Abstract
Anthracyclines such as doxorubicin are used extensively in the treatment of cancers. Anthraquinone-related angucyclines also exhibit antiproliferative properties and have been proposed to operate via similar mechanisms, including direct genome targeting. Here, we report the chemical synthesis of marmycin A and the study of its cellular activity. The aromatic core was constructed by means of a one-pot multistep reaction comprising a regioselective Diels–Alder cycloaddition, and the complex sugar backbone was introduced through a copper-catalysed Ullmann cross-coupling, followed by a challenging Friedel–Crafts cyclization. Remarkably, fluorescence microscopy revealed that marmycin A does not target the nucleus but instead accumulates in lysosomes, thereby promoting cell death independently of genome targeting. Furthermore, a synthetic dimer of marmycin A and the lysosome-targeting agent artesunate exhibited a synergistic activity against the invasive MDA-MB-231 cancer cell line. These findings shed light on the elusive pathways through which anthraquinone derivatives act in cells, pointing towards unanticipated biological and therapeutic applications.
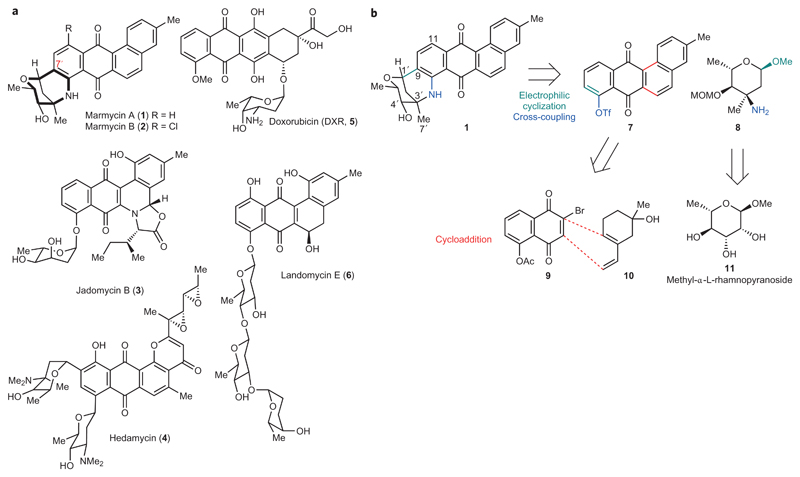
The family of angucyclines comprises a large number of natural products formally arising from polyketide biosynthesis1. This family has attracted considerable attention due to the diverse nature of its structural variants and the widespread biological activities associated with them. In particular, the angucyclines display antiproliferative, antibiotic and antiviral properties, but the underlying mechanisms are not always fully understood1. Angucyclines are characterized by an anthraquinone core, reminiscent of that of known anticancer drugs, embedded within an extended angular aromatic backbone. Typically, the aromatic core is decorated by C- and O-glycosides as well as unsaturated and oxidized functionalities. Marmycin A (1) and B (2) are novel angucyclines, recently isolated from a marine Streptomyces-related actinomycete, which exhibit relevant cytotoxic properties (Fig. 1a)2. These natural products have caught our attention partly as a result of their unique molecular architecture, which contains both C- and N-glycosidic bonds organized as a complex hexacyclic framework. Based on structural similarities with other angucyclines, pluramycins and anthracyclines, the marmycins have been proposed to kill cancer cells through DNA targeting. For instance, the angucycline-derived jadomycin B (3) promotes the cleavage of double-stranded DNA in vitro upon exposure to copper and light (Fig. 1a)3. The pluramycin derivative hedamycin (4) intercalates within double-stranded DNA in a sequence-specific manner and covalently reacts with guanine residues in vitro, indicating that the flat core is prone to π–π stacking and some degree of hydrophobic interactions (Fig. 1a)4,5. Doxorubicin (DXR, 5) triggers the production of deleterious reactive oxygen species (ROS)6, mediates the eviction of histones7,8 and interferes with topoisomerase II to promote genomic instability and cell death (Fig. 1a)9,10. The notion of direct genome targeting is, however, confounded by landomycin E (6), a potent anticancer compound known to induce mitochondrial dysfunction through as yet unidentified molecular mechanisms (Fig. 1a)11. Similarly, the exact mechanism by which the marmycins operate has remained elusive. As part of our continuing efforts towards the design and synthesis of biologically active small molecules capable of selective genome targeting12–15, we became interested in dissecting the mammalian cell biology of the marmycins.
Figure 1. Anthraquinone-containing natural products.
a, Molecular structures of marmycin A (1) and B (2), jadomycin B (3), hedamycin (4), the clinically approved anthracycline doxorubicin (DXR, 5) and landomycin E (6). b, Retrosynthetic analysis towards marmycin A precursors featuring a stepwise fusion of the aromatic and chiral sugar moieties and a regioselective cycloaddition.
Results
Chemical synthesis of the marmycins
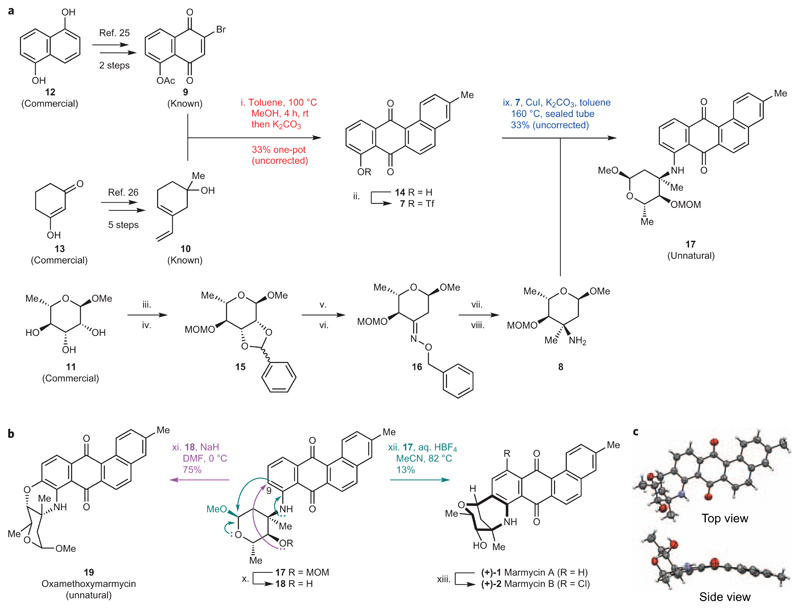
The limited supply of naturally occurring marmycin A (1) prompted us to establish a synthetic route and set up a sustainable source. Significant work in this area16–19 led to the construction of 7′-nor-marmycin A. For instance, remarkable Lewis acid-catalysed one-pot assemblies of suitable anilines and glycals have enabled the construction of the core structure of marmycin A devoid of the C7′ methyl group17,18. The presence of alkyl substituents at the junction of bicyclic systems is a ubiquitous feature of natural products, as illustrated by taxanes20 and steroids21, and represents a significant synthetic obstacle in some cases. Regardless, these pioneering studies have provided relevant clues and set important milestones towards achieving these complex structures. Previous expertise in natural product chemistry22–24 led us to consider a biomimetic route, whereby the marmycins could arise from the stepwise assembly of the aromatic core and a sugar derived from 3-epi,4-epi-vancosamine2. From a retrosynthetic standpoint (Fig. 1b), we reasoned that a seco-marmycin intermediate could be obtained by reacting the aromatic triflate 7 with the free amine 8. Such a substrate was predicted to exhibit a sufficiently high electron density at C9 to allow for a late-stage cyclization upon activation of the anomeric position, despite the presence of the electron-withdrawing quinone fragment. However, the quaternary centre carrying the amine of 8 was expected to impose a high level of steric hindrance that could preclude an effective metal-catalysed cross-coupling of the aminosugar with the anthraquinone 7. Additionally, the absence of a known natural analogue functionalized with a glycoside at C11 hinted towards a biosynthesis involving first a C–N coupling, which in turn would impose the formation of the C–C bond at C9 and yield the β-epimer as the sole reaction product. Next, we hypothesized that the aromatic building block could be built from known dienophile 925 and diene 1026 through a Diels–Alder cycloaddition. This disconnection was, however, distinct from a biomimetic step, which formally involves polyketide chemistry rather than pericyclic processes. Finally, we anticipated that the aminosugar 8 could be readily obtained from the chiral pool 11 based on previous work27,28.
With this strategy in mind, we first prepared the requisite diene25 and dienophile26 from commercially available starting materials (Fig. 2a). Dienophile 9 was synthesized in two steps from naphthalene-1,5-diol 12, whereas diene 10 was prepared in five steps from 1,3-cyclohexanedione 13 using modified procedures27. The brominated dienophile 9 and unprotected diene 10 were then reacted in hot toluene and stirred in methanolic potassium carbonate to afford a mixture of reaction intermediates and the desired cyclo-adduct 14 in good overall yields. Interestingly, we found that stirring the crude cycloaddition mixture in methanol for several hours before the addition of potassium carbonate was necessary for this multistep process to occur in one pot, suggesting that traces of hydrobromic acid may be released after cycloaddition, acting as a catalyst for dehydration and aromatization in methanol. Furthermore, using 10 as a protected tert-butyldimethylsilyl ether increased the yields of cycloaddition, but precluded dehydration/aromatization, illustrating the virtue of protecting group-free chemistry in some cases29. The corresponding triflate 7 was synthesized using a standard procedure and directly engaged in the next step. Methyl-α-l-rhamnopyranoside 11 was used as a chiral starting material, and was first reacted with benzaldehyde then chloromethyl methyl ether (MOMCl), thereby enabling us to undress the chiral sugar via a Klemer–Rodemeyer elimination30 upon exposure of the diastereomeric mixture of benzylidene 15 to sec-buthyllithium in THF. The primary amine was installed in three steps through prior conversion of the ensuing ketone into hydroxylimine 16, which was then methylated in the presence of methylcerium dichloride with high diastereoselectivity. The intermediate compound was finally subjected to hydrogenolysis to provide the target diastereoisomer 8.
Figure 2. Chemical synthesis of the marmycins.
a, Synthesis of precursors of 1. (i) 9, 10, toluene, 100 °C, 16 h, then concentrated to dryness; MeOH, rt, 4 h, then K2CO3, rt, 1 h, 33%. (ii) Tf2O, Et3N, CH2Cl2, –78 °C, 6 h, 78%. (iii) Benzaldehyde dimethylacetal, PTSA, dimethylformamide (DMF), 95 °C (reduced pressure), 12 h, 98%. (iv) (iPr)2EtN, MOMCl, CH2Cl2, rt, 72 h, 91%. (v) sec-Buthyllithium (1.6 M, hexane), THF, –40 °C, 90 min, 61%. (vi) O-benzyl hydroxylamine HCl salt, NaOAc, EtOH, rt, 2 h, 90%. (vii) Anhydrous CeCl3, methyllithium (3 M, diethoxymethane), THF, –78 °C, 1 h; then 16, –78 °C to rt, 1 h, 72%. (viii) H2, Pd/C (10 mol%), MeOH, rt, 12 h, 88%. (ix) 7, 8, CuI (20 mol%), K2CO3, toluene, 160 °C, sealed tube, 72 h, 33%. b, Assembly of aromatic and sugar moieties. (x) BF3·Et2O, CH2Cl2, –78 °C to rt, 24 h, 42%. (xi) NaH, DMF, 0 °C, 2 h, 75%. (xii) 50% wt/wt aq. HBF4, MeCN, 82 °C, 8 h, 13%. (xiii) NCS, C2H4Cl2, 84 °C, 24 h, 61%. c, ORTEP drawing of the X-ray crystal structure of 1 showing top and side views. rt, room temperature.
With the advanced building blocks 7 and 8 in hand, we explored various metal-catalysed cross-coupling conditions including classical palladium-mediated Buchwald–Hartwig chemistry31. Despite several attempts by varying ligands, solvents, bases, reaction temperatures and the use of microwave and flow chemistry, we were unable to produce sufficient quantities of the desired aniline, resulting instead in the quantitative hydrolysis of triflate 7 to regenerate phenol 14 along with unreacted amine 8. In contrast, tert-butyl amine and other less hindered primary aliphatic amines were coupled in excellent yields using palladium catalysts. Further investigations led to the identification of copper as an effective metal for this reaction32. We found that a catalytic amount of copper iodide in hot toluene led to the production of the protected seco-marmycin 17 in acceptable yields, providing sufficient material to carry on with the next step. Notably, copper-catalysed Ullmann aminations are usually conducted in the presence of aromatic halides rather than triflates. Nevertheless, these conditions were suitable to couple the electron-deficient substrate 7 with the sterically demanding amine 8, where palladium chemistry had proven ineffective.
Marmycin intermediate 17 was then exposed to a series of Brønsted and Lewis acids including p-toluenesulfonic acid (PTSA), camphorsulfonic acid, pyridinium p-toluenesulfonate, indium bromide, indium triflate and scandium triflate, varying organic solvents and reaction temperatures. These conditions led either to unreacted starting material, products of hydrolysis at C1′ and C4′, or the almost quantitative production of the free aromatic amine resulting from C3′–N bond cleavage, together with other uncharacterized by-products. Thus, we postulated that varying the nature of the protecting group of the alcohol at C4′ might provide control over conformational states in favour of a kinetically driven cyclization. To this end, the protected intermediate 17 was first treated with boron trifluoride diethyl etherate to selectively release the free alcohol 18 (Fig. 2b). To our surprise, all attempts to introduce another protecting group onto alcohol 18 under basic conditions resulted in formation of the O-cyclized product 19 (Fig. 2b), highlighting the dominant electrophilic nature of C9. This observation encouraged us to take advantage of this reactivity through the generation of the nucleophile at the anomeric position. Strikingly, analysis of the reaction mixture of 17 exposed to a salt of triphenylphosphonium hydrogen tetrafluoroborate (HPPh3·BF4) by mass spectrometry revealed the presence of a product displaying the molecular weight of 1. Hence, when 17 was subjected to aq. HBF4, we observed the formation of (+)-1 in moderate yields (Fig. 2c). This outcome was in agreement with a mechanism in which stringent acidic conditions activate the anomeric position, concurrently decreasing the electron density at C9 as a result of the protonation/chelation of the aromatic amine and/or the anthraquinone moiety. Conversely, milder acidic conditions (such as aq. HBF4) target a window of reactivity in favour of the desired intramolecular Friedel–Crafts reaction. Moreover, the use of water instead of dry organic solvents probably influenced the distribution of conformational states and thus the fate of this reaction. These results emphasize the dual electrophilic/nucleophilic nature of C9 and suggest that structures such as 19 may also be natural metabolites. As we were unable to produce marmycin B (2) from the synthetic sample of 1 using a nucleophilic reagent (for example, aq. HCl), we found that 2 could instead be obtained from 1 in the presence of N-chlorosuccinimide (NCS). This supports the notion that the aromatic amine increases the electron density at both C9 and C11, hence conferring the desired reactivity giving access to both natural small molecules. Although moderate reaction yields could be broadly considered intractable, we were able to produce appropriate quantities of rare natural products to explore the associated cell biology and did not seek to improve the reaction conditions further.
Cellular activity of marmycin A
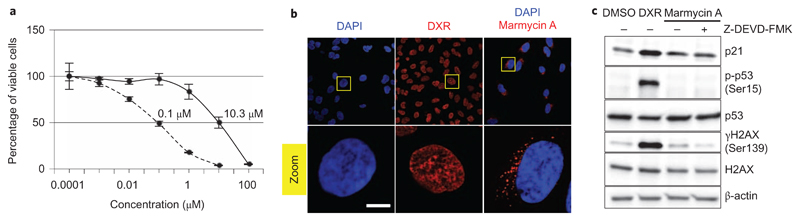
Previous work has shown that 1 exhibits cytotoxic properties against several cancer cell lines associated with a G1/S cell cycle arrest in ovarian A2780 cancer cells, consistent with the idea that 1 operates via a well-defined mechanism2. In our hands, 1 induced cell death, but no cell cycle arrest was detected by fluorescence-activated cell sorting (Supplementary Figs 1 and 2). Side-by-side comparison of 1 and the clinically approved DNA intercalating agent DXR revealed that higher doses of 1 were required to kill human osteosarcoma U2OS cells, with a half-maximum inhibitory concentration (IC50) value of ~10 μM (against ~0.1 μM for DXR) at 72 h treatment (Fig. 3a). This result, and the fact that 1 has a rather curved molecular structure (Fig. 2c), suggested that 1 may operate independently of DNA intercalation. Interestingly, visual inspection of growing cells indicated a diffuse staining of the red natural product inside the cell. This prompted us to exploit this property to identify the subcellular localization of 1 without the need to chemically label the natural product in cells, as previously done by us to detect otherwise invisible small molecules14,33. Analysis of 1 showed an excitation at ~525 nm and fluorescence emission at ~650 nm (Supplementary Fig. 3). Comparative analysis of cellular staining of the minor groove DNA binder 4,6-diamidino-2-phenylindole (DAPI), the DNA intercalating agent DXR and natural product 1 using fluorescence microscopy revealed that DAPI and DXR gave a similar nuclear staining, while 1 did not penetrate into the nucleus but accumulated instead as small vesicles in the cytosol (Fig. 3b). These data strongly challenge the notion of genome targeting by angucycline 1. To confirm this, we studied markers of the DNA damage response. Little increase of phosphorylated histone variant H2AX (γH2AX), a surrogate mark of DNA double-strand breaks, was observed upon treatment with 1, as monitored by western blotting (Fig. 3c). In line with this, no phosphorylation of p53 or significant induction of p21 could be detected, consistent with the idea that 1 does not directly promote DNA damage or a checkpoint-dependent cell cycle arrest. It is noteworthy that the low amount of γH2AX induced by 1 was prevented by co-treatment with the caspase 3 inhibitor Z-DEVD-FMK, indicating some level of caspase activation and nuclease-mediated DNA cleavage34, characteristic of an apoptotic response. In comparison, the DNA intercalating drug DXR increased the level of γH2AX and induced phosphorylation of tumour suppressor p53 and the production of p21.
Figure 3. Marmycin A kills human cancer cells independently of genome targeting.
a, Percentage of viable U2OS cells after 72 h treatment with DXR (dashed line) or 1 (black). n = 3. Error bars, s.d. b, Confocal microscopy images showing accumulation of DAPI (pan-nuclear blue), DXR (pan-nuclear red) and 1 (red puncta) in U2OS cells. Yellow boxes indicate the area of magnification of the main images. Zoomed images are ×5. Scale bar, 10 μm. c, Western blot analysis showing levels of γH2AX, p-p53 and p21 in U2OS cells treated with dimethylsulfoxide (DMSO), DXR or 1 (50 μM, 24 h) including a co-treatment with Z-DEVD-FMK.
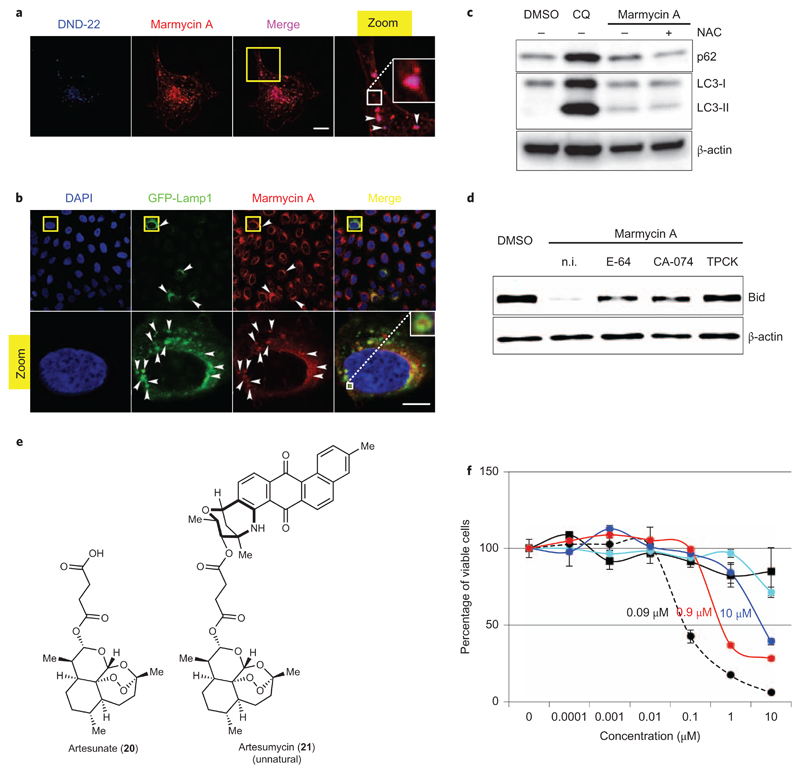
The fact that 1 is inherently fluorescent allowed us to perform live cell imaging experiments without the use of reagents that would otherwise alter the plasma membrane of cells. By doing so, we could visualize the accumulation of 1 throughout the apolar membrane, reflecting the lipophilic nature of the small molecule (Fig. 4a). Furthermore, this staining was more pronounced at some hotspots defining small vesicles of 1 sporadically disseminated throughout the plasma membrane, which suggested that 1 was, at least in part, internalized along with other components of the membrane by means of endocytosis (Fig. 4a). We therefore attempted to identify the nature of the organelles in which 1 accumulated, with particular focus on endocytic compartments, in search of putative cell death mechanisms. Simultaneous treatment of living cells with 1 (red staining) and the lysosomal marker DND-22 (blue staining) led to a significant co-localization of both substrates (merged pink staining), characteristic of a physical proximity of these compounds in the lysosomes (Fig. 4a). Conversely, pretreatment with 1 prevented the accumulation of DNA-22 in the vesicles, further indicating that marmycin A targets the lysosomal compartment. To confirm this we evaluated the localization of 1 with a biological marker of this organelle. Cells were virally infected using the BacMam system to transiently express a green fluorescent protein-fused lysosomal-associated membrane protein 1 (GFP-Lamp1). Lamp1 is involved in the structural integrity of the lysosomal compartment and the regulation of lysosome/autophagosome fusion35,36. Although the transduction efficiency was moderate, some cells contained green hollow vesicles filled with 1 (red staining), further demonstrating that 1 accumulated within the lumen of lysosomes (Fig. 4b and Supplementary Fig. 4). In contrast, marmycin A did not accumulate in the Golgi apparatus, demonstrating that the small molecule is not a generic lipophilic membrane interacting agent (Supplementary Fig. 5).
Figure 4. Marmycin A accumulates in the lysosomes and triggers programmed cell death.
a, Live imaging of U2OS cells co-treated with 1 and DND-22. The yellow box indicates the area of magnification of the main image. The zoomed image is ×3. White arrowheads indicate sites of co-localization. The white box represents a further ×3 magnification. b, Confocal microscopy images of U2OS cells expressing GFP-Lamp1 treated with 1. Yellow boxes indicate the area of magnification of the main images. Zoomed images are ×5. White arrowheads indicate sites of co-localization. The white box shows a further ×3 magnification. Scale bars, 10 μm. c, WB analysis of p62 and LC3 in U2OS cells treated with DMSO, CQ or 1 (50 µM, 24 h) including a co-treatment with NAC. d, Western blot analysis of Bid in U2OS cells treated with DMSO or 1 (10 µM, 96 h) including a co-treatment with CB or CT inhibitors. n.i., no inhibitor. e, Molecular structures of 20 and 21. f, Percentage of viable MDA-MB-231 cells after 72 h treatment with DXR (dashed line), 1 (black), 20 (cyan), 21 (red) and an equimolar mixture of 1 and 20 (blue). n = 3; error bars, s.d.
To evaluate the functional consequences of the accumulation of 1 in lysosomes, we monitored (by western blotting) the turnover of the ubiquitin-binding and oxidative stress response protein p62 as well as the autophagy marker protein LC3-II (Fig. 4c)37. We found that treatments with 1 resulted in a modest increase in p62 and LC3-II levels, indicating that 1 had little influence on the autophagic flux. In comparison, the lysosome targeting agent chloroquine (CQ) induced significantly higher levels of both p62 and LC3-II, indicating a strong inhibition of the flux. Interestingly, co-treatment of cells with 1 and the ROS scavenger N-acetyl cysteine (NAC) prevented p62 induction by 1, but did not rescue cell viability. This result indicated that 1 induced some level of ROS, as a peripheral event and not the primary cause of cell death. Additionally, while CQ is known to inhibit the autophagic flux by increasing the pH in lysosomes37, the inability of 1 to inhibit autophagy suggested that this natural product had no effect on the lysosomal pH at the concentration used.
Lipophilic small molecules such as sphingosine have been shown to induce lysosomal membrane permeabilization (LMP) in some circumstances38–40, resulting in the translocation of lysosomal proteases within the cytosol. This, in turn, can promote the cleavage and activation of specific cytosolic proteins including the pro-apoptotic BH3 interacting-domain death agonist protein (Bid), leading to mitochondrial outer membrane permeabilization (MOMP)41,42, cytochrome c release and caspases-dependent cell death. Because 1 contains a polar aminosugar fused to a lipophilic aromatic core and accumulates within the lysosomal compartment, we reasoned that 1 may also act as a detergent and trigger a similar response. Western blot analysis revealed a clear reduction of Bid protein upon treatment with 1 for 96 h (Fig. 4d). It is noteworthy that cleavage of Bid was prevented when cells were co-treated with the cathepsin B (CB) inhibitors E-64 and CA-074, or the chymotrypsin (CT) inhibitor TPCK, in strong agreement with LMP and the release of these lysosomal proteases within the cytosol. Consistent with this, 1 induced a destabilization of the lysosomal membrane, as indicated by the release of bulky FITC-dextran from lysosomes (Supplementary Fig. 6)39. Additionally, we also observed reduced levels of caspase 3, and cell viability was partially rescued when cells were co-treated with the caspase 3 inhibitors Z-DEVD-FMK or Z-VAD-FMK (Supplementary Figs 7 and 8).
The antimalarial and antiproliferative drug artesunate (20, Fig. 4e) has been shown to exert its activity by targeting the lysosomal compartment through Fenton-type chemistry43. We therefore reasoned that a synthetic dimer of 1 and 20 would combine the properties of each substrate embedded within a single scaffold. Remarkably, we found that the synthetic dimer 21 (which we named artesumycin) exhibited enhanced antiproliferative activity compared to 1 and 20 when used either independently or as a co-treatment (Fig. 4e). For example, we found that 21 displayed an IC50 value of ~0.9 μM against the metastatic MDA-MB-231 breast cancer cell line at 72 h incubation (Fig. 4f). In contrast, compounds 1 and 20 were poorly effective against this cell line at up to 10 μM when used independently, and 10 μM of each drug was required to reduce the number of viable cells by ~50%. These data indicate that 21 does not undergo hydrolysis in cells to release 1 and 20, strongly supporting a mechanistic synergy mediated by 21 against lysosomal integrity rather than independent additive effects of 1 and 20. It is noteworthy that 21 accumulated in the lysosomes, as confirmed by its co-localization with GFP-Lamp1, in line with the organelle-specific targeting capacity of 1 (Supplementary Fig. 9). Together, these data provide solid evidence for a novel model, whereby 1 induces cell death through lysosomal dysfunction, excluding direct genome targeting.
Discussion
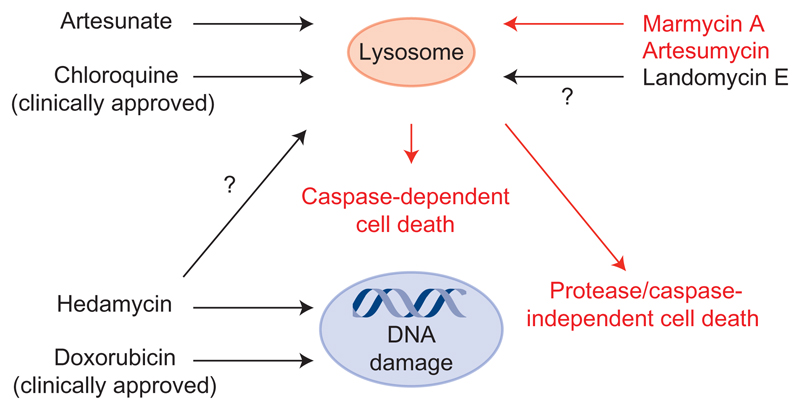
We describe an expeditious synthetic route to 1. Our strategy features a straightforward one-pot access to the angular aromatic core and its assembly with a vancosamine-derived aminosugar using a copper-catalysed cross-coupling followed by a challenging acid-mediated cyclization. This synthetic sequence lends strong support to a plausible biosynthetic route involving similar intermediates, thereby establishing the absolute configuration of the marmycins by means of chemical synthesis from known precursors. Importantly, although natural sources of 1 are scarce, this synthesis allowed the production of sufficient quantities of material to identify some of the cellular events through which the natural product exerts its activity and the preparation of the more potent derivative 21. Unbiased strategies based on quantitative proteomics44 and high-throughput sequencing14,15 have previously enabled the identification of biological targets of small molecules in cells. Here, we have used high-resolution microscopy14,33 to identify, instead, a cellular organelle as the biologically relevant target of 1. In particular, we have discovered that 1 operates as a lysosomotropic small molecule (Fig. 5), suggesting that other structurally related compounds could trigger similar responses. The biology of the lysosome is complex. Small molecules capable of targeting this organelle can induce pleiotropic responses, making it difficult to detect and quantify each single event. This includes a partial or complete LMP, the release of proteases with redundant functions, and the activation of caspase-dependent and -independent pathways, among other processes. The current study shows that 1 induces the cleavage of Bid and caspase-dependent cell death. More work is required to delineate the fine details of this response, such as the occurrence of proteases/caspase-independent cell death pathways40, which could take place on the basis that cell killing by 1 was not completely prevented by caspase inhibitors. It is also conceivable that the sudden release of the acidic content and metal ions from lysosomes upon treatment with 1 contributed to cell death in a manner that did not directly implicate the translocation of lysosomal proteases, and so explicit cell death pathways remain open to question. Regardless, our results provide substantial evidence of an alternative mechanism of action of a natural angucycline that does not involve genomic DNA. Targeting the lysosome to elicit cell death is emerging as an attractive therapeutic strategy for the treatment of cancers39,45,46. For example, the clinically approved drug CQ has been shown to prevent the relapse observed in certain cancers refractory to a DXR regimen47–50. Accordingly, marmycin A—and perhaps other angucyclines—represent valuable probes with which to study the biology of the lysosome, leaning towards unanticipated therapeutic applications for this class of natural products.
Figure 5. Antiproliferative mechanisms of anthraquinone-containing drugs.
Black arrows indicate previously established mechanisms. Red arrows highlight the cellular activity of marmycin A. Black arrows with question marks indicate putative mechanisms of action of other anthraquinone-containing small molecules.
Methods
Cell culture and proliferation assays
U2OS, A2780 and MDA-MB-231 cells were purchased from ATCC and were maintained in McCoy’s 5A or RPMI 1640, respectively, supplemented with 10% fetal bovine serum (FBS) and 1× antibiotic–antimycotic (Gibco) and incubated at 37 °C with 5% CO2. Cell viability assays were carried out by plating 2,000 cells per well in 96-well plates. Cells were treated with the relevant drug for 24 h or 72 h, then incubated with CellTiter-Blue (20 μl/well) for 1 h before recording fluorescence (560(20)Ex/590(10)Em) using a PerkinElmer Wallac 1420 Victor2 Microplate Reader. For cell cycle analysis, cells were fixed in ice-cold 70% ethanol, stained with propidium iodide (10 mg ml–1 PI, 250 mg ml–1 RNase A, 0.5% BSA, 0.02% sodium azide in 1× PBS) and analysed by FACS on a CyFlow Ploidy Analyser from Partec.
Drugs and inhibitors
Marmycin A was prepared in the laboratory according to Fig. 2. Cells were treated with 10 μM marmycin A for 24 h unless stated otherwise. Doxorubicin (Sigma) was used at 1 μM for 6 h (cell imaging) or 1 μM for 24 h (western blotting); chloroquine (Sigma) was used at 100 μM for 24 h; N-acetyl cysteine (Sigma) was used at 2 mM for 24 h (2 h pre-treatment). Z-DEVD-FMK (BD Biosciences) was used at 100 μM for 24 h (30 min pre-treatment). Z-VAD-FMK (BD Biosciences) was used at 100 μM for 24 h (30 min pretreatment). E-64 (Sigma) was used at 15 μM (added 48 h after 1, cells harvested at 96 h). CA-074 Me (Enzo Life Sciences) was used at 1 μM (added 48 h after 1, cells harvested at 96 h). N-tosyl-l-phenylalanine chloromethyl ketone (TPCK, Sigma) was used at 5 μM (added 48 h after 1, cells harvested at 96 h). Artesunate was purchased from Sigma.
Immunofluorescence analysis
U2OS cells were cultured and treated with 1 or DXR at ~40% confluence. LysoTracker Blue DND-22 (L7525, Life Technologies) and 1 were added and used without fixation in live cell imaging experiments. Fluorescence was recorded 10 min after the addition of both reagents. GFP-Lamp1 was transiently expressed in U2OS cells following the manufacturer’s instructions. Briefly, 5 μl of CellLight Lysosomes-GFP BacMam 2.0 (C10596, Life Technologies) was mixed well with 200 μM U2OS culture medium and added to each well of a 24-well plate loaded poly-l-lysine-treated coverslips. After 16 h incubation, cells were washed with PBS and treated with 1 (10 μM, 6 h). Cells were then washed with PBS, fixed for 12 min with 2% formaldehyde/PBS and permeabilized for 10 min with 0.1% Triton X-100/PBS, then washed with PBS. The Golgi apparatus was detected using an anti-RCAS1 antibody (1/100, #12290, Cell Signaling) and anti-rabbit Alexa Fluor secondary antibody (1/500, A-11008, Life Technologies). Coverslips were mounted with VectaShield mounting medium with or without DAPI (Vector Laboratories). Images were taken with Leica SP8 microscope or Nikon Te(-E) inverted confocal microscopes equipped with a spinning disk Yokogawa CXU-X1 A1 (DND-22 experiment only). Data were analysed with ImageJ software (NIH).
Western blotting
Cells were treated as indicated in Figs 3 and 4, then washed twice with PBS and lysed with 2× Laemmli buffer. Cell extracts were heated for 5 min at 95 °C, sheared through a 26-gauge needle and quantified with a Nanodrop 2000 (Thermo Scientific). Protein lysates (~100 μg) were loaded on a 4–20% Mini-PROTEAN TGX Stain-Free Gel (BioRad), resolved by SDS-PAGE electrophoresis and transferred onto a nitrocellulose membrane (Amersham). Proteins were probed with the following antibodies: anti-β-actin (ab8226, Abcam), anti-H2AX (PA1-14198, Thermo Scientific), anti-γH2AX (#2577, Cell Signaling), anti-p21 (#2947, Cell Signaling), anti-p53 (#2524, Cell Signaling), anti-p-p53 (#9284, Cell Signaling), anti-p62 (610,833, BD Transduction Laboratories), anti-LC3 (#2775, Cell Signaling), anti-Bid (#2002, Cell Signaling) and anti-Caspase 3 (#9665, Cell Signaling) and were diluted 1/1,000 in 5% BSA, 0.1% Tween-20/TBS. The secondary antibodies were anti-rabbit HRP (A120-108P, Bethyl), anti-mouse HRP (A90-116P) and anti-donkey HRP (A140-107P, Bethyl). Antigens were detected by ECL (Amersham). Imaging was performed using a ChemiDoc XRS+ System.
Supplementary Material
Acknowledgements
The authors thank the CNRS for funding, and the Imagif Cell Biology Unit, J.-F. Gallard and F. Blanchard for assistance with cell imaging, NMR spectroscopy and X-ray crystallography, respectively. R.R. thanks J.A. Yeoman, S. Müller, J.E. Moses, S.L. Buchwald, L. Johannes, M. Mehrpour and members of R.R.’s laboratory for discussions and proofreading of this manuscript. Research in R.R.’s laboratory is supported by the European Research Council (grant no. 647973), the Fondation pour la Recherche Médicale (grant no. AJE20141031486), the Emergence Ville de Paris Program and the Ligue Nationale Contre le Cancer.
Footnotes
Author contributions
R.R., with contributions from T.C., G.M. and M.M., conceived and designed the experiments. T.C., with assistance from F.G., synthesized the marmycins and artesumycin. T.T.M. performed proliferation assays, fluorescence-activated cell sorting, western blotting and cellular imaging. R.R., G.M. and M.M. supervised the research. All the authors analysed the data. R.R. wrote the manuscript. All authors commented on the manuscript. T.T.M. and F.G. contributed equally to this work.
Competing financial interests
The authors declare no competing financial interests.
Additional information
Reprints and permissions information is available online at www.nature.com/reprints.
References
- 1.Kharel MK, et al. Angucyclines: biosynthesis, mode-of-action, new natural products, and synthesis. Nat Prod Rep. 2012;29:264–325. doi: 10.1039/c1np00068c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin GDA, et al. Marmycins A and B, cytotoxic pentacyclic C-glycosides from a marine sediment-derived actinomycete related to the genus Streptomyces. J Nat Prod. 2007;70:1406–1409. doi: 10.1021/np060621r. [DOI] [PubMed] [Google Scholar]
- 3.Cottreau KM, et al. Diverse DNA-cleaving capacities of the jadomycins through precursor-directed biosynthesis. Org Lett. 2010;12:1172–1175. doi: 10.1021/ol902907r. [DOI] [PubMed] [Google Scholar]
- 4.Sun D, Hansen M, Hurley L. Molecular basis for the DNA sequence specificity of the pluramycins. A novel mechanism involving groove interactions transmitted through the helix via intercalation to achieve sequence selectivity at the covalent bonding step. J Am Chem Soc. 1995;117:2430–2440. [Google Scholar]
- 5.Hansen M, Yun S, Hurley L. Hedamycin intercalates the DNA helix and, through carbohydrate-mediated recognition in the minor groove, directs N7-alkylation of guanine in the major groove in a sequence-specific manner. Chem Biol. 1995;2:229–240. doi: 10.1016/1074-5521(95)90273-2. [DOI] [PubMed] [Google Scholar]
- 6.Singal PK, Iliskovic N. Doxorubicin-induced cardiomyopathy. N Engl J Med. 1998;339:900–905. doi: 10.1056/NEJM199809243391307. [DOI] [PubMed] [Google Scholar]
- 7.Yang F, Kemp CJ, Henikoff S. Doxorubicin enhances nucleosome turnover around promoters. Curr Biol. 2013;23:782–787. doi: 10.1016/j.cub.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pang B, et al. Drug-induced histone eviction from open chromatin contributes to the chemotherapeutic effects of doxorubicin. Nature Commun. 2013;4:1908. doi: 10.1038/ncomms2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pigram WJ, Fuller W, Hamilton LD. Stereochemistry of intercalation: interaction of daunomycin with DNA. Nature. 1972;235:17–19. doi: 10.1038/newbio235017a0. [DOI] [PubMed] [Google Scholar]
- 10.Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nature Rev Cancer. 2009;9:338–350. doi: 10.1038/nrc2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korynevska A, et al. Mechanisms underlying the anticancer activities of the angucycline landomycin E. Biochem Pharmacol. 2007;74:1713–1726. doi: 10.1016/j.bcp.2007.08.026. [DOI] [PubMed] [Google Scholar]
- 12.Müller S, Kumari S, Rodriguez R, Balasubramanian S. Small-molecule-mediated G-quadruplex isolation from human cells. Nature Chem. 2010;2:1095–1098. doi: 10.1038/nchem.842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koirala D, et al. A single-molecule platform for investigation of interactions between G-quadruplexes and small-molecule ligands. Nature Chem. 2011;3:782–787. doi: 10.1038/nchem.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez R, et al. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nature Chem Biol. 2012;8:301–310. doi: 10.1038/nchembio.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodriguez R, Miller KM. Unravelling the genomic targets of small molecules using high-throughput sequencing. Nature Rev Genet. 2012;77:5439–5444. doi: 10.1038/nrg3796. [DOI] [PubMed] [Google Scholar]
- 16.Yadav JS, et al. InBr3-catalyzed cyclization of glycals with aryl amines. Angew Chem Int Ed. 2003;42:5198–5201. doi: 10.1002/anie.200351267. [DOI] [PubMed] [Google Scholar]
- 17.Ding C, et al. Synthesis study on marmycin A: preparation of the C3′-desmethyl analogues. J Org Chem. 2009;74:6111–6119. doi: 10.1021/jo9011078. [DOI] [PubMed] [Google Scholar]
- 18.Maugel N, Snider BB. Efficient synthesis of the tetracyclic aminoquinone moiety of marmycin A. Org Lett. 2009;11:4926–4929. doi: 10.1021/ol9020496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bourcet E, Bröhmer MC, Nieger M, Bräse S. Synthetic studies towards marmycins A and B: development of the vinylogous aldol–aza-Michael domino reaction. Org Biomol Chem. 2011;9:3136–3138. doi: 10.1039/c1ob05124e. [DOI] [PubMed] [Google Scholar]
- 20.Mendoza A, Ishihara Y, Baran PS. Scalable enantioselective total synthesis of taxanes. Nature Chem. 2012;4:21–25. doi: 10.1038/nchem.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodriguez R, Chapelon A-S, Ollivier C, Santelli M. Stereoselective synthesis of CD-ring precursors of vitamin D derivatives. Tetrahedron. 2009;65:7001–7015. [Google Scholar]
- 22.Rodriguez R, Adlington RM, Moses JE, Cowley A, Baldwin JE. A new and efficient method for o-quinone methide intermediate generation: application to the biomimetic synthesis of (±)-alboatrin. Org Lett. 2004;6:3617–3619. doi: 10.1021/ol048479d. [DOI] [PubMed] [Google Scholar]
- 23.Rodriguez R, et al. Total synthesis of cyercene A and the biomimetic synthesis of (±)-9,10-deoxytridachione and (±)-ocellapyrone A. Tetrahedron. 2007;63:4500–4509. [Google Scholar]
- 24.Eade SJ, et al. Biomimetic synthesis of pyrone-derived natural products: exploring chemical pathways from a unique polyketide precursor. J Org Chem. 2008;73:4830–4839. doi: 10.1021/jo800220w. [DOI] [PubMed] [Google Scholar]
- 25.Heinzman SW, Grunwell JR. Regiospecific synthesis of bromojuglone derivatives. Tetrahedron Lett. 1980;21:4305–4308. [Google Scholar]
- 26.Carreño MC, Urbano A, Di Vitta C. Enantioselective Diels–Alder approach to C-3-oxygenated angucyclinones from (SS)-2-(p-tolylsulfinyl)-1,4-naphthoquinone. Chem Eur J. 2000;6:906–913. doi: 10.1002/(sici)1521-3765(20000303)6:5<906::aid-chem906>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 27.Brimacombe JS, Hanna R, Saeed MS, Tucker LCN. Convenient syntheses of l-digitoxose, l-cymarose, and l-ristosamine. J Chem Soc. 1982:2583–2587. [Google Scholar]
- 28.Greven R, Jütten P, Scharf H-D. A new stereoselective route to branched-chain nitro and amino sugars: synthesis of both enantiomers of decilonitrose and avidinosamine. J Org Chem. 1993;58:3742–3747. [Google Scholar]
- 29.Baran PS, Maimone TJ, Richter JM. Total synthesis of marine natural products without using protecting groups. Nature. 2007;446:404–408. doi: 10.1038/nature05569. [DOI] [PubMed] [Google Scholar]
- 30.Klemer A, Rodemeyer G. Simple synthesis of methyl 4,6-ortho-benzylidene-2-desoxy-alpha-d-erythro-hexopyranosid-3-ulose. Chem Ber. 1974;107:2612–2614. [Google Scholar]
- 31.Wolfe JP, Wagaw S, Marcoux J-F, Buchwald SL. Rational development of practical catalysts for aromatic carbon–nitrogen bond formation. Acc Chem Res. 1998;31:805–818. [Google Scholar]
- 32.Sambiagio C, Marsden SP, Blacker AJ, McGowan PC. Copper catalysed Ullmann type chemistry: from mechanistic aspects to modern development. Chem Soc Rev. 2014;43:3525–3550. doi: 10.1039/c3cs60289c. [DOI] [PubMed] [Google Scholar]
- 33.Larrieu D, Britton S, Demir M, Rodriguez R, Jackson SP. Chemical inhibition of NAT10 corrects defects of laminopathic cells. Science. 2014;344:527–532. doi: 10.1126/science.1252651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Enari M, et al. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998;391:43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- 35.Eskelinen E-L, Tanaka Y, Saftig P. At the acidic edge: emerging functions for lysosomal membrane proteins. Trends Cell Biol. 2003;13:137–145. doi: 10.1016/s0962-8924(03)00005-9. [DOI] [PubMed] [Google Scholar]
- 36.Huynh KK, et al. LAMP proteins are required for fusion of lysosomes with phagosomes. EMBO J. 2007;26:313–324. doi: 10.1038/sj.emboj.7601511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kågedal K, Zhao M, Svensson I, Brunk UT. Sphingosine-induced apoptosis is dependent on lysosomal proteases. Biochem J. 2001;359:335–343. doi: 10.1042/0264-6021:3590335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boya P, Kroemer G. Lysosomal membrane permeabilization in cell death. Oncogene. 2008;27:6434–6451. doi: 10.1038/onc.2008.310. [DOI] [PubMed] [Google Scholar]
- 40.Galluzzi L, Bravo-San Pedro JM, Kroemer G. Organelle-specific initiation of cell death. Nature Cell Biol. 2014;16:728–736. doi: 10.1038/ncb3005. [DOI] [PubMed] [Google Scholar]
- 41.Li H, Zhu H, Xu C-J, Yuan J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- 42.Chipuk JE, Bouchier-Hayes L, Green DR. Mitochondrial outer membrane permeabilization during apoptosis: the innocent bystander scenario. Cell Death Differ. 2006;13:1396–1402. doi: 10.1038/sj.cdd.4401963. [DOI] [PubMed] [Google Scholar]
- 43.Hamacher-Brady A, et al. Artesunate activates mitochondrial apoptosis in breast cancer cells via iron-catalyzed lysosomal reactive oxygen species production. J Biol Chem. 2011;286:6587–6601. doi: 10.1074/jbc.M110.210047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ziegler S, Pries V, Hedberg C, Waldmann H. Target identification for small bioactive molecules: finding the needle in the haystack. Angew Chem Int Ed. 2013;52:2744–2792. doi: 10.1002/anie.201208749. [DOI] [PubMed] [Google Scholar]
- 45.Li Y, et al. Amplification of LAPTM4B and YWHAZ contributes to chemotherapy resistance and recurrence of breast cancer. Nature Med. 2010;16:214–218. doi: 10.1038/nm.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kreuzaler P, Watson CJ. Killing a cancer: what are the alternatives? Nature Rev Cancer. 2012;12:411–424. doi: 10.1038/nrc3264. [DOI] [PubMed] [Google Scholar]
- 47.Shiraishi N, Akiyama S-I, Kobayashi M, Kuwano M. Lysosomotropic agents reverse multiple drug resistance in human cancer cells. Cancer Lett. 1986;30:251–259. doi: 10.1016/0304-3835(86)90049-2. [DOI] [PubMed] [Google Scholar]
- 48.Zamora JM, Beck WT. Chloroquine enhancement of anticancer drug cytotoxicity in multiple drug resistant human leukemic cells. Biochem Pharmacol. 1986;35:4303–4310. doi: 10.1016/0006-2952(86)90710-0. [DOI] [PubMed] [Google Scholar]
- 49.Vezmar M, Georges E. Reversal of MRP-mediated doxorubicin resistance with quinoline-based drugs. Biochem Pharmacol. 2000;59:1245–1252. doi: 10.1016/s0006-2952(00)00270-7. [DOI] [PubMed] [Google Scholar]
- 50.Savarino A, Lucia MB, Giordano F, Cauda R. Risks and benefits of chloroquine use in anticancer strategies. Lancet Oncol. 2006;7:792–793. doi: 10.1016/S1470-2045(06)70875-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.