Abstract
Mammalian haploid embryonic stem cells (haESCs) serve as a powerful tool for genetic analyses at both the cellular and organismal levels. However, spontaneous diploidization of haESCs limits their use in these analyses. Addition of small molecules to the culture medium to control the cell cycle can slow down diploidization, but cell-sorting methods such as FACS are still required to enrich haploid cells for long-term maintenance in vitro. Here, acting on our observation that haploid and diploidized cells differ in diameter, we developed a simplified filtration method to enrich haploid cells from cultured haESCs. We found that regular cell filtration with this system reliably maintained the haploidy of mouse haESCs for over 30 passages. Importantly, CRISPR/Cas9-mediated knockout and knockin were successfully achieved in the filtered cells, leading to stable haploid cell lines carrying the desired gene modifications. Of note, by injecting haESCs into metaphase II oocytes, we efficiently obtained live mice with the expected genetic traits, indicating that regular filtration maintained the functional integrity of haESCs. Moreover, this filtration system was also feasible for derivation of mouse haESCs from parthenogenetic haploid blastocysts and for human haESC maintenance. In conclusion, we have identified a reliable, efficient, and easy-to-handle technique for countering diploidization of haploid cells, a major obstacle in haESC applications.
Keywords: embryonic stem cell, CRISPR/Cas, cell sorting, cell biology, embryo, diploidization, FACS, filtration, haploid embryonic stem cells, haploidy
Introduction
HaESCs3 hold great advantages to reveal gene functions and mechanisms underlying rudimentary bioprocesses because they contain only one set of chromosomes (1–4). Advances in mammalian haESC-related techniques, from generating stable haESCs of different species (5–10) to producing semicloned (SC) mice through intracytoplasmic injection of androgenetic haESCs (AG-haESCs) (ICAHCI) into metaphase II oocytes (11–13), have been extending the utility of haESCs in a revolutionary way. Those successes bring genetic study from the cellular to the organismal level, provide more reliable and systematic analyses, and cut labor and time cost dramatically compared with regular genetic strategies. Recent thriving of CRISPR-Cas9 technology (14, 15) gives a major boost to haESC application, allowing haESCs to be used for genome-wide knockout-based screening and efficient site-specific knock-in (12, 16, 17). All of the improvements in haESCs techniques give researchers a versatile tool to study genes of interest.
Nevertheless, the major hurdle of haESC application is spontaneous diploidization of haploid cells along cell passages. Diploidization is one specific characteristic of haESCs (1) and may be related to endoreduplication (18) or prolonged metaphase during the cell cycle in haESCs compared with that in diploid ESCs (19, 20). Chemicals that accelerate G2/M transition have been successfully used to stabilize the haploid state for several weeks longer (20–22). However, detailed differences in mitosis between haploid cells and diploid cells and the molecular mechanisms underlying diploidization for generating haESCs with more stable haploidy remain to be uncovered (22, 23).
To date, haploidy maintenance in the long term still needs regular FACS to enrich haploid cells. This is a complicated procedure in which haESCs are first stained with Hoechst 33342 and then employed for haploid cell enrichment according to DNA content using a flow cytometer (24). In this study, we describe a simple but efficient filtration method for long-term haESC maintenance without hampering haESC physiology and functionality. This method provides an alternative way to maintain haploidy and will greatly promote the application of haESCs.
Results
Long-term maintenance of mouse haESCs through filtration
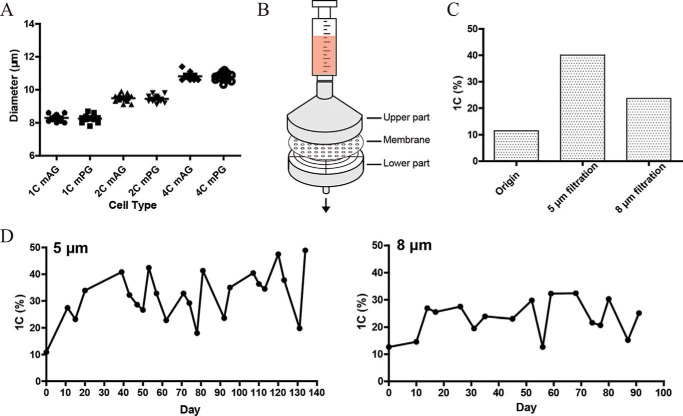
A cell flow cytometer can separate Hoechst 33342–stained haESCs into three groups according to DNA content: 1C (haploid cells in G0/G1 phase), 2C (haploid cells in G2/M phase and diploid cells in G0/G1 phase), and 4C (diploid cells in G2/M phase) (24). Cell size analyses showed that the average diameter of 1C, 2C, and 4C in mouse haESCs is 8.3 μm, 9.5 μm, and 10.8 μm, respectively (Fig. 1A), indicating an obvious size difference among cells with different DNA content. Parthenogenetic haESCs (PG-haESCs) have a similar size as AG-haESCs with same DNA content (Fig. 1A). These results suggest that 1C cells might be directly purified from a cell mixture by physical filtration based on their size. To test this, we assembled a filter using commercialized membranes with a pore size of 5 or 8 μm (Fig. 1B). A haESC suspension could be easily filtered through our device, and both 5- and 8-μm pore size membranes could be used efficiently to enrich haploid cells (Fig. 1C). Membranes with 5-μm pore size showed a higher efficiency compared with 8 μm pores, indicating that smaller pores block more large cells. To optimize the filtration conditions, we performed systematic experiments to test whether the cell concentration before filtration and pore size of the filter membrane would affect the filtration results. Cell suspensions with different concentrations were used for filtration, and filtrates were collected for analyzing the ratio of 1C cells. The results showed that haploid cells could be efficiently enriched from cell suspensions of different concentrations by using membranes with both a 5-μm and 8-μm pore size, and efficiency was higher in initial filtered drops, suggesting that if enough filtered cells were obtained for experiments, then the later drops could be discarded (Fig. S1, A–C). Meanwhile, membranes with a 5-μm pore size exhibited a relatively better enrichment efficiency compared with that 8-μm pore size, consistent with the results in Fig. 1C.
Figure 1.
Long-term maintenance of mouse haESCs through filtration. A, diameters of mouse haESCs with 1C, 2C, or 4C (C, DNA content; mAG, mouse androgenetic haESCs; mPG, mouse parthenogenetic haESCs). B, diagram of the filtration system. C, filters with 5-μm and 8-μm pore size membranes could enrich mouse AG-haESCs efficiently. D, filters with 5-μm (left panel) and 8 μm (right panel) membranes could be used to maintain the haploidy of H19▵DMR-IG▵DMR-AGH for a long term.
Next we asked whether filtration could be used to maintain the haploidy of mouse haESCs stably for long-term in vitro culturing. To test this, we used two haploid cells lines, H19▵DMR-IG▵DMR-AGH (O48) and H19▵DMR-IG▵DMR-AGH-OG3 (408), generated previously (12). As expected, the haploidy of both cell lines could be well maintained with regular filtration using membranes with 5- or 8-μm pore size for over 30 passages (Fig. 1D and S1D). Taken together, these results demonstrate that filtration can serve as an effective method to maintain mouse haESCs.
Filtrated haESCs are functional
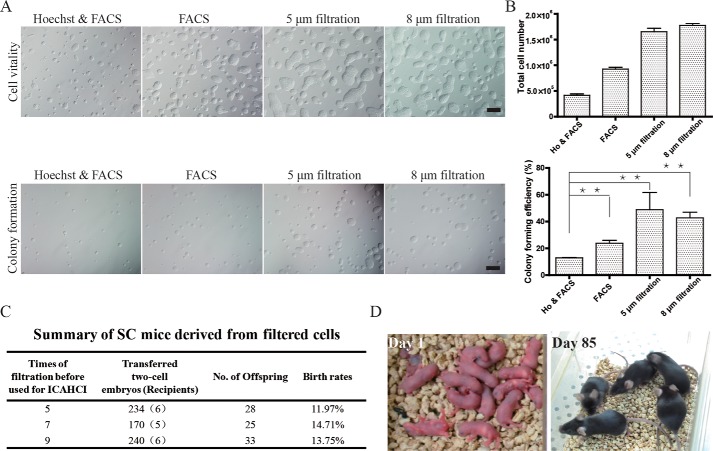
Having demonstrated that filtration can be efficiently employed to enrich haploid cells, we next examined whether haESCs maintained by regular filtration are functionally equivalent to those from regular FACS enrichment. We first tested the proliferation and colony formation abilities of the filtered cells. To do this, we plated cells obtained by different strategies and counted the total cell and colony numbers after 3 and 5 days, respectively. Compared with haploid cells enriched by FACS with or without Hoechst 33342 staining, filtered cells had better cell vitality and colony formation ability in the first passage (Fig. 2, A and B). Moreover, cells enriched by FACS need more time to recover from the damage caused by the FACS procedures, but filtered cells required less time to recover, indicating that there is less damage caused by filtration. These results imply that filtration might avoid the potential damage induced by the low cytotoxicity of Hoechst staining (25, 26) and trauma attributable to FACS purification.
Figure 2.
Filtered haESCs are functional. A, filtered cells had better cell vitality (scale bar = 200 μm) and colony-forming ability (scale bar = 500 μm) in the first passage after enrichment compared with FACS. B, summary of A. Total cell numbers and colony-forming efficiencies of filtered haESCs were significantly better than haESCs sorted by FACS. Ho, Hoechst 33342. **, p < 0.01. C, filtered haESCs of H19▵DMR-IG▵DMR-AGH with different times of filtration could efficiently support the birth of SC mice. D, newborn and adult SC mice generated using filtered haESCs from H19▵DMR-IG▵DMR-AGH.
AG-haESCs carrying both H19-DMR and IG-DMR deletions (DKO-AG-haESCs) could be used as the sperm replacement to efficiently produce SC mice through ICAHCI (12). We then tested whether DKO-AG-haESCs maintained by regular filtration are still feasible for generation of SC mice. To do this, DKO-AG-haESCs, after multiple rounds of filtration, were used as donors for ICAHCI. The results indicated that, as expected, all tested cells were capable of efficiently generating SC mice (Fig. 2, C and D), further confirming that haESCs enriched by filtration preserve normal function as those from FACS.
Generation of mutant mice using DKO-AG-haESCs enriched by filtration
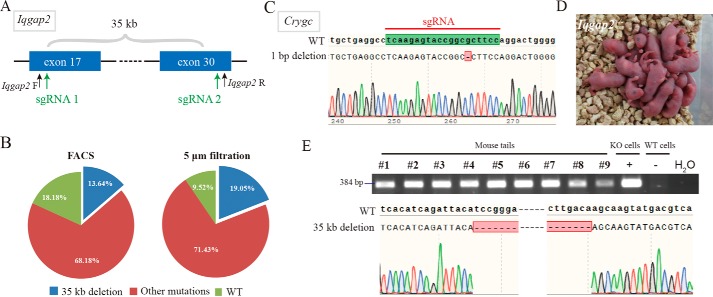
Because AG-haESCs are a feasible tool for genetic modifications using CRISPR-Cas9 and ICAHCI to give pups carrying the corresponding genetic traits (12, 16), we tested whether filtration could be used in place of FACS during this process. We first attempted induction of large deletion in a gene of interest, Iqgap2 (27, 28). We transfected constructs expressing Cas9 and two sgRNAs targeting exon 17 and exon 30, respectively, which may cause a 35-kb deletion (Fig. 3A). The results showed no significant difference in transfection efficiency and generation of mutant cell lines between cells enriched by filtration and FACS (Fig. 3B and Table S1). A total of 21 stable haploid cell lines were generated through the filtration method. DNA sequencing analyses indicated that four of 21 carried an expected mutation of 35-kb deletion. We next tested whether, through filtration, a precise point mutation could be inserted into Crygc by transfection of both CRISPR and donor DNA with the 1-bp deletion in filtered DKO-AG-hESCs (23, 29, 30) (Fig. 3C). As expected, haploid cell lines with the point mutation could be efficiently generated in filtrated cells (Table S1). Finally, we performed ICHACI using filtered haploid cells carrying a 35-kb deletion in Iqgap2 from the cell line of 2-1 as donors. Of 215 transferred SC embryos, a total of 42 live pups were born naturally (Fig. 3D), consistent with our previous observations that pregnant females carrying SC embryos from DKO-AG-haESCs could deliver pups by themselves (12, 16). Genotyping analyses indicated that all of them carried heterozygous deletion in Iqgap2 (Fig. 3E). Taken together, these point out that our enrichment method does not hamper the vitality and functional integrity of haESCs and that filtration is a reliable procedure to maintain haploidy.
Figure 3.
Generation of mutant mice using DKO-AG-haESCs enriched by filtration. A, diagram of two sgRNAs targeting two exons of Iqgap2, respectively, that may cause 35-kb deletion. Iqgap2 F and Iqgap2 R represent primers used for genotyping of Iqgap2. B, Iqgap2 with 35-kb deletion achieved efficiently in filtered H19▵DMR-IG▵DMR-AGH cells. C, diagram of 1-bp deletion in Crygc and the sequence of sgRNA targeting Crygc. D, filtered haploid cells carrying the 35-kb deletion in Iqgap2 supported the birth of SC mice. E, genetic trait analysis revealed that the SC mice generated in D all carried the heterozygous mutation in Iqgap2. KO cells, cells from the line of 2-1; WT cells, H19▵DMR-IG▵DMR-AGH.
Applications of filtration in derivation of mouse PG-haESCs and in maintaining human haESCs
We also explored the potential use of filtration in generating haESCs from haploid PG blastocysts obtained by artificial activation. PG haploid blastocysts were cultured in a standard ESC culture system supplemented with 2i (MEK and GSK inhibitors) (11, 31). Eight ESC lines were generated and subjected to filtration to enrich for haploid 1C cells, leading to the establishment of PG-haESC lines (Fig. 4A).
Figure 4.
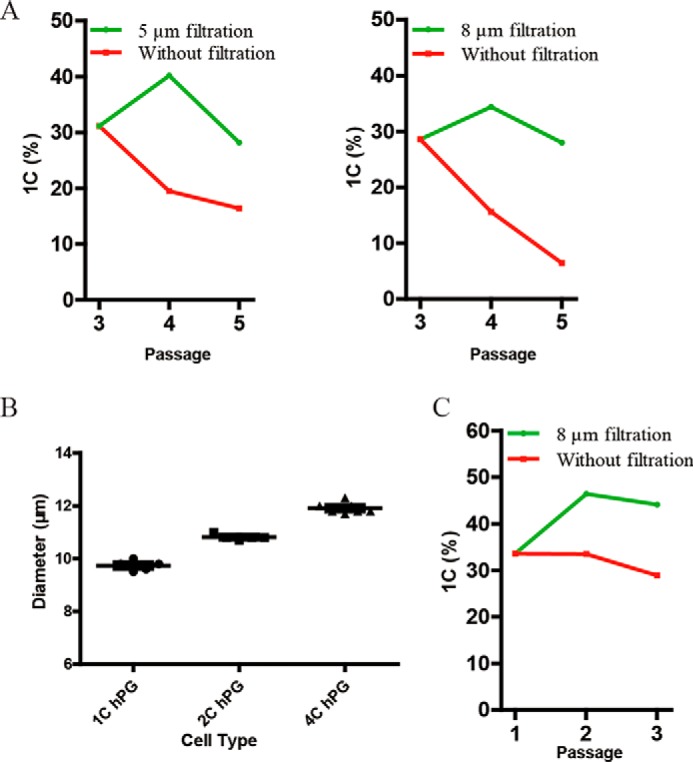
Applications of filtration in derivation of mouse PG-haESCs and maintaining human haESCs. A, two PG-haESCs were derived from mouse parthenogenetic haploid blastocysts by using filters with 5-μm or 8-μm membranes. B, diameters of human haESCs with 1C, 2C, or 4C. C, DNA content; hPG, human parthenogenetic haESCs. C, human haESCs could be enriched by using filters with 8-μm membranes.
Finally, we tested whether filtration could be adopted for maintenance of human haESCs, which we generated previously (8). To this, we first performed size analyses and found that the diameters of 1C, 2C, and 4C in human haESCs are 9.7 μm, 10.8 μm, and 11.9 μm (Fig. 4B), respectively. We then chose a filter membrane with 8-μm pore size for filtration of human haESCs. Consistently, our method could well preserve the haploidy of human haESCs (Fig. 4C). These results indicate that filtration has a broad use with haESCs from different species.
Discussion
Mammalian haESCs have been proven to be invaluable for genetic analysis (1–4, 32). However, self-diploidization, an intrinsic feature of haESCs, limits their broad application because of a requirement of frequent FACS to enrich haploid cells regularly. FACS is large device–dependent, high-cost, and complicated, which may generate some unexpected mutations by Hoechst, laser, and voltage in sorted cells (25, 26). Here we establish a new strategy to isolate haploid cells from diploidized cells through filtration, which is an easy, efficient, stable, and reliable method for long-term maintenance of haESCs. We only tested 5- and 8-μm pore membranes in this study, but considering the size and plasticity of haploid cells, with choices of 6- or 7-μm pore membranes if commercially available, we might achieve a better balance between efficiency and yield. Nevertheless, our method enriches mouse and human haploid cells at relatively high ratios, and enriched cells by filtration preserve not only their vitality well but also their functional integrity. In addition, filtration has several obvious advantages over FACS, such as convenient handling, less physical damage to cells, no cell toxicity, and economy. Our method works on both mouse haESCs and human haESCs, and with proper pore size membranes, it can also be applied to haESCs from other species.
During the preparation of this work for publication, an independent group also reported filtration-mediated enrichment of mouse PG-haESCs (33). In comparison with their separation unit using hydrostatic pressure to separate cells, our filter is assembled much easier, and the filtration process is simpler, steadier, and more reliable. All parts of the filter can be subjected to sterilization. Meanwhile, beside mouse haESCs, we also demonstrated that filtration can be used to maintain human haESCs. Moreover, we explored the use of filtration to derive haploid cell lines from blastocysts and establish gene-modified AG-haESCs, followed by generation of SC mice with corresponding genetic traits through ICAHCI. Interestingly, we showed that both 5-μm and 8-μm pore size membranes can be adopted for filtration. In contrast, Freimann and Wutz (33) reported that 5 μm did not work in their system. One potential reason could be that the pressure induced by syringe in our device is higher than their hydrostatic pressure. Actually, in our system, a 5-μm pore size membrane allows us to obtain haploid cells at high efficiency without causing detectable damage (Fig. 2, A and B).
In summary, filtration is an efficient and simple method for maintenance of haESCs. A future study needs to be done to fine-tune the filtration procedures. In the meantime, we hope that the method we have developed will facilitate the application of haESCs in genetic analysis.
Experimental procedures
Cell culture
Mouse haESCs were cultured in a standard ESC culture system supplemented with 2i (MEK and GSK inhibitors) (11, 12, 31). Cells were dissociated by 0.05% trypsin–EDTA and passaged regularly every 2–3 days. Human haESCs were cultured in mTeSRTM1 (Stem Cell) with 10 μm Y-27632 (Selleck), disassociated by Accutase, and passaged regularly every 4–8 days. Plates were coated with Matrigel matrix (Corning, with a 1:100 dilution) overnight before passage to increase attachment of cells. Two mouse AG-haESCs, H19▵DMR-IG▵DMR-AGH and H19▵DMR-IG▵DMR-AGH-OG3 (12), and one human haESC, hPGES2 (8), were used.
Fluorescence-activated cell sorting
HaESCs were dissociated into single cells and incubated with 15 μg/ml Hoechst 33342 in a 37 °C water bath for 5 min. Sorting was then conducted by using FACS Aria II (BD Biosciences). When sorting haploid cells without Hoechst staining, we used the same cells that were stained with Hoechst 33342 to help us to gate cells containing 1C in the forward-scattered light (FSC) and side-scattered light (SSC) coordinate plane system.
HaESC diameter measurement
FACS was performed to purify 1C, 2C, and 4C peaks. Cell diameters were then measured by using CountessTM (Invitrogen).
Filtration
A filter was composed of a 25-mm filter holder (Millipore, SX0002500) and a membrane with 5-μm pores (Millipore, TMTP02500) or 8-μm pores (Millipore, TETP02500). The filter holders were sterilized by autoclaving, and membranes were sterilized by 25-kilogray γ-rays. The concentrations of cell suspensions used for filtration affect the distribution of cell numbers and percentages of 1C in filtered drops (Fig. S1, A–C). For a 5-μm-pore membrane, 2 ml of cell suspension with a concentration about 1 × 106/ml was prepared, and the initial approximately 20 drops were collected for plating in a well of a 96-well plate. For an 8-μm-pore membrane, 1 ml of cell suspension with a concentration about 5 × 105/ml was prepared, and the initial approximately 10 drops were collected and plated in a well of a 96-well plate. Cells were passaged to a well of a 6-well plate 2–3 days later. When enriching human haESCs, we used 8-μm filters and collected enough filtered cells to plate in a well of a 48-well plate. Movie S1 shows the workflow of filtration.
Cell state evaluation
Haploid cells enriched by filtration and FACS were collected to evaluate cell proliferation and colony-forming abilities. For cell proliferation ability, 45,000 cells enriched by each method were cultured in a well of a 24-well plate. After 3 days, cells were dissociated and counted by CountessTM. For colony-forming efficiency, 4000 cells were plated into a well of a 12-well plate. After 5 days, the colony number in each well was estimated.
Animal use and care
All animal procedures were performed under the ethical guidelines of the Shanghai Institute of Biochemistry and Cell Biology.
Derivation of PG-haESCs from mouse PG blastocysts
Oocytes of B6D2F1 (C57BL/6 × DBA2) were obtained from superovulated females and activated for 5 h in activation medium. Embryos were then cultured in EmbryoMax® KSOM+amino acids with d-glucose (Millipore) at 37 °C under 5% CO2 in air until blastocysts formed. After removing the zona pellucida by acid Tyrode's solution, each blastocyst was transferred into a well of a 96-well plate covered with feeder cells. After around 1 week, colonies were passaged to 96-well plates. About 4 days later, cells were passaged to 24-well or 12-well plates. Filtration experiments were then performed to enrich haESCs (34).
CRISPR-Cas9-mediated gene manipulation in haESCs
SgRNAs targeting Iqgap2 were connected to pX458, which expresses green fluorescent protein. The sgRNA of Crygc was connected to the pX330-mCherry plasmid, which expresses red fluorescence protein (Addgene, 98750) (29). When constructing the double-stranded DNA donor, the sequences of the left arm and right arm were amplified from the genome and inserted into the EcoR V site of the pMD19T vector (Takara) using the Seamless Cloning Kit (Beyotime). AG-haESCs were transfected with corresponding plasmids using Lipofectamine 3000 (Thermo Fisher). 20–48 h after transfection, haploid cells expressing green or red fluorescent protein were enriched by FACS and plated into a well of a 6-well plate at a low density. Single colonies were picked and passaged to a well of a 96-well plate after 5–8 days. Filtration was performed for enrichment of haploid cells. Related sequences are listed in Table S2.
ICAHCI and embryo transfer
ICAHCI and embryo transfer were performed as described previously (12).
Author contributions
C. Q. and J. L. conceptualization; C. Q. data curation; C. Q. and M. Y. formal analysis; C. Q., S. Y., L. W., Q. Y., Y. L., and Y. C. methodology; C. Q., M. Y., and J. L. writing-original draft; C. Q., M. Y. and J. L. writing-review and editing; J. L. supervision; J. L. funding acquisition.
Supplementary Material
Acknowledgment
We thank the Genome Tagging Project Center for technical support.
This study was supported by Ministry of Science and Technology of China Grant 2014CB964803; National Natural Science Foundation of China Grants 31530048, 81672117, and 31730062; Chinese Academy of Sciences Grants XDB19010204, OYZDJ-SSW-SMC023, and KJZD-EW-L13 and the Facility-Based Open Research Program; and Shanghai Municipal Commission for Science and Technology Grants 16JC1420500, 17JC1400900, 17JC1420102, and 17411954900. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Fig. S1, Tables S1 and S2, and Movie S1.
- haESC
- haploid embryonic stem cells
- AG
- androgenetic
- PG
- parthenogenetic
- DKO
- double knockout
- SC
- semicloned
- ICAHCI
- intracytoplasmic AG-haESC injection
- sgRNA
- single-guide RNA
- MEK
- mitogen-activated protein kinase/extracellular signal-regulated kinase kinase
- GSK
- glycogen synthase kinase.
References
- 1. Bai M., Wu Y., and Li J. (2016) Generation and application of mammalian haploid embryonic stem cells. J. Int. Med. 280, 236–245 10.1111/joim.12503 [DOI] [PubMed] [Google Scholar]
- 2. Yilmaz A., Peretz M., Sagi I., and Benvenisty N. (2016) Haploid human embryonic stem cells: half the genome, double the value. Cell Stem Cell 19, 569–572 10.1016/j.stem.2016.10.009 [DOI] [PubMed] [Google Scholar]
- 3. Kokubu C., and Takeda J. (2014) When half is better than the whole: advances in haploid embryonic stem cell technology. Cell Stem Cell 14, 265–267 10.1016/j.stem.2014.02.001 [DOI] [PubMed] [Google Scholar]
- 4. Shi L., Yang H., and Li J. (2012) Haploid embryonic stem cells: an ideal tool for mammalian genetic analyses. Protein Cell 3, 806–810 10.1007/s13238-012-2096-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Leeb M., and Wutz A. (2011) Derivation of haploid embryonic stem cells from mouse embryos. Nature 479, 131–134 10.1038/nature10448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Elling U., Taubenschmid J., Wirnsberger G., O'Malley R., Demers S. P., Vanhaelen Q., Shukalyuk A. I., Schmauss G., Schramek D., Schnuetgen F., von Melchner H., Ecker J. R., Stanford W. L., Zuber J., Stark A., and Penninger J. M. (2011) Forward and reverse genetics through derivation of haploid mouse embryonic stem cells. Cell Stem Cell 9, 563–574 10.1016/j.stem.2011.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang H., Liu Z., Ma Y., Zhong C., Yin Q., Zhou C., Shi L., Cai Y., Zhao H., Wang H., Tang F., Wang Y., Zhang C., Liu X. Y., Lai D., et al. (2013) Generation of haploid embryonic stem cells from Macaca fascicularis monkey parthenotes. Cell Res. 23, 1187–1200 10.1038/cr.2013.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhong C., Zhang M., Yin Q., Zhao H., Wang Y., Huang S., Tao W., Wu K., Chen Z. J., and Li J. (2016) Generation of human haploid embryonic stem cells from parthenogenetic embryos obtained by microsurgical removal of male pronucleus. Cell Res. 26, 743–746 10.1038/cr.2016.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sagi I., Chia G., Golan-Lev T., Peretz M., Weissbein U., Sui L., Sauer M. V., Yanuka O., Egli D., and Benvenisty N. (2016) Derivation and differentiation of haploid human embryonic stem cells. Nature 532, 107–111 10.1038/nature17408 [DOI] [PubMed] [Google Scholar]
- 10. Li W., Li X., Li T., Jiang M. G., Wan H., Luo G. Z., Feng C., Cui X., Teng F., Yuan Y., Zhou Q., Gu Q., Shuai L., Sha J., Xiao Y., et al. (2014) Genetic modification and screening in rat using haploid embryonic stem cells. Cell Stem Cell 14, 404–414 10.1016/j.stem.2013.11.016 [DOI] [PubMed] [Google Scholar]
- 11. Yang H., Shi L., Wang B. A., Liang D., Zhong C., Liu W., Nie Y., Liu J., Zhao J., Gao X., Li D., Xu G. L., and Li J. (2012) Generation of genetically modified mice by oocyte injection of androgenetic haploid embryonic stem cells. Cell 149, 605–617 10.1016/j.cell.2012.04.002 [DOI] [PubMed] [Google Scholar]
- 12. Zhong C., Yin Q., Xie Z., Bai M., Dong R., Tang W., Xing Y. H., Zhang H., Yang S., Chen L. L., Bartolomei M. S., Ferguson-Smith A., Li D., Yang L., Wu Y., and Li J. (2015) CRISPR-Cas9-mediated genetic screening in mice with haploid embryonic stem cells carrying a guide RNA library. Cell Stem Cell 17, 221–232 10.1016/j.stem.2015.06.005 [DOI] [PubMed] [Google Scholar]
- 13. Li W., Shuai L., Wan H., Dong M., Wang M., Sang L., Feng C., Luo G. Z., Li T., Li X., Wang L., Zheng Q. Y., Sheng C., Wu H. J., Liu Z., et al. (2012) Androgenetic haploid embryonic stem cells produce live transgenic mice. Nature 490, 407–411 10.1038/nature11435 [DOI] [PubMed] [Google Scholar]
- 14. Mali P., Yang L., Esvelt K. M., Aach J., Guell M., DiCarlo J. E., Norville J. E., and Church G. M. (2013) RNA-guided human genome engineering via Cas9. Science 339, 823–826 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P. D., Wu X., Jiang W., Marraffini L. A., and Zhang F. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wei L., Wang X., Yang S., Yuan W., and Li J. (2017) Efficient generation of the mouse model with a defined point mutation through haploid cell-mediated gene editing. J. Genet. Genomics 44, 461–463 10.1016/j.jgg.2017.07.004 [DOI] [PubMed] [Google Scholar]
- 17. Jin L. F., and Li J. S. (2016) Generation of genetically modified mice using CRISPR/Cas9 and haploid embryonic stem cell systems. Zool. Res. 37, 205–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leeb M., Walker R., Mansfield B., Nichols J., Smith A., and Wutz A. (2012) Germline potential of parthenogenetic haploid mouse embryonic stem cells. Development 139, 3301–3305 10.1242/dev.083675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guo A., Huang S., Yu J., Wang H., Li H., Pei G., and Shen L. (2017) Single-cell dynamic analysis of mitosis in haploid embryonic stem cells shows the prolonged metaphase and its association with self-diploidization. Stem Cell Rep. 8, 1124–1134 10.1016/j.stemcr.2017.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Takahashi S., Lee J., Kohda T., Matsuzawa A., Kawasumi M., Kanai-Azuma M., Kaneko-Ishino T., and Ishino F. (2014) Induction of the G2/M transition stabilizes haploid embryonic stem cells. Development 141, 3842–3847 10.1242/dev.110726 [DOI] [PubMed] [Google Scholar]
- 21. Li H., Guo A., Xie Z., Tu W., Yu J., Wang H., Zhao J., Zhong C., Kang J., Li J., Huang S., and Shen L. (2017) Stabilization of mouse haploid embryonic stem cells with combined kinase and signal modulation. Sci. Rep. 7, 13222 10.1038/s41598-017-13471-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. He Z. Q., Xia B. L., Wang Y. K., Li J., Feng G. H., Zhang L. L., Li Y. H., Wan H. F., Li T. D., Xu K., Yuan X. W., Li Y. F., Zhang X. X., Zhang Y., Wang L., et al. (2017) Generation of mouse haploid somatic cells by small molecules for genome-wide genetic screening. Cell Rep. 20, 2227–2237 10.1016/j.celrep.2017.07.081 [DOI] [PubMed] [Google Scholar]
- 23. Zhao H., Yang L., and Cui H. (2015) SIRT1 regulates autophagy and diploidization in parthenogenetic haploid embryonic stem cells. Biochem. Biophys. Res. Commun. 464, 1163–1170 10.1016/j.bbrc.2015.07.098 [DOI] [PubMed] [Google Scholar]
- 24. Zhong C., and Li J. (2017) Efficient generation of gene-modified mice by haploid embryonic stem cell-mediated semi-cloned technology. Methods Mol. Biol. 1498, 121–133 10.1007/978-1-4939-6472-7_8 [DOI] [PubMed] [Google Scholar]
- 25. Wiezorek C. (1984) Cell cycle dependence of Hoechst 33342 dye cytotoxicity on sorted living cells. Histochemistry 81, 493–495 10.1007/BF00489756 [DOI] [PubMed] [Google Scholar]
- 26. Durand R. E., and Olive P. L. (1982) Cytotoxicity, mutagenicity and DNA damage by Hoechst 33342. J. Histochem. Cytochem. 30, 111–116 10.1177/30.2.7061816 [DOI] [PubMed] [Google Scholar]
- 27. Vaitheesvaran B., Hartil K., Navare A., Zheng C., OBroin P., Golden A., Guha C., Lee W., Kurland I. J., and Bruce J. E. (2014) Role of the tumor suppressor IQGAP2 in metabolic homeostasis: possible link between diabetes and cancer. Metabolomics 10, 920–937 10.1007/s11306-014-0639-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kumar D., Hassan M. K., Pattnaik N., Mohapatra N., and Dixit M. (2017) Reduced expression of IQGAP2 and higher expression of IQGAP3 correlates with poor prognosis in cancers. PLoS ONE 12, e0186977 10.1371/journal.pone.0186977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu Y., Liang D., Wang Y., Bai M., Tang W., Bao S., Yan Z., Li D., and Li J. (2013) Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell 13, 659–662 10.1016/j.stem.2013.10.016 [DOI] [PubMed] [Google Scholar]
- 30. Wu Y., Zhou H., Fan X., Zhang Y., Zhang M., Wang Y., Xie Z., Bai M., Yin Q., Liang D., Tang W., Liao J., Zhou C., Liu W., Zhu P., et al. (2015) Correction of a genetic disease by CRISPR-Cas9-mediated gene editing in mouse spermatogonial stem cells. Cell Res. 25, 67–79 10.1038/cr.2014.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ying Q. L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., and Smith A. (2008) The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 10.1038/nature06968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Elling U., Wimmer R. A., Leibbrandt A., Burkard T., Michlits G., Leopoldi A., Micheler T., Abdeen D., Zhuk S., Aspalter I. M., Handl C., Liebergesell J., Hubmann M., Husa A. M., Kinzer M., et al. (2017) A reversible haploid mouse embryonic stem cell biobank resource for functional genomics. Nature 550, 114–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Freimann R., and Wutz A. (2017) A fast and efficient size separation method for haploid embryonic stem cells. Biomicrofluidics 11, 054117 10.1063/1.5006326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang H., Shi L., Zhang S., Ling J., Jiang J., and Li J. (2010) High-efficiency somatic reprogramming induced by intact MII oocytes. Cell Res. 20, 1034–1042 10.1038/cr.2010.97 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.