Abstract
We have characterized two isoforms of ATP-phosphoribosyl transferase (ATP-PRT) from Arabidopsis (AtATP-PRT1 [accession no. AB025251] and AtATP-PRT2), catalyzing the first step of the pathway of hisidine (His) biosynthesis. The primary structures deduced from AtATP-PRT1 and AtATP-PRT2 cDNAs share an overall amino acid identity of 74.6% and contain N-terminal chloroplast transit peptide sequences. DNA-blot analyses indicated that the ATP-PRTs in Arabidopsis are encoded by two separate genes with a closely similar gene structural organization. Both gene transcripts were detected throughout development, and protein-blot analysis revealed predominant accumulation of the AtATP-PRT proteins in Arabidopsis leaves. The His auxotrophy of a his1 mutant of Saccharomyces cerevisiae was suppressed by the transformation with AtATP-PRT1 and AtATP-PRT2 cDNAs, indicating that both isoforms are functionally active ATP-PRT enzymes. The Km values for ATP and phosphoribosyl pyrophosphate of the recombinant AtATP-PRT proteins were comparable to those of the native ATP-PRTs from higher plants and bacteria. It was demonstrated that the recombinant AtATP-PRTs were inhibited by l-His (50% inhibition of initial activity = 40–320 μm), suggesting that His biosynthesis was regulated in plants through feedback inhibition by l-His.
The biochemistry and genetics of His biosynthesis (Fig. 1) have been extensively studied in a number of microorganisms (Winkler, 1987; Alifano et al., 1996). In eubacteria such as Escherichia coli and Salmonella typhimurium, the complete nucleotide sequences of the genes involved in the His biosynthetic pathway have been determined, and it was shown that 10 enzymatic activities are encoded by eight genes organized in a single operon (Carlomagno et al., 1988). The His biosynthetic genes have also been isolated from a variety of organisms, including lower eukaryotes such as Saccharomyces cerevisiae, Schizosaccharomyces pombe, and Neurospora crassa (Alifano et al., 1996). The rate of His biosynthesis in bacteria is primarily regulated through the attenuation control of His operon expression (Winkler, 1987; Alifano et al., 1996), which is also repressed by high intracellular His concentration (Alifano et al., 1996). His biosynthesis is also regulated at the enzyme level. For example, ATP-phosphoribosyl transferase (ATP-PRT; EC 2.4.2.17), which catalyzes the first committed step of His biosynthesis, is feedback inhibited by the pathway end-product, l-His (Winkler, 1987; Alifano et al., 1996).
Figure 1.
His biosynthetic pathway in microorganisms. The first enzyme, ATP-PRT, is feedback inhibited by the pathway end product, l-His (Winkler 1987; Alifano et al., 1996).
The first experimental evidence for His biosynthesis in higher plants was reported by Wiater et al. (1971), who detected the enzyme activities of ATP-PRT, imidazoleglycerolphosphate dehydratase (IGPD) (EC 4.2.1.19), and histidinolphosphate phosphatase (HPP) (EC 3.1.3.15) in crude extracts from the shoots of barley, oat, and pea (Wiater et al., 1971). However, His biosynthetic enzymes had never been characterized in detail until the histidinol dehydrogenase (HDH) (EC 1.1.1.23) from cabbage and the IGPD from wheat germ were purified to apparent homogeneity. The corresponding cDNAs have also been cloned (Ward and Ohta, 1998, and refs. cited therein). On the contrary, recent progress of molecular biology has considerably accelerated the elucidation of the plant His biosynthetic pathway. Thus, we have isolated and characterized other His biosynthetic genes from Arabidopsis including those for the bifunctional phosphoribosyl (PR)-ATP pyrophosphohydrolase (PRA-PH)/PR-AMP cyclohydrolase (PRA-CH) (Fujimori and Ohta, 1998a), N′-[(5′-phosphoribosyl)-formimino]-5-aminoimidazole-4-carboxamide ribonucleotide (BBM II) isomerase (Fujimori et al., 1998), and the bifunctional Gln amidotransferase (GAT)/cyclase (Fujimori and Ohta, 1998b). In addition, a cDNA for histidinolphosphate aminotransferase (HPA) (EC 2.6.1.9) has been cloned from tobacco (Nicotiana tabacum) by functional complementation of an E. coli hisC mutant (El Malki et al., 1998). These results suggested that the His biosynthetic pathway in plants is essentially the same as those operating in microorganisms. The ATP-PRT from wheat germ has been purified and partially characterized (Münzer et al., 1992), whereas the gene encoding the enzyme has not been cloned. It is still unknown whether a phosphatase specific for histidinolphosphate is present in plants, although auxotrophic mutants lacking HPP have been isolated from bacteria and yeast (Ward and Ohta, 1998).
In this paper, we report the biochemical and molecular biological characterization of two ATP-PRT isoforms (AtATP-PRT1 and AtATP-PRT2; submitted to the GenBank/EMBL/DDBJ with accession nos. AB025249 and AB025250, respectively) from Arabidopsis. These two Arabidopsis ATP-PRT cDNAs were able to suppress the His auxotrophy of the S. cerevisiae his1 mutant, indicating that both genes encoded active ATP-PRT enzymes. Gene-specific probes detected both transcripts in all tissues examined, and protein-blot analyses with polyclonal antibodies against a recombinant AtATP-PRT1 protein demonstrated that the ATP-PRT proteins accumulated throughout development. Biochemical properties of the recombinantly expressed ATP-PRT proteins indicated that His biosynthesis in plants could also be feedback regulated by l-His. These findings have demonstrated, together with other His biosynthetic genes so far reported (El Malki et al., 1998; Fujimori and Ohta, 1998a, 1998b; Fujimori et al., 1998; Ward and Ohta, 1998), that His is synthesized in plants through a similar enzymatic process as that functioning in microorganisms.
MATERIALS AND METHODS
Plant Materials and Microbial Strains
Seeds of Arabidopsis ecotype Columbia (Lehle Seeds, Round Rock, TX) were surface-sterilized and cultivated as described previously (Fujimori and Ohta, 1998a). Escherichia coli strain JM109 was used as the host for the propagation and manipulation of plasmid DNAs. The media for E. coli and Saccharomyces cerevisiae were as described previously (Fujimori and Ohta, 1998b).
Isolation of Arabidopsis ATP-PRT cDNAs
An internal amino acid sequence (YIFDEDT) was determined from the ATP-PRT protein purified from wheat germ (Münzer et al., 1992). From this amino acid sequence, we designed a primer (AR5: 5′-GTCTCCTCGTCAAA- TATGTA-3′) to amplify a partial fragment of ATP-PRT cDNA by PCR using a Lambda ZAP II (Stratagene, La Jolla, CA) cDNA library prepared from 7-d-old Arabidopsis seedlings (Mizutani et al., 1997) as the template. The primers were AR5 and SK (5′-TCTAGAACTAGTGGATC-3′), which was derived from the vector sequence. The PCR products were cloned into a pCRII vector (Invitrogen, San Diego), and 50 independent clones were selected to analyze their insert DNA fragments. A plasmid, pAR5–13, was revealed to carry an insert of approximately 500 bp, of which the sequence was homologous to the ATP-PRT genes from microorganisms. Next, we screened the Arabidopsis cDNA library used as the PCR template for full-length AtATP-PRT cDNAs. The insert DNA of pAR5–13 was labeled with [α-32P]dCTP (Amersham, Buckinghamshire, UK) by the random priming method (Feinberg and Vogelstein, 1983). Prehybridization, hybridization, and wash were performed as described previously (Fujimori and Ohta, 1998a). Twenty-six out of 6 × 105 recombinant phages were obtained through a two-round plaque purification and converted to phagemids by the in vivo excision method according to the manufacturer's instructions (Stratagene). After restriction enzyme analyses and partial DNA sequencing, these inserts were grouped into two types, AtATP-PRT1 and AtATP-PRT2, encoding two ATP-PRT isoforms in Arabidopsis.
Isolation of Arabidopsis ATP-PRT1 Gene
Approximately 5 × 105 recombinant phages of an Arabidopsis Lambda ZAP II genomic library (Stratagene) were screened using the AtATP-PRT1 cDNA as a probe. The screening was continued until pure phages were obtained, and 18 independent phage plaques were finally isolated and analyzed. The probe was labeled by the random priming method using [α-32P]dCTP (Feinberg and Vogelstein, 1983).
Determination of DNA Sequences
DNA sequences were determined from both strands using a dye terminator cycle sequencing kit (Prism, Applied Biosystems, Foster City, CA). DNA and amino acid sequences were analyzed using the software DNASIS, version 3.4 (Hitachi Software Engineering, Yokohama, Japan).
DNA-Blot and RNA-Blot Analyses
Total genomic DNA was prepared from 2-week-old Arabidopsis seedlings as described previously (Sambrook et al., 1989). The full-length cDNAs for AtATP-PRT1 and AtATP-PRT2 were labeled by the random priming method as described above and used as the probes for DNA-blot analyses. The blots were hybridized and washed under low- or high-stringency conditions.
Total RNA was extracted as described previously (Lagrimini et al., 1987), and RNA gel-blot analysis was carried out using gene-specific probes as described previously (Fujimori and Ohta, 1998a). Gene-specific probes (a region spanning −318 to +219 of the AtATP-PRT1 gene [submitted to the GenBank/EMBL/DDBJ with accession no. AB025251] and a region from −318 to +297 of the AtATP-PRT2 gene) were prepared by PCR. These gene-specific probes were able to distinguish the ATP-PRT isoforms in a preliminary DNA-gel-blot analysis with the full-length cDNAs of AtATP-PRT1 and AtATP-PRT2, and no cross-hybridization was detected (data not shown).
Construction of a his1 Defective Strain of S. cerevisiae
We constructed a S. cerevisiae strain in which the HIS1 gene was disrupted as described previously (Fujimori and Ohta, 1998b). The coding region of S. cerevisiae HIS1 gene was amplified by PCR using H1F (5′-GGAATTCGG- ATCCAGAAAAATGGATTTGGTGAACCATC-3′) and H1R (5′-GATCTAGACGTTCTATCTTATACACGACAATTAG-3′) as the primers and genomic DNA from S. cerevisiae strain S288C as the template. After initial cloning of the PCR products into a pCRII vector (Invitrogen), a full-length HIS1 coding region was obtained by digesting with EcoRI and then re-cloned into a Gal-inducible expression vector, pYES2 (Invitrogen), yielding a plasmid, pKF110. The BamHI-XhoI fragment of the S. cerevisiae LEU2 gene (Andreadis et al., 1982) was replaced with the BglII-SalI fragment of pKF110 to obtain the plasmid pKF157. A S. cerevisiae strain, SH782 (MATa ura3-52 leu2-3,102), was transformed with the 3-kb BamHI-XhoI fragment from pKF157 by the method of Ito et al. (1983). The his1 mutant strain thus obtained was designated BY1001 (MATa ura3-52 leu2-3,102 his1::LEU2). The homologous recombination event to integrate the BamHI-XhoI fragment into the gene of BY1001 was confirmed by PCR (data not shown).
Suppression of the his1 Mutation by AtATP-PRT cDNAs
The coding regions for AtATP-PRTs without the putative chloroplast transit peptide portions were amplified by PCR using gene-specific primers and either AtATP-PRT1 cDNA or AtATP-PRT2 cDNA as the template. For the AtATP-PRT1 amplification, EF3 (5′-CGGGATCCATGAAGCGTGACCAGATTCGTCTTG-3′) and ERXH (5′-GCTCTAGAAGCTTCAGCATATGCATCTTCC-3′) were used as the primers, and a set of primers, AP2F (5′-CGGGATCCCGGGAGCAGATTCGTCTT-3′) and AP2R (5′-CGAAGCTTGAGAAGCAGCATCAAAGGCCG-3′), were used for the amplification of the AtATP-PRT2 fragment. The amplified AtATP-PRT1 and AtATP-PRT2 cDNA fragments were cloned into a yeast expression plasmid, pYES2 (Invitrogen), to obtain expression plasmids, pKF251 and pKF252, respectively. The newly constructed his1 mutant, BY1001, was transformed with either pKF251 or pKF252 (Ito et al., 1983). The yeast transformants were grown for 4 d at 30°C on a synthetic medium plate containing 2% (w/v) Glc and an amino acid mixture without Leu and uracil (SC/Glc-Leu-Ura). Colonies were selected and streaked on new SC/Gal-His-Leu-Ura plates to confirm whether or not the His auxotrophy of strain BY1001 was suppressed. Strain BY1001 transformed with pKF110 (carrying the S. cerevisiae HIS1 gene) was used as a positive control.
Expression of Recombinant Arabidopsis ATP-PRT Proteins
The full-length cDNAs for AtATP-PRT isoforms were used as the template for the PCR to amplify the coding regions without the chloroplast transit peptide portions, as described above. In this PCR, a set of EF3 and ERXH were used as the primers for the AtATP-PRT1 expression, and a set of AP2F and AP2R were for the AtATP-PRT2 expression. A pMAL-c2 vector (New England Biolabs, Beverly, MA) was used to express AtATP-PRT1 and AtATP-PRT2 cDNAs in E. coli XL1-Blue. These recombinant proteins were produced as fusion proteins with a maltose binding protein (MBP). The recombinant proteins were purified employing a two-step amylose resin column chromatography according to the method provided by the manufacturer. The ATP-PRT proteins were separated from the MBP domain by digesting with factor Xa (New England Biolabs). The protein concentration was determined by the method of Bradford (1976). SDS-PAGE was performed according to the method of Laemmli (1970), and the gel was stained with Coomassie Brilliant Blue (Sigma-Aldrich, St. Louis). The purified recombinant AtATP-PRT1 was injected intradermally with a complete Freund's adjuvant into two rabbits. Boosts were carried out in an incomplete Freund's adjuvant. The anti-AtATP-PRT1 antibodies were used in protein-blot analyses.
Assay for ATP-PRT Activity
The recombinant AtATP-PRTs was assayed by the method of Martin (1963). The reaction mixture contained 111.1 mm Tris-HCl (pH 8.5), 22.2 mm MgCl2, 83.3 mm KCl, 5.6 mm ATP, 1 mm DTT, 0.56 mm phosphoribosyl pyrophosphate (PRPP), and enzyme in a final volume of 360 μL. After preincubation at 30°C for 5 min, the reaction was started by adding PRPP, and A290 increase was monitored at 30°C for 2 min. One unit of activity was defined as the enzyme amount capable of a 0.02 absorbance increase per min in a cuvette of the light-pass of 1 cm, which corresponds to the formation of 1.67 nmol of PR-ATP per min (Voll et al., 1967).
RESULTS AND DISCUSSION
Isolation of Arabidopsis ATP-PRT cDNAs
We isolated full-length cDNAs encoding two ATP-PRT isoforms from Arabidopsis. First, putative ATP-PRT cDNA fragments were amplified by PCR from a cDNA library of 7-d-old Arabidopsis seedlings made using a Lambda ZAP II system (Mizutani et al., 1997). In this PCR, we used a forward primer derived from a vector sequence (SK) and a reverse primer (AR5) of which the sequence was designed referring to the internal amino acid sequence (YIFDEDT) of the ATP-PRT protein purified from wheat germ (Münzer et al., 1992). A DNA fragment of approximately 500 bp was thus obtained and found to contain a DNA sequence encoding the amino acid sequence YIFDEDT, together with several sequences highly homologous to those found in the microbial ATP-PRT proteins. Using the 500-bp DNA fragment as a screening probe, we identified 26 positive clones carrying ATP-PRT cDNA fragments from the Arabidopsis cDNA library (6 × 105 plaques). Partial DNA sequencing and restriction enzyme analyses indicated that these clones belonged to two separate groups, AtATP-PRT1 (four clones) and AtATP-PRT2 (22 clones). The longest clones of AtATP-PRT1 and AtATP-PRT2 were 1485 and 1535 bp in length, respectively. They contained open reading frames encoding 411 and 413 amino acid residues, and the calculated molecular mass of these proteins was 44.6 and 44.8 kD, respectively (Fig. 2).
Figure 2.
Alignment of ATP-PRT protein sequences. The primary structures of Arabidopsis AtATP-PRT1 (At ATP-PRT1) and AtATP-PRT2 (At ATP-PRT2) proteins deduced from the corresponding cDNA sequences are aligned with other ATP-PRT proteins. T. goesingense ATP-PRT, T.g ATP-PRT (GenBank accession no. AF003347; X.H. Yan, U. Krämer, I. Raskin, R.D. Smith, and D.E. Salt, unpublished data); S. cerevisiae HIS1, S.c HIS1 (GenBank accession no. J01329; Hinnebusch and Fink, 1983); C. albicans HIS1, C.a HIS1 (GenBank accession no. X83871; Pla et al., 1995); E. coli hisG, E.c hisG (GenBank accession no. V00284; Carlomagno et al., 1988); M. jannaschii hisG, M.j hisG (GenBank accession no. U67562; Bult et al., 1996); and Synechocystis sp. PCC6803 hisG, S.y hisG (GenBank accession no. D64006; Kaneko et al., 1996). Dashes indicate gaps inserted to allow optimal sequence alignment. Conserved amino acid residues were shaded. Single-letter codes for amino acid residues are used, and asterisks indicate termination codons for translation. Intron positions determined from the gene structures for AtATP-PRT1 and AtATP-PRT2 are indicated by arrows above the sequence.
Characterization of AtATP-PRT Genes of Arabidopsis
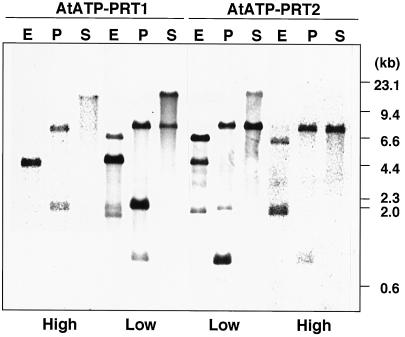
The gene for AtATP-PRT1 was isolated by screening an Arabidopsis genomic library using the AtATP-PRT1 cDNA as a probe. The entire region of AtATP-PRT2 gene was found to be present in an Arabidopsis genomic clone (F21M12, GenBank accession no. AC000132, V.S. Vysotskaia, B.I. Osborne, M. Toriumi, G. Yu, O. Oji, Y.K. Shen, R. Araujo, M. Au, E. Buehler, A.B. Conway, A.R. Conway, K. Dewar, J. Feng, C. Kim, D. Kurtz, Y. Li, P. Shinn, H. Sun, R.W. Davis, J.R. Ecker, N.A. Federspiel, and A. Theologis, unpublished data). Sequence analysis showed that both genes consisted of 11 exons and 10 introns (Fig. 2), and that all of the introns follow the GT-AG rule for the exon-intron junctions (Breathnach and Chambon, 1981). Although the nucleotide sequences of the 5′- and 3′-untranslated regions and introns were divergent, the intron positions of both genes (Fig. 2) were completely conserved with different intron lengths. Genomic DNA-blot analyses were performed using the full-lengths of AtATP-PRT1 and AtATP-PRT2 cDNAs as hybridization probes under low- or high-stringency conditions (Fig. 3). Arabidopsis genomic DNA was digested with SalI (no restriction site in both cDNAs), EcoRI (one restriction site in AtATP-PRT2 cDNA), or PstI (two restriction sites in AtATP-PRT1 cDNA and one restriction site in AtATP-PRT2 cDNA). The simple hybridization pattern detected under the high-stringency condition was in agreement with the bands identified under the low-stringency condition. The results obtained under the low-stringency condition indicated that no isoforms other than AtATP-PRT1 and AtATP-PRT2 were present in Arabidopsis genome.
Figure 3.
Genomic DNA-blot hybridization analysis. Arabidopsis genomic DNA (10 μg) was digested with EcoRI, PstI, or SalI, and subjected to hybridization analysis using the full-lengths of the AtATP-PRT1 and AtATP-PRT2 cDNAs as probes under low- or high-stringency conditions. The restriction enzymes used are indicated at the top of the figure: E, EcoRI; P, PstI; and S, SalI. Molecular size markers are shown on the right.
Steady-State Levels of AtATP-PRT mRNA in Arabidopsis
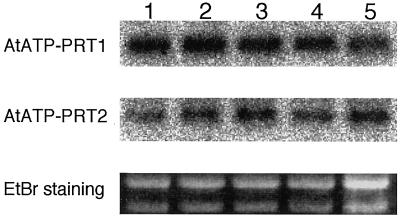
Total RNA was prepared from different tissues of Arabidopsis and used for RNA-blot hybridization analyses using gene-specific probes. Both gene-specific probes were able to detect the transcripts of approximately 1.6 kb in length (Fig. 4), indicating that both cDNA clones were representing their full-length transcripts. The mRNA levels of AtATP-PRT2 and AtATP-PRT1 were constant throughout development (Fig. 4).
Figure 4.
Steady-state levels of Arabidopsis AtATP-PRT gene transcripts. Total RNA (10 μg) was extracted from 1-week-old germinating seeds (lane 1), roots and leaves from 2-week-old seedlings (lane 2), roots and leaves of 3-week-old seedlings (lane 3), and leaves (lane 4) and inflorescence stems (lane 5) from 4-week-old plants. For hybridization, probes specific for either AtATP-PRT1 or AtATP-PRT2 were amplified by PCR (see “Materials and Methods”). The ethidium bromide (EtBr) staining of the rRNA bands for each RNA preparation is also shown.
Comparison of Amino Acid Sequences
The primary structures of AtATP-PRT1 and AtATP-PRT2 proteins deduced from the cDNA sequences were compared with those of S. cerevisiae HIS1 (Hinnebusch and Fink, 1983), Candida albicans HIS1 (Pla et al., 1995), E. coli hisG (Carlomagno et al., 1988), Synechocystis sp. PCC6803 hisG (Kaneko et al., 1996), and Methanococcus jannaschii hisG (Bult et al., 1996) proteins (Fig. 2). The ATP-PRT protein encoded by a newly isolated cDNA from a higher plant species, Thlaspi goesingense (Yan et al., GenBank accession no. AF003347) was also included in the comparison. The amino acid sequences were highly conserved among the ATP-PRT proteins of Arabidopsis and T. goesingense. It should be noted that the amino acid identity between AtATP-PRT2 and the ATP-PRT from T. goesingense (81.6%) was higher than that observed between AtATP-PRT1 and AtATP-PRT2 proteins (74.6%). On the other hand, overall similarity of the primary structures was very low among organisms, whereas sequence conservation was remarkable in several specific regions (Fig. 2). In addition, approximately 80 amino acid residues at the amino termini of the AtATP-PRT proteins showed the features as the transit peptide (von Heijne and Nishikawa, 1991). This was also true for the ATP-PRT from T. goesingense. These N-terminal extensions were rich in Ser residues and contained only a few negatively charged residues, having no homology with the N-terminal regions of the microbial ATP-PRTs (Fig. 2). We have already demonstrated that wheat IGPD and cabbage HDH proteins are localized in chloroplasts (Nagai et al., 1992; Tada et al., 1995), and the protein sequences deduced from the cDNAs for BBM II isomerase (Fujimori et al., 1998), PRA-PH/PRA-CH (Fujimori and Ohta, 1998a), GAT/cyclase (Fujimori and Ohta, 1998b), and HPA (El Malki et al., 1998) also contained putative chloroplast transit peptides at their N termini. These findings indicate that the overall His biosynthesis, which requires 41 ATP molecules for every His molecule synthesized (Alifano et al., 1996), completes within chloroplasts.
Functional Expression of AtATP-PRT cDNAs in a his1 Mutant of S. cerevisiae
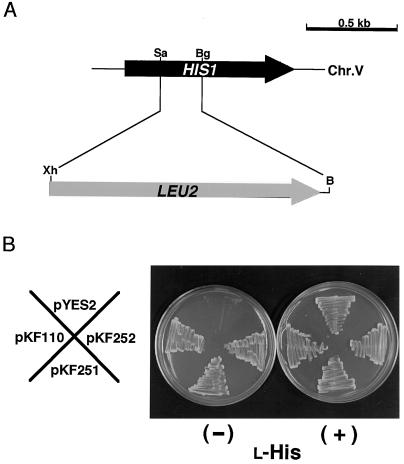
We confirmed using a his1 mutant of S. cerevisiae that both AtATP-PRT cDNAs encoded functionally active enzymes catalyzing the first step of the His biosynthetic pathway. In S. cerevisiae, ATP-PRT is encoded by HIS1 gene (Fig. 5A). A his1 mutant strain lacking ATP-PRT activity (BY1001) was constructed through homologous recombination of LEU2 gene with the coding region of HIS1 gene. Strain BY1001 was unable to grow in the absence of exogenous l-His supply (Fig. 5B). The His auxotrophy of strain BY1001 was suppressed when transformed with a plasmid, pKF110, harboring the HIS1 coding region of S. cerevisiae (Fig. 5B). We have truncated the regions corresponding to the N-terminal 74 and 76 amino acid residues from the AtATP-PRT1 and AtATP-PRT2 cDNAs, respectively, and constructed expression plasmids, pKF251 (carrying the AtATP-PRT1 cDNA) and pKF252 (harboring the AtATP-PRT2 cDNA). The his1 mutation of strain BY1001 was suppressed by transformation with either pKF251 or pKF252 (Fig. 5B), demonstrating that both Arabidopsis cDNAs indeed encoded the active ATP-PRT enzymes. Also, the truncated N-terminal regions were not essential for the AtATP-PRTs to function in the S. cerevisiae cells (Fig. 5B), supporting the idea that they were transit peptides. While the primary structures were highly divergent between the fungal proteins and the plant ATP-PRTs (Fig. 2), the successful suppression of the his1 mutation of S. cerevisiae by the AtATP-PRT cDNAs implicated a conservation of functional motifs among organisms.
Figure 5.
Suppression of a S. cerevisiae his1 null mutant, BY1001, with the Arabidopsis AtATP-PRT cDNAs. A, Construction of a his1::LEU2 null allele on chromosome 5 (Chr.V). Restriction enzyme sites are designated: B, BamHI; Bg, BglII; Sa, SalI; and Xh, XhoI. B, Growth of a S. cerevisiae his1 mutant (BY1001). Strain BY1001 was transformed with either pYES2 (empty plasmid), pKF110 (plasmid carrying S. cerevisiae HIS1 coding region), pKF251 (plasmid harboring AtATP-PRT1 cDNA truncated at the chloroplast transit peptide portion), or pKF252 (plasmid containing AtATP-PRT2 cDNA without the chloroplast transit peptide region). After the transformation, the cells were cultivated on a minimal-Gal plate supplemented with an amino acids mixture without l-His, Leu, and Ura (SC/Gal-His- Leu-Ura).
Arabidopsis ATP-PRT Proteins
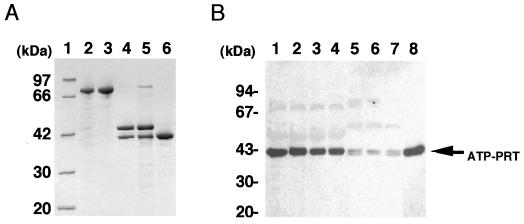
The AtATP-PRT1 (Fig. 6) and AtATP-PRT2 (data not shown) cDNAs were expressed in an E. coli expression system with pMAL-c2 vector, and the enzymatic properties of the AtATP-PRTs in the crude bacterial cell extracts were studied (Table I). No ATP-PRT activity was detectable in the crude extracts from E. coli transformed with an empty pMAL-c2 plasmid (data not shown). The endogenous ATP-PRT activity of E. coli cells was too low to be detected under our experimental conditions. The apparent Km values for PRPP and ATP of the purified AtATP-PRT1 were determined to be 0.13 and 0.60 mm, respectively (Table I). These values were similar to those of recombinant AtATP-PRT2 determined using the crude cell extracts, the native T. aestivum protein (Münzer et al., 1992), and the hisG protein of S. typhimurium (Martin, 1963; Whitfield, 1971). The Km values for PRPP and ATP of the purified recombinant AtATP-PRT1 were not consistent with those values determined using crude extracts (Table I). In the crude extracts, recombinant AtATP-PRT1 protein was present as the fusion protein with the MBP, and it is possible that the enzymatic function of the AtATP-PRT1 protein might be affected by the presence of MBP domain.
Figure 6.
Analysis of Arabidopsis ATP-PRT proteins. A, Purification of the recombinant AtATP-PRT1 protein. Sample protein (10 μg for each lane) was analyzed by SDS-PAGE and Coomassie Brilliant Blue staining. A protein size marker was applied in lane 1. Whole-cell extract from the E. coli after the isopropylthio-β-galactoside induction (lane 2) was applied to an amylose resin column. The expressed fusion protein was eluted with 10 mm maltose (lane 3). After digesting with factor Xa (lane 4), the sample was sequentially purified by a hydroxylapatite (lane 5) and a second amylose resin chromatography to remove MBP (lane 6). B, Crude extracts (containing 10 μg of protein) from Arabidopsis seedlings were separated by 10% to 20% SDS-PAGE, and the proteins were electrophoretically transferred to a PVDF membrane for immunodetection with anti-AtATP-PRT1 polyclonal antibodies. Lane 1, Leaves from 1-week-old seedlings; lane 2, leaves from 2-week-old seedlings; lane 3, leaves from 3-week-old seedlings; lane 4, leaves from 4-week-old seedlings; lane 5, roots from 2-week-old seedlings; lane 6, roots from 3-week-old seedlings; lane 7, roots from 4-week-old seedlings; and lane 8, the purified recombinant AtATP-PRT1. Molecular size markers are shown on the left. Six independent experiments were carried out, and one of the representative results is shown.
Table I.
Properties of the recombinant AtATP-PRT proteins
| Property | AtATP-PRT1 | AtATP-PRT2 | T. aestivuma | S. typhimurium |
|---|---|---|---|---|
| Km for PRPP (mm) | 37 (0.13) | 0.57 | 0.13 | 0.067b, 0.056c |
| Km for ATP (mm) | 89 (0.60) | 0.51 | 0.78 | 0.20b, 0.43c |
| IC50 of l-His (μm) | 40 (45) | 320 | 75 | 60–80b, 70–80c |
| IC50 of d-His | No inhibition | No inhibition | ND | ND |
| IC50 of 1,2,4-triazole-3-Ala (mm) | 0.47 (0.65) | 2.4 | ND | ND |
Escherichia coli strain XL-1-Blue was transformed with the expression plasmid harboring the coding region of either AtATP-PRT1 (pMA1) or AtATP-PRT2 (pMA2), and crude cell extracts were prepared for the assay of AtATP-PRT1 and AtATP-PRT2. The numbers in the parentheses are the values determined using the purified recombinant AtATP-PRT1 protein. Each value represents an average of duplicate assays of three independent experiments. ND, Not determined.
Data from Münzer et al. (1992).
Data from Martin (1963).
Data from Whitfield (1971).
The catalytic activities of the recombinant AtATP-PRT1 and AtATP-PRT2 were inhibited by l-His with the IC50 values of 40 and 320 μm, respectively (Table I). The recombinant AtATP-PRT1 activity (both the purified protein and the crude preparation) were inhibited by l-His and 1,2,4-triazole-3-Ala. These IC50 values, specifically those observed with AtATP-PRT1, were in the same range as those reported with the microbial enzymes (Table I). 1,2,4-Triazole-3-Ala, a structure analog of l-His, inhibited the Arabidopsis enzymes, while no inhibition was detected with d-His (Table I). AtATP-PRT2 was less sensitive toward the inhibition by l-His and 1,2,4-triazole-3-Ala compared with AtATP-PRT1. 1,2,4-Triazole-3-Ala has been known to have a weak herbicidal effect (Heim and Larrinua, 1989; Ward and Ohta, 1998). However, it has been controversial whether or not the phytotoxicity was solely attributable to the inhibition of ATP-PRT activity. This compound could be incorporated into newly synthesized polypeptides, leading to the production of functionally defective proteins (Ward and Ohta, 1998). Present results suggested in vitro that His biosynthesis was feedback regulated in planta by l-His and that the phytotoxicity of 1,2,4-triazole-3-Ala was at least in part ascribed to the inhibition of ATP-PRT.
The recombinant AtATP-PRT1 protein was purified to apparent homogeneity after cleavage from MBP using factor Xa (Fig. 6A), and used as the antigen to prepare polyclonal antibodies, which were found to cross-react with the recombinant AtATP-PRT2 protein (data not shown). A recombinant AtATP-PRT2 protein was also expressed as a fusion protein with the MBP of almost the same Mr as that of the AtATP-PRT1 fusion protein (data not shown), and its activity could be determined in the bacterial crude extracts as described above. However, the recombinant AtATP-PRT2 was not stable enough to retain its activity during the purification. The instability of the recombinant AtATP-PRT2 could be due to the inappropriate truncation of the N-terminal 76 amino acid residues, which might be involved in correct folding. However, the stability of the recombinant AtATP-PRT2 obtained by expressing the full-length cDNA was not significantly different from the N-terminally truncated form. Furthermore, swapping the N-terminal portions between AtATP-PRT1 and AtATP-PRT2 did not affect the stability of the recombinantly expressed enzymes (data not shown). We cannot rule out the possibility that the lower stability of the recombinant AtATP-PRT2 might be reflected in the enzymatic properties experimentally determined.
Steady-State Levels of ATP-PRT Proteins
The anti-AtATP-PRT1 antibodies strongly reacted with a protein of approximately 42 kD in leaves (Fig. 6B, lanes 1–4). Two immunoreactive bands were detected in the extracts from roots at almost the same size as that observed in leaves (Fig. 6B, lanes 5–7). The anti-AtATP-PRT1 antibodies reacted equally with both isoforms, and no differential reactivity toward any of the recombinant AtATP-PRTs was observed (data not shown). Therefore, it is thought that the immunoreactive bands in the leaf samples (Fig. 6B, lanes 1–4) were derived from two AtATP-PRT proteins with closely similar molecular size. This consideration is consistent with the RNA-blot analysis demonstrating that both gene transcripts accumulated in leaves to almost the same levels (Fig. 4). However, the ATP-PRT activity levels in these Arabidopsis tissues were too low to detect, and therefore we could not correlate the protein levels (Fig. 6B) with extractable enzyme activity levels.
Possible Mechanisms to Control His Biosynthesis in Plants
Current results have now established, together with other His biosynthetic genes so far reported (El Malki et al., 1998; Fujimori and Ohta, 1998a, 1998b; Fujimori et al., 1998a; Ward and Ohta, 1998, and refs cited therein), that His is synthesized in plants through a similar enzymatic process as that functioning in microorganisms. In microbial cells, several mechanisms are known to control the rate of His biosynthesis at the levels of both gene expression and enzyme regulation (Winkler 1987; Alifano et al., 1996). The attenuation control in S. typhimurium and E. coli and the general control in S. cerevisiae constitute the regulation of gene expression (Alifano et al., 1996), and the feedback inhibition of ATP-PRT by l-His is the biochemical regulation at the enzyme level (Alifano et al., 1996).
The properties of the recombinantly expressed ATP-PRT proteins (Table I) suggested that His biosynthesis in plants was feedback regulated by l-His. It remains to be clarified whether the inhibition of ATP-PRT by l-His actually contributes to the regulation of the pathway flux rate to reflect intracellular His levels in planta. We still know only a little about His biosynthesis in plants in terms of the gene regulation at the levels of post-transcription and post-translation. Complete elucidation of His biosynthetic pathway genes in plants provides the basis for understanding possible regulation mechanisms of His biosynthesis in plants.
LITERATURE CITED
- Alifano P, Fani R, Liu P, Lazcano A, Bazzicalupo M, Carlomagno MS, Bruni CB. Histidine biosynthetic pathway and genes: structure, regulation, and evolution. Microbiol Rev. 1996;60:44–69. doi: 10.1128/mr.60.1.44-69.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreadis A, Hsu YP, Kohlhaw GB, Schimmel P. Nucleotide sequence of yeast LEU2shows 5′-noncoding region has sequences cognate to leucine. Cell. 1982;31:319–325. doi: 10.1016/0092-8674(82)90125-8. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Breathnach R, Chambon P. Organization of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Bult CJ, White O, Olsen GJ, Zhou L, Fleischmann RD, Sutton GG, Blake JA, FitzGerald LM, Clayton RA, Gocayne JD, Kerlavage AR, Dougherty BA, Tomb J-F, Adams MD, Reich CI, Overbeek R, Kirkness EF, Weinstock KG, Merrick JM, Glodek A, Scott JL, Geoghagen NSM, Weidman JF, Fuhrmann JL, Nguyen D, Utterback TR, Kelley JM, Peterson JD, Sadow PW, Hanna MC, Cotton MD, Roberts KM, Hurst MA, Kaine BP, Borodovsky M, Klenk H-P, Fraser CM, Smith HO, Woese CR, Venter JC. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- Carlomagno MS, Chiariotti L, Alifano P, Nappo AG, Bruni CB. Structure and function of the Salmonella typhimurium and Escherichia coliK-12 histidine operons. J Mol Biol. 1988;203:585–606. doi: 10.1016/0022-2836(88)90194-5. [DOI] [PubMed] [Google Scholar]
- El Malki F, Frankard V, Jacobs M. Molecular cloning and expression of a cDNA sequence encoding histidinol phosphate aminotransferase from Nicotiana tabacum. Plant Mol Biol. 1998;37:1013–1022. doi: 10.1023/a:1006007125448. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fujimori K, Ohta D. Isolation and characterization of a histidine biosynthetic gene in Arabidopsis encoding a polypeptide with two separate domains for phosphoribosyl-ATP pyrophosphohydrolase and phosphoribosyl-AMP cyclohydrolase. Plant Physiol. 1998a;118:275–283. doi: 10.1104/pp.118.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori K, Ohta D. An Arabidopsis cDNA encoding a bifunctional glutamine amidotransferase/cyclase suppresses the histidine auxotrophy of a Saccharomyces cerevisiae his7mutant. FEBS Lett. 1998b;428:229–234. doi: 10.1016/s0014-5793(98)00535-3. [DOI] [PubMed] [Google Scholar]
- Fujimori K, Tada S, Kanai S, Ohta D. Molecular cloning and characterization of a gene encoding N′-[(5′-phosphoribosyl)-formimino]-5-aminoimidazole-4-carboxamide ribonucleotide isomerase from Arabidopsis thaliana. Mol Gen Genet. 1998;259:216–223. doi: 10.1007/s004380050807. [DOI] [PubMed] [Google Scholar]
- Heim DR, Larrinua IM. Primary site of action of amitrole in Arabidopsis thalianainvolves inhibition of root elongation but not of histidine or pigment biosynthesis. Plant Physiol. 1989;91:1226–1231. doi: 10.1104/pp.91.3.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG, Fink GR. Repeated DNA sequences upstream from HIS1 also occur at several other co-regulated genes in Saccharomyces cerevisiae. Biol Chem. 1983;258:5238–5247. [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystissp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lagrimini LM, Burkhart W, Moyer M, Rothstein S. Molecular cloning of complementary DNA encoding the lignin-forming peroxidase from tobacco: molecular analysis and tissue-specific expression. Proc Natl Acad Sci USA. 1987;84:7542–7546. doi: 10.1073/pnas.84.21.7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RG. The first enzyme in histidine biosynthesis: the nature of feedback inhibition by histidine. J Biol Chem. 1963;238:257–268. [Google Scholar]
- Mizutani M, Ohta D, Sato R. Isolation of a cDNA and a genomic clone encoding cinnamate 4-hydroxylase from Arabidopsis and its expression manner in planta. Plant Physiol. 1997;113:755–763. doi: 10.1104/pp.113.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münzer S, Hashimoto-Kumpaisal R, Scheidegger A, Ohta D. Purification and properties of ATP-phosphoribosyl transferase from wheat germ. In: Murata N, editor. Research in Photosynthesis. IV. Netherlands: Kluwer Academic Publishers; 1992. pp. 91–94. [Google Scholar]
- Nagai A, Suzuki K, Ward E, Moyer M, Mano J, Beck J, Tada S, Hashimoto M, Chang J-Y, Ryals J, Scheidegger A, Ohta D. Histidinol dehydrogenase in higher plants: purification, cloning and expression. In: Murata N, editor. Research in Photosynthesis. IV. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1992. pp. 95–98. [Google Scholar]
- Pla J, Perez-Diaz RM, Navarro-Garcia F, Sanchez M, Nombela C. Cloning of the Candida albicans HIS1 gene by direct complementation of a C. albicanshistidine auxotroph using an improved double-ARS shuttle vector. Gene. 1995;165:115–120. doi: 10.1016/0378-1119(95)00492-o. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Tada S, Hatano M, Nakayama Y, Volrath S, Guyer D, Ward E, Ohta D. Insect cell expression of recombinant imidazoleglycerolphosphate dehydratase of Arabidopsis and wheat and inhibition by triazole herbicides. Plant Physiol. 1995;109:153–159. doi: 10.1104/pp.109.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voll MJ, Appella E, Martin R. Purification and composition studies of phosphoribosyladenosine triphosphate: pyrophosphate phosphoribosyltransferase, the first enzyme of histidine biosynthesis. J Biol Chem. 1967;242:1760–1767. [PubMed] [Google Scholar]
- von Heijne G, Nishikawa K. Chloroplast transit peptides: the perfect random coil? FEBS Lett. 1991;278:1–3. doi: 10.1016/0014-5793(91)80069-f. [DOI] [PubMed] [Google Scholar]
- Ward E, Ohta D. Histidine Biosynthesis. In: Singh BJ, editor. Plant Amino Acids, Biochemistry and Biotechnology. New York: Marcel Dekker; 1998. pp. 293–303. [Google Scholar]
- Whitfield HJ., Jr Purification and properties of the wild type and a feedback-resistant phosphoribosyladenosine triphosphate. J Biol Chem. 1971;246:899–908. [PubMed] [Google Scholar]
- Wiater A, Krajewska-Grynkiewicz K, Klopotowski T. Histidine biosynthesis and its regulation in plants. Acta Biochem Pol. 1971;18:299–307. [PubMed] [Google Scholar]
- Winkler ME. Biosynthesis of histidine in Escherichia coli and Salmonella typhimurium. In: Neidhardt FC, Ingraham JL, Low KB, Magasanik B, Schaechter M, Umbarger HE, editors. Cellular and Molecular Biology. I. Washington, DC: American Society of Microbiology; 1987. pp. 395–411. [Google Scholar]