Abstract
Background/Aim: Acute myeloid leukemia (AML) represents a heterogeneous disease with varying morphologic, immunophenotypic, and genetic features, along with varying patient outcomes. The genomic tractability of AML makes it amenable for targeted next-generation sequencing (NGS) testing clinically. Materials and Methods: One hundred eights-seven unique patients with a diagnosis of acute myeloid leukemia between May 2011 and Oct 2014 and with mutational analysis by NGS were included in this study. The distribution of gene mutations was investigated in different subcategories of AML. Results: Most patients in this study (n=182) received Genoptix testing (either 5-gene panel or 21-gene panel). In 130/187 (70%) cases, there was an average of 2.3 mutations per case (range=0-7 mutations). We specifically mention mutations in 32 genes, their significance and co-occurrence as detected in different types of AML. Conclusion: The genetic heterogeneity of AML signifies the importance of taking a personalized-medicine approach to the management of patients with AML.
Keywords: AML, genomics, NGS, mutation
Acute myeloid leukemia (AML) is a deadly cancer with a 5-year relative survival of approximately 25%. However, at both the biological and the clinical level, it represents a heterogeneous disease with varying morphological, immunophenotypic, and genetic features, along with varying patient outcomes (1,2).
Molecular testing plays a major role in AML and is routinely performed as a part of the diagnostic work-up. For example, cytogenetic testing is necessary for disease subclassification with certain aberrations being definitional in AML with recurrent cytogenetic abnormalities. Furthermore, the detection of select cytogenetic markers serves as the strongest predictive marker for determining prognostic subgroups in AML (3,4). In the most recent iteration of the WHO classification of hematopoietic tumors in 2008, provisional categories for AML with certain gene mutations (NPM1 and CEPBA) were also introduced. Furthermore, the 2016 revision of the WHO classification consisted of an additional section on myeloid neoplasms with germline predisposition, identifying germline mutations in CEBPA, DDX41, RUNX1, ANKRD26, ETV6, or GATA2 (5). In clinical practice, FLT3, NPM1 and CEBPA are often assayed given their role in stratification of cases of AML with normal cytogenetics (3). More recently, several other clinically-significant genes have been identified in AML (6). In fact, comprehensive sequencing of de novo AML has identified >2,300 mutations. Thankfully, in any given tumor, the number of mutations is on average less than 15 and overall only around 20 genes are significantly mutated in AML (7). In fact, the AML genome is one of the simplest cancer genomes.
Therefore, the genomic tractability of AML makes it amenable for targeted next-generation sequencing (NGS) testing clinically. Understanding of the frequencies of key AML-related gene mutations across different diagnostic subcategories is important because it can facilitate selection of targeted therapies, help us understand potential pathways or resistance mechanism, and serve as a scaffold to discover relevant clinical correlations as our mutation database is populated. At our high-volume cancer center which is home to many refractory AML patients, we instituted a policy of routinely performing next generation sequencing testing on all new cases of AML in order to provide more refined predictive and prognostic information on our patients to guide therapy and clinical decision making. Depending on the clinical need, limited (5 genes) to extensive (>400 genes) NGS gene panels were utilized. Thus, we summarize the data collected from NGS reports on 187 AML patients.
Materials and Methods
All 187 unique patients with a diagnosis of acute myeloid leukemia between May 2011 and Oct 2014 and with mutational analysis by NGS were included in this study. The clinical history and pathologic diagnoses were retrieved from the PowerChart electronic medical records at the Moffitt Cancer Center (MCC). Pathologic diagnoses were reviewed and verified by slide review as necessary.
Next Generation Sequencing: Mutational analysis by NextGen Sequencing was performed by Genoptix, using a 5-panel gene panel initially and later, a 21-gene panel. The minimal depth of coverage was 500x. The genes tested by Genoptix panels include ASXL1, CBL, DNMT3A, ETV6, EZH2, IDH1 IDH2, JAK2, KIT, MPL, NPM1, NRAS, PHF6, RUNX1, SF3B1, SRSF2, SETBP1, TET2, TP53, U2AF1, and ZRSR2. FLT-3, CALR, and CEBPA mutations were ordered separately for some patients and identified by alternative molecular testing methods. Those data are also included. Some cases were sent for the more extensive FoundationOne panel. The 436 genes tested by this panel are listed in Table I.
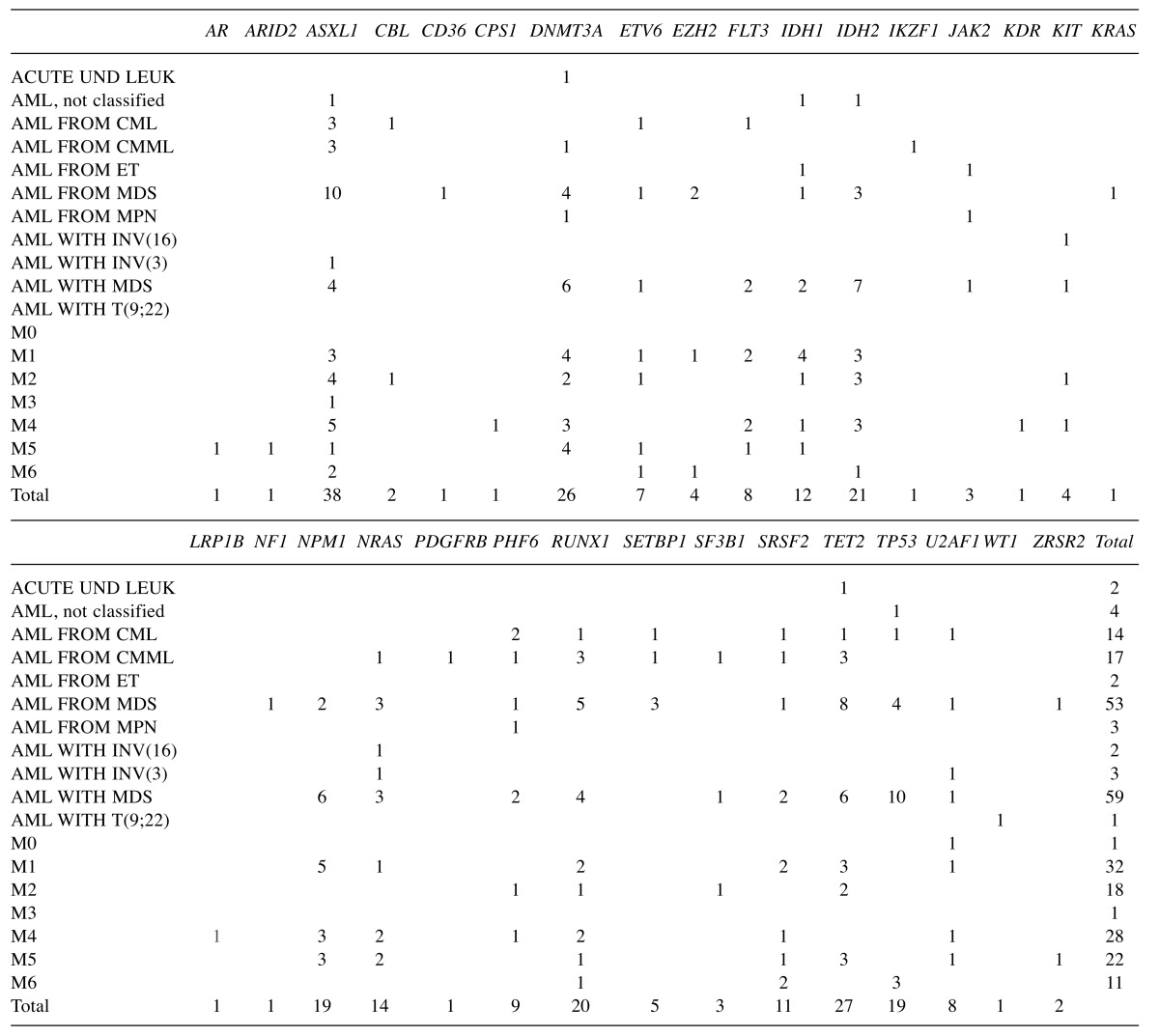
Table I. Summary of genes mutated in AML by AML subcategory.
Acute UND Leuk: Acute undifferentiated leukemia; AML, not classified: AML not otherwise classified; CML: chronic myeloid leukemia; CMML: Chronic myelomonocytic leukemia; ET: essential thrombocythemia; MDS: myelodysplastic syndrome; MPN: AML from myeloproliferative neoplasms; INV(16): inversion in chromosome 16; INV(3): inversion in chromosome 3; T(9;22): translocation between chromosomes 9 and 22; M0: AML with minimal differentiation; M1: AML without maturation; M2: AML with maturation; M3: acute promyelocytic leukemia; M4: acute myelomonocytic leukemia; M5: acute monocytic leukemia; M6: acute erythroid leukemia.
The percentage of patients with a mutation for each gene was calculated by dividing the number of patients who were positive for the mutation for each gene by the total number of patients who tested for that gene by NGS sequencing. This was done to account and compensate for the different genes tested in the different panels performed on patients. The mutation results and the diagnoses were analyzed to investigate the distribution of gene mutations in different subcategories of AML.
Results
A total of 187 unique AML patients were tested for gene mutations. Most patients in this study (n=182) received Genoptix testing (either 5-gene panel or 21-gene panel). A minority (n=8) patients were tested by FoundationOne panel, and 3 patients were tested on both platforms.
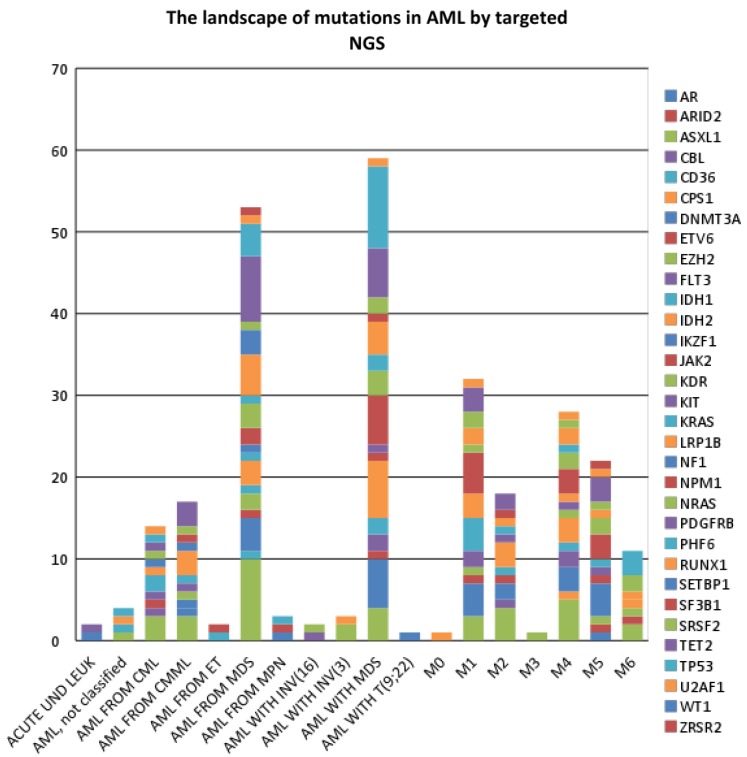
The most common type of AML was de novo AML with myelodysplasia related changes (MRC) (n=51), followed by secondary AML-MRC such as AML arising from a prior Myelodysplastic Syndrome (MDS) or MDS/Myeloprolife-rative Neoplasm (MDS/MPN) (n=29). Overall, mutations in 32 genes were detected: AR, ARID2, ASXL1, CBL, CD36, CPS1, DNMT3A, ETV6, EZH2, FLT3, IDH1, IDH2, IKZF1, JAK2, KDR, KIT, KRAS, LRP1B, NF1, NPM1, NRAS, PDGFRB, PHF6, RUNX1, SETBP1, SF3B1, SRSF2, TET2, TP53, U2AF1, WT1, and ZRSR2. When compared to the Genoptix 5 or 21 gene panel, testing with FoundationOne panel resulted in the detection of 11 additional mutated genes, namely: AR, ARID2, CD36, CPS1, IKZF1, KDR, KRAS, LRP1B, NPM1, PDGFRB, and WT1. For cases which had both panels performed (n=3), FoundationOne testing yielded detection of 4 additional mutated genes, namely: KDR, PDGFB, NRAS, IKZF1. The overall data are summarized in Table I and graphically represented in Figure 1.
Figure 1. The landscape of non-synonymous mutations in AML by AML subcategory.
At least one non-synonymous gene mutation was detected. In 130/187 (70%) cases, there was an average of 2.3 mutations per case (range=0-7 mutations). The highest number of non-synonymous mutations in any case was from a case of chronic myelogenous leukemia (CML) that transformed to AML (7 mutations). In the AML subgroup, secondary AML-MRC showed the highest mutation rate (82.8%), followed by AML M6 (77.7%), M5 (75%), M4 (68.8%), de novo AML-MRC (64.7%), and M2 (64.3%). AML with inv(16) or t(16;16) showed the lowest mutation rate (25%). Most cases lacking any detectable mutation were from the AML not otherwise specified (NOS) category and de novo AML-MRC subgroups (49% of negative cases).
ASXL1 had the highest mutation rate (20.7%) in the entire cohort, followed by TET2 (15.3%), RUNX1 (15.2%), DNMT3A (14.8%), and TP53 (14.4%), IDH2 (12%), NRAS (11.9%), FLT3 (11%), and NPM1 (11%). The remaining genes were mutated at <10% frequency. No MPL mutation was detected in the 119 samples tested. In terms of which diseases were more likely to harbor certain gene mutations, ASXL1 mutations occurred most frequently in AML arising from CML (75%) or chronic myelomonocytic leukemia (CMML) (60%). DNMT3A mutations appeared to be common in M5 (33.3%). FLT-3 mutation appeared to be more frequent in M4 and M1 subtypes and was not seen in secondary AML-MRC. IDH1 mutations are more common in M1 (19%) than other subcategories of AML, while IDH2 was more common in M2 (23.1%). NPM1 mutation was most common in M5 (25%) and M1 (23.8%). NRAS mutation was most commonly seen in secondary AML-MRC (20%). RUNX1 mutation was most commonly seen in AML arising from prior CMML (75%). SRSF2 was most common in M6 (33.3%). TET2 mutations were most common in AML from CMML (60%). TP53 was most common in M6 (42.9%).
Secondary AML-MRC and de novo AML-MRC showed different mutational profiles. The most frequently mutated genes in secondary AML-MRC were: ASXL1 (34.5%), TET2 (32%), NRAS/SETBP1/TP53 (20% each), DNMT3A (16%), IDH2 (12%), EZH2 (10%). On the other hand, the most frequently mutated genes in de novo AML-MRC with MDS were: TP53 (27%), IDH2 (14.6%), NPM1 (12.8%), DNMT3A, TET2 (12.5%), FLT-3 (11.8%), RUNX1 (10.8%). Of these genes, the mutation rate of ASXL1 was significantly higher in secondary AML-MRC than de novo AML-MRC (p=0.0051). The mutation rate of SETBP1 was also significantly higher in secondary AML-MRC (20%) than in de novo AML-MRC (0%) (p=0.0247). The other mutation rates for other genes were not statistically significantly different between secondary AML-MRC and de novo AML-MRC in this limited study.
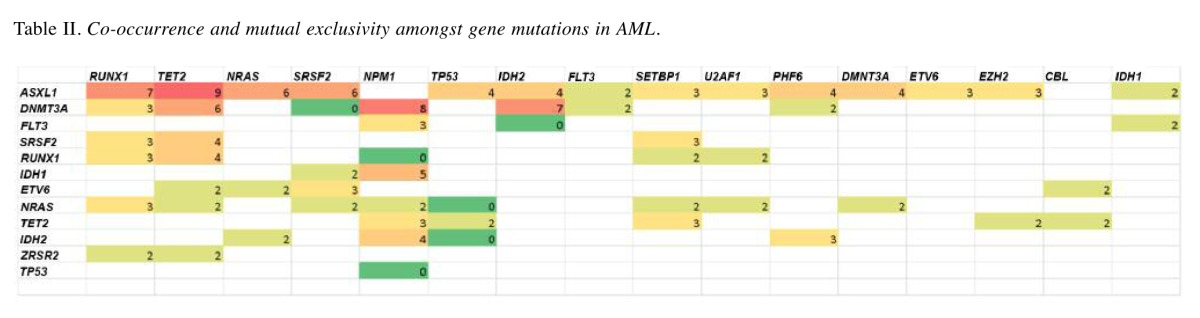
The most commonly co-mutated genes in AML were ASXL1 with RUNX1 (5.3% co-occurrence), TET2 (5.1% co-occurrence), or NRAS (5.1% co-occurrence). DNMT3A was most commonly co-mutated with NPM1 (4.6% co-occurrence), IDH2 (4% co-occurrence), or TET2 (3.4% co-occurrence). Co-occurrence was not found to be specific for or significantly enriched in any AML subcategory.
We also noted that some genes never co-occurred (Table II). For example, 176 patients were tested for both IDH2 and TET2 mutations, in which 21 patients were found to be positive for IDH2 mutations and 27 patients were found to be positive for TET2 mutations, but none of them were found to be positive for both. Therefore, IDH2 may be potentially mutually exclusive of TET2 and TP53. TP53 may be potentially mutually exclusive of IDH2, NRAS, NPM1, and SRSF2. Mutations in NPM1 and RUNX1, as well as DNMT3A and SRSF2 were also potentially mutually exclusive.
Table II. Co-occurrence and mutual exclusivity amongst gene mutations in AML.
Discussion
In our study, 70% of AML patients were found to have at least one mutation detected by targeted next-generation sequencing for genes previously described in hematopoietic malignancies or cancer in general. This figure is similar to what is previously reported in the literature for targeted NGS sequencing in AML (8). This high incidence of mutation detection further justifies the performance of NGS testing on AML cases as it can yield, at high frequency, genetic information that may be clinically actionable. For example, ASXL1 mutations were detected in approximately 1 out of every 5 cases. In MDS and AML, ASXL1 variants have been associated with worse prognosis (9,10). In CMML, ASXL1 variants are associated with poorer survival and transformation to AML (11). ASXL1 variants have also been reported in CML associated with accelerated or blast phase (12). The maximum number of genes mutated in one patient was seven, which occurred in one patient with blast phase CML. This is congruent with our current understanding of leukemogenesis which posits the sequential acquisition of driver mutations.
ASXL1 was found to have the highest mutation rate in AML patients, followed by TET2, RUNX1, DNMT3A, and TP53. Mutations of ZRSR2, CBL, KIT, JAK2, SF3B1, EZH2, SETBP1 were much less common. MPL mutation was not found in any of the AML patients tested. We detected DNMT3A mutations at a frequency of 15% which is similar to that reported by Thol et al. in 489 AMLs patients <60 years old (13). However other groups have reported frequencies ranging from 19% -36% (14). We detected IDH2 and IDH1 mutations at 12% and 7% frequency, respectively. IDH mutations have been reported at an incidence of 6%-16% for IDH1 and 8-19% for IDH2 (15), 25924101. TET2 mutations were detected at 15% and in the literature the mutation rate ranges from 13.2%-34% (16,17). ASXL1 mutations have been identified at 5-17% in the literature in sequencing studies of large cohorts (9,18,19). Our detection rate was slightly higher at 21%. This may be due to the disproportionate number of AML cases associated with MDS in our cohort or because we have more refractory AML patients in our cancer institute. It has been previously reported that ASXL1 mutations are more common in intermediate-risk karyotype AML as compared to copy number-neutral AML (19). Our data confirms the inverse relationship of ASXL1 mutations (less common in M5 and M1) with FLT3 (most common in M4 and M1) and/or NPM1 (most common in M5 and M1) mutations.
Interestingly, patients with AML-MRC that evolved from MDS showed a different mutational profile compared to de novo AML-MRC, with mutations most often involving ASXL1 and TET2 as opposed to TP53 and IDH2. This may indicate that either mutation in ASXL1 or mutation in IDH2 are sufficient to dysregulate DNA methylation patterns in AML pathogenesis (20). Furthermore, secondary AML-MRC had a somewhat higher mutation rate (82.8%) than de novo AML-MRC (64.7%). Overall, the data suggest possibly different leukemogenic pathways for these two diseases. The higher incidence of genetic mutations in MDS-related AML versus those with AML with recurrent genetic abnormalities is in line with previously reported findings (8).
We also found that some genes were more likely to be co-mutated; however, given these combinations were not specific to any subcategory of AML, this may simply be due to the higher incidence of these genes being mutated in AML. On the other hand, some genes appear to be potentially mutually exclusive (e.g., NPM1 and RUNX1; DNMT3A and SRSF2). Mutual exclusivity and co-occurrence for gene mutation in AML has previously been analyzed using Dendrix++. The previously reported exclusivity for NMP1 with TP53, NPM1 with RUNX1 was corroborated in our study (7).
In summary, clinical NGS sequencing frequently detects mutations in genes associated with myeloid disease (~70%). Importantly, different gene mutations occur at different frequencies across different subcategories of AML underscoring the genetic heterogeneity of AML and the importance of taking a personalized-medicine approach to the management of patients with AML.
References
- 1.Grimwade D, Walker H, Oliver F, Wheatley K, Harrison C, Harrison G. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children's Leukaemia Working Parties. Blood. 1998;92(7):2322–2333. [PubMed] [Google Scholar]
- 2.Horton SJ, Huntly BJ. Recent advances in acute myeloid leukemia stem cell biology. Haematologica. 2012;97(7):966–974. doi: 10.3324/haematol.2011.054734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeisig BB, Kulasekararaj AG, Mufti GJ, So CW. SnapShot: Acute myeloid leukemia. Cancer Cell. 2012;22(5):698–e1. doi: 10.1016/j.ccr.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 4.Estey EH. Acute myeloid leukemia: 2013 update on risk-stratification and management. Am J Hematol. 2013;88(4):318–327. doi: 10.1002/ajh.23404. [DOI] [PubMed] [Google Scholar]
- 5.Cazzola M. Introduction to a review series: the 2016 revision of the WHO classification of tumors of hematopoietic and lymphoid tissues. Blood. 2016;127(20):2361–2364. doi: 10.1182/blood-2016-03-657379. [DOI] [PubMed] [Google Scholar]
- 6.Naoe T, Kiyoi H. Gene mutations of acute myeloid leukemia in the genome era. Int J Hematol. 2013;97(2):165–174. doi: 10.1007/s12185-013-1257-4. [DOI] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research Network Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2014;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohgami RS, Ma L, Merker JD, Gotlib JR, Schrijver I, Zehnder JL. Next-generation sequencing of acute myeloid leukemia identifies the significance of TP53, U2AF1, ASXL1, and TET2 mutations. Mod Pathol. 2015;28(5):706–714. doi: 10.1038/modpathol.2014.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pratcorona M, Abbas S, Sanders MA, Koenders JE, Kavelaars FG, Erpelinck-Verschueren CA. Acquired mutations in ASXL1 in acute myeloid leukemia: prevalence and prognostic value. Haematologica. 2012;97(3):388–392. doi: 10.3324/haematol.2011.051532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abdel-Wahab O, Adli M, LaFave LM, Gao J, Hricik T, Shih AH. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer cell. 2012;22(2):180–193. doi: 10.1016/j.ccr.2012.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gelsi-Boyer V, Trouplin V, Roquain J, Adelaide J, Carbuccia N, Esterni B. ASXL1 mutation is associated with poor prognosis and acute transformation in chronic myelomonocytic leukaemia. Br J Haematol. 2010;151(4):365–375. doi: 10.1111/j.1365-2141.2010.08381.x. [DOI] [PubMed] [Google Scholar]
- 12.Pieri L, Spolverini A, Scappini B, Occhini U, Birtolo S, Bosi A. Concomitant occurrence of BCR-ABL and JAK2V617F mutation. Blood. 2011;118(12):3445–3346. doi: 10.1182/blood-2011-07-365007. [DOI] [PubMed] [Google Scholar]
- 13.Thol F, Damm F, Ludeking A, Winschel C, Wagner K, Morgan M. Incidence and prognostic influence of DNMT3A mutations in acute myeloid leukemia. J Clin Oncol. 2011;29(21):2889–2896. doi: 10.1200/JCO.2011.35.4894. [DOI] [PubMed] [Google Scholar]
- 14.Roller A, Grossmann V, Bacher U, Poetzinger F, Weissmann S, Nadarajah N. Landmark analysis of DNMT3A mutations in hematological malignancies. Leukemia. 2013;27(7):1573–1578. doi: 10.1038/leu.2013.65. [DOI] [PubMed] [Google Scholar]
- 15.Im AP, Sehgal AR, Carroll MP, Smith BD, Tefferi A, Johnson DE, et al. DNMT3A and IDH mutations in acute myeloid leukemia and other myeloid malignancies: associations with prognosis and potential treatment strategies. Leukemia. 2014;28(9):1774–1783. doi: 10.1038/leu.2014.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chou WC, Chou SC, Liu CY, Chen CY, Hou HA, Kuo YY. TET2 mutation is an unfavorable prognostic factor in acute myeloid leukemia patients with intermediate-risk cytogenetics. Blood. 2011;118(14):3803–3810. doi: 10.1182/blood-2011-02-339747. [DOI] [PubMed] [Google Scholar]
- 17.Grossmann V, Haferlach C, Nadarajah N, Fasan A, Weissmann S, Roller A. CEBPA double-mutated acute myeloid leukaemia harbours concomitant molecular mutations in 76.8% of cases with TET2 and GATA2 alterations impacting prognosis. Br J Haematol. 2013;161(5):649–658. doi: 10.1111/bjh.12297. [DOI] [PubMed] [Google Scholar]
- 18.Metzeler KH, Becker H, Maharry K, Radmacher MD, Kohlschmidt J, Mrozek K. ASXL1 mutations identify a high-risk subgroup of older patients with primary cytogenetically normal AML within the ELN Favorable genetic category. Blood. 2011;118(26):6920–6929. doi: 10.1182/blood-2011-08-368225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schnittger S, Eder C, Jeromin S, Alpermann T, Fasan A, Grossmann V. ASXL1 exon 12 mutations are frequent in AML with intermediate risk karyotype and are independently associated with an adverse outcome. Leukemia. 2013;27(1):82–91. doi: 10.1038/leu.2012.262. [DOI] [PubMed] [Google Scholar]
- 20.Zhang L, Padron E, Lancet J. The molecular basis and clinical significance of genetic mutations identified in myelodysplastic syndromes. Leuk Res. 2015;39(1):6–17. doi: 10.1016/j.leukres.2014.10.006. [DOI] [PubMed] [Google Scholar]