Abstract
Mycolic acids are the hallmark of the cell envelope in mycobacteria, which include the important human pathogens Mycobacterium tuberculosis and Mycobacterium leprae. Mycolic acids are very long C60–C90 α-alkyl β-hydroxy fatty acids having a variety of functional groups on their hydrocarbon chain that define several mycolate types. Mycobacteria also produce an unusually large number of putative epoxide hydrolases, but the physiological functions of these enzymes are still unclear. Here, we report that the mycobacterial epoxide hydrolase EphD is involved in mycolic acid metabolism. We found that orthologs of EphD from M. tuberculosis and M. smegmatis are functional epoxide hydrolases, cleaving a lipophilic substrate, 9,10-cis-epoxystearic acid, in vitro and forming a vicinal diol. The results of EphD overproduction in M. smegmatis and M. bovis BCG Δhma strains producing epoxymycolic acids indicated that EphD is involved in the metabolism of these forms of mycolates in both fast- and slow-growing mycobacteria. Moreover, using MALDI-TOF-MS and 1H NMR spectroscopy of mycolic acids and lipids isolated from EphD-overproducing M. smegmatis, we identified new oxygenated mycolic acid species that accumulated during epoxymycolate depletion. Disruption of the ephD gene in M. tuberculosis specifically impaired the synthesis of ketomycolates and caused accumulation of their precursor, hydroxymycolate, indicating either direct or indirect involvement of EphD in ketomycolate biosynthesis. Our results clearly indicate that EphD plays a role in metabolism of oxygenated mycolic acids in mycobacteria.
Keywords: mycobacteria, cell wall, fatty acid metabolism, lipid, mass spectrometry (MS), bacterial cell envelope, epoxide hydrolase, epoxymycolic acid, mycolic acids
Introduction
The genus Mycobacterium comprises more than 150 species, among which pathogens such as Mycobacterium tuberculosis and Mycobacterium leprae have attracted much attention as causative agents of tuberculosis and leprosy, respectively. However, a large number of mycobacterial species can also be found in various biotopes as saprophytic bacteria (1). Mycobacteria are characterized by a unique cell wall structure composed of peptidoglycan linked to a heteropolysaccharide arabinogalactan with its arabinosyl termini esterified by mycolic acids, very-long-chain fatty acids unique to mycobacteria, and related genera of the suborder Corynebacterineae (2). Arabinogalactan-bound mycolic acids represent the main lipids of the inner leaflet of the outer membrane of mycobacteria, also referred to as mycomembrane. The outer leaflet of this membrane consists of a variety of non-covalently associated glycolipids, some of the most prominent forms of which are trehalose esters of mycolates: trehalose monomycolates (TMMs)2 and trehalose dimycolates (TDMs) (3).
Mycolic acids are the hallmark of the mycobacterial cell envelope with a characteristic α-alkyl β-hydroxy structure and a very long, C60–C90 hydrocarbon chain. The main meromycolate chain is typically modified in two distinct positions along the hydrocarbon chain, with the proximal position closer to the carboxyl group and the distal position closer to the ω-end of the meromycolate (Fig. S1). These modifications include various functional groups, such as double bonds, methyl groups, cyclopropane, and oxygenated groups. The latter are present only at the distal position of the meromycolate chain and have a methyl group attached at the adjacent carbon (2). Based on the presence of functional groups, a few types of mycolic acids were defined, and interestingly, mycobacterial species synthesize distinct sets of several mycolic acid classes. For example, in M. tuberculosis, the least polar α-mycolates with two cyclopropane rings can be found, as well as methoxy- and ketomycolates (Fig. S1). The fast growing environmental strain M. smegmatis, which is a widely used model organism in mycobacteriology, produces mainly α-mycolates with two double bonds, shorter α′-mycolates and epoxymycolates. The biosynthesis of mycolic acids involves the production of (i) C22–C26 α-alkyl chain by the mycobacterial type I fatty acid synthase (FASI) system and (ii) the meromycolate, which is synthesized by elongation of C16–C18 FASI products to fatty acids up to C54 long by enzymes of the type II fatty acid synthase (FASII) system (2). These precursors then undergo a Claisen-type condensation forming a β-keto product, which is subsequently attached to trehalose, and its β-keto group is reduced (4, 5). The resulting TMM are further transported across the plasma membrane, where mycolates are transferred from TMM to arabinogalactan or another TMM molecule, resulting in the formation of TDM (6–8).
It is generally accepted that the attachment of functional groups to the proximal and distal positions of the meromycolate is initiated by desaturation of the chain, although neither the enzyme(s) nor the precise stage at which these reactions occur have been identified. In M. tuberculosis, the chains with double bonds serve as the substrates for at least six S-adenosyl methionine-dependent methyltransferases encoded by mmaA1–mmaA4, cmaA2, and pcaA genes (9). Although catalytic activities of MmaA1, MmaA2, CmaA2, and PcaA give rise to specific cyclopropane and methyl groups, the methyltransferase MmaA4 (Hma) has been shown to be required for the production of methoxy and ketomycolates (10, 11). Synthesis of oxygenated mycolates in M. tuberculosis probably occurs through a hydroxy fatty acid precursor, which can be either O-methylated by the methyltransferase MmaA3, leading to the production of methoxymycolates, or further oxidized resulting in ketomycolates (12). Studies of protein–protein interactions of core enzymes of FASII system with known meromycolate modifying enzymes from M. tuberculosis have strengthened the hypothesis that reactions of modifications of the meromycolate chain probably take place during its elongation by FASII (13, 14).
The biosynthesis of individual classes of mycolates in M. smegmatis has been extensively studied, including several important observations related to the growth of this organism at different temperatures. In this respect, it was shown that the amounts of epoxymycolates relative to α-mycolates and α′-mycolates produced by M. smegmatis, increase at lower temperatures (15). Although most α-mycolates in M. smegmatis grown at 37 °C are diunsaturated, some level of cyclopropanation was also detected, and it appears to be induced at lower growth temperatures (16, 17). Temperature-induced changes in mycolic acid composition were also observed in Mycobacterium thermoresistibile, in which cyclopropanation of α-mycolates decreased at 55 °C compared with 37 °C (18). It is therefore of significance to address the temperature dependence of mycolate composition in fast-growing mycobacteria when studying individual mycolic acid classes in these species. These data are not very surprising, given the range of growth conditions to which environmental species are exposed and must adapt to, including possibly via modification of their mycolic acid content. Although this adaptability is more pronounced in environmental mycobacteria, pathogenic species also face different hostile conditions encountered in infected macrophages, which require adaptation to maintain virulence (19, 20).
In this work we describe the involvement of a novel protein: the epoxide hydrolase EphD from M. tuberculosis H37Rv and its ortholog MSMEG_4280 from M. smegmatis mc2155, hereafter referred to as EphDtb and EphDsmeg, respectively, in the metabolism of mycolic acids in mycobacteria.
Results
Epoxide hydrolases (EHs) are enzymes that catalyze the addition of water to epoxides to yield the corresponding 1,2-diols and can use a broad range of substrates, among which are lipids such as arachidonic acid and cholesterol metabolites (Fig. 1A) (21). EphDtb is one of the seven putative EHs predicted in the genome of M. tuberculosis H37Rv, six of which (EphA–EphF) share similarity to mammalian EHs and one of which (EphG) is related to the limonene EH from Rhodococcus erythropolis (22). So far, none of these EHs was ascribed a physiological function, although mycobacterial EHs have been suggested to play a role in fatty acid metabolism (23). Contrary to all other human-like soluble EHs, proteins EphD and EphE are conserved across the Mycobacterium genus. Comparing bioinformatic data of EHs from M. tuberculosis, EphDtb shows several distinct features: (i) it is the only membrane-associated putative EH and (ii) along with having an α/β-hydrolase domain characteristic of most soluble EHs, it contains a C-terminal short-chain dehydrogenase (SDR) domain (Fig. S2).
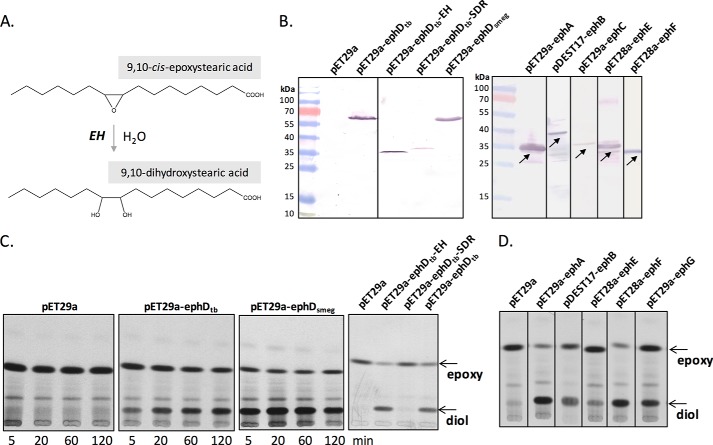
Figure 1.
Epoxide hydrolase activity of EphA–EphG proteins in vitro. A, reaction catalyzed by EH. B, analysis of recombinant protein production in E. coli BL21(DE3) pET29a, BL21(DE3) pET29a-ephDtb, BL21(DE3) pET29a-ephDtb-EH, BL21(DE3) pET29a-ephDtb-SDR, BL21(DE3) pET29a-ephDsmeg, BL21(DE3) pET29a-ephAtb, BL21(AI) pDEST17-ephBtb, BL21(DE3) pET29a-ephCtb, BL21(AI) pET28a-ephEtb, and BL21(DE3) pET28a-ephFtb. (The line with recombinant EphG is not shown because the signal was hardly visible.) SDS-PAGE analysis of 10,000 × g supernatants of cell lysates, 60 μg of proteins were loaded per lane. Recombinant proteins were detected by Western blotting analysis and immunodetection with anti-His antibodies. C, TLC analysis of the reaction products resulting from the incubation of [14C]9,10-cis-epoxystearic acid with E. coli BL21(DE3) pET29a, BL21(DE3) pET29a-ephDtb, or BL21(DE3) pET29a-ephDsmeg fractions for 5–120 min at 37 °C and E. coli BL21(DE3) pET29a, BL21(DE3) pET29a-ephDtb-EH, BL21(DE3) pET29a-ephDtb-SDR, and BL21(DE3) pET29a-ephDtb for 60 min. The plates were developed using n-hexane:diethyl ether:formic acid (70:30:2) and exposed to autoradiography film at −80 °C for 2–5 days. D, TLC analysis of the products of the EH assays containing 10,000 × g supernatants of lysates of E. coli BL21(DE3) pET29a, BL21(DE3) pET29a-ephAtb, BL21(AI) pDEST17-ephBtb, BL21 (AI) pET28a-ephEtb, BL21(DE3) pET28a-ephFtb, and BL21(DE3) pET29a-ephGtb as the enzymatic sources. The reactions ran for 1 h at 37 °C. The plates were developed and exposed as described above.
EphDtb and EphDsmeg are functional epoxide hydrolases
The epoxide hydrolase activity of EphDtb and EphDsmeg was tested using cell-free extracts of Escherichia coli strains producing these proteins, as well as recombinant proteins corresponding to the individual EH and SDR domains of EphDtb. Testing EH activity in cell-free extracts of E. coli without the need to purify individual proteins is possible due to the lack of endogenous EH activity in this microorganism (24). The ephD gene from M. smegmatis mc2155, orthologous gene from M. tuberculosis H37Rv and the EH- and SDR-encoding segments of ephDtb were individually expressed as C-terminal histidine-tagged proteins in E. coli BL21(DE3) using the expression system pET29a (Fig. 1B). Cell-free extracts from the resulting strains BL21(DE3) pET29a-ephDtb, BL21(DE3) pET29a-ephDtb-EH, BL21(DE3) pET29a-ephDtb-SDR, and BL21(DE3) pET29a-ephDsmeg together with the control strain BL21(DE3) pET29a were used as enzyme sources in an EH assay where each recombinant protein was tested for its ability to hydrolyze the generic substrate [14C]9,10-cis-epoxystearic acid (Fig. 1C) (25). TLC analyses of organic solvent-extracted lipids clearly demonstrated that cell-free extracts from the EphDtb- and EphDsmeg-producing strains, but not those from the control strain, were capable of converting 9,10-epoxystearic acid into the corresponding diol. The identity of the diol product was confirmed by LC-MS/MS (Fig. S3). Formation of the diol was also evident when a cell-free extract from BL21(DE3) pET29a-ephDtb-EH was used as an enzyme source, whereas the lipids extracted from the reaction containing BL21(DE3) pET29a-ephDtb-SDR were comparable to the negative control (Fig. 1C). We thus conclude that EphDtb and EphDsmeg are functional EHs in vitro and that the α/β-hydrolase domain of EphD alone is responsible for this activity. In the context of this study, we addressed also the epoxide hydrolase activity of all remaining putative EHs from M. tuberculosis H37Rv, and we confirmed that all of these proteins except EphC catalyze the cleavage of 9,10-epoxystearic acid (Fig. 1, B and D).
M. smegmatis overproducing EphD possesses altered lipid and mycolic acid compositions
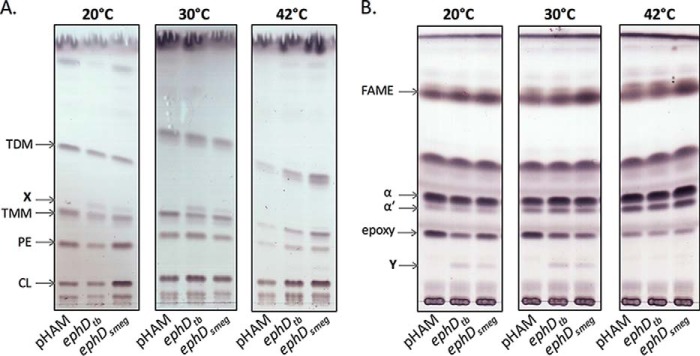
To assess the effect of ephD expression on lipids and mycolic acids, EphDtb and EphDsmeg were individually overproduced in M. smegmatis mc2155 using the inducible expression system pHAM at three different temperatures. The production of recombinant proteins was confirmed by Western blotting analysis and immunodetection with anti-His antibodies (Fig. S4). The cells were harvested, and the total lipids extracted from these cells were analyzed by TLC in different solvent systems. Compared with the negative control strain, strains overproducing EphDtb and EphDsmeg synthesized a novel lipid species (X) migrating slightly faster than TMM (Fig. 2A). Moreover, analysis of mycolic acid methyl esters (MAMEs) extracted and further derivatized from the same batch of whole cells revealed a new fatty acid (Y) correlating with depletion of epoxymycolates in the overproducing strains (Fig. 2B). Interestingly, these changes were not observed in cells grown at 42 °C.
Figure 2.
Effect of EphD overproduction on lipids and mycolic acids in M. smegmatis. A, thin-layer chromatography of extracted lipids developed in chloroform:methanol:water (20:4:0.5, v/v/v). CL, cardiolipin; PE, phosphatidylethanolamine; X, unknown lipid (see text for details). B, thin-layer chromatography of mycolic acid methyl esters isolated from whole cells developed in n-hexane:ethyl acetate (95:5, v/v, three runs). α, α′, and epoxy indicate individual types of mycolic acid methyl esters. Y, unknown fatty acid (see text for details). pHAM lane, M. smegmatis mc2155 pHAM; ephDtb lane, M. smegmatis mc2155 pHAM-ephDtb; ephDsmeg lane, M. smegmatis mc2155 pHAM-ephDsmeg.
Next, we analyzed fatty and mycolic acid compositions of the individual mycolic acid-containing molecules including TMM, TDM, delipidated cell residues, and lipid X synthesized by the overproducing strains grown at 30 °C. Individual lipids were purified by preparative TLC and further subjected to saponification, and free fatty and mycolic acids were derivatized to their corresponding methyl esters and analyzed by TLC. This unveiled that the majority of species Y is bound to populations of extractable lipids, mainly TMM and lipid X; surprisingly, the latter compound contained practically no α-, α′-, or epoxymycolates (Fig. 3). Y in the form of methyl ester was further isolated using preparative TLC and subjected to structural analysis. MALDI-TOF mass spectrometry showed that species Y represents a series of odd and even carbon number mycolic acids with mass values expected for dicarboxymycolates (m/z 925 (C57), 939 (C58), 953 (C59), 967 (C60), and 981 (C61)) (26). The presence of an ω-carboxyl group in molecules of population Y was confirmed by 1H NMR spectroscopy, with characteristic singlets at 3.65 and 3.70 ppm, corresponding to the signals of two methyl protons linked to carboxyl groups (Fig. S5). Similar dicarboxy mycolic acids were previously reported in several Mycobacterium species that synthesize wax-ester mycolates, such as Mycobacterium aurum, Mycobacterium phlei, or the Mycobacterium avium–intracellulare–scrofulaceumcomplex (27–30). Dicarboxymycolates are formed from wax-ester mycolates of these strains during the isolation of mycolic acids from their native complex forms, which involves drastic hydrolysis in alkaline conditions (saponification).
Figure 3.
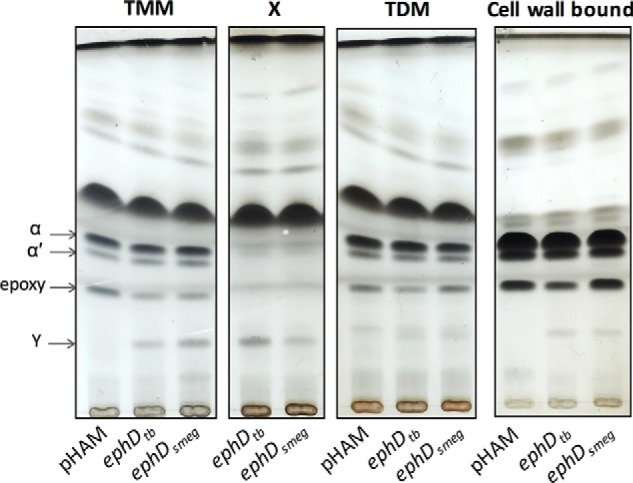
Mycolic acid composition of individual population of lipids from M. smegmatis overproducing EphD. Shown is thin-layer chromatography of mycolic acid methyl esters from isolated populations of TMMs, TDMs, and lipid X and from delipidated cell pellets developed in n-hexane:ethyl acetate (95:5, v/v, three runs). α, α′, and epoxy indicate individual types of mycolic acid methyl esters. pHAM lane, M. smegmatis mc2155 pHAM; ephDtb lane, M. smegmatis mc2155 pHAM-ephDtb; ephDsmeg lane, M. smegmatis mc2155 pHAM-ephDsmeg.
M. smegmatis overproducing EphD produces unique mycolic acids containing internal ester
To confirm whether detected dicarboxymycolates really originated from wax-ester mycolates, we attempted circumventing the need for saponification. We reasoned that the presence of wax-ester mycolates should be detectable by MALDI-TOF analysis of the purified intact lipid molecules TMM and lipid X. The comparative analysis of MALDI-TOF mass spectra of TMM from control and overproducing strains yielded the following results (Table 1): In the mass spectra of TMM from the control strain, the peaks with [M+Na]+ at m/z 1470 and 1498 were assigned to TMM with C77 and C79 α-mycolic acids and the peaks of [M+Na]+ at m/z 1514 and 1528 to the main representatives of TMM with C79 and C80 epoxymycolic acids. In the case of TMM from EphDtb- and EphDsmeg-overproducing strains, the mass values were respectively assigned to TMM with α-C77 and α-C79, as well as epoxy-C79–C80 as in the control strain, but also additional peaks at m/z 1502, 1516, 1530, and 1544 were observed. The mass values of these peaks could be interpreted as TMM molecules bearing a supplementary oxygens in the meromycolate chain (31).
Table 1.
Comparative MALDI-MS analysis of TMM
TMM was isolated from M. smegmatis mc2155 pHAM (1), M. smegmatis mc2155 pHAM-ephDtb (2), and M. smegmatis mc2155 pHAM-ephDsmeg (3). The values represent m/z of [M+Na]+.
| Type of mycolic acid | Number of carbons |
Unknown number of carbons | |||||
|---|---|---|---|---|---|---|---|
| 77 | 78 | 79 | 80 | 81 | 82 | ||
| 1. pHAM | |||||||
| α- | 1470 | 1498 | |||||
| Epoxy- | 1486 | 1500 | 1514 | 1528 | 1542 | 1556 | |
| 2. pHAM-ephDtb | |||||||
| α- | 1470 | 1498 | |||||
| Epoxy- | 1486 | 1500 | 1514 | 1528 | |||
| Unknown | 1502 | ||||||
| 1516 | |||||||
| 1530 | |||||||
| 1544 | |||||||
| 3. pHAM-ephDsmeg | |||||||
| α- | 1470 | 1498 | |||||
| Epoxy- | 1486 | 1500 | 1514 | 1528 | |||
| Unknown | 1502 | ||||||
| 1516 | |||||||
| 1530 | |||||||
| 1544 | |||||||
Each representative peak was further analyzed by MS/MS fragmentation, which confirmed the identity of trehalose esters by the appearance of a fragment at m/z 715, indicating C2–C3 cleavage of the mycolate unit releasing a C24 fragment with trehalose (Table S1). Loss of glucose in high mass values was observed in all of the samples, as well as the presence of a peak at m/z 365, assigned to trehalose.
Comparative 1H NMR studies of TMM isolated from the control and EphD-overproducing strains confirmed the presence of disaccharide trehalose with one glucose acylated at the 6 position, as well as 2-alkyl 3-hydroxy fatty acids (H-2 and H-3 at 2.35 and 3.60 ppm). Other specific signals characteristic for mycolic acids of M. smegmatis were found, including those for cis and trans double bonds and epoxy ring; this last signal was absent in TMMs from both overproducers. The signals corresponding to a wax-ester were not clearly identified. However, MALDI-MS analysis of mycolic acids released after saponification of TMM confirmed the presence of dicarboxymycolic acids in the samples from the strains overproducing EphD (Table 2). Interestingly, no diol mycolic acids were observed, but the presence of peaks of pseudomolecular [M+Na]+ species at m/z 1220, 1234, 1248, and 1262 was identified in these spectra. These mass values could be interpreted as either hydroxy-mycolic acid (1220 for the C80 homolog) or long chain dicarboxylic mycolic acid (1248 for the C80 homolog) or mycolic acid harboring an internal ester function; however, this last structure needs strong alkaline conditions for their isolation. These data together with the results of MALDI-TOF analysis of the purified intact TMMs from EphD-overproducing strains suggested that the analyzed TMM population contains mycolic acids with internal ester group.
Table 2.
Comparative MALDI-MS analysis of methylated mycolic acids
Methylated mycolic acids were obtained by saponification and methylation of TMM, isolated from M. smegmatis mc2155 pHAM (1), M. smegmatis mc2155 pHAM-ephDtb (2), and M. smegmatis mc2155 pHAM-ephDsmeg (3). The values represent m/z of [M+Na]+.
| Type of mycolic acid | Number of carbons |
Unknown number of carbons | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 58 | 59 | 60 | 61 | 77 | 78 | 79 | 80 | 81 | 82 | ||
| 1. pHAM | |||||||||||
| α- | 1174 | 1188 | 1202 | ||||||||
| Epoxy- | 1190 | 1204 | 1218 | 1232 | 1246 | ||||||
| 2. pHAM-ephDtb | 1174 | 1188 | 1202 | ||||||||
| α- | 1160 | 1188 | |||||||||
| Epoxy- | 1176 | 1204 | 1218 | 1232 | 1246 | ||||||
| Unknown | 1220 | ||||||||||
| 1234 | |||||||||||
| 1248 | |||||||||||
| 1262 | |||||||||||
| Dicarboxy- | 939 | 953 | 967 | 981 | |||||||
| 3. pHAM-ephDsmeg | |||||||||||
| α- | 1160 | 1174 | 1188 | ||||||||
| Epoxy- | 1176 | 1190 | 1204 | 1218 | 1232 | ||||||
| Unknown | 1220 | ||||||||||
| 1234 | |||||||||||
| 1248 | |||||||||||
| 1262 | |||||||||||
| 1276 | |||||||||||
| Dicarboxy- | 939 | 953 | 967 | 981 | |||||||
The MALDI-TOF mass spectra of isolated intact lipid X contained a mixture of three populations of lipids (Table S2): (i) glycopeptidolipids represented by two peaks at m/z 1232 and 1260 corresponding to one series of the species-specific lipids of M. smegmatis and characterized by their fragmentation pattern; (ii) trehalose β-keto esters with C77 and C79 diethylenic meromycolate chains, characterized by the main peaks with [M+Na]+ at m/z 1468 and 1496; These compounds, intermediates of mycolic acids synthesis, did not show C2–C3 cleavage of the mycolate by MS/MS, as the hallmark of the mycolic acid motif is absent in these molecules; and (iii) in the high mass values, at m/z from 1740 to 1894, ultra long-chain complex lipids ranging from C95 to C105 and belonging likely to the TMM series. The MS/MS fragmentation of the long chain compounds confirmed the nature of trehalose monomycolates by the occurrence of peak at m/z 715 indicating C2–C3 cleavage. Another peak resulting from this cleavage could be interpreted as a possible fragment of the meroaldehyde at m/z 1068, 1173, and 1201. The long-chain mycolic acid could explain the higher migration of this TMM fraction. Ultra long-chain mycolic acids possessing three functional groups have already been identified as minor components of M. tuberculosis (32), and a recent study has shown the involvement of the FASII hydroxyacyl dehydratase HadBC in their synthesis (33).
MALDI-TOF mass spectrometry of fatty acids released after saponification of lipid X and subsequently methylated showed the appearance of peaks at m/z from 939 to 995 corresponding to C58–C62 dicarboxymycolates, suggesting an internal ester function in these molecules. Unfortunately, the small amounts of the analyzed material did not allow carrying on their precise characterization.
Saponification products of known wax-ester mycolates, alongside dicarboxymycolates, are secondary alcohols 2-octadecanol and 2-eicosanol (29, 34, 35). To find these fragments, the lipids extracted from the control and EphD-overproducing strains were saponified, methylated, and analyzed by GC/MS (data not shown). C16 and C18 fatty acid methyl esters (FAMEs) were present in all of the strains and interestingly, also α-methyl branched FAMEs were characterized in EphD overproducers on the basis of their retention time and the base peak at m/z 88 characteristic for 2-methyl FAMEs, such as methyl-2 stearic acid and methyl 2-eicosanoic acid. In addition, minor amounts of compounds such as methyl ketone or small secondary alcohols (base peak m/z 57) were identified in the EphD overproducers, but 2-octadecanol or 2-eicosanol were not clearly detected.
Mycolic acids with internal ester are synthesized in parallel with acetate uptake
The decreased amounts of epoxymycolic acids and the formation of mycolates with internal ester in M. smegmatis overproducing EphD raise the question of whether EphD catalyzes the hydrolysis of the epoxy group on mature epoxymycolates already esterified in lipids or whether it acts during their synthesis. To distinguish between these hypotheses, we metabolically labeled M. smegmatis strains overproducing EphD with [1,2-14C]acetate by adding the label either 8 h prior to the induction of recombinant protein production or at the same time as the inducer (acetamide) was added to the culture. The cells were harvested after 3 or 24 h of further cultivation to encompass different growth stages, and their mycolic acid methyl esters obtained by saponification, extraction, and methylation were analyzed by TLC (Fig. 4). Upon detection with CuSO4, we observed the presence of dicarboxymycolates in EphD-overproducing strains of both sets of samples (data not shown). However, after autoradiography, the presence of [14C]dicarboxymycolates was detected exclusively in the cells in which [14C]acetate was added at the same time as EphD production was induced. This suggests that EphD acts on epoxymycolic acids during their synthesis.
Figure 4.
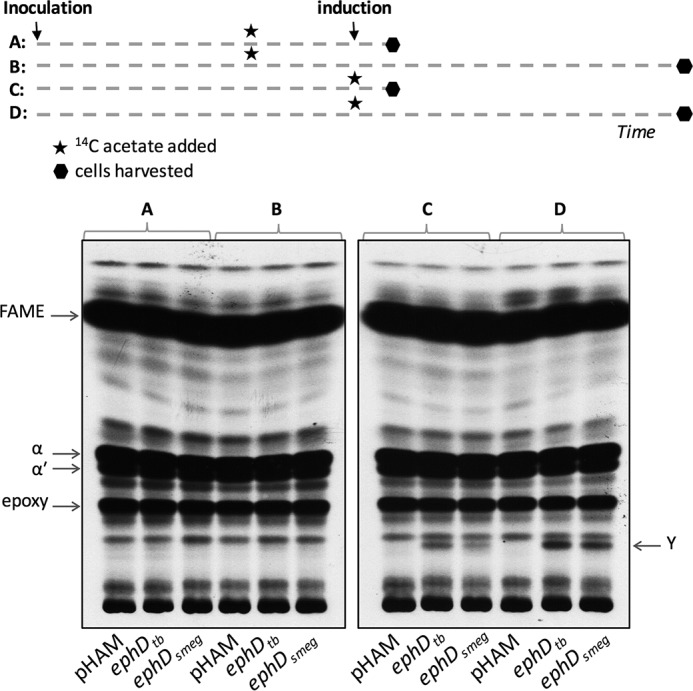
Metabolic labeling of M. smegmatis strains overproducing EphD. The top panel shows a scheme of metabolic labeling of culture sets A, B, C, and D described under “Experimental procedures”; also shown is an autoradiogram of TLC analysis of radiolabeled fatty and mycolic acid methyl esters from whole cells of cultures A and B (radioactive label added 8 h before induction of recombinant EphD production) and C and D (label added in parallel with induction of recombinant EphD production). The samples were developed in n-hexane:ethyl acetate (95:5, v/v, three runs). α, α′, and epoxy indicate individual types of mycolic acid methyl esters; pHAM lane, M. smegmatis mc2155 pHAM; ephDtb lane, M. smegmatis mc2155 pHAM-ephDtb; ephDsmeg lane, M. smegmatis mc2155 pHAM-ephDsmeg.
EphD is involved in epoxymycolic acid metabolism in slow growing mycobacteria
We demonstrated the effect of EphD overproduction on the metabolism of epoxymycolates in M. smegmatis, a fast-growing Mycobacterium species in which this type of mycolic acids naturally occurs. However, orthologs of EphD are found in slow- and fast-growing Mycobacterium species alike, most of which are not known to synthesize detectable amounts of epoxymycolates. In M. tuberculosis, the production of epoxymycolates was observed under certain conditions. Upon disruption of the gene encoding the mycolic acid methyltransferase Hma (MmaA4), which is essential for the synthesis of methoxy- and ketomycolates in both M. tuberculosis and M. bovis, an accumulation of α-mycolates was observed that accompanied the build-up of small amounts of mycolates with a cis-epoxy group at the distal position of the meromycolate chain (10). We used a strain of M. bovis BCG Δhma (36) to test, whether overproduction of EphD in this strain would affect its mycolic acid composition. Because acetamide-inducible expression systems are usually unstable in slow-growing mycobacteria (37), we attempted to constitutively express ephD using the pVV2 or pVV16 expression plasmids, but without success. We were successful, however, in generating strains of M. bovis BCG Δhma producing the individual domains of EphD: EphDtb-EH and EphDtb-SDR, using these plasmids. As expected, analysis of the mycolic acids derived from the saponification and derivatization of purified TMM, TDM, and delipidated cells from the control strain M. bovis BCG Δhma pVV16 showed the presence of α-mycolates with minor amounts of epoxymycolates and absence of ketomycolates (10) (Fig. 5). This profile was not affected by the production of the SDR domain of EphD. In sharp contrast, the same lipids obtained from M. bovis BCG Δhma producing the α/β-hydrolase domain of EphD were almost completely devoid of epoxy species. TLC analysis of those samples did not reveal detectable amounts of dicarboxymycolates, probably because of too-low amounts of epoxymycolates in these samples or because of the absence of yet unknown enzymes required for the synthesis of their precursors from epoxymycolates. Nevertheless, based on these results, we suggest that EphD is involved in the metabolism of epoxymycolates also in M. bovis.
Figure 5.
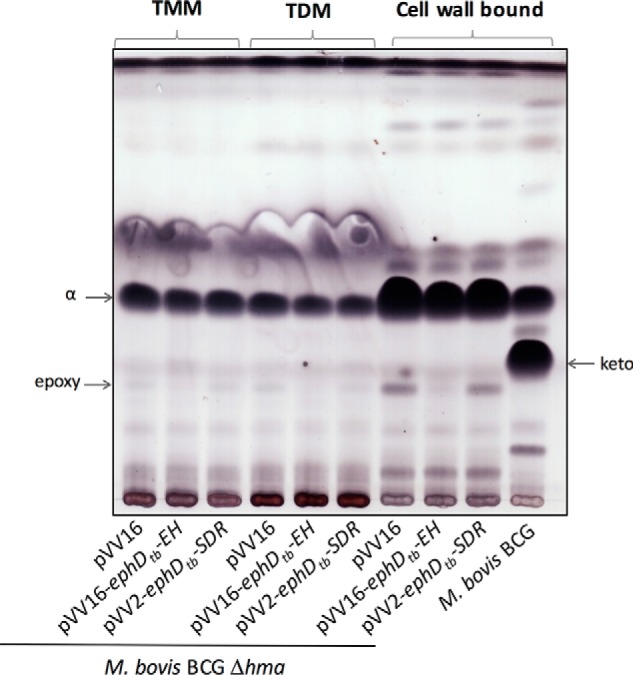
Effect of overproduction of single domains of EphD in M. bovis BCG Δhma. The results of thin-layer chromatography of mycolic acid methyl esters from isolated populations of TMMs, TDMs, and from delipidated cell pellets developed in n-hexane:ethyl acetate (95:5, v/v, three runs) are shown. α, epoxy, and keto indicate individual types of mycolic acid methyl esters.
Disruption of ephD in M. tuberculosis impairs biosynthesis of ketomycolic acids
By means of overproduction of EphD in M. smegmatis and M. bovis BCG Δhma, we show that its activity leads to the degradation/remodeling of epoxymycolates. To gain further insight into the function of EphD, we disrupted the ephD genes of M. smegmatis and M. tuberculosis and analyzed the effects of these knock-outs on mycolic acid metabolism in both species.
M. smegmatis mc2155 ΔephD was prepared by insertion of a kanamycin cassette in the ORF of ephD gene in the region encoding the α/β-hydrolase domain of the protein. The mutant strain and the wildtype parent strain, M. smegmatis mc2155, were cultivated in different media (Sauton, GAS, 7H9 + ADC, LB, and modified M63) at 20, 30, 37, or 42 °C, after which cells were harvested and saponified, and their mycolic acids were extracted, methylated, and analyzed by TLC as described above. The growth of the mutant strain was significantly impaired, and compared with the control, there was a significant clumping of mutant cells in all of the tested conditions (Fig. S6). Nevertheless, no dramatic reproducible differences were found in the fatty and mycolic acids profiles (data not shown). α-Mycolates in M. smegmatis were shown to comprise five subpopulations (α1–α5) that differ one from another in their degree of cyclopropanation and methylation at the distal and proximal positions of the meromycolate chain (16, 17). Populations of α-mycolates from total mycolic acid extracts prepared from the mutant and parent strains grown in modified M63 medium at 20, 30, or 42 °C were isolated by preparative TLC and individual subpopulations further separated by argentation TLC. Again, no reproducible differences between analyzed strains were detected (data not shown).
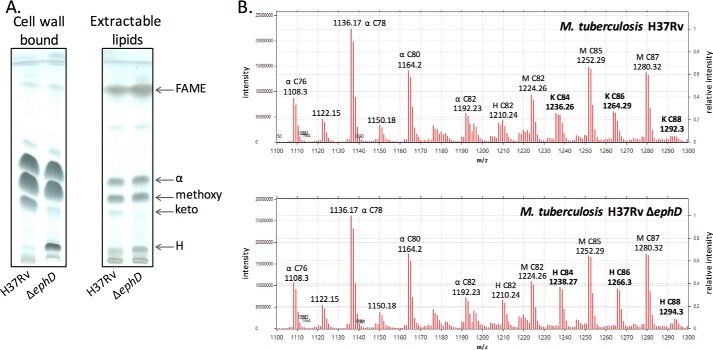
Next, we constructed the mutant strain M. tuberculosis H37Rv ΔephD harboring a hygromycin cassette insertion in the α/β-hydrolase domain of ephD. Mycolic acids were isolated from both the extractable lipids and delipidated cells of the wildtype and mutant strains and analyzed by TLC. Disruption of the ephD gene in M. tuberculosis H37Rv led to a decrease of ketomycolates and an accumulation fatty/mycolic acid H (Fig. 6A). The accumulated species H was identified via comparative LC-MS analysis of de-esterified samples of mycolic acid methyl esters prepared from M. tuberculosis H37Rv and M. tuberculosis H37Rv ΔephD as hydroxymycolic acids related to ketomycolates in the wildtype strain (Fig. 6B). Attempts to complement this phenotype by a functional copy of the ephDtb gene or fragments of this gene encoding the individual domains of EphD: EphDtb-EH and EphDtb-SDR, were not successful, probably because of the difficulty of constitutively overproducing these proteins in M. tuberculosis H37Rv strain. However, because the gene pepB (Rv2213) located in the downstream vicinity of ephD is oriented in the opposite direction, we do not expect that disruption of ephD affects downstream genes. Based on these data, we concluded that the presence of EphD is required for the synthesis of ketomycolic acids in M. tuberculosis.
Figure 6.
Effect of ephD disruption on mycolic acids in M. tuberculosis. A, thin-layer chromatography of mycolic acid methyl esters isolated from extractable lipids and delipidated cells of M. tuberculosis. H37Rv lane, M. tuberculosis H37Rv; ΔephD lane, M. tuberculosis H37Rv ΔephD. Chromatograms were developed in n-hexane:ethyl acetate (95:5, v/v, three runs). B, LC-MS comparative analysis of mycolic acids from M. tuberculosis H37Rv and M. tuberculosis H37Rv ΔephD. α, H, K, and M indicate α-, hydroxy, keto-, and methoxymycolates.
Discussion
As major lipid components of the mycomembrane, mycolic acids are essential for mycobacterial viability, which is reflected in the fact that several important and effective antimycobacterials such as isoniazid, ethionamide, isoxyl, and thiacetazone target the FASII system (38, 39). Although non-essential per se, oxygenated and cyclopropyl functionalities of the meromycolate chain in M. tuberculosis play significant roles in virulence and maintaining the integrity of the cell envelope (9, 11, 40, 41). Reports of temperature-induced changes in the mycolic acid composition of fast-growing mycobacteria such as M. smegmatis indicate that, much like changes in the fatty acid composition of phospholipids, modulation of the fine structure of mycolic acids may contribute to the adaptation of mycobacteria to environmental conditions (15, 17, 18). In this work, we provided evidence of the involvement of a novel enzyme endowed with epoxide hydrolase activity, present in both pathogenic and environmental species of mycobacteria, in the metabolism of mycolic acids. In vitro, EphD cleaves the epoxy group of the generic substrate, 9,10-cis-epoxystearic acid. Defining the physiological substrate of EphD, like that of any other mycolic acid-modifying enzyme, is complicated by the complexity of the biosynthesis of these very-long-chain fatty acids. To date, all of the mycolic acid methyltransferases of M. tuberculosis have been characterized solely on the basis of the phenotype of recombinant knock-out or knock-in strains (2). We showed that overproduction of the EphD enzyme from M. smegmatis or M. tuberculosis in M. smegmatis leads to a decrease in epoxymycolates and an accumulation of mycolic acids with internal ester, which are degraded to dicarboxylic mycolates by alkaline treatment.
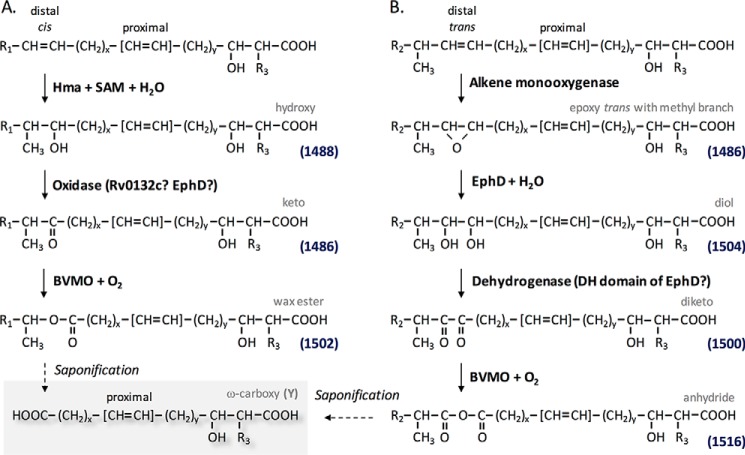
The origin of similar molecules, wax-ester mycolates, in mycobacteria has been shown to derive from the ketomycolic acids by Baeyer–Villiger oxidation, i.e. insertion of molecular oxygen into the hydrocarbon chain, thus converting a keto derivative into a wax-ester (35) (Fig. 7A). Saponification of these mycolic waxes releases the ω-carboxymycolic acids on one hand and the secondary alcohols, 2-octadecanol and 2-eicosanol, on the other. However, the origin of dicarboxymycolic acids formed in EphD-overproducing strains of M. smegmatis can be explained by another hypothetical pathway proposed on the basis of the reaction sequence postulated for the degradation of limonene in R. erythropolis (42). This reaction sequence includes the opening of the oxirane ring providing a diol, followed by oxidation of the diol, which transforms hydroxyl groups into a ketone and the introduction of molecular oxygen between these two functional groups by a Baeyer–Villiger monooxygenase (Fig. 7B).
Figure 7.
Origin of dicarboxymycolic acids in mycobacteria. A, known metabolic pathway for wax-ester mycolates. B, pathway proposed in this work. The mass values in blue are indicated for a C77 mycolyl-containing TMM. Functions between brackets in proximal position correspond to double bonds or cyclopropane.
Although the above-mentioned pathways are of a catabolic process, metabolic labeling of M. smegmatis cells overproducing EphD show that 14C-labeled dicarboxymycolates, which are probable degradation products of saponification of mycolates containing internal ester, appear simultaneously with [14C]acetate uptake, suggesting that the hydrolysis of the epoxy group and subsequent reactions occur concurrently with the synthesis of mycolates. We also found that the described modified mycolic acids are present as trehalose esters. In addition, we detected the β-keto ester product of mycolic condensation, and in the high mass values we characterized a trehalose monoester with ultra long-chain mycolic acid. Characterization of described intermediates as TMM derivatives may suggest that a certain regulatory point exists at the level of trehalose monoesters. This notion is strengthened by the identification of transient acetylation of TMM required for periplasmic export of this molecule in corynebacteria (43).
Our results further suggest that EphD is also involved in the processing of epoxymycolates in slow-growing mycobacteria. It is not clear why hma mutants of M. tuberculosis and M. bovis BCG synthesize epoxymycolates because the substrate of this methyltransferase has been proposed to contain a cis-double bond (10, 36). It is possible that epoxidation serves as a starting point for double bond degradation, although it cannot be excluded that the complete loss of oxygenated mycolates is unfavorable to these bacteria and that certain levels of epoxymycolates are being synthesized by an unknown monooxygenase to compensate for these changes and restore the integrity of the mycomembrane.
Partial but specific loss of ketomycolates in a M. tuberculosis H37Rv strain deficient in EphD that accompanied the accumulation of their hydroxy precursors indicates impaired function of the enzyme that oxidizes the hydroxylated products of Hma to their keto-derivatives (44, 45). This begs the question of whether EphD itself, or rather its SDR domain, could oxidize hydroxy-mycolates to ketomycolates. Oxidation of hydroxy-mycolates to ketomycolates was suggested to be catalyzed by coenzyme F420-dependent dehydrogenase Rv0132c (46). Co-production of this protein with methyltransferase Hma in M. smegmatis stimulated production of keto-forms. We prepared a strain of M. smegmatis co-producing both Hma and EphDtb; however, the amount of ketomycolates in this strain did not differ from the one producing Hma alone (not shown). Indeed, the Hma producing M. smegmatis strain (also described by Dubnau et al. (45)) synthesizes hydroxy- and minor amounts of ketomycolates, probably as the result of endogenous oxygenase activity in M. smegmatis. This indicates that in mycobacteria there are at least two independent pathways for the production of keto-mycolic acids. To test whether the observed production of ketomycolates is due to the activity of the endogenous EphDsmeg, we produced the Hma protein in M. smegmatis mc2155 ΔephD; however, we did not see any changes in the production of ketomycolates in this strain compared with the control (Fig. S7). Therefore, it is difficult to conclude whether EphD has a direct catalytic role in ketomycolate synthesis in M. tuberculosis.
Under the conditions tested, we did not reveal any changes in mycolic acids profile following the disruption of ephD in M. smegmatis. Hence, we can hypothesize that in M. smegmatis, the activity of EphD is physiologically induced under conditions that, for example, require changes in the amount of epoxymycolates and those were not captured during our analysis. The regulatory mechanisms underlying the subtle structural changes in mycobacterial cell envelope lipids remain essentially unknown (47). The activities of much of the FASII enzymes seem to be regulated, at least by protein phosphorylation, as was demonstrated in several studies (48–52). In silico analysis indicates a possible regulation of ephDtb by cyclic AMP, which is often a signal for environmental or metabolic changes in bacteria (53). Further efforts are needed to explore the impact of this regulation and subsequent phenotypic changes on the adaptation of mycobacteria to environmental changes.
Experimental procedures
Bacterial strains and growth conditions
E. coli strains XL1 and DH5α were used for cloning purposes and cultivated in LB (Luria–Bertani) medium (Invitrogen) at 37 °C with shaking. M. smegmatis mc2155 strains were grown in modified M63 medium (7.6 × 10−2 m (NH4)2SO4, 0.5 m KH2PO4, 1 mm MgSO4, 2% succinate, 0.025% tyloxapol, pH 7) with shaking. M. tuberculosis and M. bovis BCG strains were cultivated statically at 37 °C in Middlebrook 7H9 medium (Difco) with the addition of 10% albumin-dextrose-catalase (ADC) supplement and 0.05% Tween 80. When necessary, antibiotics were added into the medium: 20 μg·ml−1 kanamycin; 20 μg·ml−1 hygromycin for mycobacteria or 100 μg·ml−1 hygromycin for E. coli.
Gene cloning and expression
DNAs corresponding to ephAtb, ephCtb, ephDtb, ephDtb-EH, ephDtb-SDR, ephDsmeg, ephEtb, ephFtb, and ephGtb were amplified from M. tuberculosis H37Rv or M. smegmatis mc2155 chromosomal DNA by PCR using primers listed in Table S3. The resulting fragments corresponding to ephAtb, ephCtb, ephDtb, ephDtb-EH, ephDtb-SDR, ephDsmeg, and ephGtb were cloned into pET29a (Novagen) and fragments corresponding to ephEtb and ephFtb were cloned into pET28a (Novagen) using the NdeI and HindIII restriction sites. The protein EphB was produced using pDEST17-ephD construct (54). Recombinant strains of E. coli BL21(DE3) or E. coli BL21(AI) were grown in LB medium at 37 °C to an A600 of 0.4–0.6, at which point the production of recombinant proteins was induced with 0.4 mm isopropyl-β-d-1-thiogalactopyranoside. The cells producing EphD proteins were cultivated at 30 °C for an additional 4 h, and the cells producing EphA, EphB, EphC, EphE, EphF, and EphG proteins were cultivated at 16 °C for an additional 16 h. The cells were harvested, washed with 50 mm Tris-HCl, pH 8.0, and finally stored at −20 °C.
Similarly, the ephDtb and ephDsmeg ORFs were PCR-amplified from M. tuberculosis H37Rv or M. smegmatis mc2155 chromosomal DNA (Table S3) and cloned into pHAM, a derivative of pJAM2 harboring a hygromycin resistance cassette (55) using BamHI and XbaI for ephDtb or ScaI and XbaI for ephDsmeg. M. smegmatis mc2155 pHAM, M. smegmatis mc2155 pHAM-ephDtb, and M. smegmatis mc2155 pHAM-ephDsmeg were cultivated in modified M63 medium at 20, 30, or 42 °C until A600 reached 0.6–0.9. At that point, recombinant protein production was induced by adding 0.2% acetamide to the culture medium, and the cells were further grown at the given temperatures for up to 72 h.
pVV16-ephDtb-EH was prepared by excision of the ephDtb-EH fragment from pET29a-ephDtb-EH using NdeI and HindIII and ligation into pVV16 cut with the same enzymes (56). The ephDtb-SDR fragment was amplified from M. tuberculosis H37Rv chromosomal DNA by PCR (Table S3) and cloned into the NdeI and HindIII restriction sites of pVV2 (57). Constructs were introduced into M. bovis BCG Δhma (obtained from Dr. W. Jacobs (36)) by electroporation, and the resulting strains were cultivated at 37 °C as described above. Production of recombinant proteins was confirmed by SDS-PAGE and immunodetection with anti-His antibodies.
Epoxide hydrolase assay
E. coli BL21(DE3) pET29a, BL21(DE3) pET29a-ephAtb, BL21(AI) pDEST17-ephBtb, BL21(DE3) pET29a-ephCtb, BL21(DE3) pET29a-ephDtb, BL21(DE3) pET29a-ephDtb-EH, BL21(DE3) pET29a-ephDtb-SDR, BL21(DE3) pET29a-ephDsmeg, BL21(AI) pET28a-ephEtb, BL21(DE3) pET28a-ephFtb, and BL21(DE3) pET29a-ephGtb cells were suspended in 50 mm Tris-HCl, pH 8.0, and subjected to probe sonication in the form of ten 20-s pulses with 40-s cooling intervals between pulses. Sonicates were centrifuged for 10 min at 10,000 × g, and the resulting supernatants were kept frozen at −20 °C. [14C]9,10-cis-Epoxystearic acid (56 mCi mmol−1) was synthesized from radiolabeled oleic acid as described by Müller et al. (58), and the EH assay using this substrate was conducted as described by Arand et al. (25) with minor modifications. Briefly, reaction mixtures containing 10,000 × g supernatants of E. coli extracts (100 μg of proteins), [14C]9,10-cis-epoxystearic acid (5,000 dpm), and 50 mm Tris-HCl buffer, pH 8.0, in a final volume of 250 μl were incubated from 5 to 120 min at 37 °C, and the reactions were stopped by the addition of 500 μl of ethyl acetate. The samples were vortexed and centrifuged for 1 min on a tabletop centrifuge. The upper organic phase was transferred to a clean tube and dried under a stream of nitrogen, and half of the material was analyzed by TLC using n-hexane:diethyl ether:formic acid (70:30:2) as the eluent. Radiolabeled compounds were visualized by exposure of the developed silica plates to Kodak BIOMAX MR film at −80 °C.
Construction of deletion strains
The Ts-sacB method (59) was used to achieve allelic replacement at the ephD loci of M. tuberculosis H37Rv and M. smegmatis mc2155. The M. tuberculosis ephD (Rv2214c) gene and flanking regions was PCR-amplified from M. tuberculosis H37Rv genomic DNA using primers KOF (5′-actagtggactcgggttgggc-3′) and KOR (5′-actagtaacaccgacgcggag-3′) and a disrupted allele, ephD::hyg, was obtained by insertion of hygromycin resistance cassette into the KpnI restriction site of ephD, thereby disrupting the N-terminal α/β hydrolase domain of the protein. ephD::hyg was then cloned into the SpeI-cut and blunt-ended pPR27-xylE to obtain pPR27Rv2214cHX, the construct used for allelic replacement in M. tuberculosis.
The M. smegmatis ephD (MSMEG_4280; ephDsmeg) gene and flanking regions were PCR-amplified from M. smegmatis mc2155 genomic DNA using primers smg2214c.1 (5′-cccgaattcagcgggtcaccgtcccctctg-3′) and smg2214c.2 (5′-ggtctagaccacatgacgtccgacatgggtg-3′), and a disrupted allele, ephDsmeg::kan, was obtained by replacing 550 bp of the ephDsmeg ORF flanked between two PmlI restriction sites with the kanamycin resistance cassette from pUC4K (GE Healthcare). ephDsmeg::kan was then cloned into the XbaI-cut and blunt-ended pPR27-xylE to obtain pPR27ephDsmegKX, the construct used for allelic replacement in M. smegmatis. The cells of control and mutant strains of M. smegmatis used for lipid, fatty, and mycolic acids analysis were cultivated in modified M63 medium for 4 and 9 days at 20 °C, for 24 h at 30 °C, and for 24 h at 42 °C.
Lipid and mycolic acid analysis
Lipids were extracted from whole cells by 1.5 h of extraction with chloroform:methanol (1:2) at 56 °C, followed by two 1.5-h extractions with chloroform:methanol (2:1) at the same temperature. Extracts were collected into a glass tube, dried under a stream of nitrogen, and subjected to a wash in chloroform, methanol, and water (4:2:1) (60). Dried organic phases of each sample after this biphasic wash were dissolved in chloroform:methanol (2:1) and analyzed by thin-layer chromatography on silica gel 60 F254 plates (Merck) using chloroform:methanol:water (20:4:0.5) as the eluent. Lipids were visualized by spraying the plates with 10% cupric sulfate in 8% phosphoric acid and subsequent charring. Individual populations of selected lipids were isolated by preparative TLC after visualization with 0.01% rhodamine B. The silica corresponding to migration of the analyzed population was scraped off the plate, and the lipids were isolated by repeated extractions in the mixture of chloroform:methanol (2:1).
Mycolic acids were isolated from whole cells, delipidated cell pellets, or purified lipids (61). Briefly, 1 ml of 15% solution of tetrabuthylammonium hydroxide was added to each sample, and the samples were saponified at 100 °C overnight. After cooling, 1.5 ml of dichloromethane, 1 ml of demineralized water, and 150 μl of iodomethane were added, and samples were methylated for 4 h at room temperature on a rotary shaker. Methylated samples were washed twice with water, and the organic phase was transferred to clean tubes and dried under nitrogen flow. Fatty and mycolic acid methyl esters (FAMEs/MAMEs) were extracted with 2 ml of diethyl ether. Dried samples were analyzed by TLC in n-hexane:ethyl acetate (95:5, three runs) as the eluent and visualized as described above. Different types of MAMEs were isolated by preparative TLC as described above and analyzed on silver-impregnated silica plates using dichloromethane as the eluent (16).
Metabolic labeling
Four sets (A, B, C, and D) of 10-ml cultures of modified M63 medium were inoculated with the cells of M. smegmatis mc2155 pHAM, M. smegmatis mc2155 pHAM-ephDtb, and M. smegmatis mc2155 pHAM-ephDsmeg to a final A600 of ∼0.01 and cultivated at 30 °C with shaking. When A600 reached 0.07, 0.5 μCi·ml−1 [1,2-14C]acetate (American Radiolabeled Chemicals; specific activity, 106 mCi·mmol−1) was added to the sets A and B. The cultures were further cultivated until A600 = ∼0.3 when the production of recombinant proteins was induced by adding 0.2% acetamide. Sets C and D were grown in parallel, and in their case [1,2-14C]acetate was added together with acetamide at an A600 of ∼0.3. Aliquots of all of the cultures were harvested after 3 h (sets A and C) or 24 h (sets A and C) after induction.
Structural analysis
MALDI-TOF/TOF-MS and MS/MS analyses were conducted in the positive ionization and reflection mode by accumulating 10 spectra of 250 laser shots, using the 5800 MALDI TOF/TOF analyzer (Applied Biosystems/AB SCIEX) equipped with a Nd:YAG laser (349-nm wavelength). For MS and MS/MS data acquisitions, uniform, continuous, and random stage motion was selected at a fixed laser intensity of 4000 (instrument-specific units) and 400 Hz pulse rate and 6000 (instrument-specific units) and 1000 Hz, respectively. For MS/MS data acquisition, the fragmentation of selected precursor ions was performed at collision energy of 1 kV. Lipid samples were dissolved in chloroform and were directly spotted onto the target plate as 0.5-μl droplets, followed by the addition of 0.5 μl of matrix solution (2,5-dihydroxybenzoic acid, 10 mg/ml in CHCl3/CH3OH, 1:1 (v/v)). Samples were allowed to crystallize at room temperature. The spectra were externally calibrated using lipid standards.
Gas chromatography-mass spectrometry
GC-MS of volatile fatty esters was performed with 1 μl of sample solubilized into petroleum ether, which was injected into a Thermo Trace GC ultra chromatograph equipped with an Inferno ZB5HT column of 15 m for fatty acid methyl ester separation. The injector temperature was fixed at 220 °C with a split ratio of 20:1. Helium was circulated at a constant flow rate of 1.2 ml/min as a carrier gas. The oven temperature program was as follows: initial temperature at 120 °C increased at 10 °C/min to 380 °C (held for 3 min). The coupled mass spectrometer ISQ was operated in electron impact ionization of 70 eV from m/z 60 to 600, and the transfer line was maintained at 275 °C.
Nuclear magnetic resonance
1H NMR spectra of purified lipids were obtained in CDCl3 (99.9% D) at 298 K using a 600-MHz Brucker Avance III spectrometer equipped with a TCI cryoprobe. Chemical shift values (in ppm) were relative to the internal CHCl3 resonance at 7.27 ppm.
LC-MS analysis of lipids and mycolic acids
LC-MS analysis of lipids and mycolic acids was as described in Ref. 6.
LC-MS/MS analysis of cis-epoxystearic acid turnover
For the analysis of the products of the reaction by LC-MS/MS, assays using non-radiolabeled cis-epoxystearic acid were run. One microliter of 1 mm solution of cis-epoxystearic dissolved in acetonitrile was added to 100 μl of E. coli crude extracts (control and EphDtb producer). Mixtures were incubated at 37 °C for 10 min and subsequently extracted with an equal volume of ethyl acetate. After centrifugation for 3 min at 10,000 × g, 50 μl of the upper organic phase were transferred into a new tube and evaporated under a gentle stream of nitrogen at 40 °C. The remaining material was dissolved in 100 μl of acetonitrile, sonified in a water bath for 3 min, and centrifuged at 10,000 × g. The supernatant was analyzed on an Agilent 1100 liquid chromatography system using an Agilent eclipse XDB-C18 reverse phase column (4.6 × 150 mm, 5-μm pore size) equipped with a corresponding Opti-Gard precolumn as the stationary phase. The mobile phase consisted of (A) 0.1% formic acid and (B) acetonitrile containing 0.1% formic acid at a flow rate of 400 μl/min. Samples were applied in an injection volume of 20 μl. Starting conditions of 70% buffer B were maintained for 2 min, followed by a linear gradient from 70 to 100% B within 8 min. An isocratic flow of 100% buffer B was held for 8 min. Thereafter, the column was re-equilibrated for 2 min with 70% buffer B. The HPLC system was coupled to a 4000 QTRAP hybrid quadrupole linear ion trap mass spectrometer (Applied Biosystems) equipped with a TurboV source and electrospray ionization interface. Analytes were recorded using multiple reaction monitoring in the negative mode, using the following source specific parameters: IS −4500V, TEM 450 °C, curtain gas (CUR = 30 p.s.i.), nebulizer gas (GS1 = 50 p.s.i.), heater gas (GS2 = 70 p.s.i.), and collision gas (CAD = 10 p.s.i.). 9,10-dihydroxystearic acid was detected as a single homogenous peak eluting at 8.2 min using the two molecular transitions m/z 315/297 (loss of water) and m/z 315/141 (α-elimination) for specific detection. These were previously established with the molecular standard prepared by hydrolysis with recombinant human soluble epoxide hydrolase (58).
Author contributions
J. M., F. L., A. G., A. L., M. Z., M. A., M. J., M.-A. L., and J. K. investigation; J. M., M.-A. L., and J. K. methodology; J. M. and J. K. writing-original draft; A. L., M. Z., M. A., M. D., M. J., M.-A. L., and J. K. data curation; M. M., M. D., M. J., M.-A. L., and J. K. conceptualization; M. D., M. J., M.-A. L., and J. K. supervision; M. D., M. J., and J. K. funding acquisition; M. D., M. J., M.-A. L., and J. K. writing-review and editing.
Supplementary Material
Acknowledgments
We are grateful to Dr. W. R. Jacobs, Jr. (Albert Einstein College of Medicine, New York, New York) for the kind gift of M. bovis BCG Δhma, to Dr. M. James (University of Alberta, Edmonton, Canada) for the kind gift of pDEST17-ephD construct, to Dr. Gundi (Colorado State University, Fort Collins, Colorado) for help with the construction of the ephD knock-out mutant of M. tuberculosis H37Rv, to M. Kopál (Comenius University, Bratislava, Slovakia) for help with preparing the constructs, to Dr. K. Mikušová (Comenius University, Bratislava, Slovakia) for critically reading the manuscript, and to Dr. M. Neboháčová (Comenius University, Bratislava, Slovakia) and Dr. Jacques Prandi (Institut de Pharmacologie et de Biologie Structurale, Toulouse, France) for fruitful discussions. NMR experiments were performed on the PICT-Genotoul platform of Toulouse and funded by CNRS, Université Paul Sabatier, Toulouse III, Ibisa, European structural funds, and the Midi Pyrénées region.
This work was supported by Slovak Research and Development Agency Grant APVV-0441-10 (to J. K.), Slovak Grant Agency VEGA Grant 1/0284/15 (to J. K.), the project supported by the Research and Development Operational Programe funded by European Regional Development Fund Contract ITMS 26240120027 (to J. K.), and NIAID, National Institutes of Health Grant AI063054 (to M. J.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Tables S1–S3, Figs. S1–S7, and references.
- TMM
- trehalose monomycolate
- TDM
- trehalose dimycolates
- EH
- epoxide hydrolase
- FAME
- fatty acid methyl ester
- FAS
- fatty acid synthase
- MAME
- mycolic acid methyl ester
- SDR
- short-chain dehydrogenase.
References
- 1. Tortoli E. (2014) Microbiological features and clinical relevance of new species of the genus Mycobacterium. Clin. Microbiol. Rev. 27, 727–752 10.1128/CMR.00035-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Marrakchi H., Lanéelle M. A., and Daffé M. (2014) Mycolic acids: structures, biosynthesis, and beyond. Chem. Biol. 21, 67–85 10.1016/j.chembiol.2013.11.011 [DOI] [PubMed] [Google Scholar]
- 3. Rodriguez-Rivera F. P., Zhou X., Theriot J. A., and Bertozzi C. R. (2017) Visualization of mycobacterial membrane dynamics in live cells. J. Am. Chem. Soc. 139, 3488–3495 10.1021/jacs.6b12541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gavalda S., Léger M., van der Rest B., Stella A., Bardou F., Montrozier H., Chalut C., Burlet-Schiltz O., Marrakchi H., Daffé M., and Quémard A. (2009) The Pks13/FadD32 crosstalk for the biosynthesis of mycolic acids in Mycobacterium tuberculosis. J. Biol. Chem. 284, 19255–19264 10.1074/jbc.M109.006940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gavalda S., Bardou F., Laval F., Bon C., Malaga W., Chalut C., Guilhot C., Mourey L., Daffé M., and Quémard A. (2014) The polyketide synthase Pks13 catalyzes a novel mechanism of lipid transfer in mycobacteria. Chem. Biol. 21, 1660–1669 10.1016/j.chembiol.2014.10.011 [DOI] [PubMed] [Google Scholar]
- 6. Grzegorzewicz A. E., Pham H., Gundi V. A., Scherman M. S., North E. J., Hess T., Jones V., Gruppo V., Born S. E., Korduláková J., Chavadi S. S., Morisseau C., Lenaerts A. J., Lee R. E., McNeil M. R., et al. (2012) Inhibition of mycolic acid transport across the Mycobacterium tuberculosis plasma membrane. Nat. Chem. Biol. 8, 334–341 10.1038/nchembio.794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Belisle J. T., Vissa V. D., Sievert T., Takayama K., Brennan P. J., and Besra G. S. (1997) Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science 276, 1420–1422 10.1126/science.276.5317.1420 [DOI] [PubMed] [Google Scholar]
- 8. Jackson M., Raynaud C., Lanéelle M. A., Guilhot C., Laurent-Winter C., Ensergueix D., Gicquel B., and Daffé M. (1999) Inactivation of the antigen 85C gene profoundly affects the mycolate content and alters the permeability of the Mycobacterium tuberculosis cell envelope. Mol. Microbiol. 31, 1573–1587 10.1046/j.1365-2958.1999.01310.x [DOI] [PubMed] [Google Scholar]
- 9. Barkan D., Hedhli D., Yan H. G., Huygen K., and Glickman M. S. (2012) Mycobacterium tuberculosis lacking all mycolic acid cyclopropanation is viable but highly attenuated and hyperinflammatory in mice. Infect. Immun. 80, 1958–1968 10.1128/IAI.00021-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dinadayala P., Laval F., Raynaud C., Lemassu A., Laneelle M. A., Laneelle G., and Daffe M. (2003) Tracking the putative biosynthetic precursors of oxygenated mycolates of Mycobacterium tuberculosis: structural analysis of fatty acids of a mutant strain deviod of methoxy- and ketomycolates. J. Biol. Chem. 278, 7310–7319 10.1074/jbc.M210501200 [DOI] [PubMed] [Google Scholar]
- 11. Dubnau E., Chan J., Raynaud C., Mohan V. P., Lanéelle M. A., Yu K., Quémard A., Smith I., and Daffé M. (2000) Oxygenated mycolic acids are necessary for virulence of Mycobacterium tuberculosis in mice. Mol. Microbiol. 36, 630–637 [DOI] [PubMed] [Google Scholar]
- 12. Yuan Y., and Barry C. E. 3rd. (1996) A common mechanism for the biosynthesis of methoxy and cyclopropyl mycolic acids in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 93, 12828–12833 10.1073/pnas.93.23.12828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Veyron-Churlet R., Bigot S., Guerrini O., Verdoux S., Malaga W., Daffé M., and Zerbib D. (2005) The biosynthesis of mycolic acids in Mycobacterium tuberculosis relies on multiple specialized elongation complexes interconnected by specific protein–protein interactions. J. Mol. Biol. 353, 847–858 10.1016/j.jmb.2005.09.016 [DOI] [PubMed] [Google Scholar]
- 14. Cantaloube S., Veyron-Churlet R., Haddache N., Daffé M., and Zerbib D. (2011) The Mycobacterium tuberculosis FAS-II dehydratases and methyltransferases define the specificity of the mycolic acid elongation complexes. PLoS One 6, e29564 10.1371/journal.pone.0029564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baba T., Kaneda K., Kusunose E., Kusunose M., and Yano I. (1989) Thermally adaptive changes of mycolic acids in Mycobacterium smegmatis. J. Biochem. 106, 81–86 10.1093/oxfordjournals.jbchem.a122825 [DOI] [PubMed] [Google Scholar]
- 16. Laval F., Haites R., Movahedzadeh F., Lemassu A., Wong C. Y., Stoker N., Billman-Jacobe H., and Daffé M. (2008) Investigating the function of the putative mycolic acid methyltransferase UmaA: divergence between the Mycobacterium smegmatis and Mycobacterium tuberculosis proteins. J. Biol. Chem. 283, 1419–1427 10.1074/jbc.M708859200 [DOI] [PubMed] [Google Scholar]
- 17. Alibaud L., Alahari A., Trivelli X., Ojha A. K., Hatfull G. F., Guerardel Y., and Kremer L. (2010) Temperature-dependent regulation of mycolic acid cyclopropanation in saprophytic mycobacteria: role of the Mycobacterium smegmatis 1351 gene (MSMEG_1351) in CIS-cyclopropanation of α-mycolates. J. Biol. Chem. 285, 21698–21707 10.1074/jbc.M110.125724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kremer L., Guérardel Y., Gurcha S. S., Locht C., and Besra G. S. (2002) Temperature-induced changes in the cell-wall components of Mycobacterium thermoresistibile. Microbiology 148, 3145–3154 10.1099/00221287-148-10-3145 [DOI] [PubMed] [Google Scholar]
- 19. Singh A., Gupta R., Vishwakarma R. A., Narayanan P. R., Paramasivan C. N., Ramanathan V. D., and Tyagi A. K. (2005) Requirement of the mymA operon for appropriate cell wall ultrastructure and persistence of Mycobacterium tuberculosis in the spleens of guinea pigs. J. Bacteriol. 187, 4173–4186 10.1128/JB.187.12.4173-4186.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Barry C. E. 3rd, Lee R. E., Mdluli K., Sampson A. E., Schroeder B. G., Slayden R. A., and Yuan Y. (1998) Mycolic acids: structure, biosynthesis and physiological functions. Prog. Lipid Res. 37, 143–179 10.1016/S0163-7827(98)00008-3 [DOI] [PubMed] [Google Scholar]
- 21. Morisseau C. (2013) Role of epoxide hydrolases in lipid metabolism. Biochimie 95, 91–95 10.1016/j.biochi.2012.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Johansson P., Unge T., Cronin A., Arand M., Bergfors T., Jones T. A., and Mowbray S. L. (2005) Structure of an atypical epoxide hydrolase from Mycobacterium tuberculosis gives insights into its function. J. Mol. Biol. 351, 1048–1056 10.1016/j.jmb.2005.06.055 [DOI] [PubMed] [Google Scholar]
- 23. Tekaia F., Gordon S. V., Garnier T., Brosch R., Barrell B. G., and Cole S. T. (1999) Analysis of the proteome of Mycobacterium tuberculosis in silico. Tuber Lung. Dis. 79, 329–342 10.1054/tuld.1999.0220 [DOI] [PubMed] [Google Scholar]
- 24. van Loo B., Kingma J., Arand M., Wubbolts M. G., and Janssen D. B. (2006) Diversity and biocatalytic potential of epoxide hydrolases identified by genome analysis. Appl Environ Microbiol. 72, 2905–2917 10.1128/AEM.72.4.2905-2917.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Arand M., Cronin A., Adamska M., and Oesch F. (2005) Epoxide hydrolases: structure, function, mechanism, and assay. Methods Enzymol. 400, 569–588 10.1016/S0076-6879(05)00032-7 [DOI] [PubMed] [Google Scholar]
- 26. Laval F., Lanéelle M. A., Déon C., Monsarrat B., and Daffé M. (2001) Accurate molecular mass determination of mycolic acids by MALDI-TOF mass spectrometry. Anal. Chem. 73, 4537–4544 10.1021/ac0105181 [DOI] [PubMed] [Google Scholar]
- 27. Minnikin D. E., Minnikin S. M., Parlett J. H., and Goodfellow M. (1985) Mycolic acid patterns of some rapidly-growing species of Mycobacterium. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 259, 446–460 [DOI] [PubMed] [Google Scholar]
- 28. Levy-Frebault V., Daffe M., Restrepo E., Grimont F., Grimont P. A., and David H. L. (1986) Differentiation of Mycobacterium thermoresistibile from Mycobacterium phlei and other rapidly growing mycobacteria. Ann. Inst. Pasteur Microbiol. (1985) 137A, 143–151 [DOI] [PubMed] [Google Scholar]
- 29. Minnikin D. E., Minnikin S. M., Parlett J. H., Goodfellow M., and Magnusson M. (1984) Mycolic acid patterns of some species of Mycobacterium. Arch. Microbiol. 139, 225–231 [DOI] [PubMed] [Google Scholar]
- 30. Toriyama S., Yano I., Masui M., Kusunose M., and Kusunose E. (1978) Separation of C50–60 and C70–80 mycolic acid molecular species and their changes by growth temperatures in Mycobacterium phlei. FEBS Lett. 95, 111–115 10.1016/0014-5793(78)80063-5 [DOI] [PubMed] [Google Scholar]
- 31. Fujita Y., Naka T., Doi T., and Yano I. (2005) Direct molecular mass determination of trehalose monomycolate from 11 species of mycobacteria by MALDI-TOF mass spectrometry. Microbiology 151, 1443–1452 10.1099/mic.0.27791-0 [DOI] [PubMed] [Google Scholar]
- 32. Watanabe M., Aoyagi Y., Ridell M., and Minnikin D. E. (2001) Separation and characterization of individual mycolic acids in representative mycobacteria. Microbiology 147, 1825–1837 10.1099/00221287-147-7-1825 [DOI] [PubMed] [Google Scholar]
- 33. Slama N., Jamet S., Frigui W., Pawlik A., Bottai D., Laval F., Constant P., Lemassu A., Cam K., Daffé M., Brosch R., Eynard N., and Quémard A. (2016) The changes in mycolic acid structures caused by hadC mutation have a dramatic effect on the virulence of Mycobacterium tuberculosis. Mol. Microbiol. 99, 794–807 10.1111/mmi.13266 [DOI] [PubMed] [Google Scholar]
- 34. Lanéelle M. A., and Lanéelle G. (1970) [Structure of mycolic acids and an intermediate in the biosynthesis of dicarboxylic mycolic acids]. Eur. J. Biochem. 12, 296–300 10.1111/j.1432-1033.1970.tb00850.x [DOI] [PubMed] [Google Scholar]
- 35. Toriyama S., Imaizumi S., Tomiyasu I., Masui M., and Yano I. (1982) Incorporation of 18O into long-chain, secondary alcohols derived from ester mycolic acids. Biochim. Biophys. Acta 712, 427–429 10.1016/0005-2760(82)90363-0 [DOI] [Google Scholar]
- 36. Derrick S. C., Dao D., Yang A., Kolibab K., Jacobs W. R., and Morris S. L. (2012) Formulation of a mmaA4 gene deletion mutant of Mycobacterium bovis BCG in cationic liposomes significantly enhances protection against tuberculosis. PLoS One 7, e32959 10.1371/journal.pone.0032959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brown A. C., and Parish T. (2006) Instability of the acetamide-inducible expression vector pJAM2 in Mycobacterium tuberculosis. Plasmid 55, 81–86 10.1016/j.plasmid.2005.06.005 [DOI] [PubMed] [Google Scholar]
- 38. Grzegorzewicz A. E., Korduláková J., Jones V., Born S. E., Belardinelli J. M., Vaquié A., Gundi V. A., Madacki J., Slama N., Laval F., Vaubourgeix J., Crew R. M., Gicquel B., Daffé M., Morbidoni H. R., et al. (2012) A common mechanism of inhibition of the Mycobacterium tuberculosis mycolic acid biosynthetic pathway by isoxyl and thiacetazone. J. Biol. Chem. 287, 38434–38441 10.1074/jbc.M112.400994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Banerjee A., Dubnau E., Quemard A., Balasubramanian V., Um K. S., Wilson T., Collins D., de Lisle G., and Jacobs W. R. Jr. (1994) inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263, 227–230 10.1126/science.8284673 [DOI] [PubMed] [Google Scholar]
- 40. Barkan D., Liu Z., Sacchettini J. C., and Glickman M. S. (2009) Mycolic acid cyclopropanation is essential for viability, drug resistance, and cell wall integrity of Mycobacterium tuberculosis. Chem. Biol. 16, 499–509 10.1016/j.chembiol.2009.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yuan Y., Zhu Y., Crane D. D., and Barry C. E. 3rd (1998) The effect of oxygenated mycolic acid composition on cell wall function and macrophage growth in Mycobacterium tuberculosis. Mol. Microbiol. 29, 1449–1458 10.1046/j.1365-2958.1998.01026.x [DOI] [PubMed] [Google Scholar]
- 42. van der Werf M. J., Swarts H. J., and de Bont J. A. (1999) Rhodococcus erythropolis DCL14 contains a novel degradation pathway for limonene. Appl. Environ. Microbiol. 65, 2092–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yamaryo-Botte Y., Rainczuk A. K., Lea-Smith D. J., Brammananth R., van der Peet P. L., Meikle P., Ralton J. E., Rupasinghe T. W., Williams S. J., Coppel R. L., Crellin P. K., and McConville M. J. (2015) Acetylation of trehalose mycolates is required for efficient MmpL-mediated membrane transport in Corynebacterineae. ACS Chem. Biol. 10, 734–746 10.1021/cb5007689 [DOI] [PubMed] [Google Scholar]
- 44. Boissier F., Bardou F., Guillet V., Uttenweiler-Joseph S., Daffé M., Quémard A., and Mourey L. (2006) Further insight into S-adenosylmethionine-dependent methyltransferases: structural characterization of Hma, an enzyme essential for the biosynthesis of oxygenated mycolic acids in Mycobacterium tuberculosis. J. Biol. Chem. 281, 4434–4445 10.1074/jbc.M510250200 [DOI] [PubMed] [Google Scholar]
- 45. Dubnau E., Lanéelle M. A., Soares S., Bénichou A., Vaz T., Promé D., Promé J. C., Daffé M., and Quémard A. (1997) Mycobacterium bovis BCG genes involved in the biosynthesis of cyclopropyl keto- and hydroxy-mycolic acids. Mol. Microbiol. 23, 313–322 10.1046/j.1365-2958.1997.2301589.x [DOI] [PubMed] [Google Scholar]
- 46. Purwantini E., and Mukhopadhyay B. (2013) Rv0132c of Mycobacterium tuberculosis encodes a coenzyme F420-dependent hydroxymycolic acid dehydrogenase. PLoS One 8, e81985 10.1371/journal.pone.0081985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Quémard A. (2016) New Insights into the mycolate-containing compound biosynthesis and transport in Mycobacteria. Trends Microbiol. 24, 725–738 10.1016/j.tim.2016.04.009 [DOI] [PubMed] [Google Scholar]
- 48. Veyron-Churlet R., Zanella-Cléon I., Cohen-Gonsaud M., Molle V., and Kremer L. (2010) Phosphorylation of the Mycobacterium tuberculosis β-ketoacyl-acyl carrier protein reductase MabA regulates mycolic acid biosynthesis. J. Biol. Chem. 285, 12714–12725 10.1074/jbc.M110.105189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jang J., Stella A., Boudou F., Levillain F., Darthuy E., Vaubourgeix J., Wang C., Bardou F., Puzo G., Gilleron M., Burlet-Schiltz O., Monsarrat B., Brodin P., Gicquel B., and Neyrolles O. (2010) Functional characterization of the Mycobacterium tuberculosis serine/threonine kinase PknJ. Microbiology 156, 1619–1631 10.1099/mic.0.038133-0 [DOI] [PubMed] [Google Scholar]
- 50. Molle V., Gulten G., Vilchèze C., Veyron-Churlet R., Zanella-Cléon I., Sacchettini J. C., Jacobs W. R. Jr., and Kremer L. (2010) Phosphorylation of InhA inhibits mycolic acid biosynthesis and growth of Mycobacterium tuberculosis. Mol. Microbiol. 78, 1591–1605 10.1111/j.1365-2958.2010.07446.x [DOI] [PubMed] [Google Scholar]
- 51. Slama N., Leiba J., Eynard N., Daffé M., Kremer L., Quémard A., and Molle V. (2011) Negative regulation by Ser/Thr phosphorylation of HadAB and HadBC dehydratases from Mycobacterium tuberculosis type II fatty acid synthase system. Biochem. Biophys. Res. Commun. 412, 401–406 10.1016/j.bbrc.2011.07.051 [DOI] [PubMed] [Google Scholar]
- 52. Corrales R. M., Molle V., Leiba J., Mourey L., de Chastellier C., and Kremer L. (2012) Phosphorylation of mycobacterial PcaA inhibits mycolic acid cyclopropanation: consequences for intracellular survival and for phagosome maturation block. J. Biol. Chem. 287, 26187–26199 10.1074/jbc.M112.373209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Akhter Y., Yellaboina S., Farhana A., Ranjan A., Ahmed N., and Hasnain S. E. (2008) Genome scale portrait of cAMP-receptor protein (CRP) regulons in mycobacteria points to their role in pathogenesis. Gene 407, 148–158 10.1016/j.gene.2007.10.017 [DOI] [PubMed] [Google Scholar]
- 54. Biswal B. K., Garen G., Cherney M. M., Garen C., and James M. N. (2006) Cloning, expression, purification, crystallization and preliminary X-ray studies of epoxide hydrolases A and B from Mycobacterium tuberculosis. Acta Crystallogr Sect F. Struct. Biol. Cryst Commun 62, 136–138 10.1107/S1744309106000637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Triccas J. A., Parish T., Britton W. J., and Gicquel B. (1998) An inducible expression system permitting the efficient purification of a recombinant antigen from Mycobacterium smegmatis. FEMS Microbiol Lett. 167, 151–156 10.1111/j.1574-6968.1998.tb13221.x [DOI] [PubMed] [Google Scholar]
- 56. Jackson M., Crick D. C., and Brennan P. J. (2000) Phosphatidylinositol is an essential phospholipid of mycobacteria. J. Biol. Chem. 275, 30092–30099 10.1074/jbc.M004658200 [DOI] [PubMed] [Google Scholar]
- 57. Dhiman R. K., Schulbach M. C., Mahapatra S., Baulard A. R., Vissa V., Brennan P. J., and Crick D. C. (2004) Identification of a novel class of omega,E,E-farnesyl diphosphate synthase from Mycobacterium tuberculosis. J. Lipid Res. 45, 1140–1147 10.1194/jlr.M400047-JLR200 [DOI] [PubMed] [Google Scholar]
- 58. Müller F., Arand M., Frank H., Seidel A., Hinz W., Winkler L., Hänel K., Blée E., Beetham J. K., Hammock B. D., and Oesch F. (1997) Visualization of a covalent intermediate between microsomal epoxide hydrolase, but not cholesterol epoxide hydrolase, and their substrates. Eur. J. Biochem. 245, 490–496 10.1111/j.1432-1033.1997.00490.x [DOI] [PubMed] [Google Scholar]
- 59. Pelicic V., Jackson M., Reyrat J. M., Jacobs W. R. Jr., Gicquel B., and Guilhot C. (1997) Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 94, 10955–10960 10.1073/pnas.94.20.10955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Folch J., Lees M., and Sloane Stanley G. H. (1957) A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226, 497–509 [PubMed] [Google Scholar]
- 61. Phetsuksiri B., Baulard A. R., Cooper A. M., Minnikin D. E., Douglas J. D., Besra G. S., and Brennan P. J. (1999) Antimycobacterial activities of isoxyl and new derivatives through the inhibition of mycolic acid synthesis. Antimicrob. Agents Chemother. 43, 1042–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.