Abstract
The Norway rat, Rattus norvegicus, is one of the most important pest species globally and the main reservoir of leptospires causing human leptospirosis in the urban slums of tropical regions. Rodent control is a frequent strategy in those settings to prevent the disease but rapid growth from residual populations and immigration limit the long-term effectiveness of interventions. To characterize the breeding ecology of R. norvegicus and provide needed information for the level of genetic mixing, which can help identify inter-connected eradication units, we estimated the occurrence of multiple paternity, distances between mothers and sires, and inbreeding in rats from urban slum habitat in Salvador, Brazil. We genotyped 9 pregnant females, their 66 offspring, and 371 males at 16 microsatellite loci. Multiple paternity was observed in 22% (2/9) of the study litters. Of the 12 sires that contributed to the 9 litters, we identified 5 (42%) of those sires among our genotyped males. Related males were captured in close proximity to pregnant females (the mean inter-parent trapping distance per litter was 70 m, ±58 m SD). Levels of relatedness between mother–sire pairs were higher than expected and significantly higher than relatedness between all females and non-sire males. Our findings indicate multiple paternity is common, inbreeding is apparent, and that mother–sire dyads occur in close proximity within the study area. This information is relevant to improve the spatial definition of the eradication units that may enhance the effectiveness of rodent management programs aimed at preventing human leptospirosis. High levels of inbreeding may also be a sign that eradication efforts are successful.
Keywords: brown rat, genetics, mating behavior, microsatellite, multiple paternity, polyandry, Rattus norvegicus, urban slums
The Norway rat (Rattus norvegicus) is one of the most important pest species globally, which adversely affects agriculture productivity, ecosystem (native species and habitats), and public health (Capizzi et al. 2014). Although its geographic range spans 6 continents and many islands, low socioeconomic areas and slums within urban settings, most notably in developing countries, are at highest risk for rat-associated diseases (Himsworth et al. 2013).
A major example of rat-borne zoonoses with extensive global public health consequences is leptospirosis, with an estimated annual incidence of approximately 1 million people (Costa et al. 2015a; Torgerson et al. 2015). However, this figure is a gross underestimate as exemplified by studies of the etiology of acute fevers of unknown origin (Crump et al. 2013; Susilawati and McBride 2014). Long-term studies on human leptospirosis and risk factors for infection associated with rats in Salvador, Brazil, have established a unique opportunity for fine-scale studies of rat populations, genetic structure, the prevalence of leptospiral carriage, and their contribution to environmental contamination through shedding of infectious leptospires in urine (Kajdacsi et al. 2013; Costa et al. 2014a, 2014b, 2015b; Porter et al. 2015).
In Salvador and other large urban centers, rodent control programs employ rodenticide, environmental modification, and educational interventions in high-risk areas for leptospirosis transmission (Brazil 2002). However, rapid recolonization and population rebound after rat control or extensive trapping limit the effectiveness of interventions (Emlen et al. 1948; Glass et al. 2009).
Determining the mating structure and breeding ecology of R. norvegicus and in particular assessing the presence and extent of polyandry (the female production of offspring resulting from mating with multiple males) can provide important insights on levels of inbreeding in the population. Mating structure is also relevant to characterize patterns and levels of connectivity among sampling sites if the fathers come from different sites (Kennis et al. 2008; Moore et al. 2008). This information has important control and monitoring implications, as it informs on the possible contributions of residual and immigrating rats to post-control population growth and helps define the inter-connected populations to be considered as an eradication unit requiring simultaneous control effort (Abdelkrim et al. 2005). Herein, we estimate the occurrence of multiple paternity and inbreeding among Norway rats from the urban slum areas of Pau da Lima, Salvador, Brazil, to identify the minimum number of sires required to explain the offspring’s genotype.
Methods
Study Site and Sampling
Salvador, the capital city of the state of Bahia in Brazil, has 2.67 million inhabitants (Instituto Brasileiro de Geografia e Estatística 2007). The neighborhood of Pau da Lima (13°32′53.47″S; 38°43′51.10″W), an urban slum community comprised of 3 valleys with a combined area of 0.18 km2, was chosen for rodent sampling based on the abundance of Norway rats, the high Leptospira prevalence in rats (Costa et al. 2014a) and the high annual incidence of human Leptospira infection and leptospirosis cases, 3780/100000 and 20/100000 population, respectively (Felzemburgh et al. 2014).
Norway rats were captured between May and July of 2013 at 108 spatially randomized sites within the 3 valleys of Pau da Lima (Figure 1), using methods previously described (Porter et al. 2015). During this period, every point was visited 3 times with a total trap effort of 1994 trap-nights. Additionally, 14 more distant sites, 0.5–8.2 km from Pau da Lima, were sampled to identify potential male sires. Points of capture were geocoded and entered in a Geographic Information System (GIS) database. Procedures for obtaining tissue samples followed previously established protocols (Costa et al. 2014a).
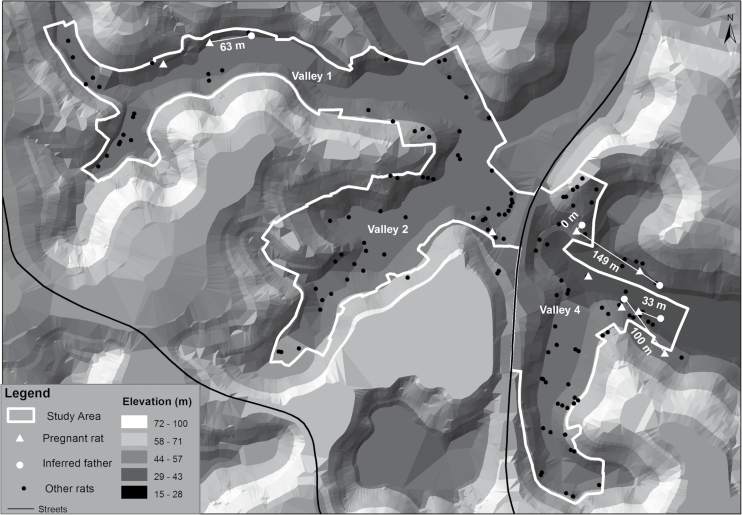
Figure 1.
The 3 valley study area of Pau da Lima. White triangles indicate the location where pregnant rats (included in this study) were caught. White dots show the location of inferred sires. Straight white lines and numbers indicate distance (m) between pregnant rats and inferred sires. Black dots indicate all other rats sampled during May and July of 2013. Black lines indicate the location of streets. Topography generated by a digital terrain model is shown in different categories of elevation.
Only pregnant rats with ≥5 fetuses were selected for this study. Fetuses from 9 rats were collected and stored in 98% alcohol until tested. The number of pregnant females, site of capture, and the number of fetuses obtained per each female are shown in Table 1. Tail snip samples from 371 (255 from the 3 valleys in Pau da Lima and 116 from the 14 distant sites) male rats were collected and genotyped following Kajdacsi et al. (2013). All animal procedures and methods were approved by IACUC committees at the Oswaldo Cruz Foundation (Salvador, Brazil, protocol number 003/2012) and Yale University (New Haven, CT; protocol number 2012-11498).
Table 1.
Inferred sire for each individual Rattus norvegicus fetus
| Offspring ID | Inferred sire | Probability of sire | Valley of female | Valley of sire | Distance between female/sire (m) |
|---|---|---|---|---|---|
| Litter 103021, individuals 1–8 | 104223.01 | 1.00 | 4 | 4 | 33.1 |
| Litter 106613, individuals 1–2, 4 | 119333.02 | 0.99 | 4 | 4 | 0 |
| Litter 106613, individuals 3, 5 | 106611.01 | 0.99 | 4 | 4 | 149.6 |
| Litter 119813, individuals 1–6 | 102512.02 | 1.00 | 4 | 4 | 100.2 |
| Litter 124431, individuals 2–4 & 6–8 | 102732.01 | 1.00 | 1 | 1 | 63.8 |
| Litter 124431, individual 1 | 226015.01 | 0.004 | 1 | 5 | 2150.6 |
| Litter 124431, individual 5 | 121531.02 | 0.14 | 1 | 1 | 209.3 |
| Litter 103031, individuals 1–9 | 122633.02 | 0.10 | 4 | 4 | 163.2 |
| Litter 111511, individuals 1–6 | 12063.11 | 0.001 | 1 | 6 | 690.4 |
The inferred sire for each individual R. norvegicus fetus is listed, along with the maximum likelihood probability of each offspring–sire paring. All offspring with inferred sires are listed, with black font denoting pairings above the 0.90 probability threshold and pairings below this threshold denoted in gray.
DNA Extractions and Genotyping
DNA from female and male rats was extracted from 3–6mm of tail tissue using the DNeasy Blood and Tissue Kit (Qiagen). Fetal DNA was similarly extracted from approximately 20–25mg of tissue. Samples were left to lyse at 56 °C overnight. We used the same PCR primers to genotype all specimens at 16 highly variable microsatellite loci to test for deviations from Hardy–Weinberg equilibrium and linkage disequilibrium, under conditions previously described (Kajdacsi et al. 2013) (Supplementary Table 1).
Paternity Analysis
We used a maximum likelihood approach to identify putative sires of the 66 embryos using the COLONY program (Wang 2004; Wang and Santure 2009). This likelihood framework evaluates levels of relatedness between individuals in the data set, including sibships and parentage (Jones and Wang 2010). Long run iterations ensured that the maximum likelihood algorithm converged by the end. Importantly, we were able to include genotypes from each rat fetus and its mother to establish known mother–offspring relationships. We then added the 371 genotypes from males as candidate sires. COLONY accommodated genetic marker error rates in each likelihood algorithm.
We set both allelic dropout frequencies and genotyping error rates at 0.05 based on preliminary runs. However, through a series of exploratory runs, there was no qualitative difference when using error rates of 0, 0.01, 0.025, and 0.05. Here, we report the results from the most conservative of these parameters, with an error rate of 5%.
Consistent with ecological evidence from this species, both male and female mating systems were treated as polygamous in each algorithm, without cloning and inbreeding. As a measure of confidence, we also estimated the probability that each sire fathered each fetus. To reduce false paternity assignment, we set 90% as the minimum assignment threshold between sire and offspring. Therefore, we did not consider paternity probabilities less than 0.9 robust enough to assign a male as the sire of a fetus.
We evaluated the precision of the likelihood algorithm from the maternal likelihood values for the known mother–offspring relationships. Each fetus was correctly paired with its mother with greater than 0.99 certainty, indicating that the maximum likelihood approach in COLONY could identify parental relationships with high accuracy. We also calculated the exclusion probability for each locus and cumulatively across loci. This metric indicates how informative loci are for eliminating a male as the putative sire and is based on allelic diversity across the sampled individuals.
Lastly, we assessed levels of inbreeding within Pau da Lima rats using genetic relatedness between known mothers and putative sires with single population pairwise relatedness in GenAlEx (Peakall and Smouse 2012). We calculated the relatedness coefficient r (Queller and Goodnight 1989) between each mother and each sire identified in the paternity analysis. This relatedness coefficient compares the proportion of alleles that are shared between 2 individuals with population-wide allele frequencies. Relatedness values range between −1 and 1, with larger r values corresponding to more closely related individuals (Burton and Krebs 2003). In order to look for correspondence between alternative approaches, we also calculated the maximum likelihood-based relatedness coefficient r using MLRelate (Kalinowski et al. 2006) to directly compare with the r relatedness values above.
Data Archiving
In fulfillment of data archiving guidelines (Baker 2013), we have deposited the primary data underlying these analyses with Dryad.
Results
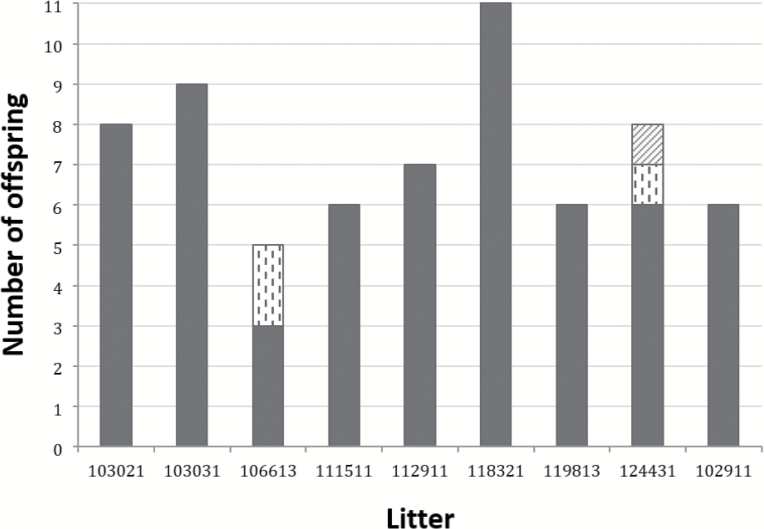
Nine mothers and 66 offspring were genotyped at 16 microsatellite loci. Litter size ranged from 5 to 11 fetuses per female with females 118321 and 106613 having the largest and smallest litter, respectively (Figure 2). The total number of alleles across all loci and litters was 117 (mean 7.3 per locus). Allele calls were not possible in less than 2% of the genotype data, meaning that missing data were very low. Locus D4Cebr2 was the most polymorphic with a total of 11 alleles, whereas locus D3Wox12 had just 3 (Supplementary Tables 2.1, 2.2, and 3).
Figure 2.
Paternal contributions to litters are indicated on the x axis and total number of offspring per litter on the y axis. Column color/pattern depicts the number of sires per litter and how many offspring each father sired.
A possible mutation was detected at locus D4Cebr2 for a fetus belonging to litter 103021. The fetus was homozygous for an allele not maternally inherited or shared by any other individual examined within this study. The fetus’ genotype was validated through repeated DNA extractions and PCR amplifications; however, COLONY flagged it as a genotyping error. COLONY also indicated a genotyping error at locus D6Cebr1 for an offspring from litter 102911. The allele in question was novel and was replicated after a repeated DNA extraction and fragment analysis. Therefore, it is more likely that it is a mutation or further instance of multiple paternity, although of all the loci examined, this was the only one that might suggest multiple paternity for this litter.
Among the 66 rat offspring, the putative sires of 25 offspring were identified among the 371 adult males in the data set with probabilities greater than 0.90 (Table 1). For 7 of the 9 litters, a single sire was most likely. However, offspring from 2 of the 9 litters were clearly attributed to multiple sires (Figure 2, Supplementary Figures 1 and 2). One of these litters (#106613) was sired by 2 males, with 1 male siring 3 offspring and the second male siring 2. Offspring from the other multiple paternity litter (#124431) were sired by 3 males. One male sired 6 of 8 fetuses in this litter, whereas the other 2 offspring were each sired by 2 separate males. The dominant male contributing to this litter (with 6 of 8 offspring) was identified among the candidate males with high likelihood (probability of 0.99). However, the other 2 males siring fetuses in litter 124431 were unlikely to have been captured during our sampling (paternity probabilities of 0.14 and 0.01). Our microsatellite data were highly informative for discriminating among paternity identities. The probability that each locus could exclude males as putative sires ranged from 0.34 to 0.7 (mean = 0.54). Only 5 loci were needed to reach an exclusion probability of 0.99; the cumulative exclusion probability across all 16 loci was optimal (i.e., 1).
Adult males contributing to parentage with probabilities greater than 0.99 were located in the same valley as the pregnant females. In one case, the male belonged to the same household (peridomestic area surrounding the household and usually limited by fences) where the female was caught. The mean inter-parent trapping distance per litter was 70 m (±58 m SD). Fetal rat samples analyzed in this study were from females captured 12–878 m apart.
Levels of inbreeding were high between mothers and the putative sires of their offspring. The mean relatedness coefficient (r) between females and their mates was significantly higher than the mean r between all mothers and other males in reproductive condition (0.31 vs. −0.10, respectively). Because relatedness coefficients were not normally distributed, we assessed significance using nonparametric permutation tests in the R statistical software (P < 0.001). Results were quantitatively similar regardless of the method used to calculate relatedness and associated estimates, with significance correspondence between values in a linear regression (r2 > 0.8, P < 0.001).
Discussion
The study of polyandry in wild-caught animals is difficult as they require capturing a sufficient number of pregnant females, males in reproductive conditions, and analysis with highly variable genetic markers to distinguish offspring from multiple males (King et al. 2014). Of the few studies of multiple paternity R. norvegicus, data from islands off the coast of New Zealand indicate that promiscuity among females provides for high levels of polyandry. On the island of Pakihi, New Zealand, a parentage analysis using 11 microsatellite loci identified polyandry in 66% (2/3) of the litters examined (Miller et al. 2010). A second study conducted on Moturemu Island, New Zealand, using 8 microsatellite markers, identified a genetic bottleneck (high levels of inbreeding; an estimate of only 30 rats inhabited the island) and breeding by a few dominant animals (Russell et al. 2009). Polyandry was high, but only 8 females were tested and no embryos were sampled (Russell et al. 2009).
Studies of other murid rodents also suggest that polyandry is significant. Among black rats, Rattus rattus, sampled from pastoral land settings in New Zealand, 94% (16/17) of females mated with multiple males as detected by 9 microsatellite loci (King et al. 2014). In Arizona (Dean et al. 2006), an average of 25% (N = 143) of wild house mouse, Mus musculus, litters were sired by multiple males, as detected by 8 microsatellites loci, although there was marked variation among populations (ranging from 6% to 43% multiple paternity).
Each of the previous studies on Norway rats were restricted by sample size and limited to island populations most likely founded by only a few rats. In this study of urban Norway rat populations multiple paternity was found in 22% (2/9) of litters. As in previous studies high levels of inbreeding were documented among dyads of females and sires. Two and 3 sires were responsible for the multiple paternity litters. All males with the highest probability of siring litters were captured from the same valley, in close proximity to pregnant females (mean inter-parent trapping distance per litter of 70 m, ±58 m SD; Figure 1). Similarly, Russell et al. (2009) found that the average distance between family group member captures was 66 m (range 0–149 m; maximum of 200 m). We previously described high levels of genetic diversity and genetic differentiation among samples from different valleys, which are only 100–400 m apart (Kajdacsi et al. 2013).
Although multiple paternity was not studied, microsatellite genotyping of rat populations in Baltimore, MD, indicated that ~95% of all Norway rats were assigned to their area of capture (62 m, within a typical alley length), indicating strong site fidelity. However, several rats were assigned to areas 2–11.5 km away, indicating some long-distance movement. Our results also indicate that population-level and polyandry genotypic patterns are most strongly associated with proximity among individuals, however gene flow among isolated and distant populations occur (Kajdacsi et al. 2013), indicating that residual and immigrant populations contribute to genetic diversity within a locale.
Polyandry among R. norvegicus is an expected finding, as females can have hundreds of sexual encounters with up to 12 males in a single night, but many may not end in male emission (Calhoun 1962). Although dominant males may attempt to protect their mates from breeding with other males, subordinate male exclusion is incomplete and female mating with multiple males is the rule (Macdonald et al. 1999). Despite this promiscuity, most of the litters in this study (7/9) were sired by a single male. This pattern is consistent with observations from laboratory experiments of Norway rats where 11 groups of 2 males and a single primiparous female produced only a single litter with 2 males contributing (Zewail-Foote et al. 2009). Female choice (some males are consistently preferred by females and have higher number of copulations) (Lovell et al. 2007), sperm competition (Breed and Taylor 2000), and colony/demographic structure (Calhoun 1962) may also influence the frequency of polyandry.
Levels of relatedness between mother–sire pairs were higher than expected and significantly higher than relatedness between all females and non-sire males (r coefficient of 0.31 vs. −0.10, respectively). The mean r of 0.31 for mating pairs is slightly higher than the expected r of 0.25 expected with half-sibling pairs (Blouin et al. 1996; Burton and Krebs 2003). This suggests that inbreeding is occurring in this population of rats within Pau da Lima. This may be due to spatial proximity of the sires and mothers in relation to the dispersal range of offspring after weaning. Previous work at this site identified genetic clusters associated with different valleys (Kajdacsi et al. 2013) suggesting, together with this data, that high levels of relatedness may result from site fidelity or geographic barriers limiting rat movement. High levels of inbreeding may also be a sign that eradication efforts are successful if genetic diversity in the mating pool is reduced, where mating with closely related individuals becomes more prevalent (Kirkpatrick and Jarne 2000; Christensen et al. 2011). In islands, eradication efforts seem to select a subsample of the rat population present before the eradication, resulting on reduction in population size, inbreeding of separated cores of survivors and leading to genetic structure (Abdelkrim et al. 2005) as observed in Salvador (Kajdacsi et al. 2013). In the slums of Salvador it is still not clear if the high levels of inbreeding are related to the separate effect of site fidelity, geographic barriers, previous eradication or is a combination of those factors. Additional studies during rodent control campaigns will be needed to answer those questions.
Genetic structure observed between different locations on the same island may be to local and subsequent local founder effects during population recovery.
Although our findings were based on only 9 pregnant females and 66 fetuses, this is a far larger number than previously studies. The large number of unassigned sires remains problematic in an attempt to define an eradication unit but breeding by residual populations and immigration from other sites, as described by Kajdacsi (Kajdacsi et al. 2013) is occurring. Additionally, the median survival time of Norway rats in urban areas (from the United States) is only 2 months (Glass et al. 1989), which reduces the chances to sample all potential sires. Studies with larger samples sizes will be required to better characterize multiple paternity patterns for R. norvegicus.
Supplementary Material
Supplementary material can be found at http://www.jhered.oxfordjournals.org/.
Funding
Oswaldo Cruz Foundation and Secretariat of Health Surveillance, Brazilian Ministry of Health; National Institutes of Health (F31 AI114245, R01 AI052473, U01 AI088752, R01 TW009504, R25 TW009338); Wellcome Trust (102330/Z/13/Z).
Data Availability
Data deposited at Dryad: http://dx.doi.org/doi:10.5061/dryad.2vh04
Supplementary Material
Acknowledgments
We would like to thank the staff of Fiocruz (Mitermayer Reis) and Zoonosis Control Center (Gorete Rodriguez) from Salvador for their assistance in conducting the study; Soledad Serrano, Gabriel Ghizzi, and Daiana Santos for their critical support during field work. We would also like to thank Priscila Machado for their assistance with database processing, GIS, and relatedness calculations. We would like to thank the Global Leptospirosis Environmental Action Network (GLEAN).
References
- Abdelkrim J, Pascal M, Calmet C, Samadi S. 2005. Importance of assessing population genetic structure before eradication of invasive species: examples from insular Norway rat populations. Conserv Biol. 19:1509–1518. [Google Scholar]
- Abdelkrim J, Pascal M, Samadi S. 2005. Island colonization and founder effects: the invasion of the Guadeloupe islands by ship rats (Rattus rattus). Mol Ecol. 14:2923–2931. [DOI] [PubMed] [Google Scholar]
- Baker CS. 2013. Journal of heredity adopts joint data archiving policy. J Hered. 104:1. [DOI] [PubMed] [Google Scholar]
- Blouin MS, Parsons M, Lacaille V, Lotz S. 1996. Use of microsatellite loci to classify individuals by relatedness. Mol Ecol. 5:393–401. [DOI] [PubMed] [Google Scholar]
- Brazil 2002. Manual de controle de roedores. Fundação Nacional de Saúde. Brasília: Funasa. [Google Scholar]
- Breed WG, Taylor J. 2000. Body mass, testes mass, and sperm size in murine rodents. J Mammal. 81:758–768. [Google Scholar]
- Burton C, Krebs C. 2003. Influence of relatedness on snowshoe hare spacing behavior. J Mammal. 84:1100–1111. [Google Scholar]
- Calhoun JB. 1962. The ecology and sociology of the Norway rat. Washington (DC): U.S. Dept. Health Educ. Welfare. [Google Scholar]
- Capizzi D, Bertolino S, Mortelliti A. 2014. Rating the rat: global patterns and research priorities in impacts and management of rodent pests. Mammal Rev. 44:148–162. [Google Scholar]
- Christensen LS, Gram-Hansen L, Chriél M, Jensen TH. 2011. Diversity and stability of Aleutian mink disease virus during bottleneck transitions resulting from eradication in domestic mink in Denmark. Vet Microbiol. 149:64–71. [DOI] [PubMed] [Google Scholar]
- Costa F, Hagan JE, Calcagno J, Kane M, Torgerson P, Martinez-Silveira MS, Stein C, Abela-Ridder B, Ko AI. 2015a. Global morbidity and mortality of leptospirosis: a systematic review. PLoS Negl Trop Dis. 9:e0003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa F, Porter FH, Rodrigues G, Farias H, de Faria MT, Wunder EA, Osikowicz LM, Kosoy MY, Reis MG, Ko AI, et al. 2014a. Infections by Leptospira interrogans, Seoul virus, and Bartonella spp. among Norway rats (Rattus norvegicus) from the urban slum environment in Brazil. Vector Borne Zoonotic Dis. 14:33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa F, Ribeiro GS, Felzemburgh RDM, Santos N, Reis RB, Santos AC, Fraga DBM, Araujo WN, Santana C, Childs JE, et al. 2014b. Influence of household rat infestation on Leptospira transmission in the urban slum environment. PLoS Negl Trop Dis. 8:e3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa F, Wunder E, Bisht V, Santos D, Ko AI, Begon M, Childs J. 2015b. Patterns in Leptospira shedding in Norway rats (Rattus norvegicus) from Brazilian slum communities at high risk of disease transmission. PLoS Negl Trop Dis. 9:e0003819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump JA, Morrissey AB, Nicholson WL, Massung RF, Stoddard RA, Galloway RL, Ooi EE, Maro VP, Saganda W, Kinabo GD, et al. 2013. Etiology of severe non-malaria febrile illness in Northern Tanzania: a prospective cohort study. PLoS Negl Trop Dis. 7:e2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean MD, Ardlie KG, Nachman MW. 2006. The frequency of multiple paternity suggests that sperm competition is common in house mice (Mus domesticus). Mol Ecol. 15:4141–4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emlen JT, Jr, Stokes AW, Winsor CP. 1948. The rate of recovery of decimated populations of brown rats in nature. Ecology. 29:133–145. [Google Scholar]
- Felzemburgh RD, Ribeiro GS, Costa F, Reis RB, Hagan JE, Melendez AX, Fraga D, Santana FS, Mohr S, dos Santos BL, et al. 2014. Prospective study of leptospirosis transmission in an urban slum community: role of poor environment in repeated exposures to the Leptospira agent. PLoS Negl Trop Dis. 8:e2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass GE, Childs JE, Korch GW, LeDuc JW. 1989. Comparative ecology and social interactions of Norway rat (Rattus norvegicus) populations in Baltimore, Maryland. Occasional Papers of the Museum of Natural History. Lawrence (KS): University of Kansas; p. 130. [Google Scholar]
- Glass GE, Gardner-Santana LC, Holt RD, Chen J, Shields TM, Roy M, Schachterle S, Klein SL. 2009. Trophic garnishes: cat-rat interactions in an urban environment. PLoS One. 4:e5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himsworth CG, Parsons KL, Jardine C, Patrick DM. 2013. Rats, cities, people, and pathogens: a systematic review and narrative synthesis of literature regarding the ecology of rat-associated zoonoses in urban centers. Vector Borne Zoonotic Dis. 13:349–359. [DOI] [PubMed] [Google Scholar]
- Instituto Brasileiro De Geografia E Estatística 2007. Contagem da População 2007. Tabela 1.1.16 - População recenseada e estimada, segundo os municípios - Bahia - 2007. [Google Scholar]
- Jones OR, Wang J. 2010. COLONY: a program for parentage and sibship inference from multilocus genotype data. Mol Ecol Resour. 10:551–555. [DOI] [PubMed] [Google Scholar]
- Kajdacsi B, Costa F, Hyseni C, Porter F, Brown J, Rodrigues G, Farias H, Reis MG, Childs JE, Ko AI, et al. 2013. Urban population genetics of slum-dwelling rats (Rattus norvegicus) in Salvador, Brazil. Mol Ecol. 22:5056–5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowski ST, Wagner AP, Taper ML. 2006. ML-Relate: a computer program for maximum likelihood estimation of relatedness and relationship. Mol Ecol Notes. 6:576–579. [Google Scholar]
- Kennis J, Sluydts V, Leirs H, Van Hooft WFP. 2008. Polyandry and polygyny in an African rodent pest species, Mastomys natalensis. Mammalia. 72:150–160. [Google Scholar]
- King C, Winstanley T, Innes J, Gleeson D. 2014. Multiple paternity and differential male breeding success in wild ship rats (Rattus rattus). N Z J Ecol. 38:76–85. [Google Scholar]
- Kirkpatrick M, Jarne P. 2000. The effects of a bottleneck on inbreeding depression and the genetic load. Am Nat. 155:154–167. [DOI] [PubMed] [Google Scholar]
- Lovell JL, Diehl A, Joyce E, Cohn J, Lopez J, Guarraci FA. 2007. “Some guys have all the luck”: mate preference influences paced-mating behavior in female rats. Physiol Behav. 90:537–544. [DOI] [PubMed] [Google Scholar]
- Macdonald DW, Mathews M, Berdoy M. 1999. The behaviour and ecology of Rattus norvegicus: from opportunities to kamikaze tendencies. In: Singleton G, Hinds L, Leirs H, Zhang Z, editors. Ecologically-based rodent management. 1st ed. Canberra: Australian Centre for International Agricultural Research; p. 49–80. [Google Scholar]
- Miller SD, Russell JC, Macinnes HE, Abdelkrim J, Fewster RM. 2010. Multiple paternity in wild populations of invasive Rattus species. N Z J Ecol. 34:360–363. [Google Scholar]
- Moore J, Nelson N, Keall S, Daugherty C. 2008. Implications of social dominance and multiple paternity for the genetic diversity of a captive-bred reptile population (tuatara). Conserv Genet. 9:1243–1251. [Google Scholar]
- Peakall R, Smouse PE. 2012. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research–an update. Bioinformatics. 28:2537–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter FH, Costa F, Rodrigue G, Farias H, Glass G, Reis MG, Ko AI, Childs JE. 2015. Morphometric and demographic differences between tropical and temperate Norway rats (Rattus norvegicus). J Mammal. 96:317–323. [Google Scholar]
- Queller D, Goodnight K. 1989. Estimating relatedness using genetic markers. Evolution. 43:258–275. [DOI] [PubMed] [Google Scholar]
- Russell J, Abdelkrim J, Fewster R. 2009. Early colonisation population structure of a Norway rat island invasion. Biol Invasions. 11:1557–1567. [Google Scholar]
- Susilawati TN, McBride WJ. 2014. Acute undifferentiated fever in Asia: a review of the literature. Southeast Asian J Trop Med Public Health. 45:719–726. [PubMed] [Google Scholar]
- Torgerson PR, Hagan JE, Costa F, Calcagno J, Kane M, Martinez-Silveira MS, Goris MG, Stein C, Ko AI, Abela-Ridder B. 2015. Global burden of leptospirosis: estimated in terms of disability adjusted life years. PLoS Negl Trop Dis. 9:e0004122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. 2004. Sibship reconstruction from genetic data with typing errors. Genetics. 166:1963–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Santure AW. 2009. Parentage and sibship inference from multilocus genotype data under polygamy. Genetics. 181:1579–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zewail-Foote M, Diehl A, Benson A, Lee KH, Guarraci FA. 2009. Reproductive success and mate choice in Long-Evans rats. Physiol Behav. 96:98–103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data deposited at Dryad: http://dx.doi.org/doi:10.5061/dryad.2vh04