Abstract
Background
Nitric oxide (NO) is released in the airway as a critical component of innate immune defense against invading pathogenic organisms. It is well documented that bacteriostatic and bactericidal effects of NO are concentration-dependent. However, few data exist comparing relative susceptibility of common pathogens to NO at physiologic concentrations. In this study we evaluated the effects of NO on 4 common airway bacteria and 1 fungus, and examined the potential implications of discrepancies in sensitivity.
Methods
Staphylococcus epidermis, Staphylococcus aureus, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Candida albicans cultures were adjusted to a uniform optical density (OD) and grown in log phase at 37°C with varying concentrations of NO formed by DETA NONOate. Both OD readings and colony forming units (CFUs) were measured at varying time-points to evaluate for inhibitory effects of NO.
Results
P aeruginosa and C albicans were significantly more sensitive to NO at physiologic concentrations typical of the human airway. S aureus was a enuated by NO to a lesser degree, and K pneumoniae and S epidermis were more resistant to NO at all concentrations tested. Air surface liquid from cultured human sinonasal epithelial cells had an additive effect in bacterial killing of P aeruginosa, but not in S aureus.
Conclusion
Common airway pathogens have varying levels of susceptibility to NO at physiologic concentrations of innate immune defense. Relative sensitivity of P aeruginosa and relative resistance of S epidermis may help explain the composition of the healthy microbiome, as well as opportunistic infection in the absence of induced NO release.
Keywords: Nitric Oxide, Bacteria Innate Immunity, Microbiome, Airway Defense, Airway Microbes, Nitric Oxide Susceptibility
A number of immune defense mechanisms function in concert to prevent infection of the sinonasal tract, a daunting task in the face of constant environmental exposure. In addition, the complexity of the nasal microbiome in health and disease supports the assertion that it is the resident microbe homeostasis that is of significance, as opposed to sterility.1 When the balance is perturbed, even commensal organisms can become pathogenic causes of inflammation or infection.2,3 Staphylococcus aureus is a frequent colonizer of healthy subjects, but it is also present at increased density in patients with chronic rhinosinusitis (CRS).4 Klebsiella pneumoniae and coagulase-negative Staphylococcus species, such as Staphylococcus epidermis, show similar patterns as representative commensal bacteria that can play a role in infectious processes.5,6 An under-standing of the host-pathogen interactions that cause these transitions allow for novel therapeutic opportunities.
Nitric oxide (NO) is a free radical compound that has potent antimicrobial effects.7 Cells that line the upper airway release NO as an innate immune mechanism,8–12 often in response to bacterial insult. Bitter secreted products from common organisms, such as S aureus, S epidermis, and Pseudomonas aeruginosa, are detected by bitter taste receptors of upper airway epithelial cells and elicit many of these downstream NO responses.8,13,14 The NO diffuses through the apical mucus into the offending organism, where it produces peroxynitrites and S-nitrosothiols that are destructive to bacterial components.15–17 DNA, replication machinery, enzymes, or virulence factors can all be affected by the NO free radical.18 In addition to being directly antimicrobial, NO also speeds up ciliary beating and resultant mucociliary clearance (MCC),19,20 with movement of trapped organisms in the sinonasal tract to the oropharynx.21
NO can have bacteriostatic to bactericidal effects that may be species-specific in magnitude. Escherichia coli and Salmonella enterica are affected in replication frequency, but not in overall cell survival, whereas other organisms, such as Burkholderia species and P aeruginosa, are more readily killed by the effects of NO.12,22–24 In this study, we aimed to characterize the susceptibility of 4 common airway bacteria and 1 fungus to NO at physiologic concentrations.15,25 In addition, we suspected that NO may display synergy with other antibacterial compounds secreted by sinonasal epithelial cells. The characterization of microbial resistance to innate immune mechanisms, including NO production, may provide critical insight into the pathogenesis of disease and the composition of the healthy microbiome.
Material and methods
Bacterial and fungal cultures
Methicillin-resistant Staphylococcus aureus (MRSA) ATCC strain 33591, Pseudomonas aeruginosa strain PA01, Klebsiella pneumoniae ATCC strain 13883, and Staphylococcus epidermis ATCC strain 14990 were obtained from frozen −80°C glycerol stocks. Samples from these stocks were inoculated into 4 mL of Luria-Bertani (LB) broth and grown to stationary phase over 24 hours at 37°C with shaking. Samples were diluted from this stock in 50% LB broth to a selected optical density prior to all experiments.
Candida albicans cultures were prepared from strain HGFP3 (provided by Katharina Ribbeck, Massachusetts Institute of Technology, Cambridge, MA, with the permission of P. Sundstrom).26 The C albicans colonies were grown on YPD agar plates (2% Bacto Peptone [Sigma-Aldrich], 2% glucose, 1% yeast extract, 2% agar) at 30°C. For preparation of liquid cultures, individual colonies were inoculated into YPD broth and grown with shaking at 30°C for 12 hours. Samples from this stock were diluted to a desired optical density in YPD broth prior to experimentation.
NO sensitivity assays
Using a spectrophotometer (Model 680, BioRad), organisms were adjusted to an optical density at 600 nm (OD600) of 0.1 in the appropriate broth at the beginning of each experiment. One-milliliter samples for each experimental condition were aliquoted into culture tubes. The cells were then incubated with increasing concentrations of DETA NONOate (Cayman Chemicals), an NO donor that has been used in previous susceptibility experiments.27 DETA NONOate was made fresh at the start of each experiment from lyophilized powder stored at −80°C, and made in stock solutions of 0.1% sodium hydroxide prior to addition to the culture media. Cultures were placed in a 37°C incubator (30°C for C albicans) with shaking at 300 rpm and grown aerobically. At predetermined time intervals, cultures were briefly removed from the incubator and 150 μL of sample was removed and placed in a 96-well plate. OD600 readings were obtained, and these samples were serially diluted and spotted on LB or YPD agar plates as appropriate. The number of colony forming units (CFUs)/mL was calculated from culture growth after 16 hours on agar at 37°C.
Determination of 50% inhibitory concentration and minimal concentration values
DETA NONOate was utilized at concentrations ranging from 0.1 to 10 mg/mL. As shown in previous work,27 a concentration of 1 mg/mL corresponds to a steady-state concentration of 6 μmol/L NO in solution. Fifty-percent inhibitory concentration (IC50) values were calculated based on nonlinear regression curve fitting of bacterial growth after 3 hours of exposure to increasing concentrations of NO. Minimal concentration (MIC) was calculated using Gompertz modeling to fit the bottom plateau of bacterial growth. Minimum bactericidal concentration (MBC) was defined as the concentration of NO that killed at least 99.9% of the inoculum relative to the start of the experiment.
Human tissue acquisition and air-liquid interface cultures
All human tissue was obtained with informed consent and institutional review board approval. Patients undergoing sinonasal surgery at the Division of Rhinology, Department of Otorhinolaryngology, University of Pennsylvania, were recruited and tissue was procured from excess clinical material. Our previous work has extensively described the culture of human nasal epithelial cells at an air-liquid interface (ALI).28,29 Briefly, human sinonasal epithelial cells were enzymatically dissociated and grown with medium containing Dulbecco’s modified Eagle medium (DMEM)/Ham’s F12 and bronchial epithelial-based medium (Clonetics), as well as 100 U/mL penicillin and 100 μg/mL streptomycin for 1 week. After this, cells were trypsinized and placed on porous polyester membranes in Transwell cell culture inserts (Transwell-clear, 12-mm diameter, 0.4-μm pores; Corning). The inserts were coated with 100 μL of coating solution (bovine serum albumin 0.1 mg/ml; Sigma-Aldrich), type 1 bovine collagen (30 μg/mL; BD Biosciences), and fibronectin (10 μg/mL; BD) in LHC basal medium (Invitrogen). After 5 days, the apical compartment was cleared and the epithelium was allowed to differentiate using a medium of 1:1 DMEM (Invitrogen) and bronchial epithelial cell basal medium (Clonetics, Cambrex), with the Clonetics complements for human epidermal growth factor (0.5 ng/mL), epinephrine (5 μg/mL), hydrocortisone (0.5 μg/mL), bovine pituitary extract (0.13 mg/mL), insulin (5 μg/mL), triiodothyronine (6.5 μg/mL), and transferrin (0.5 μg/ml), supplemented with 100 IU/mL penicillin, 100 g/mL streptomycin, 0.1 nmol/L retinoic acid (Sigma-Aldrich), and 10% fetal bovine serum (Sigma-Aldrich) in the basal compartment. Cellular differentiation was allowed to progress for 1 month. Forty-eight hours prior to experimentation, ALIs were washed on the apical and basal surfaces with phosphate-buffered saline and then placed in antibiotic-free media. Then, 48 hours later, airway surface liquid (ASL) was aspirated from the apical compartment of these cells and used in the experiments.
Statistical analysis
Stata version 13 (StataCorp, College Station, TX) software was used for statistical analysis, with p < 0.05 considered statistically significant. One-way analysis of variance (ANOVA) was used to assess differences in bacterial growth curves at 3- and 6-hour time-points, as well as in comparisons of lactate dehydrogenase (LDH) release between human cultures exposed to varying levels of NO. Two-way ANOVA was used to assess for synergistic effects of NO and ASL treatments. Data are reported as mean ± standard deviation (SD).
Results
We investigated the relative susceptibilities of 4 bacteria and 1 fungus to NO chemically generated by the NO donor DETA NONOate. DETA NONOate has a half-life of 20 hours and releases 2 mol of NO per mole of parent compound. The steady-state concentration of NO created is 6 μmol/L per milligram per milliliter of DETA NONOate used.27 OD600 readings were obtained and CFUs/mL were counted (Fig. 1) at several distinct time intervals to determine the effects of experimental manipulations on bacterial growth.
FIGURE 1.
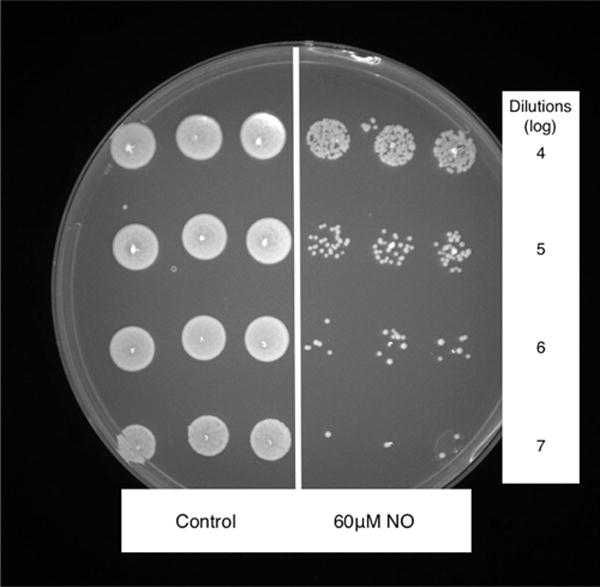
Plate of S aureus CFUs following 6 hours of growth in LB media (left) and 6 hours of growth in LB media with 60 μmol/L NO (right). CFUs = colony forming units; LB = Luria-Bertani; NO = nitric oxide.
Susceptibility of bacterial species to NO
S aureus, S epidermis, K pneumoniae, and P aeruginosa strains were grown overnight in LB broth and then diluted to an OD600 of 0.1 prior to the start of the experiment. The bacteria were exposed to increasing concentrations of NO (0, 0.6, 3, 6, 30, and 60 μmol/L) and grown at 37°C with shaking. These concentrations of NO were chosen based on the physiologic range observed in experiments demonstrating human sinonasal epithelial cell release of NO in response to microbial challenge.15,25,28,30 Specifically, the concentration range of NO observed in vivo in health and disease generally ranges from 100 to 1000 parts per billion, which corresponds to 3.3 to 33 μmol/L NO.30 CFUs were counted at 0, 3, and 6 hours and recorded for concentrations of 0, 3, 6, and 30 μmol/L (Fig. 2). OD600 was recorded at all 3 time-points for all concentrations, and these data were used to calculate IC50 and MIC for each species (nonlinear regression curve-fitting, and Gompertz modeling, respectively; Table 1).
FIGURE 2.
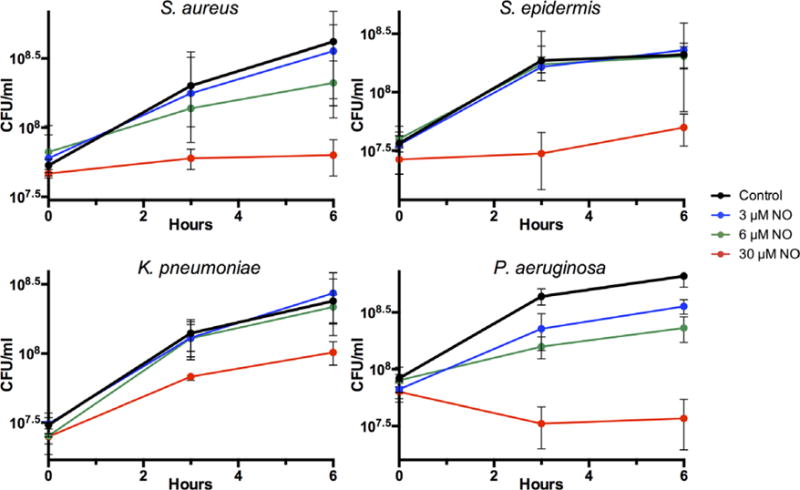
Susceptibility of log-phase S aureus, S epidermis, K pneumoniae, and P aeruginosa to varying concentrations of NO in LB media (0 μmol/L, 3 μmol/L, 6 μmol/L, and 30 μmol/L). Individual bacteria CFUs/mL were calculated at 0, 3, and 6 hours after initiation of treatment with NO. The data are mean ± standard error of 3 individual observations. CFUs = colony forming units; LB = Luria-Bertani; NO = nitric oxide.
TABLE 1.
Calculation of IC50 and MIC of NO for all organisms tested
| S aureus | S epidermis | K pneumoniae | P aeruginosa | C albicans | |
|---|---|---|---|---|---|
| IC50 (μmol/L NO) | 8.4 | 27.0 | 30.0 | 9.0 | 4.8 |
| MIC (μmol/L NO) | 42.6 | 35.4 | 55.8 | 23.4 | 6.0 |
Data points used were 3-hour OD600 for bacteria and 6-hour OD600 for fungus, in the presence of 0 μmol/L, 0.6 μmol/L, 3 μmol/L, 6 μmol/L, 30 μmol/L, and 60 μmol/L NO. IC50 was calculated using nonlinear regression curve fitting, and MIC was calculated by fitting the data to a Gompertz model. IC50 = 50% inhibitory concentration; MIC = minimum inhibitory concentration; OD600 = optical density at 600 nm; NO = nitric oxide.
Overall, S epidermis and K pneumoniae demonstrated minimal susceptibility to concentrations of 6 μmol/L NO or below, and had IC50 values of 27.0 μmol/L and 30.0 μmol/L NO, respectively. This is in contrast to S aureus (IC50 8.4 μmol/L NO) and P aeruginosa (IC50 9.0 μmol/L NO), which showed stunting of growth in response to the lower NO concentrations tested. A concentration of 30 μmol/L NO was slightly bactericidal in P aeruginosa, yet this concentration was only bacteriostatic in S aureus. This phenomenon is reflected in the calculated MICs for each bacterium, as P aeruginosa had an MIC of 23.4 μmol/L, whereas the other 3 bacteria had MICs between 30 and 60 μmol/L NO. Of interest, S aureus had a low IC50 value with a higher MIC value, indicating that, although stunting of bacterial growth occurs early, complete inhibition of bacterial growth requires a much higher concentration of NO. This is juxtaposed with the high IC50 and relatively low MIC value of S epidermis, indicating that early initial resistance to lower NO concentrations can quickly be overcome with small NO concentration increases in this species. The MBC could not be calculated, as no bacteria showed 99.9% inhibition to the highest concentration of NO tested. Of note, apical concentrations of NO from 0 to 60 μmol/L demonstrated no overt cytoxicity in human airway epithelial cells over a 6-hour time period, as assessed by LDH release (refer to Fig. S1 in the Supplementary Material online). NO of 600 μmol/L, a concentration 10-fold higher than that used in bacterial experiments, elicited a high degree of LDH release with suspected cellular death of epithelial cells in the experimental paradigm.
Susceptibility of C albicans to NO
C albicans liquid cultures were prepared from individual colonies grown on YPD agar that were inoculated into YPD broth and shaken at 30°C for 12 hours at 300 rpm, then diluted to an OD600 of 0.1. Samples from the culture were exposed to the same NO concentrations as the bacteria in the previous experiment, but CFUs were counted only at 6 hours (Fig. 3). OD600 was also recorded at 6 hours to calculate IC50 and MIC values (Table 1). C albicans demonstrated much lower IC50 (4.8) and MIC (6.0) than any of the bacteria, signifying a much higher susceptibility to NO at all concentrations.
FIGURE 3.
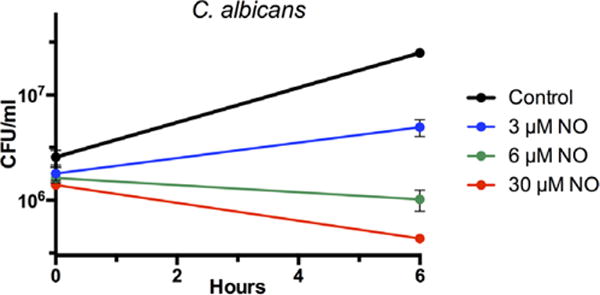
Susceptibility of log-phase C albicans to varying concentrations of NO in LB media (0 μmol/L, 3 μmol/L, 6 μmol/L, and 30 μmol/L). CFUs/mL were calculated at 0 and 6 hours after initiation of treatment with NO. The data are mean ± standard error of 3 individual observations. CFUs = colony forming units; NO = nitric oxide; LB = Luria-Bertani.
Effects of culture ASL on NO-susceptible bacteria
As many diverse innate immune defenses function together in the sinonasal tract, we were interested in a possible synergistic effect between ASL compounds and NO. Prior work has demonstrated that human airway epithelial cells release antimicrobial peptides, such as β-defensins, as well as other toxic radicals that serve as an additional layer of innate immune defense against bacteria.31,32 In addition, our previous work has shown the sensitivity of all 4 bacteria utilized in this study to some of these compounds.8 To test the hypothesis that effects of these compounds and NO would be additive, ASL was aspirated from 18 ALI cultures that were at least 1 month old, and the ASL was then diluted in 1 mL of deionized water. Seventy-five microliters of diluted ASL or deionized water was added to 75 μmol/L of full-strength LB medium with S aureus or P aeruginosa at an OD600 of 0.2, for a final OD600 of approximately 0.1. CFUs were counted at 0 and 3 hours and are shown in Figure 4. ASL was permissive of S aureus growth, and no significant effects were observed. However, P aeruginosa growth was inhibited by ASL and 6 μmol/L NO to a comparable degree, and the combination of ASL and 6 μmol/L NO caused almost complete cessation of growth (p < 0.01, 2-way ANOVA; n = 3). The effects of ASL and NO were additive, rather than synergistic, based on the absence of an observed statistical interaction between the 2 experimental manipulations.
FIGURE 4.
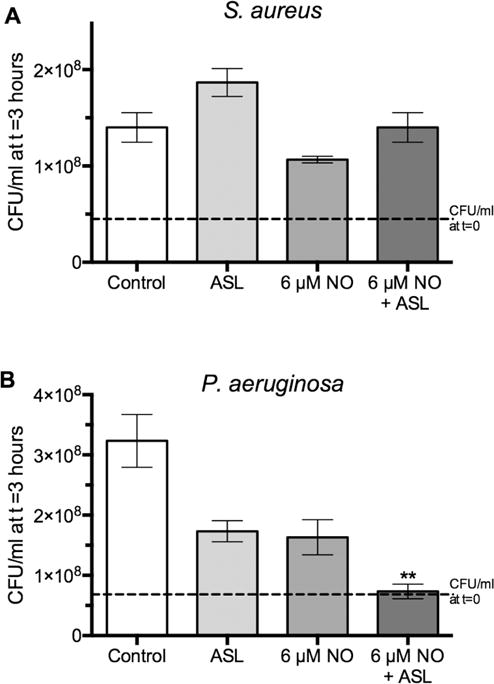
Susceptibility of log-phase S aureus and P aeruginosa to ASL from ALI cultures, with and without the addition of 6 μmol/L NO. All growth data are presented as fold increase in CFUs/mL after a 3-hour growth period. All experimental conditions were compared with the control condition (2-way ANOVA, **p < 0.01; n = 3 per condition). Dashed line demonstrates bacterial concentration at t = 0. ANOVA = analysis of variance. ALI = air-liquid interface; ANOVA = analysis of variance;ASL = airway surface liquid; CFUs = colony forming units.
Discussion
In this study we tested the relative susceptibility of common sinonasal organisms to NO, an antibacterial free radical released by airway epithelial cells as a part of innate immune defense. In bacteria, NO has toxic effects on several cellular targets, including protein thiols, iron, tyrosine residues, lipids, and DNA.18 Several discrepancies in organism sensitivity to NO were observed. P aeruginosa and C albicans were more sensitive to the effects of NO at all concentrations tested, whereas S aureus had intermediate initial sensitivity to low amounts of NO followed by resistance to complete bacteriostasis at higher concentrations. K pneumoniae and S epidermis were fairly insensitive to the effects of NO at physiologic concentrations, and very high quantities of NO were required to achieve complete bacteriostasis.
The implications for these differences in sensitivity are broad, and may influence the composition of the nasal microbiome. Species such as S aureus, S epidermis, and K pneumoniae are most commonly commensal organisms in the healthy nasopharynx, and more rarely become pathogenic when homeostasis is disrupted.33–36 These organisms are kept in check by NO production, but are not fully eradicated. Alternatively, the presence of P aeruginosa is almost always indicative of disease. The high sensitivity of P aeruginosa to NO, as suggested by its low IC50 and MIC, likely contributes to it being an uncommon presence in healthy subjects. Indeed, our earlier research has indicated that the genetic difference in the physiologic ability to release NO in response to pathogens correlates with clinical outcomes in diseased patients.37,38 In addition, work by Bommarito et al has demonstrated that there is variability in endogenous NO levels among patients, and individuals with CRS with nasal polyps have significantly lower NO concentrations in the sinonasal tract.30
Density control of bacteria by NO is not entirely predicated on bacterial killing and elimination. Rather, NO more often impairs bacterial proliferation to allow for other immune responses to act more efficiently and resolve disease with return to homeostasis.39 An example of this phenomenon is illustrated in the case of P aeruginosa, in which NO disrupts immune-resistant biofilm formation so that other immune defenses have unimpeded access to the bacterium.40,41 Although some organisms are in fact readily killed by NO,12,22 it is antimicrobial eptides like β-defensins and other related compounds that have additional potent effects.31,32 Previous experiments have also demonstrated that gram-negative bacteria have greater susceptibility than gram-positive bacteria to antimicrobial compounds, including β-defensin 2.35,42 In our study, P aeruginosa showed almost complete bacteriostasis when exposed to low levels of NO and ASL, neither of which was sufficient alone to cause the same impedance of bacterial proliferation. This additive effect was not observed in S aureus. Whereas ASL and low concentrations of NO are permissive for controlled growth of S aureus, P aeruginosa does not share the same fate, again indicating a differential response and more efficient eradication of the more virulent organism. This combination of innate immune mechanisms is also a reason that MBC of NO could not be calculated at the physiologic concentrations used in our experiment, as bacterial elimination is a result of several distinct processes.
One phenomenon of interest that we observed is the initial sensitivity of S aureus to low physiologic concentrations of NO that does not readily progress to overt bacteriostasis at higher concentrations. The exact opposite observation was made in S epidermis, with a high IC50 and low MIC indicating potent initial NO resistance, quickly progressing to bacteriostasis with small NO concentration increases beyond that point. This phenomenon is best explained by an NO-inducible LDH metabolic pathway that exists in S aureus and not in S epidermis.18 Alternative cellular machinery allows S aureus to proliferate in an attenuated fashion even at high physiologic concentrations of NO. This corroborates its role as a commensal organism that is difficult to eliminate, even when pathogenic. The differential NO sensitivity between the 2 Staphylococcus species also substantiates previous work by Carey et al, showing a less potent NO response from sinonasal epithelial cells to S epidermis when compared with that of S aureus.14 Other bacteria can upregulate NO scavengers or alter cellular respiratory function to evade the effects of NO through changes in gene expression.43 However, it is important to note that this resistance does not induce adaptive mutations in the bacteria, as expansion of cultures that initially survive NO treatment does not yield increasingly resistant cultures.27 Of additional interest is the anti-biofilm effect of high-physiologic NO on S aureus, in contrast to the biofilmenhancing effect of lower concentrations of NO on the same organism.44 This phenomenon, observed by Jardeleza et al, may help explain the low incidence of full bacterial eradication, as well as the concentration differences observed in the healthy and diseased microbiome.30 Individuals who produce low-physiologic quantities of NO may be exponentially more susceptible to overgrowth of these commensal organisms.
Beyond bacteria, C albicans is a ubiquitous fungus that is both a commensal organism and opportunistic pathogen, much like many of the bacteria we studied.24 Our experiments showed that C albicans was more sensitive to NO than any of the bacteria we tested, and had a very narrow concentration range between its IC50 and MIC. This increased responsiveness is likely multifactorial and probably related to increased cellular targets and slower adaptation mechanisms.24
The potent antimicrobial activity of NO allows for therapeutic opportunities. Ideally, harnessing the power of host signaling pathways for endogenous NO production allows for a streamlined approach. These pathways cause NO to be released steadily over time, and to minimize deleterious effects of exogenous NO on host cells. The signaling pathways that detect microbial products are still being elucidated, but the stimulation of bitter taste receptors using exogenously applied compounds shows promise, and will be a likely subject of future investigations.8 Critically, this also allows for NO to be produced at the site of interest, as opposed to in a less specific fashion.45 Other research involving the use of NO-releasing silica nanoparticles and xerogel films has also shown initial promise for candidate antimicrobials, especially against biofilm-forming bacteria.11 Further experimentation could potentially optimize other experimental variables that may have effects on NO in vivo, including nonliquid culture conditions, airflow in the sinonasal tract, and possible anaerobic environments. In addition, the identification of specific airway compounds that demonstrate additive or synergistic effects with those of NO is also an important next step.
In conclusion, there is differential NO susceptibility of common airway microbes, and these discrepancies may help explain variations in the composition of the human microbiome in health and disease. P aeruginosa was the bacterium most sensitive to NO in our study, whereas commensal organisms, such as S epidermis and K pneumoniae, were significantly more NO-resistant at physiologic NO concentrations. Tonically secreted compounds from human sinonasal epithelial cells show an additive antibacterial effect when combined with NO in P aeruginosa but not S aureus cultures. Knowledge of the relative susceptibilities of these organisms to innate immune defenses can help develop novel therapeutics that release NO when applied or act through endogenous NO pathways.
Supplementary Material
Footnotes
Additional Supporting Information may be found in the online version of this article.
Presented at the Combined Otolaryngology Spring Meeting (COSM) of the AAOA-ARS, on April 27, 2017, in San Diego, CA.
Potential conflict of interest: None provided.
References
- 1.Mahdavinia M, Keshavarzian A, Tobin MC, et al. A comprehensive review of the nasal microbiome in chronic rhinosinusitis (CRS) Clin Exp Allergy. 2016;46:21–41. doi: 10.1111/cea.12666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachert C, Gevaert P, van Cauwenberge P. Staphylococcus aureus enterotoxins: a key in airway disease? Allergy. 2002;57:480–487. doi: 10.1034/j.1398-9995.2002.02156.x. [DOI] [PubMed] [Google Scholar]
- 3.Shin SH, Ponikau JU, Sherris DA, et al. Chronic rhinosinusitis: an enhanced immune response to ubiquitous airborne fungi. J Allergy Clin Immunol. 2004;114:1369–1375. doi: 10.1016/j.jaci.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 4.Boase S, Foreman A, Cleland E, et al. The microbiome of chronic rhinosinusitis: culture, molecular diagnostics and biofilm detection. BMC Infect Dis. 2013;13:210. doi: 10.1186/1471-2334-13-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biel MA, Brown CA, Levinson RM, et al. Evaluation of the microbiology of chronic maxillary sinusitis. Ann Otol Rhinol Laryngol. 1998;107:942–945. doi: 10.1177/000348949810701107. [DOI] [PubMed] [Google Scholar]
- 6.Lau HY, Huffnagle GB, Moore TA. Host and microbiota factors that control Klebsiella pneumoniae mucosal colonization in mice. Microbes Infect. 2008;10:1283–1290. doi: 10.1016/j.micinf.2008.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parker D, Prince A. Innate immunity in the respiratory epithelium. Am J Respir Cell Mol Biol. 2011;45:189–201. doi: 10.1165/rcmb.2011-0011RT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee RJ, Kofonow JM, Rosen PL, et al. Bitter and sweet taste receptors regulate human upper respiratory innate immunity. J Clin Invest. 2014;124:1393–1405. doi: 10.1172/JCI72094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamamoto K, Ishimaru Y. Oral and extra-oral taste perception. Sem Cell Dev Biol. 2013;24:240–246. doi: 10.1016/j.semcdb.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 10.Brett PJ, Burtnick MN, Su H, et al. iNOS activity is critical for the clearance of Burkholderia mallei from infected RAW 264.7 murine macrophages. Cell Microbiol. 2008;10:487–498. doi: 10.1111/j.1462-5822.2007.01063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hetrick EM, Shin JH, Paul HS, et al. Anti-biofilm efficacy of nitric oxide-releasing silica nanoparticles. Biomaterials. 2009;30:2782–2789. doi: 10.1016/j.biomaterials.2009.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones-Carson J, Laughlin J, Hamad MA, et al. Inactivation of [Fe-S] metalloproteins mediates nitric oxide-dependent killing of Burkholderia mallei. PLoS One. 2008;3:e1976. doi: 10.1371/journal.pone.0001976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carey RM, Workman AD, Chen B, et al. Staphylococcus aureus triggers nitric oxide production in human upper airway epithelium. Int Forum Allergy Rhinol. 2015;5:808–813. doi: 10.1002/alr.21568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carey RM, Chen B, Adappa ND, et al. Human upper airway epithelium produces nitric oxide in response to Staphylococcus epidermidis. Int Forum Allergy Rhinol. 2016;6:1238–1244. doi: 10.1002/alr.21837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang FC. Perspectives series: host/pathogen interactions. Mechanisms of nitric oxide–related antimicrobial activity. J Clin Invest. 1997;99:2818–2825. doi: 10.1172/JCI119473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marcinkiewicz J. Nitric oxide and antimicrobial activity of reactive oxygen intermediates. Immunopharmacology. 1997;37:35–41. doi: 10.1016/s0162-3109(96)00168-3. [DOI] [PubMed] [Google Scholar]
- 17.Barraud N, Hassett DJ, Hwang SH, et al. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J Bacteriol. 2006;188:7344–7353. doi: 10.1128/JB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richardson AR, Libby SJ, Fang FC. A nitric oxide– inducible lactate dehydrogenase enables Staphylococcus aureus to resist innate immunity. Science. 2008;319:1672–1676. doi: 10.1126/science.1155207. [DOI] [PubMed] [Google Scholar]
- 19.Shaari J, Palmer JN, Chiu AG, et al. Regional analysis of sinonasal ciliary beat frequency. Am J Rhinol. 2006;20:150–154. [PubMed] [Google Scholar]
- 20.Salathe M. Regulation of mammalian ciliary beating. Ann Rev Physiol. 2007;69:401–422. doi: 10.1146/annurev.physiol.69.040705.141253. [DOI] [PubMed] [Google Scholar]
- 21.Hilding AC. The role of the respiratory mucosa in health and disease. Minn Med. 1967;50:915–919. [PubMed] [Google Scholar]
- 22.Yoon SS, Coakley R, Lau GW, et al. Anaerobic killing of mucoid Pseudomonas aeruginosa by acidified nitrite derivatives under cystic fibrosis airway conditions. J Clin Invest. 2006;116:436–446. doi: 10.1172/JCI24684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren B, Zhang N, Yang J, et al. Nitric oxideinduced bacteriostasis and modification of ironsulphur proteins in Escherichia coli. Mol Microbiol. 2008;70:953–964. doi: 10.1111/j.1365-2958.2008.06464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vazquez-Torres A, Jones-Carson J, Balish E. Nitric oxide production does not directly increase macrophage candidacidal activity. Infect Immunity. 1995;63:1142–1144. doi: 10.1128/iai.63.3.1142-1144.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mannick JB. Immunoregulatory and antimicrobial effects of nitrogen oxides. Proc Am Thorac Soc. 2006;3:161–165. doi: 10.1513/pats.200505-048BG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Staab JF, Bahn YS, Sundstrom P. Integrative, multifunctional plasmids for hypha-specific or constitutive expression of green fluorescent protein in Candida albicans. Microbiology. 2003;149:2977–2986. doi: 10.1099/mic.0.26445-0. [DOI] [PubMed] [Google Scholar]
- 27.Jones-Carson J, Laughlin JR, Stewart AL, et al. Nitric oxide-dependent killing of aerobic, anaerobic and persistent Burkholderia pseudomallei. Nitric Oxide Biol Chem. 2012;27:25–31. doi: 10.1016/j.niox.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee RJ, Xiong G, Kofonow JM, et al. T2R38 taste receptor polymorphisms underlie susceptibility to upper respiratory infection. J Clin Invest. 2012;122:4145–4159. doi: 10.1172/JCI64240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merkus FW, Verhoef JC, Schipper NG, et al. Nasal mucociliary clearance as a factor in nasal drug delivery. Adv Drug Deliv Rev. 1998;29:13–38. doi: 10.1016/s0169-409x(97)00059-8. [DOI] [PubMed] [Google Scholar]
- 30.Bommarito L, Guida G, Heffler E, et al. Nasal nitric oxide concentration in suspected chronic rhinosinusitis. Ann Allergy Asthma Immunol. 2008;101:358–362. doi: 10.1016/S1081-1206(10)60310-9. [DOI] [PubMed] [Google Scholar]
- 31.Ooi EH, Wormald PJ, Tan LW. Innate immunity in the paranasal sinuses: a review of nasal host defenses. Am J Rhinol. 2008;22:13–19. doi: 10.2500/ajr.2008.22.3127. [DOI] [PubMed] [Google Scholar]
- 32.Schroeder BO, Wu Z, Nuding S, et al. Reduction of disulphide bonds unmasks potent antimicrobial activity of human beta-defensin 1. Nature. 2011;469:419–423. doi: 10.1038/nature09674. [DOI] [PubMed] [Google Scholar]
- 33.Hunsaker DH, Leid JG. The relationship of biofilms to chronic rhinosinusitis. Curr Opin Otolaryngol Head Neck Surg. 2008;16:237–241. doi: 10.1097/MOO.0b013e3282fdc6d5. [DOI] [PubMed] [Google Scholar]
- 34.Niederfuhr A, Kirsche H, Riechelmann H, et al. The bacteriology of chronic rhinosinusitis with and without nasal polyps. Arch Otolaryngol Head Neck Surg. 2009;135:131–136. doi: 10.1001/archoto.2008.531. [DOI] [PubMed] [Google Scholar]
- 35.Pandak N, Pajic-Penavic I, Sekelj A, et al. Bacterial colonization or infection in chronic sinusitis. Wienerklinische Wochenschrift. 2011;123:710–713. doi: 10.1007/s00508-011-0093-x. [DOI] [PubMed] [Google Scholar]
- 36.Dlugaszewska J, Leszczynska M, Lenkowski M, et al. The pathophysiological role of bacterial biofilms in chronic sinusitis. Eur Arch Otorhinolaryngol. 2016;273:1989–1994. doi: 10.1007/s00405-015-3650-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adappa ND, Farquhar D, Palmer JN, et al. TAS2R38 genotype predicts surgical outcome in nonpolypoid chronic rhinosinusitis. Int Forum Allergy Rhinol. 2016;6:25–33. doi: 10.1002/alr.21666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adappa ND, Zhang Z, Palmer JN, et al. The bitter taste receptor T2R38 is an independent risk factor for chronic rhinosinusitis requiring sinus surgery. Int Forum Allergy Rhinol. 2014;4:3–7. doi: 10.1002/alr.21253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bogdan C. Nitric oxide synthase in innate and adaptive immunity: an update. Trends Immunol. 2015;36:161–178. doi: 10.1016/j.it.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 40.Zemke AC, Shiva S, Burns JL, et al. Nitrite modulates bacterial antibiotic susceptibility and biofilm formation in association with airway epithelial cells. Free Rad Biol Med. 2014;77:307–316. doi: 10.1016/j.freeradbiomed.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim SK, Lee JH. Biofilm dispersion in Pseudomonas aeruginosa. J Microbiol. 2016;54:71–85. doi: 10.1007/s12275-016-5528-7. [DOI] [PubMed] [Google Scholar]
- 42.Schroder JM, Harder J. Human beta-defensin-2. Int J Biochem Cell Biol. 1999;31:645–651. doi: 10.1016/s1357-2725(99)00013-8. [DOI] [PubMed] [Google Scholar]
- 43.Nunoshiba T, deRojas-Walker T, Wishnok JS, et al. Activation by nitric oxide of an oxidative-stress response that defends Escherichia coli against activated macrophages. Proc Natl Acad Sci USA. 1993;90:9993–9997. doi: 10.1073/pnas.90.21.9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jardeleza C, Foreman A, Baker L, et al. The effects of nitric oxide on Staphylococcus aureus biofilm growth and its implications in chronic rhinosinusitis. Int Forum Allergy Rhinol. 2011;1:438–444. doi: 10.1002/alr.20083. [DOI] [PubMed] [Google Scholar]
- 45.Breitbach K, Wongprompitak P, Steinmetz I. Distinct roles for nitric oxide in resistant C57BL/6 and susceptible BALB/c mice to control Burkholderia pseudomallei infection. BMC Immunol. 2011;12:20. doi: 10.1186/1471-2172-12-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


