Abstract
Hyperactivation of the PI3K pathway has been implicated in resistance to antiestrogen therapies in estrogen receptor α (ER)–positive breast cancer, prompting the development of therapeutic strategies to inhibit this pathway. Autophagy has tumor-promoting and -suppressing roles and has been broadly implicated in resistance to anticancer therapies, including antiestrogens. Chloroquine (CQ) is an antimalarial and amebicidal drug that inhibits autophagy in mammalian cells and human tumors. Herein, we observed that CQ inhibited proliferation and autophagy in ER+ breast cancer cells. PI3K inhibition with GDC-0941 (pictilisib) induced autophagy. Inhibition of autophagy using CQ or RNA interference potentiated PI3K inhibitor-induced apoptosis. Combined inhibition of PI3K and autophagy effectively induced mitochondrial membrane depolarization, which required the BH3-only proapoptotic proteins Bim and PUMA. Treatment with GDC-0941, CQ, or the combination, significantly suppressed the growth of ER+ breast cancer xenografts in mice. In an antiestrogen–resistant xenograft model, GDC-0941 synergized with CQ to provide partial, but durable, tumor regression. These findings warrant clinical evaluation of therapeutic strategies to target ER, PI3K, and autophagy for the treatment of ER+ breast cancer.—Yang, W., Hosford, S. R., Traphagen, N. A., Shee, K., Demidenko, E., Liu, S., Miller, T. W. Autophagy promotes escape from phosphatidylinositol 3-kinase inhibition in estrogen receptor–positive breast cancer.
Keywords: PI3K, GDC-0941, chloroquine, ER, BH3-only proteins
Approximately 70% of breast tumors express estrogen receptor α (ER) and/or progesterone receptor. Patients with such tumors are typically treated with antiestrogen therapies to block ER function, including tamoxifen, aromatase inhibitors (AIs), and fulvestrant (fulv). However, approximately one-third of patients initially diagnosed with early stage ER+ breast cancer ultimately develop recurrent, antiestrogen–resistant disease (1). The PI3K/AKT/mechanistic target of rapamycin (mTOR) pathway is the most frequently aberrantly activated pathway in cancer, and alterations in genes encoding PI3K pathway proteins (e.g., PIK3CA, PTEN) occur in >70% of ER+ breast cancers (2). Clinical and preclinical data suggest that hyperactivation of the PI3K/AKT/mTOR pathway promotes antiestrogen resistance (3–5). The mTOR complex I (mTORC1) inhibitor everolimus was approved for use in combination with the aromatase inhibitor (AI) exemestane for the treatment of recurrent/metastatic ER+/HER2− breast cancer based on data showing significant improvement in progression-free survival (PFS) compared with exemestane/placebo (6). However, mTORC1 inhibition blocks negative-feedback signaling loops, leading to hyperactivation of pathways upstream of mTORC1, including PI3K (7–9). In some cancer subtypes, including ER+ breast cancer, mTORC1 is tightly controlled by PI3K/AKT signaling (4, 10). Thus, PI3K inhibitors (PI3Ki) are being developed as a potentially more-effective therapeutic strategy.
Pan- and isoform-selective inhibitors of class IA PI3K (PI3Ki) are being developed clinically for the treatment of breast and other cancers. The phase II FERGI trial showed that the addition of the pan-PI3Ki pictilisib (GDC-0941) to fulv did not significantly improve PFS in postmenopausal patients with recurrent/metastatic ER+/HER2− antiestrogen–resistant breast cancer. However, a subset analysis indicated that patients with progesterone receptor-positive tumors showed significantly improved PFS with pictilisib/fulv vs. placebo/fulv (7.4 vs. 3.7 mo) (11). The phase III BELLE-2 trial testing the pan-PI3Ki buparlisib (BKM-120) in postmenopausal women with antiestrogen–resistant ER+/HER2− recurrent/metastatic breast cancer showed that buparlisib/fulv significantly improved PFS and clinical benefit compared with placebo/fulv, particularly in patients with tumors harboring activating mutations in PIK3CA (12). However, pictilisib and buparlisib showed considerable dose-limiting toxicities that prompted frequent dose reductions and treatment discontinuations, which may have limited efficacy. Clinical benefit from the PI3K/p110α isoform-selective inhibitor alpelisib (BYL-719) in combination with the AI letrozole was seen in a higher proportion of patients with PIK3CA-mutant ER+ breast tumors (44%) compared with PIK3CA–wild-type (20%) (13). Although showing clinical benefit in patients with ER+ breast cancer, antiestrogen/PI3Ki combination therapies provide a limited duration of efficacy: median PFS was 6.6 and 6.9 mo on pictilisib/fulv and buparlisib/fulv, respectively; 65% of patients treated with alpelisib/letrozole experienced disease progression within 6 mo (11–13).
Autophagy is a catabolic process that cells use to dispose of damaged organelles and misfolded/aggregated proteins. Biomolecules are recycled through lysosomal degradation in response to cellular stressors, such as starvation, hypoxia, DNA damage, and unfolded proteins (14). mTORC1 phosphorylates Ulk1 at Ser757 to disrupt Ulk1-AMPK interaction and prevent autophagy (15). mTORC1 also phosphorylates and inhibits nuclear translocation of transcription factors that drive the expression of genes encoding proteins required for lysosomal biogenesis and autophagy machinery (16). Autophagy is thought to have both tumor-promoting and -suppressing roles in cancer, and cancer cells exhibit greater dependency on autophagy for survival than noncancer cells do. Autophagy confers to resistance to antiestrogens, chemotherapy, and radiation therapy (17–21). Thus, autophagy is an attractive therapeutic target for cancer. PI3K and mTOR inhibition can induce autophagy, and inhibition of autophagy enhances the growth-suppressive and apoptotic effects of PI3K/mTOR inhibition (22–24).
Chloroquine (CQ) and hydroxychloroquine (HCQ) have long been used to treat and prevent malaria in humans, and both are known to inhibit autophagy in mammalian cells. The phosphatidylethanolamine-conjugated form of LC3 (LC3-II) is recruited to autophagosomal membranes and is used as a marker of autophagy induction (25). Autophagosomes fuse with lysosomes, which induces degradation of LC3-II. CQ/HCQ block the fusion of autophagosomes with lysosomes, inducing LC3-II accumulation; thus, LC3-II accumulation in the context of CQ treatment is a marker of autophagy inhibition (26). HCQ treatment increased LC3-II levels and autophagic vacuole accumulation in peripheral blood lymphocytes in humans (27, 28). In another study of patients with advanced solid tumors, autophagic vacuole accumulation was observed in peripheral blood mononuclear cells at 4 h after infusion of the mTORC1 inhibitor temsirolimus. After 4 wk of combination treatment with temsirolimus/HCQ, further accumulation of autophagic vacuoles in peripheral blood mononuclear cells and in tumor cells was observed. The temporal increase in autophagic vacuole accumulation was interpreted as 1) autophagy blockade by HCQ, and/or 2) induction of autophagy over longer-term exposure to temsirolimus (29).
Given the established safety profiles, CQ and HCQ are being investigated in >40 ongoing clinical trials for the treatment of cancer. In preclinical studies with cancer cells and tumors, CQ inhibits autophagic flux by accumulating in lysosomes and neutralizing pH (30). CQ has shown antitumor efficacy with antiestrogens in mice with ER+ breast tumors, potentially via macrophage activation (31). However, the potential anticancer mechanism of CQ/HCQ action in ER+ breast cancer remains unclear. We hypothesized that ER+ breast cancer resistance to PI3Ki is associated with PI3Ki-induced autophagy; blockade of which using CQ could increase efficacy of PI3Ki.
MATERIALS AND METHODS
Cell lines and RNA interference
Parental cell lines were obtained from American Type Culture Collection (Manassas, VA, USA), cultured in DMEM (CellGro, VWR International, Radnor, PA, USA) with 10% FBS (HyClone; GE Healthcare Life Sciences, Little Chalfont St. Giles, United Kingdom), and passaged for <3 mo before analysis. Fulvestrant-resistant (FR) MCF-7 (MCF-7/FR) and T47D (T47D/FR) cells were gifts from Matthew Ellis (Washington University, St. Louis, MO, USA) and were maintained in DMEM/10% FBS with 1 μM fulv (Bio-Techne, Minneapolis, MN, USA). ZR75-1/FR cells were generated through culture with 1 μM fulv for 4 mo. Cell lines were authenticated by mutational profiling using a 541-gene panel or by short tandem repeat genotyping in the University of Vermont Cancer Center DNA Analysis Facility (Burlington, VT, USA). Cells were transfected with short interfering RNA (siRNA) targeting ATG5, ATG7, Bim, PUMA, or nonsilencing control (Qiagen, Hilden, Germany; and Dharmacon, Lafayette, CO, USA) using Lipofectamine RNAiMax (Thermo Fisher Scientific, Waltham, MA, USA) per the manufacturer’s instructions. Cells were treated with GDC-0941 (kindly provided by Genentech, South San Francisco, CA, USA), CQ (Tokyo Chemical Industry, Tokyo, Japan), or fulv as indicated.
Sulforhodamine B assay
Cells were seeded in triplicate at 3000–5000/well in 96-well plates. The next day, cells were treated with CQ as indicated ≤5 d. Relative numbers of adherent cells were determined by sulforhodamine B staining (32).
Apoptosis assay
Cells were seeded in triplicate in 12-well plates and treated as indicated the next day in either low-serum (DMEM/1% FBS; GE Healthcare Life Sciences) or hormone-depleted conditions. Cells in hormone-depleted conditions were adapted to phenol red–free DMEM/10% dextran-coated charcoal–FBS (GE Healthcare Life Sciences) for 3 d before drug treatment. Apoptosis was measured by flow cytometry using the ApoScreen Annexin V apoptosis Kit (SouthernBiotech, Birmingham, AL, USA), per the manufacturer’s instructions.
Mitochondrial membrane potential assay
Cells were seeded in triplicate at 10,000–20,000/well in black, 96-well plates and treated as indicated the next day. After 24 h, cells were washed with PBS, then incubated in PBS containing 10 mg/ml JC-1 dye (Adipogen; VWR International) at 37°C for 20 min. Cells were washed twice with PBS, and fluorescence was measured using a SpectraMax M2 Multi-Mode microplate reader (Molecular Devices, Sunnyvale, CA, USA). JC-1 exhibits potential-dependent accumulation in mitochondria, as indicated by a fluorescence emission shift from red to green. The ratio of red to green fluorescence indicates relative mitochondrial membrane potential. Red fluorescence was detected at excitation/emission wavelengths of 560/595 nM, and green fluorescence was detected at excitation/emission wavelengths of 485/535 nM.
Mouse studies
Animal studies were approved by the Dartmouth College Institutional Animal Care and Use Committee. Female, NOD-scid/IL-2Rγ−/− (NSG; NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) mice (6–7 wk old; Norris Cotton Cancer Center Transgenic and Genetic Construct Shared Resource) were s.c. injected with 5–10 × 106 ZR75-1 or ZR75-1/FR cells suspended in 100 μl matrigel (BD Biosciences, Franklin Lakes, NJ, USA). Mice were also s.c. implanted on the same day with a 17β-estradiol pellet (0.72 mg, 60-d release; Innovative Research of America, Sarasota, FL, USA). In mice injected with ZR75-1/FR cells, fulv (5 mg/wk) was s.c. administered throughout the study starting on the same day as cell implantation [clinical formulation; provides ER inhibition in xenografts for ≥8 d (data not shown); kindly provided by AstraZeneca, Cambridge, United Kingdom]. Tumor volumes were measured twice weekly using calipers [volume = (length2 × width/2)]. Mice bearing tumors ∼300 mm3 were randomized to treatment with vehicle, GDC-0941 (100 mg/kg/d × 5 d/wk, or 800 mg/kg/wk, by mouth in 100 μl 0.5% methylcellulose/0.2%Tween-80; kindly provided by Genentech, South San Francisco, CA, USA), CQ [0.288 mg/ml in drinking water to reach a dose of ∼1.15–2.02 mg/d (33) or 50 mg/kg/d, i.p. in 100 μl PBS], and combinations. At the end of the study, mice were euthanized by CO2 asphyxiation and cervical dislocation, then tumors were harvested and cut in pieces for snap-freezing or formalin fixation followed by paraffin embedding.
Immunoblot analysis
Cells were lysed, and frozen tumors were homogenized and lysed in RIPA buffer [50 mM Tris pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% deoxycholic acid, 0.1% SDS plus fresh Halt protease inhibitor cocktail (Thermo Fisher Scientific and VWR International) and 1 mM Na3VO4 (New England Biolabs, Ipswich, MA, USA)]. Lysates were sonicated for 15 s and centrifuged at 17,000 g for 10 min at 4°C, and the protein in the supernatants was quantified using bicinchoninic acid assay (Thermo Fisher Scientific and VWR International). Protein extracts were denatured with NuPage (Thermo Fisher Scientific), and reduced with 1.25% β-mercaptoethanol (MilliporeSigma, Billerica, MA, USA). Proteins were separated by SDS-PAGE and transferred to nitrocellulose membrane. Blots were probed with antibodies against P-AKTT308, p62, p53, ATG5, ATG7, PARP, Bim, PUMA, vinculin, and actin (Cell Signaling Technology, Danvers, MA, USA). Horseradish peroxidase–labeled secondary antibodies (GE Healthcare Life Sciences) and ECL substrate (Thermo Fisher Scientific) were used for signal detection. Densitometry analysis was performed using QuantityOne Software (Bio-Rad Laboratories, Hercules, CA, USA).
Immunohistochemistry
For immunohistochemistry (IHC), 5 µm sections of formalin-fixed, paraffin-embedded tumor tissue were used for hematoxylin and eosin staining, with Ki67 antibody (Biocare Medical, Concord, CA, USA) and TUNEL (DeadEnd Colorimetric System; Promega, Madison, WI, USA). Proportions of positively stained cells were counted in 3 random microscopic fields (×400 magnification) in each sample.
Statistical analyses
Cell growth, IHC, and TUNEL data were analyzed by ANOVA, followed by Bonferroni multiple comparison–adjusted post hoc testing between groups. To estimate progression/regression of tumors, the following linear mixed model was employed: −tumor volumeit = ai + b × t + eit, where i represents the ith mouse, and t represents the time of tumor volume measurement, ai represents the mouse-specific log tumor volume at the baseline (t = 0), slope b represents the rate of tumor volume growth (or reduction), and eit represents the deviation of measurements from the model over time (34–36). The variance of ai is interpreted as mouse heterogeneity and b × loge(10) × 100 estimates the percentage of tumor volume increase per week. The computation was carried out in the statistical software package R (37), using function lme from library nlme. Treatment groups were compared using z test for slopes with standard errors derived from lme. Synergy was assessed using the difference of slopes (b1 − b0) + (b2 − b0) − (b12 − b0), where b1, b2, b12, and b0 are the slopes from the treatment groups 1 and 2, the combined treatment, and the control group.
RESULTS
CQ treatment blocks autophagy induced by PI3K inhibition and potentiates apoptosis
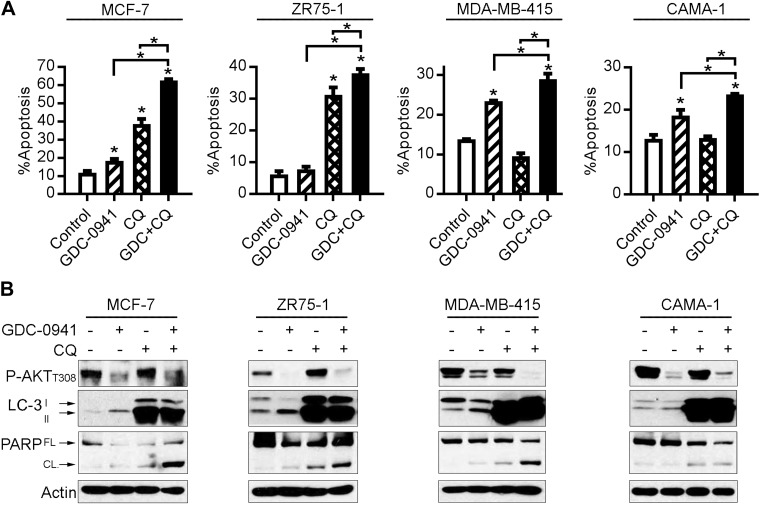
Although inhibition of oncogenic kinases (e.g., PI3K, MEK) frequently suppresses cancer-cell proliferation, apoptosis is required for regression of solid tumors (38). PI3Ki is being developed for the treatment of ER+ breast and other cancers, so we focused on harnessing the cytotoxic potential of PI3Ki. We first observed that PI3Ki with 1 μM GDC-0941 induced only limited apoptosis (control-subtracted 1.5–9.7% apoptosis) in ER+ breast cancer cells under low-serum conditions (Fig. 1A). Because PI3Ki may be more effective in combination with antiestrogens against ER+ breast cancer (39), we used hormone-depleted medium (with 10% dextran-coated charcoal–FBS) to model the estrogen deprivation induced by AI therapy in patients. In hormone-depleted conditions, 3 d of PI3Ki induced only limited apoptosis in MCF-7 (19%) and ZR75-1 (18%) ER+ breast cancer cells, but not T47D cells (Supplemental Fig. S1); T47D cells expressed low levels of the proapoptotic protein Bim (38), which may disrupt induction of apoptosis in response to drug treatment. To increase the efficacy of PI3Ki for ER+ breast cancer, we focused on strategies to potentiate apoptosis. Notably, all ER+ breast cancer cell lines investigated in this study harbor genetic alterations that activate the PI3K pathway: MCF-7 and T47D cells harbor PIK3CA mutations in exons 9 and 20, respectively, which are associated with sensitization to pan-PI3K and p110α-selective inhibitors; ZR75-1, MDA-MB-415, and CAMA-1 cells are PTEN-deficient, and PTEN loss is associated with sensitization to p110β-selective inhibitors (40–42).
Figure 1.
CQ treatment blocks autophagy and potentiates apoptosis induced by PI3K inhibition. ER+ breast cancer cells were treated with DMEM/1% FBS ± 1 μM GDC-0941 or 50 μM CQ. A) Apoptosis was measured by PI/Annexin V staining and flow cytometry after 24 h of treatment. *P < 0.05 by Bonferroni multiple comparison-adjusted post hoc test compared with control unless otherwise indicated with brackets (between GDC and GDC+CQ, or between CQ and GDC+CQ). B) Lysates were collected after 24 h of treatment and analyzed by immunoblotting using the indicated antibodies. FL, full-length; CL, cleaved.
Autophagy has been suggested to promote resistance to inhibitors of the PI3K pathway (24, 43). CQ is being investigated as an autophagy inhibitor in both preclinical and clinical studies. We observed that 50 μM CQ effectively induced accumulation of LC3-II and suppressed growth of ER+ breast cancer cells (Supplemental Fig. S2). Cells were then treated with vehicle, 1 μM GDC-0941, 50 μM CQ, or the combination for 24 h in low-serum conditions. PI3Ki alone blocked phosphorylation of AKT (a marker of PI3K activity), induced LC3-II accumulation (a marker of autophagy), and induced modest PARP cleavage and apoptosis (Fig. 1); these observations mirror those of Cook et al. (44), who found that AMPK-mediated inhibition of mTORC1 induced autophagy in ER+ breast cancer cells. Combined treatment with GDC-0941 and CQ induced greater PARP cleavage and significantly increased apoptosis compared with either single agent (Fig. 1). Similar results were obtained in hormone-depleted conditions: PI3Ki increased LC3-II and decreased levels of p62 (indicating autophagy) in 4 of 4 cell lines, CQ inhibited autophagy, and the drug combination more effectively induced apoptosis than either agent alone (Supplemental Fig. S3). CQ increased P-AKT levels in some cell lines, which was mostly mitigated by PI3Ki (Fig. 1B). We also found that PI3Ki induced autophagy in ER+ breast cancer cells with acquired resistance to the antiestrogen fulv (MCF-7/FR, T47D/FR, ZR75-1/FR). Cotreatment with GDC-0941 and CQ (in the context of fulv) induced more PARP cleavage and apoptosis than either agent alone in FR cells (Supplemental Fig. S4). These data indicate that PI3Ki induces apoptosis and autophagy, and CQ inhibits autophagy and enhances PI3Ki-induced apoptosis, suggesting that autophagy protects ER+ breast cancer cells from PI3Ki.
Genetic inhibition of autophagy enhances PI3K inhibitor–induced apoptosis
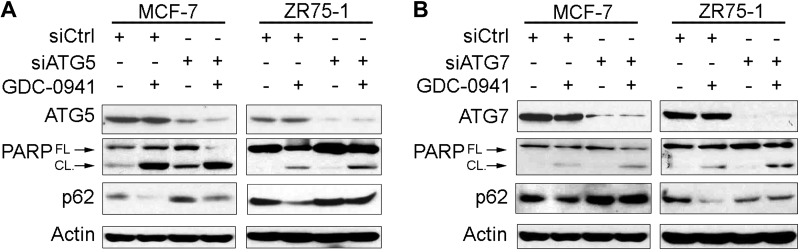
Because CQ may also affect nonautophagy pathways, we used genetic means to disrupt autophagy. ATG5 is required for the initiation of autophagy and becomes covalently conjugated to the ubiquitin-like protein ATG12 by the ubiquitin-activating enzyme ATG7 in the formation of the autophagosome (14). siRNA-induced knockdown of either ATG5 or ATG7 increased p62 levels, indicating decreased autophagy (Fig. 2 and Supplemental Fig. S5). PI3Ki induced autophagy (decreased p62), which was mitigated by siATG5 and siATG7. Knockdown of ATG5 or ATG7 combined with PI3Ki induced greater PARP cleavage than either treatment alone. Thus, autophagy hinders PI3Ki-induced apoptosis, and inhibition of autophagy can enhance the cytotoxic effects of PI3Ki.
Figure 2.
Inhibition of autophagy enhances PI3K inhibitor–induced apoptosis. Cells were transfected with siRNA targeting ATG5 (A), ATG7 (B), or nonsilencing control; 2 d after transfection, cells were treated ± 1 μM GDC-0941. Lysates were collected after 24 h of drug treatment and analyzed by immunoblotting using the indicated antibodies. FL, full-length; CL, cleaved.
Chloroquine treatment increases PI3K inhibitor-induced mitochondrial membrane depolarization
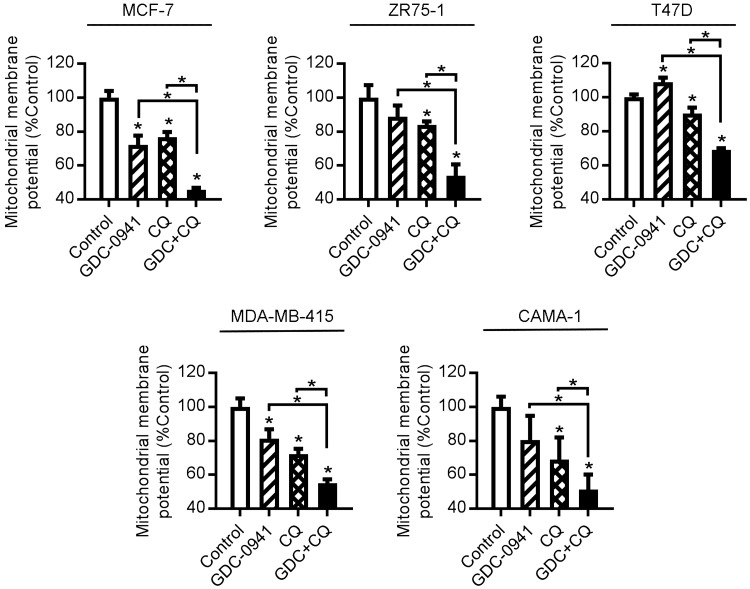
To characterize the form of apoptosis induced by combined inhibition of autophagy and PI3K, we measured changes in mitochondrial membrane potential. Twenty-four hours of GDC-0941 treatment induced significant mitochondrial membrane depolarization in 2 of 5 cell lines, and CQ induced depolarization in 5/5 cell lines. Combined CQ/PI3Ki treatment significantly decreased mitochondrial membrane potential compared to each single agent (Fig. 3). Similar effects of the drug combination were observed in 3 of 3 hormone-depleted cell lines, and in 2 of 2 FR cell lines (Supplemental Fig. S6).
Figure 3.
CQ treatment increases PI3K inhibitor–induced mitochondrial membrane depolarization. Cells were treated ± 1 μM GDC-0941 or 50 μM CQ. After 24 h of drug treatment, mitochondrial membrane potential was measured by JC-1 staining and flow cytometry. *P < 0.05 by Bonferroni multiple comparison–adjusted post hoc test compared with control unless otherwise indicated with brackets (between GDC and GDC+CQ, or between CQ and GDC+CQ).
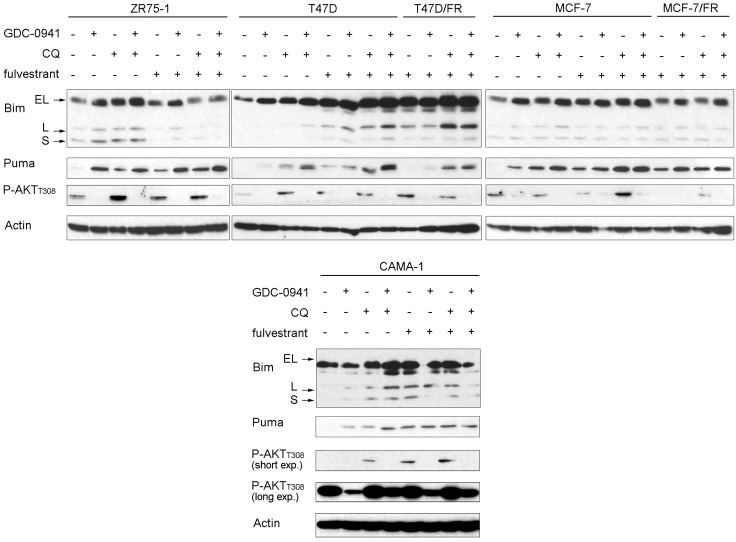
Bcl-2 family members are key regulators of mitochondrial apoptosis. The BH3-only proteins Bim and PUMA bind the proapoptotic proteins Bax and Bak, inducing Bax/Bak oligomerization in the mitochondrial outer membrane to form pores (45). In 4 of 4 ER+ breast cancer cell lines, GDC-0941 or CQ increased Bim and/or PUMA levels, which were often further elevated by the drug combination (Fig. 4). Fulvestrant had variable effects on Bim and Puma levels, increasing levels as a single agent, but not appreciably enhancing levels induced by GDC-0941 and/or CQ. The basal levels of Bim in T47D/FR and MCF-7/FR cells were higher than in respective parental T47D and MCF-7 cells, in line with the Bim-inducing activity of fulv in parental cells. MCF-7/FR and T47D/FR cells (in the presence of fulv) showed further induction of Bim/Puma in response to GDC-0941 and CQ, respectively (Fig. 4). The data suggest that inhibition of autophagy increases the levels of BH3-only proteins and potentiates PI3Ki-induced mitochondrial apoptosis.
Figure 4.
Bim and Puma are elevated by PI3K inhibition and CQ treatment. Cells were pretreated ± 1 μM fulv for 24 h, then treated without or with 1 μM GDC-0941, 50 μM CQ, or 1 μM fulv for 24 h. Lysates were analyzed by immunoblot using the indicated antibodies. EL, extra-long; L, long; S, short.
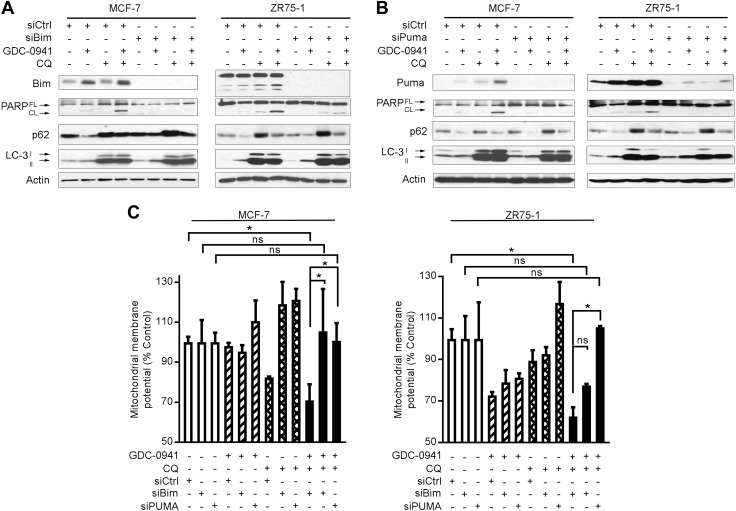
Bim and PUMA regulate apoptosis induced by inhibition of PI3K and autophagy
To functionally implicate Bim and PUMA in apoptosis induced by PI3Ki and CQ, MCF-7 and ZR75-1 cells were transfected with siRNA against Bim, PUMA, or control. Cells were then treated without or with GDC-0941 or CQ for 24 h. Knockdown of either Bim or PUMA had little effect on drug-induced autophagy but hindered apoptosis (Fig. 5A, B and Supplemental Fig. S7). In line with those results, the GDC-0941/CQ combination treatment significant decreased mitochondrial membrane potential in siControl-transfected cells, which was mitigated by knockdown of Bim or PUMA (Fig. 5C). Thus, Bim and PUMA have critical roles in the mitochondrial apoptosis induced by PI3Ki and CQ without apparent interference with drug-induced autophagy.
Figure 5.
Bim and Puma are required for apoptosis induced by inhibition of PI3K and autophagy. Cells were transfected with siRNA targeting Bim, PUMA, or nonsilencing control; 2 d after transfection, cells were treated ± 1 μM GDC-0941 or 50 μM CQ. A, B) Lysates were collected after 24 h of drug treatment and analyzed by immunoblot using the indicated antibodies. FL, full-length; CL, cleaved. C) Mitochondrial membrane potential was measured after 24 h of drug treatment by JC-1 staining and flow cytometry. *P < 0.05 by Bonferroni multiple comparison–adjusted post hoc test.
Inhibition of autophagy enhances the antitumor effects of PI3K inhibition
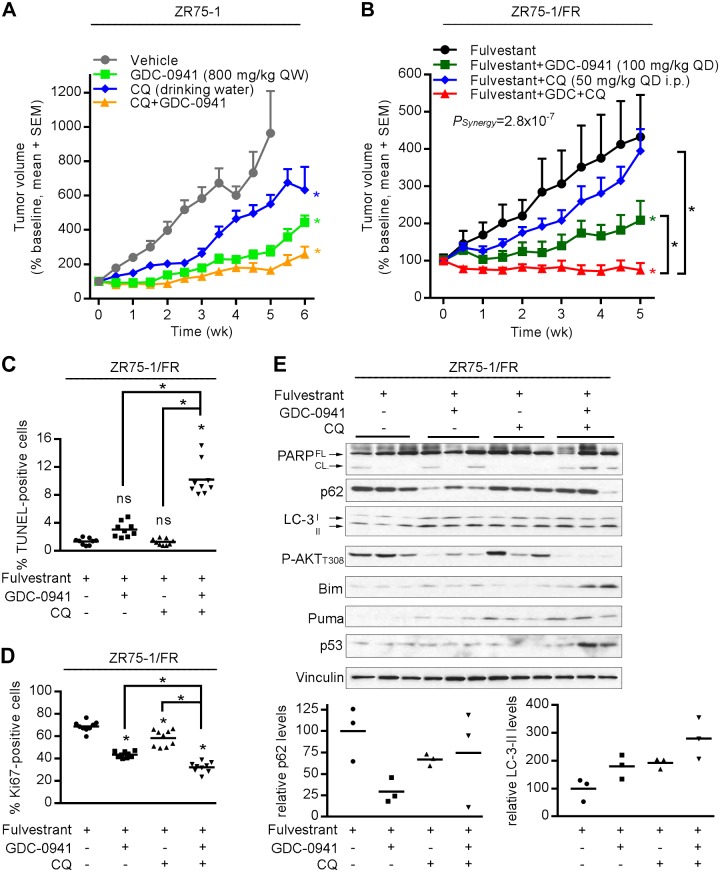
To determine whether combined inhibition of PI3K and autophagy was effective against established ER+ breast tumors, mice bearing ZR75-1 xenografts were treated with vehicle, GDC-0941, CQ (via drinking water), or the combination. Both single-agent and combination treatments significantly slowed tumor growth compared with the vehicle control (all P < 0.05; Fig. 6A and Supplemental Fig. S8).
Figure 6.
Inhibition of autophagy enhances the antitumor effects of PI3K inhibition. A) Mice bearing ZR75-1 xenografts were randomized to treatment with vehicle (n = 7), CQ (0.288 mg/ml in drinking water; n = 7), GDC-0941 [800 mg/kg per week (QW); n = 8], or the combination (n = 8). B) Mice bearing ZR75-1/FR xenografts were treated with a fulv (5 mg QW) backbone, and randomized to vehicle (n = 5), CQ (50 mg/kg i.p. QD; n = 5), GDC-0941 [100 mg/kg per day (QD); n = 5], or the combination (n = 7). Means ± sem are shown. *P < 0.05 compared with control unless otherwise indicated with brackets. C–E) ZR75-1/FR tumors were harvested at 4 h after the final dose of GDC-0941 or CQ on d 3. Tumor sections were analyzed by TUNEL to assess apoptosis (C) and by IHC for Ki67 to assess cell proliferation (D). *P < 0.05 by Bonferroni multiple comparison–adjusted post hoc test compared with control unless otherwise indicated with brackets. Ns, not significant. E) Tumor lysates were analyzed by immunoblot using the indicated antibodies. FL, full-length; CL, cleaved. Densitometry analysis was performed on p62, LC-3-II, and vinculin. p62 and LC-3-II signal intensities were normalized to vinculin control, and data are shown as the percentage of vehicle-treated controls.
Because PI3Ki, being developed for the treatment of ER+ breast cancer, would be first drug approved for use in the recurrent/metastatic setting in which many patients have antiestrogen-resistant disease, we also tested the effects of GDC-0941 and CQ in fulv-treated mice bearing antiestrogen-resistant ZR75-1/FR tumors. The addition of CQ to fulv slowed tumor growth compared with fulv/vehicle (Fig. 6B); that effect did not reach statistical significance, possibly because of the increased intertumor variability in volume compared with ZR75-1 tumors in Fig. 6A. Treatment with fulv/GDC-0941 appreciably delayed tumor growth compared with fulv/vehicle (P < 0.05). Triplet treatment with fulv/GDC-0941/CQ induced partial regression, followed by tumor stasis, and statistical analysis indicated that GDC-0941 synergized with CQ (P = 2.8 × 10−7; Fig. 6B and Supplemental Fig. S8).
ZR75-1/FR tumors were harvested 4 h after the last dose of drug(s) on d 3. TUNEL analysis revealed that triplet treatment with GDC-0941/CQ/fulv significantly increased tumor cell apoptosis compared with each other treatment group (all P < 0.05), whereas doublet treatments of GDC-0941/fulv and CQ/fulv did not increase apoptosis beyond vehicle/fulv control levels (Fig. 6C and Supplemental Fig. S9A). Ki67 IHC analysis indicated that triplet and doublet treatments significantly decreased tumor cell proliferation compared with vehicle/fulv control, and triplet treatment was significantly more effective than each doublet treatment (all P < 0.05; Fig. 6D and Supplemental Fig. S9B). Thus, the combination of GDC-0941/CQ/fulv completely suppressed ZR75-1/FR tumor growth through inhibition of tumor cell survival and proliferation, whereas GDC-0941/fulv or CQ/fulv only suppressed proliferation.
Immunoblot analysis of lysates from the same tumors as above showed that combined GDC-0941/CQ/fulv treatment induced PARP cleavage more effectively than other drug treatments (Fig. 6E), in line with results from the TUNEL assay (Fig. 6C). Compared with vehicle/fulv control, GDC-0941/fulv treatment decreased p62 levels and increased LC3-II, indicating induction of autophagy. CQ/fulv induced LC3-II up-regulation compared with vehicle/fulv, and blocked the GDC-0941–induced decrease in p62, confirming inhibition of PI3Ki-induced autophagy. Triplet drug treatment induced up-regulation of Bim and PUMA compared with other treatment groups (Fig. 6E), which may be due, in part, to induction of p53 (46). These results collectively suggest that inhibition of autophagy with CQ potentiates PI3K inhibitor–induced apoptosis and proliferative inhibition in antiestrogen-resistant ER+ breast tumors.
DISCUSSION
Herein, we demonstrated that PI3Ki induced autophagy in ER+ breast cancer cells and tumors. Inhibition of autophagy by genetic (RNAi) or pharmacologic (CQ) means suppressed growth and survival of ER+ breast cancer cells, enhancing the anticancer effects of PI3Ki. Similar effects of inhibition of autophagy and PI3K occurred in the presence and absence of estrogen, and in antiestrogen-sensitive and -resistant cells and tumors. Combined PI3Ki and CQ induced expression of the BH3-only proteins Bim and Puma to trigger mitochondrial membrane depolarization and apoptosis. In antiestrogen-resistant ER+ breast tumors, the triplet combination of fulv/GDC-0941/CQ provided durable tumor regression, whereas fulv/GDC-0941 and fulv/CQ only slowed tumor growth.
Bim and PUMA are required for apoptosis induced by inhibition of EGFR or HER2, which lie upstream of PI3K/AKT (47). Herein, we showed that PI3Ki induced up-regulation of Bim, PUMA, and apoptosis in ER+ breast cancer cells (Figs. 4 and 5A, B). Bim is destabilized by MAPK phosphorylation (48). We and others showed previously that PI3K drives MAPK activation via PI3K-dependent activation of Rac in ER+ breast cancer cells (49, 50), which may underlie PI3Ki-induced stabilization of Bim. PUMA can be induced by FOXO transcription factors, which are inhibited by AKT (51, 52). In ZR75-1/FR tumors, triplet PI3Ki/CQ/antiestrogen treatment up-regulated p53 (Fig. 6E), which has also been shown to induce PUMA expression (53). Inhibition of autophagy or AKT can induce p53 (54, 55), suggesting that PUMA may be transcriptionally induced via FOXO- and p53-dependent mechanisms in ER+ breast cancer. Our demonstration that Bim and PUMA are implicated in the apoptotic response of ER+ breast cancer cells and tumors to inhibition of PI3K and autophagy (Figs. 5 and 6B, C, E) aligns with the findings of Vaillant et al. (56), who showed that Bcl-2 inhibition (which can release BH3-only proteins to trigger apoptosis) enhances the response of ER+ breast tumors to combined PI3Ki/mTORi/antiestrogen therapy.
CQ and HCQ have been tested in phase I/II clinical trials for the treatment of cancer at doses ranging from 200 to 1200 mg/d, which is ∼3- to 18-fold less than the dose administered to mice herein. Although clinical studies reported CQ/HCQ concentrations ranging from 1 to 3 μg/ml (∼3–10 μM) in blood and plasma (29, 57–59), intratumoral CQ/HCQ concentrations were rarely evaluated. In dogs bearing spontaneously occurring lymphoma, significant accumulations of HCQ and its metabolite in tumor tissues was apparent after only 3 d of treatment, with an ∼100- to 150-fold increase in tumor levels compared with plasma levels (60). Because the period of CQ/HCQ treatment in humans typically lasts weeks to months, CQ/HCQ is predicted to accumulate within tumors. Because cultured cancer cells were only treated for a short time in our studies, we used 50 μM CQ in vitro to mimic potential drug accumulation that is predicted to occur in tumors. Future studies are needed to determine the concentrations of CQ/HCQ achievable in tumors in patients treated with safe and accepted doses of CQ/HCQ and whether such doses and concentrations are sufficient to block autophagy. Alternatively, other agents that inhibit autophagy are being developed [reviewed in reference (61)], which may ultimately prove less toxic and more selective than CQ/HCQ.
We observed that CQ (0.288 mg/ml in drinking water) modestly but significantly slowed the growth of antiestrogen-sensitive ZR75-1 ER+ breast tumors in NSG mice, but CQ (50 mg/kg per day i.p.) did not significantly slow the growth of antiestrogen-resistant ZR75-1/FR tumors in the context of a fulv (5 mg/wk) backbone therapy (Fig. 6A, B). Cook et al. (31) found that single-agent CQ (same dose via drinking water) induced partial regression of pan-antiestrogen–resistant (MCF-7-derived) LCC9 xenografts without affecting growth of tamoxifen-resistant MCF7-RR xenografts in athymic, nude mice. Combined CQ/tamoxifen treatment inhibited the growth of MCF7-RR tumors. Interestingly, fulv (0.5 mg/wk) partly hindered the effects of CQ on LCC9 tumors, which may have been caused by fulv-suppressed recruitment of macrophages to tumors (31). It is conceivable that cotreatment with fulv also hindered the effects of CQ on ZR75-1/FR tumors (Fig. 6B), but such effects are unlikely to have been macrophage-dependent because NSG mice are macrophage-deficient. Our collective results suggest that CQ and CQ/antiestrogen combinations have mixed effects on antiestrogen-sensitive and -resistant tumors. It is conceivable that PI3K/AKT/mTOR inhibition, which induces autophagy, is required to sensitize some ER+ breast tumors to CQ (Fig. 6B, E). Several clinical studies are ongoing to test the effects of CQ/HCQ in combination with an AKTi or an mTORC1i in patients with cancer, but none include cotreatment with an antiestrogen for ER+ breast cancer.
In summary, we provided herein the first demonstration, to our knowledge, that a clinically validated PI3K-selective inhibitor (GDC-0941) synergizes with CQ to induce regression of antiestrogen-resistant ER+ breast tumors (Fig. 6B). These results warrant consideration of clinical testing of an autophagy inhibitor (e.g., CQ/HCQ) with antiestrogen/PI3Ki combination therapy for the treatment of ER+ breast cancer.
Supplementary Material
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
ACKNOWLEDGMENTS
The authors thank the following Norris Cotton Cancer Center Shared Resources for assistance: Flow Cytometry, Microscopy, Pathology, and the Center for Comparative Medicine and Research. This work was supported by the American Cancer Society (Grant RSG-13-292-01-TBE to T.W.M.) and U.S. National Institutes of Health, National Cancer Institute (Grant P30CA023108 to Dartmouth College, Norris Cotton Cancer Center Support). The authors declare no conflicts of interest.
Glossary
- AI
aromatase inhibitor
- CQ
chloroquine
- ER
estrogen receptor α
- FR
fulvestrant-resistant
- fulv
fulvestrant
- HCQ
hydroxychloroquine
- IHC
immunohistochemistry
- mTOR
mechanistic target of rapamycin
- mTORC1
mTOR complex I
- NSG
NOD-scid/IL-2Rγ−/−
- PFS
progression-free survival
- PI3Ki
PI3K inhibitor
- siRNA
short interfering RNA
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
W. Yang and T. W. Miller designed the research; W. Yang, S. R. Hosford, N. A. Traphagen, and S. Liu performed experiments; W. Yang, K. Shee, E. Demidenko, and T. W. Miller analyzed data; W. Yang and T. W. Miller wrote the manuscript; and all authors reviewed and approved the manuscript.
REFERENCES
- 1.Davies C., Godwin J., Gray R., Clarke M., Cutter D., Darby S., McGale P., Pan H. C., Taylor C., Wang Y. C., Dowsett M., Ingle J., Peto R.; Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) . (2011) Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet 378, 771–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas N.; Cancer Genome Atlas Network . (2012) Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller T. W., Rexer B. N., Garrett J. T., Arteaga C. L. (2011) Mutations in the phosphatidylinositol 3-kinase pathway: role in tumor progression and therapeutic implications in breast cancer. Breast Cancer Res. 13, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang W., Hosford S. R., Dillon L. M., Shee K., Liu S. C., Bean J. R., Salphati L., Pang J., Zhang X., Nannini M. A., Demidenko E., Bates D., Lewis L. D., Marotti J. D., Eastman A. R., Miller T. W. (2016) Strategically timing inhibition of phosphatidylinositol 3-kinase to maximize therapeutic index in estrogen receptor α-positive, PIK3CA-mutant breast cancer. Clin. Cancer Res. 22, 2250–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller T. W., Hennessy B. T., González-Angulo A. M., Fox E. M., Mills G. B., Chen H., Higham C., García-Echeverría C., Shyr Y., Arteaga C. L. (2010) Hyperactivation of phosphatidylinositol-3 kinase promotes escape from hormone dependence in estrogen receptor-positive human breast cancer. J. Clin. Invest. 120, 2406–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tartarone A., Lerose R., Aieta M. (2012) Everolimus in HR-positive advanced breast cancer. N. Engl. J. Med. 366, 1739–1740, author reply 1739–1740 [DOI] [PubMed] [Google Scholar]
- 7.Miller T. W., Forbes J. T., Shah C., Wyatt S. K., Manning H. C., Olivares M. G., Sanchez V., Dugger T. C., de Matos Granja N., Narasanna A., Cook R. S., Kennedy J. P., Lindsley C. W., Arteaga C. L. (2009) Inhibition of mammalian target of rapamycin is required for optimal antitumor effect of HER2 inhibitors against HER2−overexpressing cancer cells. Clin. Cancer Res. 15, 7266–7276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Reilly K. E., Rojo F., She Q. B., Solit D., Mills G. B., Smith D., Lane H., Hofmann F., Hicklin D. J., Ludwig D. L., Baselga J., Rosen N. (2006) mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 66, 1500–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carracedo A., Ma L., Teruya-Feldstein J., Rojo F., Salmena L., Alimonti A., Egia A., Sasaki A. T., Thomas G., Kozma S. C., Papa A., Nardella C., Cantley L. C., Baselga J., Pandolfi P. P. (2008) Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J. Clin. Invest. 118, 3065–3074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hosford S. R., Dillon L. M., Bouley S. J., Rosati R., Yang W., Chen V. S., Demidenko E., Morra R. P., Jr., Miller T. W. (2017) Combined inhibition of both p110α and p110β isoforms of phosphatidylinositol 3-kinase is required for sustained therapeutic effect in PTEN-deficient, ER+ breast cancer. Clin. Cancer Res. 23, 2795–2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krop I. E., Mayer I. A., Ganju V., Dickler M., Johnston S., Morales S., Yardley D. A., Melichar B., Forero-Torres A., Lee S. C., de Boer R., Petrakova K., Vallentin S., Perez E. A., Piccart M., Ellis M., Winer E., Gendreau S., Derynck M., Lackner M., Levy G., Qiu J., He J., Schmid P. (2016) Pictilisib for oestrogen receptor-positive, aromatase inhibitor-resistant, advanced or metastatic breast cancer (FERGI): a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol. 17, 811–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baselga J., Im S. A., Iwata H., Clemons M., Ito Y., Awada A., Chia S., Jagiello-Gruszfeld A., Pistilli B., Tseng L. M., Hurvitz S., Masuda N., Cortés J., De Laurentiis M., Arteaga C. L., Jiang Z., Jonat W., Hachemi S., Le Mouhaër S., Di Tomaso E., Urban P., Massacesi C., Campone M. (2016) PIK3CA status in circulating tumor DNA (ctDNA) predicts efficacy of buparlisib (BUP) plus fulvestrant (FULV) in postmenopausal women with endocrine-resistant HR+/HER2− advanced breast cancer (BC): first results from the randomized, phase III BELLE-2 trial. Cancer Res 76 (4 Suppl), S6-01 (abstr.) [Google Scholar]
- 13.Mayer I. A., Abramson V. G., Formisano L., Balko J. M., Estrada M. V., Sanders M. E., Juric D., Solit D., Berger M. F., Won H. H., Li Y., Cantley L. C., Winer E., Arteaga C. L. (2017) A phase Ib study of alpelisib (BYL719), a PI3Kα-specific inhibitor, with letrozole in ER+/HER2− metastatic breast cancer. Clin. Cancer Res. 23, 26–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kreuzaler P., Watson C. J. (2012) Killing a cancer: what are the alternatives? Nat. Rev. Cancer 12, 411–424 [DOI] [PubMed] [Google Scholar]
- 15.Kim J., Kundu M., Viollet B., Guan K. L. (2011) AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13, 132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martina J. A., Chen Y., Gucek M., Puertollano R. (2012) MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 8, 903–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo J. Y., White E. (2016) Autophagy, metabolism, and cancer. Cold Spring Harb. Symp. Quant. Biol. 81, 73–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Green D. R., Levine B. (2014) To be or not to be? how selective autophagy and cell death govern cell fate. Cell 157, 65–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang C., Hu Q., Shen H. M. (2016) Pharmacological inhibitors of autophagy as novel cancer therapeutic agents. Pharmacol. Res. 105, 164–175 [DOI] [PubMed] [Google Scholar]
- 20.Lui A., New J., Ogony J., Thomas S., Lewis-Wambi J. (2016) Everolimus downregulates estrogen receptor and induces autophagy in aromatase inhibitor-resistant breast cancer cells. BMC Cancer 16, 487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schoenlein P. V., Periyasamy-Thandavan S., Samaddar J. S., Jackson W. H., Barrett J. T. (2009) Autophagy facilitates the progression of ERα-positive breast cancer cells to antiestrogen resistance. Autophagy 5, 400–403 [DOI] [PubMed] [Google Scholar]
- 22.Ji Y., Di W., Yang Q., Lu Z., Cai W., Wu J. (2015) Inhibition of autophagy increases proliferation inhibition and apoptosis induced by the PI3K/mTOR inhibitor NVP-BEZ235 in breast cancer cells. Clin. Lab. 61, 1043–1051 [DOI] [PubMed] [Google Scholar]
- 23.Wang Y., Liu J., Qiu Y., Jin M., Chen X., Fan G., Wang R., Kong D. (2016) ZSTK474, a specific class I phosphatidylinositol 3-kinase inhibitor, induces G1 arrest and autophagy in human breast cancer MCF-7 cells. Oncotarget 7, 19897–19909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Echeverry N., Ziltener G., Barbone D., Weder W., Stahel R. A., Broaddus V. C., Felley-Bosco E. (2015) Inhibition of autophagy sensitizes malignant pleural mesothelioma cells to dual PI3K/mTOR inhibitors. Cell Death Dis. 6, e1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimura S., Fujita N., Noda T., Yoshimori T. (2009) Monitoring autophagy in mammalian cultured cells through the dynamics of LC3. Methods Enzymol. 452, 1–12 [DOI] [PubMed] [Google Scholar]
- 26.Klionsky D. J., Abdelmohsen K., Abe A., Abedin M. J., Abeliovich H., Arozena A. A., Adachi H., Adams C. M., Adams P. D., Adeli K., Adhihetty P. J., Adler S. G., Agam G., Agarwal R., Aghi M. K., Agnello M., Agostinis P., Aguilar P. V., Aguirre-Ghiso J., Airoldi E. M., Ait-Si-Ali S., Akematsu T., Akporiaye E. T., Al-Rubeai M., Albaiceta G. M., Albanese C., Albani D., Albert M. L., Aldudo J., Algul H., Alirezaei M., Alloza I., Almasan A., Almonte-Beceril M., Alnemri E. S., Alonso C., Altan-Bonnet N., Altieri D. C., Alvarez S., Alvarez-Erviti L., Alves S., Amadoro G., Amano A., Amantini C., Ambrosio S., Amelio I., Amer A. O., Amessou M., Amon A., An Z. Y., Anania F. A., Andersen S. U., Andley U. P., Andreadi C. K., Andrieu-Abadie N., Anel A., Ann D. K., Anoopkumar-Dukie S., Antonioli M., Aoki H., Apostolova N., Aquila S., Aquilano K., Araki K., Arama E., Aranda A., Araya J., Arcaro A., Arias E., Arimoto H., Ariosa A. R., Armstrong J. L., Arnould T., Arsov I., Asanuma K., Askanas V., Asselin E., Atarashi R., Atherton S. S., Atkin J. D., Attardi L. D., Auberger P., Auburger G., Aurelian L., Autelli R., Avagliano L., Avantaggiati M. L., Avrahami L., Awale S., Azad N., Bachetti T., Backer J. M., Bae D. H., Bae J. S., Bae O. N., Bae S. H., Baehrecke E. H., Baek S. H., Baghdiguian S., Bagniewska-Zadworna A., Bai H., Bai J., Bai X. Y., Bailly Y., Balaji K. N., Balduini W., Ballabio A., Balzan R., Banerjee R., Banhegyi G., Bao H. J., Barbeau B., Barrachina M. D., Barreiro E., Bartel B., Bartolome A., Bassham D. C., Bassi M. T., Bast R. C., Basu A., Batista M. T., Batoko H., Battino M., Bauckman K., Baumgarner B. L., Bayer K. U., Beale R., Beaulieu J. F., Beck G. R., Becker C., Beckham J. D., Bedard P. A., Bednarski P. J., Begley T. J., Behl C., Behrends C., Behrens G. M. N., Behrns K. E., Bejarano E., Belaid A., Belleudi F., Benard G., Berchem G., Bergamaschi D., Bergami M., Berkhout B., Berliocchi L., Bernard A., Bernard M., Bernassola F., Bertolotti A., Bess A. S., Besteiro S., Bettuzzi S., Bhalla S., Bhattacharyya S., Bhutia S. K., Biagosch C., Bianchi M. W., Biard-Piechaczyk M., Billes V., Bincoletto C., Bingol B., Bird S. W., Bitoun M., Bjedov I., Blackstone C., Blanc L., Blanco G. A., Blomhoff H. K., Boada-Romero E., Bockler S., Boes M., Boesze-Battaglia K., Boise L. H., Bolino A., Boman A., Bonaldo P., Bordi M., Bosch J., Botana L. M., Botti J., Bou G., Bouche M., Bouchecareilh M., Boucher M. J., Boulton M. E., Bouret S. G., Boya P., Boyer-Guittaut M., Bozhkov P. V., Brady N., Braga V. M. M., Brancolini C., Braus G. H., Bravo-San Pedro J. M., Brennan L. A., Bresnick E. H., Brest P., Bridges D., Bringer M. A., Brini M., Brito G. C., Brodin B., Brookes P. S., Brown E. J., Brown K., Broxmeyer H. E., Bruhat A., Brum P. C., Brumell J. H., Brunetti-Pierri N., Bryson-Richardson R. J., Buch S., Buchan A. M., Budak H., Bulavin D. V., Bultman S. J., Bultman S. J., Bumbasirevic V., Burelle Y., Burke R. E., Burmeister M., Butikofer P., Caberlotto L., Cadwell K., Cahova M., Cai D. S., Cai J. J., Cai Q., Calatayud S., Camougrand N., Campanella M., Campbell G. R., Campbell M., Campello S., Candau R., Caniggia I., Cantoni L., Cao L. Z., Caplan A. B., Caraglia M., Cardinali C., Cardoso S. M., Carew J. S., Carleton L. A., Carlin C. R., Carloni S., Carlsson S. R., Carmona-Gutierrez D., Carneiro L. A. M., Carnevali O., Carra S., Carrier A., Carroll B., Casas C., Casas J., Cassinelli G., Castets P., Castro-Obregon S., Cavallini G., Ceccherini I., Cecconi F., Cederbaum A. I., Cena V., Cenci S., Cerella C., Cervia D., Cetrullo S., Chaachouay H., Chae H. J., Chagin A. S., Chai C. Y., Chakrabarti G., Chamilos G., Chan E. Y. W., Chan M. T. V., Chandra D., Chandra P., Chang C. P., Chang R. C. C., Chang T. Y., Chatham J. C., Chatterjee S., Chauhan S., Che Y. S., Cheetham M. E., Cheluvappa R., Chen C. J., Chen G., Chen G. C., Chen G. Q., Chen H. Z., Chen J. W., Chen J. K., Chen M., Chen M. Z., Chen P. W., Chen Q., Chen Q., Chen S. D., Chen S., Chen S. S. L., Chen W., Chen W. J., Chen W. Q., Chen W. L., Chen X. M., Chen Y. H., Chen Y. G., Chen Y., Chen Y. Y., Chen Y. S., Chen Y. J., Chen Y. Q., Chen Y. J., Chen Z., Chen Z., Cheng A., Cheng C. H. K., Cheng H., Cheong H. S., Cherry S., Chesney J., Cheung C. H. A., Chevet E., Chi H. C., Chi S. G., Chiacchiera F., Chiang H. L., Chiarelli R., Chiariello M., Chieppa M., Chin L. S., Chiong M., Chiu G. N. C., Cho D. H., Cho S. G., Cho W. C., Cho Y. Y., Cho Y. S., Choi A. M. K., Choi E. J., Choi E. K., Choi J. Y., Choi M. E., Choi S. I., Chou T. F., Chouaib S., Choubey D., Choubey V., Chow K. C., Chowdhury K., Chu C. T., Chuang T. H., Chun T., Chung H. W., Chung T. J., Chung Y. L., Chwae Y. J., Cianfanelli V., Ciarcia R., Ciechomska I. A., Ciriolo M. R., Cirone M., Claerhout S., Clague M. J., Claria J., Clarke P. G. H., Clarke R., Clementi E., Cleyrat C., Cnop M., Coccia E. M., Cocco T., Codogno P., Coers J., Cohen E. E. W., Colecchia D., Coletto L., Coll N. S., Colucci-Guyon E., Comincini S., Condello M., Cook K. L., Coombs G. H., Cooper C. D., Cooper J. M., Coppens I., Corasaniti M. T., Corazzari M., Corbalan R., Corcelle-Termeau E., Cordero M. D., Corral-Ramos C., Corti O., Cossarizza A., Costelli P., Costes S., Costes S., Coto-Montes A., Cottet S., Couve E., Covey L. R., Cowart L. A., Cox J. S., Coxon F. P., Coyne C. B., Cragg M. S., Craven R. J., Crepaldi T., Crespo J. L., Criollo A., Crippa V., Cruz M. T., Cuervo A. M., Cuezva J. M., Cui T. X., Cutillas P. R., Czaja M. J., Czyzyk-Krzeska M. F., Dagda R. K., Dahmen U., Dai C. S., Dai W. J., Dai Y., Dalby K. N., Valle L. D., Dalmasso G., D’Amelio M., Damme M., Darfeuille-Michaud A., Dargemont C., Darley-Usmar V. M., Dasarathy S., Dasgupta B., Dash S., Dass C. R., Davey H. M., Davids L. M., Davila D., Davis R. J., Dawson T. M., Dawson V. L., Daza P., de Belleroche J., de Figueiredo P., de Figueiredo R. C. B. Q., de la Fuente J., De Martino L., De Matteis A., De Meyer G. R. Y., De Milito A., De Santi M., de Souza W., De Tata V., De Zio D., Debnath J., Dechant R., Decuypere J. P., Deegan S., Dehay B., Del Bello B., Del Re D. P., Delage-Mourroux R., Delbridge L. M. D., Deldicque L., Delorme-Axford E., Deng Y. Z., Dengjel J., Denizot M., Dent P., Der C. J., Deretic V., Derrien B., Deutsch E., Devarenne T. P., Devenish R. J., Di Bartolomeo S., Di Daniele N., Di Domenico F., Di Nardo A., Di Paola S., Di Pietro A., Di Renzo L., DiAntonio A., Diaz-Araya G., Diaz-Laviada I., Diaz-Meco M. T., Diaz-Nido J., Dickey C. A., Dickson R. C., Diederich M., Digard P., Dikic I., Dinesh-Kumar S. P., Ding C., Ding W. X., Ding Z. F., Dini L., Distler J. H. W., Diwan A., Djavaheri-Mergny M., Dmytruk K., Dobson R. C. J., Doetsch V., Dokladny K., Dokudovskaya S., Donadelli M., Dong X. C., Dong X. N., Dong Z., Donohue T. M., Doran K. S., D’Orazi G., Dorn G. W., Dosenko V., Dridi S., Drucker L., Du J., Du L. L., Du L. H., du Toit A., Dua P., Duan L., Duann P., Dubey V. K., Duchen M. R., Duchosal M. A., Duez H., Dugail I., Dumit V. I., Duncan M. C., Dunlop E. A., Dunn W. A., Dupont N., Dupuis L., Duran R. V., Durcan T. M., Duvezin-Caubet S., Duvvuri U., Eapen V., Ebrahimi-Fakhari D., Echard A., Eckhart L., Edelstein C. L., Edinger A. L., Eichinger L., Eisenberg T., Eisenberg-Lerner A., Eissa N. T., El-Deiry W. S., El-Khoury V., Elazar Z., Eldar-Finkelman H., Elliott C. J. H., Emanuele E., Emmenegger U., Engedal N., Engelbrecht A. M., Engelender S., Enserink J. M., Erdmann R., Erenpreisa J., Eri R., Eriksen J. L., Erman A., Escalante R., Eskelinen E. L., Espert L., Esteban-Martinez L., Evans T. J., Fabri M., Fabrias G., Fabrizi C., Facchiano A., Faergeman N. J., Faggioni A., Fairlie W. D., Fan C. H., Fan D. P., Fan J., Fang S. Y., Fanto M., Fanzani A., Farkas T., Faure M., Favier F. B., Fearnhead H., Federici M., Fei E., Felizardo T. C., Feng H., Feng Y. B., Feng Y. C., Ferguson T. A., Fernandez A. F., Fernandez-Barrena M. G., Fernandez-Checa J. C., Fernandez-Lopez A., Fernandez-Zapico M. E., Feron O., Ferraro E., Ferreira-Halder C. V., Fesus L., Feuer R., Fiesel F. C., Filippi-Chiela E. C., Filomeni G., Fimia G. M., Fingert J. H., Finkbeiner S., Finkel T., Fiorito F., Fisher P. B., Flajolet M., Flamigni F., Florey O., Florio S., Floto R. A., Folini M., Follo C., Fon E. A., Fornai F., Fortunato F., Fraldi A., Franco R., Francois A., Francois A., Frankel L. B., Fraser I. D. C., Frey N., Freyssenet D. G., Frezza C., Friedman S. L., Frigo D. E., Fu D. X., Fuentes J. M., Fueyo J., Fujitani Y., Fujiwara Y., Fujiya M., Fukuda M., Fulda S., Fusco C., Gabryel B., Gaestel M., Gailly P., Gajewska M., Galadari S., Galili G., Galindo I., Galindo M. F., Galliciotti G., Galluzzi L., Galluzzi L., Galy V., Gammoh N., Gandy S., Ganesan A. K., Ganesan S., Ganley I. G., Gannage M., Gao F. B., Gao F., Gao J. X., Nannig L. G., Vescovi E. G., Garcia-Macia M., Garcia-Ruiz C., Garg A. D., Garg P. K., Gargini R., Gassen N. C., Gatica D., Gatti E., Gavard J., Gavathiotis E., Ge L., Ge P. F., Ge S. F., Gean P. W., Gelmetti V., Genazzani A. A., Geng J. F., Genschik P., Gerner L., Gestwicki J. E., Gewirtz D. A., Ghavami S., Ghigo E., Ghosh D., Giammarioli A. M., Giampieri F., Giampietri C., Giatromanolaki A., Gibbings D. J., Gibellini L., Gibson S. B., Ginet V., Giordano A., Giorgini F., Giovannetti E., Girardin S. E., Gispert S., Giuliano S., Gladson C. L., Glavic A., Gleave M., Godefroy N., Gogal R. M., Gokulan K., Goldman G. H., Goletti D., Goligorsky M. S., Gomes A. V., Gomes L. C., Gomez H., Gomez-Manzano C., Gomez-Sanchez R., Goncalves D. A. P., Goncu E., Gong Q. Q., Gongora C., Gonzalez C. B., Gonzalez-Alegre P., Gonzalez-Cabo P., Gonzalez-Polo R. A., Goping I. S., Gorbea C., Gorbunov N. V., Goring D. R., Gorman A. M., Gorski S. M., Goruppi S., Goto-Yamada S., Gotor C., Gottlieb R. A., Gozes I., Gozuacik D., Graba Y., Graef M., Granato G. E., Grant G. D., Grant S., Gravina G. L., Green D. R., Greenhough A., Greenwood M. T., Grimaldi B., Gros F., Grose C., Groulx J. F., Gruber F., Grumati P., Grune T., Guan J. L., Guan K. L., Guerra B., Guillen C., Gulshan K., Gunst J., Guo C. Y., Guo L., Guo M., Guo W. J., Guo X. G., Gust A. A., Gustafsson A. B., Gutierrez E., Gutierrez M. G., Gwak H. S., Haas A., Haber J. E., Hadano S., Hagedorn M., Hahn D. R., Halayko A. J., Hamacher-Brady A., Hamada K., Hamai A., Hamann A., Hamasaki M., Hamer I., Hamid Q., Hamid Q., Han F., Han W. D., Handa J. T., Hanover J. A., Hansen M., Harada M., Harhaji-Trajkovic L., Harper J. W., Harrath A. H., Harris J., Hasler U., Hasselblatt P., Hasui K., Hawley R. G., Hawley T. S., He C. C., He C. Y., He F. T., He G., He R. R., He X. H., He Y. W., He Y. Y., Heath J. K., Hebert M. J., Heinzen R. A., Helgason G. V., Hensel M., Henske E. P., Her C. T., Herman P. K., Hernandez A., Hernandez C., Hernandez-Tiedra S., Hetz C., Hiesinger P. R., Higaki K., Hilfiker S., Hill B. G., Hill J. A., Hill W. D., Hino K., Hofius D., Hofman P., Hoglinger G. U., Hohfeld J., Holz M. K., Hong Y. G., Hood D. A., Hoozemans J. J. M., Hoppe T., Hsu C., Hsu C. Y., Hsu L. C., Hu D., Hu G. C., Hu H. M., Hu H. B., Hu M. C., Hu Y. C., Hu Z. W., Hua F., Hua Y., Huang C. H., Huang H. L., Huang K. H., Huang K. Y., Huang S. L., Huang S. Q., Huang W. P., Huang Y. R., Huang Y., Huang Y. F., Huber T. B., Huebbe P., Huh W. K., Hulmi J. J., Hur G. M., Hurley J. H., Husak Z., Hussain S. N. A., Hussain S., Hwang J. J., Hwang S. M., Hwang T. I. S., Ichihara A., Imai Y., Imbriano C., Inomata M., Into T., Iovane V., Iovanna J. L., Iozzo R. V., Ip N. Y., Irazoqui J. E., Iribarren P., Isaka Y., Isakovic A. J., Ischiropoulos H., Isenberg J. S., Ishaq M., Ishida H., Ishii I., Ishmael J. E., Isidoro C., Isobe K. I., Isono E., Issazadeh-Navikas S., Itahana K., Itakura E., Ivanov A. I., Iyer A. K. V., Izquierdo J. M., Izumi Y., Izzo V., Jaattela M., Jaber N., Jackson D. J., Jackson W. T., Jacob T. G., Jacques T. S., Jagannath C., Jain A., Jana N. R., Jang B. K., Jani A., Janji B., Jannig P. R., Jansson P. J., Jean S., Jendrach M., Jeon J. H., Jessen N., Jeung E. B., Jia K. L., Jia L. J., Jiang H., Jiang H. C., Jiang L. W., Jiang T., Jiang X. Y., Jiang X. J., Jiang X. J., Jiang Y., Jiang Y. J., Jimenez A., Jin C., Jin H. C., Jin L., Jin M. Y., Jin S. K., Jinwal U. K., Jo E. K., Johansen T., Johnson D. E., Johnson G. V. W., Johnson J. D., Jonasch E., Jones C., Joosten L. A. B., Jordan J., Joseph A. M., Joseph B., Joubert A. M., Ju D. W., Ju J. F., Juan H. F., Juenemann K., Juhasz G., Jung H. S., Jung J. U., Jung Y. K., Jungbluth H., Justice M. J., Jutten B., Kaakoush N. O., Kaarniranta K., Kaasik A., Kabuta T., Kaeffer B., Kagedal K., Kahana A., Kajimura S., Kakhlon O., Kalia M., Kalvakolanu D. V., Kamada Y., Kambas K., Kaminskyy V. O., Kampinga H. H., Kandouz M., Kang C., Kang R., Kang T. C., Kanki T., Kanneganti T. D., Kanno H., Kanthasamy A. G., Kantorow M., Kaparakis-Liaskos M., Kapuy O., Karantza V., Karim M. R., Karmakar P., Kaser A., Kaushik S., Kawula T., Kaynar A. M., Ke P. Y., Ke Z. J., Kehrl J. H., Keller K. E., Kemper J. K., Kenworthy A. K., Kepp O., Kern A., Kesari S., Kessel D., Ketteler R., Kettelhut I. D., Khambu B., Khan M. M., Khandelwal V. K. M., Khare S., Kiang J. G., Kiger A. A., Kihara A., Kim A. L., Kim C. H., Kim D. R., Kim D. H., Kim E. K., Kim H. Y., Kim H. R., Kim J. S., Kim J. H., Kim J. C., Kim J. H., Kim K. W., Kim M. D., Kim M. M., Kim P. K., Kim S. W., Kim S. Y., Kim Y. S., Kim Y., Kimchi A., Kimmelman A. C., Kimura T., King J. S., Kirkegaard K., Kirkin V., Kirshenbaum L. A., Kishi S., Kitajima Y., Kitamoto K., Kitaoka Y., Kitazato K., Kley R. A., Klimecki W. T., Klinkenberg M., Klucken J., Knaevelsrud H., Knecht E., Knuppertz L., Ko J. L., Kobayashi S., Koch J. C., Koechlin-Ramonatxo C., Koenig U., Koh Y. H., Kohler K., Kohlwein S. D., Koike M., Komatsu M., Kominami E., Kong D. X., Kong H. J., Konstantakou E. G., Kopp B. T., Korcsmaros T., Korhonen L., Korolchuk V. I., Koshkina N. V., Kou Y. J., Koukourakis M. I., Koumenis C., Kovacs A. L., Kovacs T., Kovacs W. J., Koya D., Kraft C., Krainc D., Kramer H., Kravic-Stevovic T., Krek W., Kretz-Remy C., Krick R., Krishnamurthy M., Kriston-Vizi J., Kroemer G., Kruer M. C., Kruger R., Ktistakis N. T., Kuchitsu K., Kuhn C., Kumar A. P., Kumar A., Kumar A., Kumar D., Kumar D., Kumar R., Kumar S., Kundu M., Kung H. J., Kuno A., Kuo S. H., Kuret J., Kurz T., Kwok T., Kwon T. K., Kwon Y. T., Kyrmizi I., La Spada A. R., Lafont F., Lahm T., Lakkaraju A., Lam T., Lamark T., Lancel S., Landowski T. H., Lane D. J. R., Lane J. D., Lanzi C., Lapaquette P., Lapierre L. R., Laporte J., Laukkarinen J., Laurie G. W., Lavandero S., Lavie L., LaVoie M. J., Law B. Y. K., Law H. K. W., Law K. B., Layfield R., Lazo P. A., Le Cam L., Le Roch K. G., Le Stunff H., Leardkamolkarn V., Lecuit M., Lee B. H., Lee C. H., Lee E. F., Lee G. M., Lee H. J., Lee H., Lee J. K., Lee J., Lee J. H., Lee J. H., Lee M., Lee M. S., Lee P. J., Lee S. W., Lee S. J., Lee S. J., Lee S. Y., Lee S. H., Lee S. S., Lee S. J., Lee S., Lee Y. R., Lee Y. J., Lee Y. H., Leeuwenburgh C., Lefort S., Legouis R., Lei J. Z., Lei Q. Y., Leib D. A., Leibowitz G., Lekli I., Lemaire S. D., Lemasters J. J., Lemberg M. K., Lemoine A., Leng S. L., Lenz G., Lenzi P., Lerman L. O., Barbato D. L., Leu J. I. J., Leung H. Y., Levine B., Lewis P. A., Lezoualc’h F., Li C., Li F. Q., Li F. J., Li J., Li K., Li L., Li M., Li M., Li Q., Li R., Li S., Li W., Li W., Li X. T., Li Y. M., Lian J. Q., Liang C. Y., Liang Q. R., Liao Y. L., Liberal J., Liberski P. P., Lie P., Lieberman A. P., Lim H. J., Lim K. L., Lim K., Lima R. T., Lin C. S., Lin C. F., Lin F., Lin F. M., Lin F. C., Lin K., Lin K. H., Lin P. H., Lin T. W., Lin W. W., Lin Y. S., Lin Y., Linden R., Lindholm D., Lindqvist L. M., Lingor P., Linkermann A., Liotta L. A., Lipinski M. M., Lira V. A., Lisanti M. P., Liton P. B., Liu B., Liu C., Liu C. F., Liu F., Liu H. J., Liu J. X., Liu J. J., Liu J. L., Liu K., Liu L. Y., Liu L., Liu Q. T., Liu R. Y., Liu S. M., Liu S. W., Liu W., Liu X. D., Liu X. G., Liu X. H., Liu X. F., Liu X., Liu X. Q., Liu Y., Liu Y. L., Liu Z. X., Liu Z., Liuzzi J. P., Lizard G., Ljujic M., Lodhi I. J., Logue S. E., Lokeshwar B. L., Long Y. C., Lonial S., Loos B., Lopez-Otin C., Lopez-Vicario C., Lorente M., Lorenzi P. L., Lorincz P., Los M., Lotze M. T., Lovat P. E., Lu B. F., Lu B., Lu J., Lu Q., Lu S. M., Lu S. Y., Lu Y. Y., Luciano F., Luckhart S., Lucocq J. M., Ludovico P., Lugea A., Lukacs N. W., Lum J. J., Lund A. H., Luo H. L., Luo J., Luo S. Q., Luparello C., Lyons T., Ma J. J., Ma Y., Ma Y., Ma Z. Y., Machado J., Machado-Santelli G. M., Macian F., MacIntosh G. C., MacKeigan J. P., Macleod K. F., MacMicking J. D., MacMillan-Crow L. A., Madeo F., Madesh M., Madrigal-Matute J., Maeda A., Maeda T., Maegawa G., Maellaro E., Maes H., Magarinos M., Maiese K., Maiti T. K., Maiuri L., Maiuri M. C., Maki C. G., Malli R., Malorni W., Maloyan A., Mami-Chouaib F., Man N., Mancias J. D., Mandelkow E. M., Mandell M. A., Manfredi A. A., Manie S. N., Manzoni C., Mao K., Mao Z. X., Mao Z. W., Marambaud P., Marconi A. M., Marelja Z., Marfe G., Margeta M., Margittai E., Mari M., Mariani F. V., Marin C., Marinelli S., Marino G., Markovic I., Marquez R., Martelli A. M., Martens S., Martin K. R., Martin S. J., Martin S., Martin-Acebes M. A., Martin-Sanz P., Martinand-Mari C., Martinet W., Martinez J., Martinez-Lopez N., Martinez-Outschoorn U., Martinez-Velazquez M., Martinez-Vicente M., Martins W. K., Mashima H., Mastrianni J. A., Matarese G., Matarrese P., Mateo R., Matoba S., Matsumoto N., Matsushita T., Matsuura A., Matsuzawa T., Mattson M. P., Matus S., Maugeri N., Mauvezin C., Mayer A., Maysinger D., Mazzolini G. D., McBrayer M. K., McCall K., McCormick C., McInerney G. M., McIver S. C., McKenna S., McMahon J. J., McNeish I. A., Mechta-Grigoriou F., Medema J. P., Medina D. L., Megyeri K., Mehrpour M., Mehta J. L., Mei Y. D., Meier U. C., Meijer A. J., Melendez A., Melino G., Melino S., de Melo E. J. T., Mena M. A., Meneghini M. D., Menendez J. A., Menezes R., Meng L. S., Meng L. H., Meng S. S., Menghini R., Menko A. S., Menna-Barreto R. F. S., Menon M. B., Meraz-Rios M. A., Merla G., Merlini L., Merlot A. M., Meryk A., Meschini S., Meyer J. N., Mi M. T., Miao C. Y., Micale L., Michaeli S., Michiels C., Migliaccio A. R., Mihailidou A. S., Mijaljica D., Mikoshiba K., Milan E., Miller-Fleming L., Mills G. B., Mills I. G., Minakaki G., Minassian B. A., Ming X. F., Minibayeva F., Minina E. A., Mintern J. D., Minucci S., Miranda-Vizuete A., Mitchell C. H., Miyamoto S., Miyazawa K., Mizushima N., Mnich K., Mograbi B., Mohseni S., Moita L. F., Molinari M., Molinari M., Moller A. B., Mollereau B., Mollinedo F., Monick M. M., Monick M. M., Montagnaro S., Montell C., Moore D. J., Moore M. N., Mora-Rodriguez R., Moreira P. I., Morel E., Morelli M. B., Moreno S., Morgan M. J., Moris A., Moriyasu Y., Morrison J. L., Morrison L. A., Morselli E., Moscat J., Moseley P. L., Mostowy S., Motori E., Mottet D., Mottram J. C., Moussa C. E. H., Mpakou V. E., Mukhtar H., Levy J. M. M., Muller S., Munoz-Moreno R., Munoz-Pinedo C., Munz C., Murphy M. E., Murray J. T., Murthy A., Mysorekar I. U., Nabi I. R., Nabissi M., Nader G. A., Nagahara Y., Nagai Y., Nagata K., Nagelkerke A., Nagy P., Naidu S. R., Nair S., Nakano H., Nakatogawa H., Nanjundan M., Napolitano G., Naqvi N. I., Nardacci R., Narendra D. P., Narita M., Nascimbeni A. C., Natarajan R., Navegantes L. C., Nawrocki S. T., Nazarko T. Y., Nazarko V. Y., Neill T., Neri L. M., Netea M. G., Netea-Maier R. T., Neves B. M., Ney P. A., Nezis I. P., Nguyen H. T. T., Nguyen H. P., Nicot A. S., Nilsen H., Nilsson P., Nishimura M., Nishino I., Niso-Santano M., Niu H., Nixon R. A., Njar V. C. O., Noda T., Noegel A. A., Nolte E. M., Norberg E., Norga K. K., Noureini S. K., Notomi S., Notterpek L., Nowikovsky K., Nukina N., Nurnberger T., O’Donnell V. B., O’Donovan T., O’Dwyer P. J., Oehme I., Oeste C. L., Ogawa M., Ogretmen B., Ogura Y., Oh Y. J., Ohmuraya M., Ohshima T., Ojha R., Okamoto K., Okazaki T., Oliver F. J., Ollinger K., Olsson S., Orban D. P., Ordonez P., Orhon I., Orosz L., O’Rourke E. J., Orozco H., Ortega A. L., Ortona E., Osellame L. D., Oshima J., Oshima S., Osiewacz H. D., Otomo T., Otsu K., Ou J. H. J., Outeiro T. F., Ouyang D. Y., Ouyang H. J., Overholtzer M., Ozbun M. A., Ozdinler P. H., Ozpolat B., Pacelli C., Paganetti P., Page G., Pages G., Pagnini U., Pajak B., Pak S. C., Pakos-Zebrucka K., Pakpour N., Palkova Z., Palladino F., Pallauf K., Pallet N., Palmieri M., Paludan S. R., Palumbo C., Palumbo S., Pampliega O., Pan H. M., Pan W., Panaretakis T., Pandey A., Pantazopoulou A., Papackova Z., Papademetrio D. L., Papassideri I., Papini A., Parajuli N., Pardo J., Parekh V. V., Parenti G., Park J. I., Park J., Park O. K., Parker R., Parlato R., Parys J. B., Parzych K. R., Pasquet J. M., Pasquier B., Pasumarthi K. B. S., Patschan D., Patterson C., Pattingre S., Pattison S., Pause A., Pavenstadt H., Pavone F., Pedrozo Z., Pena F. J., Penalva M. A., Pende M., Peng J. X., Penna F., Penninger J. M., Pensalfini A., Pepe S., Pereira G. J. S., Pereira P. C., la Cruz V. P., Perez-Perez M. E., Perez-Rodriguez D., Perez-Sala D., Perier C., Perl A., Perlmutter D. H., Perrotta I., Pervaiz S., Pesonen M., Pessin J. E., Peters G. J., Petersen M., Petrache I., Petrof B. J., Petrovski G., Phang J. M., Piacentini M., Pierdominici M., Pierre P., Pierrefite-Carle V., Pietrocola F., Pimentel-Muinos F. X., Pinar M., Pineda B., Pinkas-Kramarski R., Pinti M., Pinton P., Piperdi B., Piret J. M., Platanias L. C., Platta H. W., Plowey E. D., Poggeler S., Poirot M., Polcic P., Poletti A., Poon A. H., Popelka H., Popova B., Poprawa I., Poulose S. M., Poulton J., Powers S. K., Powers T., Pozuelo-Rubio M., Prak K., Prange R., Prescott M., Priault M., Prince S., Proia R. L., Proikas-Cezanne T., Prokisch H., Promponas V. J., Przyklenk K., Puertollano R., Pugazhenthi S., Puglielli L., Pujol A., Puyal J., Pyeon D., Qi X., Qian W. B., Qin Z. H., Qiu Y., Qu Z. W., Quadrilatero J., Quinn F., Raben N., Rabinowich H., Radogna F., Ragusa M. J., Rahmani M., Raina K., Ramanadham S., Ramesh R., Rami A., Randall-Demllo S., Randow F., Rao H., Rao V. A., Rasmussen B. B., Rasse T. M., Ratovitski E. A., Rautou P. E., Ray S. K., Razani B., Reed B. H., Reggiori F., Rehm M., Reichert A. S., Rein T., Reiner D. J., Reits E., Ren J., Ren X. C., Renna M., Reusch J. E. B., Revuelta J. L., Reyes L., Rezaie A. R., Richards R. I., Richardson D. R., Richetta C., Riehle M. A., Rihn B. H., Rikihisa Y., Riley B. E., Rimbach G., Rippo M. R., Ritis K., Rizzi F., Rizzo E., Roach P. J., Robbins J., Roberge M., Roca G., Roccheri M. C., Rocha S., Rodrigues C. M. P., Rodriguez C. I., de Cordoba S. R., Rodriguez-Muela N., Roelofs J., Rogov V. V., Rohn T. T., Rohrer B., Romanelli D., Romani L., Romano P. S., Roncero M. I. G., Rosa J. L., Rosello A., Rosen K. V., Rosenstiel P., Rost-Roszkowska M., Roth K. A., Roue G., Rouis M., Rouschop K. M., Ruan D. T., Ruano D., Rubinsztein D. C., Rucker E. B., Rudich A., Rudolf E., Rudolf R., Ruegg M. A., Ruiz-Roldan C., Ruparelia A. A., Rusmini P., Russ D. W., Russo G. L., Russo G., Russo R., Rusten T. E., Ryabovol V., Ryan K. M., Ryter S. W., Sabatini D. M., Sacher M., Sachse C., Sack M. N., Sadoshima J., Saftig P., Sagi-Eisenberg R., Sahni S., Saikumar P., Saito T., Saitoh T., Sakakura K., Sakoh-Nakatogawa M., Sakuraba Y., Salazar-Roa M., Salomoni P., Saluja A. K., Salvaterra P. M., Salvioli R., Samali A., Sanchez A. M. J., Sanchez-Alcazar J. A., Sanchez-Prieto R., Sandri M., Sanjuan M. A., Santaguida S., Santambrogio L., Santoni G., dos Santos C. N., Saran S., Sardiello M., Sargent G., Sarkar P., Sarkar S., Sarrias M. R., Sarwal M. M., Sasakawa C., Sasaki M., Sass M., Sato K., Sato M., Satriano J., Savaraj N., Saveljeva S., Schaefer L., Schaible U. E., Scharl M., Schatzl H. M., Schekman R., Scheper W., Schiavi A., Schipper H. M., Schmeisser H., Schmidt J., Schmitz I., Schneider B. E., Schneider E. M., Schneider J. L., Schon E. A., Schonenberger M. J., Schonthal A. H., Schorderet D. F., Schroder B., Schuck S., Schulze R. J., Schwarten M., Schwarz T. L., Sciarretta S., Scotto K., Scovassi A. I., Screaton R. A., Screen M., Seca H., Sedej S., Segatori L., Segev N., Seglen P. O., Segui-Simarro J. M., Segura-Aguilar J., Seiliez I., Seki E., Sell C., Semenkovich C. F., Semenza G. L., Sen U., Serra A. L., Serrano-Puebla A., Sesaki H., Setoguchi T., Settembre C., Shacka J. J., Shajahan-Haq A. N., Shapiro I. M., Sharma S., She H., Shen C. K. J., Shen C. C., Shen H. M., Shen S. B., Shen W. L., Sheng R., Sheng X. Y., Sheng Z. H., Shepherd T. G., Shi J. Y., Shi Q., Shi Q. H., Shi Y. G., Shibutani S., Shibuya K., Shidoji Y., Shieh J. J., Shih C. M., Shimada Y., Shimizu S., Shin D. W., Shinohara M. L., Shintani M., Shintani T., Shioi T., Shirabe K., Shiri-Sverdlov R., Shirihai O., Shore G. C., Shu C. W., Shukla D., Sibirny A. A., Sica V., Sigurdson C. J., Sigurdsson E. M., Sijwali P. S., Sikorska B., Silveira W. A., Silvente-Poirot S., Silverman G. A., Simak J., Simmet T., Simon A. K., Simon H. U., Simone C., Simons M., Simonsen A., Singh R., Singh S. V., Singh S. K., Sinha D., Sinha S., Sinicrope F. A., Sirko A., Sirohi K., Sishi B. J. N., Sittler A., Siu P. M., Sivridis E., Skwarska A., Slack R., Slaninova I., Slavov N., Smaili S. S., Smalley K. S. M., Smith D. R., Soenen S. J., Soleimanpour S. A., Solhaug A., Somasundaram K., Son J. H., Sonawane A., Song C. J., Song F. Y., Song H. K., Song J. X., Song W., Soo K. Y., Sood A. K., Soong T. W., Soontornniyomkij V., Sorice M., Sotgia F., Soto-Pantoja D. R., Sotthibundhu A., Sousa M. J., Spaink H. P., Span P. N., Spang A., Sparks J. D., Speck P. G., Spector S. A., Spies C. D., Springer W., St Clair D., Stacchiotti A., Staels B., Stang M. T., Starczynowski D. T., Starokadomskyy P., Steegborn C., Steele J. W., Stefanis L., Steffan J., Stellrecht C. M., Stenmark H., Stepkowski T. M., Stern S. T., Stevens C., Stockwell B. R., Stoka V., Storchova Z., Stork B., Stratoulias V., Stravopodis D. J., Strnad P., Strohecker A. M., Strom A. L., Stromhaug P., Stulik J., Su Y. X., Su Z. L., Subauste C. S., Subramaniam S., Sue C. M., Suh S. W., Sui X. B., Sukseree S., Sulzer D., Sun F. L., Sun J. R., Sun J., Sun S. Y., Sun Y., Sun Y., Sun Y. J., Sundaramoorthy V., Sung J., Suzuki H., Suzuki K., Suzuki N., Suzuki T., Suzuki Y. J., Swanson M. S., Swanton C., Sward K., Swarup G., Sweeney S. T., Sylvester P. W., Szatmari Z., Szegezdi E., Szlosarek P. W., Taegtmeyer H., Tafani M., Taillebourg E., Tait S. W. G., Takacs-Vellai K., Takahashi Y., Takats S., Takemura G., Takigawa N., Talbot N. J., Tamagno E., Tamburini J., Tan C. P., Tan L., Tan M. L., Tan M., Tan Y. J., Tanaka K., Tanaka M., Tang D. L., Tang D. Z., Tang G. M., Tanida I., Tanji K., Tannous B. A., Tapia J. A., Tasset-Cuevas I., Tatar M., Tavassoly I., Tavernarakis N., Taylor A., Taylor G. S., Taylor G. A., Taylor J. P., Taylor M. J., Tchetina E. V., Tee A. R., Teixeira-Clerc F., Telang S., Tencomnao T., Teng B. B., Teng R. J., Terro F., Tettamanti G., Theiss A. L., Theron A. E., Thomas K. J., Thome M. P., Thomes P. G., Thorburn A., Thorner J., Thum T., Thumm M., Thurston T. L. M., Tian L., Till A., Ting J. P. Y., Titorenko V. I., Toker L., Toldo S., Tooze S. A., Topisirovic I., Torgersen M. L., Torosantucci L., Torriglia A., Torrisi M. R., Tournier C., Towns R., Trajkovic V., Travassos L. H., Triola G., Tripathi D. N., Trisciuoglio D., Troncoso R., Trougakos I. P., Truttmann A. C., Tsai K. J., Tschan M. P., Tseng Y. H., Tsukuba T., Tsung A., Tsvetkov A. S., Tu S. P., Tuan H. Y., Tucci M., Tumbarello D. A., Turk B., Turk V., Turner R. F. B., Tveita A. A., Tyagi S. C., Ubukata M., Uchiyama Y., Udelnow A., Ueno T., Umekawa M., Umemiya-Shirafuji R., Underwood B. R., Ungermann C., Ureshino R. P., Ushioda R., Uversky V. N., Uzcategui N. L., Vaccari T., Vaccaro M. I., Vachova L., Vakifahmetoglu-Norberg H., Valdor R., Valente E. M., Vallette F., Valverde A. M., Van den Berghe G., Van Den Bosch L., van den Brink G. R., van der Goot F. G., van der Klei I. J., van der Laan L. J. W., van Doorn W. G., van Egmond M., van Golen K. L., Van Kaer L., Campagne M. V., Vandenabeele P., Vandenberghe W., Vanhorebeek I., Varela-Nieto I., Vasconcelos M. H., Vasko R., Vavvas D. G., Vega-Naredo I., Velasco G., Velentzas A. D., Velentzas P. D., Vellai T., Vellenga E., Vendelbo M. H., Venkatachalam K., Ventura N., Ventura S., Veras P. S. T., Verdier M., Vertessy B. G., Viale A., Vidal M., Vieira H. L. A., Vierstra R. D., Vigneswaran N., Vij N., Vila M., Villar M., Villar V. H., Villarroya J., Vindis C., Viola G., Viscomi M. T., Vitale G., Vogl D. T., Voitsekhovskaja O. V., von Haefen C., von Schwarzenberg K., Voth D. E., Vouret-Craviari V., Vuori K., Vyas J. M., Waeber C., Walker C. L., Walker M. J., Walter J., Wan L., Wan X. B., Wang B., Wang C. H., Wang C. Y., Wang C. S., Wang C. R., Wang C. H., Wang D., Wang F., Wang F. X., Wang G. H., Wang H. J., Wang H. C., Wang H. G., Wang H. M., Wang H. D., Wang J., Wang J. J., Wang M., Wang M. Q., Wang P. Y., Wang P., Wang R. C., Wang S., Wang T. F., Wang X., Wang X. J., Wang X. W., Wang X., Wang X. J., Wang Y., Wang Y., Wang Y., Wang Y. J., Wang Y. P., Wang Y., Wang Y. T., Wang Y. Q., Wang Z. N., Wappner P., Ward C., Ward D. M., Warnes G., Watada H., Watanabe Y., Watase K., Weaver T. E., Weekes C. D., Wei J. W., Weide T., Weihl C. C., Weindl G., Weis S. N., Wen L. P., Wen X., Wen Y. F., Westermann B., Weyand C. M., White A. R., White E., Whitton J. L., Whitworth A. J., Wiels J., Wild F., Wildenberg M. E., Wileman T., Wilkinson D. S., Wilkinson S., Willbold D., Williams C., Williams K., Williamson P. R., Winklhofer K. F., Witkin S. S., Wohlgemuth S. E., Wollert T., Wolvetang E. J., Wong E., Wong G. W., Wong R. W., Wong V. K. W., Woodcock E. A., Wright K. L., Wu C. L., Wu D. F., Wu G. S., Wu J., Wu J. F., Wu M., Wu M., Wu S. Z., Wu W. K. K., Wu Y. H., Wu Z. L., Xavier C. P. R., Xavier R. J., Xia G. X., Xia T., Xia W. L., Xia Y., Xiao H. Y., Xiao J., Xiao S., Xiao W. H., Xie C. M., Xie Z. P., Xie Z. L., Xilouri M., Xiong Y. Y., Xu C. S., Xu C. F., Xu F., Xu H. X., Xu H. W., Xu J., Xu J. Z., Xu J. X., Xu L., Xu X. L., Xu Y. Q., Xu Y., Xu Z. X., Xu Z. H., Xue Y., Yamada T., Yamamoto A., Yamanaka K., Yamashina S., Yamashiro S., Yan B., Yan B., Yan X., Yan Z., Yanagi Y., Yang D. S., Yang J. M., Yang L., Yang M. H., Yang P. M., Yang P., Yang Q., Yang W. N., Yang W. Y., Yang X. S., Yang Y., Yang Y., Yang Z. F., Yang Z. H., Yao M. C., Yao P. J., Yao X. F., Yao Z. Y., Yao Z. Y., Yasui L. S., Ye M. X., Yedvobnick B., Yeganeh B., Yeh E. S., Yeyati P. L., Yi F., Yi L., Yin X. M., Yip C. K., Yoo Y. M., Yoo Y. H., Yoon S. Y., Yoshida K. I., Yoshimori T., Young K. H., Yu H. M., Yu J. J., Yu J. T., Yu J., Yu L., Yu W. H., Yu X. F., Yu Z. P., Yuan J. Y., Yuan Z. M., Yue B. Y. J. T., Yue J. B., Yue Z. Y., Zacks D. N., Zacksenhaus E., Zaffaroni N., Zaglia T., Zakeri Z., Zecchini V., Zeng J. S., Zeng M., Zeng Q., Zervos A. S., Zhang D. D., Zhang F., Zhang G., Zhang G. C., Zhang H., Zhang H., Zhang H., Zhang H. B., Zhang J., Zhang J., Zhang J. W., Zhang J. H., Zhang J. P., Zhang L., Zhang L., Zhang L., Zhang L., Zhang M. Y., Zhang X. N., Zhang X. D., Zhang Y., Zhang Y., Zhang Y. J., Zhang Y. M., Zhang Y. J., Zhao M., Zhao W. L., Zhao X. N., Zhao Y. G., Zhao Y., Zhao Y. C., Zhao Y. X., Zhao Z. D., Zhao Z. Z. J., Zheng D. X., Zheng X. L., Zheng X. X., Zhivotovsky B., Zhong Q., Zhou G. Z., Zhou G. F., Zhou H. P., Zhou S. F., Zhou X. J., Zhu H. X., Zhu H., Zhu W. G., Zhu W. H., Zhu X. F., Zhu Y. H., Zhuang S. M., Zhuang X. H., Ziparo E., Zois C. E., Zoladek T., Zong W. X., Zorzano A., Zughaier S. M. (2016) Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolpin B. M., Rubinson D. A., Wang X., Chan J. A., Cleary J. M., Enzinger P. C., Fuchs C. S., McCleary N. J., Meyerhardt J. A., Ng K., Schrag D., Sikora A. L., Spicer B. A., Killion L., Mamon H., Kimmelman A. C. (2014) Phase II and pharmacodynamic study of autophagy inhibition using hydroxychloroquine in patients with metastatic pancreatic adenocarcinoma. Oncologist 19, 637–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosenfeld M. R., Ye X., Supko J. G., Desideri S., Grossman S. A., Brem S., Mikkelson T., Wang D., Chang Y. C., Hu J., McAfee Q., Fisher J., Troxel A. B., Piao S., Heitjan D. F., Tan K. S., Pontiggia L., O’Dwyer P. J., Davis L. E., Amaravadi R. K. (2014) A phase I/II trial of hydroxychloroquine in conjunction with radiation therapy and concurrent and adjuvant temozolomide in patients with newly diagnosed glioblastoma multiforme. Autophagy 10, 1359–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rangwala R., Chang Y. C., Hu J., Algazy K. M., Evans T. L., Fecher L. A., Schuchter L. M., Torigian D. A., Panosian J. T., Troxel A. B., Tan K. S., Heitjan D. F., DeMichele A. M., Vaughn D. J., Redlinger M., Alavi A., Kaiser J., Pontiggia L., Davis L. E., O’Dwyer P. J., Amaravadi R. K. (2014) Combined MTOR and autophagy inhibition: phase I trial of hydroxychloroquine and temsirolimus in patients with advanced solid tumors and melanoma. Autophagy 10, 1391–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kimura T., Takabatake Y., Takahashi A., Isaka Y. (2013) Chloroquine in cancer therapy: a double-edged sword of autophagy. Cancer Res. 73, 3–7 [DOI] [PubMed] [Google Scholar]
- 31.Cook K. L., Wärri A., Soto-Pantoja D. R., Clarke P. A., Cruz M. I., Zwart A., Clarke R. (2014) Hydroxychloroquine inhibits autophagy to potentiate antiestrogen responsiveness in ER+ breast cancer. Clin. Cancer Res. 20, 3222–3232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vichai V., Kirtikara K. (2006) Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat. Protoc. 1, 1112–1116 [DOI] [PubMed] [Google Scholar]
- 33.Lewis M. D., Pfeil J., Mueller A. K. (2011) Continuous oral chloroquine as a novel route for plasmodium prophylaxis and cure in experimental murine models. BMC Res. Notes 4, 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Demidenko E. (2006) The assessment of tumour response to treatment. Appl. Stat. 55, 365–377 [Google Scholar]
- 35.Demidenko E. (2010) Three endpoints of in vivo tumour radiobiology and their statistical estimation. Int. J. Radiat. Biol. 86, 164–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Demidenko E. (2013) Mixed Models: Theory and Applications with R, Wiley, New York, NY, USA [Google Scholar]
- 37.Team R. C. (2014) R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 38.Faber A. C., Corcoran R. B., Ebi H., Sequist L. V., Waltman B. A., Chung E., Incio J., Digumarthy S. R., Pollack S. F., Song Y., Muzikansky A., Lifshits E., Roberge S., Coffman E. J., Benes C. H., Gómez H. L., Baselga J., Arteaga C. L., Rivera M. N., Dias-Santagata D., Jain R. K., Engelman J. A. (2011) BIM expression in treatment-naive cancers predicts responsiveness to kinase inhibitors. Cancer Discov. 1, 352–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller T. W., Balko J. M., Arteaga C. L. (2011) Phosphatidylinositol 3-kinase and antiestrogen resistance in breast cancer. J. Clin. Oncol. 29, 4452–4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garner F., Brown J., Katzenellenbogen J., Lyttle C.R., Hattersley G. (2016) Abstract P3-05-07: RAD1901, a novel oral, selective estrogen receptor degrader (SERD) with single agent efficacy in an ER+ primary patent derived ESR1 mutant xenograft model. Cancer Res. 76, Abstract nr P3-05-07 [Google Scholar]
- 41.O’Brien C., Wallin J. J., Sampath D., GuhaThakurta D., Savage H., Punnoose E. A., Guan J., Berry L., Prior W. W., Amler L. C., Belvin M., Friedman L. S., Lackner M. R. (2010) Predictive biomarkers of sensitivity to the phosphatidylinositol 3′ kinase inhibitor GDC-0941 in breast cancer preclinical models. Clin. Cancer Res. 16, 3670–3683 [DOI] [PubMed] [Google Scholar]
- 42.Ni J., Liu Q., Xie S., Carlson C., Von T., Vogel K., Riddle S., Benes C., Eck M., Roberts T., Gray N., Zhao J. (2012) Functional characterization of an isoform-selective inhibitor of PI3K-p110β as a potential anticancer agent. Cancer Discov. 2, 425–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lamoureux F., Thomas C., Crafter C., Kumano M., Zhang F., Davies B. R., Gleave M. E., Zoubeidi A. (2013) Blocked autophagy using lysosomotropic agents sensitizes resistant prostate tumor cells to the novel Akt inhibitor AZD5363. Clin. Cancer Res. 19, 833–844 [DOI] [PubMed] [Google Scholar]
- 44.Cook K. L., Shajahan A. N., Wärri A., Jin L., Hilakivi-Clarke L. A., Clarke R. (2012) Glucose-regulated protein 78 controls cross-talk between apoptosis and autophagy to determine antiestrogen responsiveness. Cancer Res. 72, 3337–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Happo L., Strasser A., Cory S. (2012) BH3-only proteins in apoptosis at a glance. J. Cell Sci. 125, 1081–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Villunger A., Michalak E. M., Coultas L., Müllauer F., Böck G., Ausserlechner M. J., Adams J. M., Strasser A. (2003) p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science 302, 1036–1038 [DOI] [PubMed] [Google Scholar]
- 47.Bean G. R., Ganesan Y. T., Dong Y., Takeda S., Liu H., Chan P. M., Huang Y., Chodosh L. A., Zambetti G. P., Hsieh J. J., Cheng E. H. (2013) PUMA and BIM are required for oncogene inactivation-induced apoptosis. Sci. Signal. 6, ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ley R., Balmanno K., Hadfield K., Weston C., Cook S. J. (2003) Activation of the ERK1/2 signaling pathway promotes phosphorylation and proteasome-dependent degradation of the BH3-only protein, Bim. J. Biol. Chem. 278, 18811–18816 [DOI] [PubMed] [Google Scholar]
- 49.Dillon L. M., Bean J. R., Yang W., Shee K., Symonds L. K., Balko J. M., McDonald W. H., Liu S., Gonzalez-Angulo A. M., Mills G. B., Arteaga C. L., Miller T. W. (2015) P-REX1 creates a positive feedback loop to activate growth factor receptor, PI3K/AKT and MEK/ERK signaling in breast cancer. Oncogene 34, 3968–3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ebi H., Costa C., Faber A. C., Nishtala M., Kotani H., Juric D., Della Pelle P., Song Y., Yano S., Mino-Kenudson M., Benes C. H., Engelman J. A. (2013) PI3K regulates MEK/ERK signaling in breast cancer via the Rac-GEF, P-Rex1. Proc. Natl. Acad. Sci. USA 110, 21124–21129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.You H., Pellegrini M., Tsuchihara K., Yamamoto K., Hacker G., Erlacher M., Villunger A., Mak T. W. (2006) FOXO3a-dependent regulation of Puma in response to cytokine/growth factor withdrawal. J. Exp. Med. 203, 1657–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Amente S., Zhang J., Lavadera M. L., Lania L., Avvedimento E. V., Majello B. (2011) Myc and PI3K/AKT signaling cooperatively repress FOXO3a-dependent PUMA and GADD45a gene expression. Nucleic Acids Res. 39, 9498–9507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakano K., Vousden K. H. (2001) PUMA, a novel proapoptotic gene, is induced by p53. Mol. Cell 7, 683–694 [DOI] [PubMed] [Google Scholar]
- 54.White E. (2015) The role for autophagy in cancer. J. Clin. Invest. 125, 42–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mayo L. D., Donner D. B. (2001) A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc. Natl. Acad. Sci. USA 98, 11598–11603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vaillant F., Merino D., Lee L., Breslin K., Pal B., Ritchie M. E., Smyth G. K., Christie M., Phillipson L. J., Burns C. J., Mann G. B., Visvader J. E., Lindeman G. J. (2013) Targeting BCL-2 with the BH3 mimetic ABT-199 in estrogen receptor-positive breast cancer. Cancer Cell 24, 120–129 [DOI] [PubMed] [Google Scholar]
- 57.Molenaar R. J., Coelen R. J., Khurshed M., Roos E., Caan M. W., van Linde M. E., Kouwenhoven M., Bramer J. A., Bovée J. V., Mathôt R. A., Klümpen H. J., van Laarhoven H. W., van Noorden C. J., Vandertop W. P., Gelderblom H., van Gulik T. M., Wilmink J. W. (2017) Study protocol of a phase IB/II clinical trial of metformin and chloroquine in patients with IDH1-mutated or IDH2-mutated solid tumours. BMJ Open 7, e014961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosenfeld M. R., Ye X., Supko J. G., Desideri S., Grossman S. A., Brem S., Mikkelson T., Wang D., Chang Y. C., Hu J., McAfee Q., Fisher J., Troxel A. B., Piao S., Heitjan D. F., Tan K. S., Pontiggia L., O’Dwyer P. J., Davis L. E., Amaravadi R. K. (2014) A phase I/II trial of hydroxychloroquine in conjunction with radiation therapy and concurrent and adjuvant temozolomide in patients with newly diagnosed glioblastoma multiforme. Autophagy 10, 1359–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rangwala R., Leone R., Chang Y. C., Fecher L. A., Schuchter L. M., Kramer A., Tan K. S., Heitjan D. F., Rodgers G., Gallagher M., Piao S., Troxel A. B., Evans T. L., DeMichele A. M., Nathanson K. L., O’Dwyer P. J., Kaiser J., Pontiggia L., Davis L. E., Amaravadi R. K. (2014) Phase I trial of hydroxychloroquine with dose-intense temozolomide in patients with advanced solid tumors and melanoma. Autophagy 10, 1369–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barnard R. A., Wittenburg L. A., Amaravadi R. K., Gustafson D. L., Thorburn A., Thamm D. H. (2014) Phase I clinical trial and pharmacodynamic evaluation of combination hydroxychloroquine and doxorubicin treatment in pet dogs treated for spontaneously occurring lymphoma. Autophagy 10, 1415–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chude C. I., Amaravadi R. K. (2017) Targeting autophagy in cancer: update on clinical trials and novel inhibitors. Int. J. Mol. Sci. 18, 1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.