Abstract
The Wnt/β-catenin pathway is essential for embryonic development and homeostasis, but excessive activation of this pathway is frequently observed in various human diseases, including cancer. Current therapeutic drugs targeting the Wnt pathway often lack sufficient efficacy, and new compounds targeting this pathway are therefore greatly needed. Here we report that the plant-derived natural product parthenolide (PTL), a sesquiterpene lactone, inhibits Wnt signaling. We found that PTL dose-dependently inhibits Wnt3a- and CHIR99021-induced transcriptional activity assessed with the T-cell factor (TCF)/lymphoid enhancer factor (LEF) firefly luciferase (TOPFlash) assay in HEK293 cells. Further investigations revealed that PTL decreases the levels of the transcription factors TCF4/LEF1 without affecting β-catenin stability or subcellular distribution. Moreover, this effect of PTL on TCF4/LEF1 was related to protein synthesis rather than to proteasome-mediated degradation. Of note, siRNA-mediated knockdown of RPL10, a ribosome protein PTL binds, substantially decreased TCF4/LEF1 protein levels and also Wnt3a-induced TOPFlash activities, suggesting a potential mechanism by which PTL may repress Wnt/β-catenin signaling. In summary, PTL binds RPL10 and thereby potently inhibits the Wnt/β-catenin pathway.
Keywords: inhibitor, protein synthesis, small molecule, T-cell factor (TCF), Wnt signaling
Introduction
Since the first Wnt protein was identified 35 years ago, the Wnt/β-catenin pathway has been well known for its essential role in both embryo development and adult tissue homeostasis (1). In the absence of Wnt signals, cytoplasm β-catenin is phosphorylated by the Ser/Thr kinases GSK3 and CK1. The phosphorylated β-catenin is then recognized and ubiquitylated by β-TRCP and subjected to proteasome-mediated degradation. Upon binding of the Wnt proteins with the membrane receptor Frizzled and LRP5/6, β-catenin is stabilized and shuttled into the nucleus, where it binds to the transcriptional factor TCFs/LEF14 and activates expression of various target genes. Excessive activation of Wnt/β-catenin signaling is frequently observed in a wide range of tumors, such as colorectal cancer, liver cancer, melanoma, prostate cancer, and breast cancer (2), making this pathway an attractive target for cancer therapy. Many efforts have been made to identify and develop drugs that can modulate Wnt/β-catenin signaling, especially by targeting β-catenin and its downstream players (3). Methyl 3-{[(4-methylphenyl)sulfonyl]amino}benzoate is a selective inhibitor of Wnt/β-catenin signaling that has been found to bind to β-catenin and induce its proteasome-mediated degradation (4). PNU-74654 and iCRT3 are two other small molecules that inhibit Wnt signaling by disturbing TCF4–β-catenin interaction (5, 6). ICG-001 has been found to compete with CREB-binding protein for binding β-catenin, thereby inhibiting Wnt signaling (7). Our laboratory recently reported NC043 as a Wnt signaling inhibitor by targeting CARF (collaborator of ARF) to disturb TCF4 and β-catenin interaction (8, 9).
PTL, a sesquiterpene lactone, was first purified from the shoots of the medicinal plant feverfew (Tanacetum parthenium) (10, 11), which is a Chinese folk medical plant and has been used to treat fever, headache, and arthritis for many years (12). In recent years, it has attracted wide attention for its considerable pharmacological activities, including antimicrobial, anti-inflammatory, and especially anticancer effects (13). PTL is well known for its activity of inhibiting NF-κB signaling and is commonly used as an NF-κB inhibitor (14). However, the mechanisms behind the anti-cancer effects of PTL have not been completely understood. Some studies indicate that PTL inhibits tumor growth by increasing reactive oxygen species (ROS) (15, 16). PTL was also found to target epigenetic factors, such as DNMT1 and HDAC1, to inhibit tumor growth (17, 18). Of note, in 2005, PTL was identified as the first small molecule that selectively kills cancer stem cells while sparing normal stem cells in acute and chronic myelogenous leukemia stem cell models (19). After that, the selective killing of cancer stem cells by PTL was also observed in a number of other cancers, such as bone, breast, melanoma, mesenchyme, and prostate cancer (20–25). On the other hand, Wnt/β-catenin signaling is well recognized for its essential role in the maintenance and self-renewal of embryonic and adult stem cells. However, it remains unknown whether PTL may act on Wnt signaling to inhibit tumor growth. In this work, through high-throughput screening, we identified that PTL potently inhibits Wnt3a-induced TOPFlash activity. Further studies indicated that PTL acts through reducing TCF4/LEF1 synthesis via targeting RPL10.
Results
PTL inhibits Wnt/β-catenin signaling in HEK293 cells
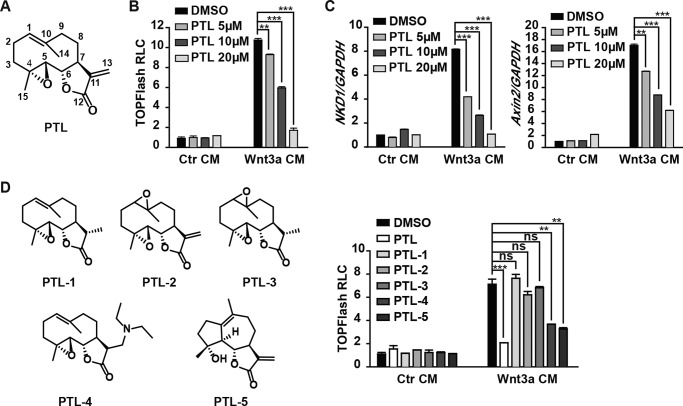
In our high-throughput screening effort for small-molecule compounds modulating Wnt signaling, we identified the small molecule PTL (Fig. 1A). Our results showed that PTL dose-dependently inhibited Wnt3a conditioned medium (CM)–induced TOPFlash activity in HEK293cells (Fig. 1B). Next we examined the effect of PTL on the expression of Wnt target genes. We treated HEK293 cells with PTL and then carried out real-time PCR. As expected, PTL inhibited expression of the endogenous Wnt target genes Axin2 and NKD1 in a dose-dependent manner (Fig. 1C). To further explore the structure–activity relationship of PTL, we synthesized a series of PTL derivatives and tested their effects on TOPFlash activity. We found that PTL-4 and PTL-5 exhibited similar inhibitory effects as PTL, whereas PTL-1, PTL-2, and PTL-3 lost the inhibitory effect on TOPFlash reporter activity (Fig. 1D). These results indicated that the α,β-unsaturated lactone and the 1(10) double bond of PTL might be the functional groups that are responsible for PTL inhibitory activity against Wnt signaling. Because PTL-4 can undergo retro-Michael additions to regenerate the parent molecule that is PTL, it was not surprising to find that PTL-4 inhibits Wnt/β-catenin signaling, as does PTL.
Figure 1.
PTL inhibits Wnt/β-catenin signaling. A, chemical structure of PTL. B, PTL inhibits Wnt/β-catenin signaling in a dose-dependent manner. HEK293 cells were transfected with TOPFlash plasmids. After 18 h of transfection, the cells were treated with DMSO or PTL for 1 h, followed by Wnt3a CM or control (Ctr) CM plus the same dose of PTL for another 6 h and then lysed for luciferase assays. RLC, relative luciferase count. C, PTL inhibits Wnt target gene expression. Expression of the Wnt target genes NKD1 and Axin2 was determined by quantitative real-time PCR and normalized to GAPDH expression. D, distinct effects of PTL and its derivates on TOPFlash reporter activity. HEK293 cells were treated with DMSO or 20 μm PTL, PTL-1, PTL-2, PTL-3, PTL-4, or PTL-5 in the presence of either the Ctr CM or Wnt3a CM. Data represent the mean ± S.D. from one experiment. Each experiment was repeated at least three times. **, p < 0.01; ***, p < 0.001; significant relative to vehicle control; ns, no statistical significance.
PTL does not affect β-catenin stability or subcellular distribution
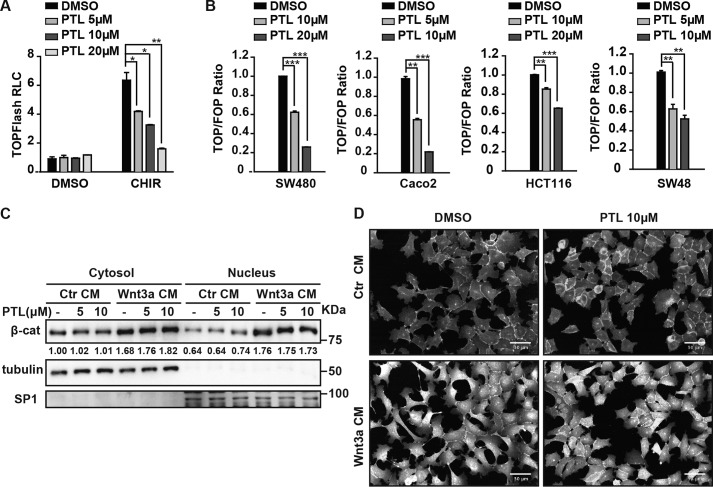
As mentioned before, β-catenin is the central player of canonical Wnt signaling, and its accumulation and nuclear translocation are hallmarks for activation of the signaling pathway. Thus, we wanted to ask whether PTL affects Wnt signaling through regulating the accumulation of β-catenin. To answer this question, we first tested the effect of PTL on TOPFlash activity induced by a glycogen synthase kinase (GSK) inhibitor, CHIR99021 (26). We transfected HEK293 cells with TOPFlash plasmids, and then we treated cells with 2 μm CHIR, followed by PTL treatment for the TOPFlash activity test. As shown in Fig. 2A, PTL showed similar inhibitory effects on TOPFlash luciferase reporter activity regardless of CHIR treatment. This result indicates that PTL functions downstream of the “β-catenin destruction complex”. Next we wanted to investigate the activity of PTL in the case of constant β-catenin activation. For this purpose, we used the colon cancer cell lines Caco2 and SW480, which harbor adenomatous polyposis coli (APC) mutations, as well as HCT116 and SW48, which harbor β-catenin mutations. We transfected the plasmids of TOPFlash/FOPFlash to those colon cancer cells, followed by treatment with PTL. As shown in Fig. 2B, PTL efficiently inhibited TOPFlash reporter activity in all four cell lines, indicating that PTL acts downstream of β-catenin accumulation. To further examine the effects of PTL on β-catenin activation, we treated HEK293 cells with two doses of PTL in the presence of either control conditioned medium or Wnt3a conditioned medium. We then separated the cytosol and nucleus of the cells and subjected them to Western blot analysis using β-catenin/tubulin/SP1 antibodies. As shown in Fig. 2C, PTL had no effect on endogenous β-catenin protein levels or its subcellular distribution. Consistent with this result, in an immunofluorescence assay with a β-catenin-specific antibody, β-catenin levels and its subcellular distribution showed no differences between the PTL-treated and control groups (Fig. 2D). These results further confirm that PTL does not affect β-catenin stability or subcellular distribution.
Figure 2.
PTL does not affect β-catenin stability and subcellular distribution. A, PTL inhibits CHIR-induced Wnt/β-catenin signaling in a dose-dependent manner. HEK293 cells were treated with increasing doses of PTL with DMSO or 2 μm CHIR. RLC, relative luciferase count. B, PTL inhibits TOPFlash activity in a dose-dependent manner in SW480, Caco2, HCT116, and SW48 cells. Cells were treated with PTL for 24 h. TOPFlash (TOP) responds to Wnt signaling. FOPFlash (FOP) does not respond to Wnt singling and was used as a control. C, PTL has no effect on β-catenin (β-cat) stability and subcellular distribution. HEK293 cells were incubated with DMSO or two doses of PTL. After 1 h, cells were treated with control or Wnt3a CM containing the same dose of PTL for an additional 2 h. The cells were then fractioned as described under “Experimental procedures,” and the samples were analyzed by Western blot analysis. Tubulin was used as a cytosolic marker, and SP1 was used as a nuclear marker. The immunoblots were quantified by densitometry, and the values are given beneath each band. D, immunofluorescence staining shows that PTL does not influence β-catenin protein levels and distribution. HEK293 cells were treated with 10 μm of PTL for 1 h, and then cells were incubated with control or Wnt3a CM containing the same dose of PTL for an additional 2 h. The cells were then fixed as described under “Experimental procedures,” and the images were collected and analyzed by Opera. Scale bars = 50 μm. Data represent the mean ± S.D. from one experiment. Each experiment was repeated at least three times. *, p < 0.05; **, p < 0.01; ***, p < 0.001; significant relative to vehicle control.
PTL decreases TCF4/LEF1 protein levels
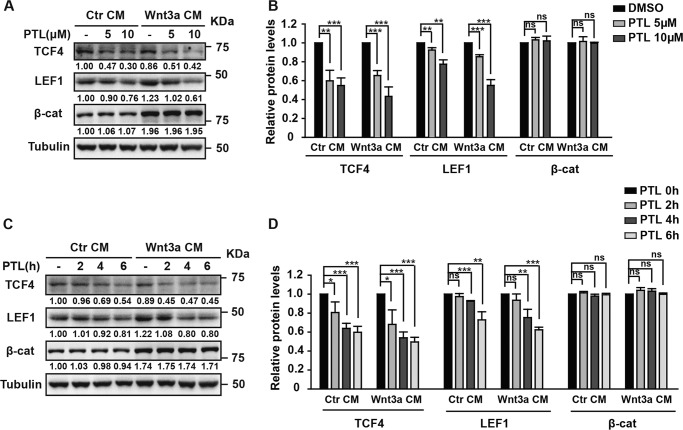
Our results above suggest that PTL acts downstream of β-catenin accumulation and nuclear localization. Thus, PTL very likely acts on the TCF/LEF1 transcriptional factors in the nucleus. To test this possibility, we first examined whether PTL affects TCF/LEF1 protein levels. The human TCF family has four members: TCF1, TCF3, TCF4 and LEF1. Before testing the effect of PTL on TCF family proteins, we measured the mRNA levels of TCF1, TCF3, TCF4, and LEF1 in HEK293 cells. We found that the mRNA levels of TCF4 and LEF1 are much higher (nearly 40- to 50-fold) than that of TCF1 and TCF3 (Fig. S2). Therefore we carried out our further studies on TCF4 and LEF1. HEK293 cells were cultured in control CM or Wnt3a CM with 5 or 10 μm PTL for 6 h. As shown in Fig. 3, A and B, both TCF4 and LEF1 protein levels were reduced by PTL in a dose-dependent manner regardless of Wnt stimulation. PTL also reduced TCF4/LEF1 levels in a time-dependent manner (Fig. 3, C and D). In view of the significant differences in mRNA levels of TCF family members, although the TCF1 and TCF3 protein levels in HEK293 cells were also decreased after PTL treatment, PTL inhibition of Wnt signaling was mainly due to the decrease of TCF4 and LEF1 (Fig. S3). However, there was no difference between the levels of TCF4/LEF1 mRNAs before and after 10 μm PTL treatment in the control conditioned medium, indicating that PTL could not function by regulating the transcription of TCF4/LEF1. Consistent with this, upon Wnt treatment, TCF4 mRNA levels were also not affected by PTL treatment, supporting that PTL does not function through affecting gene transcription (Fig. S2). However, LEF1 mRNA levels were apparently reduced by PTL. This is due to the fact that LEF1 is a target gene of Wnt signaling. Because PTL treatment leads to inhibition of Wnt signaling, it was not surprising to find a decrease of LEF1 mRNA levels by PTL treatment. Thus, this decrease of LEF1 mRNA levels is most likely due to inhibition of Wnt signaling rather than a direct effect on LEF1 transcription by PTL.
Figure 3.
PTL decreases TCF4/LEF1 protein levels. A, PTL reduces TCF4 and LEF1 protein levels in a dose-dependent manner. HEK293 cells were incubated with DMSO or two doses of PTL. After 1 h, cells were treated with control or Wnt3a CM containing the same dose of PTL for an additional 6 h. The cells were then harvested and analyzed by Western blot analysis. The immunoblots were quantified by densitometry, and the values are given beneath each band. β-cat, β-catenin. B, statistics of three independent experiments of the protein quantification in A. For each protein, the control group was normalized to 1 in both control and Wnt3a CM stimulation. C, PTL reduces TCF4 and LEF1 protein levels in a time-dependent manner. HEK293 cells were stimulated by control or Wnt3a CM for 6 h. During this time, control or Wnt3a CM containing 10 μm PTL was exchanged for the cells every 2 h. PTL treatment time is indicated. The immunoblots were quantified by densitometry, and values are given beneath each band. D, statistics of three independent experiments of the protein quantification in C. For each protein, the control group was normalized to 1 in both control and Wnt3a CM stimulation. ns, no statistical significance. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
PTL decreases TCF4/LEF1 levels by blocking protein synthesis
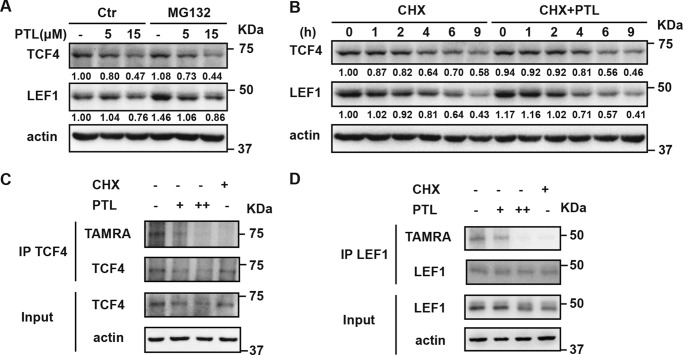
Because our data suggest that PTL does not act through affecting gene transcription, we asked how PTL reduces TCF4/LEF1 protein levels. One possibility is that PTL may facilitate proteasome-mediated degradation of TCF4/LEF1 proteins. Therefore, we blocked proteasome-induced degradation by using MG132. We treated HEK293 cells with 20 μm MG132 followed by PTL treatment. We found that blocking proteasome-induced degradation with MG132 does not affect PTL activity in decreasing TCF4/LEF1 protein levels (Fig. 4A), indicating that PTL does not act through promoting TCF4/LEF1 degradation. Next we tested whether blocking protein synthesis by using CHX affects the PTL activity of inhibiting Wnt signaling. As shown in Fig. 4B, when protein synthesis process is blocked by CHX, treatment with PTL could not further decrease TCF4/LEF1 levels (Fig. 4B). To further address whether PTL inhibits protein synthesis of TCF4 and LEF1, we carried out a nonradioactive metabolic labeling assay. As shown in Fig. 4, C and D, PTL significantly inhibits the protein synthesis of both TCF4 and LEF1. Thus, we conclude that PTL decreases TCF4 and LEF1 by blocking protein synthesis rather than affecting TCF4/LEF1 protein stability.
Figure 4.
PTL decreases TCF4/LEF1 by blocking protein synthesis. A, PTL can still decrease TCF4 and LEF1 when blocking proteasome-induced degradation with MG132. HEK293 cells were treated with DMSO or 20 μm MG132 containing different doses of PTL for 6 h. Then cells were harvested and analyzed by Western blot analysis. B, PTL could not further decrease TCF4/LEF1 levels when blocking protein synthesis using CHX. HEK293 cells were treated with 100 μg/ml CHX or 100 μg/ml CHX containing 15 μm PTL for the indicated time. Then cells were harvested and analyzed by Western blot analysis. The immunoblots were quantified by densitometry, and the corresponding values are given beneath each band. C, PTL significantly reduces nascent TCF4. Nascent TCF4 in HEK293 cells was labeled after DMSO, 7.5 μm PTL, 15 μm PTL, or 100 μg/ml CHX treatment, which was detected by TAMRA antibody. IP, immunoprecipitation. D, PTL significantly reduces nascent LEF1. Nascent LEF1 in HEK293 cells was labeled after DMSO, 7.5 μm PTL, 15 μm PTL, or 100 μg/ml CHX treatment, which was detected by TAMRA antibody.
RPL10 knockdown decreases TCF4/LEF1 protein levels
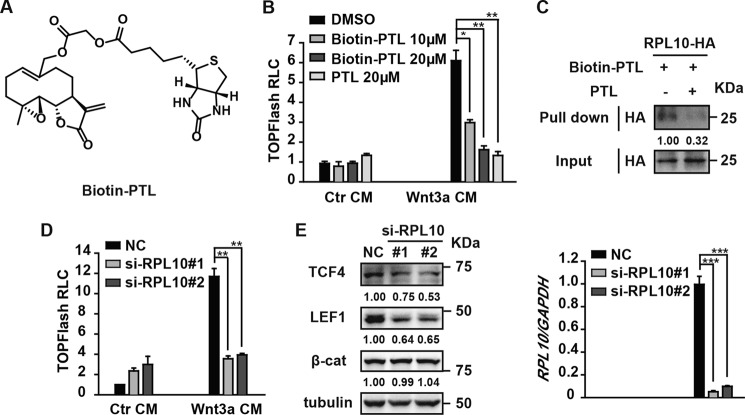
RPL10, a ribosome protein, was recently reported as a target of DMAPT, which is a water-soluble analogue of PTL (27). Chen et al. uncovered that DMAPT targets and decreases RPL10, leading to reduced protein levels of p65 and IKKγ (27). This mechanism is responsible at least partially for DMAPT inhibitory activity against NF-κB (27). To test whether PTL could bind to RPL10, we first synthesized biotinylated PTL (Fig. 5A) and confirmed that biotin-PTL dose-dependently inhibits Wnt signaling, as does PTL (Fig. 5B). Next we overexpressed HA-tagged human RPL10 in HEK293T cells and carried out a pulldown assay using biotinylated PTL as the bait with or without addition of unlabeled PTL. As shown in Fig. 5C, biotin-PTL captured RPL10, and this binding could be competed off by free PTL, supporting PRL10 as the target of PTL. To examine the role of PRL10 in Wnt signaling, we deprived HEK293 cells of RPL10 using the method of RNAi. As shown in Fig. 5D, knockdown of RPL10 significantly reduced TOPFlash reporter activity. Moreover, RPL10 deprivation led to a marked decrease in TCF4 and LEF1 levels (Fig. 5E). Our results, combined with the findings of Chen et al., confirmed that PTL reduces TCF4/LEF1 protein levels by targeting the ribosome protein RPL10.
Figure 5.
RPL10 knockdown decreases TCF4/LEF1 protein levels. A, chemical structure of biotin-PTL. B, biotin-PTL inhibits Wnt/β-catenin signaling, as does PTL. HEK293 cells were transfected with TOPFlash plasmids. After 18 h of transfection, the cells were treated with biotin-PTL or PTL for 1 h, followed by Wnt3a CM or Ctr CM plus the same dose of biotin-PTL or PTL for another 6 h. The cells were then lysed for luciferase assays. RLC, relative luciferase count. C, PTL specifically binds to RPL10. HEK293T cells were transfected with HA-tagged RPL10 plasmids. After 24 h of transfection, the cell lysates were used for a biotin pulldown assay with biotin-PTL (10 μm) as the bait. PTL (20 μm) was added to compete with biotin-PTL for binding to RPL10. The immunoblots were quantified by densitometry, and the values are given beneath each band. D, knockdown of RPL10 inhibits Wnt3a-induced TOPFlash activity. HEK293 cells were transfected with the indicated siRNA on the first day using RNAiMAX, and 24 h later, cells were retransfected with TOPFlash plasmids using Lipofectamine 2000. After 18 h, cells were treated with Wnt3a CM or Ctr CM for another 6 h and then lysed for the luciferase assay. E, RPL10 knockdown significantly reduces TCF4 and LEF1 levels. HEK293 cells transfected with the indicated siRNA were harvested. Half of the cells were evaluated for expression of endogenous proteins by Western blot analysis using antibodies as indicated, and the other half of the cells were evaluated for mRNA levels of RPL10 by quantitative real-time PCR. The immunoblots were quantified by densitometry, and the values are given beneath each band. Data represent the mean ± S.D. from one experiment. Each experiment was repeated at least three times. *, p < 0.05; **, p < 0.01; ***, p < 0.001; significant relative to vehicle control. NC, negative control.
PTL inhibits proliferation of colon cancer cells
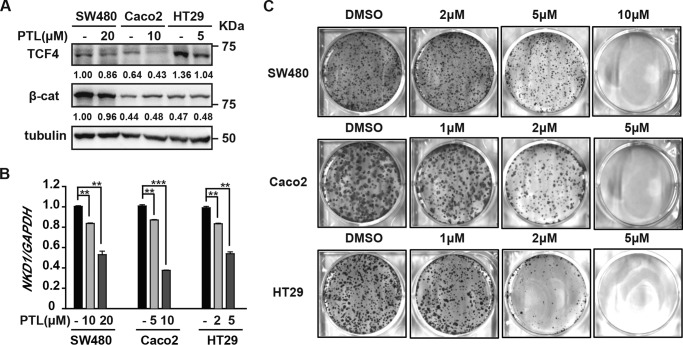
As mentioned before, overactivation of Wnt/β-catenin signaling is closely related to tumor formation and progression. One anecdote is colon cancer. More than 80% of colon cancers harbor mutations in the scaffold protein APC and have overactivated Wnt signaling (28). Thus, we used human colon cancer cells to test the potential of PTL in cancer therapy. We found that PTL significantly reduced TCF4 levels in the colon cancer cell lines SW480, Caco2, and HT29 but has no apparent effects on β-catenin levels in these cells (Fig. 6A). Consistently, PTL inhibited expression of the Wnt target gene NKD1 in these three colon cancer cell lines in a dose-dependent manner (Fig. 6B). A colony-forming assay also showed that PTL inhibited colon cancer cell proliferation at a level similar to its inhibitory activity toward Wnt signaling (Fig. 6C). These results demonstrate the potential of PTL in the treatment of colon cancers that are mostly driven by overactivated Wnt signaling.
Figure 6.
PTL inhibits proliferation of colon cancer cells. A, PTL reduces TCF4 levels in colon cancer cells. Colon cancer, SW480, Caco2, and HT29 cells were treated with the indicated doses of PTL for 24 h, and then cells were harvested and analyzed by Western blot analysis. The immunoblots were quantified by densitometry, and the values are given beneath each band. β-cat, β-catenin. B, PTL dose-dependently down-regulates NKD1 mRNA in colon cancer cells. Colon cancer cells were treated with the indicated doses of PTL for 24 h, and then the mRNA levels of NKD1 were evaluated by quantitative real-time PCR. C, PTL inhibits colony formation of colon cancer cells. Cells were treated with the indicated doses of PTL, with medium changes every 3 days until visible colonies formed. Data represent the mean ± S.D. from one experiment. Each experiment was repeated at least three times. **, p < 0.01; ***, p < 0.001; significant relative to vehicle control.
Discussion
The Wnt/β-catenin signaling pathway is one of the most important pathways in development and is involved in many aspects of tumorigenesis. Considerable efforts have been made to identify and develop effective inhibitors against the Wnt signaling pathway. However, successes in finding inhibitors of this pathway are very limited, and so far, no drugs specific for Wnt signaling have been approved for clinical applications. In this study, we found that PTL, a sesquiterpene lactone, exhibits potent inhibitory activities against Wnt/β-catenin signaling. Further mechanistic study showed that PTL reduced TCF4/LEF1 synthesis by targeting RPL10 (Fig. 7).
Figure 7.
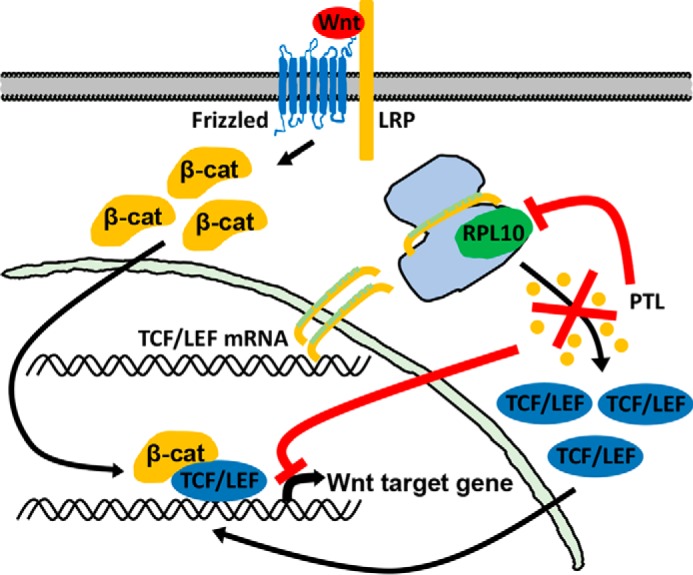
A model showing that PTL inhibits Wnt/β-catenin signaling. Binding of Wnt protein with its receptors induces cytoplasmic β-catenin (β-cat) to aggregate and translocate into the nucleus. In the nucleus, β-catenin binds to TCF/LEF to induce Wnt target gene expression. When PTL binds to RPL10, TCF/LEF levels are reduced, leading to inhibition of Wnt/β-catenin signaling.
TCF/LEF1 transcription factors are the downstream effectors of Wnt/β-catenin signaling. Recent genome-wide studies in mammals and Drosophila suggest that all direct activation of β-catenin target gene expression requires TCF/LEF1 as the final effectors (29, 30). Moreover, the levels of TCF/LEF1 in colon cancer cells are substantially increased compared with normal tissues, and TCF4 expression is positively correlated with a poor prognosis in resectable esophageal squamous cell carcinoma (31, 32). Therefore, targeting TCF/LEF1 would be a potentially effective way in reversing excessive activation of Wnt/β-catenin signaling in these cancer cells. However, few of the compounds have so far been reported to target TCF/LEF1. Thus, our findings provide a valuable prototype for new therapeutic agents in treating tumors driven by overactivated Wnt signaling.
RPL10, a component of the 60S ribosome, functions in protein synthesis, and its mutations have been found in T-cell acute lymphoblastic leukemia patients (33). Here we found that TCF4/LEF1 levels were significantly reduced by deprivation of RPL10, but β-catenin levels were not affected. An interesting question is whether the ribosome has selectivity in mediating protein translation. Recently, Shi et al. reported that heterogeneous ribosomes tend to translate distinct subpools of mRNAs genome-wide (34). Sendoel et al. reported that, during tumor initiation, the translational apparatus is redirected toward non-canonical upstream initiation sites, enhancing the translational efficiency of oncogenic mRNAs (35). Thus, it is possible that RPL10 may selectively regulate TCF4/LEF1 synthesis without affecting β-catenin translation. Further studies are required to investigate this possibility and address the mechanisms behind how RPL10 regulates TCF/LEF synthesis.
PTL has been reported to specially inhibit cancer stem cell growth without killing normal tissue cells in leukemia and solid tumors (13). Wnt/β-catenin signaling is known to be essential for the maintenance of stem cells (1). Our work shows that PTL inhibits colon cancer cell growth. Whether this inhibition of colon cancer cell growth is caused by inhibition of Wnt/β-catenin signaling in cancer stem cells also warrants further study.
In summary, our study identifies PTL as a new inhibitor of Wnt/β-catenin signaling by targeting RPL10 and blocking TCF4/LEF1 synthesis. Given that DMAPT, a more hydrophilic form of PTL, has been approved for clinical trial for hematologic malignancies, our findings provide proof of concept for the use of PTL as a potential drug for treating Wnt-driven tumors.
Experimental procedures
Isolation of PTL, synthesis of biotin-PTL, and PTL derivatives
For isolation of PTL, air-dried powdered leaves of Magnolia grandiflora (10.0 kg) were extracted with MeOH (20 liters × 3) under reflux three times (4, 3, and 3 h, respectively) at 80 °C. The combined MeOH extracts were concentrated in a vacuum to give a crude residue (600 g) that was suspended in water. The water layer was successively partitioned with ethyl acetate (EtOAc) (10 liters × 3). The EtOAc portion (220 g) was repeatedly subjected to a silica gel column to get parthenolide (2.0 g). The purity of parthenolide was greater than 95% as determined by TLC and NMR spectra. Biotin-PTL and PTL derivatives were synthesized as described in Fig. S1.
Antibodies and reagents
Antibodies for β-catenin (BD Transduction Laboratories), TCF4 (Millipore), LEF1 (Cell Signaling Technology), SP1 (Sigma), tubulin (Sigma), actin (Santa Cruz Biotechnology), and HA (Covance) were utilized in this study. Wnt3a CM and control medium were described previously (36). CHIR-99021 was purchased from Selleckchem. MG132 and CHX were purchased from Sigma-Aldrich. The full-length complementary DNA of human RPL10 was generated by PCR from HEK293 complementary DNA libraries, cloned into the mammalian expression vector pCMV tagged with HA, and verified by DNA sequencing. The siRNA sequences of RPL10 #1 and #2 were 5′-GGCCAAGUUAUCAUGUCCA-3′ and 5′-CUGAUGCCAAGAUUCGCAU-3′, respectively. The negative control siRNA sequence was 5′-UUCUCCGAACGUGUCACGU-3′.
Cell culture, transfection, and reporter gene assay
HEK293 cells were cultured in Dulbecco's modified Eagle's medium containing 10% FBS. HT29 and HCT116 cells were maintained in McCoy's 5A medium with 10% FBS. SW480 and SW48 cells were maintained in Leibovitz's medium with 10% FBS. Caco2 cells were maintained in minimum Eagle's medium with 20% FBS. Mycoplasma contamination tests were performed for all cells used in this study.
HEK293 cells were transfected by Lipofectamine 2000 (Invitrogen) or RNAiMAX (Invitrogen) according to the manufacturer's instructions, whereas SW480, Caco2, HCT116, and SW48 cells were transfected by Lipofectamine 3000 (Invitrogen). For reporter gene assays, cells were seeded in 24-well plates. Each well of HEK293 cells was transfected with 250 ng of plasmids in total, including 30 ng of TOPFlash and 25 ng of GFP. The LacZ plasmid was added to equalize the total amount of transfected DNA. After 18 h, the cells were treated with Wnt3a conditioned medium or control medium for another 6 h and then lysed for luciferase assays. Each well of SW480, Caco2, HCT116, and SW48 cells was transfected with 250 ng of plasmids in total, including 50 ng of TOPFlash or FOPFlash, 50 ng of GFP, and 150 ng of LacZ. The cells were lysed for luciferase assays 24 h after transfection. The GFP expression levels were determined for normalization as described previously (37).
Drug treatment and Western blot analysis
Cells were treated with the indicated dose of drugs and harvested with 2× loading buffer (0.3 m Tris-HCl, 10% SDS, 6% β-mercaptoethanol, 50% glycerol, and 0.05% bromphenol blue). Then proteins were separated by SDS-PAGE and blotted onto polyvinylidene difluoride membranes (Millipore). Membranes were blocked with 5% nonfat dry milk for 1 h and then incubated with primary antibodies for 1 h at room temperature or overnight at 4 °C. After being washed, membranes were incubated for 1 h at room temperature with the appropriate horseradish peroxidase–conjugated secondary antibodies (Thermo Scientific) for 1 h. Results were visualized with Tanon 5200.
Cytosol and nucleus fractionation
HEK293 cells were grown in 6-well plates, harvested with a cell scraper into 1.5 ml of PBS, and spun at 700 × g for 10 min. Then the cytosol and nucleus fractions were separated with a nuclear and cytoplasmic extraction kit (CW0199S, CWBIO) according to the manufacturer's instructions.
Immunofluorescence staining
HEK293 cells were seeded into a 96-well plate to make 6000 cells/well. 48 h later, the indicated dose of PTL was added to the wells for 1 h, followed by Wnt3a conditioned medium or control conditioned medium plus the same dose of PTL for an additional 2 h. Then cells were fixed with 4% paraformaldehyde (Alfa Aesar, 30525894) for 15 min and permeabilized with PBST (PBS and 0.1% Triton-X100) for 20 min at room temperature. The fixed samples were incubated with a primary antibody against β-catenin overnight at 4 °C, followed by Cy3-conjugated anti-mouse secondary antibody and 4′,6-diamidino-2-phenylindole. For each well, 10 random-field fluorescence images were captured by the Opera LX high-content confocal imaging system with a ×20 Air-LUCPLFLN objective (numerical aperture = 0.45, PerkinElmer Life Sciences) (38).
RT-PCR and quantitative real-time PCR
The total RNA was isolated with a TRIzol kit (Invitrogen). Reverse transcription of purified RNA was performed using oligo(dT) priming and Superscript III reverse transcriptase according to the manufacturer's instructions (Invitrogen). Quantitative real-time PCR for Axin2, NKD1, and GAPDH was performed with the TaKaRa SYBR Premix Ex Taq kit on the ABI PRISM 7500 system (Applied Biosystems). The primers used were human-specific as follows: Axin2, 5′-AGTGTGAGGTCCACGGAAAC-3′ (forward) and 5′-CTTCACACTGCGATGCATTT-3′ (reverse); NKD1, 5′-GTCAACCACTCCCCAACATC-3′ (forward) and 5′-AATGGTGGTAGCAGCCAGAC-3′ (reverse); GAPDH, 5′-AGGTCGGAGTCAACGGATTTG-3′ (forward) and 5′-TGTAAACCATGTAGTTGAGGTCA-3′ (reverse); RPL10, 5′-AGCTGCAGAACAAG GAGCAT-3′ (forward) and 5′-GTGAAGCCCC ACTTCTTTGA-3′ (reverse).
Nonradioactive metabolic labeling assay
The nonradioactive metabolic labeling assay mainly includes four parts: nascent protein labeling, immunoprecipitation of the protein of interest (for example, TCF4), Click-iT reaction of the immunoprecipitated protein, and detection of the nascent protein of interest by immunoblotting (39). For nascent protein labeling, HEK293 cells at 80–90% confluency were incubated in complete normal medium with the indicated dose of PTL or CHX for 1 h. Then HEK293 cells were washed once with warm PBS and incubated with methionine-free medium (Thermo Fisher Scientific) together with PTL or CHX at 37 °C for 1 h to deplete methionine reserves. Cells were further incubated with methionine-free medium plus 40 μm methionine analog l-homopropargylglycine (Thermo Fisher Scientific) with PTL or CHX for 4 h instead of the traditional [35S]methionine. 4 h later, cells were harvested in lysis buffer (1% SDS in 50 mm Tris-HCl [pH 8.0]) with protease inhibitors. The immunoprecipitation experiment was performed using protein A/G Plus–agarose (Santa Cruz Biotechnology) and antibody against TCF4 (Millipore) or LEF1 (Cell Signaling Technology), following the recommended procedures. The immunoprecipitated TCF4 or LEF1 was further labeled with tetramethylrhodamine (TAMRA) azide (Thermo Fisher Scientific) via a copper-catalyzed reaction between azide and l-homopropargylglycine using the Click-iT reaction buffer kit (Thermo Fisher Scientific). The TAMRA-labeled protein was detected by standard immunoblotting using an antibody against TAMRA azide (Thermo Fisher Scientific). Of note, the molecular mass of detected protein should be a bit larger than expected, and the increased molecular mass should be the molecular mass of TAMRA azide (555 Da) multiplied by the number of methionine residues in the labeled protein, and this labeled protein is probably not recognized by the normal antibody because of the integration of TAMRA azide.
Biotin pulldown assay
HEK293T cells were transfected with HA-tagged RPL10 plasmids. After 24 h of transfection, the cells were lysed with 500 μl of lysis buffer (50 mm Tris-HCl, 150 mm NaCl, 1% (v/v) Triton X-100, and 5 mm EDTA [pH 7.4]) containing proteinase inhibitors and centrifuged at 16,000 × g for 15 min at 4 °C. 20 μl of the supernatant was used as the input by adding 20 μl of 2× loading buffer, and the remaining 480 μl of supernatant was mixed with biotin-PTL (10 μm) together with streptavidin–agarose (Novex, Carlsbad, CA) at 4 °C for 1 h. For competition experiments, PTL (20 μm) was added to the mixture. The beads were washed three times and resuspended in 40 μl of 2× loading buffer.
Colony-forming assays
For colony formation assays, cells were seeded in 6-well plates (1500 cells/well for Caco2 cells, 2000 cells/well for HT29 cells, and 1000 cells/well for SW480 cells). The cells were treated with control (DMSO) or PTL with medium changes every 3 days until visible colonies formed. At the end of the experiment, cells were washed with PBS twice and fixed with cold methanol for 30 min. After fixation, cells were stained with 2% crystal violet (25% methanol) at room temperature for 1 h. After staining, the plates were rinsed with distilled water and dried.
Statistical analysis
An unpaired Student's t test was used to evaluate the difference between the two treatments. p < 0.05 was considered to be statistically significant, and a p < 0.01 was considered to be extremely statistically significant. Statistical analyses were performed using SYSTAT SigmaPlot 10.0 statistics software.
Author contributions
X. Z., C. Y., C. T., C. L., and F. N. conducted the experiments. X. S., R. Z., X. H., D. W., and L. L. analyzed the results. X. Z., C. Y., X. S., and L. L. wrote the manuscript with comments from all authors. L. L. and X. H. guided all aspects of this study.
Supplementary Material
Acknowledgments
We thank the Core Facility of Molecular Biology (Shanghai Institute of Biochemistry and Cell Biology) for providing instrument access and technological backup.
This work was supported by National Key R&D Program of China Grant 2017YFA0503600), National Natural Science Foundation of China Grants 31530094 and 31571456, and Strategic Priority Research Program of the Chinese Academy of Sciences Grants XDB19000000 and XDA12010301. This work was also supported by the Chinese Academy of Sciences/State Administration of Foreign Experts Affairs International Partnership Program for Creative Research Teams. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S3 and information.
- LEF
- lymphoid enhancer factor
- TCF
- T-cell factor
- CREB
- cAMP-response element-binding protein
- PTL
- parthenolide
- CM
- conditioned medium
- CHIR
- CHIR99021
- CHX
- cycloheximide
- HA
- hemagglutinin
- FBS
- fetal bovine serum
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- TAMRA
- tetramethylrhodamine
- Ctr
- control
- DMAPT
- dimethylamino-parthenolide.
References
- 1. Nusse R., and Clevers H. (2017) Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell 169, 985–999 10.1016/j.cell.2017.05.016 [DOI] [PubMed] [Google Scholar]
- 2. Clevers H., and Nusse R. (2012) Wnt/β-catenin signaling and disease. Cell 149, 1192–1205 10.1016/j.cell.2012.05.012 [DOI] [PubMed] [Google Scholar]
- 3. Anastas J. N., and Moon R. T. (2013) WNT signalling pathways as therapeutic targets in cancer. Nat. Rev. Cancer 13, 11–26 10.1038/nrc3419 [DOI] [PubMed] [Google Scholar]
- 4. Hwang S. Y., Deng X., Byun S., Lee C., Lee S. J., Suh H., Zhang J., Kang Q., Zhang T., Westover K. D., Mandinova A., and Lee S. W. (2016) Direct targeting of β-catenin by a small molecule stimulates proteasomal degradation and suppresses oncogenic Wnt/β-catenin signaling. Cell Rep. 16, 28–36 10.1016/j.celrep.2016.05.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Trosset J. Y., Dalvit C., Knapp S., Fasolini M., Veronesi M., Mantegani S., Gianellini L. M., Catana C., Sundström M., Stouten P. F., and Moll J. K. (2006) Inhibition of protein-protein interactions: the discovery of druglike β-catenin inhibitors by combining virtual and biophysical screening. Proteins 64, 60–67 10.1002/prot.20955 [DOI] [PubMed] [Google Scholar]
- 6. Gonsalves F. C., Klein K., Carson B. B., Katz S., Ekas L. A., Evans S., Nagourney R., Cardozo T., Brown A. M., and DasGupta R. (2011) An RNAi-based chemical genetic screen identifies three small-molecule inhibitors of the Wnt/wingless signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 108, 5954–5963 10.1073/pnas.1017496108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Emami K. H., Nguyen C., Ma H., Kim D. H., Jeong K. W., Eguchi M., Moon R. T., Teo J. L., Oh S. W., Kim H. Y., Moon S. H., Ha J. R., and Kahn M. (2004) A small molecule inhibitor of β-catenin/cyclic AMP response element-binding protein transcription. Proc. Natl. Acad. Sci. U.S.A. 101, 12682–12687 10.1073/pnas.0404875101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang W., Liu H., Wang S., Hao X., and Li L. (2011) A diterpenoid derivative 15-oxospiramilactone inhibits Wnt/β-catenin signaling and colon cancer cell tumorigenesis. Cell Res 21, 730–740 10.1038/cr.2011.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. He X., Zhang W., Yan C., Nie F., Li C., Liu X., Fei C., Li S., Song X., Jia Y., Zeng R., Wu D., Pan W., Hao X., and Li L. (2017) Chemical biology reveals CARF as a positive regulator of canonical Wnt signaling by promoting TCF/β-catenin transcriptional activity. Cell Discov. 3, 17003 10.1038/celldisc.2017.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bohlmann F., and Zdero C. (1982) Sesquiterpene lactones and other constituents from Tanacetum parthenium. Phytochemistry 21, 2543–2549 10.1016/0031-9422(82)85253-9 [DOI] [Google Scholar]
- 11. Stojakowska A., and Kisiel W. (1997) Production of parthenolide in organ cultures of feverfew. Plant Cell Tiss. Org. 47, 159–162 10.1007/BF02318952 [DOI] [Google Scholar]
- 12. Knight D. W. (1995) Feverfew: chemistry and biological activity. Nat. Prod. Rep. 12, 271–276 10.1039/np9951200271 [DOI] [PubMed] [Google Scholar]
- 13. Ghantous A., Sinjab A., Herceg Z., and Darwiche N. (2013) Parthenolide: from plant shoots to cancer roots. Drug. Discov. Today 18, 894–905 10.1016/j.drudis.2013.05.005 [DOI] [PubMed] [Google Scholar]
- 14. Bork P. M., Schmitz M. L., Kuhnt M., Escher C., and Heinrich M. (1997) Sesquiterpene lactone containing Mexican Indian medicinal plants and pure sesquiterpene lactones as potent inhibitors of transcription factor NF-κB. FEBS Lett. 402, 85–90 10.1016/S0014-5793(96)01502-5 [DOI] [PubMed] [Google Scholar]
- 15. Wen J., You K. R., Lee S. Y., Song C. H., and Kim D. G. (2002) Oxidative stress-mediated apoptosis: the anticancer effect of the sesquiterpene lactone parthenolide. J. Biol. Chem. 277, 38954–38964 10.1074/jbc.M203842200 [DOI] [PubMed] [Google Scholar]
- 16. Yang C., Yang Q. O., Kong Q. J., Yuan W., and Ou Yang Y. P. (2016) Parthenolide induces reactive oxygen species-mediated autophagic cell death in human osteosarcoma cells. Cell Physiol. Biochem. 40, 146–154 10.1159/000452532 [DOI] [PubMed] [Google Scholar]
- 17. Liu Z., Liu S., Xie Z., Pavlovicz R. E., Wu J., Chen P., Aimiuwu J., Pang J., Bhasin D., Neviani P., Fuchs J. R., Plass C., Li P. K., Li C., Huang T. H., et al. (2009) Modulation of DNA methylation by a sesquiterpene lactone parthenolide. J. Pharmacol. Exp. Ther. 329, 505–514 10.1124/jpet.108.147934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gopal Y. N., Arora T. S., and Van Dyke M. W. (2007) Parthenolide specifically depletes histone deacetylase 1 protein and induces cell death through ataxia telangiectasia mutated. Chem. Biol. 14, 813–823 10.1016/j.chembiol.2007.06.007 [DOI] [PubMed] [Google Scholar]
- 19. Guzman M. L., Rossi R. M., Karnischky L., Li X., Peterson D. R., Howard D. S., and Jordan C. T. (2005) The sesquiterpene lactone parthenolide induces apoptosis of human acute myelogenous leukemia stem and progenitor cells. Blood 105, 4163–4169 10.1182/blood-2004-10-4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zuch D., Giang A. H., Shapovalov Y., Schwarz E., Rosier R., O'Keefe R., and Eliseev R. A. (2012) Targeting radioresistant osteosarcoma cells with parthenolide. J. Cell. Biochem. 113, 1282–1291 10.1002/jcb.24002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou J. B., Zhang H., Gu P. H., Bai J. N., Margolick J. B., and Zhang Y. (2008) NF-κB pathway inhibitors preferentially inhibit breast cancer stem-like cells. Breast Cancer Res. Tr. 111, 419–427 10.1007/s10549-007-9798-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Czyz M., Koprowska K., and Sztiller-Sikorska M. (2013) Parthenolide reduces the frequency of ABCB5-positive cells and clonogenic capacity of melanoma cells from anchorage independent melanospheres. Cancer Biol. Ther. 14, 135–145 10.4161/cbt.22952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Uchibori R., Tsukahara T., Mizuguchi H., Saga Y., Urabe M., Mizukami H., Kume A., and Ozawa K. (2013) NF-κB activity regulates mesenchymal stem cell accumulation at tumor sites. Cancer Res. 73, 364–372 10.1158/0008-5472.CAN-12-0088 [DOI] [PubMed] [Google Scholar]
- 24. Kawasaki B. T., Hurt E. M., Kalathur M., Duhagon M. A., Milner J. A., Kim Y. S., and Farrar W. L. (2009) Effects of the sesquiterpene lactone parthenolide on prostate tumor-initiating cells: an integrated molecular profiling approach. Prostate 69, 827–837 10.1002/pros.20931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Birnie R., Bryce S. D., Roome C., Dussupt V., Droop A., Lang S. H., Berry P. A., Hyde C. F., Lewis J. L., Stower M. J., Maitland N. J., and Collins A. T. (2008) Gene expression profiling of human prostate cancer stem cells reveals a pro-inflammatory phenotype and the importance of extracellular matrix interactions. Genome Biol. 9, R83 10.1186/gb-2008-9-5-r83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang S. M., Mishina Y. M., Liu S., Cheung A., Stegmeier F., Michaud G. A., Charlat O., Wiellette E., Zhang Y., Wiessner S., Hild M., Shi X., Wilson C. J., Mickanin C., Myer V., et al. (2009) Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461, 614–620 10.1038/nature08356 [DOI] [PubMed] [Google Scholar]
- 27. Shi C., Wang Y., Guo Y., Chen Y., and Liu N. (2017) Cooperative down-regulation of ribosomal protein L10 and NF-κB signaling pathway is responsible for the anti-proliferative effects by DMAPT in pancreatic cancer cells. Oncotarget 8, 35009–35018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Giles R. H., van Es J. H., and Clevers H. (2003) Caught up in a Wnt storm: Wnt signaling in cancer. Biochim. Biophys. Acta 1653, 1–24 [DOI] [PubMed] [Google Scholar]
- 29. Schuijers J., Mokry M., Hatzis P., Cuppen E., and Clevers H. (2014) Wnt-induced transcriptional activation is exclusively mediated by TCF/LEF. EMBO J. 33, 146–156 10.1002/embj.201385358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Franz A., Shlyueva D., Brunner E., Stark A., and Basler K. (2017) Probing the canonicity of the Wnt/Wingless signaling pathway. PLoS Genet. 13, e1006700 10.1371/journal.pgen.1006700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hrckulak D., Kolar M., Strnad H., and Korinek V. (2016) TCF/LEF transcription factors: an update from the internet resources. Cancers (Basel) 8, 70 10.3390/cancers8070070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ishiguro H., Wakasugi T., Terashita Y., Sakamoto N., Tanaka T., Sagawa H., Okubo T., and Takeyama H. (2016) Nuclear expression of TCF4/TCF7L2 is correlated with poor prognosis in patients with esophageal squamous cell carcinoma. Cell. Mol. Biol. Lett. 21, 5 10.1186/s11658-016-0006-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Goudarzi K. M., and Lindström M. S. (2016) Role of ribosomal protein mutations in tumor development (review). Int. J. Oncol. 48, 1313–1324 10.3892/ijo.2016.3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shi Z., Fujii K., Kovary K. M., Genuth N. R., Röst H. L., Teruel M. N., and Barna M. (2017) Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genome-wide. Mol. Cell 67, 71–83.e7 10.1016/j.molcel.2017.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sendoel A., Dunn J. G., Rodriguez E. H., Naik S., Gomez N. C., Hurwitz B., Levorse J., Dill B. D., Schramek D., Molina H., Weissman J. S., and Fuchs E. (2017) Translation from unconventional 5′ start sites drives tumour initiation. Nature 541, 494–499 10.1038/nature21036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mao J., Wang J., Liu B., Pan W., Farr G. H. 3rd, Flynn C., Yuan H., Takada S., Kimelman D., Li L., and Wu D. (2001) Low-density lipoprotein receptor-related protein-5 binds to Axin and regulates the canonical Wnt signaling pathway. Mol. Cell 7, 801–809 10.1016/S1097-2765(01)00224-6 [DOI] [PubMed] [Google Scholar]
- 37. Li L., Yuan H., Xie W., Mao J., Caruso A. M., McMahon A., Sussman D. J., and Wu D. (1999) Dishevelled proteins lead to two signaling pathways: regulation of LEF-1 and c-Jun N-terminal kinase in mammalian cells. J. Biol. Chem. 274, 129–134 10.1074/jbc.274.1.129 [DOI] [PubMed] [Google Scholar]
- 38. Mao L., Liu C., Wang Z., Niu X., Xue L., Zhou Z., Cai Z., Yu M., Li Y., Wu D., and Li L. (2016) A genome-wide loss-of-function screening method for minimizing false-negatives caused by functional redundancy. Cell Res. 26, 1067–1070 10.1038/cr.2016.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Qu Y., Olsen J. R., Yuan X., Cheng P. F., Levesque M. P., Brokstad K. A., Hoffman P. S., Oyan A. M., Zhang W., Kalland K. H., and Ke X. (2018) Small molecule promotes β-catenin citrullination and inhibits Wnt signaling in cancer. Nat. Chem. Biol. 14, 94–101 10.1038/nchembio.2510 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.