Abstract
Ephrin (Eph) receptor tyrosine kinases have evolutionarily diverged from other tyrosine kinases to respond to specific activation and regulatory signals that require close coupling of kinase catalytic and regulatory functions. However, the evolutionary basis for such functional coupling is not fully understood. We employed an evolutionary systems approach involving statistical mining of large sequence and structural data sets to define the hallmarks of Eph kinase evolution and functional specialization. We found that some of the most distinguishing Eph-specific residues structurally tether the flanking juxtamembrane and sterile α motif (SAM) linker regions to the kinase domain, and substitutions of these residues in EphA3 resulted in faster kinase activation. We report for the first time that the SAM domain linker is functionally coupled to the juxtamembrane through co-conserved residues in the kinase domain and that together these residues provide a structural framework for coupling catalytic and regulatory functions. The unique organization of Eph-specific tethering networks and the identification of other Eph-specific sequence features of unknown functions provide new hypotheses for future functional studies and new clues to disease mutations altering Eph kinase–specific functions.
Keywords: allosteric regulation, receptor tyrosine kinase, structure-function, protein sequence, bioinformatics, protein evolution
Introduction
The ephrin (Eph)2 family of receptor tyrosine kinases comprises the largest family of tyrosine kinases and controls critical developmental processes by modulating cell adhesion, cell migration, and cytoskeletal organization in a variety of cell types (1, 2). The human Eph family includes 14 members, which are further divided into two subclasses, EphA and EphB, based on sequence similarity and ligand affinity. Two members, EphA10 and EphB6, are classified as pseudokinases based on their divergence in sequence motifs essential for catalytic activity (3–5). However, all Eph members share a common domain organization, characterized by an extracellular ligand-binding region and an intracellular region that includes the juxtamembrane segment, tyrosine kinase domain, a sterile α motif (SAM) domain, and a PDZ-binding motif (Fig. S1) (1, 2). The implication of Eph family kinases in a broad array of diseases from Alzheimer's disease, viral pathogenesis, and multiple types of cancer (2, 6) underscores the biomedical significance of these enzymes and rationalizes the interest and investment in investigating their molecular mechanisms.
Like other receptor tyrosine kinases, the enzymatic activity of Eph kinases is tightly regulated by molecular mechanisms that allow specific responses to ligand binding, post-translational modifications, and other molecular events. Activation of the receptor is achieved when the extracellular domain binds membrane-anchored ephrin ligands, which triggers the autophosphorylation of tyrosines in the cytoplasmic kinase domain (1, 7). Autophosphorylation on two conserved tyrosines in the juxtamembrane (Tyr596 and Tyr602 in EphA3) is a prerequisite for catalytic activity (8–13) and precedes the autophosphorylation of the activation loop tyrosine (Tyr779 in EphA3) (Fig. S1) (8). Activation loop phosphorylation increases catalytic efficiency as seen in many protein kinases (14); however, unlike the juxtamembrane tyrosines, mutation of the activation loop tyrosine to phenylalanine decreases but does not abolish catalytic or autophosphorylation activity (10, 12). Additionally, phosphorylation of these three major sites as well as other minor sites serves to bind SH2 domain–containing proteins such as Src, Crk, Nck, and RasGAP (15–17). Thus, once the juxtamembrane becomes autophosphorylated, further autophosphorylation on additional sites as well as direct substrate phosphorylation propagates signaling through downstream pathways.
Although the roles of juxtamembrane and activation loop autophosphorylation in Eph kinase functions are well recognized, little is known about how other molecular events are involved in Eph regulation and autophosphorylation. For example, regulation via the formation of higher order complexes is well understood for some receptor tyrosine kinases, for example, in the epidermal growth factor receptor family of receptor tyrosine kinases (18, 19); however, the mechanisms of Eph kinase regulation by oligomerization are not well established. In particular, the difficulty of determining the structure of oligomeric complexes and the lack of full-length crystal structures for the large Eph family, whose members are thought to hetero-oligomerize (20), have contributed to the difficulty in understanding the intricate details of Eph oligomerization. Despite the determination of several dimeric structures of isolated SAM domains from Eph kinases (20, 21), the contribution of the SAM domain in oligomerization or other forms of regulation is yet to be fully characterized. Conflicting reports on the effects of SAM domain deletion mutations on dimerization and activity in EphA3 (22), EphA2 (23), and EphB2 (10) further confound our understanding of the Eph SAM domain. Furthermore, although the SAM domain linker is resolved in multiple crystal structures (9), whether it plays any functional role has never been explored.
Inferring mechanisms from protein sequences provides an alternative and complementary approach to experimental and biochemical characterization methods. In particular, currently unanswered questions such as the evolutionary bases for Eph kinase functional divergence through oligomerization or interactions with regulatory domains and protein segments can be inferred through statistical mining of protein sequences. Indeed, previous statistical analyses of evolutionary constraints acting on protein kinase sequences have provided important insights into protein kinase evolution and allosteric regulation (24–29). The Eph family serves as an ideal system for evolutionary sequence studies because it is a monophyletic family with detectable orthologs throughout extremely diverse metazoan phyla, from chordates to nematodes, poriferans, and choanoflagellates (30–32). The same domain structure is conserved across most metazoans, indicating that the overall structure and function of Eph receptors are likely well conserved throughout metazoan evolution (33). The Eph kinase domain conserves prototypical features of the protein kinase domain, which comprises the N-terminal ATP-binding lobe (N-lobe) and the C-terminal substrate-binding lobe (C-lobe) (9, 11, 34).
In this study, we use a Bayesian statistical framework (35) to identify sequence features most characteristic of the Eph family. We show for the first time that nearly all residues that distinguish the Eph family from other tyrosine kinases occur on the surface of the protein and that some of the most distinguishing residues tether flanking protein segments to the kinase domain. The selective conservation of these residues within the Eph family suggests that they play important roles in Eph kinase functions. Single mutations of juxtamembrane and SAM linker tethering residues both resulted in more rapid activation of EphA3, for the first time highlighting the SAM domain linker as a critical functional component in the activation of Eph kinases. Simultaneous mutation of both networks demonstrated that the SAM linker plays a regulatory role via allosteric coupling to the juxtamembrane. The emerging model of Eph evolution spotlights the coevolution of the juxtamembrane and the SAM linker with the kinase domain to precisely coordinate kinase catalytic and regulatory functions. We also identify highly distinct Eph-specific motifs of unknown functions that warrant further investigation.
Results
Unique sequence features distinguish the Eph family from other tyrosine kinases
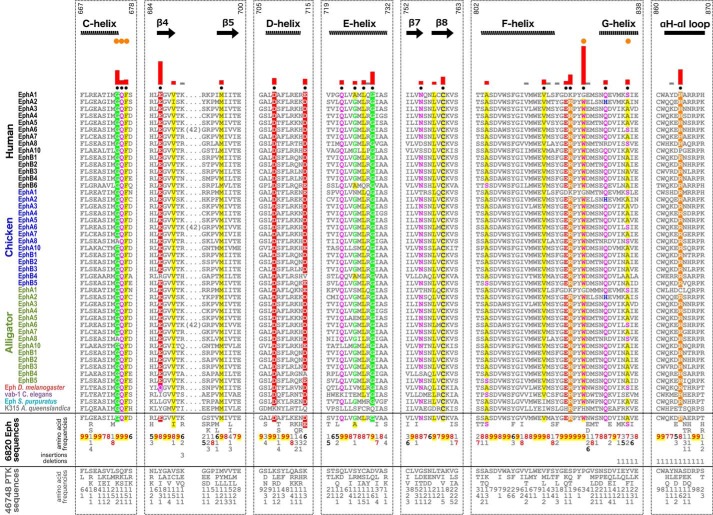
To identify sequence features that most distinguish Eph kinases from other tyrosine kinases, we systematically compared large and diverse sequence sets of Eph kinases and non-Eph tyrosine kinases using computational methods described previously (35). We note that the regions flanking the Eph kinase domain, namely the N-terminal juxtamembrane and the C-terminal SAM domain linker, are unique to the Eph family in that they share detectable sequence similarity within the Eph family but share no similarity with kinase sequences outside of the Eph family (Fig. S2). The juxtamembrane and SAM domain linker are both well conserved, sharing 47.9 and 45.9% sequence similarity, respectively, within the Eph family. Eph kinases are the only family of tyrosine kinases with a conserved SAM domain; therefore, the SAM domain and the SAM domain linker are unique to Eph kinase sequences.
To identify sequence constraints that distinguish the Eph kinase domain from other tyrosine kinase domains, we used a Bayesian pattern partitioning procedure, which classifies multiply aligned sequences based on patterns of amino acid conservation and variation (35). Residues that are highly conserved in Eph kinase sequences but non-conserved and/or biochemically dissimilar in non-Eph tyrosine kinase sequences are highlighted in Fig. 1 where the histograms represent the relative strengths of the constraints imposed on these residues. Sequence constraints identified across the full kinase domain alignment are shown in Fig. S3. Some Eph-specific residues occur in the N-lobe, specifically in the αC helix, β4 strand, and β5 strand. Additionally, highly distinguishing Eph-specific residues are scattered throughout the C-lobe and located in the αD/αE/αF/αG helices, β7/β8 strands, αF-αG loop, and αH-αI loop.
Figure 1.
Unique sequence features defining the Eph family kinase domain. Columns are highlighted where amino acids are highly conserved in Eph family sequences and non-conserved and/or biochemically dissimilar in other tyrosine kinase (PTK) sequences. Histograms quantify the degree of divergence between Eph and other tyrosine kinase sequences. Column-wise amino acid and insertion/deletion frequencies are indicated in integer tenths where a “5” indicates an occurrence of 50–60% in the given (weighted) sequence set. Columns used by the Bayesian partitioning procedure to sort Eph sequences from other tyrosine kinase sequences are marked with black dots. Kinase secondary structures are annotated above the alignment. Eph-specific residues that participate in juxtamembrane and SAM linker tethering networks are marked by orange dots. Sequence numbering above the alignment corresponds to the human EphA3 sequence.
In the following sections, we first provide a brief overview of the structural locations of Eph-specific residues. Next, we report our detailed structural and experimental analyses of Eph-specific residues that take part in well conserved networks involving the unique N- and C-terminal segments flanking the kinase domain. Eph-specific residues of unknown functions were also investigated and are reported below.
Structural location of Eph-specific residues
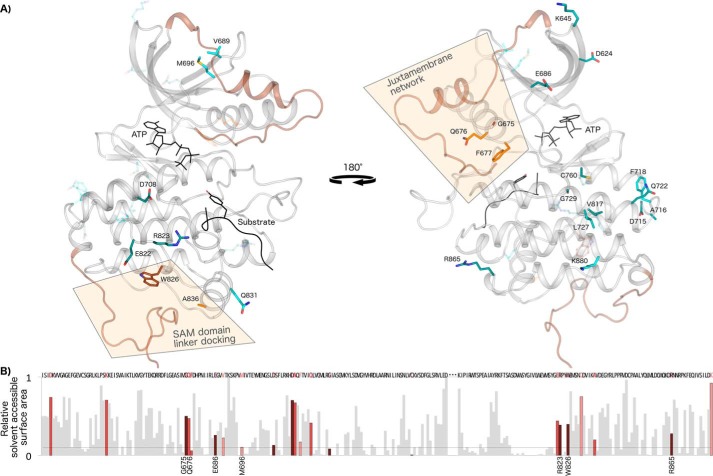
Sequence features distinguishing the Eph kinase domain are dispersed in primary sequence; however, a structural mapping of these residues onto crystal structures of EphA3 shows that Eph-specific residues cluster into networks that are largely located on the surface of the protein (Fig. 2). One of these networks has previously been noted to be significant for Eph function and tethers the unique juxtamembrane segment to the kinase domain (Fig. 2A). However, many other Eph-specific residues form networks dispersed across the N-lobe, hinge region, and C-lobe of the kinase domain whose functions are yet to be determined. To shed light on the roles of these conserved residues, we performed detailed structural analyses and mutational studies using human EphA3 as a model Eph family kinase. In our experimental analysis, we mutated Eph-specific residues to those observed in other tyrosine kinases and determined the impact of mutations on kinase activation via autophosphorylation. We also determined the effects on substrate phosphorylation using enolase as a generic protein substrate.
Figure 2.
Networks of Eph-specific residues in distinct functional regions of the kinase tertiary structure. A, key distinguishing residues of the Eph family mapped onto a model structure of human EphA3. ATP and peptide substrate (Protein Data Bank code 3FY2) are shown in black. The juxtamembrane (modeled from Protein Data Bank code 2QO2) and the SAM domain linker (modeled from Protein Data Bank code 2QOC) are shown in brown. The activation loop was modeled using active EphA2 (Protein Data Bank code 4TRL) and active human Lck as a template (Protein Data Bank code 3LCK). The 25 most distinguishing Eph-specific residues (columns with black dot in Fig. 1) are shown as sticks, which are colored darker to lighter based on the greater or lesser contrast between Ephs and other tyrosine kinases at that position. B, relative solvent-accessible surface area of residues across the kinase domain calculated from an active EphA3 structure (Protein Data Bank code 2QOC). Eph-specific residues are highlighted in dark to light red relative to the strength of the sequence constraint, with darker red indicating more highly constrained residues. Data for the disordered activation loop is omitted and marked by ellipses.
Eph-specific networks tether unique flanking segments onto the kinase domain
Eph-specific interactions between the kinase domain and juxtamembrane
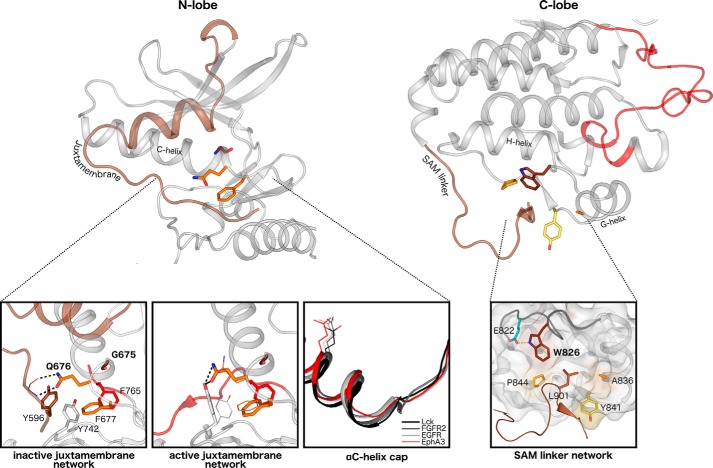
The “GQF” motif in the αC-β4 loop (Fig. 1) forms a network of critical interactions with the juxtamembrane, and these interactions have been highlighted in previous biochemical and structural studies on various Eph members (8–13). In crystal structures of autoinhibited Eph kinase domains, the N-terminal juxtamembrane phosphorylation site, Tyr596 (EphA3 numbering), docks into a pocket formed by Eph-specific residues Phe677 and Gln676. The side chain of Gln676 also forms two hydrogen bonds to the backbone of the juxtamembrane (Fig. 3) (9). This autoinhibitory configuration prohibits the activation loop from adopting an active conformation due to steric hindrance by another important tyrosine residue in the network, Tyr742 (9). In comparison, the active structure of EphA3 exhibits shifts in the side chain orientations of residues in this network. For example, the hydrogen bonds by Gln676 to the juxtamembrane backbone are instead satisfied by the side chain of Tyr742, which is rotated away from the active cleft in a manner that allows the open conformation of the activation loop (Fig. 3). Gly675 is the most highly distinguishing residue in the GQF motif (Fig. 1) and has not yet been recognized for its role in juxtamembrane function in previous studies. This glycine residue caps the C-terminal end of the αC helix, causing the αC helix to be a half to full turn shorter than observed in other tyrosine kinases (Fig. 3). Interestingly, a variation of the GQF motif is observed in chicken and alligator EphA8 sequences, which harbor a biochemically similar “AQF” variation, while all other canonical residues of the juxtamembrane network are conserved (Fig. 1). Through further taxonomic analyses of EphA8 sequences, we have determined that the alanine variant of the GQF motif is selectively conserved in non-mammalian vertebrate EphA8 sequences.
Figure 3.
Structural interactions in the juxtamembrane network and SAM domain linker network. Left, key interactions among the juxtamembrane, GQF motif, and kinase domain residues are highlighted in the autoinhibited (Protein Data Bank code 2QO2) and active (Protein Data Bank code 2QOC) structures of EphA3. The juxtamembrane tyrosine is shown in brown, the tethering GQF motif is in orange, and the DFG phenylalanine is in red. Different side chain occupancies of residues observed in the active structure are shown as lines. An alignment of αC helices of various tyrosine kinases reveals the earlier helix capping observed in Eph structures. The conserved ATP-binding αC helix glutamate is shown as a reference for the structural alignment. Right, interactions between the SAM domain linker and the C-lobe of the kinase domain are highlighted in an active structure of EphA3 (Protein Data Bank code 2QOC). The hydrophobic pocket in the C-lobe is depicted in surface representation, and the surface of Eph-specific residues that make up the pocket is colored. The αF-αG loop backbone is highlighted in dark gray.
Eph-specific interactions between the kinase domain and SAM domain linker
Another group of Eph-specific residues maps to the basal face of the C-lobe and forms a network of interactions tethering the SAM domain linker to the kinase domain. Although these tethering interactions have been observed in multiple inactive and active EphA3 structures (9), the functional significance of these interactions has not been explored. In crystal structures where the SAM domain linker is ordered, a leucine in the linker (Leu901 in EphA3) is tethered onto the C-lobe of the kinase domain between the αF-αG loop, αG helix, and αG-αH loop (9). The side chain of Trp826, which is the most distinguishing Eph-specific residue in this network, forms a hydrophobic tethering pocket along the αF-αG loop. The αF-αG loop is observed in a unique conformation that is divergent from other tyrosine kinase structures (see Fig. 6), and this conformation is associated with a hydrogen bond between the indole nitrogen of Trp826 to the side chain oxygen of another Eph-specific residue in the αF-αG loop, Glu822. The hydrophobic tethering pocket is additionally formed by other Eph-specific residues (Ala836 in αG helix and Pro844 and Tyr841 in the αG-αH loop).
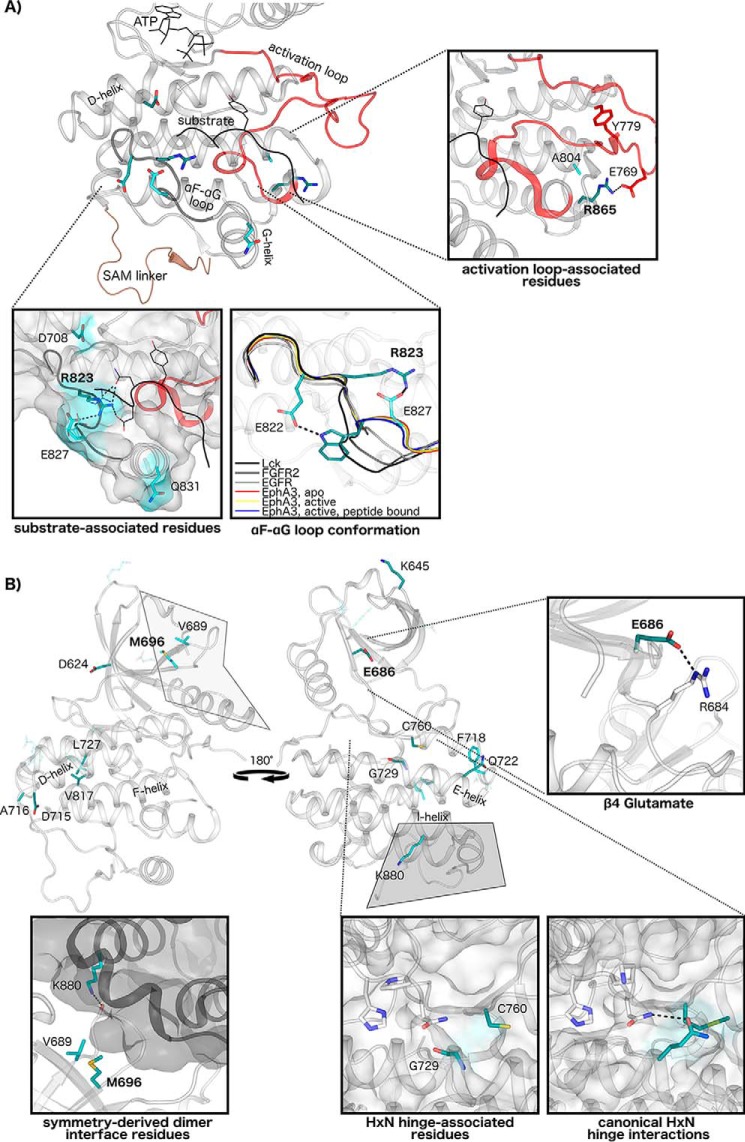
Figure 6.
Structural interactions of Eph-specific residues of unknown function. A, Eph-specific residues contribute to unique interactions in the substrate-binding region and activation loop. Key conserved residues in this region are mapped to structures of human EphA3, with darker shading indicating greater sequence constraints at that position. ATP and peptide substrate are shown in black, the activation loop is in red, and the SAM domain linker is in brown. Interactions of Eph-specific residues and peptide substrate are shown, with different rotamer conformations between peptide-bound (shown as sticks) (Protein Data Bank code 3FXX) and peptide-unbound structures (shown as lines) (Protein Data Bank code 2QOC). The unique conformation of the αF-αG loop in Eph structures is shown relative to other tyrosine kinase structures: EphA3, apo (Protein Data Bank code 2QO2); EphA3, active (Protein Data Bank code 2QOC); EphA3, active, peptide-bound (Protein Data Bank code 3FXX), Lck (Protein Data Bank code 3LCK); FGFR2 (Protein Data Bank code 2PVF); and epidermal growth factor receptor (EGFR) (Protein Data Bank code 2GS2). Interactions of Eph-specific residues Arg865 and Ala804 with the activation loop are shown (EphA2, Protein Data Bank code 4TRL). B, Eph-specific residues in the N-lobe and hinge region. Interactions of the β4 glutamate are shown (Protein Data Bank code 2QOC). The N-lobe-to-C-lobe interface in a symmetry-derived dimer observed in several EphA3 structures is shown with key Eph-specific residues in the interface (Protein Data Bank code 2QOC) highlighted. Lys880 of this network forms a hydrogen bond to a backbone residue in the β4-β5 loop in the dimeric partner. Comparisons between canonical HXN hinge network interactions, which are conserved across diverse protein kinases, and the Eph family HXN hinge network are shown from representative structures (FGFR2, Protein Data Bank code 2PVF; EphA3, Protein Data Bank code 2QOC).
Mutation of N- and C-lobe tethering networks results in more rapid activation of EphA3
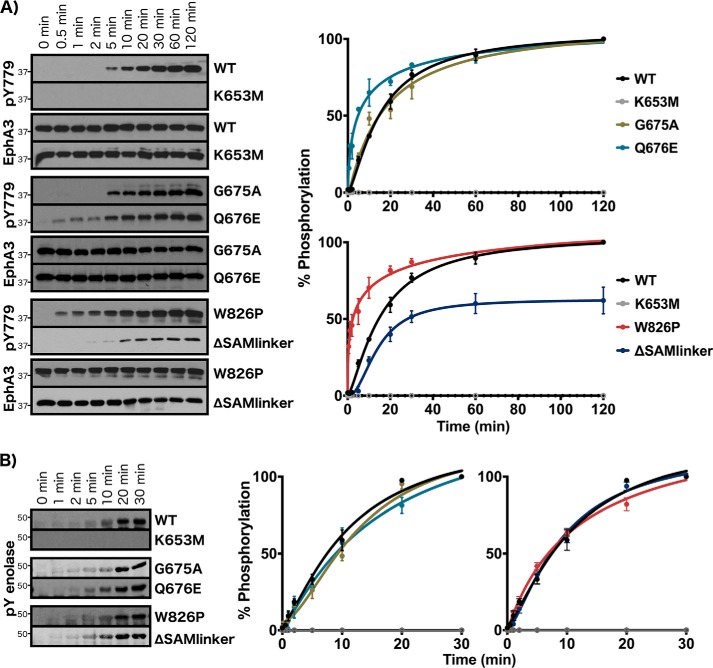
To shed light on the roles of these N-lobe and C-lobe tethering networks, we mutated select Eph-specific residues in these networks to residues observed at the equivalent position in other tyrosine kinases and determined the impact of mutations on autophosphorylation and enolase phosphorylation. As seen in Fig. 4, WT EphA3 begins to exhibit activation loop autophosphorylation (pTyr779) after 5 min of incubation with MgATP and approaches maximum phosphorylation levels at 1 h (Fig. 4). To validate the role of the GQF motif from the juxtamembrane network, we mutated the GQF glutamine (Gln676) to glutamate, which is found at this position in other tyrosine kinase sequences such as Abl and Musk. We also examined the functional impact of the AQF variant by mutating the GQF glycine (Gly675) to alanine, which is observed in select EphA8 sequences. The glycine mutant, G675A, exhibited similar activation time to WT EphA3, whereas the glutamine mutant, Q676E, achieved much faster activation with autophosphorylation detected as early as 30 s (Fig. 4A). In contrast, neither the Q676E nor G675A mutant exhibited any differences compared with WT with respect to rates of enolase phosphorylation (Fig. 4B). In the SAM domain linker network, we mutated the tryptophan (Trp826) to proline, which is the residue observed in over 30% of other tyrosine kinase sequences. We also perturbed this network via a truncation mutant that removes the SAM domain linker at position Thr892 (ΔSAMlinker). The W826P mutant exhibited significantly increased rates of autophosphorylation with high levels of autophosphorylation detected at 30 s. In contrast, the ΔSAMlinker mutation reduced rates of autophosphorylation, with activation loop phosphorylation detected only after 10 min, and decreased total autophosphorylation levels to about 60% compared with WT (Fig. 4A). Neither of the SAM linker network mutations caused any observable effect on enolase phosphorylation (Fig. 4B). We additionally characterized the rates of activation and peptide phosphorylation for WT and W826P proteins using an NADH-coupled assay, which also showed that the W826P mutant achieves faster rates of activation compared with WT (Table S1).
Figure 4.
Experimental analysis of juxtamembrane and SAM linker network residues. A, autophosphorylation time series of juxtamembrane and SAM linker network mutants. Phosphorylation levels of the activation loop site were detected (pTyr779). B, enolase phosphorylation time series. Phosphorylation levels in A and B were quantified by densitometry and normalized against total protein levels. Error bars represent standard deviation between three independent experiments.
SAM linker tethering residue is coupled to juxtamembrane autoregulatory function
Mutational analysis of the highly distinct SAM linker tethering residue (W826P) resulted in significantly more rapid activation similar to that seen when directly mutating the autoinhibitory juxtamembrane network (Q676E). Because the most critical step in Eph activation is autophosphorylation of the juxtamembrane, which is believed to precede any other autophosphorylation event, we sought to determine whether the mutation directly affected juxtamembrane autophosphorylation. We used MALDI peptide mass fingerprinting and LC-MS/MS to identify phosphorylated juxtamembrane residues (pTyr596 and pTyr602) for WT and W826P samples after 30 s and after 10 min of incubation with MgATP (Table S2 and Figs. S4 and S5). For the W826P mutant, we detected the singly phosphorylated juxtamembrane peptide for the 30-s sample, and we detected the doubly phosphorylated juxtamembrane peptide for the 10-min sample. In contrast, only the singly phosphorylated juxtamembrane peptide was readily detected for WT for the 10-min sample. Further LC-MS/MS and CID analysis of the W826P 30-s sample was used to distinguish between phosphorylated N-terminal and C-terminal tyrosine residues in the juxtamembrane. Our data showed that the juxtamembrane peptide phosphorylated on the C-terminal tyrosine is readily detected in contrast to the N-terminally phosphorylated or doubly phosphorylated peptide (Figs. S6–S8), corroborating that the C-terminal tyrosine is phosphorylated first in the sequence of autophosphorylation events as has been determined in a previous study (8).
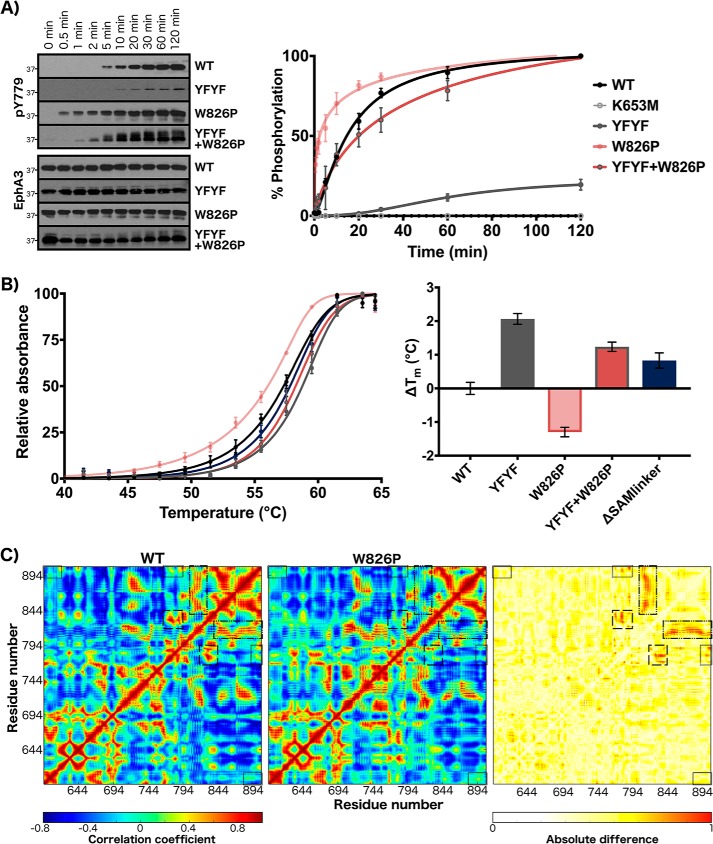
In addition, we produced double and triple mutations at sites in the juxtamembrane and SAM linker networks to investigate the cooperativity of these networks. We produced a YFYF mutant with double tyrosine-to-phenylalanine mutations in the juxtamembrane as well as a triple mutant containing the W826P mutation in addition to the YFYF mutations. Substitution of both juxtamembrane tyrosines to phenylalanine was previously found to completely abolish catalytic activity in various Eph family kinases (8–13). We observed that the juxtamembrane double mutant (YFYF) showed almost no autophosphorylation activity with minor levels of autophosphorylation detected from 20 min to 2 h (Fig. 5A). Interestingly, the W826P mutation, which significantly increased the rate of EphA3 activation when examined as a single mutant, was able to rescue activity when added in the YFYF double mutant background (Fig. 5A). The triple YFYF+W826P mutant exhibited significant levels of autophosphorylation by 10–20 min similar to WT EphA3 but lesser than levels exhibited by the single mutant alone (W826P).
Figure 5.
Characterization of coupled behavior between the juxtamembrane and SAM domain linker. A, autophosphorylation time series of WT, YFYF, W826P, and YFYF+W826P mutants. Phosphorylation levels of the activation loop site were detected (pTyr779). B, thermal stability analysis. The change in Tm for the mutants was calculated with respect to WT. Error bars represent standard deviation between three independent experiments. C, cross-correlation analysis for 500-ns WT and W826P molecular dynamics trajectories. Correlation coefficient values are colored from red (positive) to blue (negative). Cross-correlations between the juxtamembrane (residues 595–620) and SAM linker (residues 892–906) are highlighted in a black box. Also highlighted are cross-correlations of the αF-αG loop (residues 820–829) with the αG-αH loop (residues 840–849), αH helix (residues 850–860), αH-αI loop (residues 861–869), αI helix (residues 870–891), and SAM linker in the large dashed-dotted box; cross-correlations between the activation loop (residues 764–794) and αF-αG loop/αG helix (residues 820–839) in the long-dashed box; and cross-correlations between the activation loop and SAM linker in the short-dashed box. A difference map of cross-correlation coefficients between WT and W826P simulations (right) shows that these highlighted regions exhibit the greatest difference.
To determine whether the mutations may be disrupting autoinhibition by destabilizing the autoinhibited state, we examined the global protein stability of the inactive, dephosphorylated forms of WT EphA3 and its mutants through a dye-based thermal shift assay. As shown in Fig. 5B, the W826P mutation decreased global stability and resulted in a Tm 1.5 °C lower than the Tm of WT. The addition of the YFYF double mutation had a stabilizing effect on both the WT protein and the W826P mutant. The YFYF mutant exhibited a Tm 2 °C higher than the Tm of WT, and the YFYF+W826P triple mutant resulted in a 2.5 °C increase in Tm relative to the W826P single mutant. We also examined the thermal stability of the SAM domain linker deletion mutant, ΔSAMlinker, which was slightly more stable than WT with a ΔTm of +0.83 °C. The ΔSAMlinker mutant was also much more stable than the W826P mutant by 2.1 °C. In addition, we used molecular dynamics simulations to analyze the dynamics and stability of autoinhibited models of WT and W826P EphA3. As shown in Fig. 5C, the W826P mutation increases the positive correlative motions between the juxtamembrane (residues 595–620) and SAM linker (residues 892–906). The regions that show the most difference in cross-correlated motions between WT and W826P are also highlighted in Fig. 5C and include the activation loop (residues 764–794), αF-αG loop (residues 820–829), SAM linker, and other regions in the C-lobe. Notably, the positive correlations between the αF-αG loop and other regions of the C-lobe (αG-αH loop, αH and αI helices, and SAM linker) are markedly decreased in the W826P mutant. Similarly, positive correlations between the activation loop and the substrate-binding regions (αF-αG loop/αG helix) and between the activation loop and SAM linker are decreased in the W826P mutant as well (Fig. S9). We also note that the W826P mutation produces long-range effects that increase the flexibility of the juxtamembrane backbone (Fig. S9). This increased juxtamembrane flexibility is also mirrored in the destabilization of key autoinhibitory interactions between the juxtamembrane backbone and Eph-specific Gln676. Whereas the distance between interacting nitrogen and oxygen atoms remains stable between 2.5 and 3 Å through the entire duration of the WT simulation, the distance between interacting atoms in the W826P simulation fluctuates above the ideal distance for electrostatic interactions after 250 ns (Fig. S9).
Eph-specific networks of unknown function
Eph-specific residues of unknown functions are located in the β1, β3, β4, and β5 strands of the N-lobe and in the αD-αE loop, αE helix, and β7/β8 strands in the hinge region of the kinase domain. Several Eph-specific residues additionally map to the C-lobe, spanning the αF helix, αF-αG loop, αG helix, αH-αI loop, and αI helix (Fig. 6). Here, we describe the structural interactions observed for these residues in light of their uniqueness relative to other tyrosine kinase structures; however, the precise roles of these conserved residues are not readily apparent from available crystal structures alone.
Eph-specific residues in the C-lobe contribute to several unique structural interactions in the substrate-binding region, which includes the αD and αG helices, αF-αG loop, and activation loop (Fig. 6A). Two residues that appear to be involved in substrate binding, Arg823 and Glu827, may be connected to the SAM linker tethering network via the αF-αG loop, which contains the SAM linker docking residue, Trp826, and is observed to be in a highly unique conformation relative to the αF-αG loop conformations seen in other tyrosine kinase structures (Fig. 6A). Many distinguishing Eph-specific residues also broadly map to the N-lobe and hinge region of the kinase domain (Fig. 6B). The most distinctive residue networks include a charged three-residue network in the N-lobe comprising Asp624, Lys645, and Glu686; a hydrophobic network in the N-lobe comprising Met696 and Val689; and a large network spanning the hinge region to the αD, αE, and αF helices in the C-lobe, including the highly distinctive residues Cys760 and Gly729 (Fig. 6B).
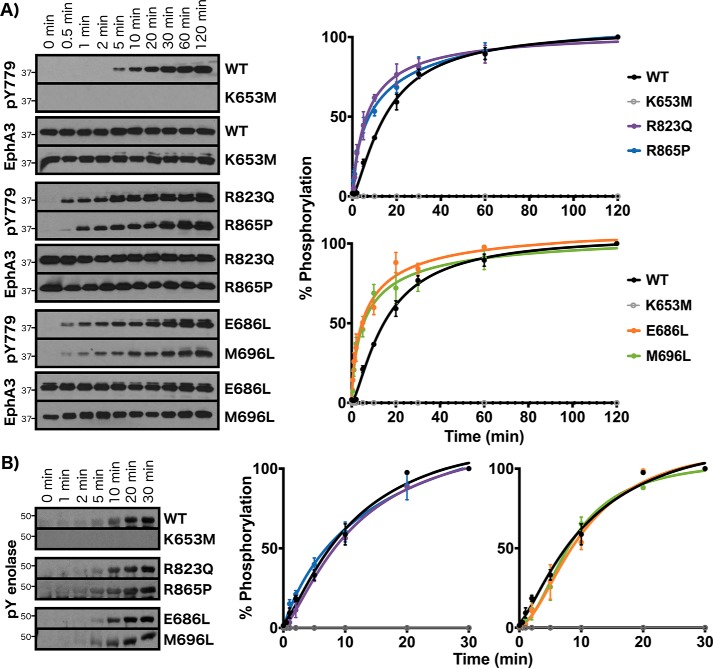
We performed an initial characterization of Eph-specific residues of unknown function by mutating conserved residues in distinct regions of the C-lobe and N-lobe in EphA3 and determining the effects on autophosphorylation and enolase phosphorylation. Mutation of the substrate-associated residue Arg823 to a glutamine (R823Q), which is conserved at the equivalent position in other receptor tyrosine kinases (i.e. Ror and discoidin domain–containing receptor families), resulted in faster autophosphorylation relative to WT (Fig. 7A). Similar to the juxtamembrane and SAM linker tethering mutants (Q676E and W826P), significant levels of autophosphorylation were detected for the R823Q mutant after just 30 s of incubation with ATP. Similarly, mutation of the activation loop–associated Arg865 to proline, which is observed in over 60% of other tyrosine kinase sequences, caused an increase in the rate of autophosphorylation relative to WT, although this mutant was activated slightly less quickly than the R823Q mutant (significant autophosphorylation levels detected after 1 min). In the N-lobe, both the β4 strand glutamate and the β5 strand methionine were mutated to leucine (E686L and M696L), which is the most common residue found at the equivalent positions in other tyrosine kinases. Similar to mutations in the C-lobe, both mutations (E686L and M696L) caused increases in the rate of autophosphorylation with respect to WT (Fig. 7A). Interestingly, although R823Q, R865P, E686L, and M696L mutants all exhibited increased rates of autophosphorylation, no effects were observed on enolase phosphorylation compared with WT (Fig. 7B).
Figure 7.
Experimental analysis of Eph-specific residues of unknown functions. A, autophosphorylation time series of WT, kinase-dead (K653M), R823Q, R865P, E686L, and M696L mutants. Phosphorylation levels of the activation loop site were detected (pTyr779). B, enolase phosphorylation time series. Phosphorylation levels in A and B were quantified by densitometry and normalized against total protein levels. Error bars represent standard deviation between three independent experiments.
Discussion
Eph kinases have evolutionarily diverged from other tyrosine kinases to respond to specific temporal and cellular signals in signaling pathways. Here, we have identified strikingly conserved sequence features shared among Eph kinases and across extremely diverse evolutionary phyla that tether the flexible juxtamembrane and SAM domain linker to the kinase domain, and we have also discovered novel Eph-specific residues of unknown function. Mutation of the most highly distinguishing Eph-specific residues from various regions of the kinase domain resulted in significantly reduced activation times with negligible effects on enolase phosphorylation. We also show for the first time that the SAM linker and its tethering interactions are essential for the regulation of activation through allosteric coupling of the SAM linker to the critical autoregulatory juxtamembrane.
Linker regions in multidomain proteins have been commonly observed in protein kinases and other diverse proteins to play autoregulatory roles (24, 27, 36–38), promote protein interactions (27, 28, 36, 39, 40), and serve as evolutionary hot spots for neofunctionalization (41, 42). The identification of highly distinct Eph-specific residues that tether the N- and C-terminal flanking segments indicates that these flexible segments have evolved to play important and conserved functional roles unique to this family of tyrosine kinases. Mutation of the juxtamembrane tethering GQF motif corroborated the autoinhibitory role of the juxtamembrane, which has previously been characterized in several biochemical and structural studies (8–13). The replacement of the GQF glutamine with glutamate caused faster activation, indicating that the glutamine residue and its electrostatic interactions with the juxtamembrane backbone are indispensable for proper autoinhibition. Although the role of the glutamine has been noted in previous studies (11), we identify for the first time the GQF glycine as a critical part of the juxtamembrane network. Despite it being the most uniquely conserved residue of the network, we identified an alanine variant of this residue in non-mammalian, vertebrate EphA8 sequences (Fig. 1). We observed that producing this variation in EphA3 (G675A) resulted in an activation time similar to that of WT EphA3 (Fig. 4). These results reflect that the AQF variant is well tolerated with respect to autoregulatory function; however, this variation could reflect functional specialization of mechanisms other than autophosphorylation. We note that the GQF glycine is also a recurrent hot spot for cancer-associated variants. The glycine has been observed to be mutated in seven different cancer samples across three different Eph members (EphA7, EphA8, and EphB3) to arginine, glutamate, valine, and cysteine (supporting Data Set 1).
In addition, we investigated the unique network of Eph-specific residues that tether the C-terminal SAM domain linker to the C-lobe of the kinase domain. Although the functional relevance of the SAM linker has remained elusive, the unique conservation of residues that tether the linker reflects an evolutionary pressure to maintain the SAM linker tethering pocket and indicates that the linker and its interaction with the kinase domain play an important role. Through mutational analysis of the SAM linker network, we discovered that the SAM domain linker is important for activation via autophosphorylation but dispensable for enolase phosphorylation. Mutation of the SAM linker tethering tryptophan residue, Trp826, which is the most distinguishing residue of the Eph family, significantly increased the speed of activation. Structurally, we expect that the W826P mutation would cause ineffective tethering of the SAM linker by reorienting the αF-αG loop portion of the interface and by widening the tethering pocket on the basal face of the C-lobe. The weaker tethering of the SAM linker via the W826P mutation is corroborated by our thermal stability studies, which showed significantly decreased global stability of the W826P mutant with respect to WT (Fig. 5), and by our molecular dynamics studies, which showed increased SAM linker dynamics in the W826P mutant (Fig. S9). Further studies of the SAM linker network via a SAM linker deletion mutant surprisingly showed that the ΔSAMlinker mutant exhibited diminished autophosphorylation activity. Thermal stability analysis of this mutant showed increased global stability compared with WT, which suggests that the SAM linker, when present, contributes to the general flexibility of the WT protein. Interestingly, we note that the SAM linker tethering tryptophan is substituted with glycine in EphA1 sequences (Fig. 1), which reflects a drastic variation of the SAM linker network. We predict that this variation would have dramatic effects on Eph-specific interactions in the αF-αG loop and would prevent effective tethering of the SAM linker. Given the effects of the W826P mutation on EphA3 activation, this variation may allow EphA1 kinases to autophosphorylate and be activated more rapidly than other Eph members. However, the possible functional consequences of this variation and whether there are EphA1-specific variations that compensate for the tryptophan-to-glycine substitution warrant further investigation. Furthermore, the tryptophan in EphA7 is observed to be mutated to leucine in two cancer samples (supporting Data Set 1). Whether this disease variant of EphA7 contributes to abnormal signaling and/or cancer progression should also be explored.
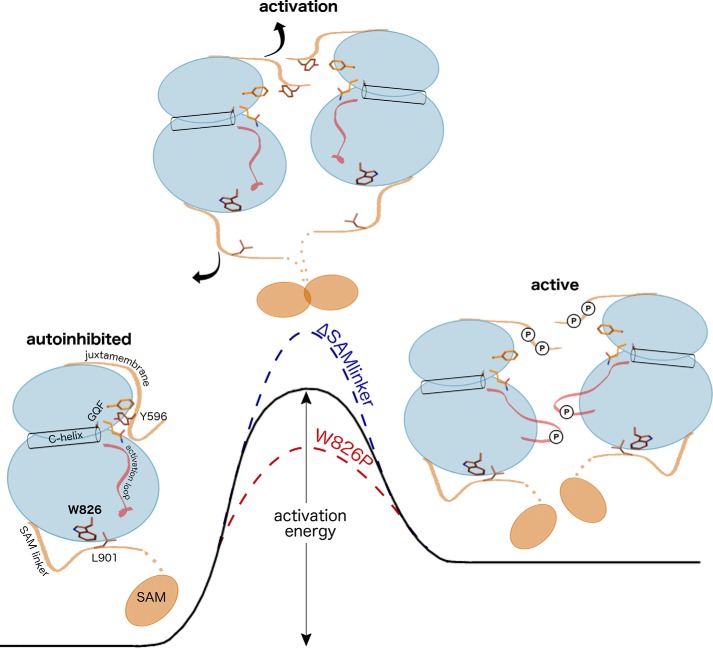
The strikingly faster activation by the W826P mutant was unexpected given the distal location of the SAM linker network to the activation loop or to the juxtamembrane, which to this point have been the only identified autoinhibitory regulators of Eph activity. We confirmed through mass spectrometry studies that the effects of the W826P mutation on activation were directly linked to changes in the rate of juxtamembrane autophosphorylation. In addition, by examining the effects of coupled mutations between the juxtamembrane and SAM linker networks, we found that the SAM linker network adds an additional layer of regulation to the autoinhibitory juxtamembrane to regulate autophosphorylation. Specifically, the W826P mutation was able to rescue activity when added to the nearly inactive YFYF juxtamembrane mutant, which demonstrates that this mutation is able to communicate to the distal juxtamembrane and disrupt its autoinhibitory function despite the lack of structural and biochemical changes induced by phosphorylation. In light of these results, we propose a model where the C-terminal SAM domain linker plays an essential and cooperative role in disengaging the conserved juxtamembrane autoinhibitory mechanism during activation (Fig. 8). Given the faster activation by the W826P mutant, we believe that the untethered, flexible SAM domain linker is able to allosterically disrupt the juxtamembrane from its autoinhibitory conformation, as supported by our molecular dynamics studies (Fig. S9), and cause the autophosphorylation of the juxtamembrane to occur more readily. We also conclude that the SAM domain linker is essential for juxtamembrane autophosphorylation and kinase domain activation to occur because the SAM domain deletion mutant exhibited decreased autophosphorylation efficiency. The co-conservation of the coupled SAM linker and juxtamembrane networks highlights the evolution of an allosteric communication network that uses both flanking segments to regulate the Eph kinase domain.
Figure 8.
Proposed energetic model for Eph activation by the juxtamembrane and SAM domain linker. The initial step of Eph activation occurs with the displacement of the juxtamembrane from its autoinhibitory conformation. Displacement of the autoinhibitory conformation occurs with an increase in entropy and is facilitated by 1) autophosphorylation of the juxtamembrane and/or 2) untethering of the SAM domain linker. This allows for further autophosphorylation to occur (i.e. autophosphorylation on the activation loop shown in red) to complete the activation process. SAM domain network mutants affect the dynamics of the autoinhibitory juxtamembrane, altering the activation energy required for activation.
Conserved allosteric communication networks have been observed in many protein kinases that couple distal protein regions and molecular events such as substrate binding, post-translational modifications, and protein interactions to the active site in ways that impact catalytic activity (25, 27, 29, 43–46). We believe that the SAM linker comprises part of an allosteric communication network in Eph kinases that allows the coupling of other important functional regions to the SAM linker and juxtamembrane. For example, oligomerization by the SAM domain may trigger the untethering of the SAM domain linker from the kinase domain and subsequently prime the juxtamembrane for autophosphorylation. Furthermore, as suggested by our molecular dynamics analysis (Fig. 5C and Fig. S9), the untethering of the SAM linker may allosterically affect the conformation of other functionally important regions such as the activation loop or substrate-binding region through other Eph-specific residues such as Arg865 and Arg823 (Fig. 6). Although it was unexpected that the R823Q mutation did not negatively affect autophosphorylation or substrate phosphorylation despite its location in the substrate-binding pocket, whether this result is attributed to changes in substrate recognition or other conformational changes remains unknown. It is also worth noting that Eph-specific residues, most of which are located on the protein surface distal to known juxtamembrane-, SAM linker–, or active site–associated regions, may serve as protein–protein interaction sites that can communicate protein interaction events to regulatory networks within the kinase domain. In light of this view, we have observed Eph-specific residues that comprise the interface in a symmetry-related dimer, although the biological relevance of this dimer remains unknown. We also note that the same position of the β4 glutamate, which is the second most distinguishing Eph-specific residue, has been observed in other tyrosine kinases such as Abl and Src to be important in regulatory inter- and intramolecular interactions (47, 48); this common docking site may have been refashioned as a unique regulatory interaction site in Eph kinases as well. In this study, mutation of Eph-specific residues resulted in faster activation in all but one case (G675A); therefore, further investigating how these residues may allosterically affect catalysis and regulation and how they may be involved in protein interactions and pathways will be important directions for future studies.
Materials and methods
Identification of Eph-specific sequence features
We used a data set of 53,568 tyrosine protein kinase sequences identified from the NCBI-nr protein database using curated hierarchical sequence profiles generated using the MAPGAPS suite (49). The 53,568 sequences were aligned using MAPGAPS, and this alignment was used as an input for mcBPPS (35). N-terminal juxtamembrane and C-terminal SAM linker and SAM domain sequences were identified using Pfam domain (50) and TMHMM transmembrane (51) predictions and then aligned using Clustal Omega (52).
Expression and purification of EphA3 WT and mutant proteins
WT and mutant EphA3 proteins were expressed and purified as described previously (25). The EphA3 construct used in the study corresponds to residues Asp577–Ser947 with an N-terminal His6 tag. Additional details are provided in the supporting information.
Autophosphorylation assays
To obtain starting samples for autophosphorylation assays, we produced inactive, dephosphorylated EphA3 by incubating 25–50 mg of purified protein samples with 100 units of CIP (New England Biolabs) overnight at 4 °C. The CIP was removed by purification on nickel-nitrilotriacetic acid-agarose columns. The complete dephosphorylation and removal of CIP was verified using Western blotting with anti-pTyr779 (Cell Signaling Technology, 8862) and anti-CIP (Fitzgerald, 20C-CR2110RP) antibodies. For autophosphorylation reactions, reactions were set up with a final reaction volume of 20 μl and with the following final concentrations: 0.375 mg/ml EphA3, 5 mm ATP, and 10 mm MgCl2. To quench the reaction at each time point, 1 μl of the reaction was diluted into 74 μl of SDS-PAGE sample buffer, and then the sample was boiled for 5 min. Additional details of Western blotting methods are provided as supporting information.
Enolase phosphorylation assays
To obtain starting samples for enolase phosphorylation assays, we produced fully active EphA3 by incubating 25–50 mg of purified protein samples with ATP and MgCl2 at final concentrations of 10 and 20 mm, respectively, for 6 h at 4 °C. Excess MgATP/ADP was dialyzed out overnight. For enolase phosphorylation reactions, rabbit muscle enolase (Sigma) was denatured in 25 mm acetic acid for 15 min. Reactions were set up with a final reaction volume of 250 μl containing 70 μg of denatured enolase and the following at the indicated final concentrations: 0.1 mg/ml EphA3, 50 μm ATP, and 25 mm MgCl2. To quench the reaction at each time point, 30 μl of the reaction was mixed into 10 μl of SDS-PAGE sample buffer, and then the sample was boiled for 5 min.
Western blotting
Samples were separated by 10% SDS-PAGE and transferred to PVDF membranes. EphA3 phosphorylation levels were detected by Western blotting using anti-pTyr779 antibody (Cell Signaling Technology, 8862), and total EphA3 levels were detected using anti-EphA3 antibody (Cell Signaling Technology, 8793). Enolase phosphorylation levels were detected by Western blotting using anti-pTyr100 antibody (Cell Signaling Technology, 9411).
Thermal melt assays
Thermal melt assays were performed as described previously (25). In brief, thermal melt assays were performed by quantifying SYPRO Orange dye (Sigma) fluorescence as a reporter for protein unfolding. Reactions were set up with a final volume of 100 μl with final EphA3 concentrations of 0.05–0.1 mg/ml. SYPRO Orange dye was used at a final dilution of 1:8000. These assays were performed in a Synergy H4 microplate reader with increasing temperature from 25 to 65 °C with a step size of 2 °C. Fluorescence was measured with excitation at 470 nm and emission at 570 nm. For all experiments, Tm values were calculated by fitting data to a Boltzmann sigmoidal curve.
Mass spectrometry analysis of autophosphorylated samples
WT and W826P 30-s and 10-min autophosphorylation samples were analyzed by in-gel trypsin digestion followed by peptide mass fingerprinting by MALDI-TOF mass spectrometry. The W826P 30-s sample was further analyzed by LC-MS/MS, CID, and HCD MS/MS. Further details are provided in the supporting information.
Molecular dynamics simulations
All-atom MD simulations were performed using GROMACS (5.0.2) (53). Structural models of autoinhibited WT and W826P EphA3 were generated using Rosetta (3.8) (54) and Modeller (9.19) (55) and used as starting structures. Simulation data were acquired for 500 ns. Analysis of MD trajectories was done using packages from the GROMACS suite, and the cross-correlation matrices of Cα–Cα motions were generated using ProDy (56). Further details are provided in the supporting information.
Author contributions
A. K. and N. K. conceptualization; A. K. data curation; A. K. and Z. R. formal analysis; A. K. validation; A. K. investigation; A. K. visualization; A. K. methodology; A. K. writing-original draft; A. K. and N. K. writing-review and editing; M. J. resources; N. K. supervision; N. K. funding acquisition; M. J. contributed to obtaining protein samples; Z. R. generated molecular dynamics data.
Supplementary Material
Acknowledgments
Members of N. K.'s laboratory are acknowledged for helpful discussions. We also acknowledge the Proteomics and Mass Spectrometry Facility at the University of Georgia and the staff for help in generating and analyzing mass spectrometry data.
This work was supported by National Institutes of Health Grant 5RO1GM114409 (to N. K). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S9, Tables S1 and S2, Data Set 1, and supporting methods.
- Eph
- ephrin
- SAM
- sterile α motif
- SH2
- Src homology 2
- GAP
- GTPase-activating protein
- N-lobe
- N-terminal ATP-binding lobe
- C-lobe
- C-terminal substrate-binding lobe
- CID
- collision-induced dissociation
- CIP
- calf intestinal alkaline phosphatase
- HCD
- higher-energy collisional dissociation.
References
- 1. Kullander K., and Klein R. (2002) Mechanisms and functions of Eph and ephrin signalling. Nat. Rev. Mol. Cell Biol. 3, 475–486 10.1038/nrm856 [DOI] [PubMed] [Google Scholar]
- 2. Pasquale E. B. (2005) Eph receptor signalling casts a wide net on cell behaviour. Nat. Rev. Mol. Cell Biol. 6, 462–475 10.1038/nrm1662 [DOI] [PubMed] [Google Scholar]
- 3. Murphy J. M., Zhang Q., Young S. N., Reese M. L.., Bailey F. P., Eyers P. A., Ungureanu D., Hammaren H., Silvennoinen O., Varghese L. N., Chen K., Tripaydonis A., Jura N., Fukuda K., Qin J., et al. (2014) A robust methodology to subclassify pseudokinases based on their nucleotide-binding properties. Biochem. J. 457, 323–334 10.1042/BJ20131174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zeqiraj E., and van Aalten D. M. (2010) Pseudokinases-remnants of evolution or key allosteric regulators? Curr. Opin. Struct. Biol. 20, 772–781 10.1016/j.sbi.2010.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boudeau J., Miranda-Saavedra D., Barton G. J., and Alessi D. R. (2006) Emerging roles of pseudokinases. Trends Cell Biol. 16, 443–452 10.1016/j.tcb.2006.07.003 [DOI] [PubMed] [Google Scholar]
- 6. Barquilla A., and Pasquale E. B. (2015) Eph receptors and ephrins: therapeutic opportunities. Annu. Rev. Pharmacol. Toxicol. 55, 465–487 10.1146/annurev-pharmtox-011112-140226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Himanen J. P., Rajashankar K. R., Lackmann M., Cowan C. A., Henkemeyer M., and Nikolov D. B. (2001) Crystal structure of an Eph receptor-ephrin complex. Nature 414, 933–938 10.1038/414933a [DOI] [PubMed] [Google Scholar]
- 8. Singla N., Erdjument-Bromage H., Himanen J. P., Muir T. W., and Nikolov D. B. (2011) A semisynthetic Eph receptor tyrosine kinase provides insight into ligand-induced kinase activation. Chem. Biol. 18, 361–371 10.1016/j.chembiol.2011.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Davis T. L., Walker J. R., Loppnau P., Butler-Cole C., Allali-Hassani A., and Dhe-Paganon S. (2008) Autoregulation by the juxtamembrane region of the human ephrin receptor tyrosine kinase A3 (EphA3). Structure 16, 873–884 10.1016/j.str.2008.03.008 [DOI] [PubMed] [Google Scholar]
- 10. Binns K. L., Taylor P. P., Sicheri F., Pawson T., and Holland S. J. (2000) Phosphorylation of tyrosine residues in the kinase domain and juxtamembrane region regulates the biological and catalytic activities of Eph receptors. Mol. Cell. Biol. 20, 4791–4805 10.1128/MCB.20.13.4791-4805.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wybenga-Groot L. E., Baskin B., Ong S. H., Tong J., Pawson T., and Sicheri F. (2001) Structural basis for autoinhibition of the Ephb2 receptor tyrosine kinase by the unphosphorylated juxtamembrane region. Cell 106, 745–757 10.1016/S0092-8674(01)00496-2 [DOI] [PubMed] [Google Scholar]
- 12. Shi G., Yue G., and Zhou R. (2010) EphA3 functions are regulated by collaborating phosphotyrosine residues. Cell Res 20, 1263–1275 10.1038/cr.2010.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wiesner S., Wybenga-Groot L. E., Warner N., Lin H., Pawson T., Forman-Kay J. D., and Sicheri F. (2006) A change in conformational dynamics underlies the activation of Eph receptor tyrosine kinases. EMBO J. 25, 4686–4696 10.1038/sj.emboj.7601315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Adams J. A. (2003) Activation loop phosphorylation and catalysis in protein kinases: is there functional evidence for the autoinhibitor model? Biochemistry 42, 601–607 10.1021/bi020617o [DOI] [PubMed] [Google Scholar]
- 15. Zisch A. H., Pazzagli C., Freeman A. L., Schneller M., Hadman M., Smith J. W., Ruoslahti E., and Pasquale E. B. (2000) Replacing two conserved tyrosines of the EphB2 receptor with glutamic acid prevents binding of SH2 domains without abrogating kinase activity and biological responses. Oncogene 19, 177–187 10.1038/sj.onc.1203304 [DOI] [PubMed] [Google Scholar]
- 16. Lawrenson I. D., Wimmer-Kleikamp S. H., Lock P., Schoenwaelder S. M., Down M., Boyd A. W., Alewood P. F., and Lackmann M. (2002) Ephrin-A5 induces rounding, blebbing and de-adhesion of EphA3-expressing 293T and melanoma cells by CrkII and Rho-mediated signalling. J. Cell Sci. 115, 1059–1072 [DOI] [PubMed] [Google Scholar]
- 17. Holland S. J., Gale N. W., Gish G. D., Roth R. A., Songyang Z., Cantley L. C., Henkemeyer M., Yancopoulos G. D., and Pawson T (1997) Juxtamembrane tyrosine residues couple the Eph family receptor EphB2/Nuk to specific SH2 domain proteins in neuronal cells. EMBO J. 16, 3877–3888 10.1093/emboj/16.13.3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang X., Gureasko J., Shen K., Cole P. A., and Kuriyan J. (2006) An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell 125, 1137–1149 10.1016/j.cell.2006.05.013 [DOI] [PubMed] [Google Scholar]
- 19. Huang Y., Bharill S., Karandur D., Peterson S. M., Marita M., Shi X., Kaliszewski M. J., Smith A. W., Isacoff E. Y., and Kuriyan J. (2016) Molecular basis for multimerization in the activation of the epidermal growth factor receptor. Elife 5, e14107 10.7554/eLife.14107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thanos C. D., Goodwill K. E., and Bowie J. U. (1999) Oligomeric structure of the human EphB2 receptor SAM domain. Science 283, 833–836 10.1126/science.283.5403.833 [DOI] [PubMed] [Google Scholar]
- 21. Stapleton D., Balan I., Pawson T., and Sicheri F. (1999) The crystal structure of an Eph receptor SAM domain reveals a mechanism for modular dimerization. Nat. Struct. Biol. 6, 44–49 10.1038/4917 [DOI] [PubMed] [Google Scholar]
- 22. Singh D. R., Cao Q., King C., Salotto M., Ahmed F., Zhou X. Y.., Pasquale E. B., and Hristova K. (2015) Unliganded EphA3 dimerization promoted by the SAM domain. Biochem. J. 471, 101–109 10.1042/BJ20150433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Singh D. R., Ahmed F., Paul M. D., Gedam M., Pasquale E. B., and Hristova K. (2017) The SAM domain inhibits EphA2 interactions in the plasma membrane. Biochim. Biophys. Acta 1864, 31–38 10.1016/j.bbamcr.2016.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mirza A., Mustafa M., Talevich E., and Kannan N. (2010) Co-conserved features associated with cis regulation of ErbB tyrosine kinases. PLoS One 5, e14310 10.1371/journal.pone.0014310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mohanty S., Oruganty K., Kwon A., Byrne D. P., Ferries S., Ruan Z., Hanold L. E., Katiyar S., Kennedy E. J., Eyers P. A., and Kannan N. (2016) Hydrophobic core variations provide a structural framework for tyrosine kinase evolution and functional specialization. PLoS Genet. 12, e1005885 10.1371/journal.pgen.1005885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Oruganty K., Talathi N. S., Wood Z. A., and Kannan N. (2013) Identification of a hidden strain switch provides clues to an ancient structural mechanism in protein kinases. Proc. Natl. Acad. Sci. U.S.A. 110, 924–929 10.1073/pnas.1207104110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kannan N., Haste N., Taylor S. S., and Neuwald A. F. (2007) The hallmark of AGC kinase functional divergence is its C-terminal tail, a cis-acting regulatory module. Proc. Natl. Acad. Sci. U.S.A. 104, 1272–1277 10.1073/pnas.0610251104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Eyers P. A., Keeshan K., and Kannan N. (2017) Tribbles in the 21st century: the evolving roles of tribbles pseudokinases in biology and disease. Trends Cell Biol. 27, 284–298 10.1016/j.tcb.2016.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nguyen T., Ruan Z., Oruganty K., and Kannan N. (2015) Co-conserved MAPK features couple D-domain docking groove to distal allosteric sites via the C-terminal flanking tail. PLoS One 10, e0119636 10.1371/journal.pone.0119636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brunet F. G., Volff J. N., and Schartl M. (2016) Whole genome duplications shaped the receptor tyrosine kinase repertoire of jawed vertebrates. Genome Biol. Evol. 8, 1600–1613 10.1093/gbe/evw103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fairclough S. R., Chen Z., Kramer E., Zeng Q., Young S., Robertson H. M., Begovic E., Richter D. J., Russ C., Westbrook M. J., Manning G., Lang B. F., Haas B., Nusbaum C., and King N. (2013) Premetazoan genome evolution and the regulation of cell differentiation in the choanoflagellate Salpingoeca rosetta. Genome Biol. 14, R15 10.1186/gb-2013-14-2-r15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Srivastava M., Simakov O., Chapman J., Fahey B., Gauthier M. E., Mitros T., Richards G. S., Conaco C., Dacre M., Hellsten U., Larroux C., Putnam N. H., Stanke M., Adamska M., Darling A., et al. (2010) The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466, 720–726 10.1038/nature09201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Drescher U. (2002) Eph family functions from an evolutionary perspective. Curr. Opin. Genet. Dev. 12, 397–402 10.1016/S0959-437X(02)00316-7 [DOI] [PubMed] [Google Scholar]
- 34. Knighton D. R., Zheng J. H., Ten Eyck L. F., Ashford V. A., Xuong N. H., Taylor S. S., and Sowadski J. M. (1991) Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253, 407–414 10.1126/science.1862342 [DOI] [PubMed] [Google Scholar]
- 35. Neuwald A. F. (2010) Bayesian classification of residues associated with protein functional divergence: Arf and Arf-like GTPases. Biol. Direct 5, 66 10.1186/1745-6150-5-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dyson H. J., and Wright P. E. (2005) Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6, 197–208 10.1038/nrm1589 [DOI] [PubMed] [Google Scholar]
- 37. Hubbard S. R. (2004) Juxtamembrane autoinhibition in receptor tyrosine kinases. Nat. Rev. Mol. Cell Biol. 5, 464–471 10.1038/nrm1399 [DOI] [PubMed] [Google Scholar]
- 38. Plaza-Menacho I., Barnouin K., Barry R., Borg A., Orme M., Chauhan R., Mouilleron S., Martínez-Torres R. J., Meier P., and McDonald N. Q. (2016) RET functions as a dual-specificity kinase that requires allosteric inputs from juxtamembrane elements. Cell Rep. 17, 3319–3332 10.1016/j.celrep.2016.11.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gajiwala K. S. (2013) EGFR: tale of the C-terminal tail. Protein Sci. 22, 995–999 10.1002/pro.2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ma B., Tsai C. J., Haliloğlu T., and Nussinov R. (2011) Dynamic allostery: linkers are not merely flexible. Structure 19, 907–917 10.1016/j.str.2011.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brown C. J., Johnson A. K., Dunker A. K., and Daughdrill G. W. (2011) Evolution and disorder. Curr. Opin. Struct. Biol. 21, 441–446 10.1016/j.sbi.2011.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Harrison S. C. (2003) Variation on an Src-like theme. Cell 112, 737–740 10.1016/S0092-8674(03)00196-X [DOI] [PubMed] [Google Scholar]
- 43. Kornev A. P., and Taylor S. S. (2015) Dynamics-driven allostery in protein kinases. Trends Biochem. Sci. 40, 628–647 10.1016/j.tibs.2015.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Laine E., Auclair C., and Tchertanov L. (2012) Allosteric communication across the native and mutated KIT receptor tyrosine kinase. PLoS Comput. Biol. 8, e1002661 10.1371/journal.pcbi.1002661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McClendon C. L., Kornev A. P., Gilson M. K., and Taylor S. S. (2014) Dynamic architecture of a protein kinase. Proc. Natl. Acad. Sci. U.S.A. 111, E4623–E4631 10.1073/pnas.1418402111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schulze J. O., Saladino G., Busschots K., Neimanis S., Süß E., Odadzic D., Zeuzem S., Hindie V., Herbrand A. K., Lisa M. N., Alzari P. M., Gervasio F. L., and Biondi R. M. (2016) Bidirectional allosteric communication between the ATP-binding site and the regulatory PIF pocket in PDK1 protein kinase. Cell Chem. Biol. 23, 1193–1205 10.1016/j.chembiol.2016.06.017 [DOI] [PubMed] [Google Scholar]
- 47. Chen S., Brier S., Smithgall T. E., and Engen J. R. (2007) The Abl SH2-kinase linker naturally adopts a conformation competent for SH3 domain binding. Protein Sci. 16, 572–581 10.1110/ps.062631007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xi G., Shen X., and Clemmons D. R. (2010) p66shc inhibits insulin-like growth factor-I signaling via direct binding to Src through its polyproline and Src homology 2 domains, resulting in impairment of Src kinase activation. J. Biol. Chem. 285, 6937–6951 10.1074/jbc.M109.069872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Neuwald A. F. (2009) Rapid detection, classification and accurate alignment of up to a million or more related protein sequences. Bioinformatics 25, 1869–1875 10.1093/bioinformatics/btp342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bateman A., Coin L., Durbin R., Finn R. D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E. L., Studholme D. J., Yeats C., and Eddy S. R. (2004) The Pfam protein families database. Nucleic Acids Res. 32, D138–D141 10.1093/nar/gkh121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Krogh A., Larsson B., von Heijne G., and Sonnhammer E. L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- 52. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Abraham M. J., Murtola T., Schulz R., Páll S., Smith J. C., Hess B., and Lindahl E. (2015) GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2, 19–25 10.1016/j.softx.2015.06.001 [DOI] [Google Scholar]
- 54. Rohl C. A., Strauss C. E., Misura K. M., and Baker D. (2004) Protein structure prediction using Rosetta. Methods Enzymol. 383, 66–93 10.1016/S0076-6879(04)83004-0 [DOI] [PubMed] [Google Scholar]
- 55. Fiser A., Do R. K., and Sali A. (2000) Modeling of loops in protein structures. Protein Sci. 9, 1753–1773 10.1110/ps.9.9.1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bakan A., Meireles L. M., and Bahar I. (2011) ProDy: protein dynamics inferred from theory and experiments. Bioinformatics 27, 1575–1577 10.1093/bioinformatics/btr168 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.