvation has also been shown to increase alka/ene production in certain strains (74). Current production of FFAs and related compounds may be limited more by the negative physiological effects that overproduction has on the host rather than metabolism (76, 77). Overexpression damages the thylakoid and plasma membranes, leading to compromised photosynthesis, increased cell permeability, and sensitivity to mechanical shock (49, 76, 77). The use of alternative cyanobacterial production hosts where toxicity is reduced offers one means to overcome the current limits (30).
Fuel-like isoprenoid products
Cyanobacteria encode the methylerythritol phosphate (MEP) pathway to synthesize isoprenoid compounds. The MEP pathway is initiated from pyruvate and GAP. Cyanobacteria use this pathway to generate precursors for carotenoids, phytol, sterols, and other pigments (78). Because of the low natural carbon flux into this pathway in cyanobacteria and inherent regulation (79–81), it has thus far been challenging to generate high yields of isoprenoid molecules. In some strains, flux through the MEP pathway can be increased by raising light intensity (79). C10–C20 isoprenoids are attractive as “green” jet fuels due to their chemical similarity to petroleum-derived fuels. A variety of C5–C30 isoprenoids has now been produced, most often by integration of a single enzyme. Cyanobacteria do not naturally encode an isoprene synthase, and integration of this gene from plants results in heterologous formation of isoprene that evaporates from the growth medium (78). Gao et al. (80) used in silico metabolic modeling to simulate flux and optimize carbon flow through the complete MEP pathway, leading to significantly increased production of isoprene. Engineering of the whole pathway represents a method to overcome the inherent regulation of native pathways that may limit product yields. Production of larger isoprenoid compounds has been accomplished by introducing terpene synthases from plant or tree species to redirect isoprenoid intermediates to limonene, β-phellandrene, caryophyllene, bisabolene, farnesene, and squalene (63, 82–86).
High-value natural products
Both plants and cyanobacteria synthesize thousands of diverse secondary metabolites, or “natural products” (NPs) (16, 17, 87, 88). Many NPs have potent bioactivities that are valuable to medicine and agriculture (17, 89). However, synthetic biology and metabolic engineering have only rarely been applied to NP pathways, due to the large size of the biosynthetic gene clusters and the complex enzymatic transformations that are involved (16, 90, 91). Fortunately, new tools should now allow large gene clusters to be more easily cloned (92). Because of their metabolic similarity to plants and capacity for production of NPs, cyanobacteria are attractive production platforms for high-value NPs, including isoprenoids and those synthesized by non-ribosomal peptide synthetases (NRPS) and polyketide synthases (PKS). In contrast, heterotrophic expression of NRPS, polyketide synthase, or cytochrome P450s from plants or cyanobacteria is challenging due to genetic and biochemical incompatibilities (93–96). Furthermore, the biosynthesis of plant terpenoids often relies on cytochrome P450 monooxygenases that use NADPH, a cofactor that is in abundance in cyanobacteria but not in heterotrophs (97). Successful expression of a membrane-bound plant cytochrome P450 in a cyanobacterium led to the production of caffeic acid from p-coumarate (98). Protein engineering has also been used to channel photosynthesis-derived electrons from cytochrome P450s to produce plant NPs (97, 99). Englund et al. (79) reported integrating a partial plant pathway to produce manoyl oxide, a precursor to the diterpenoid forskolin. Production was accomplished by integrating two stereospecific diterpene cyclases from Coleus (79). Production yields were improved by overexpressing a heterologous geranyl pyrophosphate synthase and the first enzyme in the MEP pathway (79). Videau et al. (100) engineered lyngbyatoxin production by cloning and transferring the biosynthetic NRPS genes from a slow-growing marine cyanobacterium to a laboratory strain. This study illustrates that model strains can serve as heterologous production hosts for marine cyanobacterial NPs.
Concluding comments
Significant progress has been made in the last 2 decades toward metabolic engineering of cyanobacteria. These advances are due, in part, to improvements in synthetic biology tools and our increased understanding of the underlying cyanobacterial metabolism. However, although many successful proof of concept studies have been carried out, little work is currently being performed to commercialize the technology. The determinants of commercial success lie in the productivity, titer, and stability that can ultimately be attained by engineered strains in an industrial setting. In this quest, there is a need to experimentally determine the theoretical limit of the production rates. Much more work is needed in the area of metabolic engineering to direct a larger portion of the fixed carbon into the desired end products. Most of the proof of concept studies were performed in slow-growing laboratory strains that may not be suitable for outdoor cultivation. Thus, identification and development of robust, fast-growing strains that grow well in high light and temperature and in salt or wastewater are critical. Finally, there has been little work performed on scaling up laboratory systems to production scale. The biology has come a long way, but the engineering efforts that are required for industrialization have not received enough interest. Low-cost bioreactors or other systems such as open ponds need to be improved, and technology to harvest the end products needs to be developed. As cyanobacterial productivity improves, investments should aim to acquire the technology and infrastructure to scale up production. Although we may still be years away from commercial cyanobacterial cell factories, the great potential of these organisms as sustainable green production systems should attract continued interest from metabolic engineers for years to come.
This work was supported by Indo-U.S. Science and Technology Forum for Indo-U.S. Advanced Bioenergy Consortium (IUABC) (to H. B. P. and P. P. W.), Washington University (to H. B. P.), and the Office of Science, Department of Energy–Biological and Environmental Research (to H. B. P.). This is the fourth article in the Thematic Minireview series “Green biological chemistry.” The authors declare that they have no conflicts of interest with the contents of this article.
- CBC
- Calvin-Benson cycle
- TCA
- tricarboxylic acid
- DHAP
- dihydroxyacetone phosphate
- GAP
- glyceraldehyde 3-phosphate
- 2,3-BD
- 2,3-butanediol
- 1,2-PD
- 1,2-propanediol
- 3-HP
- 3-hydroxypropionate
- 3-HB
- 3-hydroxybutyrate
- PHB
- polyhydroxybutyrate
- ACP
- acyl-carrier protein
- MEP
- methylerythritol phosphate
- NP
- natural product
- BCAA
- branched-chain amino acid
- FAS
- fatty-acid synthase
- FFA
- free fatty acid
- NRPS
- non-ribosomal peptide synthetase
- gDW
- grams dry cell weight.
References
- 1. Anastas P. T., and Warner J. C. (1998) Green Chemistry: Theory and Practice, Oxford University Press, Oxford, UK [Google Scholar]
- 2. Whitton B. A., and Potts M. (2002) in The Ecology of Cyanobacteria: Their Diversity in Time and Space (Whitton B. A., and Potts M., eds) pp. 1–11, Springer, Dordrecht, the Netherlands [Google Scholar]
- 3. Hamilton T. L., Bryant D. A., and Macalady J. L. (2016) The role of biology in planetary evolution: cyanobacterial primary production in low-oxygen proterozoic oceans. Environ. Microbiol. 18, 325–340 10.1111/1462-2920.13118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Flombaum P., Gallegos J. L., Gordillo R. A., Rincón J., Zabala L. L., Jiao N., Karl D. M., Li W. K., Lomas M. W., Veneziano D., Vera C. S., Vrugt J. A., and Martiny A. C. (2013) Present and future global distributions of the marine cyanobacteria Prochlorococcus and Synechococcus. Proc. Natl. Acad. Sci. U.S.A. 110, 9824–9829 10.1073/pnas.1307701110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bullerjahn G. S., and Post A. F. (2014) Physiology and molecular biology of aquatic cyanobacteria. Front. Microbiol. 5, 359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hu Q., Sommerfeld M., Jarvis E., Ghirardi M., Posewitz M., Seibert M., and Darzins A. (2008) Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J. 54, 621–639 10.1111/j.1365-313X.2008.03492.x [DOI] [PubMed] [Google Scholar]
- 7. Hankamer B., Lehr F., Rupprecht J., Mussgnug J. H., Posten C., and Kruse O. (2007) Photosynthetic biomass and H2 production by green algae: from bioengineering to bioreactor scale-up. Physiol. Plant 131, 10–21 10.1111/j.1399-3054.2007.00924.x [DOI] [PubMed] [Google Scholar]
- 8. Dismukes G. C., Carrieri D., Bennette N., Ananyev G. M., and Posewitz M. C. (2008) Aquatic phototrophs: efficient alternatives to land-based crops for biofuels. Curr. Opin. Biotechnol. 19, 235–240 10.1016/j.copbio.2008.05.007 [DOI] [PubMed] [Google Scholar]
- 9. Branco Dos Santos F., Du W., and Hellingwerf K. J. (2014) Synechocystis: not just a plug-bug for CO2, but a green E. coli. Front. Bioeng. Biotechnol. 2, 36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chi Z., O'Fallon J. V., and Chen S. (2011) Bicarbonate produced from carbon capture for algae culture. Trends Biotechnol. 29, 537–541 10.1016/j.tibtech.2011.06.006 [DOI] [PubMed] [Google Scholar]
- 11. Gimpel J. A., Henríquez V., and Mayfield S. P. (2015) In metabolic engineering of eukaryotic microalgae: potential and challenges come with great diversity. Front. Microbiol. 6, 1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pate R., Klise G., and Wu B. (2011) Resource demand implications for US algae biofuels production scale-up. Appl. Energy 88, 3377–3388 10.1016/j.apenergy.2011.04.023 [DOI] [Google Scholar]
- 13. Sharma N. K., and Stal L. J. (2014) Cyanobacteria, pp. 167–180 John Wiley & Sons, Ltd., Chichester, UK [Google Scholar]
- 14. Markou G., and Georgakakis D. (2011) Cultivation of filamentous cyanobacteria (blue-green algae) in agro-industrial wastes and wastewaters: a review. Applied Energy 88, 3389–3401 10.1016./j.apenergy.2010.12.042 [DOI] [Google Scholar]
- 15. Deng M.-D., and Coleman J. R. (1999) Ethanol synthesis by genetic engineering in cyanobacteria. Appl. Environ. Microbiol. 65, 523–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dittmann E., Gugger M., Sivonen K., and Fewer D. P. (2015) Natural product biosynthetic diversity and comparative genomics of the cyanobacteria. Trends Microbiol. 23, 642–652 10.1016/j.tim.2015.07.008 [DOI] [PubMed] [Google Scholar]
- 17. Burja A. M., Banaigs B., Abou-Mansour E., Grant Burgess J., and Wright P. C. (2001) Marine cyanobacteria—a prolific source of natural products. Tetrahedron 57, 9347–9377 10.1016/S0040-4020(01)00931-0 [DOI] [Google Scholar]
- 18. Stephanopoulos G. (1999) Metabolic fluxes and metabolic engineering. Metab. Eng. 1, 1–11 10.1006/mben.1998.0101 [DOI] [PubMed] [Google Scholar]
- 19. Melis A. (2013) Carbon partitioning in photosynthesis. Curr. Opin. Chem. Biol. 17, 453–456 10.1016/j.cbpa.2013.03.010 [DOI] [PubMed] [Google Scholar]
- 20. Lan E. I., and Liao J. C. (2011) Metabolic engineering of cyanobacteria for 1-butanol production from carbon dioxide. Metab. Eng. 13, 353–363 10.1016/j.ymben.2011.04.004 [DOI] [PubMed] [Google Scholar]
- 21. Lan E. I., and Liao J. C. (2012) ATP drives direct photosynthetic production of 1-butanol in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 109, 6018–6023 10.1073/pnas.1200074109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li H., and Liao J. C. (2013) Engineering a cyanobacterium as the catalyst for the photosynthetic conversion of CO2 to 1,2-propanediol. Microb. Cell Fact. 12, 4 10.1186/1475-2859-12-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Angermayr S. A., Paszota M., and Hellingwerf K. J. (2012) Engineering a cyanobacterial cell factory for production of lactic acid. Appl. Environ. Microbiol. 78, 7098–7106 10.1128/AEM.01587-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lassen L. M., Nielsen A. Z., Ziersen B., Gnanasekaran T., Møller B. L., and Jensen P. E. (2014) Redirecting photosynthetic electron flow into light-driven synthesis of alternative products including high-value bioactive natural compounds. ACS Synth. Biol. 3, 1–12 10.1021/sb400136f [DOI] [PubMed] [Google Scholar]
- 25. Hirokawa Y., Maki Y., Tatsuke T., and Hanai T. (2016) Cyanobacterial production of 1,3-propanediol directly from carbon dioxide using a synthetic metabolic pathway. Metab. Eng. 34, 97–103 10.1016/j.ymben.2015.12.008 [DOI] [PubMed] [Google Scholar]
- 26. Stal L. J., and Moezelaar R. (1997) Fermentation in cyanobacteria. FEMS Microbiol. Rev. 21, 179–211 10.1111/j.1574-6976.1997.tb00350.x [DOI] [Google Scholar]
- 27. Zhang S., and Bryant D. A. (2011) The tricarboxylic acid cycle in cyanobacteria. Science 334, 1551–1553 10.1126/science.1210858 [DOI] [PubMed] [Google Scholar]
- 28. Steinhauser D., Fernie A. R., and Araújo W. L. (2012) Unusual cyanobacterial TCA cycles: not broken just different. Trends Plant Sci. 17, 503–509 10.1016/j.tplants.2012.05.005 [DOI] [PubMed] [Google Scholar]
- 29. Anfelt J., Kaczmarzyk D., Shabestary K., Renberg B., Rockberg J., Nielsen J., Uhlén M., and Hudson E. P. (2015) Genetic and nutrient modulation of acetyl-CoA levels in Synechocystis for n-butanol production. Microb. Cell Fact. 14, 167 10.1186/s12934-015-0355-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ruffing A. M. (2014) Improved free fatty acid production in cyanobacteria with Synechococcus sp. PCC 7002 as host. Front. Bioeng. Biotechnol. 2, 17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Song K., Tan X., Liang Y., and Lu X. (2016) The potential of Synechococcus elongatus UTEX 2973 for sugar feedstock production. Appl. Microbiol. Biotechnol. 100, 7865–7875 10.1007/s00253-016-7510-z [DOI] [PubMed] [Google Scholar]
- 32. Hendry J. I., Prasannan C. B., Joshi A., Dasgupta S., and Wangikar P. P. (2016) Metabolic model of Synechococcus sp. PCC 7002: Prediction of flux distribution and network modification for enhanced biofuel production. Bioresour. Technol. 213, 190–197 10.1016/j.biortech.2016.02.128 [DOI] [PubMed] [Google Scholar]
- 33. Young J. D., Shastri A. A., Stephanopoulos G., and Morgan J. A. (2011) Mapping photoautotrophic metabolism with isotopically nonstationary 13C flux analysis. Metab. Eng. 13, 656–665 10.1016/j.ymben.2011.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jazmin L. J., Xu Y., Cheah Y. E., Adebiyi A. O., Johnson C. H., and Young J. D. (2017) Isotopically nonstationary 13C flux analysis of cyanobacterial isobutyraldehyde production. Metab. Eng. 42, 9–18 10.1016/j.ymben.2017.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hendry J. I., Prasannan C., Ma F., Möllers K. B., Jaiswal D., Digmurti M., Allen D. K., Frigaard N.-U., Dasgupta S., and Wangikar P. P. (2017) Rerouting of carbon flux in a glycogen mutant of cyanobacteria assessed via isotopically non-stationary 13C metabolic flux analysis. Biotechnol. Bioeng. 114, 2298–2308 10.1002/bit.26350 [DOI] [PubMed] [Google Scholar]
- 36. Xiong W., Morgan J. A., Ungerer J., Wang B., Maness P.-C., and Yu J. (2015) The plasticity of cyanobacterial metabolism supports direct CO2 conversion to ethylene. Nat. Plants 1, 15053 10.1038/nplants.2015.53 [DOI] [Google Scholar]
- 37. Atsumi S., Higashide W., and Liao J. C. (2009) Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat. Biotechnol. 27, 1177–1180 10.1038/nbt.1586 [DOI] [PubMed] [Google Scholar]
- 38. Oliver J. W., Machado I. M., Yoneda H., and Atsumi S. (2013) Cyanobacterial conversion of carbon dioxide to 2,3-butanediol. Proc. Natl. Acad. Sci. U.S.A. 110, 1249–1254 10.1073/pnas.1213024110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hirokawa Y., Maki Y., and Hanai T. (2017) Improvement of 1,3-propanediol production using an engineered cyanobacterium, Synechococcus elongatus by optimization of the gene expression level of a synthetic metabolic pathway and production conditions. Metab. Eng. 39, 192–199 10.1016/j.ymben.2016.12.001 [DOI] [PubMed] [Google Scholar]
- 40. Shen C. R., and Liao J. C. (2012) Photosynthetic production of 2-methyl-1-butanol from CO2 in cyanobacterium Synechococcus elongatus PCC 7942 and characterization of the native acetohydroxyacid synthase. Energy Environ. Sci. 5, 9574–9583 10.1039/c2ee23148d [DOI] [Google Scholar]
- 41. Kopka J., Schmidt S., Dethloff F., Pade N., Berendt S., Schottkowski M., Martin N., Dühring U., Kuchmina E., Enke H., Kramer D., Wilde A., Hagemann M., and Friedrich A. (2017) Systems analysis of ethanol production in the genetically engineered cyanobacterium Synechococcus sp. PCC 7002. Biotechnol. Biofuels 10, 56 10.1186/s13068-017-0741-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Niederholtmeyer H., Wolfstädter B. T., Savage D. F., Silver P. A., and Way J. C. (2010) Engineering cyanobacteria to synthesize and export hydrophilic products. Appl. Environ. Microbiol. 76, 3462–3466 10.1128/AEM.00202-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li X., Shen C. R., and Liao J. C. (2014) Isobutanol production as an alternative metabolic sink to rescue the growth deficiency of the glycogen mutant of Synechococcus elongatus PCC 7942. Photosynth. Res. 120, 301–310 10.1007/s11120-014-9987-6 [DOI] [PubMed] [Google Scholar]
- 44. Oliver J. W., and Atsumi S. (2015) A carbon sink pathway increases carbon productivity in cyanobacteria. Metab. Eng. 29, 106–112 10.1016/j.ymben.2015.03.006 [DOI] [PubMed] [Google Scholar]
- 45. Wang Y., Tao F., Ni J., Li C., and Xu P. (2015) Production of C3 platform chemicals from CO2 by genetically engineered cyanobacteria. Green Chem. 17, 3100–3110 10.1039/C5GC00129C [DOI] [Google Scholar]
- 46. Hirokawa Y., Suzuki I., and Hanai T. (2015) Optimization of isopropanol production by engineered cyanobacteria with a synthetic metabolic pathway. J. Biosci. Bioeng. 119, 585–590 10.1016/j.jbiosc.2014.10.005 [DOI] [PubMed] [Google Scholar]
- 47. Kusakabe T., Tatsuke T., Tsuruno K., Hirokawa Y., Atsumi S., Liao J. C., and Hanai T. (2013) Engineering a synthetic pathway in cyanobacteria for isopropanol production directly from carbon dioxide and light. Metab. Eng. 20, 101–108 10.1016/j.ymben.2013.09.007 [DOI] [PubMed] [Google Scholar]
- 48. Wang B., Pugh S., Nielsen D. R., Zhang W., and Meldrum D. R. (2013) Engineering cyanobacteria for photosynthetic production of 3-hydroxybutyrate directly from CO2. Metab. Eng. 16, 68–77 10.1016/j.ymben.2013.01.001 [DOI] [PubMed] [Google Scholar]
- 49. Liu X., Sheng J., and Curtiss R. 3rd (2011) Fatty acid production in genetically modified cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 108, 6899–6904 10.1073/pnas.1103014108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhou J., Zhang H., Zhang Y., Li Y., and Ma Y. (2012) Designing and creating a modularized synthetic pathway in cyanobacterium Synechocystis enables production of acetone from carbon dioxide. Metab. Eng. 14, 394–400 10.1016/j.ymben.2012.03.005 [DOI] [PubMed] [Google Scholar]
- 51. Wang W., Liu X., and Lu X. (2013) Engineering cyanobacteria to improve photosynthetic production of alka (e) nes. Biotechnol. Biofuels 6, 69 10.1186/1754-6834-6-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang Y., Sun T., Gao X., Shi M., Wu L., Chen L., and Zhang W. (2016) Biosynthesis of platform chemical 3-hydroxypropionic acid (3-HP) directly from CO2 in cyanobacterium Synechocystis sp. PCC 6803. Metab. Eng. 34, 60–70 10.1016/j.ymben.2015.10.008 [DOI] [PubMed] [Google Scholar]
- 53. Chwa J.-W., Kim W. J., Sim S. J., Um Y., and Woo H. M. (2016) Engineering of a modular and synthetic phosphoketolase pathway for photosynthetic production of acetone from CO2 in Synechococcus elongatus PCC 7942 under light and aerobic condition. Plant Biotechnol. J. 14, 1768–1776 10.1111/pbi.12536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lan E. I., Chuang D. S., Shen C. R., Lee A. M., Ro S. Y., and Liao J. C. (2015) Metabolic engineering of cyanobacteria for photosynthetic 3-hydroxypropionic acid production from CO2 using Synechococcus elongatus PCC 7942. Metab. Eng. 31, 163–170 10.1016/j.ymben.2015.08.002 [DOI] [PubMed] [Google Scholar]
- 55. Ungerer J., Tao L., Davis M., Ghirardi M., Maness P.-C., and Yu J. (2012) Sustained photosynthetic conversion of CO2 to ethylene in recombinant cyanobacterium Synechocystis 6803. Energy Environ. Sci. 5, 8998–9006 10.1039/c2ee22555g [DOI] [Google Scholar]
- 56. Lan E. I., and Wei C. T. (2016) Metabolic engineering of cyanobacteria for the photosynthetic production of succinate. Metab. Eng. 38, 483–493 10.1016/j.ymben.2016.10.014 [DOI] [PubMed] [Google Scholar]
- 57. McNeely K., Xu Y., Bennette N., Bryant D. A., and Dismukes G. C. (2010) Redirecting reductant flux into hydrogen production via metabolic engineering of fermentative carbon metabolism in a cyanobacterium. Appl. Environ. Microbiol. 76, 5032–5038 10.1128/AEM.00862-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tiruveedula G. S. S., and Wangikar P. P. (2017) Gene essentiality, conservation index and co-evolution of genes in cyanobacteria. PLoS ONE 12, e0178565 10.1371/journal.pone.0178565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Aikawa S., Nishida A., Ho S.-H., Chang J.-S., Hasunuma T., and Kondo A. (2014) Glycogen production for biofuels by the euryhaline cyanobacteria Synechococcus sp. strain PCC 7002 from an oceanic environment. Biotechnol. Biofuels 7, 88 10.1186/1754-6834-7-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Blumwald E., Mehlhorn R. J., and Packer L. (1983) Ionic osmoregulation during salt adaptation of the cyanobacterium Synechococcus 6311. Plant Physiol. 73, 377–380 10.1104/pp.73.2.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ducat D. C., and Silver P. A. (2012) Improving carbon fixation pathways. Curr. Opin. Chem. Biol. 16, 337–344 10.1016/j.cbpa.2012.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jacobsen J. H., and Frigaard N. U. (2014) Engineering of photosynthetic mannitol biosynthesis from CO2 in a cyanobacterium. Metab. Eng. 21, 60–70 10.1016/j.ymben.2013.11.004 [DOI] [PubMed] [Google Scholar]
- 63. Davies F. K., Work V. H., Beliaev A. S., and Posewitz M. C. (2014) Engineering limonene and bisabolene production in wild type and a glycogen-deficient mutant of Synechococcus sp. PCC 7002. Front. Bioeng. Biotechnol. 2, 21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ducat D. C., Avelar-Rivas J. A., Way J. C., and Silver P. A. (2012) Rerouting carbon flux to enhance photosynthetic productivity. Appl. Environ. Microbiol. 78, 2660–2668 10.1128/AEM.07901-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rock C. O., and Jackowski S. (2002) Forty years of bacterial fatty acid synthesis. Biochem. Biophys. Res. Commun. 292, 1155–1166 10.1006/bbrc.2001.2022 [DOI] [PubMed] [Google Scholar]
- 66. Winters K., Parker P. L., and van Baalen C. (1969) Hydrocarbons of blue-green algae: geochemical significance. Science 163, 467–468 10.1126/science.163.3866.467 [DOI] [PubMed] [Google Scholar]
- 67. Schirmer A., Rude M. A., Li X., Popova E., and del Cardayre S. B. (2010) Microbial biosynthesis of alkanes. Science 329, 559–562 10.1126/science.1187936 [DOI] [PubMed] [Google Scholar]
- 68. Mendez-Perez D., Begemann M. B., and Pfleger B. F. (2011) Modular synthase-encoding gene involved in α-olefin biosynthesis in Synechococcus sp. strain PCC 7002. Appl. Environ. Microbiol. 77, 4264–4267 10.1128/AEM.00467-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kaczmarzyk D., and Fulda M. (2010) Fatty acid activation in cyanobacteria mediated by acyl-acyl carrier protein synthetase enables fatty acid recycling. Plant Physiol. 152, 1598–1610 10.1104/pp.109.148007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ruffing A. M. (2013) RNA-Seq analysis and targeted mutagenesis for improved free fatty acid production in an engineered cyanobacterium. Biotechnol. Biofuels 6, 113 10.1186/1754-6834-6-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ruffing A. M. (2013) Borrowing genes from Chlamydomonas reinhardtii for free fatty acid production in engineered cyanobacteria. J. Appl. Phycol. 25, 1495–1507 10.1007/s10811-013-9993-7 [DOI] [Google Scholar]
- 72. Tan X., Yao L., Gao Q., Wang W., Qi F., and Lu X. (2011) Photosynthesis driven conversion of carbon dioxide to fatty alcohols and hydrocarbons in cyanobacteria. Metab. Eng. 13, 169–176 10.1016/j.ymben.2011.01.001 [DOI] [PubMed] [Google Scholar]
- 73. Kaiser B. K., Carleton M., Hickman J. W., Miller C., Lawson D., Budde M., Warrener P., Paredes A., Mullapudi S., Navarro P., Cross F., and Roberts J. M. (2013) Fatty aldehydes in cyanobacteria are a metabolically flexible precursor for a diversity of biofuel products. PLoS ONE 8, e58307 10.1371/journal.pone.0058307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kageyama H., Waditee-Sirisattha R., Sirisattha S., Tanaka Y., Mahakhant A., and Takabe T. (2015) Improved alkane production in nitrogen-fixing and halotolerant cyanobacteria via abiotic stresses and genetic manipulation of alkane synthetic genes. Curr. Microbiol. 71, 115–120 10.1007/s00284-015-0833-7 [DOI] [PubMed] [Google Scholar]
- 75. Jiménez-Díaz L., Caballero A., Pérez-Hernandez N., and Segura A. (2017) Microbial alkane production for jet fuel industry: Motivation, state of the art and perspectives. Microb. Biotechnol. 10, 103–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ruffing A. M., and Jones H. D. (2012) Physiological effects of free fatty acid production in genetically engineered Synechococcus elongatus PCC 7942. Biotechnol. Bioeng. 109, 2190–2199 10.1002/bit.24509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Liu X., Fallon S., Sheng J., and Curtiss R. 3rd. (2011) CO2-limitation-inducible green recovery of fatty acids from cyanobacterial biomass. Proc. Natl. Acad. Sci. U.S.A. 108, 6905–6908 10.1073/pnas.1103016108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lindberg P., Park S., and Melis A. (2010) Engineering a platform for photosynthetic isoprene production in cyanobacteria, using Synechocystis as the model organism. Metab. Eng. 12, 70–79 10.1016/j.ymben.2009.10.001 [DOI] [PubMed] [Google Scholar]
- 79. Englund E., Andersen-Ranberg J., Miao R., Hamberger B., and Lindberg P. (2015) Metabolic engineering of Synechocystis sp. PCC 6803 for production of the plant diterpenoid manoyl oxide. ACS Synth. Biol. 4, 1270–1278 10.1021/acssynbio.5b00070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gao X., Gao F., Liu D., Zhang H., Nie X., and Yang C. (2016) Engineering the methylerythritol phosphate pathway in cyanobacteria for photosynthetic isoprene production from CO2. Energy Environ. Sci. 9, 1400–1411 10.1039/C5EE03102H [DOI] [Google Scholar]
- 81. Wang X., Liu W., Xin C., Zheng Y., Cheng Y., Sun S., Li R., Zhu X.-G., Dai S. Y., Rentzepis P. M., and Yuan J. S. (2016) Enhanced limonene production in cyanobacteria reveals photosynthesis limitations. Proc. Natl. Acad. Sci. U.S.A. 113, 14225–14230 10.1073/pnas.1613340113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bentley F. K., García-Cerdán J. G., Chen H.-C., and Melis A. (2013) Paradigm of monoterpene (β-phellandrene) hydrocarbons production via photosynthesis in cyanobacteria. Bioenergy Res. 6, 917–929 10.1007/s12155-013-9325-4 [DOI] [Google Scholar]
- 83. Formighieri C., and Melis A. (2015) A phycocyanin-phellandrene synthase fusion enhances recombinant protein expression and β-phellandrene (monoterpene) hydrocarbons production in Synechocystis (cyanobacteria). Metab. Eng. 32, 116–124 10.1016/j.ymben.2015.09.010 [DOI] [PubMed] [Google Scholar]
- 84. Reinsvold R. E., Jinkerson R. E., Radakovits R., Posewitz M. C., and Basu C. (2011) The production of the sesquiterpene β-caryophyllene in a transgenic strain of the cyanobacterium Synechocystis. J. Plant Physiol. 168, 848–852 10.1016/j.jplph.2010.11.006 [DOI] [PubMed] [Google Scholar]
- 85. Englund E., Pattanaik B., Ubhayasekera S. J., Stensjö K., Bergquist J., and Lindberg P. (2014) Production of squalene in Synechocystis sp. PCC 6803. PLoS ONE 9, e90270 10.1371/journal.pone.0090270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Halfmann C., Gu L., Gibbons W., and Zhou R. (2014) Genetically engineering cyanobacteria to convert CO2, water, and light into the long-chain hydrocarbon farnesene. Appl. Microbiol. Biotechnol. 98, 9869–9877 10.1007/s00253-014-6118-4 [DOI] [PubMed] [Google Scholar]
- 87. Marienhagen J., and Bott M. (2013) Metabolic engineering of microorganisms for the synthesis of plant natural products. J. Biotechnol. 163, 166–178 10.1016/j.jbiotec.2012.06.001 [DOI] [PubMed] [Google Scholar]
- 88. Leonard E., Runguphan W., O'Connor S., and Prather K. J. (2009) Opportunities in metabolic engineering to facilitate scalable alkaloid production. Nat. Chem. Biol. 5, 292–300 10.1038/nchembio.160 [DOI] [PubMed] [Google Scholar]
- 89. Newman D. J., and Cragg G. M. (2016) Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 79, 629–661 10.1021/acs.jnatprod.5b01055 [DOI] [PubMed] [Google Scholar]
- 90. Fischbach M. A., and Walsh C. T. (2006) Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem. Rev. 106, 3468–3496 10.1021/cr0503097 [DOI] [PubMed] [Google Scholar]
- 91. Kehr J. C., Gatte Picchi D., and Dittmann E. (2011) Natural product biosyntheses in cyanobacteria: a treasure trove of unique enzymes. Beilstein J. Org. Chem. 7, 1622–1635 10.3762/bjoc.7.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Shao Z., Zhao H., and Zhao H. (2009) DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 37, e16 10.1093/nar/gkn991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Trauger J. W., and Walsh C. T. (2000) Heterologous expression in Escherichia coli of the first module of the nonribosomal peptide synthetase for chloroeremomycin, a vancomycin-type glycopeptide antibiotic. Proc. Natl. Acad. Sci. U.S.A. 97, 3112–3117 10.1073/pnas.97.7.3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Ziemert N., Ishida K., Liaimer A., Hertweck C., and Dittmann E. (2008) Ribosomal synthesis of tricyclic depsipeptides in bloom-forming cyanobacteria. Angew. Chem. Int. Ed. Engl. 47, 7756–7759 10.1002/anie.200802730 [DOI] [PubMed] [Google Scholar]
- 95. Jones A. C., Ottilie S., Eustáquio A. S., Edwards D. J., Gerwick L., Moore B. S., and Gerwick W. H. (2012) Evaluation of Streptomyces coelicolor A3(2) as a heterologous expression host for the cyanobacterial protein kinase C activator lyngbyatoxin A. FEBS J. 279, 1243–1251 10.1111/j.1742-4658.2012.08517.x [DOI] [PubMed] [Google Scholar]
- 96. Chang M. C., Eachus R. A., Trieu W., Ro D.-K., and Keasling J. D. (2007) Engineering Escherichia coli for production of functionalized terpenoids using plant P450s. Nat. Chem. Biol. 3, 274–277 10.1038/nchembio875 [DOI] [PubMed] [Google Scholar]
- 97. Lassen L. M., Nielsen A. Z., Olsen C. E., Bialek W., Jensen K., Møller B. L., and Jensen P. E. (2014) Anchoring a plant cytochrome P450 via PsaM to the thylakoids in Synechococcus sp. PCC 7002: Evidence for light-driven biosynthesis. PLoS ONE 9, e102184 10.1371/journal.pone.0102184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Xue Y., Zhang Y., Grace S., and He Q. (2014) Functional expression of an Arabidopsis p450 enzyme, p-coumarate-3-hydroxylase, in the cyanobacterium Synechocystis PCC 6803 for the biosynthesis of caffeic acid. J. Appl. Phycol. 26, 219–226 10.1007/s10811-013-0113-5 [DOI] [Google Scholar]
- 99. Wlodarczyk A., Gnanasekaran T., Nielsen A. Z., Zulu N. N., Mellor S. B., Luckner M., Thøfner J. F., Olsen C. E., Mottawie M. S., Burow M., Pribil M., Feussner I., Møller B. L., and Jensen P. E. (2016) Metabolic engineering of light-driven cytochrome P450 dependent pathways into Synechocystis sp. PCC 6803. Metab. Eng. 33, 1–11 10.1016/j.ymben.2015.10.009 [DOI] [PubMed] [Google Scholar]
- 100. Videau P., Wells K. N., Singh A. J., Gerwick W. H., and Philmus B. (2016) Assessment of Anabaena sp. strain PCC 7120 as a heterologous expression host for cyanobacterial natural products: production of Lyngbyatoxin A. ACS Synth. Biol. 5, 978–988 10.1021/acssynbio.6b00038 [DOI] [PubMed] [Google Scholar]