Abstract
Excess circulating insulin is associated with obesity in humans and in animal models. However, the physiologic causality of hyperinsulinemia in adult obesity has rightfully been questioned because of the absence of clear evidence that weight loss can be induced by acutely reversing diet-induced hyperinsulinemia. Herein, we describe the consequences of inducible, partial insulin gene deletion in a mouse model in which animals have already been made obese by consuming a high-fat diet. A modest reduction in insulin production/secretion was sufficient to cause significant weight loss within 5 wk, with a specific effect on visceral adipose tissue. This result was associated with a reduction in the protein abundance of the lipodystrophy gene polymerase I and transcript release factor (Ptrf; Cavin) in gonadal adipose tissue. RNAseq analysis showed that reduced insulin and weight loss also associated with a signature of reduced innate immunity. This study demonstrates that changes in circulating insulin that are too fine to adversely affect glucose homeostasis nonetheless exert control over adiposity.—Page, M. M., Skovsø, S., Cen, H., Chiu, A. P., Dionne, D. A., Hutchinson, D. F., Lim, G. E., Szabat, M., Flibotte, S., Sinha, S., Nislow, C., Rodrigues, B., Johnson, J. D. Reducing insulin via conditional partial gene ablation in adults reverses diet-induced weight gain.
Keywords: obesity, adipose tissue, Ptrf/Cavin, high-fat diet, innate immunity
Obesity and related diseases burden both society and the individual, increasing the prevalence and risk of comorbidities including diabetes, heart disease, and cancer (1). It is clear that obesity and hyperinsulinemia are closely related, although their causal relationship remains poorly defined, obscuring the molecular mechanisms underlying possible treatments for obesity. The common belief is that obesity precedes insulin resistance, which subsequently causes a compensatory increase in β-cell insulin secretion to prevent hyperglycemia (2). However, elevated circulating insulin levels have been reported prior to the onset of obesity (3, 4), and increasing evidence suggests that hyperinsulinemia is not simply an adaptive response to obesity (5). In support of this concept, adipose tissue–specific impairment of insulin signaling prevents obesity in mice (6–10). Furthermore, we recently reported that mice with lifelong prevention of diet-induced hyperinsulinemia by partial insulin gene deletion were protected against high-fat diet (HFD)–induced obesity (11, 12). These results provided the first evidence in mammals that hyperinsulinemia itself plays a causal role in obesity, yet this preventative approach could not determine whether insulin reduction could serve as a viable treatment in those individuals who are already obese. Suppressing hyperinsulinemia with drugs has been used as an obesity treatment in animal models and in some human studies (13–15). However, these results must be interpreted with caution, because these drugs directly affect other organs that are also implicated in weight regulation, including white adipose tissue (WAT) and the hypothalamus (13, 16). Herein, we report, that an adult-onset partial reduction of insulin (Ins)-2 gene dose in obese male mice caused significant weight loss within 5 wk, with specific effects on visceral adipose depots. The results demonstrate that adiposity and obesity can be controlled by modest changes in circulating insulin.
MATERIALS AND METHODS
Animals and in vivo physiology
Animal protocols were performed in accordance with the University of British Columbia Animal Care Committee. Ins1−/−:Ins2fl/+ mice have been described in Duvillié et al. (17). Pdx1CreERT mice (024968) and mice carrying the lineage-tracing marker membrane-targeted tdTomato/membrane-targeted enhanced green fluorescent protein; (mTmG; 007675) were purchased from The Jackson Laboratories (Bar Harbor, ME, USA). Before they were randomized into different groups, they were fed a 19% protein extruded rodent diet (Teklad; Envigo, Madison, WI, USA). At 6 wk of age, male mice were randomized and placed on 1 of 3 diets: 1) a 10% fat diet [total calories = 3.85 kcal/g; 10% calories from fat, 20% from protein, and 70% from carbohydrate; D12450B (Open Source/Research Diets, New Brunswick, NJ, USA)], 2) a 25% fat diet [total calories = 3.70 kcal/g; 25% calories from fat, 20% from protein, and 55% from carbohydrate; 5LJ5 (Lab Diet, St. Louis. MO, USA)], 3) a 58% fat diet [total calories = 5.56 kcal/g; 58.0% calories from fat, 16.4% calories from protein, and 25.5% calories from carbohydrate; D12330 (Open Source Diets/Research Diets)]. Mice that were randomized were born from 5 breeding pairs, and we ensured that pups of subsequent litters were placed on each of the three different diets before repeating any diet twice. The body mass of the mice before randomization ranged from 21.4 to 33.2 g. Because our study primarily focused on the physiologic changes in mice fed an HFD, we can report that, immediately upon randomization, the average body weights were as follows: vehicle, 26.5 ± 0.5 g; control, 26.8 ± 0.3 g; and experimental, 26.1 ± 0.3 g. Twelve weeks after the initiation of this diet, control (Ins1−/−:Ins2fl/+:mTmG) and experimental (Ins1−/−:Ins2fl/+:Pdx1CreERT:mTmG) mice were injected intraperitoneally with tamoxifen (3 mg/40 g body weight) dissolved in corn oil for 4 consecutive days. Body mass was measured between 6 and 23 wk of age between 2 and 4 pm, and daily food intake was measured 2 wk before and 4 wk after tamoxifen injections. Basal blood glucose measurements and blood collection to measure circulating insulin were performed after the mice remained unfed for 4 h. Glucose tolerance and insulin sensitivity were assessed in mice injected with 20% glucose and 1.5 U/kg body weight insulin, respectively, after 4 h without food. Glucose-stimulated insulin secretion was assessed in mice injected with 20% glucose after 4 h without food. Insulin content from in vivo isolated islet samples was measured with ELISA kits from Alpco (Salem, NH, USA). Leptin, resistin, ghrelin, glucose-dependent insulinotropic peptide (GIP), glucagon-like protein (GLP)-1, IL-6, and pancreatic polypeptide (PYY) were measured with a mouse magnetic bead panel assay and Luminex technology (Millipore-Sigma, Billerica, MA, USA). IL-1β, TNF-α, Il-2, IL-6, and IL-12p70 levels were measured with a mouse magnetic bead panel (Bio-Rad, Hercules, CA, USA). Nonesterified fatty acids (NEFAs; Wako, Richmond, VA, USA), free glycerol and glycerides (Millipore-Sigma) were measured according to the manufacturer’s protocol in serum collected from a cohort of mice fed an HFD 5 wk after tamoxifen injections and not fed for 16 h and again for 15 min after insulin stimulation (2 U/kg body weight). Adipose mass was measured from this cohort of mice.
Fluorescence microscopy
Pancreata from PBS-perfused mice were harvested and fixed in 4% paraformaldehyde for 24 h, washed, and stored in 70% ethanol, before paraffin embedding. Sections (5 μm) were taken from at least 3 different regions of the pancreas, 100 μm apart. Sections were deparaffinized, hydrated with decreasing concentrations of ethanol, and rinsed in PBS, then exposed to a 15 min heat-induced epitope retrieval at 95°C with 10 mM citrate buffer (pH 6.0). Sections were blocked and incubated with primary anti-insulin (ab7842; Abcam, Cambridge, MA, USA) and anti-green fluorescent protein (GFP) (A11122; Thermo Fisher Scientific, Waltham, MA, USA) overnight in a humid chamber at 4°C. Primary antibodies were visualized after incubation with secondary antibodies conjugated to AlexaFluor 488, 555, 594, or 647, as required (1:1000; Thermo Fisher Scientific). Gonadal and perirenal adipose tissue was harvested from mice left unfed for 4 h, and tissues were fixed in 4% paraformaldehyde. Adipocyte size was measured by staining for perilipin in 5 mm-thick sections (Cell Signaling Technology, Danvers, MA, USA). All images were taken on ImageXpressmicro with a ×10 high (NA 0.3) objective.
Immunoblot analyses and protein detection
In Western blot analyses, 30–40 μg of protein was used to probe for polymerase I and transcript release factor (PTRF; ab48824; Abcam); the ratio of phospho-caveolin-1 (3251) to total caveolin-1 (3267, clathrin (4796), insulin receptor (3020), ratio of phospho-hormone–sensitive lipase (Hsl;4139) to total Hsl (4107), ratio of phospho-Akt (4060) to total Akt (9272), and ratio of phospho-p44/42 MAPK (4370) to total p44/42 MAPK (4695; all from Cell Signaling Technology); and lipoprotein lipase (Lpl; sc-32885; Santa Cruz Biotechnology, Dallas, TX, USA) in gonadal tissue from mice that were left unfed overnight and during a 15-min insulin-stimulation (2 U/kg body weight). Actin (NB600-501; Novusbio, Littleton, CO, USA) and vinculin (13901; Cell Signaling Technology) were used as loading references.
Islet isolation, culture, and perifusion
Pancreatic islets were isolated by using collagenase, filtration, and manual picking (18). Islets were cultured overnight (37°C, 5% CO2) in RPMI 1640 medium (Thermo Fisher Scientific) with 11 mM glucose (Millipore-Sigma), 100 U/ml penicillin, 100 μg/ml streptomycin (Thermo Fisher Scientific), and 10% fetal bovine serum (Thermo Fisher Scientific). In vitro insulin secretion was measured by perifusion and radioimmunoassay (19).
RNA isolation, real-time quantitative PCR, and transcriptome analysis
Total RNA was isolated from islets with an RNeasy Mini Kit (Qiagen, Mississauga, ON, Canada). Reverse transcription was used to generate cDNA (Superscript III; Thermo Fisher Scientific). TaqMan probes (Integrated DNA Technologies, Coralville, IA, USA) were used to measure Ins2 gene expression, with actin as a reference gene. Gonadal adipose tissue was dissected from a cohort of 23-wk-old (corresponding to 5 wk after tamoxifen injection) control littermate (n = 7) and experimental (n = 8) mice euthanized after remaining unfed for 4 h. RNA isolation was performed with Trizol reagent (Thermo Fisher Scientific, Waltham, MA, USA) and Qiagen RNeasy Mini Kit (Qiagen). In brief, tissue was homogenized in Trizol and centrifuged for 10 min at 4°C. Chloroform was added to the supernatant and centrifuged for 15 min at 4°C. An equal volume of 70% ethanol was added to the aqueous layer after chloroform extraction, and this mixture was transferred to the spin column. The remainder of the protocol was followed as in the manufacturer’s instructions. cDNA synthesis and real-time quantitative PCR was conducted as published (11). For RNA sequencing, libraries were prepared with the TruSeq RNA Sample Preparation v.2 kit (Illumina, San Diego, CA, USA), from 400 ng of total RNA. Sample quality was assessed with a BioAnalyzer (Agilent Technologies, Santa Clara, CA, USA), and RNA was quantified with Qubit (Thermo Fisher Scientific). Libraries were multiplexed and sequenced over two rapid-run lanes on HiSeq2500, and raw data were converted to fastq format with bcl2fastq-v1.8.4 (Illumina). Kallisto software (https://pachterlab.github.io/kallisto/) was used for read alignment and expression quantification and the R package DESeq2 software (http://bioconductor.org/packages/release/bioc/html/DESeq.html) for differential expression analysis. In brief, kallisto (version 0.42.3) was used to build an index file for the mouse reference transcriptome GRCm38 [downloaded from Ensembl (www.ensembl.org)]. The sequence reads for each sample were then quantified with the quant function of kallisto. In-house Perl scripts were used to sum the read counts at the transcript level for each gene and to create a matrix comprising the read counts for all the genes for all the samples. Differential expression analysis was then performed on the data from that matrix by using DESeq2. Each sample was assessed with the quality control software RNA-SeQC (The Broad Institute, Cambridge, MA, USA; http://archive.broadinstitute.org/cancer/cga/rna-sequc) and the PtR script from the Trinity Suite (http://TrinityRNASeq.sourceforge.net). Quality control showed one control and two experimental outliers, and these samples were removed from differential expression analyses. Therefore, included in the analyses are expression from 6 control and 6 experimental mice. RNA sequencing transcriptome data were analyzed with GeneMania (http://genemania.org/), to construct a predicted protein–protein networking model which was manually arranged into groups based on predicted tissue origin.
Micro-computed tomography and dual-energy X-ray absorptiometry
Scans were performed with a TriFoil Micro-Computed Tomography (CT) scanner (Northridge Tri-Modality Imaging, Inc., Chatsworth, CA, USA) with the following settings: resolution, 50 μm; scan time,∼4 min. Each mouse was scanned 1 wk before and again 5 wk after tamoxifen injections. The mice were positioned on their backs in the scanner with a straight spine and a minimum 90° angle between the spine and the femur bone, which was confirmed by a scout scan. Animals were scanned while anesthetized (O2 at 1.0 L/min; 5% isoflurane). Analysis of the scanned images was performed with Amira 6.0.1 software (FEI, Hillsboro, OR, USA). The first transsectional scan showing the pelvis was set as the reference scan for each mouse. The inner layers of the abdominal wall were applied to distinguish visceral from subcutaneous fat depot volumes (µm3). Lean mass was determined in vivo by dual-energy X-ray absorptiometry (Lunar Piximus; no longer manufactured) densitometer in mice 5–7 wk after tamoxifen (from 15 control and 12 experimental mice).
Statistical analysis
Data are expressed as means ± sem unless otherwise indicated. Results were considered statistically significant at P < 0.05, by 2-tailed, unpaired Student’s t test. Statistical analyses were performed with Microsoft Excel (Redmond, WA, USA).
RESULTS AND DISCUSSION
Inducible, partial Ins2 gene deletion as an experimental model to reduce insulin production
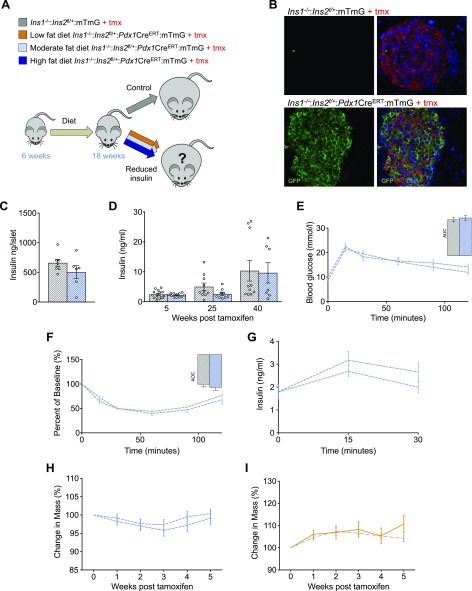
To directly assess the sustained effects of hyperinsulinemia on obesity and weight loss, we generated a mouse model in which the insulin gene dose could be reduced by using an inducible Cre-loxP system (Fig. 1A). At 6 wk of age, mice were fed 1 of 3 diets; low fat (10%), moderate fat (25%), or high fat (58%). After 12 wk of consuming these diets, littermate control (Ins1−/−:Ins2f/+:mTmG) and experimental (Ins1−/−:Ins2f/+:Pdx1CreERT:mTmG) male mice were injected intraperitoneally with tamoxifen, resulting in a near complete recombination on this genetic background (Fig. 1B) (20).
Figure 1.
Adult reduction of Ins2 gene dose in mice consuming low- and moderate-fat diets does not reverse weight gain. A) Schematic of our hypothesis that reduced insulin would reverse weight gain and obesity in adult male mice fed a low-fat (10%, yellow), moderate-fat (25%, light blue), or high-fat (58%, dark blue) diet. B) Tamoxifen induction of Pdx1CreERT led to near complete, as evidenced by membrane GFP expression (bottom). C) Insulin content in mice (n = 7, 6; control n is listed first throughout) fed a moderate-fat diet were measured from isolated islets 40 wk after tamoxifen injections. For insulin content measurements, n represents individual mice. D) Circulating insulin at 5, 25, and 40 wk after tamoxifen injection in control littermates (n = 10–15) and experimental mice (n = 7–12) fed a moderate-fat diet. E, F) Glucose tolerance (E) and insulin sensitivity (F) in mice (n = 15, 12). Insets: area under the curve (AUC) (E) and area over the curve (AOC) (F). G) Glucose-stimulated insulin secretion (n = 15, 12). H) Percentage change in body mass of male mice fed a moderate-fat diet; control littermates (n = 20; Ins1−/−:Ins2f/+:mTmG; gray dashed line) and experimental mice (n = 20; Ins1−/−:Ins2f/+:Pdx1CreERT:mTmG; blue dashed line). I) Percentage change in body mass of male mice fed a low-fat diet; control littermates (n = 3; Ins1−/−:Ins2f/+:mTmG; gray dashed line) and experimental mice (n = 2; Ins1−/−:Ins2f/+:Pdx1CreERT:mTmG; yellow line). Unless otherwise indicated, measurements were conducted from samples collected from mice between 5 and 7 wk after tamoxifen injection. Data are means ± sem.
Inducible partial loss of Ins2 does not affect the weight of mice fed a moderate- or low-fat diet
Manipulation of Ins2 gene dose in experimental mice fed a moderate-fat diet did not significantly affect islet insulin levels (Fig. 1C), nor did this manipulation lead to a statistically significant alteration in circulating insulin in experimental mice throughout the study (Fig. 1D). Consistent with these findings, glucose tolerance, insulin sensitivity, and glucose-stimulated insulin secretion were not impaired in experimental mice when compared to control littermates in the context of this diet (Fig. 1E–G). Reduction of Ins2 gene dose in male mice did not affect body mass in mice fed a moderate-fat diet (Fig. 1H). Similarly, body mass was unchanged after a reduction in the Ins2 gene dose in experimental mice fed a low-fat diet (Fig. 1I). These results are in line with previous evidence from other genetic manipulations that do not affect body mass in mice fed low- or moderate-fat diets (21).
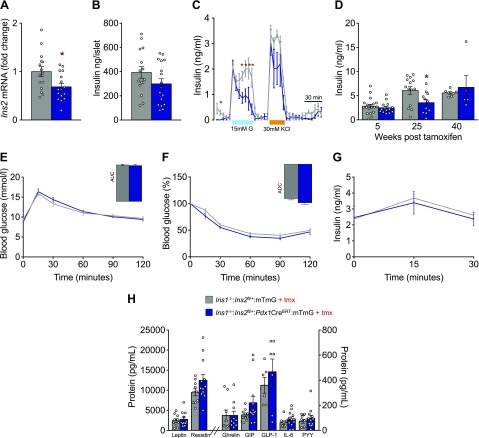
Inducible Ins2 gene reduction modestly reduces islet insulin secretion in mice fed an HFD
Ins2 gene reduction in male mice fed an HFD resulted in a significant (30%) reduction of Ins2 expression (Fig. 2A) and a similar magnitude, but nonsignificant, reduction in insulin protein content from isolated islets (Fig. 2B) compared to littermate controls. The modest reduction in insulin gene expression in experimental mice led to a significant reduction in the second phase of high glucose-stimulated insulin secretion (Fig. 2C). We detected significantly lower levels of fasting insulin in vivo at 25 wk after tamoxifen injection, although we noted high variability in the fasting insulin measurements at all ages (Fig. 2D). These modest reductions in insulin secretion were not sufficient to alter glucose tolerance, insulin sensitivity, or glucose-stimulated insulin secretion (Fig. 2E–G). We were also unable to detect any differences in the levels of several other circulating hormones, including leptin, resistin, ghrelin, GIP, GLP-1, IL-6, and PYY (Fig. 2H). Thus, in these experimental conditions, partial ablation of the Ins2 gene in adult mice results in small but significant reductions in basal and glucose-stimulated second-phase insulin release that remain within the normal range for glucose homeostasis. The maintenance of normal glucose homeostasis within our model allowed us to test for the causality of hyperinsulinemia in obesity, without confounding changes in glucose tolerance or insulin sensitivity.
Figure 2.
Adult reduction of Ins2 gene dose in mice fed an HFD impairs the second phase of high-glucose–stimulated insulin secretion without altering glucose tolerance and insulin sensitivity. A) Islet Ins2 transcript level in control littermates (n = 15, Ins1−/−:Ins2f/+:mTmG; gray) and experimental mice (n = 16; Ins1−/−:Ins2f/+:Pdx1CreERT:mTmG; blue) fed an HFD were measured from isolated islets of mice 5 wk after tamoxifen injections. B) Islet insulin content of mice (n = 15, 17; control n is listed first throughout) fed an HFD were measured from isolated islets of mice 5 wk after tamoxifen injections. For insulin content and transcript level measurements, n represents individual mice. C) In vitro basal, glucose- (15 mM) and KCl- (30 mM) stimulated insulin release from isolated islets (n = 3, 3). D) Fasting circulating insulin 5, 25, and 40 wk after tamoxifen injection in control littermate (n = 7–19) and experimental mice (n = 5–14). E, F) Glucose tolerance (E) and insulin sensitivity (F) in mice (n = 14, 19). Insets: area under the curve (AUC) (E) and area over the curve (AOC) (F). G) Glucose-stimulated insulin secretion in mice (n = 26, 19). H) Leptin, resistin, ghrelin, GIP, GLP-1, IL-6, PYY levels in plasma extracted from control littermates (n = 6–10) and experimental mice (n = 7–12). Unless otherwise indicated, measurements were conducted from samples collected from mice between 5 and 7 wk after tamoxifen injection. Data are means ± sem. *P < 0.05.
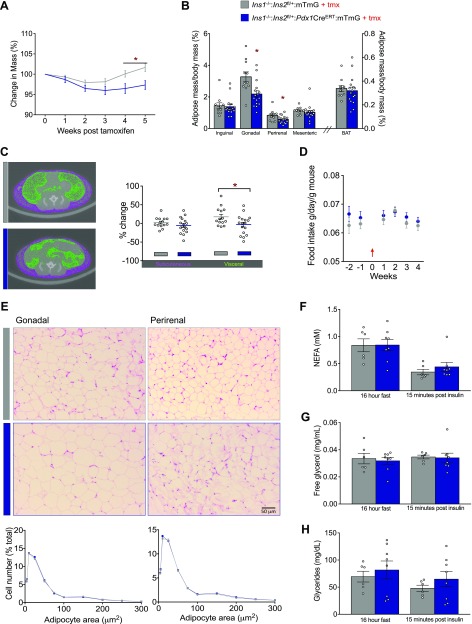
Inducible insulin reduction causes weight loss in mice fed an HFD
To test our hypothesis that obesity could be reversed by acutely suppressing insulin, we examined the body weight of HFD-fed control littermates and experimental mice 5 wk after tamoxifen injections. In a pilot study, we found that tamoxifen injection resulted in a slight reduction in body mass compared to that in vehicle-treated mice (Supplemental Fig. S1). To be conservative, we felt it was most appropriate to compare groups of mice that were both injected with tamoxifen. Using this conservative approach, we found that acute reduction in Ins2 gene dose resulted in an ∼5% reduction in body mass (Fig. 3A). Control mice had body weights ranging from 42 to 44 g, whereas experimental mice weighed between 39 and 41 g 5 wk after the tamoxifen injection. The weight loss we observed in the experimental mice was primarily related to the reduced mass of gonadal and perirenal fat pads, which were significantly lighter than those fat pads isolated from controls (Fig. 3B). Micro-CT imaging confirmed reduced visceral fat accumulation in mice after partial Ins2 gene ablation (Fig. 3C). The masses of liver, heart, and brain (data not shown), as well as brown adipose tissue (BAT) (Fig. 3C), were not affected by reducing the Ins2 gene dose. In addition, mean lean mass measurements in dual-energy X-ray absorptiometry scans did not differ between control littermate and experimental mice (30.9 ± 0.6 vs. 29.7 ± 0.7 g). There were no differences in daily food intake before or after recombination (Fig. 3D), suggesting the absence of a robust action on the brain. Analysis of gonadal and perirenal fat pad cross sections did not reveal robust differences in adipocyte morphology between control littermate and experimental mice (Fig. 3E). Together, these data demonstrate that a modest acute reduction of circulating insulin can lead to weight loss. Our data are consistent with the reduced obesity in HFD-fed mice with β-cell specific loss of glutamate dehydrogenase 1 (Glud1), an enzyme required for complete insulin secretion (22).
Figure 3.
Adult reduction of Ins2 gene dose reverses obesity in mice fed an HFD, primarily because of reduced mass of gonadal and perirenal fat pads. A) Percentage change in mass of control littermate (n = 39) and experimental (n = 29) mice 5 wk after tamoxifen injection. B) Inguinal, gonadal, perirenal, and mesenteric WAT depots and BAT from mice were weighed after 16 h without food and again at 15 min after an insulin stimulation (n = 16, 11). C) Representative transsectional images obtained by micro-CT (left) and quantification (right) of percentage change in subcutaneous and visceral fat depots after tamoxifen injection (n = 14, 17). D) Daily food intake was measured for 2 wk before, and 4 wk after, tamoxifen injection (n = 14, 19; control n is listed first throughout). Arrow: the week of tamoxifen injection. E) Representative sections of gonadal (n = 7, 10) and perirenal (n = 8, 11) adipose tissue (top; hematoxylin and eosin staining) and analysis of adipocyte size distribution (bottom; perilipin staining). F–H) NEFA (F), free glycerol (G), and glyceride (H) levels (n = 6, 8) were measured after food was withheld for 16 h and again 15 min after insulin stimulation. Unless otherwise indicated, measurements were conducted in samples collected from mice between 5 and 7 wk after tamoxifen injection. Data are means ± sem. *P < 0.05.
Markers of lipid mobilization are altered after induced insulin reduction
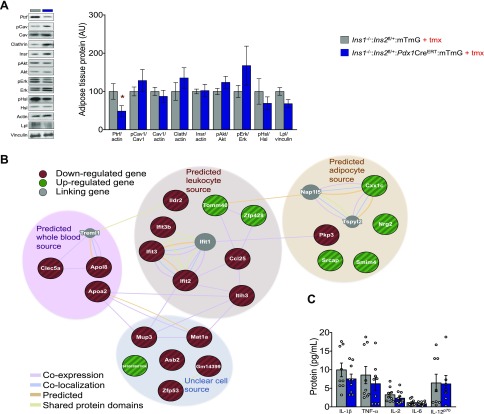
Insulin regulates lipid metabolism at multiple steps, including by inhibiting lipolysis (23). Basal levels of NEFA, free glycerol, or glycerides (Fig. 3F–H) did not differ between control littermates and experimental mice, indicating that the reduction of circulating insulin in the experimental mice did not grossly impair lipid metabolism and that insulin sensitivity remained similar between these two groups of mice. Insulin is well known to regulate WAT lipid accumulation (24). This, along with the reduction of fat mass in the experimental mice, led us to explore proteins involved in adipocyte energy storage and lipid mobilization. First, we examined the protein abundance of Ptrf/Cavin, given that both mice and humans with loss-of-function mutations in this gene display a lipodystrophic phenotype (25, 26). Indeed, Ptrf protein levels were significantly lower in experimental mice after reduction of Ins2 gene dose (Fig. 4A). Ptrf is known for its role in the formation and organization of caveolae, together with caveolin proteins, which have also been implicated in lipodystrophy (27–29). In addition to clathrin, phosphorylated caveolin-1 has been implicated recently in insulin uptake and insulin receptor internalization (30, 31). However, we did not observe significant differences in the protein abundances of clathrin, caveolin 1 (Cav1), Y14 phosphorylated Cav1, insulin receptor, phosphorylated-Akt, or phosphorylated-Erk between control littermates and experimental mice. This result suggests that Ptrf either acts downstream of reduced insulin independent from Cav1 or in a manner that does not necessitate changes in Cav1 protein levels or phosphorylation at the Y14 site. It is also consistent with our observations that insulin sensitivity was not influenced by our mild and acute reductions in insulin production. Nevertheless, caveolin proteins have been implicated in the activation of lipolysis (32) and Hsl (33). Phospho-Hsl did not differ statistically between control littermates and experimental mice, consistent with the similarities in plasma levels of free fatty acids and glycerol (Fig. 3F, G). We observed a slight reduction (P = 0.06) in Lpl in our experimental mice compared with their control littermates, suggesting another mechanism associated with altered lipid uptake into adipose tissue in the context of reduced insulin production and secretion. Given the role of Lpl as an important marker for adipogenesis (34), the reduced levels in our study may reflect impaired adipogenesis related to reduced insulin production and secretion (35).
Figure 4.
Network analysis of gonadal fat pad from adult mice with reduced Ins2 gene dose. A) Representative immunoblots (left) and quantification (right) of key proteins involved in lipid metabolism. B) Analysis of a protein–protein interaction network assembled from RNAseq data. Node color reflects whether mRNA is increased (green) or decreased (red); gray represents linking genes. C) IL-1β, TNF-α, Il-2, IL-6, and IL-12p70 levels in control littermate (n = 8–9) and experimental (n = 7–12) mice.
Inducible insulin reduction is associated with an altered immune profile in adipose tissue
We performed RNA sequencing to further examine the effects of inducible insulin reduction on the transcriptome of gonadal fat tissue 5 d after tamoxifen injection. After correction for multiple testing, we did not find any individual genes that were significantly differentially expressed when the Ins2 gene dose was reduced. However, ordering the genes by P-value before correction for multiple testing revealed that some gene families were overrepresented at the top of that list (Supplemental File S1). We therefore selected the top 24 genes, all with uncorrected P > 0.01, for protein–protein network modeling, to visualize the relationships between the expressed mRNAs; these networks were manually arranged to reflect the most likely cell source, based on the GTEx (http://www.gtexportal.org) resource. Six immune-associated genes, including chemokine ligand (Ccl)25, interferon-induced protein with tetratricopeptide repeats (Ifit)-2, Ifit3, Ifit3b, Ig-like domain-containing receptor (Ildr)-2, and inter-α-trypsin inhibitor heavy chain (Itih)-3 were down-regulated in the experimental mice compared to control littermates within the subnetwork of a predicted leukocyte source (Fig. 4B). Enriched pathways in the protein–protein network highlighted changes in immune function as well as lipid transport (Table 1). Ccl25 is primarily involved in leukocyte migration and its elevated expression is associated with chronic inflammatory conditions (36). Inflammatory conditions linked to the onset of diabetes in nonobese Goto-kakizaki rats are also associated with elevated expression of Ifit genes (37) and Itih3 and Itih4 genes have been linked to obesity (38) and diabetes (39) in mouse genetic studies. These genes bind to locally synthesized hyaluronan and, as a complex, have been reported to be involved in inflammatory diseases (40). Levels of hyaluronan, an extracellular matrix component, have been reported to increase in adipose tissue (41) as well as insulin-resistant skeletal muscle in mice with diet-induced obesity (42). Our control littermates are both obese and hyperinsulinemic compared with our experimental mice, which is consistent with altered expression of proinflammatory genes. The down-regulation we observed of immune-related genes in mice with reduced insulin, however, did not correspond to statistically significant changes in individual cytokines measured within gonadal tissue homogenates (Fig. 4C). Although we did not observe the stereotypical increase in uncoupled protein 1 expression (Supplemental File S1) in our experimental mice, we noted differential expression of Ildr2, a gene associated with white adipose browning. Specifically, the expression level of Ildr2 has been noted to be significantly lower in BAT than in subcutaneous and visceral WAT depots (43). Furthermore, we noted an increase in neuregulin (Nrg)-2 gene expression within the predicted adipocyte source. Nrg2 belongs to the neuregulin family of proteins (Nrg1–4) which shares N-terminal epidermal growth factor (EGF)-like domains, which activates membrane-associated tyrosine kinases related to the EGF receptors (44). Nrg4 has been characterized as a WAT browning adipokine (43, 45), and its overexpression has been linked to reduced chronic inflammation in preventative and treatment studies (46). Nrg2, like Nrg4, has been implicated in neurite outgrowth and may increase adipose tissue innervation, which is necessary for the initiation of WAT lipolysis (47). The RNA-sequencing data and network analysis suggest the potential for interesting differences in adipose tissue function and composition, although, clearly, additional targeted loss-of-function studies are needed to establish if any of these gene products mediate the effects of reduced insulin on body weight.
TABLE 1.
Enriched pathways in the protein–protein network in gonadal fat from mice with reduced insulin
| Gene feature | FDR |
|---|---|
| Blood microparticle | 4.24E−09 |
| Lipid transporter activity | 1.73E−02 |
| Lipid transport | 2.62E−02 |
| Steroid binding | 2.62E−02 |
| Triglyceride-rich lipoprotein particle | 4.78E−02 |
| Very-low-density lipoproteins particle | 4.78E−02 |
| Neutral lipid metabolic process | 6.20E−02 |
| Response to virus | 6.20E−02 |
| Extracellular matrix | 6.20E−02 |
| Cyclin-dependent protein kinase holoenzyme complex | 6.20E−02 |
| Acylglycerol metabolic process | 6.20E−02 |
| Lipid localization | 6.20E−02 |
| Plasma lipoprotein particle | 6.51E−02 |
| Phospholipid binding | 6.97E−02 |
| Protein–lipid complex | 7.08E−02 |
| Intestinal absorption | 7.74E−02 |
| Adenylyltrasnferase activity | 7.74E−02 |
| Lipoprotein particle receptor binding | 8.96E−02 |
Our data provide the first molecular genetic evidence that obesity can be reversed by acutely and modestly reducing insulin production alone. The results of this treatment study, combined with our previous prevention studies (11, 12), clearly demonstrate the causal control of obesity by hyperinsulinemia in a genetically tractable mammalian system. The magnitude of insulin gene dose reduction achieved in our current study (∼30%) does not cause changes in glucose homeostasis and is attainable in human populations as a preventative measure or weight-loss strategy. For example, intermittent and continuous energy restriction over a 6-mo period resulted in similar reductions in circulating insulin in obese patients, coupled with a 5–10% loss in body weight (48). Although such dietary restriction interventions are often associated with high dropout rates (49), isocaloric restriction of daily fructose intake from 28 to 10% is an alternate approach that may mitigate high dropout rates and has successfully been shown to achieve similar reductions in circulating insulin and weight loss in obese adolescent patients (50). The magnitude of weight loss in our study, when translated to humans, has been associated with improvements to risk factors linked to cardiovascular disease, as well as reductions in the risk of developing type 2 diabetes by more than 50% (51).
CONCLUSIONS
Our results have profound implications for nutritional guidelines and therapeutic efforts to combat obesity and its comorbidities. Our findings support the concept that an individual’s hyperinsulinemia status should be the target for lifestyle modifications, macronutrient dietary composition, and drug development for the purposes of weight loss. Studies where individualized dietary responses are assessed (52) and mechanistically defined will be useful for the design of personalized nutrigenomic strategies. Approaches that physiologically modulate insulin levels may also have therapeutic utility beyond weight loss, including in the improvement of insulin sensitivity, and perhaps even longevity (53).
Supplementary Material
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
ACKNOWLEDGMENTS
The authors thank Farnaz Taghizadeh and Xiaoke (Betty) Hu [both with the University of British Columbia (UBC)] for assistance with islet isolations, perifusions, and animal work; the UBC Sequencing and Bioinformatics Consortium for RNA sequencing; the UBC Centre for High-Throughput Phenogenomics for micro-CT imaging; and John Schipilow and Dr. Nancy Ford (UBC) for technical support and assistance. The Centre is supported by the Canadian Foundation for Innovation, British Columbia Knowledge Development Foundation, and the UBC Faculty of Dentistry. Financial support was provided by an operating grant from the Canadian Institutes of Health Research; Canadian Diabetes Association postdoctoral fellowship awards (to M.M.P. and M.S.); a Canadian Diabetes Association Doctoral Student Research Award (to A.P.C.); a Benzon Foundation Fellowship (to S.S.); and the Juvenile Diabetes Research Foundation and Michael Smith Foundation for Health Research Postdoctoral Fellowship Awards (to G.E.L.). The authors declare no conflicts of interest.
Glossary
- BAT
brown adipose tissue
- Ccl
chemokine ligand
- DXA
dual-energy X-ray absorptiometry
- GFP
green fluorescent protein
- GIP
glucose-dependent insulinotropic peptide
- GLP
glucagon-like protein
- HFD
high-fat diet
- Hsl
hormone-sensitive lipase
- Ildr
Ig-like domain-containing receptor
- Ins
insulin
- Itih
inter-α-trypsin inhibitor heavy chain
- Lpl
lipoprotein lipase
- mTmG
membrane-targeted tdTomato/membrane-targeted enhanced green fluorescent protein
- NEFA
nonesterified fatty acid
- Nrg
neuregulin
- Pdx
pancreatic and duodenal homeobox, PYY, pancreatic polypeptide
- Ptrf
polymerase I and transcript release factor
- WAT
white adipose tissue
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
M. M. Page designed and performed experiments, analyzed results, and wrote the manuscript; S. Skovsø performed micro-CT scans and analysis; H. Cen, A. P. Chiu, and G. E. Lim performed Western blot analyses; D. A. Dionne and D. F. Hutchinson performed experiments; M. Szabat performed imaging experiments and analysis; S. Flibotte performed RNA sequencing analysis; S. Sinha performed RNA sequencing; and J. D. Johnson designed experiments, analyzed results, edited the manuscript, and is the guarantor of this work; and all other authors reviewed and edited the manuscript.
REFERENCES
- 1.World Health Organization . (2000) Obesity: preventing and managing the global epidemic. WHO Technical Report Series 894 [PubMed] [Google Scholar]
- 2.Nolan C. J., Ruderman N. B., Kahn S. E., Pedersen O., Prentki M. (2015) Insulin resistance as a physiological defense against metabolic stress: implications for the management of subsets of type 2 diabetes. Diabetes 64, 673–686 https://doi.org/10.2337/db14-0694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kitamura T., Kahn C. R., Accili D. (2003) Insulin receptor knockout mice. Annu. Rev. Physiol. 65, 313–332 https://doi.org/10.1146/annurev.physiol.65.092101.142540 [DOI] [PubMed] [Google Scholar]
- 4.Gray S. L., Donald C., Jetha A., Covey S. D., Kieffer T. J. (2010) Hyperinsulinemia precedes insulin resistance in mice lacking pancreatic beta-cell leptin signaling. Endocrinology 151, 4178–4186 https://doi.org/10.1210/en.2010-0102 [DOI] [PubMed] [Google Scholar]
- 5.Templeman N. M., Skovsø S., Page M. M., Lim G. E., Johnson J. D. (2017) A causal role for hyperinsulinemia in obesity. J. Endocrinol. 232, R173–R183 https://doi.org/10.1530/JOE-16-0449 [DOI] [PubMed] [Google Scholar]
- 6.Blüher M., Michael M. D., Peroni O. D., Ueki K., Carter N., Kahn B. B., Kahn C. R. (2002) Adipose tissue selective insulin receptor knockout protects against obesity and obesity-related glucose intolerance. Dev. Cell 3, 25–38 https://doi.org/10.1016/S1534-5807(02)00199-5 [DOI] [PubMed] [Google Scholar]
- 7.Blüher M., Kahn B. B., Kahn C. R. (2003) Extended longevity in mice lacking the insulin receptor in adipose tissue. Science 299, 572–574 https://doi.org/10.1126/science.1078223 [DOI] [PubMed] [Google Scholar]
- 8.Boucher J., Mori M. A., Lee K. Y., Smyth G., Liew C. W., Macotela Y., Rourk M., Bluher M., Russell S. J., Kahn C. R. (2012) Impaired thermogenesis and adipose tissue development in mice with fat-specific disruption of insulin and IGF-1 signalling. Nat. Commun. 3, 902 https://doi.org/10.1038/ncomms1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boucher J., Softic S., El Ouaamari A., Krumpoch M. T., Kleinridders A., Kulkarni R. N., O’Neill B. T., Kahn C. R. (2016) Differential roles of insulin and IGF-1 receptors in adipose tissue development and function. Diabetes 65, 2201–2213 https://doi.org/10.2337/db16-0212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Softic S., Boucher J., Solheim M. H., Fujisaka S., Haering M. F., Homan E. P., Winnay J., Perez-Atayde A. R., Kahn C. R. (2016) Lipodystrophy due to adipose tissue-specific insulin receptor knockout results in progressive NAFLD. Diabetes 65, 2187–2200 https://doi.org/10.2337/db16-0213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mehran A. E., Templeman N. M., Brigidi G. S., Lim G. E., Chu K. Y., Hu X., Botezelli J. D., Asadi A., Hoffman B. G., Kieffer T. J., Bamji S. X., Clee S. M., Johnson J. D. (2012) Hyperinsulinemia drives diet-induced obesity independently of brain insulin production. Cell Metab. 16, 723–737 https://doi.org/10.1016/j.cmet.2012.10.019 [DOI] [PubMed] [Google Scholar]
- 12.Templeman N. M., Clee S. M., Johnson J. D. (2015) Suppression of hyperinsulinaemia in growing female mice provides long-term protection against obesity. Diabetologia 58, 2392–2402 https://doi.org/10.1007/s00125-015-3676-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alemzadeh R., Karlstad M. D., Tushaus K., Buchholz M. (2008) Diazoxide enhances basal metabolic rate and fat oxidation in obese Zucker rats. Metabolism 57, 1597–1607 https://doi.org/10.1016/j.metabol.2008.06.017 [DOI] [PubMed] [Google Scholar]
- 14.Alemzadeh R., Langley G., Upchurch L., Smith P., Slonim A. E. (1998) Beneficial effect of diazoxide in obese hyperinsulinemic adults. J. Clin. Endocrinol. Metab. 83, 1911–1915 [DOI] [PubMed] [Google Scholar]
- 15.Lustig R. H., Greenway F., Velasquez-Mieyer P., Heimburger D., Schumacher D., Smith D., Smith W., Soler N., Warsi G., Berg W., Maloney J., Benedetto J., Zhu W., Hohneker J. (2006) A multicenter, randomized, double-blind, placebo-controlled, dose-finding trial of a long-acting formulation of octreotide in promoting weight loss in obese adults with insulin hypersecretion. Int. J. Obes. (Lond). 30, 331–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pocai A., Lam T. K., Gutierrez-Juarez R., Obici S., Schwartz G. J., Bryan J., Aguilar-Bryan L., Rossetti L. (2005) Hypothalamic K(ATP) channels control hepatic glucose production. Nature 434, 1026–1031 https://doi.org/10.1038/nature03439 [DOI] [PubMed] [Google Scholar]
- 17.Duvillié B., Cordonnier N., Deltour L., Dandoy-Dron F., Itier J. M., Monthioux E., Jami J., Joshi R. L., Bucchini D. (1997) Phenotypic alterations in insulin-deficient mutant mice. Proc. Natl. Acad. Sci. USA 94, 5137–5140 https://doi.org/10.1073/pnas.94.10.5137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szabat M., Johnson J. D., Piret J. M. (2010) Reciprocal modulation of adult beta cell maturity by activin A and follistatin. Diabetologia 53, 1680–1689 https://doi.org/10.1007/s00125-010-1758-0 [DOI] [PubMed] [Google Scholar]
- 19.Dror V., Nguyen V., Walia P., Kalynyak T. B., Hill J. A., Johnson J. D. (2007) Notch signalling suppresses apoptosis in adult human and mouse pancreatic islet cells. Diabetologia 50, 2504–2515 https://doi.org/10.1007/s00125-007-0835-5 [DOI] [PubMed] [Google Scholar]
- 20.Szabat M., Page M. M., Panzhinskiy E., Skovsø S., Mojibian M., Fernandez-Tajes J., Bruin J. E., Bround M. J., Lee J. T., Xu E. E., Taghizadeh F., O’Dwyer S., van de Bunt M., Moon K. M., Sinha S., Han J., Fan Y., Lynn F. C., Trucco M., Borchers C. H., Foster L. J., Nislow C., Kieffer T. J., Johnson J. D. (2016) Reduced insulin production relieves endoplasmic reticulum stress and induces β cell proliferation. Cell Metab. 23, 179–193 https://doi.org/10.1016/j.cmet.2015.10.016 [DOI] [PubMed] [Google Scholar]
- 21.Nasteska D., Harada N., Suzuki K., Yamane S., Hamasaki A., Joo E., Iwasaki K., Shibue K., Harada T., Inagaki N. (2014) Chronic reduction of GIP secretion alleviates obesity and insulin resistance under high-fat diet conditions. Diabetes 63, 2332–2343 https://doi.org/10.2337/db13-1563 [DOI] [PubMed] [Google Scholar]
- 22.Vetterli L., Carobbio S., Frigerio F., Karaca M., Maechler P. (2016) The amplifying pathway of the β-cell contributes to diet-induced obesity. J. Biol. Chem. 291, 13063–13075 https://doi.org/10.1074/jbc.M115.707448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaworski K., Sarkadi-Nagy E., Duncan R. E., Ahmadian M., Sul H. S. (2007) Regulation of triglyceride metabolism, IV: hormonal regulation of lipolysis in adipose tissue. Am. J. Physiol. Gastrointest. Liver Physiol. 293, G1–G4 https://doi.org/10.1152/ajpgi.00554.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Czech M. P., Tencerova M., Pedersen D. J., Aouadi M. (2013) Insulin signalling mechanisms for triacylglycerol storage. Diabetologia 56, 949–964 https://doi.org/10.1007/s00125-013-2869-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu L., Brown D., McKee M., Lebrasseur N. K., Yang D., Albrecht K. H., Ravid K., Pilch P. F. (2008) Deletion of Cavin/PTRF causes global loss of caveolae, dyslipidemia, and glucose intolerance. Cell Metab. 8, 310–317 https://doi.org/10.1016/j.cmet.2008.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayashi Y. K., Matsuda C., Ogawa M., Goto K., Tominaga K., Mitsuhashi S., Park Y. E., Nonaka I., Hino-Fukuyo N., Haginoya K., Sugano H., Nishino I. (2009) Human PTRF mutations cause secondary deficiency of caveolins resulting in muscular dystrophy with generalized lipodystrophy. J. Clin. Invest. 119, 2623–2633 https://doi.org/10.1172/JCI38660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hill M. M., Bastiani M., Luetterforst R., Kirkham M., Kirkham A., Nixon S. J., Walser P., Abankwa D., Oorschot V. M., Martin S., Hancock J. F., Parton R. G. (2008) PTRF-Cavin, a conserved cytoplasmic protein required for caveola formation and function. Cell 132, 113–124 https://doi.org/10.1016/j.cell.2007.11.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Briand N., Prado C., Mabilleau G., Lasnier F., Le Lièpvre X., Covington J. D., Ravussin E., Le Lay S., Dugail I. (2014) Caveolin-1 expression and cavin stability regulate caveolae dynamics in adipocyte lipid store fluctuation. Diabetes 63, 4032–4044 https://doi.org/10.2337/db13-1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parton R. G., Hanzal-Bayer M., Hancock J. F. (2006) Biogenesis of caveolae: a structural model for caveolin-induced domain formation. J. Cell Sci. 119, 787–796 https://doi.org/10.1242/jcs.02853 [DOI] [PubMed] [Google Scholar]
- 30.Azizi P. M., Zyla R. E., Guan S., Wang C., Liu J., Bolz S. S., Heit B., Klip A., Lee W. L. (2015) Clathrin-dependent entry and vesicle-mediated exocytosis define insulin transcytosis across microvascular endothelial cells. Mol. Biol. Cell 26, 740–750 https://doi.org/10.1091/mbc.E14-08-1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boothe T., Lim G. E., Cen H., Skovsø S., Piske M., Li S. N., Nabi I. R., Gilon P., Johnson J. D. (2016) Inter-domain tagging implicates caveolin-1 in insulin receptor trafficking and Erk signaling bias in pancreatic beta-cells. Mol. Metab. 5, 366–378 https://doi.org/10.1016/j.molmet.2016.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen A. W., Schubert W., Brasaemle D. L., Scherer P. E., Lisanti M. P. (2005) Caveolin-1 expression is essential for proper nonshivering thermogenesis in brown adipose tissue. Diabetes 54, 679–686 https://doi.org/10.2337/diabetes.54.3.679 [DOI] [PubMed] [Google Scholar]
- 33.Haemmerle G., Zimmermann R., Strauss J. G., Kratky D., Riederer M., Knipping G., Zechner R. (2002) Hormone-sensitive lipase deficiency in mice changes the plasma lipid profile by affecting the tissue-specific expression pattern of lipoprotein lipase in adipose tissue and muscle. J. Biol. Chem. 277, 12946–12952 https://doi.org/10.1074/jbc.M108640200 [DOI] [PubMed] [Google Scholar]
- 34.Björntorp P., Karlsson M., Pertoft H., Pettersson P., Sjöström L., Smith U. (1978) Isolation and characterization of cells from rat adipose tissue developing into adipocytes. J. Lipid Res. 19, 316–324 [PubMed] [Google Scholar]
- 35.Klemm D. J., Leitner J. W., Watson P., Nesterova A., Reusch J. E., Goalstone M. L., Draznin B. (2001) Insulin-induced adipocyte differentiation: activation of CREB rescues adipogenesis from the arrest caused by inhibition of prenylation. J. Biol. Chem. 276, 28430–28435 https://doi.org/10.1074/jbc.M103382200 [DOI] [PubMed] [Google Scholar]
- 36.Svensson M., Agace W. W. (2006) Role of CCL25/CCR9 in immune homeostasis and disease. Expert Rev. Clin. Immunol. 2, 759–773 https://doi.org/10.1586/1744666X.2.5.759 [DOI] [PubMed] [Google Scholar]
- 37.Xue B., Sukumaran S., Nie J., Jusko W. J., Dubois D. C., Almon R. R. (2011) Adipose tissue deficiency and chronic inflammation in diabetic Goto-Kakizaki rats. PLoS One 6, e17386 https://doi.org/10.1371/journal.pone.0017386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheverud J. M., Lawson H. A., Fawcett G. L., Wang B., Pletscher L. S., R Fox A., Maxwell T. J., Ehrich T. H., Kenney-Hunt J. P., Wolf J. B., Semenkovich C. F. (2011) Diet-dependent genetic and genomic imprinting effects on obesity in mice. Obesity (Silver Spring) 19, 160–170 https://doi.org/10.1038/oby.2010.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lawson H. A., Lee A., Fawcett G. L., Wang B., Pletscher L. S., Maxwell T. J., Ehrich T. H., Kenney-Hunt J. P., Wolf J. B., Semenkovich C. F., Cheverud J. M. (2011) The importance of context to the genetic architecture of diabetes-related traits is revealed in a genome-wide scan of a LG/J × SM/J murine model. Mamm. Genome 22, 197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ebana Y., Ozaki K., Inoue K., Sato H., Iida A., Lwin H., Saito S., Mizuno H., Takahashi A., Nakamura T., Miyamoto Y., Ikegawa S., Odashiro K., Nobuyoshi M., Kamatani N., Hori M., Isobe M., Nakamura Y., Tanaka T. (2007) A functional SNP in ITIH3 is associated with susceptibility to myocardial infarction. J. Hum. Genet. 52, 220–229 https://doi.org/10.1007/s10038-006-0102-5 [DOI] [PubMed] [Google Scholar]
- 41.Zhu Y., Crewe C., Scherer P. E. (2016) Hyaluronan in adipose tissue: beyond dermal filler and therapeutic carrier. Sci. Transl. Med. 8, 323ps4 https://doi.org/10.1126/scitranslmed.aad6793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang L., Lantier L., Kennedy A., Bonner J. S., Mayes W. H., Bracy D. P., Bookbinder L. H., Hasty A. H., Thompson C. B., Wasserman D. H. (2013) Hyaluronan accumulates with high-fat feeding and contributes to insulin resistance. Diabetes 62, 1888–1896 https://doi.org/10.2337/db12-1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosell M., Kaforou M., Frontini A., Okolo A., Chan Y. W., Nikolopoulou E., Millership S., Fenech M. E., MacIntyre D., Turner J. O., Moore J. D., Blackburn E., Gullick W. J., Cinti S., Montana G., Parker M. G., Christian M. (2014) Brown and white adipose tissues: intrinsic differences in gene expression and response to cold exposure in mice. Am. J. Physiol. Endocrinol. Metab. 306, E945–E964 https://doi.org/10.1152/ajpendo.00473.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buonanno A., Fischbach G. D. (2001) Neuregulin and ErbB receptor signaling pathways in the nervous system. Curr. Opin. Neurobiol. 11, 287–296 https://doi.org/10.1016/S0959-4388(00)00210-5 [DOI] [PubMed] [Google Scholar]
- 45.Christian M. (2014) Transcriptional fingerprinting of “browning” white fat identifies NRG4 as a novel adipokine. Adipocyte 4, 50–54 https://doi.org/10.4161/adip.29853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma Y., Gao M., Liu D. (2016) Preventing high fat diet-induced obesity and improving insulin sensitivity through neuregulin 4 gene transfer. Sci. Rep. 6, 26242 https://doi.org/10.1038/srep26242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bartness T. J., Liu Y., Shrestha Y. B., Ryu V. (2014) Neural innervation of white adipose tissue and the control of lipolysis. Front. Neuroendocrinol. 35, 473–493 https://doi.org/10.1016/j.yfrne.2014.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harvie M. N., Pegington M., Mattson M. P., Frystyk J., Dillon B., Evans G., Cuzick J., Jebb S. A., Martin B., Cutler R. G., Son T. G., Maudsley S., Carlson O. D., Egan J. M., Flyvbjerg A., Howell A. (2011) The effects of intermittent or continuous energy restriction on weight loss and metabolic disease risk markers: a randomized trial in young overweight women. Int. J. Obes. (Lond.) 35, 714–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwingshackl L., Dias S., Hoffmann G. (2014) Impact of long-term lifestyle programmes on weight loss and cardiovascular risk factors in overweight/obese participants: a systematic review and network meta-analysis. Syst. Rev. 3, 130 https://doi.org/10.1186/2046-4053-3-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lustig R. H., Mulligan K., Noworolski S. M., Tai V. W., Wen M. J., Erkin-Cakmak A., Gugliucci A., Schwarz J. M. (2016) Isocaloric fructose restriction and metabolic improvement in children with obesity and metabolic syndrome. Obesity (Silver Spring) 24, 453–460 https://doi.org/10.1002/oby.21371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wharton S. (2016) Current perspectives on long-term obesity pharmacotherapy. Can. J. Diabetes 40, 184–191 https://doi.org/10.1016/j.jcjd.2015.07.005 [DOI] [PubMed] [Google Scholar]
- 52.Zeevi D., Korem T., Zmora N., Israeli D., Rothschild D., Weinberger A., Ben-Yacov O., Lador D., Avnit-Sagi T., Lotan-Pompan M., Suez J., Mahdi J. A., Matot E., Malka G., Kosower N., Rein M., Zilberman-Schapira G., Dohnalová L., Pevsner-Fischer M., Bikovsky R., Halpern Z., Elinav E., Segal E. (2015) Personalized nutrition by prediction of glycemic responses. Cell 163, 1079–1094 https://doi.org/10.1016/j.cell.2015.11.001 [DOI] [PubMed] [Google Scholar]
- 53.Templeman N. M., Flibotte S., Chik J. H. L., Sinha S., Lim G. E., Foster L. J., Nislow C., Johnson J. D. (2017) Reduced circulating insulin enhances insulin sensitivity in old mice and extends lifespan. Cell Rep. 20, 451–463 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.