Abstract
The restrictive element-1 silencing transcription factor)/NRSF (neuron-restrictive silencing factor (NRSF) is a transcriptional repressor which acts via epigenetic remodeling to silence target genes. Emerging evidence indicates that REST is a master transcriptional regulator of neuron-specific genes not only in neurogenesis and neuronal differentiation, but also in differentiated neurons during the critical period in postnatal brain development, where it plays a role in fine-tuning of genes involved in synaptic plasticity, and in normal aging, where it promotes neuroprotection by repressing genes involved in oxidative stress and β-amyloid toxicity. This review focuses on recent findings that dysregulation of REST and REST-dependent epigenetic remodeling provide a central mechanism critical to the progressive neurodegeneration associated with neurologic disorders and diseases including global ischemia, stroke, epilepsy, Alzheimer’s and Huntington’s disease.
Introduction
REST (also called NRSF) is a gene silencing transcription factor that is widely expressed during embryogenesis and plays a strategic role in end-stage neuronal differentiation [1,2]. In pluripotent stem cells and neural progenitors, REST acts via epigenetic remodeling to actively repress a vast number of coding and noncoding neuron-specific genes involved in synaptogenesis, axonal path-finding, synaptic plasticity and structural remodeling, including synaptic vesicle proteins, channels, receptors, transporters, and neuron-specific microRNAs that regulate networks of non-neuronal genes [3–6]. In silico analysis identifies close to 2000 putative REST targets in the mammalian genome, including both coding and noncoding genes [7,8]. During the final stages of neuronal differentiation, loss of REST is essential for acquisition of the neuronal phenotype [9].
Whereas REST was initially thought to function as a master regulator of neuronal genes involved in neurogenesis in undifferentiated neurons and stem cells. Recent findings indicate that REST is expressed in differentiated neurons during the critical period, a time of heightened sensitivity to plasticity in brain development, where it plays a role in fine-tuning of genes important to synaptic plasticity [10] and in normal aging, where it suppresses genes involved in neuronal death, thereby affording neuroprotection [11•]. Dysregulation of REST is implicated in a number of neurodegenerative disorders and diseases. REST is activated in selectively vulnerable mature hippocampal neurons in response to ischemic insults [12•,13,14•,15•,16] and seizures [17,18,19•,20]. In Huntington’s disease, REST aberrantly accumulates in the nuclei of selectively vulnerable striatal neurons [21,22]. In aging neurons, loss of REST is associated with the onset of Alzheimer’s disease in humans [11•] (Figure 1).
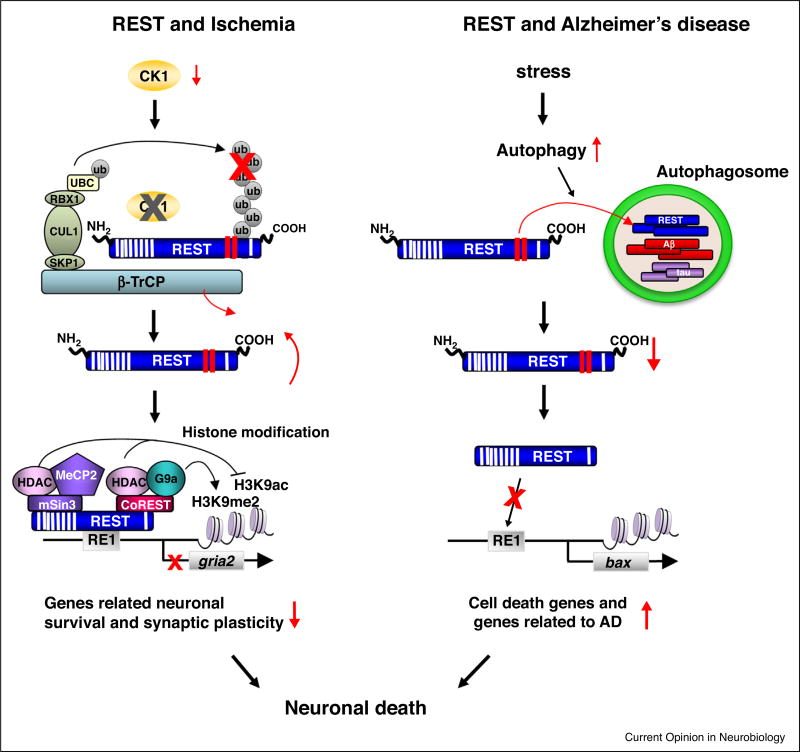
Figure 1.
Regulation of REST in global ischemia and AD. Left, global ischemia reduces abundance of CK1 and E3 ligase β-TrCP, resulting in an increase in REST in the hippocampal CA1. REST binds to the RE1 element within the promoter of target genes such as gria2 and orchestrates the assembly of mSin3A and CoREST, HDACs 1 and 2, G9a and MeCP2. The REST-corepressor complex promotes epigenetic remodeling of core histone proteins at the promoter of target genes and represses transcription of genes important to synaptic plasticity and neuronal survival. Right, In AD brains oxidative stress activates autophagy and formation of autophagosome. REST is engulfed in autophagosomes, together with misfolded proteins, such as Aβ and tau, which, in turn, reduces REST abundance in the nucleus. Loss of REST in the nucleus causes an increase in expression of genes involved in neuronal death and AD pathology.
REST-dependent epigenetic remodeling
A fundamental mechanism by which REST regulates target genes is that of epigenetic remodeling [9,10,12•,13,14•,16]. Epigenetic modifications, such as DNA methylation and hydroxymethylation and histone modifications reflect environmental influences that are not ‘hard-wired’ into the DNA sequence and represent a mechanism through which environmental cues, adverse or enriched experience, drugs of abuse, and neuronal insults such as ischemia and seizures can modify synaptic connections, synaptic efficacy and structural remodeling [23–25]. It is now well established that epigenetic modifications are critical to genome reprogramming during brain development, tissue-specific gene expression, higher cognitive function such as learning and memory, and to neuronal survival in aging neurons [23–28]. It is postulated that changes in the epigenome could be an important mechanism for enduring changes in transcription in adult neurons.
In neural progenitors [9], differentiated neurons during postnatal development [10], and insulted adult neurons [12•,14•,16], REST binds at RE1/NRSE sites within target genes and recruits CoREST [29,30] and mSin3A [31–33], corepressor platforms which in turn recruit histone deacetylases (HDACs)-1 and 2. HDACs remove acetyl moieties from core histone proteins and induce dynamic and reversible gene silencing by tightening of the core chromatin complex, thereby restricting access of the transcriptional machinery required for gene activation to the promoters of target genes [6,23–28,34]. REST mediates long-term gene silencing by association with site-specific histone methyltransferases such as G9a, which promotes trimethylation of histone 3 at lysine 9 (H3K9me3) through CoREST-dependent [9,35] and independent [36] mechanisms and the site-specific histone demethylase LSD1, which removes methyl groups from H3 at K4 (H3K4) [37,38]. In addition, REST associated with methyl-CpG binding protein 2 (MeCP2), a transcriptional repressor that reads epigenetic marks and is recruited to methylated CpGs [39]. Histone acetylation and methylation are primarily marks of dynamic gene expression. Although originally thought to be an enduring mark of gene silencing [40], we now know that changes in DNA methylation can be rapid and reversible [41].
REST not only silences, but also recruits other proteins and, which together activate REST target genes. For example, recent studies indicate that full-length REST can activate gene transcription by recruiting two proteins, TET3 hydroxylase and NSD3, chromatin remodeling proteins [42] that typically activate gene transcription. A recent study revealed a mechanism by which this occurs [42]. REST directly binds the short splice variant of TET3 (the predominant isoform found in neurons) and directs it to the promoters of REST target genes, where TET3 promotes conversion of methyl cytosine to 5-hyodroxy methylcytosine, resulting in context-specific gene transcription [42]. In addition, the REST — TET3 complex recruits methyltransferases such as NSD3, which confers a trimethyl moiety to core histone protein H3 at lysine residue 36 to generate H3K36me3, a strong and enduring mark of gene activation [43]. In addition, the short splice variant of REST, REST4, which contains the N-terminal, but lacks the C-terminal repressor domain, can activate REST target genes by coordinating with the glucocorticoid receptor (a ligand-dependent transcription factor) to recruit Brahma (Brm), a chromatin remodeling protein that also activates gene transcription [44]. These findings reveal novel mechanisms by which REST activates expression of neuronal genes.
Role of REST in differentiated neurons under physiological conditions
In addition to its role as master regulator of neurogenesis, REST plays an important role in the shaping of the synaptic output of adult neurons. During normal postnatal development, REST orchestrates the developmental switch in several key synaptic and extrasynaptic proteins including the NMDA receptor (NMDAR) and K-Cl cotransporter KCC2. NMDARs are critical to synaptogenesis, neural circuitry and information flow. REST binds and epigenetically remodels the grin2b promoter (gene encoding the NMDAR subunit GluN2B) and thereby silences GluN2B expression. This, in turn, promotes the switch between the immature (primarily GluN2B-containing) and mature (primarily GluN2A-containing) NMDAR phenotype at hippocampal synapses [10]. This is significant in that GluN2B expression restricts synaptic incorporation of AMPARs, reduces the threshold for and enhances the magnitude of LTP, and promotes hippocampal- dependent learning, plasticity-induced spine growth and dendritic patterning critical to information processing [45]. Interestingly, the switch can be disrupted by brief bouts of adverse experience in the form of maternal deprivation, indicating that the synaptic NMDAR phenotype can be regulated by REST-dependent epigenetic remodeling [10].
In addition, REST regulates chloride conductance in neurons during postnatal development. In neonatal neurons, the chloride transporter NKCC1 is robustly expressed and the K-Cl cotransporter KCC2 is repressed by binding of REST at two distinct sites within the kcc2b promoter [46]. This is significant in that NKCC1 promotes high intracellular Cl−, and drives an outward Cl− flux, resulting in depolarizing GABA currents. During early postnatal development (~P8), NKCC1 expression decreases and loss of REST from the Kcc2b promoter induces upregulation of KCC2. This is significant in that the K-Cl cotransporter KCC2 maintains the low intracellular chloride required for the hyperpolarizing actions of the inhibitory neurotransmitters GABA and glycine in mature neurons [46]. Thus, REST regulates the sign of GABA (and glycine) synaptic transmission.
Specificity of REST target genes
Given ~2000 putative REST target genes that contain the canonical RE1-NRSE element within the mammalian genome [7,47], and that noncanonical RE1 motifs in the genome [48] account for an even broader array of dynamically regulated, but lower affinity, REST targets, the question arises as to what determines the specificity of interaction between REST and its targets. Considerable evidence indicates that the ensemble of target genes responsive to REST (‘transcriptionally-responsive’ genes) varies in a cell-type-dependent and context-dependent manner. The subset of REST targets that exhibit altered expression in the hippocampal CA1 after global ischemia [14•] differs from that altered in the hippocampal CA3 in response to seizures [20], that in prefrontal cortex of humans with Huntington Disease [22] and that in the prefrontal cortex of healthy aged humans [49]. In addition, the subset of transcriptionally responsive genes identified by unbiased, genome-wide studies involving REST chromatin immunoprecipitation and deep sequencing (ChIP-seq) in a neuroblastoma cell line [49] differs from that identified by large-scale ChIP-seq in Jurkat cells [48] and by ChIP-on-chip in mouse neural stem cells [50]. These findings demonstrate that in different cell types, at different ages and disease states, REST controls expression of different ensembles of target genes. It should also be noted that experiments performed in cell lines in vitro do not reflect the impact of the cellular environment, which is known to influence the epigenetic landscape and alter the binding of REST and other transcription factors/chromatin remodeling proteins to target genes.
An attractive scenario is that the epigenetic landscape, which varies with developmental stage, cell type, brain region and disease state, determines the enrichment of REST at the promoter of a given target gene. A case in point is the polycomb group proteins, which serve as gene silencers in various cells types including neurons [51]. The polycomb complex-2 (PRC2) is recruited to RE1/NRSE elements within the promoters of REST target genes via the long noncoding (lncRNA) HOTAIR [52]. Whereas HOTAIR binds through a motif in its 50 domain to the polycomb complex 2 (PRC2), it binds through a motif in its 3′ domain to the LSD1/CoREST/REST complex, consistent with the concept that lncRNAs serve as scaffolds by providing binding platforms to assemble chromatin remodeling proteins, and thereby regulate transcription [52]. Recent studies show that the epigenetic mark H3K27me3, a functional readout of the polycomb protein EZH2, is enriched at the grin2b gene (gene encoding the NMDA receptor subunit GluN2B) during normal postnatal development [10]. It is also possible that other transcription factors and epigenetic marks also influence the affinity for REST of a given gene or set of target genes.
Another factor is that of binding affinity of target genes for REST. Using large-scale transcriptome arrays, Baram and colleagues [20] found that only a small subset (~10%) of putative REST targets are silenced by REST in response to seizures despite the fact that REST is increased several-fold. Unexpectedly, the impact of REST on target genes was greatest for genes that bound the transcription factor with an intermediate affinity, a property that renders them sensitive to modest alterations in REST abundance. Whereas genes that bind REST tightly are already switched off under physiological conditions, genes that bind REST weakly would require more REST in order to be switched off.
Regulation of REST abundance in neurons
REST abundance is bidirectionally regulated in pluripotent stem cells and cancer cells via SCF (Skp1 — Cul1 — F-box protein)/β-TrCP-dependent, ubiquitin-based proteasomal degradation [53–55] and HAUSP-dependent deubiquitination [56]. β-TrCP is an E3 ligase, which binds and initiates ubiquitination of target proteins (substrates) such as the transcription factors REST, β-catenin [57] and NFκβ2 [58]. REST harbors two neighboring, but distinct, noncanonical degron motifs in its carboxy-terminal domain [59]. A recent study identified the serine/threonine kinase casein kinase-1 (CK1) as an upstream signal that regulates REST stability. CK1 phosphorylates REST at serine residues within its degron motifs, enabling the E3 ligase β-TrCP to recognize and bind REST (the substrate) through phospho-degron motifs [15•]. A recent study by Mandel and colleagues identified a proline-rich sequence upstream of the phospho-degron motifs which, when phosphorylated by ERK1/2, facilitates loss of REST at the end stage of neural differentiation [60].
There findings are consistent with a model whereby in differentiated neurons under physiological conditions, CK1 is activate and maintains REST at low, constitutive levels. In response to neuronal insults such as global ischemia, CK1 and β-TrCP abundance are decreased, and REST rises in vulnerable hippocampal neurons [14•,16]. Once activated, REST binds to the RE1 element in the promoters of a subset of target genes including the AMPAR subunit GluA2, and assembles in a large corepressor complex (see above) that orchestrates epigenetic modifications and gene silencing in a cell-specific and context-specific manner [13,14•,16].
REST can also be degraded via the lysosomal/autophagy pathway as occurs, for example, in AD, frontotemporal dementia and dementia with Lewy bodies [49]. Activation of autophagy by serum deprivation results in translocation of REST from the nucleus to the cytoplasm of neuron-derived SH-SY5Y cells, where it colocalizes to punctate structures identified as autophagosomes [49]. Moreover, REST colocalizes with Aβ in a subset of autophagosomes [49]. A possible scenario is that the ubiquitin-proteasome system is overloaded or impaired in neurodegenerative diseases such as AD. It is well known that ubiquitinated proteins associate with the cargo adaptor p62 which target them to LC3-II (+) autophagosomes [61].
In addition to regulation of REST abundance by the ubiquitin-based, proteasomal and lysosomal pathways, REST activity can be regulated by translocation of REST into or out of the nucleus. Cattaneo and colleagues found that wild-type huntingtin resides in the cytoplasm, where it forms a complex with HAP1 and REST-interacting LIM domain protein (RILP), which together sequester REST in the cytoplasm, away from target genes [21,22]. In cell and mouse models of Huntington’s disease, mutant huntingtin disrupts the complex, liberating RILP, which directly binds REST/NRSF and promotes its translocation into the nucleus [62]. Once in the nucleus, REST assembles to form a corepressor complex, which represses the transcription of an ensemble of genes including the gene encoding brain-derived neurotrophic factor (BDNF), a member of the neurotrophin family of growth factors implicated in synaptogenesis, dendritogenesis, and neuronal survival [63].
REST and neurodegenerative disease
REST dysregulation is implicated in diseases of the nervous system. Perturbation of REST expression during embryogenesis elicits cellular apoptosis, aberrant differentiation, patterning and lethality [64,65]. In differentiated neurons, REST is quiescent, but can be activated in response to neuronal insults such as ischemia [12•,13,14•,15•] and seizures [17,18]. In these disorders, a rise in REST and REST-dependent epigenetic remodeling are causally linked to neuronal death. Increased expression of REST in response to neuronal insults would be expected to repress not only postsynaptic receptors and transporters, but also presynaptic vesicle proteins such as synapsin I, synaptophysin, synaptotagmins II, IV, VI and VII, synaptobrevin II, and the SNARE protein SNAP25 [66]. Thus, not only neurotransmission in adult neurons, but also stimulus-induced exocytosis in PC12 cells and pancreatic β-cells, relies on exceedingly low levels of REST expression [66], with exceedingly brief exceptions to enable, for example, transitions between receptor (or transporter) phenotypes. In contrast, in aging neurons, low levels of REST are neuroprotective, and loss of REST is associated with Alzheimer’s disease [11•]. Dysregulation of REST and its target genes is also implicated in the pathogenesis of epilepsy [19•,20,67], Huntington’s disease [21], Parkinson’s disease [68], and SMCX, a form of X-linked mental retardation [69]. We focus on global ischemia and Alzheimer’s disease, for which the evidence in strongest.
REST and ischemia
Recent findings demonstrate that REST expression is activated in mature, differentiated neurons in response to neuronal insults and that REST-dependent silencing of target genes is essential to neuronal death. Within 16–24 hours after global ischemia induced by 4 VO model in rats, CK1 and β — TrCP are decreased [15•] and REST protein abundance is increased, presumably due to enhanced stability in selectively vulnerable hippocampal CA1 neurons (Figure 1) [12•,13,14•,15•,16]. In the CA1 pyramidal cell layer, REST is recruited to the proximal promoter of a subset of target genes where it assembles with a corepressor complex and orchestrates epigenetic remodeling. These findings implicate epigenetic dysregulation in neurodegeneration [6]. The HDAC inhibitor TSA affords substantial protection against ischemia-induced neuronal death [14•], indicating a causal relation between epigenetic remodeling and ischemia-induced neuronal death. Expression of shRNA directed to REST or dominant-negative REST expressed in the brain of living animals afforded robust protection of CA1 neurons in a clinically relevant model of global ischemia. These findings provide strong evidence that activation of REST in excitatory neurons of hippocampal neurons is casually related to neuronal death. A similar role for REST in ischemia-induced neuronal death has been demonstrated in transient middle cerebral artery occlusion (tMCAO), a clinically relevant model of ischemic stroke [70].
ChIP-on-chip profiling and bioinformatics analysis indicate that in post ischemic neurons, the REST orchestrates the silencing of an ensemble of ‘transcriptionally responsive’ target genes, including those encoding the AMPA receptor subunit GluA2, the NMDAR subunit NR1, neuronal acetylcholine receptor subunit β2, the muscarinic acetylcholine receptor M4, NF-κB subunit 2, TRPV1, and the SNARE protein synaptotagmin 6, of which the gene encoding GluA2 was highly ranked. This is significant in that the GluA2 subunit is the ‘ion gate-keeper’ of AMPARs [71]. Whereas GluA2-lacking AMPARs are permeable to Ca2+, and Zn2+ and exhibit pronounced inward rectification, the presence of GluA2 in heteromeric AMPA receptors renders the channel impermeable to Ca2+ and Zn2+ and electrically linear [71]. The presence of GluA2 also influences channel kinetics, conductance, and targeting to and from synaptic sites [71]. This is significant in that Ca2+-permeable AMPA receptors are casually related to ischemia-induced neuronal death [12]. Genes with enhanced REST binding exhibited reduced mRNA and protein expression in the selectively vulnerable hippocampal CA1 after ischemia [14•].
REST and Alzheimer’s disease
A landmark paper by Yankner and colleagues reports that REST is neuroprotective in the aging brain [49]. The authors show that whereas REST expression, activated by the Wnt signaling pathway, is a prominent feature of normal aging, loss of REST is associated with mild or severe cognitive impairment and AD (Figure 1). Examination of autopsy tissue from normal aging human subjects revealed a striking increase in REST in the nuclei of neurons of the prefrontal cortex and the hippocampal CA1 and CA3 that was lost in subjects with mild or severe cognitive impairment AD. ChIP-seq experiments in SH-SY5Y cells [49] revealed occupancy of REST primarily at targets involved in neuronal death (p38 MAPK, FAS, FADD, TRADD, BAX, BID, BBC3 (also known as PUMA), mitochondrial permeability transition pore proteins and cytochrome c) and AD pathology (γ-secretase, presenilin 2, presenilin enhancer-2, and CDK5R1). Accordingly, aging mice with reduced REST exhibited enhanced vulnerability to oxidative stress [49]. Consistent with this, a missense variant of REST which elevates REST expression is protective against hippocampal atrophy associated with Alzheimer’s disease and neurodegeneration in patients with mild cognitive impairment [72]. The neuroprotection afforded by REST in aging brain transcends AD in that REST is also depleted in tissue from subjects with frontotemporal dementia and dementia with Lewy bodies [49]. Moreover, elevated REST expression in neurons of the substantia nigra is protective in an animal model of Parkinson’s disease [73]. These findings demonstrate that REST expression in aging neurons confers neuroprotection and implicate REST as a potential therapeutic target in both mild and severe cognitive impairment AD.
Conclusion
In summary, the past decade has witnessed new findings which collectively point to a role for dysregulation of REST in neurodegenerative disease. Whereas REST is causally to the neuronal death of post ischemic, mature neurons, in aging neurons, it affords neuroprotection. An emerging concept that warrants further exploration is that the ensemble of target genes regulated by REST varies in a cell-type-dependent and context-dependent manner. These findings drive home the concept of REST as a multifaceted regulator of neuronal genes in normal and pathological conditions. Although significant progress has been made in our understanding of how REST functions in neurons under physiological and pathological conditions, many questions remain unanswered. For example, is assembly with TET3 and NSD3 the only mechanism by which REST activates gene transcription or are there other proteins that assemble with REST to promote gene expression? Are NMDA receptors and chloride transporters the only gene targets regulated by REST-dependent epigenetic remodeling during the critical period in postnatal development or are there other genes involved in synaptic plasticity that are regulated by REST at this time in development? Are there as yet undiscovered small molecule inhibitors or activators of REST that might open the door for development of novel therapeutic strategies to ameliorate the neuronal death and impaired cognition associated with neurodegenerative disorders and diseases? In aging neurons, REST affords neuroprotection by suppressing genes involved in apoptosis and oxidative stress including many Bcl2 family members [11•]. Is there any other context in which REST suppresses this ensemble of genes?. In summary, the many unanswered questions suggest many years of fruitful research on REST and epigenetic remodeling of neuronal genes lie ahead.
Acknowledgments
This work was supported by National Institutes of Health grant NS46742, HD083828, and MH092877, and the generous grant from the F.M. Kirby Foundation to RSZ, AHA Scientist Development Grant 16SDG31500001 and NARSAD Young Investigator Grant 25369 to JYH. RSZ is the F. M. Kirby Professor in Neural Repair and Protection.
Footnotes
Conflict of interest statement
None declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, Mandel G. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- 2.Schoenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- 3.Ballas N, Mandel G. The many faces of REST oversee epigenetic programming of neuronal genes. Curr Opin Neurobiol. 2005;15:500–506. doi: 10.1016/j.conb.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 4.Ooi L, Wood IC. Chromatin crosstalk in development and disease: lessons from REST. Nat Rev Genet. 2007;8:544–554. doi: 10.1038/nrg2100. [DOI] [PubMed] [Google Scholar]
- 5.Qureshi IA, Mehler MF. Regulation of non-coding RNA networks in the nervous system – what’s the REST of the story? Neurosci Lett. 2009;466:73–80. doi: 10.1016/j.neulet.2009.07.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hwang JY, Aromolaran KA, Zukin RS. The emerging field of epigenetics in neurodegeneration and neuroprotection. Nat Rev Neurosci. 2017;18:347–361. doi: 10.1038/nrn.2017.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruce AW, Donaldson IJ, Wood IC, Yerbury SA, Sadowski MI, Chapman M, Gottgens B, Buckley NJ. Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc Natl Acad Sci U S A. 2004;101:10458–10463. doi: 10.1073/pnas.0401827101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci U S A. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 10.Rodenas-Ruano A, Chavez AE, Cossio MJ, Castillo PE, Zukin RS. REST-dependent epigenetic remodeling promotes the developmental switch in synaptic NMDA receptors. Nat Neurosci. 2012;15:1382–1390. doi: 10.1038/nn.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11•.Lu T, Aron L, Zullo J, Pan Y, Kim H, Chen Y, Yang TH, Kim HM, Drake D, Liu XS, Bennett DA, Colaiacovo MP, Yankner BA. REST and stress resistance in ageing and Alzheimer’s disease. Nature. 2014;507:448–454. doi: 10.1038/nature13163. This was the first paper to show that dysregulation of REST in AD and to show that REST is neuroprotective in ageing neurons. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12•.Calderone A, Jover T, Noh K-M, Tanaka H, Yokota H, Lin Y, Grooms S, Regis R, Bennett MV, Zukin RS. Ischemic insults de-repress the gene silencer rest in neurons destined to die. J Neurosci. 2003;23:2112–2121. doi: 10.1523/JNEUROSCI.23-06-02112.2003. This was the first paper to show that REST is activated in differentiated neurons. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Formisano L, Noh KM, Miyawaki T, Mashiko T, Bennett MV, Zukin RS. Ischemic insults promote epigenetic reprogramming of mu opioid receptor expression in hippocampal neurons. Proc Natl Acad Sci U S A. 2007;104:4170–4175. doi: 10.1073/pnas.0611704104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14•.Noh KM, Hwang JY, Follenzi A, Athanasiadou R, Miyawaki T, Greally JM, Bennett MV, Zukin RS. Repressor element-1 silencing transcription factor (REST)-dependent epigenetic remodeling is critical to ischemia-induced neuronal death. Proc Natl Acad Sci U S A. 2012;109:E962–E971. doi: 10.1073/pnas.1121568109. This was the first paper to show that REST assembles in a large corepressor complex at the promoter of a target gene and to document that REST is casually related to ischemia-induced neuronal death. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15•.Kaneko N, Hwang JY, Gertner M, Pontarelli F, Zukin RS. Casein kinase 1 suppresses activation of REST in insulted hippocampal neurons and halts ischemia-induced neuronal death. J Neurosci. 2014;34:6030–6039. doi: 10.1523/JNEUROSCI.4045-13.2014. This was the first paper to identify casein kinase 1 as an upstream signal that activates REST in insulted hippocampal neurons. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang JY, Kaneko N, Noh KM, Pontarelli F, Zukin RS. The gene silencing transcription factor REST represses miR-132 expression in hippocampal neurons destined to die. J Mol Biol. 2014;426:3454–3466. doi: 10.1016/j.jmb.2014.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palm K, Belluardo N, Metsis M, Timmusk T. Neuronal expression of zinc finger transcription factor REST/NRSF/XBR gene. J Neurosci. 1998;18:1280–1296. doi: 10.1523/JNEUROSCI.18-04-01280.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garriga-Canut M, Schoenike B, Qazi R, Bergendahl K, Daley TJ, Pfender RM, Morrison JF, Ockuly J, Stafstrom C, Sutula T, Roopra A. 2-Deoxy-D-glucose reduces epilepsy progression by NRSF-CtBP-dependent metabolic regulation of chromatin structure. Nat Neurosci. 2006;9:1382–1387. doi: 10.1038/nn1791. [DOI] [PubMed] [Google Scholar]
- 19•.McClelland S, Flynn C, Dube C, Richichi C, Zha Q, Ghestem A, Esclapez M, Bernard C, Baram TZ. Neuron-restrictive silencer factor-mediated hyperpolarization-activated cyclic nucleotide gated channelopathy in experimental temporal lobe epilepsy. Ann Neurol. 2011;70:454–464. doi: 10.1002/ana.22479. This paper was the first to show that REST represses a subset of its target genes in selectively vulnerable hippocampal neurons in response to seizures and to identify a mechanism responsible for target specificity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McClelland S, Brennan GP, Dube C, Rajpara S, Iyer S, Richichi C, Bernard C, Baram TZ. The transcription factor NRSF contributes to epileptogenesis by selective repression of a subset of target genes. Elife. 2014;3:e01267. doi: 10.7554/eLife.01267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuccato C, Tartari M, Crotti A, Goffredo D, Valenza M, Conti L, Cataudella T, Leavitt BR, Hayden MR, Timmusk T, Rigamonti D, Cattaneo E. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet. 2003;35:76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]
- 22.Zuccato C, Belyaev N, Conforti P, Ooi L, Tartari M, Papadimou E, MacDonald M, Fossale E, Zeitlin S, Buckley N, Cattaneo E. Widespread disruption of repressor element-1 silencing transcription factor/neuron-restrictive silencer factor occupancy at its target genes in Huntington’s disease. J Neurosci. 2007;27:6972–6983. doi: 10.1523/JNEUROSCI.4278-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nat Rev Neurosci. 2007;8:355–367. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- 24.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 25.Borrelli E, Nestler EJ, Allis CD, Sassone-Corsi P. Decoding the epigenetic language of neuronal plasticity. Neuron. 2008;60:961–974. doi: 10.1016/j.neuron.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levenson JM, Sweatt JD. Epigenetic mechanisms in memory formation. Nat Rev Neurosci. 2005;6:108–118. doi: 10.1038/nrn1604. [DOI] [PubMed] [Google Scholar]
- 27.Day JJ, Sweatt JD. DNA methylation and memory formation. [2010/10/27];Nat Neurosci. 2010 :1319–1323. doi: 10.1038/nn.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abel T, Zukin RS. Epigenetic targets of HDAC inhibition in neurodegenerative and psychiatric disorders. Curr Opin Pharmacol. 2008;8:57–64. doi: 10.1016/j.coph.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andres ME, Burger C, Peral-Rubio MJ, Battaglioli E, Anderson ME, Grimes J, Dallman J, Ballas N, Mandel G. CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc Natl Acad Sci U S A. 1999;96:9873–9878. doi: 10.1073/pnas.96.17.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ballas N, Battaglioli E, Atouf F, Andres ME, Chenoweth J, Anderson ME, Burger C, Moniwa M, Davie JR, Bowers WJ, Federoff HJ, Rose DW, Rosenfeld MG, Brehm P, Mandel G. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 2001;31:353–365. doi: 10.1016/s0896-6273(01)00371-3. [DOI] [PubMed] [Google Scholar]
- 31.Grimes JA, Nielsen SJ, Battaglioli E, Miska EA, Speh JC, Berry DL, Atouf F, Holdener BC, Mandel G, Kouzarides T. The co-repressor mSin3A is a functional component of the REST-CoREST repressor complex. J Biol Chem. 2000;275:9461–9467. doi: 10.1074/jbc.275.13.9461. [DOI] [PubMed] [Google Scholar]
- 32.Naruse Y, Aoki T, Kojima T, Mori N. Neural restrictive silencer factor recruits mSin3 and histone deacetylase complex to repress neuron-specific target genes. Proc Natl Acad Sci USA. 1999;96:13691–13696. doi: 10.1073/pnas.96.24.13691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang Y, Myers SJ, Dingledine R. Transcriptional repression by REST: recruitment of Sin3A and histone deacetylase to neuronal genes. Nat Neurosci. 1999;2:867–872. doi: 10.1038/13165. [DOI] [PubMed] [Google Scholar]
- 34.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 35.Lunyak VV, Burgess R, Prefontaine GG, Nelson C, Sze SH, Chenoweth J, Schwartz P, Pevzner PA, Glass C, Mandel G, Rosenfeld MG. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science. 2002;298:1747–1752. doi: 10.1126/science.1076469. [DOI] [PubMed] [Google Scholar]
- 36.Roopra A, Qazi R, Schoenike B, Daley TJ, Morrison JF. Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol Cell. 2004;14:727–738. doi: 10.1016/j.molcel.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 37.Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- 38.Shi YJ. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19:1–8. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 39.Lombardi LM, Baker SA, Zoghbi HY. MECP2 disorders: from the clinic to mice and back. J Clin Invest. 2015;125:2914–2923. doi: 10.1172/JCI78167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 41.Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 42.Perera A, Eisen D, Wagner M, Laube SK, Kunzel AF, Koch S, Steinbacher J, Schulze E, Splith V, Mittermeier N, Muller M, Biel M, Carell T, Michalakis S. TET3 is recruited by REST for context-specific hydroxymethylation and induction of gene expression. Cell Rep. 2015;11:283–294. doi: 10.1016/j.celrep.2015.03.020. [DOI] [PubMed] [Google Scholar]
- 43.Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol. 2012;13:115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abramovitz L, Shapira T, Ben-Dror I, Dror V, Granot L, Rousso T, Landoy E, Blau L, Thiel G, Vardimon L. Dual role of NRSF/REST in activation and repression of the glucocorticoid response. J Biol Chem. 2008;283:110–119. doi: 10.1074/jbc.M707366200. [DOI] [PubMed] [Google Scholar]
- 45.Philpot BD, Zukin RS. Synapse-specific metaplasticity: to be silenced is not to silence 2B. Neuron. 2010;66:814–816. doi: 10.1016/j.neuron.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yeo M, Berglund K, Augustine G, Liedtke W. Novel repression of Kcc2 transcription by REST-RE-1 controls developmental switch in neuronal chloride. J Neurosci. 2009;29:14652–14662. doi: 10.1523/JNEUROSCI.2934-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mortazavi A, Thompson EC, Garcia ST, Myers RM, Wold B. Comparative genomics modeling of the NRSF/REST repressor network: from single conserved sites to genome-wide repertoire. Genome Res. 2006;16:1208–1221. doi: 10.1101/gr.4997306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein–DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 49.Lu T, Aron L, Zullo J, Pan Y, Kim H, Chen Y, Yang TH, Kim HM, Drake D, Liu XS, Bennett DA, Colaiacovo MP, Yankner BA. REST and stress resistance in ageing and Alzheimer’s disease. Nature. 2014;507:448–454. doi: 10.1038/nature13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abrajano JJ, Qureshi IA, Gokhan S, Molero AE, Zheng D, Bergman A, Mehler MF. Corepressor for element-1-silencing transcription factor preferentially mediates gene networks underlying neural stem cell fate decisions. Proc Natl Acad Sci U S A. 2010;107:16685–16690. doi: 10.1073/pnas.0906917107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zukin RS. Eradicating the mediators of neuronal death with a fine-tooth comb. Sci Signal. 2010;3:e20. doi: 10.1126/scisignal.3125pe20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guardavaccaro D, Frescas D, Dorrello NV, Peschiaroli A, Multani AS, Cardozo T, Lasorella A, Iavarone A, Chang S, Hernando E, Pagano M. Control of chromosome stability by the beta-TrCP-REST-Mad2 axis. Nature. 2008;452:365–369. doi: 10.1038/nature06641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Westbrook TF, Hu G, Ang XL, Mulligan P, Pavlova NN, Liang A, Leng Y, Maehr R, Shi Y, Harper JW, Elledge SJ. SCFbeta-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature. 2008;452:370–374. doi: 10.1038/nature06780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh A, Rokes C, Gireud M, Fletcher S, Baumgartner J, Fuller G, Stewart J, Zage P, Gopalakrishnan V. Retinoic acid induces REST degradation and neuronal differentiation by modulating the expression of SCF(beta-TRCP) in neuroblastoma cells. Cancer. 2011;117:5189–5202. doi: 10.1002/cncr.26145. [DOI] [PubMed] [Google Scholar]
- 56.Huang Z, Wu Q, Guryanova OA, Cheng L, Shou W, Rich JN, Bao S. Deubiquitylase HAUSP stabilizes REST and promotes maintenance of neural progenitor cells. Nat Cell Biol. 2011;13:142–152. doi: 10.1038/ncb2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheong JK, Virshup DM. Casein kinase 1: complexity in the family. Int J Biochem Cell Biol. 2011;43:465–469. doi: 10.1016/j.biocel.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 58.Chen H, Ma H, Inuzuka H, Diao J, Lan F, Shi YG, Wei W, Shi Y. DNA damage regulates UHRF1 stability via the SCF(beta-TrCP) E3 ligase. Mol Cell Biol. 2013;33:1139–1148. doi: 10.1128/MCB.01191-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weissman AM. How much REST is enough? Cancer Cell. 2008;13:381–383. doi: 10.1016/j.ccr.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 60.Nesti E, Corson GM, McCleskey M, Oyer JA, Mandel G. C-terminal domain small phosphatase 1 and MAP kinase reciprocally control REST stability and neuronal differentiation. Proc Natl Acad Sci U S A. 2014;111:E3929–E3936. doi: 10.1073/pnas.1414770111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, Padmanabhan R, Hild M, Berry DL, Garza D, Hubbert CC, Yao TP, Baehrecke EH, Taylor JP. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 62.Shimojo M, Hersh LB. Characterization of the REST/NRSF-interacting LIM domain protein (RILP): localization and interaction with REST/NRSF. J Neurochem. 2006;96:1130–1138. doi: 10.1111/j.1471-4159.2005.03608.x. [DOI] [PubMed] [Google Scholar]
- 63.Park H, Poo MM. Neurotrophin regulation of neural circuit development and function. Nat Rev Neurosci. 2013;14:7–23. doi: 10.1038/nrn3379. [DOI] [PubMed] [Google Scholar]
- 64.Chen ZF, Paquette AJ, Anderson DJ. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat Genet. 1998;20:136–142. doi: 10.1038/2431. [DOI] [PubMed] [Google Scholar]
- 65.Paquette AJ, Perez SE, Anderson DJ. Constitutive expression of the neuron-restrictive silencer factor (NRSF)/REST in differentiating neurons disrupts neuronal gene expression and causes axon pathfinding errors in vivo. Proc Natl Acad Sci U S A. 2000;97:12318–12323. doi: 10.1073/pnas.97.22.12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thiel G, Ekici M, Rossler OG. RE-1 silencing transcription factor (REST): a regulator of neuronal development and neuronal/endocrine function. Cell Tissue Res. 2015;359:99–109. doi: 10.1007/s00441-014-1963-0. [DOI] [PubMed] [Google Scholar]
- 67.Patterson KP, Barry JM, Curran MM, Singh-Taylor A, Brennan G, Rismanchi N, Page M, Noam Y, Holmes GL, Baram TZ. Enduring memory impairments provoked by developmental febrile seizures are mediated by functional and structural effects of neuronal restrictive silencing factor. J Neurosci. 2017;37:3799–3812. doi: 10.1523/JNEUROSCI.3748-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chuang DM, Leng Y, Marinova Z, Kim HJ, Chiu CT. Multiple roles of HDAC inhibition in neurodegenerative conditions. Trends Neurosci. 2009;32:591–601. doi: 10.1016/j.tins.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tahiliani M, Mei P, Fang R, Leonor T, Rutenberg M, Shimizu F, Li J, Rao A, Shi Y. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature. 2007;447:601–605. doi: 10.1038/nature05823. [DOI] [PubMed] [Google Scholar]
- 70.Formisano L, Guida N, Valsecchi V, Pignataro G, Vinciguerra A, Pannaccione A, Secondo A, Boscia F, Molinaro P, Sisalli MJ, Sirabella R, Casamassa A, Canzoniero LM, Di RG, Annunziato L. NCX1 is a new rest target gene: role in cerebral ischemia. Neurobiol Dis. 2013;50:76–85. doi: 10.1016/j.nbd.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 71.Liu SJ, Zukin RS. Ca2+-permeable AMPA receptors in synaptic plasticity and neuronal death. Trends Neurosci. 2007;30:126–134. doi: 10.1016/j.tins.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 72.Nho K, Kim S, Risacher SL, Shen L, Corneveaux JJ, Swaminathan S, Lin H, Ramanan VK, Liu Y, Foroud TM, Inlow MH, Siniard AL, Reiman RA, Aisen PS, Petersen RC, Green RC, Jack CR, Jr, Weiner MW, Baldwin CT, Lunetta KL, Farrer LA, Furney SJ, Lovestone S, Simmons A, Mecocci P, Vellas B, Tsolaki M, Kloszewska I, Soininen H, McDonald BC, Farlow MR, Ghetti B, Huentelman MJ, Saykin AJ. Protective variant for hippocampal atrophy identified by whole exome sequencing. Ann Neurol. 2015;77:547–552. doi: 10.1002/ana.24349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu M, Suo H, Liu M, Cai L, Liu J, Huang Y, Xu J, Wang Y, Zhu C, Fei J, Huang F. NRSF/REST neuronal deficient mice are more vulnerable to the neurotoxin MPTP. Neurobiol Aging. 2013;34:916–927. doi: 10.1016/j.neurobiolaging.2012.06.002. [DOI] [PubMed] [Google Scholar]