Abstract
Discovery of monoclonal antibodies is most commonly performed using phage or yeast display but mammalian cells are used for production because of the complex antibody structure, including the multiple disulfide bonds and glycosylation, required for function. As this transition between host organisms is often accompanied by impaired binding, folding or expression, development pipelines include laborious plate-based screening or engineering strategies to adapt an antibody to mammalian expression. To circumvent these problems, we developed a plasmid-based Fab screening platform on Chinese hamster ovary (CHO) cells which allows for antibody selection in the production host and in the presence of the same post-translational modifications as the manufactured product. A hu4D5 variant with low affinity for the human epidermal growth factor receptor (HER2) growth factor receptor was mutagenized and this library of ~106 unique clones was screened to identify variants with up to 400-fold enhanced HER2 binding. After two rounds of fluorescence activated cell sorting (FACS), four unique clones exhibited improved antigen binding when expressed on the CHO surface or as purified human IgG. Three of the four clones contained free cysteines in third complementarity determining region of the antibody heavy chain, which did not impair expression or cause aggregation. The improved clones had similar yields and stabilities as hu4D5 and similar sub-nanomolar affinities as measured by equilibrium binding to target cells. The limited size of mammalian libraries restricts the utility of this approach for naïve antibody library screening, but it is a powerful approach for antibody affinity maturation or specificity enhancement and is readily generalizable to engineering other surface receptors, including T-cell receptors and chimeric antigen receptors.
Keywords: antibody engineering, expression engineering, hu4D5, mammalian display, Trastuzumab
Introduction
The ability of antibodies to provide targeted therapies with low immunogenicity has driven impressive growth in the number of monoclonal antibody (mAb) therapeutics in clinical trials and approved by the United States Food and Drug Administration (FDA) (Ecker et al., 2015). High-throughput screening of antibody candidates for binding to an antigen of interest has been possible for over 25 years (McCafferty et al., 1990) but screening and optimization for large-scale manufacturing suitability remains low-throughput (Tabrizi et al., 2012). Selection of clones suitable for manufacturing often involves laboriously sub-cloning, expressing and characterizing hundreds of antibodies in the mammalian hosts used for production. Mammalian expression remains the industry standard, as full-length mAbs can be produced at levels reaching 25 g/l (Chon and Zarbis-Papastoitsis, 2011) with human-like post-translational modifications, and the host meets stringent regulatory requirements.
However, simpler organisms such as phage, bacteria and yeast are traditionally used for antibody engineering (Hoogenboom, 2005). These hosts are popular since they support high-throughput screening of large libraries (up to 1010 unique clones; Weaver-Feldhaus et al., 2004) yet they also impose constraints on the antibody sequences that can be expressed and selected. For instance, bacteria lack and yeast differ in many of the sophisticated quality control mechanisms present in higher eukaryotes, including chaperones, endoplasmic reticulum-associated degradation proteins, thio-oxidoreductases and peptidyl prolyl isomerases (Ni and Lee, 2007). Yeast also modify proteins with significantly different N-glycosylated sugar structures than mammalian cells, while Escherichia coli do not perform these reactions at all (Wildt and Gerngross, 2005). The popular single-chain variable fragment (scFv) format exposes hydrophobic residues normally buried at the variable region–constant domain interface (Nieba et al., 1997) and reduces folding efficiency in many cases. Taken together, these hosts can select for diversity that is not necessarily compatible with reformatting as an IgG. Even clones that express well in an scFv format can suffer from poor expression and/or activity after conversion back to full-length antibody constructs (Hoogenboom, 2005; Bradbury et al., 2011).
In an effort to streamline antibody discovery and development, several groups have developed methods for screening libraries on the mammalian cell surface using various approaches (Ho et al., 2006; Schenk et al., 2007; Beerli et al., 2008; Bowers et al., 2011, 2013, 2014; Li et al., 2012; McConnell et al., 2012; Horlick et al., 2013; King et al., 2014). Many use human embryonic kidney (HEK) cells, which are less relevant for antibody production than Chinese hamster ovary (CHO) cells (Frenzel et al., 2013) and/or viral infection techniques, which are cumbersome. We aimed to develop a simple, widely-accessible, plasmid transfection-based CHO cell surface display system for screening antibody Fab variants with several key features. To maintain the library in the CHO cells during the selection process, the Epi-CHO (Kunaparaju et al., 2005) (Acyte Biotech) semi-stable episomal expression system utilizing Epstein–Barr and polyoma virus elements was used. The light and heavy chain coding regions were included on the same episome, separated by a 2A peptide for simple recovery of the entire coding sequence and maintenance of proper pairing throughout screening. Finally, the antibody Fab format was displayed because they retain a structure more similar to full-length IgGs than scFvs, while avoiding the bivalent nature of IgG binding.
We used this system to identify Fab variants with efficient expression in the production host and high affinity ligand binding in the context of post-translational modifications that would exist in a final therapeutic product. A low affinity variant of the well-characterized hu4D5 antibody (also known as Trastuzumab or Herceptin), bD1 (Bostrom et al., 2009), was used as a model antibody for mutagenesis and screening. After generating a large library of ~106 unique clones with residues altered at nine heavy chain CDR positions, we used the CHO cell platform to identify bD1 variants with similar expression levels, stabilities and binding affinities as hu4D5 but distinct CDR sequences. The free-cysteine containing sequences of several of the improved variants and the previously undescribed glycosylation of the bD1 parent (isolated by phage display) highlight the differences between antibodies produced in mammalian and other cells types as well as the benefits of screening in the CHO production host. Due to the culture volumes required for mammalian cell growth, this library and most mammalian libraries are limited in size to ~106 variants, reducing their utility for screening of completely naïve antibody libraries. However, the library sizes are sufficient for affinity maturation as presented here, specificity redirection, or screening of libraries preselected using phage or magnetic panning. In addition to screening antibodies, the system is readily adaptable to other biomolecules best expressed in mammalian systems such as T-cell receptors, chimeric antigen receptors and other membrane-bound receptors.
Materials and Methods
Cloning of hu4D5 Fab for display
The hu4D5 light chain variable (VL) and heavy chain variable (VH) amino acid sequences were obtained from literature (Cho et al., 2003) and submitted to Life Technologies for Cricetulus griseus codon optimization and DNA synthesis. Simultaneously, an IgG1 Fab display construct was synthesized by integrated DNA technologies with the consensus Kozak sequence (CCACCATGG), a mouse IgK secretion signal (Smith et al., 2009), a dummy VL sequence, human IgK CL sequence without a stop codon, a F2A sequence with a furin cleavage site (Szymczak-Workman et al., 2012), a dummy VH sequence, human IgG1 CH1 sequence, a glycine–serine linker and a transmembrane region from platelet derived growth factor receptor (PDGFR; Gronwald et al., 1988), and flanking KpnI and BamHI sites for cloning into pPyEBV (Kunaparaju et al., 2005; Acyte Biotech). The sequence was also codon optimized for C. griseus and codon usage was adjusted slightly to remove extended regions of sequence repetition. Useful restriction sites were integrated into the sequence to allow cloning of VL sequences with AgeI and AflII restriction sites and cloning of VH sequences with HindIII and NheI. This dummy display sequence was cloned into pPyEBV with KpnI and BamHI. The hu4D5 VL sequence was amplified with appropriate primers and cloned into the display vector via AgeI and AflII restriction sites. After sequencing, the hu4D5 VH sequence was added by digestion and ligation between the HindIII and NheI restriction sites. The hu4D5 display construct, pPy4D5disp, design is outlined in Fig. 1A and provided in full in Supplementary Fig. 1.
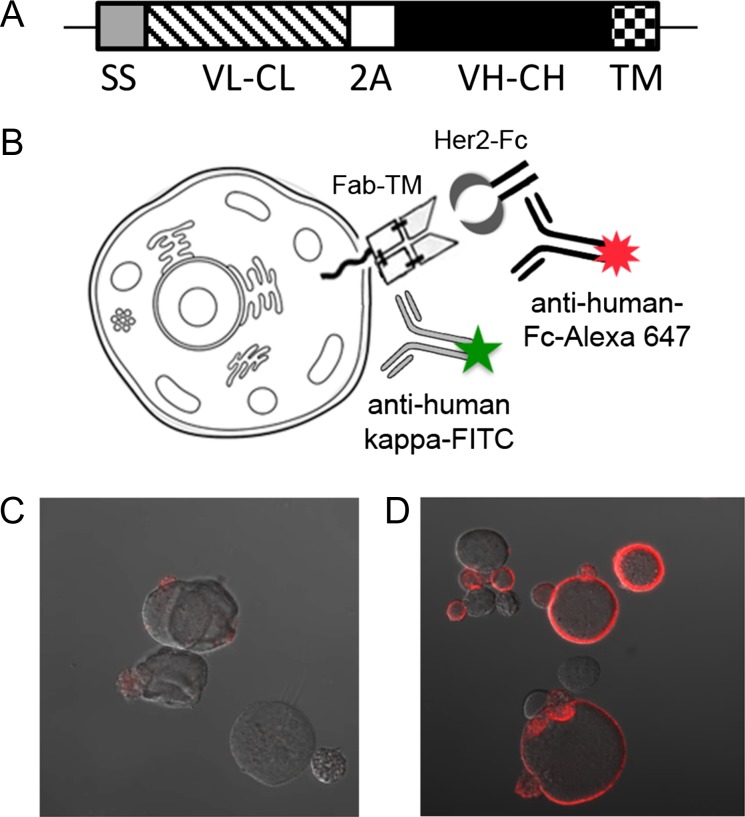
Fig. 1.
The hu4D5 Fab was effectively displayed on the surface of CHO cells. (A) Schematic of the Fab CHO expression construct. The open reading frame inserted into pPyEBV consisted of a murine IgΚ secretion signal (SS) fused to the hu4D5 IgΚ chain (VL–CL) DNA sequence, followed by DNA encoding a furin cleavage site and F2A peptide (2 A). The hu4D5 VH and CH1 coding sequence (VH–CH) fused to a short glycine–serine linker and the PDGFR transmembrane domain (TM) immediately followed the 2A peptide. (B) Schematic of the Fab CHO display and staining system. The displayed Fab was stained with HER2-Fc, then anti-human Fc-Alexa Fluor 647, and anti-human IgK-FITC was added for flow cytometric detection. For microscopy visual confirmation of the Fab CHO expression and staining procedure, CHO cells were transfected with either (C) blank pPyEBV or (D) pPyhu4D5disp and stained for HER2 binding, then imaged by confocal microscopy.
Confocal imaging of hu4D5 Fab display
CHO-T (Acyte Biotech) cells were transfected either with blank pPyEBV or pPy4D5disp using Lipofectamine 2000 (Life Technologies) as described in the Epi-CHO manual. Growth media was CHO-S-SFM-II media (Life Technologies) supplemented with 2× Glutamax (Life Technologies). Two days after transfection, cells were washed with phosphate buffered saline (PBS), 0.05% tween-20, and 1% fetal bovine serum (FBS), spun at 200× g for 2 min, and incubated for 30 min with 1 μg/ml human epidermal growth factor receptor (HER2)-Fc (R&D Systems) in wash buffer. Samples were then washed, incubated with a 1:125 dilution of goat–anti-human Fc-Alexa Fluor 647 (Jackson ImmunoResearch) in wash buffer for 30 min, washed again and mounted on slides with Fluormount G (Southern Biotech). Images were obtained on a Zeiss LSM 710 Confocal microscope (Fig. 1B–D).
Cloning of EGFP into pPyEBV
EGFP was cloned directly from pEGFP-C1 (Clontech) to pPyEBV using XhoI and NheI restriction digestion and ligation.
Flow cytometric scanning and sorting of CHO-T cells displaying Fab
For flow cytometric screening, ~106 cells were harvested and spun at 200× g for 2 min. Cells were then washed in 1 ml wash buffer (PBS with 1% bovine serum albumin (BSA, Fisher Scientific), filter sterilized), incubated for 20 min at room temperature in primary stain (specified below) in wash buffer. The samples were then washed and incubated with secondary stain (specified below) in wash buffer 20 min at room temperature. Finally, the samples were washed in wash buffer, resuspended in wash buffer and scanned by flow cytometry on a BD Fortessa instrument. Alexa Fluor 647 was excited with a 640-nm laser and emission was detected through a 670/30-bandpass filter. EGFP and FITC were excited with a 488-nm laser and emission was detected through a 530/30-bandpass filter. Cy3 was excited with a 561-nm laser and emission was detected through a 582/15-bandpass filter. There was no crosstalk between Cy3, Alexa Fluor 647 and EGFP/FITC in these channels.
Similarly, FACS of the library was performed with increased cell numbers (the larger of five times the number of library members remaining or 106 cells) on a BD FACS Aria with a 100 μm nozzle. For sorting, all wash and staining steps were performed with filter sterilized Optimem (ThermoFisher Scientific) with 1% BSA.
Determining the number of plasmids per cell
Following the Epi-CHO transfection protocol, pPy4D5disp and pPyEGFP were mixed at ratios of 0:1, 1:1, 4:1, 9:1 and 19:1, and transfected to CHO-T cells in duplicate. Two days later, the cells were harvested and analyzed by flow cytometry as described above with 0.4 μg/ml HER2-Fc as the primary stain and 1:250 anti-human Fc-Alexa Fluor 647 as the secondary stain (Fig. 2A–F). Based on negative cell populations, the percentage of cells expressing both EGFP and hu4D5 Fab was determined. These percentages were normalized by the transfection efficiency and compared to the predicted percentage of mixed fluorescence based on a Poisson distribution of plasmids introduced per cell. The average number of plasmids per cell was allowed to vary to minimize the difference between the data and the Poisson-based prediction, giving a fit with a Pearson’s correlation coefficient of 0.99 and an average number of plasmids per cell of 2.07. Based on this average number of plasmids per cell, the ratio of blank carrier plasmid to pPy4D5 was optimized such that 75% of transfected cells were predicted to have a single pPy4D5disp plasmid. The fit suggested 74% blank carrier plasmid and 26% antibody encoding plasmid was optimal, which was used for all remaining experiments.
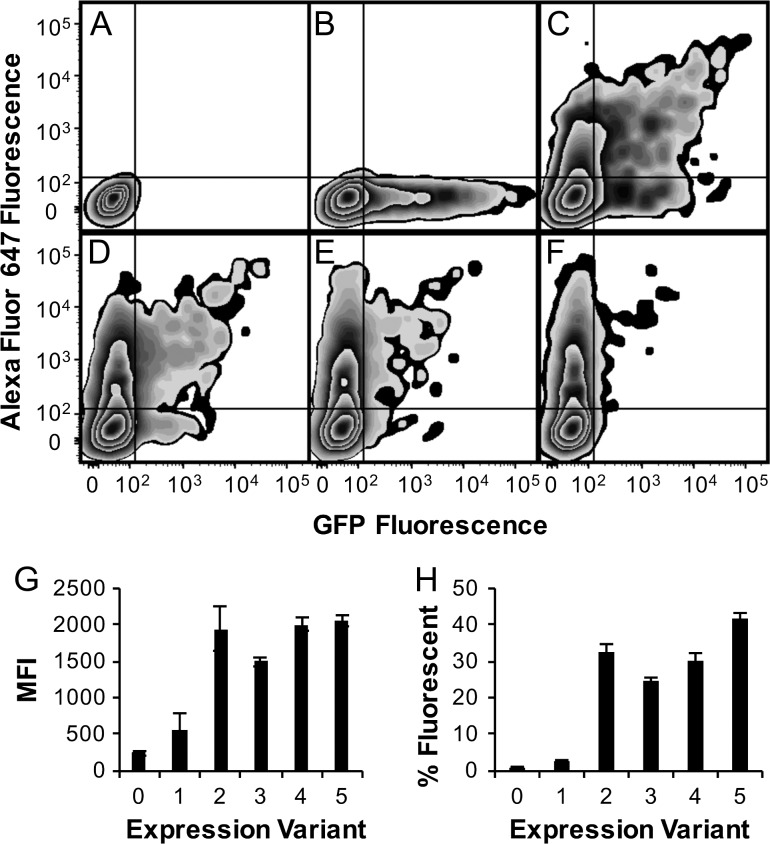
Fig. 2.
Fab display was optimized to limit the number of plasmids per cell and modulate expression level. Epi-CHO cells were transfected with: (A) no plasmid or pPy4D5disp and pPyEGFP at a ratio of (B) 0:1, (C) 1:1, (D) 4:1, (E) 9:1 or (F) 19:1. The percent in quadrant I, positive for both GFP and HER2-Fc/anti-human Fc-AF 647, were 0%, 0.6%, 19.5%, 12.5%, 8.2% and 3.8%, respectively. Plasmids encoding expression level Kozak variants pPy4D5disp0 (0), pPy4D5disp1 (1), pPy4D5disp2 (2), pPy4D5disp3 (3), pPy4D5disp4 (4) and unmodified pPy4D5disp (5) were transfected to CHO-T cells, stained with HER2-Fc/anti-human Fc- Alexa Fluor 647, and scanned by flow cytometry. Data was collected for (G) mean fluorescence intensity (MFI) of Alexa Fluor 647 in cells displaying Fab and H, the percentage of total cells expressing Fab.
Testing of display with altered Kozak sequences
The region upstream of the start codon and a large portion of the pPy4D5disp light chain sequence was amplified with primers to modify the Kozak sequence and add synthetic open reading frames in some cases. The forward primers were 5′-AAGCTGGGTACCGCGCGCCTTTTTTGGCTGGTCCTGCATCATCCTG-3′ for pPy4D5disp0, 5′-AAGCTGGGTACCGCGCGCCGGGATGGGTTGATTTATGGGCTGGTCCTGCATCAT-3′ for pPy4D5disp1, 5′-AAGCTGGGTACCGCGCGCCTAAATGGGTTGAACCATGGGCTGGTCCTGCATCAT-3′ for pPy4D5disp2, 5′-AAGCTGGGTACCGCGCGCCTTGATGGGCTGGTCCTGCATCAT-3′ for pPy4D5disp3, 5′-AAGCTGGGTACCGCGCGCCTAAATGGGCTGGTCCTGCATCAT-3′ for pPy4D5disp4 and all amplifications used the reverse primer 5′-GACTTCGCATGCGTAGACTTTGTG-3′. After polymerase chain reaction (PCR) amplification, all products were cloned into pPy4D5disp using KpnI and SphI restriction sites. Two days after transfecting pPy4D5disp, and pPy4D5disp0, 1, 2, 3 and 4 to CHO-T cells with carrier plasmid as described above, a sample of each transfection cells was harvested and stained for cytometry as described above. Live cells were analyzed for fluorescence intensity and percent fluorescent (Fig. 2G and H).
Cloning bD1
Overlapping 40-mer DNA oligomers were ordered (Sigma) to span the bD1 VL region in the forward and reverse directions. Oligos used for assembly are listed in Supplementary Table 2. Assembly PCR (Stemmer et al., 1995) was used to create the bD1 VL region DNA fragment and it was inserted into pPy4D5disp2 between the AflII and KpnI restriction sites by digestion and ligation. The resulting plasmid is pPybD1disp.
bD1 Heavy chain library creation
The crystal structure of hu4D5 complexed with HER2, PDB IN8Z (Cho et al., 2003), was examined and nine residues within 5 Å of HER2 or the light chain residues that were altered in bD1 were considered for mutagenesis. The majority of these residues were in CDRs H2 and H3, and those CDRs were compared to ~100 similar CDR sequences found in a GenBank blast search. Residues were targeted for mutagenesis based on the level of variability found in similar antibodies at that position. A designed bD1 heavy chain library was created using assembly mutagenesis (Bessette et al., 2003) with the primers listed in Supplementary Table 3. The targeted amino acids in CDRH2 according to the Kabat numbering scheme were R50 with the codon AGM (coding for R/S), Y52 with the codon TMT (coding for S/Y), T53 with the codon WMT (coding for N/S/T/Y) and N54 with the codon ART (coding for N/S). More diversity was introduced in third complementarity determining region of the antibody heavy chain (CDRH3) where there were more residues near areas of interest including encoding W95 with BDK (coding for C/D/E/F/G/H/L/Q/R/V/W/Y/stop), D98 with RVT (coding for A/D/G/N/S/T), F100 with TNK (coding for C/F/S/W/Y/stop), A100b with KVT (coding for A/C/D/G/S/Y) and M100c with NBK (coding for A/C/F/G/I/L/M/P/R/S/T/V/W). After assembly, the PCR product was cloned into pPybD1disp between the HindIII and NheI restriction sites. In all, the possible diversity of the library was 4.0 × 106 DNA sequences and 1.3 × 106 protein sequences, with 13% predicted to contain premature stop codons. The actual number of E.coli transformants was 1.0 × 106. Sequencing of 10 clones showed diversity at the targeted positions, suggesting a successful generation of the designed library (data not shown).
bD1 HC library screening
Library plasmid DNA, blank pPyEBV, pPy4D5disp2 and pPybD1disp were transfected to CHO-T cells with a similarly sized blank carrier plasmid as described above. The transformed cells were grown at 37°C overnight, spun at 200× g for 2 min and resuspended in fresh media without antibiotic. On Day 2, cells were collected again and resuspended in fresh media containing 150 μg/ml Hygromycin B (ThermoFisher Scientific) for selection. Five days after transfection, the media was replaced again, but the Hygromycin B concentration was increased to 300 μg/ml and maintained at that concentration for the duration of the experiment. The cells were carefully cultured and split throughout the first two weeks of growth to allow the Hygromycin B to kill cells not transfected with pPyEBV vector, while maintaining at least five times the library size in pPyEBV transfected cells. Two weeks after transfection, the cells were collected and stained for FACS as described above. Cells were first stained with 0.4 μg/ml HER2-Fc, 1:250 goat–anti-human Ig Κ-FITC (Sigma) and 1:250 goat–anti-rabbit IgG (H + L)-Cy3 (Life Technologies) in Optimem with 1% BSA. After washing, the cells were then incubated with 1:250 goat–anti-human kappa-FITC and 1:250 anti-human Fc-Alexa Fluor 647 in Optimem with 1% BSA. In two rounds of FACS, the 1–2% of cells with the highest Alexa Fluor 647 signal for a given FITC signal were sorted into warm media. The cells were grown for 7 days between sorts and scanned for enrichment after the final round of sorting and growth (Fig. 3). After each round of cell sorting and growth, cells were collected for DNA purification and storage.
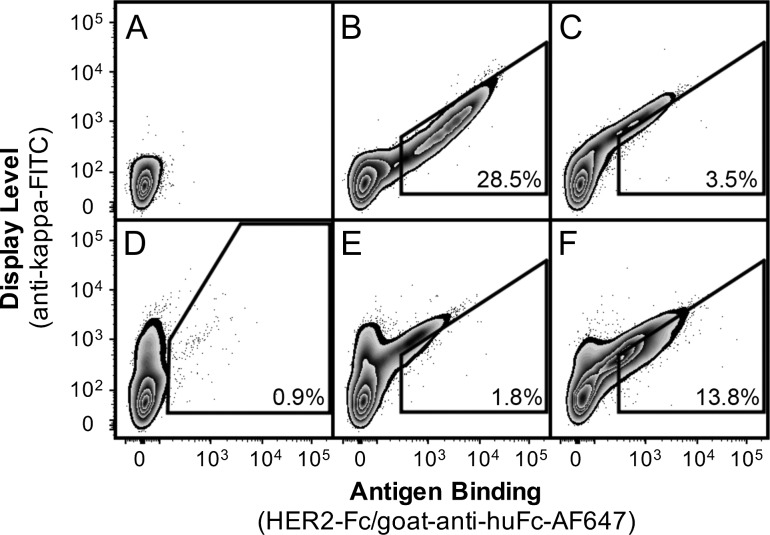
Fig. 3.
The bD1 Fab display library was enriched by FACS for increased HER2 binding. CHO-T cells transfected with (A) blank pPyEBV, (B) pPy4D5Disp2 and (C) pPybD1disp plasmid were compared to (D) Round 0 (unsorted), (E) Round 1 and (F) Round 2 sorts. Panels B, C, E and F show the Round 1 sort gate, while Panel D shows the initial sort gate. The percent of the population falling into the gate is indicated.
Isolation and screening of individual clones
A Genomic DNA Purification Kit (Invitrogen) was used to purify DNA from 106 sorted cells. The DNA was used as a template to amplify the VH regions with the forward primer 5′-ATCAGGCGTGAAGCAGACACTCAAC-3′ and the reverse primer 5′-CTGTGCCTCCTGACGTACTCTTG-3′, which were then cloned into pPybD1disp as described above. Twenty post-round 2 clones were sequenced. Four unique, non-background sequences were transfected to CHO-T cells and scanned by flow cytometry as described above (Fig. 4). For soluble, full-length human IgG1 expression, the hu4D5 and variant VH regions and the hu4D5 and bD1 VL regions were cloned into the AbVec antibody expression vectors (Smith et al., 2009), expressed transiently and purified as described previously (Nguyen et al., 2015). Antibodies were also expressed side-by-side in small scale to compare expression levels (Table 1). R2J variants were cloned by amplification of the R2J VH region with the reverse primer 5′- gcccttggtcgacgcAGAGGACACGGTCACGAGGGTTCCCTGGCCCCAATAGTCAAAAGAGTAAAAGCCAGCGCC-3′ for R2J-S and 5′- gcccttggtcgacgcAGAGGACACGGTCACGAGGGTTCCCTGGCCCCAATAGTCAAAATAGTAAAAGCCAGCGCC-3′ for R2J-Y, and the same AbVec cloning protocol. Parent hu4D5 and R2J were also cloned as His-tagged scFvs, expressed in E. coli, purified by IMAC and evaluated by SDS-PAGE for assessment of the suitability of these antibodies for prokaryotic expression (Supplementary Fig. 2).
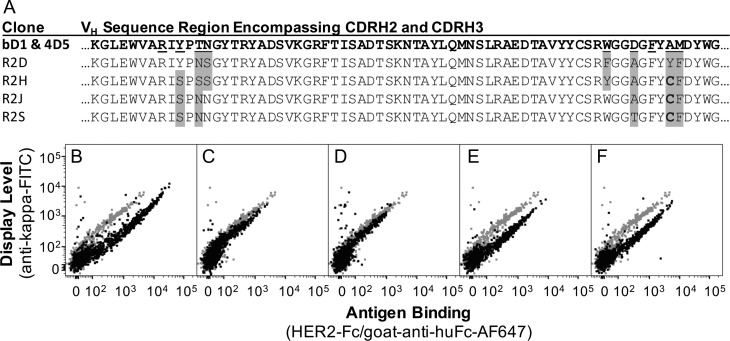
Fig. 4.
Displayed antibody variants have increased HER2 binding relative to parent bD1. Four unique variants of bD1 were sequenced (A) and scanned by flow cytometry in the Fab display format (B–F). CHO-T cells displaying bD1 (gray in panels B–F) were compared to the (B) hu4D5 control or the following variants shown in black: (C) R2D, (D) R2H, (E) R2J and (F) R2S.
Table I.
Comparison of hu4D5, bD1 and bD1 variants as purified IgG1*
| Clone | Expression level (relative to bD1) | Second transition Tm (°C) | ELISA EC50 (nM) | Effective Kd (nM) [R2] |
|---|---|---|---|---|
| hu4D5 | 1.1 | 84.6 ± 0.4 | 0.08 ± 0.01 | 0.17 ± 0.06 [0.89] |
| bD1 | 1.0 | 85.1 ± 0.1 | 5 ± 2 | 90 ± 60 [0.89] |
| R2D | 0.9 | 84.8 ± 0.1 | 0.41 ± 0.08 | 3 ± 2 [0.88] |
| R2H | 0.9 | 86.0 ± 0.1 | 0.34 ± 0.04 | ND |
| R2J | 0.8 | 86.2 ± 0.1 | 0.10 ± 0.02 | 0.22 ± 0.07 [0.91] |
| R2S | 0.9 | 86.2 ± 0.1 | 0.11 ± 0.01 | 0.22 ± 0.06 [0.92] |
| R2J-S | nd | 84.7 ± 0.1 | 0.11 ± 0.01 | ND |
| R2J-Y | nd | 87.4 ± 0.1 | 0.12 ± 0.02 | ND |
ND: no data.
*Errors are standard deviation for Tm, range of duplicates for EC50 and standard error for Kd.
Variant characterization
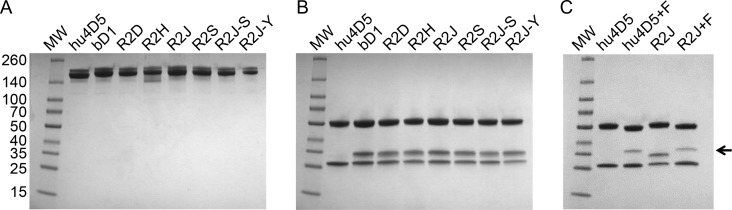
To assess antibody purity, 3 μg of each antibody variant and hu4D5 and bD1 controls were mixed with reducing or non-reducing sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer. Reduced samples were boiled for 5 min, while non-reduced samples were warmed to 42°C for 2 min, then all samples were loaded on a 4–20% acrylamide gradient gel (Bio-Rad) alongside SpectraBR pre-stained ladder (ThermoFisher Scientific). Gels were stained with GelCode Blue (ThermoFisher Scientific) and destained with water prior to imaging (Fig. 5A and B). PNGase F (NEB) digestion of 10 μg of purified hu4D5 and bD1 antibody was performed with and without PNGase F enzyme as described in the PNGase F protocol. After digestion, one-third of each product was run on a reducing SDS-PAGE gel as described above (Fig. 5C). PNGase F enzyme has a molecular weight of 36 kDa and appears in the lanes containing the enzyme.
Fig. 5.
Purified antibody variants behave as expected in SDS-PAGE as full-length antibodies and in the heavy chain, but have double light chain bands. The bD1 and hu4D5 controls and selected bD1 variants were run on SDS-PAGE in (A) non-reducing and (B and C) reducing conditions. For panel C, the labels hu4D5+F and R2J+F indicate the addition of the deglycosylating enzyme PNGase F to the protein. The arrow in panel C indicates the band corresponding to PNGase F (36 kDa).
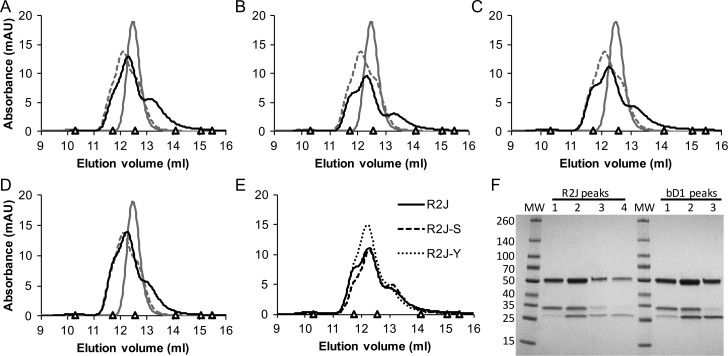
Each purified antibody was loaded on an Åkta Pure (GE Healthcare) fast protein liquid chromatography system (FPLC) and analyzed by size exclusion chromatography (SEC) over a Superdex 200 Increase 10/300 GL column (GE Healthcare) (Fig. 6A–E). In each case, PBS was used as the running buffer, and 150 μg of purified antibody in 200 μl was loaded onto a 100 μl loop to ensure consistent injection of 75 μg of antibody over the column per run. Fractions of bD1 and R2J eluting at 11.5–12 ml, 12–12.5 ml, 12.5–13 ml and 13–13.5 ml were concentrated using centrifugal filter units (Amicon), reduced and run on SDS-PAGE (Fig. 6F) and evaluated by HER2 enzyme-linked immunosorbent assay (ELISA; Supplementary Fig. 3) as described.
Fig. 6.
Full-length antibody variants were compared to bD1 and hu4D5 by SEC. Equal masses (75 μg) of purified, full-length hu4D5 (gray, solid), bD1 (gray, dashed) and variant (black, solid) antibodies were analyzed by SEC using a Superdex 200 column on an Åkta Pure FPLC. The variants shown are (A) R2D, (B) R2H, (C) R2J and (D) R2S. In panel (E), SEC profiles for R2J (black, solid), R2J-S (black, dashed) and R2J-Y (black, dotted) were compared. Standards in each SEC plot are indicated by a triangle and include: ferritin (10.3 ml, 440 kDa), beta amylase (11.7 ml, 200 kDa), aldolase (12.6 ml, 158 kDa), conalbumin (14.1 ml, 75 kDa), ovalbumin (15.1 ml, 44 kDa) and carbonic Anhydrase (16.5 ml, 29 kDa). (F) The eluted protein from each of the four R2J peak fractions (Peak 1, 11.5–12.0 ml; Peak 2, 12.0–12.5 ml; Peak 3, 12.5–13.0 ml and Peak 4, 13.0–13.5 ml) and three bD1 peak fractions (Peak 1, 11.5–12.0 ml; Peak 2, 12.0–12.5 ml and Peak 3, 12.5–13.0) were precipitated and analyzed by SDS-PAGE for comparison to molecular weight standards.
To evaluate the effect of the selected residue changes on stability, the purified antibodies were evaluated using the Protein Thermal Shift Dye Kit (Applied Biosystems) according to the kit instructions. Each antibody was evaluated in triplicate at 200 μg/ml (Table 1 and Supplementary Fig. 4).
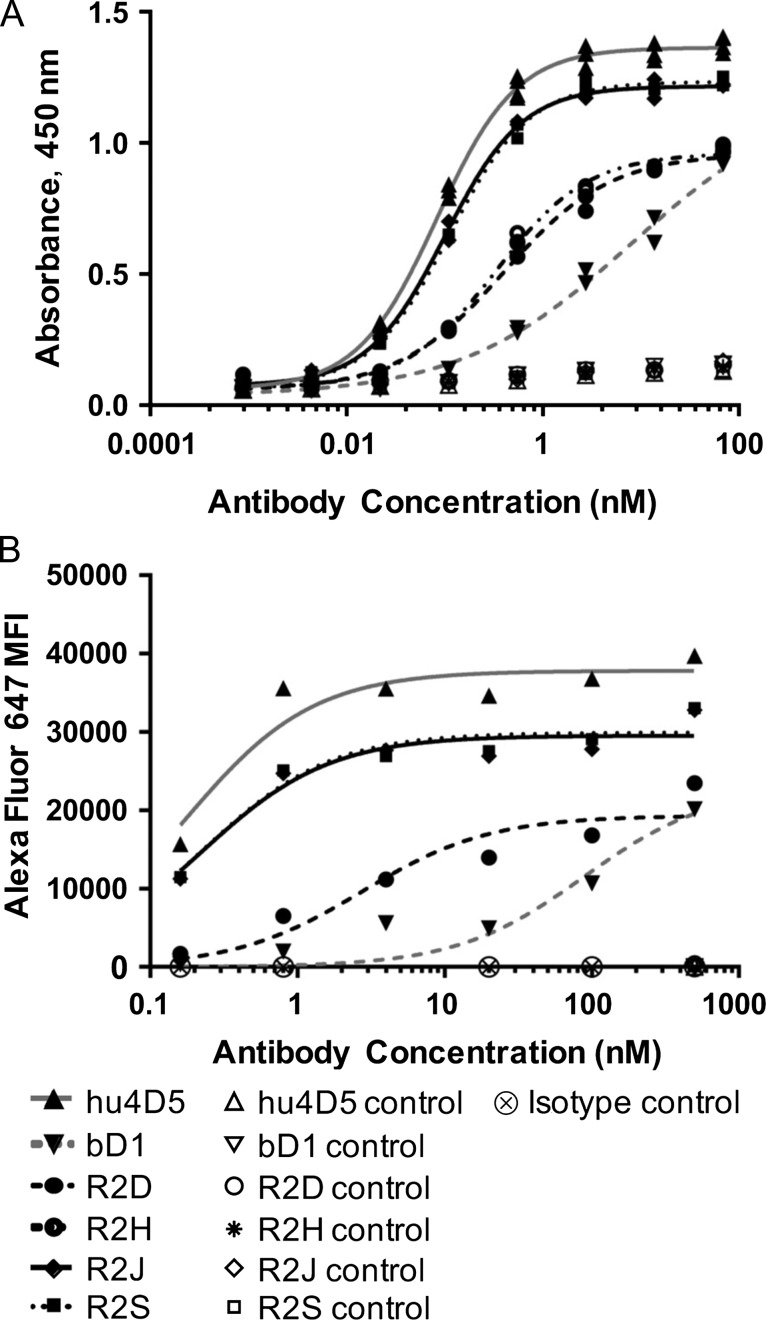
Binding of the antibodies to HER2-Fc was evaluated using ELISA with a 0.2-μg/ml HER2-Fc coat and goat–anti-human IgΚ-HRP (Southern Biotech) secondary antibody. PBS with 0.05% Tween-20 and 5% dry milk was used for blocking and all dilutions. Washes were performed with PBS with 0.05% Tween-20. ELISAs were developed and analyzed with a TMB Substrate Kit (Thermo Scientific) according to the kit instructions. The EC50 was determined using a four-parameter logistic regression, and the ranges of duplicate EC50s are reported (Fig. 7A and Table 1).
Fig. 7.
Full-length variants of bD1 have improved binding to both the soluble and membrane-bound form of HER2. (A) Each purified antibody was analyzed by ELISA to determine relative binding to soluble HER2-Fc or milk-coated (control) wells. (B) SK-OV-3 cells were incubated with purified antibody and stained with goat-anti-Fc-Alexa Fluor 647, then analyzed by flow cytometry. Control samples were antibody incubated with CHO-K1 cells and SK-OV-3 cells incubated with an irrelevant antibody isotype control.
The SK-OV-3 (ATCC) ovarian cancer cell line expresses HER2 at ~106 molecules per cell (Onsum et al., 2013). To determine effective Kd, equilibrium binding of purified antibody to SK-OV-3 cells was tested using goat–anti-human Fc-Alexa Fluor 647 secondary antibody and fit according to the protocol reported by Feldhaus and Siegel (Feldhaus and Siegel, 2004). Antibody concentrations ranged from 0.8 to 500 nM and staining volumes were adjusted to avoid ligand depletion at low concentrations. The geometric mean of the goat–anti-human Fc-Alexa Fluor 647 fluorescence (MFI) of the forward/side scatter gated cells was fit to a one site specific binding equation using GraphPad Prism 7.03 (Fig. 7B). Values reported are the Kd, standard error and R2 of the fit (Table 1).
Results
Developing the CHO display system
The Fab portion of the anti-HER2 antibody hu4D5 was cloned into the pPyEBV vector for semi-stable plasmid maintenance and display in CHO-T cells. Acyte Biotech’s CHO-T cell line contains the Polyoma large T antigen coding sequence stably integrated into the CHO cell genome. This cell line combined with additional Epstein–Barr and Polyoma viral components and a Hygromycin B resistance gene encoded on the pPyEBV vector (Kunaparaju et al., 2005) allowed plasmid maintenance and Fab expression for ~8 weeks. The expression cassette was designed with a 2 A peptide between the light chain and Fab portion of the heavy chain to allow for Fab expression from a single transcript (Fig. 1A and Supplementary Fig. 1). To anchor the Fab to the cell surface, DNA encoding a short glycine–serine linker and the PDGFR transmembrane region was appended to the 3′ end of the CH1 sequence. Expression of this construct in CHO-T cells resulted in detectable binding of soluble HER2-Fc (R&D Systems) to the cell surface by confocal microscopy (Fig. 1B–D). The Fab was also robustly displayed with the PDGFR transmembrane region appended to both the CH1 and CL regions simultaneously (data not shown), but efforts to use the native human IgG1 transmembrane region resulted in minimal display on CHO-T cells (Supplementary Fig. 5). Prior reports have employed the PDGFR (Beerli et al., 2008; Zhou et al., 2010) and IgG1 (Bowers et al., 2014) transmembrane domains (TM) to mediate full-length antibody display on mammalian cells. We evaluated both approaches, enjoying immediate success with the PDGFR domain and thus moved forward with this design.
For use in library screening, it was important that each transfected CHO-T cell carry a single antibody expressing plasmid. Lipofection can result in hundreds to thousands of plasmids entering some cell types (Cohen et al., 2009) but the average number of functional plasmids introduced into this CHO-T system with a large, 12.3 kbp plasmid was unknown. The hu4D5 Fab displaying plasmid, pPy4D5, was mixed with a green fluorescent protein expressing plasmid, pPyEGFP, at various ratios and transfected into CHO-T cells. The percentage of cells expressing both detectable hu4D5 antibody and EGFP were evaluated (Fig. 2A–F). A predictive model based on an expected Poisson distribution of plasmids per cell was fit to the data, providing an estimated average number of plasmids per cell of 2.1. Based on this information, a mixture of 74% blank carrier plasmid without a Hygromycin B resistance gene to 26% antibody expressing plasmid was predicted to result in an approximate average of one plasmid per cell. Accordingly, this ratio was used in all subsequent experiments. Two days following transfection, ~17% of cells expressed antibody, with 75% of those expected to contain a single plasmid. Growth in selective media containing 300 μg/ml Hygromycin B supported expansion of cells containing Fab-encoding plasmids, while selected against the 83% of cells that either did not internalize plasmid or internalized only blank carrier plasmid.
Another important consideration for library screening was to minimize growth bias by modulating the Fab display level. While CMV promoter-based plasmids with optimal Kozak sequences provide robust transcription and expression of soluble proteins, high expression levels may be unnecessary or even detrimental for display systems in which the cell’s surface area limits the number of molecules that may be simultaneously displayed. Lower display levels may also reduce avidity effects, making it easier to discriminate between clones with similar affinities.
To reduce the translational load, characterized modifications to the Kozak sequence were introduced to pPy4D5disp and evaluated to determine which expression-reducing modifications provided robust display. Five modifications were chosen from the ~50 tested in Ferreira et al. (2013) that ranged from 0% to 80% expression of green fluorescent protein in their system. These were cloned into pPy4D5disp, creating pPy4D5disp0, 1, 2, 3 and 4 (Supplementary Table 1). While completely disrupting the Kozak sequence, as in pPy4D5disp0, resulted into no display and pPy4D5disp1 and three had reduced display, the two other variants had equivalent mean fluorescence and similar percent fluorescent values as unmodified pPy4D5disp 2 days after transfection (Fig. 2G and H). Further development used pPy4D5disp2 as it had the lowest predicted expression level with an equivalent detected display as unmodified pPy4D5disp. In our hands, pPy4D5disp2 expressing cells exhibited more consistent growth during Hygromycin B selection and appeared qualitatively healthier during antibiotic selection than those transformed with pPy4D5disp.
bD1 library creation and screening
In order to optimize the CHO surface display screening method, a low affinity hu4D5 variant, bD1, was targeted for affinity maturation. Compared to its parent hu4D5, which has an estimated Kd of 0.1 nM, the 10 mutations introduced into the bD1 VL region were reported to reduce affinity 2000-fold (Bostrom et al., 2009). Display of bD1 indicated a similar Fab expression level and reduced HER2 binding as compared to hu4D5 by flow cytometry (Fig. 3A–C).
We hypothesized that bD1 could be improved to an affinity nearer its hu4D5 parent, and accordingly generated a VH CDR library to avoid recovery of the original hu4D5. Analysis of the hu4D5–HER2 complex crystal structure revealed several CDRH2 and CDRH3 residues in close proximity to HER2 and the bD1 light chain mutations. A list of candidate residues likely amenable to mutagenesis was compiled by comparison to similar CDRs found in a GenBank blast search, resulting in a designed bD1 VH library with degeneracies in nine CDR codons. These sequences were assembled and cloned into pPybD1disp in E. coli to create a library of ~1.0 × 106 unique transformants, which was midi-prepped and transfected into CHO cells to yield 107 transfectants.
The CHO library was subjected to two rounds of FACS (Fig. 3D–F), with selection based on Fab display level, as indicated by goat–anti-human IgΚ-FITC staining, and improved HER2-Fc affinity, as indicated binding by goat–anti-human Fc-Alexa Fluor 647 secondary staining. The cells were pre-gated to exclude goat–anti-rabbit IgG (H + L)-Cy3 binding cells as a marker of non-specific binding directly to the goat secondary antibody. The library was oversampled by 10-fold during sorting and ~105 cells (1–2% of total cells) were sorted per round and allowed to grow in selective media for 1 week before repeated sorting and/or cell collection. After growth, 106 cells from each sort were collected and total DNA was purified, amplified by PCR, sub-cloned back into pPybD1disp, and sequenced to monitor Fab sequence diversity. The total time from initial transfection to isolation of Round 2 DNA was 1 month.
Variant characterization
The VH region was amplified from the purified post-Round 2 DNA and cloned back into pPybD1disp for individual clone analysis. Because the pPybD1disp population and Round 2 population overlap, 9 of 20 sequenced clones contained unmodified pPybD1disp background. This was somewhat anticipated because the digested pPybD1disp vector was used as the vector backbone for cloning of the library, so the incidence of this sequence in the unsorted library was significantly higher than any variant (~3% vs 0.0001%). This frequency could be reduced by various methods including using a cloning vector with completely non-functional variable regions. Despite the presence of unmodified background, enrichment of improved variants was rapid. Further purification of the improved population from background would require an additional week of growth and sorting which could be associated with reduced diversity, therefore we characterized clones selected after two rounds of sorting.
Of the 11 of 20 post-Round 2 sequences that were not pPybD1disp, four unique clones (R2D, R2H, R2J and R2S) exhibited improved HER2 binding to display level fluorescence ratio as single clones in the display format (Fig. 4). Accordingly, these were transferred to vectors for transient expression of soluble full-length human IgG1. Three of these four variants contained a cysteine at the 100c position, creating a free cysteine in CDRH3. To further evaluate the effect of this free cysteine, it was replaced in R2J with a serine (R2J-S) or a tyrosine (R2J-Y). Antibody sequences were transfected and expressed at larger scale (several T-150 flasks) for purification as well as side-by-side at small scale for comparison of expression levels (Table 1). Free cysteines are predicted to be problematic in prokaryotic expression systems used in phage display and are generally avoided in library design. To evaluate the effect of the cysteine at position 100c on bacterial expression, hu4D5 and R2J were expressed as soluble scFvs appended with a hexa-histidine tag in E. coli, purified by immobilized metal affinity chromatography, and analyzed by reducing and non-reducing SDS-PAGE. The R2J variant was expressed at much lower levels with noticeable covalent and non-covalent oligomers as compared to hu4D5 in E. coli (Supplementary Fig. 2). This impaired production and folding suggest that clones with a free cysteine at 100c are unlikely to have been selected from a prokaryotic library.
The antibody variants were >90% pure, as monitored by reducing and non-reducing SDS-PAGE (Fig. 5A and B). Hu4D5 showed typical bands at ~25 and ~50 kDa in reducing SDS-PAGE, but bD1 and the variants, all of which contain the identical bD1 light chain, showed a doublet near the expected light chain molecular weight. This was also true for the R2J cysteine variants (Fig. 5A and B). After PNGase F digestion, the larger bD1 light chain band collapsed into the smaller band, which migrated at the same size as the hu4D5 light chain. This suggests that partial N-linked glycosylation was responsible for the larger light chain band (Fig. 5C). Inspection of the bD1 light chain sequence revealed an NVS motif in bD1 created by the D28N substitution relative to hu4D5. This sequence matches the NXS/T consensus sequence for N-linked glycosylation, and is a computationally predicted N-linked glycosylation site (Blom et al., 2004) (http://www.cbs.dtu.dk/services/NetNGlyc/).
SEC analysis of the purified antibodies gave no indication of large aggregates despite the free cysteine in clones R2H, R2J and R2S (Fig. 6A–D). The parent bD1 and all selected variants, including the R2J-S and R2J-Y clones lacking the free cysteine (Fig. 6E), resolved into three overlapping peaks near the elution volume corresponding to hu4D5 monomer. SDS-PAGE analysis of the four peak fractions from R2J and three peak fractions from bD1 showed that the peak eluting at the smallest volume contains only glycosylated light chain, the next peak contains a mix of glycosylated and non-glycosylated light chain and the peak eluting at the largest volume contains only non-glycosylated light chain (Fig. 6F). When each of these fractions was tested for binding to HER2-Fc by ELISA, the doubly glycosylated bD1 light chain peak exhibited reduced binding (as measured by EC50 and maximum absorbance) relative to the other peaks, consistent with the interpretation that light chain glycosylation impedes HER2 binding. In contrast, all R2J peaks exhibit similar HER2-Fc binding, supporting the idea that the selected heavy chain mutations abrogated the negative effects of light chain glycosylation (Supplementary Fig. 3).
The selected variants all possess high thermostability. Measurement of the thermal melting curve showed that the first transition occurred consistently around 71°C for all purified antibodies, corresponding to the expected human IgG1 CH2 melting temperature (Ionescu et al., 2008). The second transition, which typically reflects CH3 and Fab stability, was ~85°C for hu4D5, bD1, R2D and R2J-S. All of the variants with free cysteines had second transition temperatures that were increased by one degree to 86°C, while R2J-Y had a substantially increased second transition temperature at 87.4°C (Table 1).
The effective HER2 binding affinities of the variant antibodies were compared to bD1 and hu4D5 by two complimentary methods. First, a HER2-Fc ELISA showed that all variants bound immobilized HER2-Fc with EC50 values improved over bD1 and approaching that of hu4D5 for variants R2J, R2S, R2J-S and R2J-Y (Fig. 7A and Table 1). None of the antibodies bound control wells only blocked with milk. In an experiment more reflective of the in vivo binding of a therapeutic antibody, SK-OV-3 ovarian cancer cells that over express the HER2 receptor on their surface were cultured, incubated with purified bD1 variants in the IgG format in concentrations from 500 nM to 0.8 nM, and analyzed by flow cytometry (Fig. 7B). These results were consistent with ELISA and showed similar effective Kd values for hu4D5 and variants R2J and R2S, which represented a 400-fold improvement over bD1 in this assay. Clone R2D exhibited a 30-fold improved effective affinity over bD1 (Table 1). No binding of these antibodies to CHO cells or of an isotype control antibody to SK-OV-3 cells was detected (Fig. 7B).
Discussion
The display and screening of a bD1 library on the surface of CHO cells identified several antibodies with improved effective affinities and favorable biophysical properties indicating their suitability for manufacturing. The library of 106 unique variants was introduced to CHO-T cells by transient transfection under conditions optimized for library screening. After antibiotic selection of transfected cells, the library was sorted twice, allowing 1 week for cell recovery in between sorts. Of the resulting improved clones, two of the characterized variants, R2J and R2S, were improved by 400-fold over the bD1 parent in a flow cytometry-based equilibrium binding assay with effective Kd values of 0.22 nM. Two other variants, R2D and R2H, each showed a >10-fold improvement in HER2 binding by ELISA and 30-fold improvement of R2D by flow cytometry. The variant antibodies all expressed at levels similar to hu4D5 and bD1 with similarly low aggregation and equivalent or better thermostabilities.
Interestingly, three of the four variants contained a free cysteine in the CDRH3 position 100 c. A cysteine is also present at this position in OKT3 (Muromonab-CD3) (Arakawa et al., 1996), a mouse anti-CD3 antibody developed to treat transplant rejection and the first FDA approved mAb. In both the OKT3 (Kjer-Nielsen et al., 2004) and hu4D5 crystal structures, this residue is buried between the heavy and light chains. It may be that a cysteine in this position provides additional stability by optimal packing of the protein core since it does not form a disulfide bond in OKT3. While free cysteines are typically avoided in antibody CDR libraries, especially in phage and bacterial systems where they often result in improper disulfide bond formation, they are present in naturally occurring antibody repertoires (Zemlin et al., 2003). The R2J-S and R2J-Y variants, in which the free cysteine in R2J was replaced by serine or tyrosine, respectively, exhibited similar stabilities and HER2 binding properties as R2J, indicating that cysteine may not be the uniquely optimal amino acid at this position, but also demonstrating that the presence of a free cysteine in this position is not deleterious. When R2J was expressed as a soluble scFv in E.coli, the expression level was low and a large fraction of the protein appeared in a dimer band that was reduced to monomer upon heating and treatment with β-mercaptoethanol (Supplementary Fig. 2). The free cysteine mutation is not well-tolerated by bacterial expression and clones containing the mutation would likely not be enriched in a phage display format. The appearance of this cysteine in CHO display affinity-selected clones indicates that screening in a mammalian expression system expands the allowable sequence repertoire to be more representative of naturally occurring sequences compared to other expression systems.
Commercial and therapeutic antibody development processes typically exclude sequences containing free cysteines due to potential folding and heterogeneity issues. We neither observe any evidence of product heterogeneity nor misfolding in R2J vs R2J-S nor R2J-Y after expression in CHO cells, but we neither test for long-term stability nor cysteine adducts that may occur. It is known that free thiol groups may become modified by glutathionylation in antibodies engineered to have solvent exposed free cysteines such as THIOMABs (Chen et al., 2009). In the case of OKT3, though, no such modifications appeared in the crystal structure, indicating that these modifications are possibly less likely in the buried location the free cysteine is found in OKT3 and our hu4D5 variants. In fact, statistical analysis of free cysteines in protein crystal structures has lead researchers to consider it a hydrophobic residue when buried in the protein core (Nagano et al., 1999). Though antibody development pipelines are not likely to incorporate free cysteines in new antibodies from the perspective of minimizing risk, they are naturally occurring and it is not clear that they are always problematic. Library design can preclude introducing cysteines, avoiding the issue entirely in any display format.
The partial glycosylation of the bD1 light chain was an unexpected but interesting characteristic of bD1 that was uncovered in this work. The bD1 antibody was a hu4D5 variant discovered using phage display and expressed as a full-length IgG in HEK cells but there is no mention of glycosylation of the bD1 light chain in the original publication (Bostrom et al., 2009). This work aimed to create dual binding specificity within a single antibody binding site by maintaining heavy chain binding to HER2 and screening the light chain for variants with affinity for a second antigen. Their efforts to simultaneously target HER2 and the vascular epidermal growth factor were successful and they obtained Kd values of the variant bH1 to these antigens of 26 nM and 300 nM, respectively. However, in the case of bD1, the antibody was intended to target HER2 and death receptor 5 (DR5) simultaneously, but the full-length antibody exhibited a reported 2000-fold decrease in HER2 affinity relative to hu4D5 and only 14 μM Kd for DR5, as measured by surface plasmon resonance. The two reported monospecific antibodies targeting DR5 alone through light chain mutagenesis did not include an NXS/T N-linked glycosylation consensus sequence, and had DR5 affinities of 120 nM and 150 nM. The reason for poor HER2 and DR5 binding in bD1 compared to other variants is not clear, but screening in a phage format could have masked glycosylation and other post-translational modification sites that become relevant during mammalian expression.
Our results for bD1 binding to HER2-Fc in ELISA showed that the bD1 SEC fractions containing predominantly doubly glycosylated light chain had reduced binding relative to fractions with single or no light chain glycosylation, while R2J had equivalent binding with regardless of the level of light chain glycosylation (Supplementary Fig. 3). We were able to recover strong HER2 binding affinity in the presence of these post-translational modifications, exemplifying the benefit of screening in the same system used for downstream expression. While antibodies are unlikely to benefit from introduced glycosylation sites in the CDR region, CHO display accounts for these modifications during the initial antibody discovery step, as opposed discovering and excluding these clones later in development. Together, the unexpected glycosylation site uncovered when bD1 was expressed in mammalian cells and the enrichment of free cysteine containing sequences from the mammalian display library highlight the two main problems with screening for antibodies in hosts not used for production: (1) other systems may select for sequences that have decreased function after mammalian processing (as in the glycosylation site in bD1) and (2) other systems may miss perfectly functional sequences because they cannot process them properly (as in the free cysteines in R2J and R2S).
A growing body of literature describes diverse approaches to screening antibody libraries on the surface of mammalian cells. For example, research articles from AnaptysBio, Inc. over the previous 6 years have detailed libraries comprised of full-length human antibodies derived from pooled human repertoires expressed on the HEK293 cell surface. After in vitro somatic hypermutation and screening, affinity matured variants with sub-nanomolar affinities were identified (Bowers et al., 2011, 2013, 2014; McConnell et al., 2012; Horlick et al., 2013; King et al., 2014). The papers indicate that library screening in an episomal system involved 5–12 rounds of selection. This very long process can be attributed to the presence of only very weak binders in the naïve library and the lack of a crystal structure to guide library design, but may also have been impacted by avidity effects from the bivalent displayed IgG, separate heavy and light chain episomes requiring later deconvolution, and reliance on activation-induced cytidine deaminase (AID) for diversification of low affinity binders. While AID expression results in somatic hypermutation effects similar to those occurring during B-cell maturation, the rate of mutation is typically 10-fold lower than in vivo (Martin and Scharff, 2002). In addition, the expression host, HEK293, is less commonly used in therapeutic protein manufacturing processes and results in lower average antibody yields than CHO cells (Frenzel et al., 2013).
Another corporate venture into mammalian display, HuTARG™ (Innovative Targeting Solutions, Inc.), is less well described in the literature and highly proprietary, but seems to rely on induced RAG1, RAG2 and recombinant signal sequences much like the process used in native B-cell antibody sequence recombination. It is not clear what cell type is used for antibody display, how long the library generation process takes or how flexible the platform may be for mutagenesis strategies apart from antibody CDR recombination. While large libraries can be created using this method of V(D)J recombination of human germline sequences, the type of diversity, and more importantly, the ability to directly target residues of interest is limited relative to recombinant methods. While this method may be attractive for de novo discovery of new antibodies, it is not suitable for engineering an existing antibody through targeted or random mutagenesis, such as the project described here.
Other groups have used mammalian surface display for engineering antibody-like molecules, but these methods have been limited by poor scaffold choices such as scFvs (Ho et al., 2006; Schenk et al., 2007; Beerli et al., 2008), which often do not reflect full-length antibody behavior (Nieba et al., 1997; Hoogenboom, 2005; Bradbury et al., 2011), or full-length IgGs (Zhou et al., 2010; Li et al., 2012; Tomimatsu et al., 2013), which may complicate screening due to avidity effects. On a cell surface, numerous displayed full-length antibodies with two binding sites per molecule create a membrane saturated with binding sites in close proximity and resulting in antigen rebinding even at lower monovalent affinity levels. Thus, without careful antigen titration, it can be challenging to detect all but large differences in antibody binding affinity by flow cytometry, especially given the heterogeneity of mammalian cell populations. In contrast, engineering projects often rely on identifying a series of antibody variants, each of which represents a modest affinity increase relative to the prior clone. Our scaffold choice, the Fab, retains a structure more similar to full-length IgGs than scFvs while avoiding the bivalent nature of IgG binding, minimizing these avidity effects as long as surface density is controlled. Existing technologies also rely on genomic integration (Beerli et al., 2008; Zhou et al., 2010; Tomimatsu et al., 2013) or unstable transient expression (Ho et al., 2006; Schenk et al., 2007) for mammalian cell expression, which introduces various obstacles including the requirement for genome editing expertize, introducing considerable screening complications and/or raising biosafety concerns.
Challenges for mammalian display compared to well-established phage, bacterial and yeast systems include the difficulty of precisely controlling gene copy number to ensure a single, retrievable DNA sequence is responsible for the phenotype observed and the inability to modulate expression level to prevent growth inhibition and cell death. Published mammalian display technologies used genomic integration in an attempt to tightly control copy number or accepted multiple plasmids per cell as unavoidable and screened various heavy–light chain combinations to tease out relevant pairs. By simply determining the average number of plasmids introduced per cell and using blank carrier DNA as diluent, we largely avoided this problem. Combined with the Epi-CHO system, this approach allowed for antibiotic selection and maintenance of only those cells containing antibody encoding sequences. While strategies for mammalian transcriptional control are not highly developed for recombinant gene expression, translation modulation through Kozak sequence modifications and the addition of small open reading frames upstream of the gene of interest helped regulate expression in these experiments. Further development of mammalian plasmids and CHO cell lines that are engineered to maintain a single plasmid or contain robust inducible expression systems would be helpful for this approach but are not required.
Because mammalian cells are large and heterogeneous compared to other cell types, mammalian flow cytometry populations are often characterized by relatively high coefficients of variation in fluorescence. This phenomenon resulted in overlapping populations even for the large affinity differences between hu4D5 and bD1 (Fig. 3B and C). Thus, sorting of the bD1 library required sort gates that also encompassed the parent bD1 population. While this overlap and the bD1 cloning background resulted in 9 of the 20 sequenced clones being identified as the parental bD1 clone, the importance of collecting this population was evident in the substantial effective affinity increase seen in R2D and R2H compared to bD1 with a slight change in fluorescence profile by flow cytometry (Fig. 4C and D and Table 1). The challenges in discriminating variants with similar affinities could be exacerbated in highly avid systems due to surface crowding or multivalent antibody display formats.
Another consideration for mammalian libraries is the large culture volumes required for mammalian cell growth. The CHO display platform described here is limited by convenience to a size of ~106 library members. While this is a small fraction of the library screening capability of other systems and is not suitable for screening completely naïve libraries, the possible diversity is sufficient for many antibody engineering applications. The technology is well-suited for adjusting or enhancing existing binding or optimizing binding for unique environments.
CHO display could also displace individual colony screening approaches for testing clones after screening a naïve phage or yeast library. After an initial screen to reduce the library size in another system, an enriched population could be transferred to the CHO display format to thoroughly screen for desired binding and manufacturing suitability. While mammalian cell growth rates result in a comparatively slow sorting process (~1 month here), considerably less time is required to improve prospective antibodies for manufacturing.
Many groups still use animal immunization to discover antibody candidates. B-cells can then be harvested for the slow process of hybridoma creation, or VL and VH genes may be randomly combined to create a scFv phage library and screened to identify combinations recapitulating the original antigen binding capability. The rapid polarization and amplification of antigen binding B-cell populations after immunization as described by Reddy et al. (2010) may allow for much smaller mammalian display libraries to isolate highly functional antibodies without potential losses due to the poor behavior of some clones on phage. CHO display libraries are also well-suited for screening humanization libraries after animal immunization as the system allows for screening on the basis of retained binding, CHO expression, and ‘human-like’ staining profiles.
Finally, the CHO display format described here is not limited to antibody libraries, but can easily be adapted for the display, mutagenesis, and screening of other biomolecules. Extracellular T-cell receptor (TCR) domains, which are notoriously challenging to express in heterologous hosts (Maynard et al., 2005), are easily displayed in this format and can be screened for increased peptide-MHC affinity (unpublished data). By comparison, yeast display of TCRs often requires screening a single-chain TCR library to identify variants with increased solubility and stability that allow for display of properly folded proteins before even beginning affinity maturation (Kieke et al., 1999). Many other molecules best produced in mammalian cells may be engineered with the CHO display system, including endogenous receptors and engineered proteins such as chimeric antigen receptors.
Supplementary data
Supplementary data are available at Protein Engineering, Design and Selection online.
Funding
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health [F32GM111018 to A.W.N.] and the Welch Foundation [F-1767 to J.A.M.]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Supplementary Material
Acknowledgements
The authors would like to thank Luciano Posada for assistance with cloning pPyEGFP and hu4D5 expression variants.
References
- Arakawa F., Kuroki M., Kuwahara M., Senba T., Ozaki H., Matsuoka Y., Misumi Y., Kanda H. and Watanabe T. (1996) J. Biochem., 120, 657–662. [DOI] [PubMed] [Google Scholar]
- Beerli R.R., Bauer M., Buser R.B., Gwerder M., Muntwiler S., Maurer P., Saudan P. and Bachmann M.F. (2008) Proc. Natl. Acad. Sci. U. S. A., 105, 14336–14341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessette P.H., Mena M.A., Nguyen A.W. and Daugherty P.S. (2003) Methods Mol. Biol., 231, 29–37. [DOI] [PubMed] [Google Scholar]
- Blom N., Sicheritz-Ponten T., Gupta R., Gammeltoft S. and Brunak S. (2004) Proteomics, 4, 1633–1649. [DOI] [PubMed] [Google Scholar]
- Bostrom J., Yu S.F., Kan D., Appleton B.A., Lee C.V., Billeci K., Man W., Peale F., Ross S., Wiesmann C. et al. (2009) Science, 323, 1610–1614. [DOI] [PubMed] [Google Scholar]
- Bowers P.M., Horlick R.A., Kehry M.R., Neben T.Y., Tomlinson G.L., Altobell L., Zhang X., Macomber J.L., Krapf I.P., Wu B.F. et al. (2014) Methods, 65, 44–56. [DOI] [PubMed] [Google Scholar]
- Bowers P.M., Horlick R.A., Neben T.Y., Toobian R.M., Tomlinson G.L., Dalton J.L., Jones H.A., Chen A., Altobell L. 3rd, Zhang X. et al. (2011) Proc. Natl. Acad. Sci. U. S. A., 108, 20455–20460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers P.M., Neben T.Y., Tomlinson G.L., Dalton J.L., Altobell L., Zhang X., Macomber J.L., Wu B.F., Toobian R.M., McConnell A.D. et al. (2013) J. Biol. Chem., 288, 7688–7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury A.R., Sidhu S., Dubel S. and McCafferty J. (2011) Nat. Biotechnol., 29, 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.N., Nguyen M., Jacobson F. and Ouyang J. (2009) MAbs, 1, 563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H.S., Mason K., Ramyar K.X., Stanley A.M., Gabelli S.B., Denney D.W. Jr. and Leahy D.J. (2003) Nature, 421, 756–760. [DOI] [PubMed] [Google Scholar]
- Chon J.H. and Zarbis-Papastoitsis G. (2011) N Biotechnol, 28, 458–463. [DOI] [PubMed] [Google Scholar]
- Cohen R.N., van der Aa M.A., Macaraeg N., Lee A.P. and Szoka F.C. Jr. (2009) J Control Release, 135, 166–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker D.M., Jones S.D. and Levine H.L. (2015) MAbs, 7, 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldhaus M. and Siegel R. (2004) Methods Mol. Biol., 263, 311–332. [DOI] [PubMed] [Google Scholar]
- Ferreira J.P., Overton K.W. and Wang C.L. (2013) Proc. Natl. Acad. Sci. U. S. A., 110, 11284–11289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenzel A., Hust M. and Schirrmann T. (2013) Front Immunol, 4, 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronwald R.G., Grant F.J., Haldeman B.A., Hart C.E., O’Hara P.J., Hagen F.S., Ross R., Bowen-Pope D.F. and Murray M.J. (1988) Proc. Natl. Acad. Sci. U. S. A., 85, 3435–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho M., Nagata S. and Pastan I. (2006) Proc. Natl. Acad. Sci. U. S. A., 103, 9637–9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenboom H.R. (2005) Nat. Biotechnol., 23, 1105–1116. [DOI] [PubMed] [Google Scholar]
- Horlick R.A., Macomber J.L., Bowers P.M., Neben T.Y., Tomlinson G.L., Krapf I.P., Dalton J.L., Verdino P. and King D.J. (2013) J. Biol. Chem., 288, 19861–19869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionescu R.M., Vlasak J., Price C. and Kirchmeier M. (2008) J. Pharm. Sci., 97, 1414–1426. [DOI] [PubMed] [Google Scholar]
- Kieke M.C., Shusta E.V., Boder E.T., Teyton L., Wittrup K.D. and Kranz D.M. (1999) Proc. Natl. Acad. Sci. U. S. A., 96, 5651–5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King D.J., Bowers P.M., Kehry M.R. and Horlick R.A. (2014) Curr Drug Discov Technol, 11, 56–64. [DOI] [PubMed] [Google Scholar]
- Kjer-Nielsen L., Dunstone M.A., Kostenko L., Ely L.K., Beddoe T., Mifsud N.A., Purcell A.W., Brooks A.G., McCluskey J. and Rossjohn J. (2004) Proc. Natl. Acad. Sci. U. S. A., 101, 7675–7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunaparaju R., Liao M. and Sunstrom N.A. (2005) Biotechnol. Bioeng., 91, 670–677. [DOI] [PubMed] [Google Scholar]
- Li C.Z., Liang Z.K., Chen Z.R., Lou H.B., Zhou Y., Zhang Z.H., Yu F., Liu S., Zhou Y., Wu S. et al. (2012) Cell Mol Immunol, 9, 184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A. and Scharff M.D. (2002) Proc. Natl. Acad. Sci. U. S. A., 99, 12304–12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard J., Adams E.J., Krogsgaard M., Petersson K., Liu C.W. and Garcia K.C. (2005) J. Immunol. Methods, 306, 51–67. [DOI] [PubMed] [Google Scholar]
- McCafferty J., Griffiths A.D., Winter G. and Chiswell D.J. (1990) Nature, 348, 552–554. [DOI] [PubMed] [Google Scholar]
- McConnell A.D., Do M., Neben T.Y., Spasojevic V., MacLaren J., Chen A.P., Altobell L. 3rd, Macomber J.L., Berkebile A.D., Horlick R.A. et al. (2012) PLoS One, 7, e49458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano N., Ota M. and Nishikawa K. (1999) FEBS Lett., 458, 69–71. [DOI] [PubMed] [Google Scholar]
- Nguyen A.W., Wagner E.K., Laber J.R., Goodfield L.L., Smallridge W.E., Harvill E.T., Papin J.F., Wolf R.F., Padlan E.A., Bristol A. et al. (2015) Sci. Transl. Med., 7, 316ra195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M. and Lee A.S. (2007) FEBS Lett., 581, 3641–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieba L., Honegger A., Krebber C. and Pluckthun A. (1997) Protein Eng., 10, 435–444. [DOI] [PubMed] [Google Scholar]
- Onsum M.D., Geretti E., Paragas V., Kudla A.J., Moulis S.P., Luus L., Wickham T.J., McDonagh C.F., Macbeath G. and Hendriks B.S. (2013) Am. J. Pathol., 183, 1446–1460. [DOI] [PubMed] [Google Scholar]
- Reddy S.T., Ge X., Miklos A.E., Hughes R.A., Kang S.H., Hoi K.H., Chrysostomou C., Hunicke-Smith S.P., Iverson B.L., Tucker P.W. et al. (2010) Nat. Biotechnol., 28, 965–969. [DOI] [PubMed] [Google Scholar]
- Schenk J.A., Sellrie F., Bottger V., Menning A., Stocklein W.F. and Micheel B. (2007) Biochimie, 89, 1304–1311. [DOI] [PubMed] [Google Scholar]
- Smith K., Garman L., Wrammert J., Zheng N.Y., Capra J.D., Ahmed R. and Wilson P.C. (2009) Nat. Protoc., 4, 372–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmer W.P., Crameri A., Ha K.D., Brennan T.M. and Heyneker H.L. (1995) Gene, 164, 49–53. [DOI] [PubMed] [Google Scholar]
- Szymczak-Workman A.L., Vignali K.M. and Vignali D.A. (2012) Cold Spring Harb Protoc, 2012, 199–204. [DOI] [PubMed] [Google Scholar]
- Tabrizi M.A., Bornstein G.G. and Klakamp S.L. (2012) Development of antibody-based therapeutics: translational considerations. Springer, New York. [Google Scholar]
- Tomimatsu K., Matsumoto S.E., Tanaka H., Yamashita M., Nakanishi H., Teruya K., Kazuno S., Kinjo T., Hamasaki T., Kusumoto K. et al. (2013) Biochem. Biophys. Res. Commun., 441, 59–64. [DOI] [PubMed] [Google Scholar]
- Weaver-Feldhaus J.M., Lou J., Coleman J.R., Siegel R.W., Marks J.D. and Feldhaus M.J. (2004) FEBS Lett., 564, 24–34. [DOI] [PubMed] [Google Scholar]
- Wildt S. and Gerngross T.U. (2005) Nat. Rev. Microbiol., 3, 119–128. [DOI] [PubMed] [Google Scholar]
- Zemlin M., Klinger M., Link J., Zemlin C., Bauer K., Engler J.A., Schroeder H.W. Jr. and Kirkham P.M. (2003) J. Mol. Biol., 334, 733–749. [DOI] [PubMed] [Google Scholar]
- Zhou C., Jacobsen F.W., Cai L., Chen Q. and Shen W.D. (2010) MAbs, 2, 508–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.