Abstract
The development of novel neuroprotective treatments for acute stroke has been fraught with failures, which supports the view of ischemic brain damage as a highly complex multifactorial process. Post-translational modifications such as small ubiquitin-like modifier (SUMO)ylation have emerged as critical molecular regulatory mechanisms in states of both homeostasis and ischemic stress, as evidenced by our previous work. Accordingly, the clinical significance of the selective control of the global SUMOylation process has become apparent in studies of ischemic pathobiology and pathophysiology. Herein, we describe a process capable of identifying and characterizing small molecules with the potential of targeting the SUMO system through inhibition of SUMO deconjugation in an effort to develop novel stroke therapies.—Bernstock, J. D., Ye, D., Smith, J. A., Lee, Y.-J., Gessler, F. A., Yasgar, A., Kouznetsova, J., Jadhav, A., Wang, Z., Pluchino, S., Zheng, W., Simeonov, A., Hallenbeck, J. M., Yang, W. Quantitative high-throughput screening identifies cytoprotective molecules that enhance SUMO-conjugation via the inhibition of SUMO-specific protease (SENP)2.
Keywords: drug repurposing, ischemia, neuroprotection, stroke, SUMOylation
Cerebrovascular accidents continue to be a leading cause of both morbidity and mortality worldwide, posing an increasingly severe burden on society (1). Despite the many basic and translational studies and clinical trials (2, 3), therapeutic approaches to modulating the evolving pathobiology of ischemic stroke beyond acute thrombolysis and mechanical thrombectomy have not found their way into treatment strategies (4–6). The limited success of such massive research investments demands a reevaluation of the underlying pathobiology in an effort to identify novel therapeutic targets and translational approaches.
The pathology of stroke is complex and involves many cellular processes. Ischemic stroke, most often caused by a thromboembolism that occludes a cerebral artery, can lead to focal brain ischemia, cell death, and, ultimately, sensory, motor, and cognitive impairments (7). Treatments targeting neuroprotection and intending to prevent salvageable neurons from dying have uniformly failed in clinical trials despite showing efficacy in experimental stroke studies (8, 9). Furthermore, no treatment or combination of treatments yet explored have been able to address the entirety of damaging processes that occur during or after ischemic stroke (e.g., excitotoxicity, inflammation, and apoptosis) (7, 9).
The complexity of ischemic stroke demands therapies that simultaneously engage multiple axes involved in the pathobiology and pathophysiology of ischemia (2, 10). Accordingly, a body of evidence has arisen to support that small ubiquitin-like modifier (SUMO)ylation, a dynamic post-translational modification, may be a relevant therapeutic target in ischemic stroke (11, 12). Protein SUMOylation has been documented to play a role in numerous homeostatic processes throughout the cell, including signal transduction, chromatin remodeling, gene expression, and protein quality control (13–15). In brief, SUMO in mammals is found as 4 distinct systemically distributed paralogues: SUMO-2 and -3, sharing 96% homology; SUMO-1, sharing only 45% homology with SUMO-2 and -3; and the recently identified but ill-characterized SUMO-4, sharing 86% homology with SUMO-2 (16, 17). Like its namesake ubiquitin, SUMO is synthesized as an inactive precursor that is maturated by the endopeptidase activity of SUMO-specific proteases (SENPs) to expose a C-terminal di-glycine (GG) motif (14). Conjugation begins when the heterodimeric E1 enzyme (SUMO-activating enzyme-1/2) adenylates SUMO in an ATP-dependent process to form a covalent thioester E1-SUMO intermediate, which is then transferred to the catalytic cysteine of Ubc9, the sole E2 conjugase. Ubc9, often with the assistance of a target-specific E3 ligase, catalyzes the formation of an isopeptide linkage between the C-terminal GG motif of SUMO and the ε-amino-group lysine of the target substrate (14, 15). The consequences of SUMO conjugation include the modulation of protein interactions (e.g., providing SUMO interacting motifs), alterations in target protein stability (e.g., recruiting SUMO-targeted ubiquitin ligases or hindering ubiquitin conjugation), and regulation of activity (e.g., inducing changes in protein conformation) (13, 15).
Based on a substantial body of evidence, it is now believed that SUMOylation is an endogenous neuroprotective mechanism (12, 18–27). For example, in vitro studies have shown that silencing SUMOs sensitizes primary neurons to oxygen-glucose deprivation (OGD)-induced damage, whereas overexpression of SUMO in cortical neurons induced OGD tolerance (12). Furthermore, compared to wild-type mice, mice overexpressing Ubc9 demonstrated improved resistance to focal ischemia–induced damage (23), and transgenic mice in which SUMO-1–3 were knocked down in neurons displayed worse functional outcomes after transient forebrain ischemia (24). SUMOylation is increased during animal hibernation and clinically relevant hypothermia (12, 18, 25, 26), and it has been proposed that increased global SUMOylation is a key mechanism underlying hypothermia-induced protection (12, 18, 21, 27). Taken together, increasing global SUMOylation is a promising new clinical strategy for protecting the brain against ischemic damage.
Accordingly, we recently explored alternative approaches in an effort to increase SUMOylation, and identified specific inhibitors of miR-182 or -183 (28). The rationale for this study was based on the fact that levels of the miR-200 family and the miR-182 family decrease within the squirrel brain during the torpor phase when global SUMOylation is increased, and further, that inhibiting these microRNAs serves to increase global SUMOylation in cell culture (28, 29). A number of compounds were identified that could increase global SUMOylation and ultimately provide protective effects after OGD/restoration of oxygen and glucose in vitro (28).
Conjugation of SUMO, however, is not the only axis by which SUMOylation may be targeted; increases in global SUMOylation can also be achieved by inhibiting SUMO-deconjugation; SUMOylation is a highly dynamic process and is balanced through deconjugating activity (i.e., isopeptidase activity) of the various SENPs, comprising SENP1–3 and -5–7 in mammals (15, 30). In vitro, SENP1 and -2 demonstrate broad specificity for all SUMO paralogues, with the latter bearing a preference for SUMO-2/3; both also process SUMO precursors and do not edit SUMO-2/3 chains (31). However, it has been suggested that SENP1 may be specific for SUMO-1-conjugated proteins in vivo (32). SENP3 and -5–7 have a preference for SUMO-2/3, with only SENP5 being able to process SUMO precursors; and SENP6 and -7 being capable of chain editing (31). Among the SENPs, SENP1 and -2 have the highest catalytic activity (33).
It is prudent to note that the role of the SENPs in neuroprotection remains controversial, and that although the current body of knowledge suggests that SENP3 may be an ideal target for therapies centered on decreasing SUMO deconjugation (34–36), technical challenges preclude obtaining sufficient quantities of stable or active SENP3 catalytic domain for use in high-throughput screening (37). Given this technical limitation and the knowledge that deletion of SENP2 in vivo dramatically increases both SUMO-1 and -2/3 conjugation (38), we chose to use SENP2 and its catalytic domain as our screening targets.
Accordingly, herein we describe a process capable of identifying and characterizing approved agents that inhibit SENP2, thus bearing the potential to modulate SUMO pathways in ischemic stroke. The validity of the quantitative high-throughput screen (qHTS), a titration-based approach for the rapid testing of small-molecule libraries, was confirmed using orthogonal assays comprising immunoblot analysis, target engagement, and the induction of neuroprotection during OGD.
MATERIALS AND METHODS
Compounds
All compounds were initially sourced as screening libraries from the National Center for Advancing Translational Sciences (NCATS)/U.S. National Institutes of Health (NIH). For follow-up studies, 3,3′-methylene-bis(4-hydroxycoumarin) (dicumarol), protocatechuic acid ethyl ester (ethyl protocatechuate), tenatoprazole, isoprenaline hydrochloride, acriflavine hydrochloride, and DMSO were purchased from Millipore-Sigma (Billerica, MA, USA), and ebselen was procured from Enzo Life Sciences (Farmingdale, NY, USA).
Antibodies
Anti-SUMO-2/3 and anti-SUMO-1 rabbit polyclonal antibodies were developed in-house (NIH) (39). Anti-SENP2 (H-300) rabbit polyclonal IgG was purchased from Santa Cruz Biotechnology (Dallas, TX, USA). Anti-β-actin mouse monoclonal IgG was purchased from Millipore-Sigma.
qHTS AlphaScreen assay miniaturization, primary screen, and hit validation
The AlphaScreen (PerkinElmer, Waltham, MA, USA) assay used in this study was established within our laboratory and has been described in detail (40). The assay uses a recombinant substrate, SS3HS2 [His-SUMO2(K11R) conjugated to Strep-SUMO3ΔGG], that resembles a physiologically relevant SUMOylated protein (SUMO-2 conjugated to SUMO-3) (Fig. 1A). The spatial proximity between the His-tag and Strep-tag within the intact SS3HS2 recombinant protein allows for excitation at 680 nm and the generation of an emission signal at 520–600 nm via the transfer of singlet oxygen [∆·O2] from the Strep-tactin donor beads to the nickel chelate acceptor beads. If substrate SS3HS2 is ultimately cleaved by an SENP, the distance between the His-tag and Strep-tag is increased, thereby resulting in a dramatic decrease in emission signal. A small-molecule inhibitor of SENP prevents substrate cleavage, maintaining signal and allowing the identification of putative hits and lead compounds. In this study, our AlphaScreen assay was further optimized for use in low-volume settings (384- and 1536-well, white, solid-bottom plates; Greiner Bio-One, Monroe, NC, USA). The human SENP2 catalytic domain (hSENP2c) was purchased from Enzo Life Sciences (BML-UW9765-0100), and the substrate SS3HS2 was produced as has been described (40). The final concentrations of enzyme and substrate were determined by performing a substrate titration in the 384-well format at a fixed bead concentration of 5 µg/ml (Supplemental Fig. 1A; mixture of Strep-Tactin donor and (His)6-nickel chelate acceptor beads). Based on these results, we moved forward with a 100 nM concentration of substrate and performed an enzyme titration and time course in the 1536-well format (Supplemental Fig. 1B). An enzyme concentration of 10 nM and an incubation time of 30 min were chosen to balance the need for a low concentration of reagents and a large signal window. The miniaturized assay (final volume, 4 µl) used the following protocol. First, 2 µl of hSENP2c (final concentration 10 nM) or buffer [25 mM hydroxyethyl piperazineethanesulfonic acid HEPES (pH 7.5) and 100 mM NaCl with 0.01% Tween 20] were dispensed into a 1536-well, solid-bottom, white plate (Greiner Bio One). Then, SUMO2-aldehyde (an irreversible SENP inhibitor; final concentration 65.4–143 nM) control, compounds, or DMSO neutral control were pin transferred (23 nl; Wako Automation, Richmond, VA, USA) and incubated for 15 min at room temperature, followed by addition of 1 µl of SS3HS2 substrate (final concentration 100 nM). After a 15 min incubation, a 1 µl mixture of Strep-Tactin donor and (His)6-nickel chelate acceptor beads (final concentration 5 µg/ml; PerkinElmer) was dispensed, and samples were incubated for additional 30 min at room temperature, followed by a read on an EnVision plate reader with the 1536 plate HTS AlphaScreen aperture (AlphaScreen optics; both from PerkinElmer). The assay showed minimal well-to-well variation and a Z′ factor > 0.5. Further, the assay reagents, as formulated at the working concentrations, were stable overnight, thereby allowing an unattended robotic screening (Supplemental Fig. 1C). SUMO2-aldehyde exhibited a concentration-dependent inhibition with an IC50 of ∼11 nM (Supplemental Fig. 1D), consistent with our previous results (40). The above assay was used in a screen against a collection of 4096 samples contained within both the Library of Pharmaceutically Active Compounds (LOPAC1280; Millipore-Sigma) and the NIH Chemical Genomics Center (NCGC) Pharmaceutical Collection (NPC) compound library (41). Each compound was tested as a 5- or 7-point dilution series, with concentrations ranging from 3.6 nM to 57.2 µM.
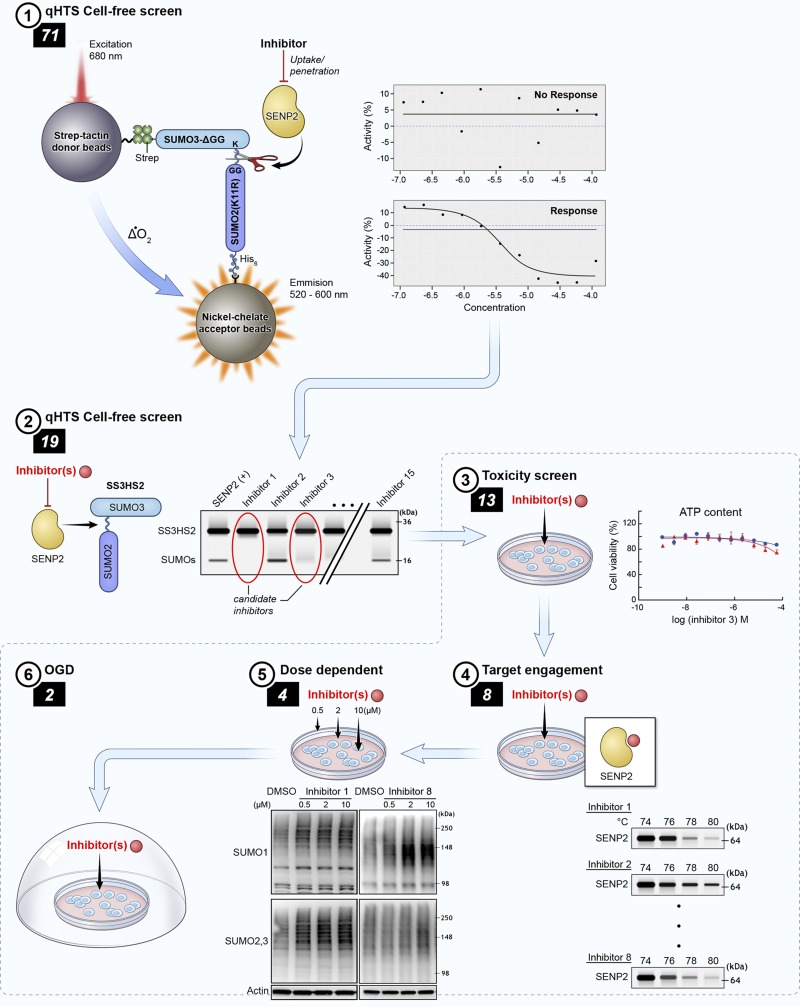
Figure 1.
Compound triaging paradigm. A qHTS (AlphaScreen) (1) identified 71 putative inhibitors of SENP2 from the NPC and LOPAC1280 libraries. These compounds were taken into a cell-free hSENP2 catalytic activity assay (2), and 19 were identified as being total inhibitors of SENP2 catalytic activity. A toxicity screen (3) of those 19 compounds in B35 rat neuroblastoma cells identified 6 that were cytotoxic at 18 h. The 13 remaining nontoxic compounds were tested for target engagement with SENP2 using CETSA in total cell lysates (4); 8 were determined to be engagers. B35 cells were treated with those 8 compounds at doses of 0.5, 2, and 10 μM, and the levels of SUMO-1 and -2/3 conjugation were measured with Western blot analysis (5) to confirm in vitro upregulation of SUMO conjugation via inhibition of SENP2 activity by 4 compounds that were then used to treat rat B35 cells in an in vitro stroke model (OGD) (6).
Cell-free SENP activity assays
A cell-free SENP2 activity assay was used to confirm the inhibitory efficacy of the 71 compounds selected from the AlphaScreen on the isopeptidase activity of SENP2 (40). hSENP2c at 2 nM was incubated with compounds at 100 μM before the addition of SS3HS2 at 400 nM. Results were visualized via immunoblot analysis. Total inhibition of hSENP2c activity was defined as the absence of a cleaved SUMO band at ∼18 kDa. The same method was used to explore the capacity of those compounds identified as noncytotoxic total inhibitors (i.e., 13 compounds) of hSENP2c with regard to the human SENP1 catalytic domain (hSENP1c) (Boston Biochem, Cambridge, MA, USA). Glutathione S-transferase controls were not used in our cell-free SENP activity assay.
Cell culture
The rat neuroblastoma cell line B35 was cultured in DMEM (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Atlanta Biologicals, Flowery Branch, GA, USA), 100 U/ml penicillin, and 100 mg/ml streptomycin in 5% CO2 at 37°C. Of note, the B35 cell line was confirmed to be Mycoplasma negative with the MycoAlert Mycoplasma Detection Kit (Lonza, Basel, Switzerland).
Cytotoxicity
An ATP-content assay using a luminescent readout was used to measure the cytotoxicity of the 19 total inhibitors of hSENP2c in B35 neuroblastoma cells. Compounds identified as hits in the primary assay were dissolved in DMSO as 10 mM stock solutions, and plated into a 1536-well plate as a 3-fold, 11-point serial dilution, ranging from 57.5 µM to 236.6 nM (final concentration). B35 neuroblastoma cells were dispensed into 1536-well, white, collagen-coated plates (Corning, Corning, NY, USA), at a total of 2000 cells per well in 4 µl medium with a Multidrop Combi dispenser (Thermo Fisher Scientific). After a 16 h incubation at 5% CO2 and 37°C, 23 nl of compounds were transferred to the assay plates via a NX-TR pintool station (Wako Scientific Solutions, San Diego, CA, USA), resulting in a 173.9-fold dilution. Cells were incubated with the compounds for an additional 18 or 24 h in a humidified incubator at 5% CO2 and 37°C. Cellular ATP levels were quantitated by adding 4 µl of detection reagent (CellTiter-Glo; Promega, Madison, WI, USA), and the resultant luminescence was detected on the ViewLux plate reader (PerkinElmer). The activity of each compound was normalized to vehicle control wells (no killing) and 100 µM tamoxifen (100% killing). IC50 values were calculated from dose-response curves using Prism software (GraphPad Prism, La Jolla, CA, USA).
CETSA
CETSA was used to investigate target engagement of the 13 nontoxic compounds with SENP2, as described by Almqvist et al. (42), with some modifications. In brief, 5 × 106 cells/ml were suspended in DMEM without glucose and phenol red (Thermo Fisher Scientific), lysed via ultrasonication on ice, and treated with 100 μM of compound in 0.1% DMSO for 1 h at 37°C with mixing every 10–15 min. The cell suspensions were then aliquoted into 100-μl volumes, which were heated for 3.5 min at the appropriate temperatures in a 96-well Veriti thermal cycler (Thermo Fisher Scientific). Samples were then centrifuged at 15,000 g for 15 min at 4°C to separate stable and denatured proteins. The supernatants were transferred and mixed with an equal volume of 2× Laemmli loading buffer and 10% 2-ME, and incubated for 10 min at 90°C. Proteins were separated on 4–20% Tris-glycine gels (Thermo Fisher Scientific) and transferred to PVDF membranes (Thermo Fisher Scientific). Images were taken with the FluorChem system (Protein Simple, San Jose, CA, USA). Protein expression levels were determined via densitometric analysis of the bands corresponding to the proteins of interest by using ImageJ (NIH). Target engagement of compounds was evaluated quantitatively as a significant increase in remaining SENP2 protein at temperatures equal to or greater than the melting temperature vs. the DMSO control.
Molecular docking calculations
Ligand models were developed using Avogadro (v1.2.0) (43) and converted to a protein data bank, partial charge (q), and atom type (T) format using AutoDockTools (v.1.5.6) (44). Crystal structure 2IO2, human SENP2 in complex with RanGAP1-SUMO1 (45), was obtained from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (46). The RanGAP1-SUMO1 conjugate was deleted before docking. Docking calculations were performed using AutoDock Vina (v.1.1.2) (47) with the Vina Control and Post Processing Tools extension (http://www.biochemsolutions.com). A cubic search space of 30 × 30 × 30 Å was used, centered on the reported catalytic site of the target enzyme (delimited by residues Trp410, His478, Gln542, and C548S). Lowest-energy ligand poses were rendered using PyMOL (v.0.99rc6; DeLano Scientific, Palo Alto, CA, USA) and intermolecular interactions analyzed using Discovery Studio Visualizer (v.4.5; Biovia, San Diego, CA, USA).
Western blot analysis
The 8 compounds that were determined to be engagers by CETSA were assessed in vitro in the B35 cell line. In brief, B35 cells were plated at 5 × 105 cells per well into 6-well collagen-coated tissue culture plates and allowed to adhere and grow overnight before treatment with compounds in 0.2% DMSO for 18 h. Growth medium was removed, and the cells were gently washed with PBS before being lysed [100 mM Tris-Cl (pH 7.4), 2% SDS, 50 mM EDTA, and 20% glycerol] and boiled for 10 min at 95°C. Samples were subsequently sonicated on ice and incubated at 95°C for 5 min. Protein concentrations were normalized using the Pierce Bicinchoninic Protein Assay (Thermo Fisher Scientific) before separation on 4–20% Tris-Glycine gels and transfer to PVDF membranes. Images were taken with the FluorChem system (Protein Simple). Protein expression levels were determined via densitometric analysis of the bands corresponding to the proteins of interest using ImageJ. All densities were normalized to the corresponding β-actin levels and expressed as the ratio to the control (DMSO alone).
Oxygen/glucose deprivation and the assessment of cell death
The 4 compounds that were determined to be engagers by CETSA and were capable of upregulating SUMOylation were assessed for their ability to protect cells from OGD–induced cell death. OGD was performed as previously described, with slight modifications (48). We subjected B35 neuroblastoma cells that had been plated at 7.5 × 105 cells/well to 20 h of OGD; cells were incubated with compounds in 0.2% DMSO throughout the 4 h preconditioning and 20 h OGD periods. Induction of OGD was executed by replacing the basal culture medium with DMEM lacking both glucose and sodium pyruvate (Thermo Fisher Scientific) followed by incubation in a hypoxic incubator containing 95% N2, 5% CO2, and <0.1% O2. Oxygen levels were maintained at 0.1% or less using a ProOx 110 compact oxygen controller (BioSpherix, Parish, NY, USA). After hypoxic incubation, cells were detached with papain after washing with 1 mM EDTA in PBS. Cell death was assessed via nuclear staining with Hoechst 33342 and propidium iodide followed by fluorescence-activated cell sorting analysis; typically, 5 × 104 events were captured. The percentages of vital or apoptotic and necrotic cells in compound-treatment conditions were normalized to those in the DMSO condition to derive a fold difference for survival.
Animals
Animal experiments were approved by the Duke University Animal Care and Use Committee. In brief, 1 h after C57/BL6 mice were injected with ebselen (at 12 mg/kg, i.p.) or vehicle DMSO, their cortical brain tissue was extracted and snap frozen and ultimately evaluated via Western blotting for levels of global SUMOylation.
Statistical analyses
Unless otherwise noted, Student’s t test, or 1-way ANOVA, or both, with Dunnett’s post hoc correction as appropriate, were used to test the significance of differences between conditions, as detailed in figure legends. Values of P ≤ 0.05 were deemed statistically significant.
RESULTS
qHTS identifies inhibitors of SENP2
We set out to identify compounds capable of upregulating SUMOylation through the inhibition of the deconjugating SUMO-specific isopeptidase SENP2. To accomplish this identification, we used the experimental workflow and compound triage outlined in Fig. 1, which was centered on hits obtained with a primary qHTS assay with the AlphaScreen format (40, 49). We screened the NCATS NPC and Sigma LOPAC1280 libraries, which consisted of 4096 compounds, where 166 compounds were identified as hits (Supplemental Table 1). Of the 166 compounds, 99 exhibited inhibition in at least 1 of the incubation times used during confirmatory screening, and of those 71 were inactive within the counterscreen and were therefore chosen for further testing.
Select compounds inhibit hSENP2c and are nontoxic
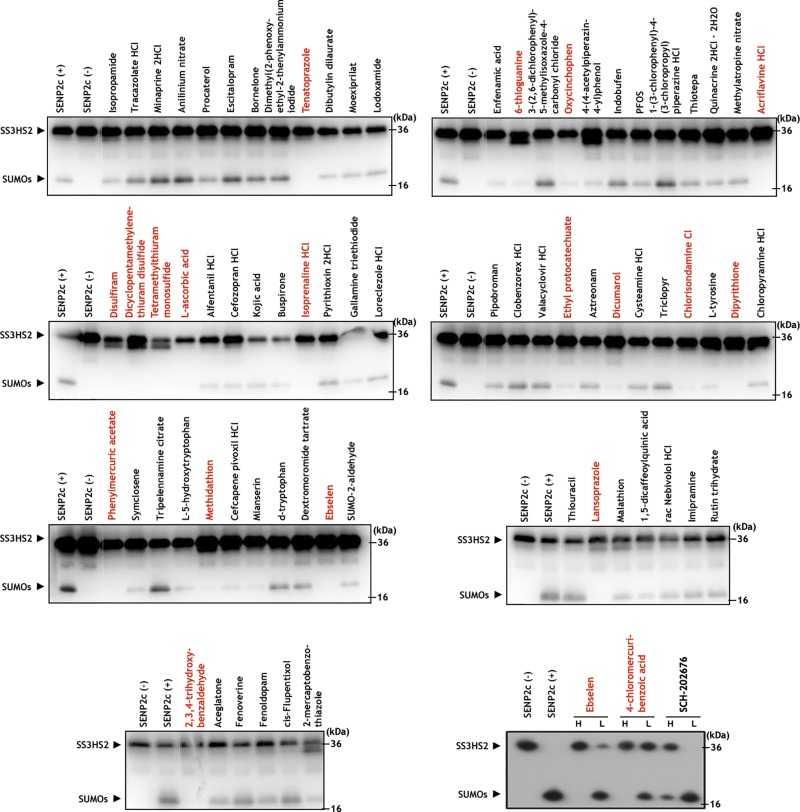
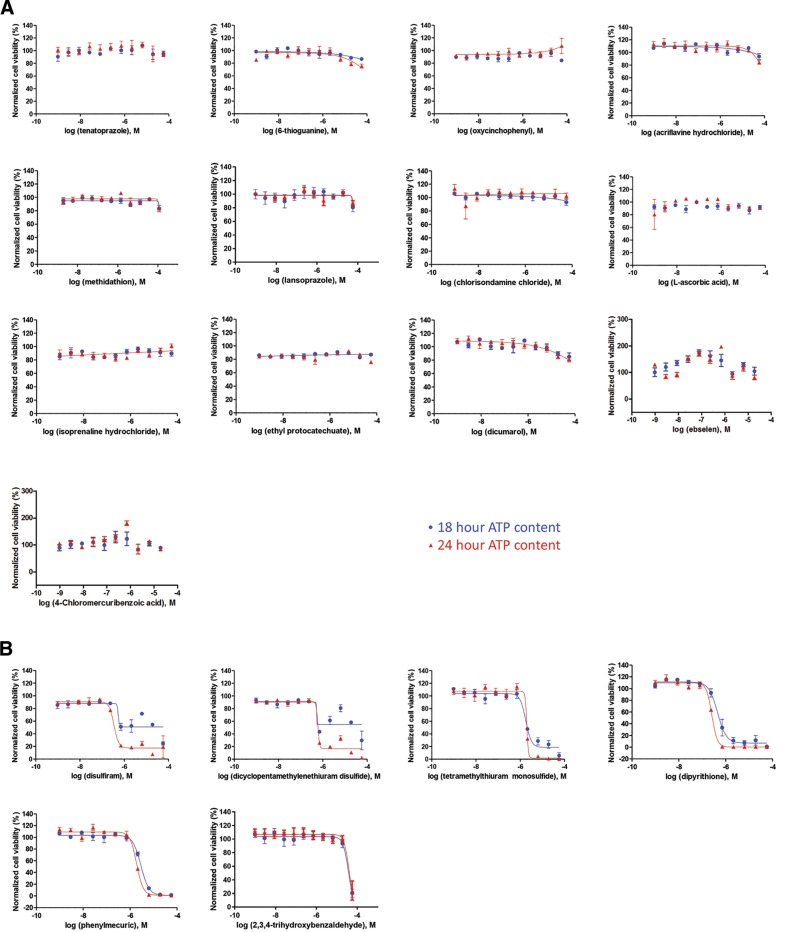
To determine the effect of the active compounds identified via the primary screen on the inhibition of SENP2 activity, the 71 compounds were tested in an orthogonal cell-free assay with the recombinant substrates SS3HS2 and hSENP2c. hSENP2c was incubated with compounds at 100 μM before the addition of SS3HS2 and results of the experiment were visualized by Western blot analysis with a SUMO-2/3 antibody. Total inhibition of hSENP2c activity was defined as the absence of a cleaved SUMO band at ∼18 kDa. We demonstrated that 19 of the 71 hits effected complete inhibition of the catalytic domain (Fig. 2). To exclude the confounding contributions of compound cytotoxicity downstream, a cell-viability assay was performed in parallel by using measurement of ATP levels. We found that 6 compounds displayed cytotoxicity at either 18 or 24 h of incubation (Fig. 3). Cells were treated with the compounds for an equivalent amount of time when the levels of global SUMOylation were ultimately explored via Western blot analysis. Of note, six of the compounds shown to inhibit SENP2 were also determined to be inhibitors of the human SENP1 catalytic domain, suggesting that this screening approach can be used to identify inhibitors of all SENPs (i.e., pan-SENP inhibitors) (Supplemental Fig. 2). This finding is most likely attributable to partial conservation of the C-terminal catalytic regions among the SENPs, being that the SENP2 catalytic fragment was used as the screening target during the qHTS (31).
Figure 2.
A cell-free assay for inhibition of SENP2 catalytic activity. Final concentration of all compounds was 100 μM. The band corresponding to the SS3HS2 conjugate was visualized at ∼36 kDa. Total inhibition of hSENP2c was defined as the qualitatively determined near-total absence of the band corresponding to cleaved free SUMO products, visualized at ∼18 kDa. The following compounds were identified as total inhibitors of hSENP2c activity: tenatoprazole, oxycinchophen, acriflavine hydrochloride, 6-thioguanine, disulfiram, dicyclopentamethylenethiuram disulfide, tetramethythiuram monosulfide, l-ascorbic acid, isoprenaline hydrochloride, ethyl protocatechuate, dicumarol, chlorisondamine chloride, dipyrithione, phenylmercuric acetate, methidathion, lansoprazole, 2,3,4-trihydroxybenzaldehyde, ebselen, and 4-chloromercuribenzoic acid.
Figure 3.
Determination of compound toxicity. Cell viability curves for the 19 compounds identified as total inhibitors of hSENP2 catalytic activity in the cell-free assay. Viability of rat B35 neuroblastoma cells at a given dose of compound was determined through measurement of ATP content at 18 and 24 h after treatment, and response curves were generated by plotting the negative logarithm of dose concentration against percent cell viability. A) Nontoxic compounds. Toxic compounds (B) that were removed according to the triage paradigm: disulfiram, dicyclopentamethylenethiuram disulfide, tetramethylthiuram monosulfide, dipyrithione, phenylmercuric acetate, and 2,3,4-trihydroxybenzaldehyde.
CETSA demonstrates compound target engagement with SENP2 within the cell
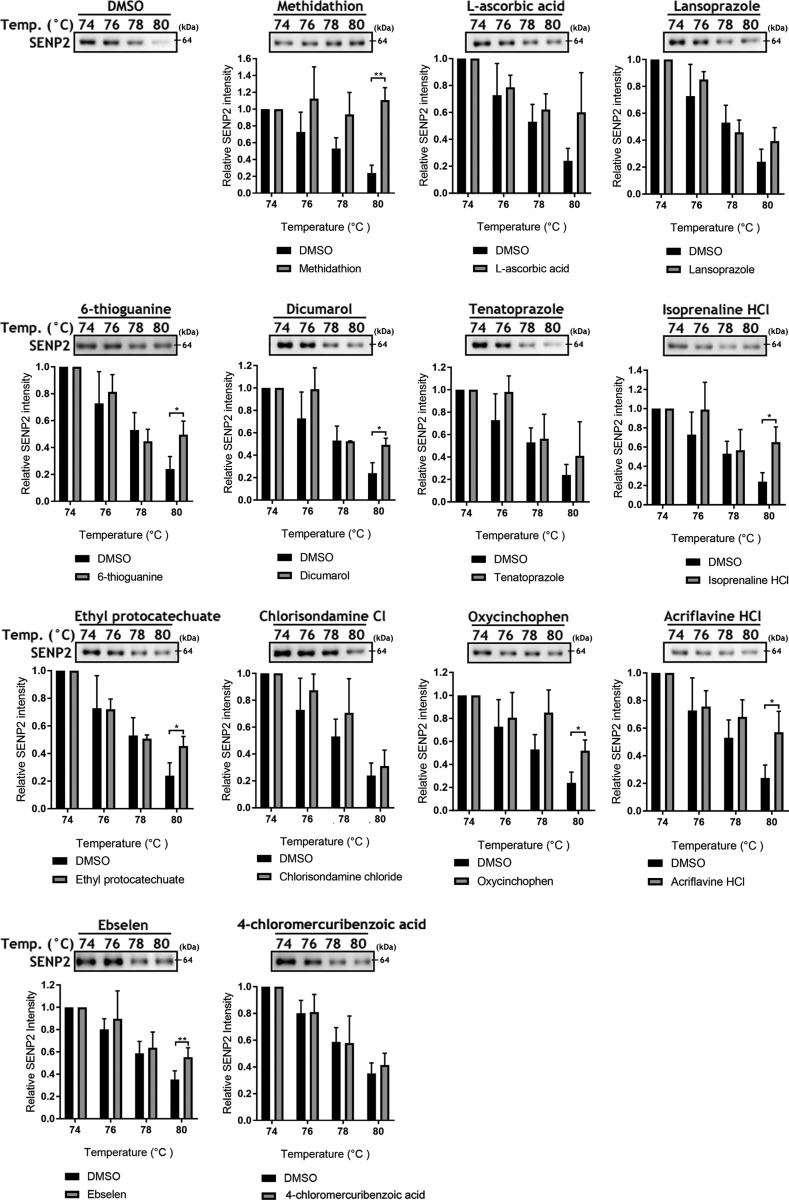
As thermal shifts have been shown capable of revealing thermal stabilization of proteins upon ligand binding (50), we performed CETSA to confirm target engagement in a more physiologic scenario. The assay involves treatment of cells or cell lysates with a compound of interest, heating to denature and precipitate proteins, and the separation of cell debris and aggregates from the soluble protein fraction (42, 50). Via a series of pilot experiments (Supplemental Fig. 3) we determined that the melting temperature of SENP2 was ∼76°C. Of the 13 compounds identified as inhibitors of hSENP2c in the cell-free system that were also nontoxic (Figs. 2 and 3), CETSA analysis indicated engagement with endogenous SENP2 in 8 tested compounds (Fig. 4).
Figure 4.
Cellular thermal shift assay for target engagement with SENP2. Thirteen nontoxic total inhibitors of hSENP2 catalytic activity were subjected to CETSA at 4 temperatures from 74 to 80°C. Eight compounds showed significant increases in stable SENP2 protein at a temperature above the melting point, indicating ligand-binding-induced stabilization; representative immunoblots shown. The band corresponding to SENP2 (detected at ∼64 kDa; expected, ∼68 kDa) was cropped in each lane and the total intensities were measured and expressed as the fold difference relative to the DMSO control at the same temperature. Means ± sd (n = 3). *P < 0.05, **P < 0.01 (by Student’s t test).
Molecular docking calculations position the lowest-energy poses of the most potent inhibitors at the active site of a human SENP2 crystal structure
Models of each of the 8 best candidate inhibitors (as determined via CETSA) were subjected to docking calculations with a crystal structure-derived model of human SENP2 (45) (Fig. 5A, in complex with RanGAP1-SUMO-1), to establish their lowest energy binding poses (Fig. 5C–J). In each instance, the lowest energy configuration situated the ligand at the entrance to a tunnel through the solvent-accessible surface of the enzyme (delineated by Trp410, His474, Lys476, Val477, and Trp479). This tunnel guides the C-terminal tail of SUMO-1 toward the catalytic residues of the SENP2 structure (Fig. 5B), including His478 and Cys548 (Ser548 in this inactivated mutant), where deconjugation would occur upon exit. Each of the putative inhibitors would appear to occlude the entrance to the tunnel, suggesting a mode of action whereby these ligands impede the proper approach of the SUMO C-terminal tail to the catalytic site of the SENP. Indeed, isoprenaline and ethyl protocatechuate (Fig. 5F, G, respectively) appear to fully occupy the tunnel enclosed by Trp410, whereas each of the other ligands coincide spatially with several of the C-terminal residues of the overlaid structure of complexed SUMO-1. Binding of the inhibitors to SENP2 is promoted by a variety of electrostatic and hydrophobic interactions with residues at the entrance to the tunnel, as well as van der Waals contacts with the surfaces of the enzyme. Interactions with Trp410 or His474 or both are common to all ligands, involving a mixture of hydrogen bonding and π–π interactions. 6-Thioguanine demonstrated a particularly elaborate network of hydrogen bonding to multiple residues around the tunnel entrance, including both Trp410 and His474.
Figure 5.
Low-energy binding poses of putative inhibitors from molecular docking calculations. A) The 8 compounds identified by CETSA as SENP2 engagers were subjected to molecular docking calculations with the structure (PDB ID: 2IO2) of human SENP2 (cyan ribbons/surfaces) in complex with a RanGAP1-SUMO-1 conjugate (green and pink ribbons, respectively). Yellow: catalytic residues (Trp410, His478, Asp495, and C548S) and their respective surfaces. B–J) Close-up of the tunnel in the surface of SENP2 directing the C-terminal tail of SUMO-1 to the catalytic site (B), shown with the low-energy binding poses of dicumarol (C), oxycinchophen (D), ebselen (E), isoprenaline (F), ethyl protocatechuate (G), acriflavine (H), 6-thioguanine (I), and methidathion (J) (ligands depicted as spheres color-coded by element). Each low-energy binding pose is also depicted from a different angle as transparent spheres illustrating proximity and contacts to residues Trp410 (yellow) or His474 (green) or both, and as stick models illustrating their orientation relative to the SENP2 catalytic tunnel (surfaces arising from catalytic residues in yellow).
SENP2 inhibitors increase SUMO conjugation in B35 cells
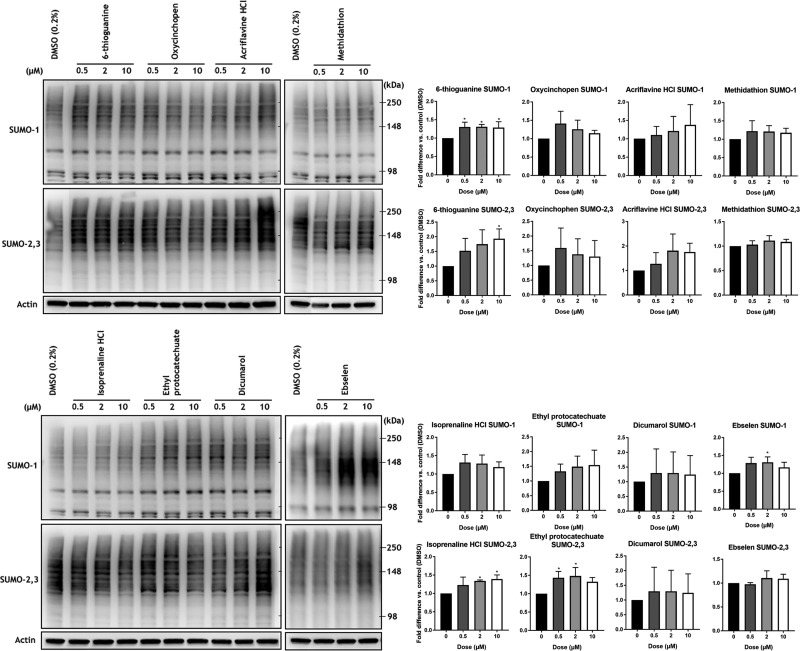
To determine their effects on the induction of SUMO-conjugation, the remaining 8 compounds were tested in orthogonal cell-based assays again using B35 cells. The compounds were incubated with the neuroblastoma cells for 18 h at 3 concentrations. Immunoblot analysis effectively demonstrated that 4 of these compounds were indeed capable of upregulating SUMO-1 and/or SUMO-2/3 conjugation, thereby confirming the biologic validity of the positive hits resulting from the screen (Fig. 6).
Figure 6.
Effects of compounds on SUMOylation in rat B35 neuroblastoma cells. The 8 compounds identified by CETSA as SENP2 engagers were tested at 0.5, 2, and 10 μM in the B35 rat neuroblastoma cell line. Four compounds significantly increased SUMO-1 and -2/3 conjugation with at least 1 dose: 6-thioguanine increased SUMO-1 conjugation at all doses and SUMO-2/3 conjugation at 10 μM; isoprenaline hydrochloride increased SUMO-2/3 conjugation at 10 μM; ethyl protocatechuate increased SUMO-2/3 conjugation at 0.5 and 2 μM; and ebselen increased SUMO-1 conjugation at 2 μM. High molecular mass (>100 kDa) SUMO-1 and -2/3 conjugates were cropped in each lane and the total intensities were measured. Densitometries were normalized to the β-actin loading control and expressed as fold difference relative to the DMSO control. Data are means ± sd (n ≥ 3). *P < 0.05 (1-way ANOVA with Dunnett’s post hoc correction).
Select compounds are capable of inducing protection against oxygen and glucose deprivation in B35 cells, and, of these, ebselen upregulates global SUMOylation within the brains of mice after intraperitoneal injection
The ultimate goal of this process is to identify small molecules that might be used for the treatment of ischemic stroke via the inhibition of SENPs. The 4 compounds that had shown increased SUMOylation in B35 cells, were consequently tested for their putative efficacy in protecting cells from OGD in vitro. Two compounds (ebselen and 6-thioguanine) were protective against OGD-induced cell death in B35 cells (Fig. 7A, B). Last, we tested ebselen in vivo. Here, ebselen administered by IP injection significantly increased the level of global SUMOylation within the brain in vivo (Fig. 7C).
Figure 7.
Pretreatment and OGD in B35 neuroblastoma cells. A) Ebselen and 6-thioguanine protected neuroblastoma cells from OGD-induced cell death, with significantly greater vital populations and significantly lesser necrotic and apoptotic populations, as compared to the DMSO control, was used to determine significance. Data are means ± sd (n ≥ 3). *P < 0.05, **P < 0.01, ***P < 0.005, ****P < 0.001 (1-way ANOVA with Dunnett’s post hoc correction). B) Representative fluorescence-activated well sorting plots showing the cell population gated into vital, necrotic, and apoptotic populations. The population of dead cells is the aggregate of the necrotic and apoptotic populations. C) Ebselen significantly increases SUMO-1 and -2/3 conjugation in mouse brains after intraperitoneal injection. Data are means ± sd (n = 3). *P < 0.05 (by Student’s t test).
DISCUSSION
In this study, we sought to describe a process capable of identifying and characterizing small molecules with the potential to be developed or repurposed as clinical therapies for ischemic stroke through the inhibition of the SENPs.
Brain ischemia is a complex pathologic process involving modulation of numerous signaling pathways (51). In the past, however, development of neuroprotective therapies has focused on single signaling pathways; all such clinical trials have failed (52). New therapeutic strategies targeting a key regulatory switch that affects many downstream pathways in brain ischemia should therefore be considered (51, 53). Notably, mounting evidence indicates that SUMOylation has the potential to be such a switch, because of its numerous targets and overall neuroprotective effects in brain ischemia and hypothermia. Accordingly, small molecules that inhibit SENP2 and thus increase SUMOylation must be identified.
Major efforts have been invested in screening for specific SENP inhibitors via both in silico and conventional high-throughput screening methods (54, 55). However, few of these screens have identified candidates with medicinal properties and have made use of physiologically relevant substrates (54). Accordingly, in this study, we miniaturized an AlphaScreen-based qHTS assay to a 1536-well format designed to screen for inhibitors of the isopeptidase activity of SENPs and used an endogenous SUMO-conjugated protein substrate and the conserved SENP catalytic domain (40). Understanding the requirements for a drug or compound to bear translational relevance, compounds from the NPC and LOPAC1280 libraries (i.e., a myriad of approved drugs) were first tested in a cell-free screen with recombinant proteins, and then in a cytotoxicity screen. Only compounds that demonstrated inhibition of SENP2 activity and displayed a lack of cytotoxicity were approved to continue in this triaging process. Subsequently, those compounds were tested for their ability to definitively engage with SENP2 in an in silico computer model of binding energies and poses, as well as the CETSA, which is capable of definitely demonstrating target engagement and direct binding to a protein target of interest within a cell (50). Neuroblastoma cells were then treated with these compounds to validate that they were indeed capable of increasing SUMO-1 and -2/3 conjugation in vitro via the inhibition of SENP2. The select final group of compounds was then tested against OGD, an in vitro model of ischemia.
Some of the compounds identified or triaged ultimately were found capable of inducing protection against OGD-induced cell death. We recently confirmed the validity of such an approach, having demonstrated for the first time that increasing global SUMOylation via the use of an SENP inhibitor (i.e., quercetin) is in fact neuroprotective in cell culture (48). Of the 2 compounds identified that were ultimately shown to provide protection in the face of OGD, it is prudent to note that 6-thioguanine (56), a thiol analogue of guanine used as a chemotherapeutic, is far from an ideal candidate but could ultimately represent a core molecular structure for future medicinal chemistry explorations designed to engineer ideal clinical-grade inhibitors of SENP2. In line with the inhibition of SENP2, purine analogues, such as 6-thioguanine and 2-aminopurine, have been found to inhibit protein kinase N in vitro (57).
The identification of ebselen (a synthetic organo-selenium compound) supports the validity of this screening paradigm’s ability to identify inhibitors with potential for repurposing as stroke therapeutics. Ebselen is an organo-selenium compound that was originally found to reduce hydroperoxides owing to its glutathione peroxidase–like activity. This antioxidant activity was later attributed to the neuroprotective effects of ebselen observed in various experimental stroke models (58, 59). Concerns related to potential selenium toxicity and insufficient efficacy reported in clinical trials have thus far precluded its clinical development (58, 59). However, it is important to note that the clinical trial performed did identify benefits for those patients treated early (i.e., before 24 h) (60), thereby highlighting the potential of this compound as a prophylactic for those patients at increased risk of a cerebrovascular accident (e.g., undergoing carotid endarterectomy, having a diagnosis of cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy or Binswanger’s disease) or have been shown by current brain-imaging protocols to have salvageable tissue after a stroke (61). The increased level of global SUMOylation demonstrated within the brain by ebselen is similar to hypothermia-induced levels that have been shown to protect brains from ischemic damage (27). However, whether this SUMOylation-activator activity of ebselen contributes to its protective effect will need further clarification. For such compounds, additional target identification and profiling techniques, such as thermal proteome profiling may be useful toward fully elucidating the mechanism by which neuroprotection is conferred.
As both in vitro and in vivo data have illustrated that increased global SUMOylation is neuroprotective in brain ischemia, we have established a platform for pursuing small molecules capable of increasing global SUMOylation. It should be noted that this neuroprotective effect arises from the summed effect of SUMOylated proteins on overall outcome, as the SUMO proteome comprises diverse proteins reported to be involved in both prosurvival and prodeath pathways. For example, dynamin-related protein 1 SUMOylation suppresses cytochrome c release and protects the cell from apoptosis (34), whereas hypoxia-inducible factor (HIF)-1α SUMOylation may promote HIF-1α degradation and thereby impair HIF-1α–mediated protective effects after ischemia (62). It must be noted that current knowledge with regard to specific SUMO targets involved in brain ischemia is highly limited, and the potential effects of individual SUMOylated proteins within complex pathologies such as brain ischemia are primarily speculative (63).
Although our results validated our approach, only 2 of our identified compounds showed neuroprotective efficacy in cells. Considering the great potential of SENP inhibitors for neuroprotection in brain ischemia, there is a clear need to rapidly identify compounds of interest, thereby suggesting that larger libraries should in fact be interrogated. Our platform represents a strong approach to screening large libraries of small molecules to identify new classes of compounds that inhibit SENP isopeptidase activity. Once more ideal candidate compounds are identified in future projects, they should first be subjected to extensive medicinal chemistry optimization to improve potency, specificity, and drug-like properties. The optimized drug candidates can then be tested in various brain ischemia models, including global cerebral ischemia and focal ischemia (transient and permanent) animal models, using both pre- and posttreatment paradigms. Ultimately, the most promising compounds would be expected to advance to preclinical development with the final goal of translation into clinical interventions to prevent brain ischemia-induced damage.
Finally, beyond stroke, this platform may also be used in future qHTS studies to screen for new classes of inhibitors that target SENP isopeptidase activity. For example, SENP1 has been reported to be an oncogene (64) in various cancers (e.g., prostate, kidney, bladder, neuroblastoma, and multiple myeloma), and a strategy targeting SENP1 could identify adjuvant treatments relevant to these malignancies.
CONCLUSIONS
Therapeutic options for ischemic brain injury remain extremely limited. During the past decade, substantial evidence has accumulated to support increases in the levels of global SUMOylation as a mechanism to protect the brain from ischemic damage. As such, it is encouraging to speculate that pharmaceutically increased global SUMOylation under normothermic conditions may provide similar neuroprotection without the adverse effects associated with hypothermia. To investigate this possibility, small molecules that can increase global SUMOylation in cells and can cross the blood–brain barrier have been actively sought. In this study, we have presented a streamlined process that effectively identified SENP2 inhibitors with the ability to increase global SUMOylation in vitro and in vivo. It is our hope that this finding will ultimately lead to advanced treatments for patients who have ischemic brain damage.
Supplementary Material
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
ACKNOWLEDGMENTS
The authors thank Dr. Gregory K. Friedman (University of Alabama at Birmingham) and Dr. Jorn Karhausen (Duke University Medical Center) for critical reading and editing of the manuscript. The authors also thank Dr. Dragan Maric [Flow Cytometry Core Facility, U.S. National Institutes of Health (NIH) National Institute of Neurological Disorders and Stroke Sciences (NINDS)]. This research was supported by the Intramural Research Program of the NIH/NINDS; the National Center for Advancing Translational Sciences; NIH/NINDS Grant R01 NS099590 and American Heart Association Grant 16GRNT30270003 (to W.Y.); and an NIH/OxCam Fellowship (to J.D.B.). The authors declare no conflicts of interest.
Glossary
- CETSA
cellular thermal shift assay
- GG
di-glycine
- HIF
hypoxia-inducible factor
- hSENPc
human SENP catalytic domain
- LOPAC1280
Library of Pharmaceutically Active Compounds
- NCATS
National Center for Advancing Translational Sciences
- NCGC
National Center for Advancing Translational Sciences
- NPC
NCGC Pharmaceutical Collection
- OGD
oxygen-glucose deprivation
- qHTS
quantitative high-throughput screen
- SENP
small ubiquitin-like modifier-specific protease
- SUMO
small ubiquitin-like modifier
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
J. D. Bernstock, D. Ye, Y.-J. Lee, F. A. Gessler, A. Yasgar, A. Jadhav, S. Pluchino, W. Zheng, A. Simeonov, J. M. Hallenbeck, and W. Yang designed the research; J. D. Bernstock, D. Ye, J. A. Smith, A. Yasgar, J. Kouznetsova, and Z. Wang performed the research; J. A. Smith, A. Yasgar, A. Jadhav, W. Zheng, A. Simeonov, and J. M. Hallenbeck contributed critical reagents or analytical tools; J. D. Bernstock, D. Ye, J. A. Smith, A. Yasgar, J. Kouznetsova, and W. Yang analyzed the data; and J. D. Bernstock, D. Ye, and W. Yang wrote and revised the manuscript.
REFERENCES
- 1.Mozaffarian D., Benjamin E. J., Go A. S., Arnett D. K., Blaha M. J., Cushman M., Das S. R., de Ferranti S., Despres J. P., Fullerton H. J., Howard V. J., Huffman M. D., Isasi C. R., Jimenez M. C., Judd S. E., Kissela B. M., Lichtman J. H., Lisabeth L. D., Liu S., Mackey R. H., Magid D. J., McGuire D. K., Mohler E. R., III, Moy C. S., Muntner P., Mussolino M. E., Nasir K., Neumar R. W., Nichol G., Palaniappan L., Pandey D. K., Reeves M. J., Rodriguez C. J., Rosamond W., Sorlie P. D., Stein J., Towfighi A., Turan T. N., Virani S. S., Woo D., Yeh R. W., Turner M. B. (2015) Heart disease and stroke statistics-2016 update: a report from the American Heart Association. Circulation 133, e38–360 [DOI] [PubMed] [Google Scholar]
- 2.O’Collins V. E., Macleod M. R., Donnan G. A., Horky L. L., van der Worp B. H., Howells D. W. (2006) 1,026 experimental treatments in acute stroke. Ann. Neurol. 59, 467–477 https://doi.org/10.1002/ana.20741 [DOI] [PubMed] [Google Scholar]
- 3.Ginsberg M. D. (2008) Neuroprotection for ischemic stroke: past, present and future. Neuropharmacology 55, 363–389 https://doi.org/10.1016/j.neuropharm.2007.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang P., Prabhakaran S. (2017) Recent advances in the management of acute ischemic stroke. F1000 Res. 6, 484 https://doi.org/10.12688/f1000research.9191.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.National Institute of Neurological Disorders and Stroke rt-PA Stroke Study Group . (1995) Tissue plasminogen activator for acute ischemic stroke. N. Engl. J. Med. 333, 1581–1587 https://doi.org/10.1056/NEJM199512143332401 [DOI] [PubMed] [Google Scholar]
- 6.Goyal M., Demchuk A. M., Menon B. K., Eesa M., Rempel J. L., Thornton J., Roy D., Jovin T. G., Willinsky R. A., Sapkota B. L., Dowlatshahi D., Frei D. F., Kamal N. R., Montanera W. J., Poppe A. Y., Ryckborst K. J., Silver F. L., Shuaib A., Tampieri D., Williams D., Bang O. Y., Baxter B. W., Burns P. A., Choe H., Heo J. H., Holmstedt C. A., Jankowitz B., Kelly M., Linares G., Mandzia J. L., Shankar J., Sohn S. I., Swartz R. H., Barber P. A., Coutts S. B., Smith E. E., Morrish W. F., Weill A., Subramaniam S., Mitha A. P., Wong J. H., Lowerison M. W., Sajobi T. T., Hill M. D.; ESCAPE Trial Investigators . (2015) Randomized assessment of rapid endovascular treatment of ischemic stroke. N. Engl. J. Med. 372, 1019–1030 https://doi.org/10.1056/NEJMoa1414905 [DOI] [PubMed] [Google Scholar]
- 7.Dirnagl U., Iadecola C., Moskowitz M. A. (1999) Pathobiology of ischaemic stroke: an integrated view. Trends Neurosci. 22, 391–397 https://doi.org/10.1016/S0166-2236(99)01401-0 [DOI] [PubMed] [Google Scholar]
- 8.Minnerup J., Sutherland B. A., Buchan A. M., Kleinschnitz C. (2012) Neuroprotection for stroke: current status and future perspectives. Int. J. Mol. Sci. 13, 11753–11772 https://doi.org/10.3390/ijms130911753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chamorro Á., Dirnagl U., Urra X., Planas A. M. (2016) Neuroprotection in acute stroke: targeting excitotoxicity, oxidative and nitrosative stress, and inflammation. Lancet Neurol. 15, 869–881 https://doi.org/10.1016/S1474-4422(16)00114-9 [DOI] [PubMed] [Google Scholar]
- 10.Hallenbeck J. M. (2012) Tracks of a non-main path traveler: 2011 Thomas Willis Lecture. Stroke 43, 585–590 https://doi.org/10.1161/STROKEAHA.111.643668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tempé D., Piechaczyk M., Bossis G. (2008) SUMO under stress. Biochem. Soc. Trans. 36, 874–878 https://doi.org/10.1042/BST0360874 [DOI] [PubMed] [Google Scholar]
- 12.Lee Y. J., Hallenbeck J. M. (2013) SUMO and ischemic tolerance. Neuromolecular Med. 15, 771–781 https://doi.org/10.1007/s12017-013-8239-9 [DOI] [PubMed] [Google Scholar]
- 13.Geiss-Friedlander R., Melchior F. (2007) Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8, 947–956 https://doi.org/10.1038/nrm2293 [DOI] [PubMed] [Google Scholar]
- 14.Gareau J. R., Lima C. D. (2010) The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11, 861–871 https://doi.org/10.1038/nrm3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flotho A., Melchior F. (2013) Sumoylation: a regulatory protein modification in health and disease. Annu. Rev. Biochem. 82, 357–385 https://doi.org/10.1146/annurev-biochem-061909-093311 [DOI] [PubMed] [Google Scholar]
- 16.Tatham M. H., Jaffray E., Vaughan O. A., Desterro J. M. P., Botting C. H., Naismith J. H., Hay R. T. (2001) Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276, 35368–35374 https://doi.org/10.1074/jbc.M104214200 [DOI] [PubMed] [Google Scholar]
- 17.Henley J. M., Craig T. J., Wilkinson K. A. (2014) Neuronal SUMOylation: mechanisms, physiology, and roles in neuronal dysfunction. Physiol. Rev. 94, 1249–1285 https://doi.org/10.1152/physrev.00008.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee Y. J., Miyake S., Wakita H., McMullen D. C., Azuma Y., Auh S., Hallenbeck J. M. (2007) Protein SUMOylation is massively increased in hibernation torpor and is critical for the cytoprotection provided by ischemic preconditioning and hypothermia in SHSY5Y cells. J. Cereb. Blood Flow Metab. 27, 950–962 https://doi.org/10.1038/sj.jcbfm.9600395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang W., Sheng H., Warner D. S., Paschen W. (2008) Transient focal cerebral ischemia induces a dramatic activation of small ubiquitin-like modifier conjugation. J. Cereb. Blood Flow Metab. 28, 892–896 https://doi.org/10.1038/sj.jcbfm.9600601 [DOI] [PubMed] [Google Scholar]
- 20.Yang W., Sheng H., Warner D. S., Paschen W. (2008) Transient global cerebral ischemia induces a massive increase in protein sumoylation. J. Cereb. Blood Flow Metab. 28, 269–279 https://doi.org/10.1038/sj.jcbfm.9600523 [DOI] [PubMed] [Google Scholar]
- 21.Lee Y. J., Castri P., Bembry J., Maric D., Auh S., Hallenbeck J. M. (2009) SUMOylation participates in induction of ischemic tolerance. J. Neurochem. 109, 257–267 https://doi.org/10.1111/j.1471-4159.2009.05957.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Datwyler A. L., Lättig-Tünnemann G., Yang W., Paschen W., Lee S. L., Dirnagl U., Endres M., Harms C. (2011) SUMO2/3 conjugation is an endogenous neuroprotective mechanism. J. Cereb. Blood Flow Metab. 31, 2152–2159 https://doi.org/10.1038/jcbfm.2011.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee Y. J., Mou Y., Maric D., Klimanis D., Auh S., Hallenbeck J. M. (2011) Elevated global SUMOylation in Ubc9 transgenic mice protects their brains against focal cerebral ischemic damage. PLoS One 6, e25852 https://doi.org/10.1371/journal.pone.0025852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L., Liu X., Sheng H., Liu S., Li Y., Zhao J. Q., Warner D. S., Paschen W., Yang W. (2017) Neuron-specific SUMO knockdown suppresses global gene expression response and worsens functional outcome after transient forebrain ischemia in mice. Neuroscience 343, 190–212 https://doi.org/10.1016/j.neuroscience.2016.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L., Ma Q., Yang W., Mackensen G. B., Paschen W. (2012) Moderate hypothermia induces marked increase in levels and nuclear accumulation of SUMO2/3-conjugated proteins in neurons. J. Neurochem. 123, 349–359 https://doi.org/10.1111/j.1471-4159.2012.07916.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang W., Ma Q., Mackensen G. B., Paschen W. (2009) Deep hypothermia markedly activates the small ubiquitin-like modifier conjugation pathway; implications for the fate of cells exposed to transient deep hypothermic cardiopulmonary bypass. J. Cereb. Blood Flow Metab. 29, 886–890 https://doi.org/10.1038/jcbfm.2009.16 [DOI] [PubMed] [Google Scholar]
- 27.Lee Y. J., Mou Y., Klimanis D., Bernstock J. D., Hallenbeck J. M. (2014) Global SUMOylation is a molecular mechanism underlying hypothermia-induced ischemic tolerance. Front. Cell. Neurosci. 8, 416 https://doi.org/10.3389/fncel.2014.00416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernstock J. D., Lee Y. J., Peruzzotti-Jametti L., Southall N., Johnson K. R., Maric D., Volpe G., Kouznetsova J., Zheng W., Pluchino S., Hallenbeck J. M. (2016) A novel quantitative high-throughput screen identifies drugs that both activate SUMO conjugation via the inhibition of microRNAs 182 and 183 and facilitate neuroprotection in a model of oxygen and glucose deprivation. J. Cereb. Blood Flow Metab. 36, 426–441 https://doi.org/10.1177/0271678X15609939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee Y. J., Johnson K. R., Hallenbeck J. M. (2012) Global protein conjugation by ubiquitin-like-modifiers during ischemic stress is regulated by microRNAs and confers robust tolerance to ischemia. PLoS One 7, e47787 https://doi.org/10.1371/journal.pone.0047787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukhopadhyay D., Dasso M. (2007) Modification in reverse: the SUMO proteases. Trends Biochem. Sci. 32, 286–295 https://doi.org/10.1016/j.tibs.2007.05.002 [DOI] [PubMed] [Google Scholar]
- 31.Hickey C. M., Wilson N. R., Hochstrasser M. (2012) Function and regulation of SUMO proteases. Nat. Rev. Mol. Cell Biol. 13, 755–766 https://doi.org/10.1038/nrm3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma P., Yamada S., Lualdi M., Dasso M., Kuehn M. R. (2013) Senp1 is essential for desumoylating Sumo1-modified proteins but dispensable for Sumo2 and Sumo3 deconjugation in the mouse embryo. Cell Rep. 3, 1640–1650 https://doi.org/10.1016/j.celrep.2013.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mendes A. V., Grou C. P., Azevedo J. E., Pinto M. P. (2016) Evaluation of the activity and substrate specificity of the human SENP family of SUMO proteases. Biochim. Biophys. Acta 1863, 139–147 https://doi.org/10.1016/j.bbamcr.2015.10.020 [DOI] [PubMed] [Google Scholar]
- 34.Guo C., Hildick K. L., Luo J., Dearden L., Wilkinson K. A., Henley J. M. (2013) SENP3-mediated deSUMOylation of dynamin-related protein 1 promotes cell death following ischaemia. EMBO J. 32, 1514–1528 https://doi.org/10.1038/emboj.2013.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fanis P., Gillemans N., Aghajanirefah A., Pourfarzad F., Demmers J., Esteghamat F., Vadlamudi R. K., Grosveld F., Philipsen S., van Dijk T. B. (2012) Five friends of methylated chromatin target of protein-arginine-methyltransferase[prmt]-1 (chtop), a complex linking arginine methylation to desumoylation. Mol. Cell. Proteomics 11, 1263–1273 https://doi.org/10.1074/mcp.M112.017194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo J., Gurung S., Lee L., Henley J. M., Wilkinson K. A., Guo C. (2017) Increased SUMO-2/3-ylation mediated by SENP3 degradation is protective against cadmium-induced caspase 3-dependent cytotoxicity. J. Toxicol. Sci. 42, 529–538 https://doi.org/10.2131/jts.42.529 [DOI] [PubMed] [Google Scholar]
- 37.Mikolajczyk J., Drag M., Békés M., Cao J. T., Ronai Z., Salvesen G. S. (2007) Small ubiquitin-related modifier (SUMO)-specific proteases: profiling the specificities and activities of human SENPs. J. Biol. Chem. 282, 26217–26224 https://doi.org/10.1074/jbc.M702444200 [DOI] [PubMed] [Google Scholar]
- 38.Kang X., Qi Y., Zuo Y., Wang Q., Zou Y., Schwartz R. J., Cheng J., Yeh E. T. (2010) SUMO-specific protease 2 is essential for suppression of polycomb group protein-mediated gene silencing during embryonic development. Mol. Cell 38, 191–201 https://doi.org/10.1016/j.molcel.2010.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bernstock J. D., Ye D., Gessler F. A., Lee Y. J., Peruzzotti-Jametti L., Baumgarten P., Johnson K. R., Maric D., Yang W., Kögel D., Pluchino S., Hallenbeck J. M. (2017) Topotecan is a potent inhibitor of SUMOylation in glioblastoma multiforme and alters both cellular replication and metabolic programming. Sci. Rep. 7, 7425 https://doi.org/10.1038/s41598-017-07631-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang W., Wang L., Paschen W. (2013) Development of a high-throughput screening assay for inhibitors of small ubiquitin-like modifier proteases. J. Biomol. Screen. 18, 621–628 https://doi.org/10.1177/1087057113479971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang R., Southall N., Wang Y., Yasgar A., Shinn P., Jadhav A., Nguyen D. T., Austin C. P. (2011) The NCGC pharmaceutical collection: a comprehensive resource of clinically approved drugs enabling repurposing and chemical genomics. Sci. Transl. Med. 3, 80ps16 https://doi.org/10.1126/scitranslmed.3001862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Almqvist H., Axelsson H., Jafari R., Dan C., Mateus A., Haraldsson M., Larsson A., Martinez Molina D., Artursson P., Lundbäck T., Nordlund P. (2016) CETSA screening identifies known and novel thymidylate synthase inhibitors and slow intracellular activation of 5-fluorouracil. Nat. Commun. 7, 11040 https://doi.org/10.1038/ncomms11040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanwell M. D., Curtis D. E., Lonie D. C., Vandermeersch T., Zurek E., Hutchison G. R. (2012) Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 4, 17 https://doi.org/10.1186/1758-2946-4-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanner M. F. (1999) Python: a programming language for software integration and development. J. Mol. Graph. Model. 17, 57–61 [PubMed] [Google Scholar]
- 45.Reverter D., Lima C. D. (2006) Structural basis for SENP2 protease interactions with SUMO precursors and conjugated substrates. Nat. Struct. Mol. Biol. 13, 1060–1068 https://doi.org/10.1038/nsmb1168 [DOI] [PubMed] [Google Scholar]
- 46.Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. (2000) The protein data bank. Nucleic Acids Res. 28, 235–242 https://doi.org/10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trott O., Olson A. J. (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee Y. J., Bernstock J. D., Nagaraja N., Ko B., Hallenbeck J. M. (2016) Global SUMOylation facilitates the multimodal neuroprotection afforded by quercetin against the deleterious effects of oxygen/glucose deprivation and the restoration of oxygen/glucose. J. Neurochem. 138, 101–116 https://doi.org/10.1111/jnc.13643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yasgar A., Jadhav A., Simeonov A., Coussens N. P. (2016) AlphaScreen-based assays: ultra-high-throughput screening for small-molecule inhibitors of challenging enzymes and protein-protein interactions. Methods Mol. Biol. 1439, 77–98 https://doi.org/10.1007/978-1-4939-3673-1_5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jafari R., Almqvist H., Axelsson H., Ignatushchenko M., Lundbäck T., Nordlund P., Martinez Molina D. (2014) The cellular thermal shift assay for evaluating drug target interactions in cells. Nat. Protoc. 9, 2100–2122 https://doi.org/10.1038/nprot.2014.138 [DOI] [PubMed] [Google Scholar]
- 51.Fisher M. (2011) New approaches to neuroprotective drug development. Stroke 42(1 Suppl), S24–S27 https://doi.org/10.1161/STROKEAHA.110.592394 [DOI] [PubMed] [Google Scholar]
- 52.Moretti A., Ferrari F., Villa R. F. (2015) Neuroprotection for ischaemic stroke: current status and challenges. Pharmacol. Ther. 146, 23–34 https://doi.org/10.1016/j.pharmthera.2014.09.003 [DOI] [PubMed] [Google Scholar]
- 53.Savitz S. I., Fisher M. (2007) Future of neuroprotection for acute stroke: in the aftermath of the SAINT trials. Ann. Neurol. 61, 396–402 https://doi.org/10.1002/ana.21127 [DOI] [PubMed] [Google Scholar]
- 54.Yang W., Sheng H., Wang H. (2016) Targeting the SUMO pathway for neuroprotection in brain ischaemia. Stroke Vasc Neurol 1, 101–107 https://doi.org/10.1136/svn-2016-000031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumar A., Zhang K. Y. (2015) Advances in the development of SUMO specific protease (SENP) inhibitors. Comput. Struct. Biotechnol. J. 13, 204–211 https://doi.org/10.1016/j.csbj.2015.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nelson J. A., Carpenter J. W., Rose L. M., Adamson D. J. (1975) Mechanisms of action of 6-thioguanine, 6-mercaptopurine, and 8-azaguanine. Cancer Res. 35, 2872–2878 [PubMed] [Google Scholar]
- 57.Volonté C., Greene L. A. (1992) 6-Methylmercaptopurine riboside is a potent and selective inhibitor of nerve growth factor-activated protein kinase N. J. Neurochem. 58, 700–708 https://doi.org/10.1111/j.1471-4159.1992.tb09774.x [DOI] [PubMed] [Google Scholar]
- 58.Parnham M. J., Sies H. (2013) The early research and development of ebselen. Biochem. Pharmacol. 86, 1248–1253 https://doi.org/10.1016/j.bcp.2013.08.028 [DOI] [PubMed] [Google Scholar]
- 59.Noguchi N. (2016) Ebselen, a useful tool for understanding cellular redox biology and a promising drug candidate for use in human diseases. Arch. Biochem. Biophys. 595, 109–112 https://doi.org/10.1016/j.abb.2015.10.024 [DOI] [PubMed] [Google Scholar]
- 60.Yamaguchi T., Sano K., Takakura K., Saito I., Shinohara Y., Asano T., Yasuhara H.; Ebselen Study Group . (1998) Ebselen in acute ischemic stroke: a placebo-controlled, double-blind clinical trial. Stroke 29, 12–17 https://doi.org/10.1161/01.STR.29.1.12 [DOI] [PubMed] [Google Scholar]
- 61.Tymianski M. (2017) Combining neuroprotection with endovascular treatment of acute stroke: is there hope? Stroke 48, 1700–1705 https://doi.org/10.1161/STROKEAHA.117.017040 [DOI] [PubMed] [Google Scholar]
- 62.Gu J., Fan Y., Liu X., Zhou L., Cheng J., Cai R., Xue S. (2014) SENP1 protects against myocardial ischaemia/reperfusion injury via a HIF1α-dependent pathway. Cardiovasc. Res. 104, 83–92 https://doi.org/10.1093/cvr/cvu177 [DOI] [PubMed] [Google Scholar]
- 63.Peters M., Wielsch B., Boltze J. (2017) The role of SUMOylation in cerebral hypoxia and ischemia. Neurochem. Int. 107, 66–77 https://doi.org/10.1016/j.neuint.2017.03.011 [DOI] [PubMed] [Google Scholar]
- 64.Seeler J. S., Dejean A. (2017) SUMO and the robustness of cancer. Nat. Rev. Cancer 17, 184–197 https://doi.org/10.1038/nrc.2016.143 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.