Abstract
Background
Mov10 is an RNA helicase that modulates access of Argonaute 2 to microRNA recognition elements in mRNAs. We examined the role of Mov10 in Xenopus laevis development and show a critical role for Mov10 in gastrulation and in the development of the central nervous system.
Results
Knockdown of maternal Mov10 in Xenopus embryos using a translation blocking morpholino led to defects in gastrulation and the development of notochord and paraxial mesoderm, and a failure to neurulate. RNA sequencing of the Mov10 knockdown embryos showed significant upregulation of many mRNAs when compared to controls at stage 10.5 (including those related to the cytoskeleton, adhesion, and extracellular matrix, which are involved in those morphogenetic processes). Additionally, the degradation of the miR-427 target mRNA, cyclin A1, was delayed in the Mov10 knockdowns. These defects suggest that Mov10’s role in miRNA-mediated regulation of the maternal to zygotic transition could lead to pleiotropic effects that cause the gastrulation defects. Additionally, the knockdown of zygotic Mov10 showed that it was necessary for normal head, eye and brain development in Xenopus consistent with a recent study in the mouse.
Conclusions
Mov10 is essential for gastrulation and normal CNS development.
Keywords: RNA helicase, RISC, brain, embryonic development, Mov10, gastrulation
Introduction
The RNA helicase Mov10 was originally described as a cofactor for RNA-induced silencing complex (RISC) component Argonaute 2 (Ago2) that was required for microRNA (miRNA)-guided cleavage of a reporter (Meister et al., 2005). Mov10 binds to G-rich secondary structures in mRNAs and unwinds RNA in a 5′-3′ direction in an ATP-dependent manner (Gregersen et al., 2014; Kenny et al., 2014). Mov10 also associates with nonsense-mediated decay factor UPF1 (Gregersen et al., 2014). In addition, Mov10 suppresses viral RNAs and retrotransposition in cultured cells (Burdick et al., 2010; Goodier et al., 2012). We recently showed that Mov10 is required during embryonic development because the Mov10 knockout mouse is embryonic lethal. As we were unable to identify the early developmental defects associated with this lethality in the mouse model (Skariah et al., 2017), we sought to determine the function of Mov10 in another well-established vertebrate model system, Xenopus laevis.
Here, we demonstrate a conserved role for Mov10 during embryonic development in X. laevis. Blocking translation of maternal Mov10 in X. laevis embryos leads to a severe gastrulation defect and failure of the embryo to undergo neurulation. This may be due to the misregulation of the maternal to zygotic transition (MZT), where one proposed mechanism involves the degradation of maternal mRNAs by RISC to permit zygotic transcription (Tadros and Lipshitz, 2009; Langley et al., 2014). Loss of zygotic Mov10 using a splice-blocking morpholino in X. laevis leads to defects in the differentiation of the retina and abnormalities in brain development. These data agree with our findings in mice where, in addition to being essential for early development, Mov10 expression was found to be significantly elevated in the brain shortly after birth through adolescence (Skariah et al., 2017). We propose that Mov10 plays a vital role in gastrulation and normal CNS development.
Results
Mov10 is required for normal gastrulation and neural tube formation
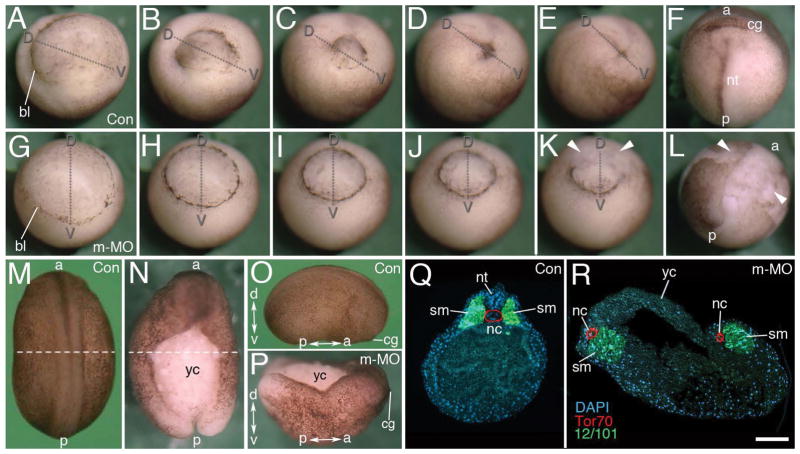
Based on our finding that Mov10 knockouts show early embryonic lethality in mice, we used X. laevis to study the cause of this lethality because of its accessible, external mode of development. We introduced either a control- or a Mov10 translation blocking morpholino into one-cell X. laevis embryos (Fig. 1), which targeted maternal, as well as zygotic mRNA transcripts of Mov10 to prevent them from being translated. In contrast to the control embryos (Fig. 1A–F), the translation blocking morpholino (m-MO) disrupted the completion of gastrulation (Fig. 1G–L). Time-lapse imaging of the m-MO injected embryos showed that gastrulation is initiated, but epiboly appeared to proceed in a more uniform, symmetrical fashion around the blastopore, as indicated by a complete ring of dark pigmentation, when compared to the asymmetrical appearance found in control embryos (Supplemental videos 1 and 2, Fig. 1A–L). Sections revealed that there is no distinct formation of Brachet’s cleft (compare Fig. 2B to 2C) and that vegetal rotation is delayed and limited in extent. This symmetrical blastopore formation may be driven primarily by convergent thickening (Fig. 1, 2C). Ultimately, the blastopore remains open in the m-MO injected embryos to generate a central mass of exposed endodermal cells (Fig. 1K–L, N, P, Supplemental videos 1 and 2). Some yolky vegetal cells are expelled from these embryos and cells located along the dorsal lip also lose their integrity. The blastopore remains open and as a consequence, the neural plate and neural tube fail to form in these embryos (Fig. 1L,N, R and quantified in Fig. 2A, m-Mo injected).
Figure 1. Maternal Mov10 is required for gastrulation and neural tube formation.
A–F) Successive time-lapse images of whole-mount control embryo undergoing normal gastrulation. G–L) Successive time-lapse images of whole-mount maternal Mov10 knockdown (m-MO) embryo that fails to complete gastrulation. Dorso-ventral and anterior-posterior axes are as labeled. White arrowheads in K and L point to yolky debris and loose cells of the blastopore and dorsal lip. M–P) Dorsal, whole-mount images from Control (Con) and m-MO injected embryos. M) dorsal view of stage 21 control embryo. N) Dorsal view of sibling m-MO injected embryo. Note large opening with exposed yolky mass of cells on the dorsal surface. The loose yolky debris has washed away from this hatched embryo. O) Lateral view of a Con embryo like that shown in M. P) Dorsal view from anterior end of m-MO injected embryo showing the “boat-shaped” phenotype. The dotted lines in M and N show the plane of sections for Q and R, respectively. Q) Section through a stage 21 control embryo showing a single notochord and two rows of somites united along the dorsal midline. Antibody to Tor70 recognizes notochord cells (in red); Antibody 12/101 recognizes somitic mesoderm (in green); DAPI in blue R) Section through a typical m-MO embryo showing failure to complete gastrulation and neurulation. This embryo has two separated files of notochord and somitic mesoderm. The dorsal side is located towards the top in Q and R. a, anterior; bl, blastopore lip; cg, cement gland; d, dorsal; nt, neural tube; nc, notochord; nt, neural tube; p, posterior; sm, somite; v, ventral; yc, yolk cells. Scale bar in R equals 120μm (for A–L), 450μm (for M–N), 440μm (for O–P), and 80μm (for Q–R).
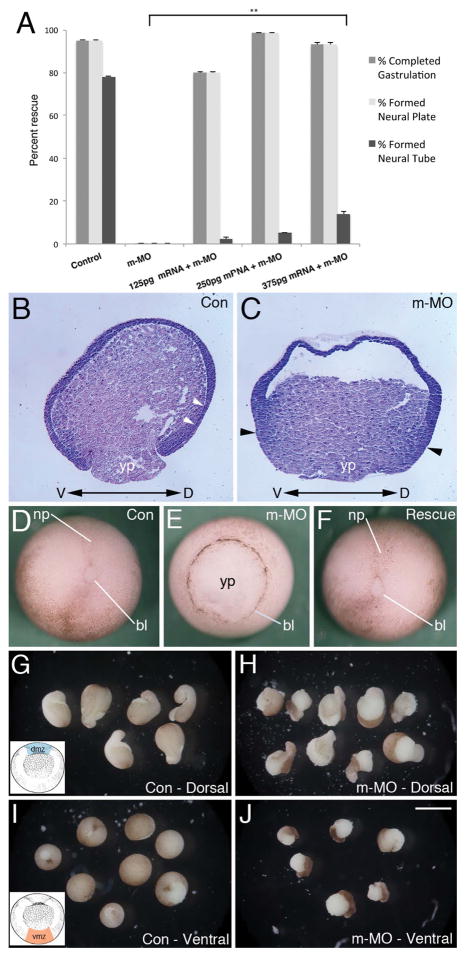
Figure 2. mRNA rescue and MO effects on convergent extension.
A) Partial rescue of m-MO phenotype by introduction of Mov10 mRNA. Error bars represent SEM, **p<0.01 (Student’s t-test, two-tailed). Mov10 mRNA rescued cases that underwent gastrulation also formed a neural plate, but those cases typically disintegrate a few hours later so they are unable to complete neurulation and form a neural tube. B–C) X. laevis stage 10 sagittal sections stained with Hematoxylin and Eosin. Dorsal (D) and ventral (V) sides are as noted. B) Representative section of a control (Con) embryo, showing Brachet’s cleft (white arrowheads), which separates the outer ectoderm from the underlying mesendoderm. C) Representative section of a sibling m-MO injected embryo, which lacks a visible delineation between ectodermal and mesendodermal layers (Brachet’s cleft). Note that cells along the dorsal and ventral sides of the blastopore and yolk plug, denoted with black arrowheads, appear to be thicker, having mainly undergone convergent thickening (compare B to C). D) Whole-mount image of representative stage 13–14 Control (Con) embryo with closed blastopore. E) Representative sibling m-MO embryo with unclosed blastopore. F) Rescued embryo co-injected with m-MO and Mov10 mRNA (250pg) with closed blastopore. G–J). Development of stage 10 dorsal (dmz) and ventral marginal zone (vmz) explants harvested from the region shown in the inset included in G and I, respectively. G) Control dmz explants eventually form mesoderm that undergoes convergent extension, as revealed by the formation of elongated projections. H) dmz explants from m-MO injected embryos also show signs of convergent extension and elongation. Note, however, that the pigmented ectoderm does not cover the mesodermal tissue as completely as seen in control explants (compare with G). I) vmz explants harvested from stage 10 control embryos do not exhibit signs of elongation and form spherical embryoids. J) Likewise, m-MO injected embryos also form spherical embryoids. Note that the pigmented ectoderm has not covered the surface of these explants. bl, lip of blastopore; np, neural plate; yp, yolk plug. Scale bar in J equals 180μm for B–C, 250μm for D–F, 850μm for G–J.
During normal development, cells of the notochord undergo convergent extension (CE) and the notochord shears posteriorly along the anterior-posterior axis with respect to the adjacent paraxial mesoderm. Tissue separation occurs to permit involution of marginal zone tissues, and the notochord acts as a zipper to pull together the paraxial somitic files at the midline. The location of these cells can be visualized by immunofluorescence using the Tor70 and 12/101 antibodies, which stain the notochord and somites, respectively (Fig. 1Q–R) (Keller et al., 1985; Davidson and Keller, 1999). In the m-MO injected embryos convergent extension of notochord cells still took place; however, the notochord and paraxial, somitic mesoderm were spilt into bilateral files located on both sides of the exposed dorsal opening (Fig. 1R). The behavior of these tissues likely enlarges and elongates the persistent opening of the blastopore, which ends up being located on the dorsal side of the embryo. As a result, the defective embryos exhibited an elongated “boat-shaped” phenotype (Fig. 1N, P). Together, these findings indicate that Mov10 is required early during development for normal gastrulation and neural tube formation. We were able to partially rescue defects caused by m-MO, by co-injecting Mov10 mRNA designed to be resistant to m-MO action (Fig. 2A,D–F). These embryos were able to complete gastrulation and form the neural plate. We saw a dose dependent increase in the rescued phenotypes as we injected a greater amount of Mov10 mRNA (Fig. 2A) suggesting that the gastrulation defect is a result of the loss of Mov10. Typically, most of these embryos disintegrate a few hours later and do not complete neurulation (e.g., form a neural tube). Why these cases ultimately do not survive is unclear.
To examine the behavior of blastopore lip tissues and look for further evidence of convergent extension, we prepared dorsal and ventral lip explants (Fig. 2G–J). Dorsal lip explants from both control and m-MO underwent convergent extension as evidenced by their elongated morphology (Fig. 2G–H). One difference was noted in that the ectoderm did not cover the endoderm and mesoderm as completely in the m-MO explants. In contrast, control ventral lip explants do not elongate and simply form a rounded ball of tissue, as expected, where the pigmented epithelium expands to cover most of the exposed surface (Fig. 2I). Ventral lip explants from m-MO injected embryos, formed similar rounded balls of tissue (Fig. 2J) though these exhibited a failure of the ectoderm to cover the exposed surface. The failure of the ectoderm to cover these m-MO explants is consistent with the gastrulation defects we saw in the intact embryos, where gastrulation appears to be driven mainly by convergent thickening.
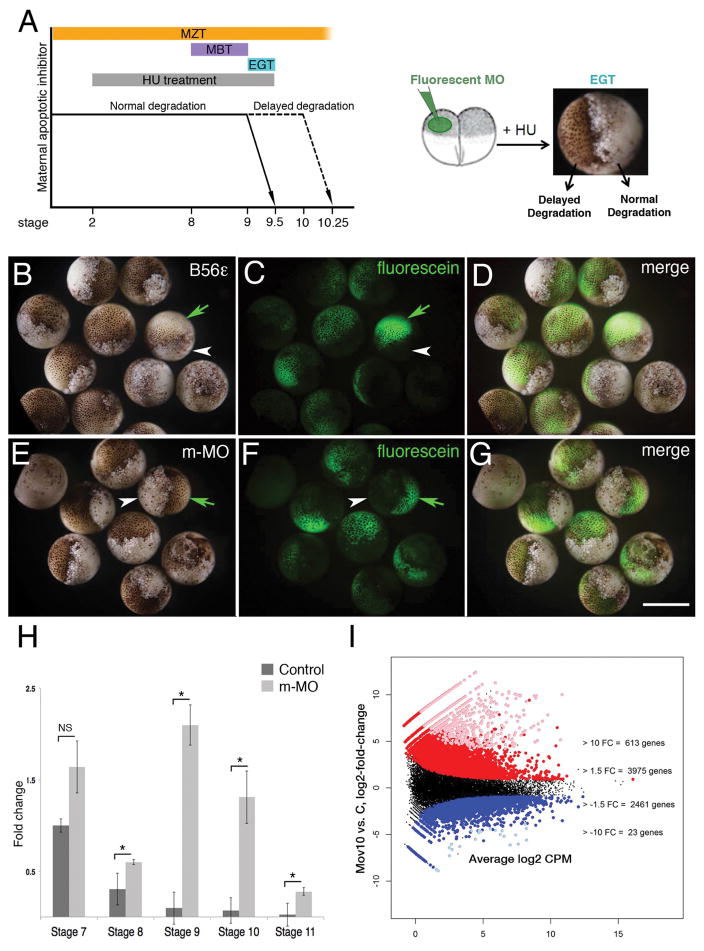
The early stages of embryonic development in metazoans mainly occur in the absence of de novo transcription and are directed by maternally deposited transcripts in the egg (Tadros and Lipshitz, 2009; Langley et al., 2014). In X. laevis embryos, the first stages following fertilization are characterized by fairly rapid cleavage divisions, synchronous cell cycles and the onset of maternal mRNA degradation. After twelve synchronous cleavage divisions, the cell cycles become asynchronous and large-scale zygotic transcription is detected for the first time. This developmental time-period is referred to as the Mid-Blastula Transition (MBT) and occurs between stages 8 and 9 (Fig. 3A). However, cell cycle progression at MBT is still under the control of maternal transcripts, and becomes dependent on zygotic transcripts following three more divisions at the onset of gastrulation. At this point, there are also large-scale cell movements, and the cells become susceptible to apoptosis due to the degradation of maternal apoptotic inhibitors. This developmental time period occurring after MBT is called the Early Gastrulation Transition (EGT, stage 9 – 9.5, see Fig. 3A; (Howe et al., 1995; Langley et al., 2014). Both MBT and EGT lie within the larger time period of embryonic development termed the Maternal to Zygotic Transition (MZT). MZT starts just after fertilization at stage 1 when maternal transcripts are beginning to be degraded, spanning the onset of zygotic transcription, and ending after the point when the cells have become susceptible to apoptosis [Fig. 3A, (Stack and Newport, 1997; Tadros and Lipshitz, 2009)]. Thus, zygotic transcription is coupled with the degradation of maternal mRNA transcripts during MZT, as exemplified by the miRNA-mediated degradation of cyclin A1 (Lund et al., 2009). Additionally, the key component of the RNA-Induced Silencing Complex (RISC), Ago2, has been shown to regulate MZT in mice (Lykke-Andersen et al., 2008). Since Mov10 and Ago2 cooperate to regulate transcripts through the RISC pathway (Sievers et al., 2012; Kenny et al., 2014), we examined whether Mov10 may be involved in MZT using the Hydroxyurea (HU) Assay (Fig. 3). HU induces apoptosis in embryos after the maternally-encoded apoptotic inhibitors are degraded during MZT [Fig. 3A, (Stack and Newport, 1997)]. Thus, if a protein involved in the degradation of maternal mRNAs is absent, the embryo is protected from HU-induced apoptosis at EGT, as shown previously for B56-epsilon mRNA [green arrows in Fig. 3B–D, (Jin et al., 2010)]. In the case of Mov10, m-MO injected into one cell of the two-cell embryo the fluorescently-labeled progeny showed a normal pigmented appearance (green arrows in Fig. 3E,F), suggesting that the maternal apoptotic inhibitors were not degraded; thus, MZT was blocked. In contrast, the uninjected side underwent HU-induced apoptosis to form a mass of yolky debris (white arrowheads in Fig. 3E,F). The apoptotic-resistance of the m-MO and B56-epsilon morpholino injected embryos showed that MZT is delayed and implicates Mov10 in MZT. We verified this by examining the effect of m-MO on cyclin A1 mRNA, a target that contains a single miR-427 binding site and that undergoes miRNA-mediated degradation at the onset of zygotic transcription (Lund et al., 2009). In m-MO injected embryos, cyclin A1 mRNA levels remained high at stages 8–11, in contrast to controls, where the levels decreased drastically as zygotic transcription begins (Fig. 3H). We conclude that Mov10 participates in MZT, possibly by mediating RISC activity.
Figure 3. Mov10 regulates MZT through RISC.
A) Schematic of developmental time periods and the Hydroxyurea (HU) assay. The Maternal to Zygotic Transition (MZT) spans between stage 1 and stage 10. The Mid-Blastula Transition (MBT) lies between stages 8 to 9. Early Gastrula Transition (EGT) occurs between stage 9 to 9.5. Hydroxyurea (HU) treatments lasted from the two-cell stage to stage 9.5. At the two-cell stage one of the blastomeres was injected with fluorescein-tagged morpholinos and the embryos were treated with HU until stage 9.5. B–D) Images of stage 9.5 embryos, where one cell had been injected with morpholino, targeting a positive regulator of MZT (B56ε) at the 20-cell stage. A total of 22 embryos were injected, and 21 of those showed the expected phenotype. Green arrows point to the live progeny of the injected cell in one of the cases shown. White arrowheads point to the dead progeny of the uninjected cell. E–G) Images of stage 9.5 embryos, where only one cell was injected with m-MO. A total of 22 embryos were injected and all of them showed the expected phenotype. H) qRT-PCR of cyclin A1 levels in control and m-MO embryos at indicated stages. Error bars represent SEM. NS- Not significant, p *<0.05 (Student’s t-test, two-tailed). I) Differential expression results for maternal Mov10 morpholino (m-MO) injected (Mov10) vs. control embryos (C), X-axis-average expression value (TMM-normalized Counts Per Million, log2 scale) of each X. laevis gene and y-axis is log2 (Mov10/C). Each point is a single gene: groups of genes colored red or blue had significantly greater than or less than 1.5 FC difference (unlogged), respectively. Groups of genes colored pink or light blue also had significantly greater than or less than 10 FC difference (unlogged), respectively. The numbers of genes listed at the +/− 1.5 FC level include the genes with +/− 10 FC. Scale bar in G equals 100μm for A an 300μm for B–G. Gene identities and statistical analysis are in Supplemental Table 2 (accessible through NCBI GEO accession number GSE86382).
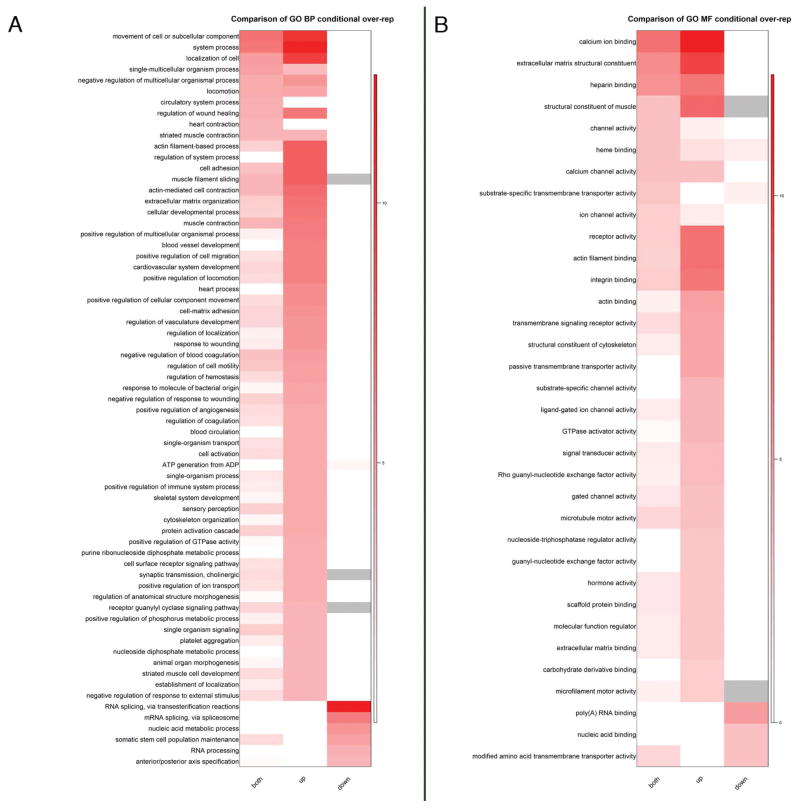
To examine the global effect of Mov10 loss on mRNAs at this stage, we isolated total RNA from stage 10.5 control and sibling m-MO injected embryos for RNA-seq. As expected for a miRNA-mediated regulatory event, Mov10 knockdown embryos showed a significant upregulation of mRNAs compared to controls both at a 1.5-fold and a 10-fold cut-off (Fig. 3I, gene identities and statistics are included in Supplementary Tables 1 and 2 at NCBI GEO GSE86382). Gene ontology analysis of the upregulated genes included RNAs involved in cellular movements, extracellular matrix, actin-filament based processes and cell adhesion under the category of “Biological Processes” or “BP” (Fig. 4). Misregulation of RNAs involved in these processes may explain the loss of tissue integrity on the dorsal side of the Mov10 m-Mo injected embryos (Fig. 1K,L). Under the category “Molecular Function” (MF), RNAs for calcium binding and actin filament binding are also significantly increased. Since activation of the egg at the onset of fertilization causes an increase in intracellular calcium levels, perturbation of these RNAs should be expected to affect signaling cascades necessary for multiple events including cytoskeletal rearrangements (Tadros and Lipshitz, 2009). The misregulated genes could thus contribute to the defects seen in gastrulation. These data suggest that Mov10 affects MZT through the RISC pathway, which subsequently causes pleiotropic effects leading to defective gastrulation and neurulation during embryonic development.
Figure 4. Gene ontology analysis (GO) of significantly changed RNAs between mMO injected and control embryos.
A) Heat maps of –log10 (p-values) showing the comparison of all significantly changed (both), up- or down-regulated gene sets from X. laevis for Biological Process (BP) category. Due to the larger number of BP terms, only those with raw p-values < 0.0001 in any gene set were included in the heatmap. B) Heat maps of –log10 (p-values) showing the comparison of all (both), up or down gene sets from X. laevis for Molecular Function (MF) category. GO terms with raw p-values < 0.001 in any gene set were included for MF.
Zygotic Mov10 is required for normal CNS development
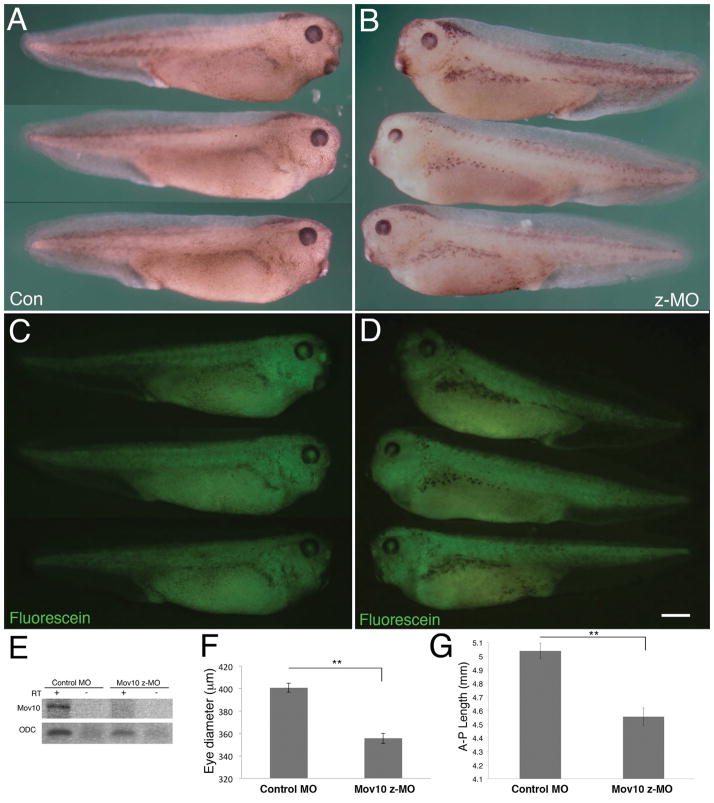
We also examined the effect of zygotic Mov10 knockdown using a splice-blocking morpholino (z-MO) that targets the 5′ splice junction between the third and fourth exon, resulting in a downstream frameshift and degradation of the zygotic pre-mRNA (Fig. 5). X. laevis embryos were injected with control morpholino or z-MO at the one-cell stage, which allows them to progress past MZT, and then examined at the tadpole stage, where the effect of a zygotic knockdown might be seen (stage 36; Fig. 5A–D). Mov10 targeting by this morpholino was confirmed by RT-PCR (Fig. 5E). The z-MO injected embryos hatch normally and show muscle contractility (data not shown). Interestingly, Mov10 z-MO injected embryos showed a significantly smaller eye diameter compared to control morpholino injected embryos (Fig. 5A–B,F). This may be relevant to Mov10’s role in the CNS since retinal tissue is derived from the diencephalon, which is a component of the forebrain. In a recent study, we showed that Mov10 plays a role in the development of the CNS in the mouse (Skariah et al., 2017). Our data agree with that study and another where whole-mount in situ hybridization of Mov10 in X. laevis tail-bud stage showed expression in the eye anlage and nervous system (Owens et al., 2017). In addition to these defects, we also observed that the z-MO injected embryos had a significantly smaller anterior-posterior body length compared to controls (Fig. 5G).
Figure 5. Knockdown of zygotic Mov10 causes decreases in eye and body size.
A) Whole-mount images of control morpholino (Con) injected tadpoles at stage 36. A total of 46 tadpoles were analyzed, all of which exhibited this normal phenotype. B) Whole-mount images of zygotic Mov10 knockdown (z-MO) tadpoles at stage 36. A total of 48 tadpoles were analyzed and 45 of them showed a small eye phenotype. (C, D) Fluorescein images of the tadpoles from A and B, respectively, showing the distribution of the fluorescein-tagged morpholinos. E) RT-PCR using Mov10 primers for control and z-MO injected embryo cDNA showing effective splicing blocking and absence of Mov10 splice junction (PCR product expected size is 197nt, + reaction included cDNA, - reactions did not include cDNA). As a positive control, ODC (Ornithine Decarboxylase) was found to be present in both samples (PCR product expected size is 221nt). F) Graph showing eye diameter in μm from control morpholino (con) injected and z-MO injected embryos measured at stage 36. A total of 67 tadpoles were analyzed. G) Graph showing overall anterior-posterior (AP) length in mm from control morpholino (con) injected and z-MO injected embryos measured at stage 36. A total of 67 tadpoles were analyzed. Error bars represent SEM, **p<0.01 (Student’s t-test, two-tailed). Scale bar in D equals 800μm.
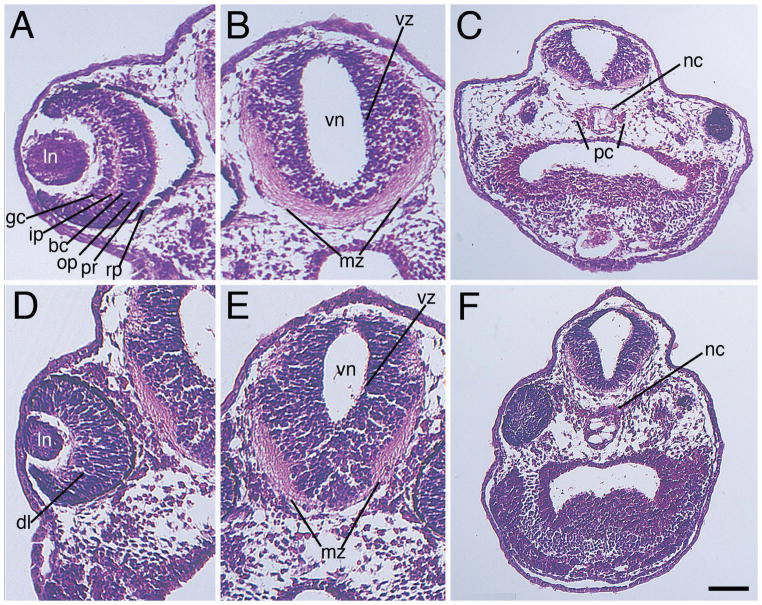
To further characterize the eye size, as well as query for internal brain defects, we sectioned and stained control-morpholino and Mov10 z-MO-injected embryos (Fig. 6). Unlike the control, there was diminished differentiation of the various retinal layers (compare Fig. 6A to 6D). Within the developing forebrain, we observed a smaller mass of peripheral (marginal) axons in the z-MO specimens, and what appeared to be an expanded ventricular zone (compare Fig. 6B to 6E, and see Sox3 expression discussed below, and in Fig. 7). We also saw reduced parachordal cartilages surrounding the notochord in the z-MO injected embryos (compare Fig. 6C to 6F) (Bernardini, 1999; Wiechmann, 2003). Parachordal cartilages expand and form the basal plate of the chondrocranium, and are important for craniofacial development (McBratney-Owen et al., 2008).
Figure 6. Knockout of zygotic Mov10 leads to defects in the eye and brain structure.
A–C) Hematoxylin and Eosin staining of control (Con) morpholino injected embryos, as labeled. A) Representative section of a control eye showing distinct, well-organized layers of the retina in a control embryo, as labeled. B) Forebrain region displaying a well-developed ventricular zone and marginal zone. Note large mass of axonal fibers within the ventral marginal zone. C) Lower magnification image showing the notochord and parachordal cartilage (D–F) Hematoxylin and Eosin stain of z-MO injected embryos. D) Section reveals a smaller eye with disorganized retinal layers. E) Section of the brain with a reduced marginal zone area. Note a reduced mass of axonal fibers within the ventral marginal zone. F) Lower magnification image revealing an enlarged notochord and no distinguishable parachordal cartilage. Dorsal is located towards the top of these images. bc, bipolar cell layer; dl, disorganized layers; gc, ganglion cell layer; ip, inner plexiform layer; mz, marginal zone; nc, notochord; op, outer plexiform layer; pc, parachordal cartilage; pr, photoreceptor; rp, retinal pigment epithelium; vz, ventricular zone. Scale bar in F equals 100μm (for A–B and D–E), and 170μm (for C and F).
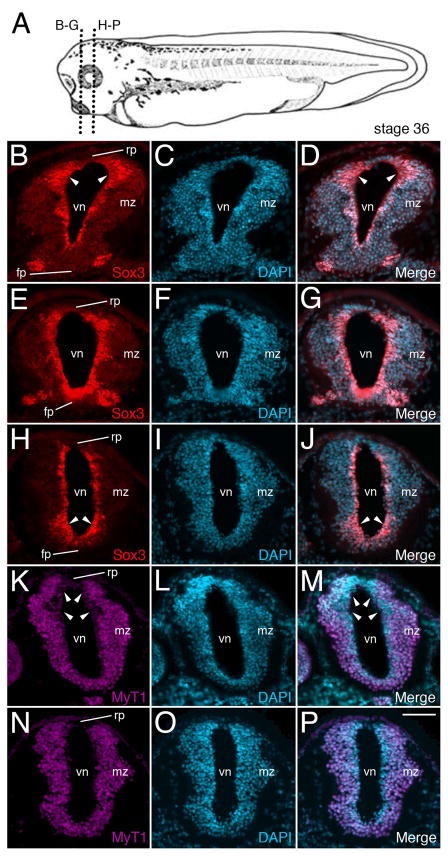
Figure 7. Knockout of zygotic Mov10 shows abnormal staining of neuronal precursors in the ventricular zone.
A) A stage 36 tadpole showing the plane of sectioning for B through P. B) Representative fluorescence image of control embryo shows the forebrain region with Sox3 positive precursors (in red) surrounding the lumen of the ventricle. Note the expanded dorsal Sox3 labeling in the ventricular zone, denoted by white arrowheads. C) DAPI staining (in blue) of the same section shown in B. D) Merged images from B and C. E–G) Representative fluorescence images from a similar region, as shown in B, from representative z-MO injected tadpoles. E) Sox3 positive neuronal precursor cells are located in the ventricular zone. F) DAPI staining of the same section shown in E. G) Merged images from E and F. Notice the enhanced overall staining of Sox3, including in the more ventral regions of the ventricular zone, denoted by white arrowheads (compare to the control embryo shown in B). H–J) More posterior section from the forebrain region of a z-MO injected tadpole. H) Sox3 neuronal precursors. Note the expanded ventral labeling denoted by arrowheads. I) Corresponding DAPI staining for H. J) Merge of Sox3 and DAPI stains. K–M) MyT1 staining for differentiated neurons in a control embryo. K) MyT1 positive differentiated neurons. Note expanded dorsal area devoid of MyT1 expression within the ventricular zone (denoted by white arrowheads). L) DAPI staining of the same section shown in K. M) Merged images from K and L. N–P) Representative sections from a z-MO injected tadpoles. N) Wide distribution of MyT1 positive differentiated neurons. Note uniform MyT1 staining in the dorsal ventricular zone (compare with K). O) DAPI staining of the same section shown in N. P) Merged images from N and O. fp, floor plate; mz, marginal zone; rp, roof plate; vn, ventricle. Scale bar in P equals 40μm
The ventricular zone, which, at this stage (NP36) is one to four cells thick, encloses the ventricle of the CNS, and contains neural precursor cells (NPCs) (Noctor et al., 2001; Rakic, 2009; Thuret et al., 2015), which can be identified by expression of Sox3 [SRY (sex determining region Y)-box 3] (Koyano et al., 1997; Macedoni-Luksic et al., 2009; Zhang et al., 2016) (Fig. 7). In control MO-injected cases, we observed Sox3 staining in cells around the ventricle, which is more pronounced in the dorsal region (Fig. 7B–D). In contrast, there is expanded, intense expression of Sox3 throughout the ventricular zone in the z-MO injected embryos (Fig. 7E–J), and this appears to be more pronounced in the ventral region (Fig. 7H), as compared to controls (Fig. 7B).
We also examined the localization of MyT1 (Myelin Transcription factor 1), a marker for primary differentiated neurons (Bellefroid et al., 1996; Zhang et al., 2016), which are located more distally to the ventricle and are non-overlapping with the Sox3 positive NPCs (Thuret et al., 2015). In control embryos, neurons are localized outside of the ventricular zone and in particular, seem to be more lateral on the dorsal side (Fig. 7K, arrowheads). This appears to match the expanded expression of Sox3, which is seen in the dorsal region of the neural tube, as described above (Fig. 7B). These differences appear to be lost in the z-MO injected cases (Fig. 7N) as compared to the control (Fig. 7K).
Discussion
The embryonic lethality observed in X. laevis following the knockdown of Mov10 is consistent with the developmental increase in Mov10 mRNA levels seen at stage 8 in both X. laevis and X. tropicalis, suggesting an important role for Mov10 during this period of development (Yanai et al., 2011). Elimination of maternal Mov10 leads to severe gastrulation defects and failure to complete neurulation (Fig. 1). Like other RISC components (Dicer, Drosha and Ago2), loss of Mov10 causes embryonic lethality in mouse (Bernstein et al., 2003; Liu et al., 2004; Morita et al., 2007; Skariah et al., 2017). The regulation of MZT by Mov10 in X. laevis suggests a role similar to that of Ago2 at the two-cell stage in mouse embryos (Lykke-Andersen et al., 2008). Additionally, our data show that the loss of Mov10 at the onset of development results in an overall increase in mRNA levels around MZT consistent with impaired RISC function (Fig. 3I). This increase in mRNA levels could be due to misregulation of multiple maternal transcripts or a few key regulatory transcripts that then leads to large-scale defects at gastrulation.
Mov10 has low expression levels in the adult mouse brain (Skariah et al., 2017) similar to what is reported in the Allen brain atlas (Nagase et al., 2000, 2008); however, there is an approximately 40-fold increase in Mov10 levels in P0-P3 mouse brain (Skariah et al., 2017), suggesting an important role in brain development. As the neural stem cells become depleted through their differentiation into neurons, the ventricular zone becomes diminished (Dehay and Kennedy, 2007). In X. laevis, there is abnormal organization of the brain when Mov10 is reduced, which we confirmed by observing disorganized retinal patterning (a derivative of the forebrain), and an expanded ventricular zone with abnormal NPC distribution, as evidenced by changes in Sox3 expression. The increased presence of MyT1+ cells near the dorsal ventricular zone of Mov10 z-Mo injected stage 36 embryos, which are absent in WT sections, suggests either increased differentiation into neurons or a defect in migration of neurons (Figs. 6 and 7). These findings suggest a conserved role for Mov10 in normal embryonic and CNS development, as observed in mice (Skariah et al., 2017).
In summary, we show a key role for Mov10 in gastrulation and in the central nervous system during embryonic development in X. laevis, which supports our earlier study in mouse (Skariah et al., 2017). Knockdown of maternal Mov10 in X. laevis embryos leads to gastrulation defects, which are likely a result of the large scale changes in gene expression. The disregulation of mRNA expression, particularly for those encoding cytoskeletal, cell adhesion, and extracellular matrix proteins could be due to Mov10’s role in miRNA-mediated regulation of MZT. In addition, zygotic Mov10 is necessary for normal head and eye development in X. laevis consistent with its function in mouse (Skariah et al., 2017).
Experimental Procedures
Morpholino Oligonucleotide Design
The Mov10 RISC complex RNA helicase sequence is publicly available from Xenbase in the X. laevis J-strain version 9.1 genome data. Following analysis, this sequence data was used to design a translation blocking morpholino (m-MO, Gene Tools, LLC) that targets bases -17 to 9 of the X. laevis Mov10 transcript. Splice blocking morpholinos (z-MO) were also designed to target the intron/exon 4 region. These morpholinos also include an incorporated 5′-fluorescein tag (green) to allow imaging of cells containing the morpholino. Additionally, a standard fluorescein-tagged random control morpholino (Con) was used as a negative control (targeting the human globin intron). This control morpholino is not known to target any X. laevis sequences and has been used in previous experiments to assay for potential non-specific effects from the injections or possible toxicity (Perry et al., 2010).
Generation of X. laevis Mov10 mRNA
An altered full-length synthetic RNA of X. laevis Mov10 was generated for injection into X. laevis embryos. The sequence immediately downstream of the Mov10 start site was altered by changing the third/wobble amino acid while preserving the original protein coding sequence to prevent Mov10 translation blocking Morpholino hybridization (IDT gBlocks, Coralville, IA). The full-length altered cDNA was synthesized in two pieces (fragment 1 = 1221nt; fragment 2 = 1987nt) and Gibson assembly (New England Biolabs, Ipswich, MA) was used to directionally ligate the gBlocks into the pCS2+ vector between the ClaI and XhoI restriction sites. Sequences were verified by the Roy J. Carver Biotechnology Center (University of Illinois, Urbana, IL). Rescue RNA was made from PCR-amplified template (SP6 and T3 primers). The SP6 mMessage mMachine kit (Thermo/Ambion) was used to transcribe capped mRNA, followed by purification of the RNA with the RNeasy MinElute Cleanup Kit (Qiagen). PCR was performed as described (Skariah et al., 2017). PCR primers are listed in Supplemental Table 4.
Microinjection of X. laevis embryos
Morpholinos were dissolved in RNase-free dH20 to a stock concentration of 2mM to facilitate co-injection with RNA at various concentrations. Depending on the experiment, either zygotes or embryos at the two-cell stage [stage 2, all stages follow (Nieuwkoop, 1956)] were immersed in 5% Ficoll solution (diluted with 1/20X Normal Amphibian Medium; Slack, 1984) and immobilized in rounded pits made in clay-lined petri dishes (Elkins and Henry, 2006; Wolfe and Henry, 2006). Graded concentrations of morpholinos or RNAs were injected using glass microinjection needles with a Narishige micromanipulator (Narishige USA, East Meadow, NY) and Harvard Apparatus pressure injector (World Precision Instruments, Inc., Sarasota, FL) into either fertilized eggs or unilaterally into single cells at the two-cell stage for the hydroxyurea assay (described below). Following injections, embryos were transferred to 1/20X NAM solution to recover and cultured at room temperature or 16°C with daily 1/20X NAM changes until various time points were reached for analysis. The experiments were carried out with embryos from at least 6 biological replicates (i.e., embryos from six mating pairs).
Dorsal and Ventral Lip Explants
Stage 10 control and m-MO injected embryos were collected and placed into ¾ NAM solution. Embryos were removed from their vitelline envelope with sharpened forceps and immobilized in small pits a clay-lined dish with the blastopore oriented upwards and visible for surgeries. A square patch of tissue was removed from either the dorsal (as shown in the Fig. 2G inset) or ventral (as shown in the Fig. 2I inset) lip of the blastopore, making sure to avoid the yolky cells of the blastopore itself and free of any deeper yolky vegetal cells. In the m-MO injected cases the dorsal side can be distinguished by a slight depression at the dorsal lip, which had been verified by continuous examination of the development of m-MO injected embryos. These blastopore lip explants were transferred to a small petri dish containing ¾ NAM solution and placed with the outermost, pigmented ectodermal layer facing downward, towards the bottom surface of the dish. These dishes were left undisturbed for three to four hours at room temperature, and then imaged for analysis.
Hydroxyurea Assay
Two-cell X. laevis embryos were rinsed in 1X MMR and one cell was injected with m-MO using glass microinjection needles. The embryos were transferred to 0.25X MMR containing a final concentration of 30mM hydroxyurea (Sigma Aldrich, St. Louis, MO) and incubated at 16°C. The uninjected cell served as an internal control. The embryos were monitored closely until they reached stage 9.5 and imaged. The experiment was conducted three times using embryos from independent mating pairs.
RNA-Seq Analysis
Total RNA was isolated from three biological replicates of stage 10.5 embryos from WT and m-MO injected embryos using TRIZOL reagent (Ambion, Thermo Fisher Scientific, Waltham, MO). The samples were sequenced at the High-Throughput Sequencing and Genotyping Unit of the Carver Biotechnology Center at the University of Illinois. Strand-specific single-end libraries were prepared using the TruSeq Stranded mRNAseq Sample Prep kit (Illumina, San Diego, CA). The libraries were quantitated by qPCR, pooled in equimolar amounts and sequenced on 2 lanes of a HiSeq 4000 (Illumina; sequencing kit version 1), generating over 780 million 100 bp single-end reads. Fastq files were generated and demultiplexed per sample with the bcl2fastq v2.17.1.14 Conversion Software (Illumina), which also trims Illumina adapters from the reads and removes any resulting sequences shorter than 35 bp. All bases across the reads showed quality scores greater than Q30 (FASTQC version 0.11.4) so quality trimming was not performed. The X. laevis reference genome version 9.1 was downloaded from Xenbase along with the version 1.8.3.1 gene models containing 45,099 genes. The gene models were converted from gff3 to gtf format using the gff_read program from cufflinks (version 2.2.1) (Trapnell et al., 2010). Reads were aligned to the genome using STAR (version 2.5.2a) (Dobin et al., 2013) using parameters --sjdbGTFfeatureExon CDS, --sjdbGTFtagExonParentGene gene_id and –quantMode GeneCounts, the last of which outputs read counts per gene_id, summing over all coding sequences of the gene.
The read counts were input into R 3.3.1 (Team, 2011) for data pre-processing and statistical analysis using packages from Bioconductor (Huber et al., 2015), as indicated below. Initial quality control analysis indicated that one of the three Mov10 m-MO injected replicates had many fewer reads and looked very different, and therefore, it was removed from the analysis. Genes without 1 Count Per Million mapped reads in at least two of the 5 samples were filtered out; 19,368 of the 45,099 genes passed this filter and were analyzed using edgeR 3.16.0.(Robinson and Oshlack, 2010) using the quasi-likelihood pipeline (Chen et al., 2016; Lun et al., 2016) that also accounted for the total library size for each sample and an extra TMM normalization factor (Robinson and Oshlack, 2010) for any biases due to changes in total RNA composition of the samples. Due to the large number of genes with very small fold-changes, we tested for differential expression of at least 1.5 fold and at least 10 fold change up or down, using the TREAT method (McCarthy and Smyth, 2009) adapted for quasi-likelihood F-tests (Chen et al., 2016). Adjustment for multiple testing was done using the False Discovery Rate method (Benjamini and Hochberg, 1995).
Additional annotation information for the X. laevis genes was drawn from two sources that were published concurrently: blast mappings to human genes done by (Ding et al., 2016) and gene symbols assigned by Xenbase in gene model annotation 1.8.3, which was a major update to the annotated gene symbols by merging two databases and human curation by three experts (Fortriede). Supplemental Table S1 [based on (Ding et al., 2016)] in NCBI’s Gene Expression Omnibus (GSE75278) contains X. laevis IDs from “genome assembly JGI9.1”, human protein IDs, human gene symbols and descriptions. These X. laevis IDs are actually the Xenopus Gene Nomenclature Committee’s symbols, but for the Xenbase version 1.8.0 gene models (Name attribute in Xlaevisv1.8.Named.gene.gff3.gz). The major annotation update to Xenbase version 1.8.3 only changed the symbols, not the number or locations of genes, so the internal ID attribute is the same for all 1.8 gene model sets and was used to link the human gene symbols to our differential expression results. Xenbase recently published a small version upgrade (1.8.3.2) to the gene model set we had used (1.8.3.1), which corrected gene symbols for five gene models. We used the gene symbols from 1.8.3.2, which are based on human gene nomenclature. Both sets of gene symbols, (Ding et al., 2016) and Xenbase, needed minor correcting to remove leading/trailing white spaces, X. laevis’ sub-genome extensions, and to fix capitalization differences with the symbols in Bioconductor’s org.Hs.eg.db_3.4.0 annotation package. Both sets of gene symbols also contained older symbols that org.Hs.eg.db_3.4.0 listed as retired aliases; we updated the older symbols to current symbols, as needed, also resolving a few aliases that mapped to more than one current symbol by inspecting the gene description (Ding et al., 2016). Comparisons of (Ding et al., 2016) and Xenbase annotations showed overwhelming agreement between the two with 73.8% of genes annotated to the same symbol or to no symbol (see Supplemental Table 1 accessible through NCBI GEO record GSE86382). Only 2.6% of genes were annotated to different symbols by the two sources, and many of these were just different members of the same gene family (e.g., DOCK6 vs. DOCK7). In these cases we preferentially used the (Ding et al., 2016) annotation because it annotated more genes overall and had descriptions listed as well. The 23.6% of genes annotated with a symbol in only one source were assigned that symbol. The selected gene symbols were then used to pull NCBI Entrez Gene IDs, gene names and Gene Ontology terms from org.Hs.eg.db_3.4.0.
Of the 19,368 X. laevis gene models that were expressed in our samples, only 1,470 were not annotated to a human symbol and the rest were annotated to only 10,935 unique human genes due to the tetraploid genome of X. laevis. To do over-representation testing on GO term pathways based on human annotations, each unique human symbol was deemed “significant” if the X. laevis genes mapping to it had FDR p < 0.05 for the > 1.5 FC test. 4,596 human genes were called significant under these criteria, irrespective of direction of change. When directionality is taken into account, 140 human genes had significant X. laevis genes changing in both directions; this is not surprising given the nature of mapping to human genes and both results could be true if the gene has multiple splice variants. We did not try to resolve these discrepancies, but instead counted the genes as both significantly up (2,826 genes) and down (1,883) when conducting GO testing separately based on direction. Gene identities and statistical analysis are in Supplemental Table 2 accessible through NCBI GEO accession number GSE86382.
Over-representation testing was done separately for the all-significant, up-significant and down-significant gene sets using the 10,935 unique genes as the background. The GOstat package’s (v 2.40.0) conditional hypergeometric testing was performed to reduce the redundancy of related GO terms. Comparison of GO terms’ raw p-values between the all, up and down gene sets was done to see which GO terms were specific to the direction of change and which included genes changing in both directions. Heatmaps of –log10 (p-values) across the 3 gene sets were made separately for BP, MF and CC categories; GO terms with raw p-values < 0.001 in any gene set were included for Molecular Function (MF) and Cellular Compartment (CC), but due to the larger number of Biological Processes (BP) terms, only those with raw p-values < 0.0001 in any gene set were included in the heatmap. All of the R codes necessary for the above analyses are in Supplemental File 3 (Skariah_GSE86382_Bioc3.4.txt). No additional files are needed because the codes show how to download the necessary sample counts and annotations from GEO and Xenbase. The codes produce the results presented in (Skariah et al., 2017) when run with R 3.3.1/Bioconductor 3.4. Links to this X. laevis RNAseq data set can be found with the following NCBI accession number: GSE86382.
X. laevis histology
Stage 36 tadpoles were fixed in 4% Paraformaldehyde in 1X PBS for 1 hour at room temperature (RT), followed by three washes in 1X PBS for 15 min each. The tadpoles were serially dehydrated in 30%, 50% and 70% ethanol for an hour each at RT and stored in 70% ethanol at 4°C. Body length and eye diameter were measured using ImageJ software. For whole mount histology, the tadpoles were incubated in 95% ethanol for 15min followed by three 100% ethanol washes for 15 min each. The tadpoles were then moved to xylene and incubated for 30min with fresh changes of xylene every 10 min. This was followed by a 6-hour incubation in 50% Paraplast plus xylene solution at 60°C. The tadpoles were then moved to fresh Paraplast plus twice for 6 hours each at 60°C before embedding in plastic boats. Sections were prepared using a Spencer 820 rotary microtome, affixed to subbed slides and dried overnight. The sections were deparaffinized and stained with Hematoxylin 7211 and Eosin Y reagents (ThermoScientific, Waltham, MA) according to the manufacturer’s protocol. The images were captured using a Spot digital camera (Diagnostic Instruments, Inc., Sterling Heights, MI). A biological replicate of 25 – 30 stage 36 tadpoles from three different matings were scored.
X. laevis embryo immunohistochemistry
Embryos were devitellinized and fixed in 4% paraformaldehyde in PBS for 15 min at RT, followed by three washes of 1X PBS for 15min each. The embryos were sequentially dehydrated using 25%, 50%, 75%, 95% and 100% ethanol for 15 min each at RT. The embryos were prepared for paraffin embedding and sectioned, as described in the X. laevis histology methods, above. For immunohistochemistry, the sections were deparaffinized using xylene and rehydrated before Heat-induced Antigen retrieval in 1X citrate buffer (pH-6.0). The slides were stained using the following primary antibodies at 0.5ug/ml- Tor-70 (notochord cells, a kind gift from Prof. Ray Keller), 12/101 (somitic mesoderm, Developmental Studies Hybridoma Bank, Iowa), at 1:500 dilution for Sox3 (neural precursor cells) and MyT1 (differentiated neurons, kind gift from Prof. Klymkowsky and Prof. Papalapoulo respectively) (Zhang et al., 2016). The secondary antibodies were Alexa Flour 488 (Jackson Immunoresearch, West Grove, PA) and RITC-conjugated IgM specific antibody (Jackson Immuoresearch) used at a concentration of 1:200. The experiment was done using 6 biological replicates (i.e., embryos from six mating pairs for each condition).
Supplementary Material
Supplemental Table 1 (.xls). Annotation of Xenopus RNA seq data (accessible through NCBI GEO accession number GSE86382).
Supplemental Table 2 (.xls). Statistical analysis of Mov10 morpholino injected embryos compared to control RNA seq data (accessible through NCBI GEO accession number GSE86382).
Supplemental File 3 (.txt). R codes for RNA seq analysis
Supplemental File 4. PCR primers used in this study
Time-lapse imaging of control morpholino injected X. laevis embryos
Time-lapse imaging of Mov10 m-MO injected X. laevis embryos
Acknowledgments
Grant Sponsor and number - NIH/NIMH MH093661 (to SC), Spastic Paralysis Research Foundation of Illinois-Eastern Iowa District of Kiwanis International (to SC), NIH/NEI EY023979 (to JJH).
We thank Dr. Ray Keller for advice in interpreting X. laevis gastrulation phenotypes and for the Tor-70 antibody, Dr. Sayee Anakk, Dr. Auinash Kalsotra, Dr. Jing Yang for advice on this manuscript and the HU assays. We thank Dr. Mike Klymkowsky for the Sox3 antibody and Dr. Nancy Papalopulu and Dr. Enrique Amaya for the MyT1 antibody. The authors declare that they have no competing interests.
References
- Allen developing Mouse Brain Atlas [Online] Allen Brain Institute; Available: http://developingmouse.brain-map.org/[Accessed] [Google Scholar]
- Bellefroid EJ, Bourguignon C, Hollemann T, Ma Q, Anderson DJ, Kintner C, Pieler T. X-MyT1, a Xenopus C2HC-Type Zinc Finger Protein with a Regulatory Function in Neuronal Differentiation. Cell. 87:1191–1202. doi: 10.1016/s0092-8674(00)81815-2. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- Bernardini G, Prati M, Bonetti F, Scarì G. Atlas of xenopus development. New York: Springer; 1999. [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Burdick R, Smith JL, Chaipan C, Friew Y, Chen J, Venkatachari NJ, Delviks-Frankenberry KA, Hu W-S, Pathak VK. P Body-Associated Protein Mov10 Inhibits HIV-1 Replication at Multiple Stages. J Virol. 2010;84:10241–10253. doi: 10.1128/JVI.00585-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Lun AT, Smyth GK. From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Res. 2016;5:1438. doi: 10.12688/f1000research.8987.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson LA, Keller RE. Neural tube closure in Xenopus laevis involves medial migration, directed protrusive activity, cell intercalation and convergent extension. Development. 1999;126:4547–4556. doi: 10.1242/dev.126.20.4547. [DOI] [PubMed] [Google Scholar]
- Dehay C, Kennedy H. Cell-cycle control and cortical development. Nature Reviews Neuroscience. 2007;8:438. doi: 10.1038/nrn2097. [DOI] [PubMed] [Google Scholar]
- Ding Y, Colozza G, Zhang K, Moriyama Y, Ploper D, Sosa EA, Benitez MD, De Robertis EM. Genome-wide analysis of dorsal and ventral transcriptomes of the Xenopus laevis gastrula. Dev Biol. 2016 doi: 10.1016/j.ydbio.2016.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkins MB, Henry JJ. Isolation and characterization of a novel gene, xMADML, involved in Xenopus laevis eye development. Developmental Dynamics. 2006;235:1845–1857. doi: 10.1002/dvdy.20824. [DOI] [PubMed] [Google Scholar]
- Fortriede J. Available: ( ftp://ftp.xenbase.org/pub/Genomics/JGI/Xenla9.1/1.8.3.1/ReadMe.txt). [Accessed]
- Goodier JL, Cheung LE, Kazazian HH., Jr MOV10 RNA helicase is a potent inhibitor of retrotransposition in cells. PLoS Genet. 2012;8:e1002941. doi: 10.1371/journal.pgen.1002941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregersen LH, Schueler M, Munschauer M, Mastrobuoni G, Chen W, Kempa S, Dieterich C, Landthaler M. MOV10 Is a 5′ to 3′ RNA helicase contributing to UPF1 mRNA target degradation by translocation along 3′ UTRs. Mol Cell. 2014;54:573–585. doi: 10.1016/j.molcel.2014.03.017. [DOI] [PubMed] [Google Scholar]
- Howe JA, Howell M, Hunt T, Newport JW. Identification of a developmental timer regulating the stability of embryonic cyclin A and a new somatic A-type cyclin at gastrulation. Genes & Development. 1995;9:1164–1176. doi: 10.1101/gad.9.10.1164. [DOI] [PubMed] [Google Scholar]
- Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, Gottardo R, Hahne F, Hansen KD, Irizarry RA, Lawrence M, Love MI, MacDonald J, Obenchain V, Oles AK, Pages H, Reyes A, Shannon P, Smyth GK, Tenenbaum D, Waldron L, Morgan M. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Wallace L, Harper SQ, Yang J. PP2A:B56{epsilon}, a substrate of caspase-3, regulates p53-dependent and p53-independent apoptosis during development. J Biol Chem. 2010;285:34493–34502. doi: 10.1074/jbc.M110.169581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller RE, Danilchik M, Gimlich R, Shih J. The function and mechanism of convergent extension during gastrulation of Xenopus laevis. J Embryol Exp Morphol. 1985;89(Suppl):185–209. [PubMed] [Google Scholar]
- Kenny PJ, Zhou H, Kim M, Skariah G, Khetani RS, Drnevich J, Arcila ML, Kosik KS, Ceman S. MOV10 and FMRP regulate AGO2 association with microRNA recognition elements. Cell Rep. 2014;9:1729–1741. doi: 10.1016/j.celrep.2014.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyano S, Ito M, Takamatsu N, Takiguchi S, Shiba T. The Xenopus Sox3 gene expressed in oocytes of early stages. Gene. 1997;188:101–107. doi: 10.1016/s0378-1119(96)00790-1. [DOI] [PubMed] [Google Scholar]
- Langley AR, Smith JC, Stemple DL, Harvey SA. New insights into the maternal to zygotic transition. Development. 2014;141:3834–3841. doi: 10.1242/dev.102368. [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Lun ATL, Chen Y, Smyth GK. It’s DE-licious: A Recipe for Differential Expression Analyses of RNA-seq Experiments Using Quasi-Likelihood Methods in edgeR. In: Mathé E, Davis S, editors. Statistical Genomics: Methods and Protocols. New York, NY: Springer New York; 2016. pp. 391–416. [DOI] [PubMed] [Google Scholar]
- Lund E, Liu M, Hartley RS, Sheets MD, Dahlberg JE. Deadenylation of maternal mRNAs mediated by miR-427 in Xenopus laevis embryos. Rna. 2009;15:2351–2363. doi: 10.1261/rna.1882009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen K, Gilchrist MJ, Grabarek JB, Das P, Miska E, Zernicka-Goetz M. Maternal Argonaute 2 is essential for early mouse development at the maternal-zygotic transition. Mol Biol Cell. 2008;19:4383–4392. doi: 10.1091/mbc.E08-02-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macedoni-Luksic M, Greiss-Hess L, Rogers SJ, Gosar D, Lemons-Chitwood K, Hagerman R. Imitation in fragile X syndrome. Implications for autism. Autism. 2009;13:599–611. doi: 10.1177/1362361309337850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBratney-Owen B, Iseki S, Bamforth SD, Olsen BR, Morriss-Kay GM. Development and Tissue Origins of the Mammalian Cranial Base. Developmental biology. 2008;322:121–132. doi: 10.1016/j.ydbio.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Smyth GK. Testing significance relative to a fold-change threshold is a TREAT. Bioinformatics. 2009;25:765–771. doi: 10.1093/bioinformatics/btp053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Luhrmann R, Tuschl T. Identification of novel argonaute-associated proteins. Curr Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- Morita S, Horii T, Kimura M, Goto Y, Ochiya T, Hatada I. One Argonaute family member, Eif2c2 (Ago2), is essential for development and appears not to be involved in DNA methylation. Genomics. 2007;89:687–696. doi: 10.1016/j.ygeno.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Nagase T, Kikuno R, Hattori A, Kondo Y, Okumura K, Ohara O. Prediction of the coding sequences of unidentified human genes. XIX. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 2000;7:347–355. doi: 10.1093/dnares/7.6.347. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PDFJ. Normal table of Xenopus laevis. Amsterdam: North Holland Publishing; 1956. [Google Scholar]
- Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- Owens DA, Butler AM, Aguero TH, Newman KM, Van Booven D, King ML. High-throughput analysis reveals novel maternal germline RNAs crucial for primordial germ cell preservation and proper migration. Development. 2017;144:292–304. doi: 10.1242/dev.139220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry KJ, Johnson VR, Malloch EL, Fukui L, Wever J, Thomas AG, Hamilton PW, Henry JJ. The G-protein-coupled receptor, GPR84, is important for eye development in Xenopus laevis. Dev Dyn. 2010;239:3024–3037. doi: 10.1002/dvdy.22446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. Evolution of the neocortex: Perspective from developmental biology. Nature reviews. Neuroscience. 2009;10:724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers C, Schlumpf T, Sawarkar R, Comoglio F, Paro R. Mixture models and wavelet transforms reveal high confidence RNA-protein interaction sites in MOV10 PAR-CLIP data. Nucleic Acids Res. 2012;40:e160. doi: 10.1093/nar/gks697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skariah G, Seimetz J, Norsworthy M, Lannom MC, Kenny PJ, Elrakhawy M, Forsthoefel C, Drnevich J, Kalsotra A, Ceman S. Mov10 suppresses retroelements and regulates neuronal development and function in the developing brain. BMC Biology. 2017;15:54. doi: 10.1186/s12915-017-0387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack JH, Newport JW. Developmentally regulated activation of apoptosis early in Xenopus gastrulation results in cyclin A degradation during interphase of the cell cycle. Development. 1997;124:3185–3195. doi: 10.1242/dev.124.16.3185. [DOI] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136:3033–3042. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- Team RDC. R: A Language and Environment for Statistical Computing. Vienna, Austria: the R Foundation for Statistical Computing; 2011. [Google Scholar]
- Thuret R, Auger H, Papalopulu N. Analysis of neural progenitors from embryogenesis to juvenile adult in Xenopus laevis reveals biphasic neurogenesis and continuous lengthening of the cell cycle. Biology Open. 2015;4:1772–1781. doi: 10.1242/bio.013391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xenbase [Online] 2008 Available: ( http://www.xenbase.org/, RRID:SCR_003280)[Accessed]
- Wiechmann AF, Wirsig-Wiechmann Celeste E. Color atlas of Xenopus laevis histology. Boston: Kluwer Academic Publishers; 2003. [Google Scholar]
- Wolfe AD, Henry JJ. Neuronal leucine-rich repeat 6 (XlNLRR-6) is required for late lens and retina development in Xenopus laevis. Dev Dyn. 2006;235:10270–11041. doi: 10.1002/dvdy.20691. [DOI] [PubMed] [Google Scholar]
- Yanai I, Peshkin L, Jorgensen P, Kirschner MW. Mapping gene expression in two Xenopus species: evolutionary constraints and developmental flexibility. Dev Cell. 2011;20:483–496. doi: 10.1016/j.devcel.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Li J, Lea R, Amaya E. Assessing Primary Neurogenesis in Xenopus Embryos Using Immunostaining. Journal of Visualized Experiments: JoVE:53949. 2016 doi: 10.3791/53949. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1 (.xls). Annotation of Xenopus RNA seq data (accessible through NCBI GEO accession number GSE86382).
Supplemental Table 2 (.xls). Statistical analysis of Mov10 morpholino injected embryos compared to control RNA seq data (accessible through NCBI GEO accession number GSE86382).
Supplemental File 3 (.txt). R codes for RNA seq analysis
Supplemental File 4. PCR primers used in this study
Time-lapse imaging of control morpholino injected X. laevis embryos
Time-lapse imaging of Mov10 m-MO injected X. laevis embryos