SUMMARY
Long noncoding RNAs (lncRNAs) are involved in diverse cellular processes through multiple mechanisms. Here, we describe a previously uncharacterized human lncRNA, CONCR (cohesion regulator noncoding RNA), that is transcriptionally activated by MYC and is upregulated in multiple cancer types. The expression of CONCR is cell cycle regulated, and it is required for cell-cycle progression and DNA replication. Moreover, cells depleted of CONCR show severe defects in sister chromatid cohesion, suggesting an essential role for CONCR in cohesion establishment during cell division. CONCR interacts with and regulates the activity of DDX11, a DNA-dependent ATPase and helicase involved in DNA replication and sister chromatid cohesion. These findings unveil a direct role for an lncRNA in the establishment of sister chromatid cohesion by modulating DDX11 enzymatic activity.
Graphical abstract
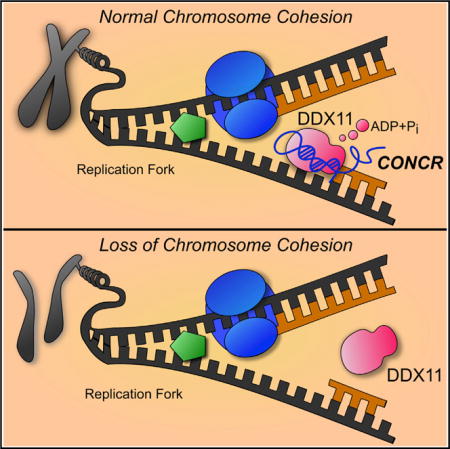
INTRODUCTION
The human genome is known to encode thousands of RNA transcripts, the majority of which do not produce proteins and are referred to as noncoding RNAs (ncRNAs). Among this class of gene products, the largest group is represented by RNAs longer than 200 nt and with mRNA-like characteristics (5′-cap and 3′-polyA), known as long noncoding RNAs (lncRNAs) (Harrow et al., 2012). Long noncoding RNAs represent more than 25% of all human genes (GENCODE v24), although only a small number of them have been functionally characterized to date. LncRNAs described until now have been shown to be involved in the most diverse cellular processes, such as cell growth and apoptosis, cell pluripotency, and differentiation, through multiple and diverse mechanisms (Bonasio and Shiekhattar, 2014; Fatica and Bozzoni, 2014; Rinn and Chang, 2012). Moreover, our work and that of others has shown that lncRNAs are key regulators of cell transformation and, in some cases, direct transcriptional targets of well-known tumor suppressor and oncogenic factors, such as p53 (Huarte, 2015; Sánchez et al., 2014) and MYC (Hart et al., 2014; Kim et al., 2015).
Faithful DNA replication and proper sister chromatid cohesion ensure the correct propagation of the genetic material to daughter cells during cell division. A large number of factors involved in these processes have been identified and characterized (Masai et al., 2010; Peters and Nishiyama, 2012), as well as their alterations associated with genome instability and eventually tumorigenesis (Gaillard et al., 2015; Losada, 2014; Skibbens et al., 2013). Among these factors is DEAD/H box protein 11 (DDX11), a DNA-dependent ATPase and helicase involved in the processing of the lagging strand during DNA replication and in the maintenance of the fork structure for the establishment of cohesion (Bharti et al., 2014; Parish et al., 2006). Mutations in DDX11 have been in fact associated with a rare pathological condition known as Warsaw breakage syndrome, a syndrome characterized at the cellular level by sister chromatid cohesion defects (Capo-Chichi et al., 2013; van der Lelij et al., 2010).
Although we have a large knowledge of the proteins involved in DNA replication and cohesion, the involvement of lncRNAs in these processes remains undetermined. Here, we describe a human lncRNA, which regulates DNA replication and sister chromatid cohesion by modulating the activity of the helicase DDX11. The expression of the lncRNA is directly linked with the ability of tumor cells to proliferate, conferring them with increased malignancy.
RESULTS
CONCR Is Negatively Regulated by p53 and Activated by MYC
p53 is a master regulator of cellular homeostasis that inhibits uncontrolled cell proliferation. Consistently with the known function of p53, p53−/− cells bypass cell-cycle arrest caused by DNA damage (Kaeser et al., 2004). In order to identify lncRNAs involved in this process, we searched for those with altered expression in cells with impaired p53. To do this, we performed polyA+ RNA sequencing of p53−/− and p53+/+ HCT116 cells either untreated or treated with the DNA-damaging drug 5-FU. By comparing the expression values of each transcript identified in HCT116 p53−/− and p53+/+ cells, we ranked those transcripts in which expression levels were significantly greater in the absence of p53 both in the presence and absence of DNA damage. We identified 4,143 mRNAs that showed increased expression in p53−/− compared to p53+/+ cells, with functions related to cell cycle, mitosis, DNA repair, and DNA replication (Table S1). Similarly, we identified 81 lncRNAs with induced expression in p53−/− cells (Table S1), and we hypothesized their involvement in cell-cycle progression.
Among the lncRNAs identified by our analysis, we found an lncRNA previously annotated as DDX11 antisense RNA 1 (DDX11-AS1), which is a divergent non-overlapping transcript of the protein-coding gene DDX11, that we named “cohesion regulator noncoding RNA,” or CONCR (Figure 1A). CONCR showed greater levels in HCT116 p53−/− compared to wild-type cells (Figure 1B and Table S1). This anti-correlation between p53 and CONCR was confirmed in A549 cells depleted of p53 by RNAi both in the presence or absence of treatment with DNA-damaging drugs, 5-FU or doxorubicin (Figures 1C, S1A, and S1B), suggesting that absence of p53 causes an increase in CONCR expression.
Figure 1. CONCR Is a Nuclear lncRNA Negatively Regulated by p53 and Activated by MYC.
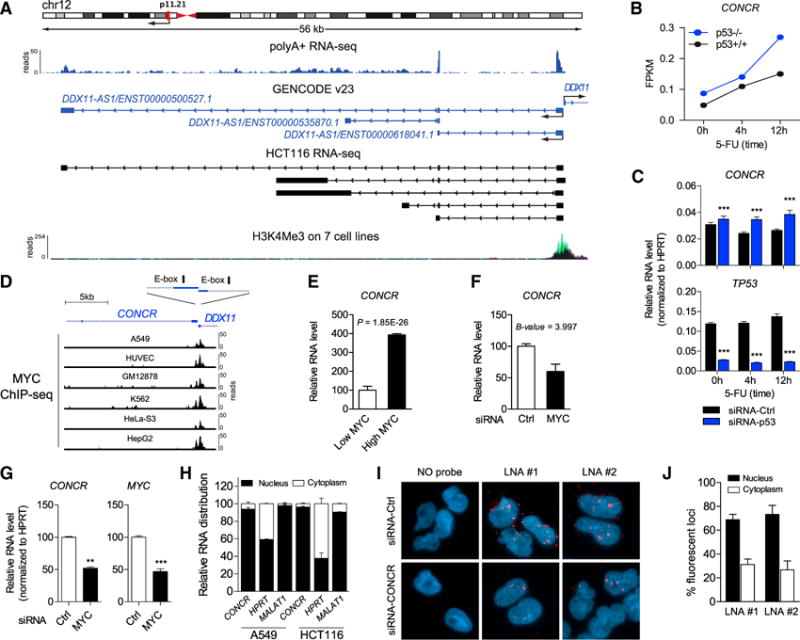
(A) CONCR genomic locus. Ideogram of location on chromosome 12; RNA expression detected by whole-cell polyA+ RNA-seq of A549 from ENCODE/CSHL; structure and directionality of CONCR (DDX11-AS1) and its neighbor gene DDX11 as annotated in GENCODE v23; RNA structures identified by RNA-seq in HCT116; and H3K4me3 mark on seven cell lines from ENCODE defining active transcription.
(B) CONCR expression level determined by polyA+ RNA-seq of p53−/− and p53+/+ HCT116 cells either untreated or treated with the DNA-damaging drug 5-FU for 4 and 12 hr.
(C) CONCR and TP53 expression levels determined by qRT-PCR of A549 cells transfected with a control siRNA (siRNA-Ctrl) or with a siRNA-targeting TP53 (siRNA-p53), either untreated or treated with the DNA-damaging drug 5-fluorouracil (5-FU), for 4 and 12 hr. Graph shows mean ± SEM of three independent experiments.
(D) Schematic of the location of the E-box CACGTG motifs in the promoter region of CONCR-DDX11; MYC binding to the promoter region determined by ChIP-seq in different cell lines (ENCODE/SYDH and ENCODE/OpenChrom-UTA).
(E) CONCR relative expression level determined by polyA+ RNA-seq in the human B cell line P493-6 expressing either low or high levels of MYC (Hart et al., 2014). Graph shows mean ± SEM of values and significance as reported in the original study (Hart et al., 2014).
(F) CONCR relative expression level determined by microarray in HCT116 depleted of MYC by RNAi (Kim et al., 2015). Graph shows mean ± SEM of values and significance as reported in the original study (Kim et al., 2015).
(G) CONCR and MYC RNA levels determined by qRT-PCR in A549 depleted or not of MYC by RNAi.
(H) Relative subcellular localization of CONCR and control RNAs, i.e., HPRT and MALAT1, determined by nucleus/cytoplasm fractionation and qRT-PCR of A549 and HCT116. Graph shows mean ± SD of two independent experiments.
(I) RNA FISH of A549 transfected with a control siRNA (siRNA-Ctrl) or with a combination of two siRNAs targeting CONCR (siRNA-CONCR) using two independent LNA probes (LNA #1 and #2) or a no probe condition as control.
(J) Percentage of fluorescent foci detected by RNA FISH in the nucleus or in the cytoplasm of siRNA-Ctrl cells as in (I). Fluorescent foci were quantified by imaging and counting approximately 100 cells per condition. Graph shows mean ± SD of two independent experiments.
p53 is known to regulate gene expression both by direct transcriptional activation and by indirect effects on cellular pathways and transcription factors that in turn become active or inactive (Fischer et al., 2014; Rinn and Huarte, 2011). Analysis of the promoter region of CONCR failed to identify a p53-binding motif. Consistently, we could not find any evidence of binding in the promoter region by p53 chromatin immunoprecipitation sequencing (ChIP-seq) analysis (Sánchez et al., 2014), suggesting that the lower level of expression of CONCR observed in p53+/+ cells may be the result of indirect p53-dependent repression. In contrast, the analysis of the promoter region of CONCR identified two canonical E-box CACGTG binding motifs for the transcription factor MYC (Sabò and Amati, 2014) (Figure 1D), which has been described as transcriptionally repressed in a p53-dependent manner (Ho et al., 2005; Sachdeva et al., 2009). Indeed, ChIP-seq data from ENCODE showed that MYC is bound to CONCR promoter region in multiple cell types (Figure 1D). Furthermore, CONCR was significantly identified by RNA sequencing (RNA-seq) as upregulated in response to MYC overexpression in the human B cell line P493-6 (Hart et al., 2014) (Figure 1E), while inhibition of MYC by RNAi in HCT116 and array analysis (Kim et al., 2015) showed downregulation of CONCR (Figure 1F). Similarly, when we silenced Mcs in CONCR levels were indirectly due to cell-cycle deregulation, we silenced E2F1, a transcription factor involved in the control of cell-cycle progression from G1 to S-phase (Biswas and Johnson, 2012), observing, as expected, that cell cycle was affected, while CONCR levels were not (Figures S1C and S1D). Although it remains difficult to discriminate between the causes and consequences of perturbations of the cell cycle, altogether these data suggest that CONCR is transcriptionally regulated by MYC, and the greater levels of CONCR following p53 depletion may be ascribed to the transcriptional activity of MYC.
CONCR is ubiquitously expressed in a panel of different human cell lines (Figure S1E). We confirmed by qRT-PCR the presence of three different transcriptional isoforms of CONCR, as annotated in GENCODE v23, although the isoform comprising the first two exons (ENST00000618041.1) is the most abundant in the cell (>10-fold), and its 5′ and 3′ ends are confirmed by 5′ cap gene expression (CAGE) analysis and the presence of a polyadenylation signal, respectively (Figures S1F and S1G). The noncoding nature of CONCR lncRNA was confirmed by the lack of significant open reading frames (Table S1) and its low coding potential (Figure S1H). CONCR is a predominantly nuclear lncRNA, shown by sub-cellular fractionation (Figure 1H) and RNA FISH with two independent oligonucleotide probes (Figures 1I and 1J). To control for the specificity of the RNA FISH probes, we silenced CONCR by RNAi and quantified the number of fluorescent foci. Results showed significant reduction in the number of CONCR foci following knockdown of the lncRNA, confirming specific binding of the probes to CONCR (Figures 1I and S1I–S1L).
CONCR Is Upregulated in Multiple Cancer Types
The relationship observed for CONCR with p53 and MYC suggests an implication of the lncRNA in cancer. We determined the levels of CONCR across hundreds of tumors and adjacent normal tissues from different cancer types using publicly available data derived from the computational analysis of RNA sequencing (Iyer et al., 2015). CONCR expression was significantly greater in the majority of cancer types analyzed (9 out of 12) when comparing tumor specimens with healthy tissue-paired samples (Figure 2A and Table S2). Moreover, when the mutational status of p53 was taken into account, the expression level of CONCR appeared significantly greater in tumors with mutations in the TP53 gene compared to tumors presenting the wild-type gene (Figure 2B). Therefore, CONCR presents greater levels of expression in cancer as a result of the mutational status and impaired functionality of p53. We then investigated the ability of cells to form tumors in a mouse xenograft model dependent on the presence or absence of CONCR. For this, an equal number of HCT116 cells depleted of CONCR or control cells were injected subcutaneously in immunocompromised mice and tumor growth was followed for the indicated time (Figure 2C). Results showed that CONCR knockdown affects the ability of cells to form tumors when comparing tumor sizes of mice injected with cells transfected with a control siRNA or CONCR-targeting siRNAs (Figures 2C and 2D), suggesting that CONCR contributes to tumor growth.
Figure 2. CONCR Is Upregulated in Multiple Cancer Types.
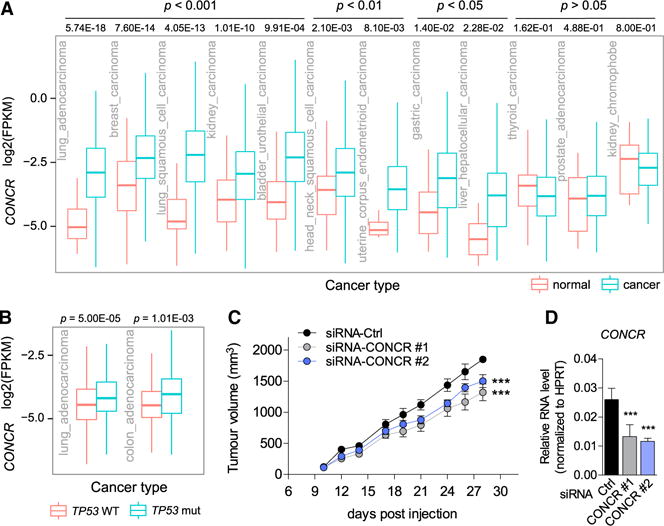
(A) CONCR expression level in paired normal-cancer samples of different cancer types. Data were obtained from http://mitranscriptome.org (Iyer et al., 2015). Significance was determined by Welch’s t test (Table S2).
(B) CONCR expression level determined in lung adenocarcinoma and colon adenocarcinoma RNA-seq data available through the TCGA database (https://gdc-portal.nci.nih.gov), i.e., LUAD and COAD datasets (Table S2). Expression was compared in each cancer type between tumor samples from individuals bearing mutations in TP53 and those with the wild-type gene. Significance was determined by unpaired Student’s t test.
(C) HCT116 cells transfected with CONCR-targeting siRNAs (#1 and #2) or with a control siRNA subcutaneously injected in immunodeficient mice (n = 6 per experimental condition). Tumor volume was measured at the indicated times. Graph shows mean ± SEM of n = 6 mice per experimental condition. Significance was determined by one-way ANOVA and Bonferroni Multiple Comparison Test comparing to siRNA-Ctrl.
(D) CONCR knockdown efficiencies in cells used in (C) were determined by qRT-PCR.
See also Table S2.
Expression of CONCR Is Periodic in the Cell Cycle and Is Required for Efficient Cell Division and Survival
To investigate the biological function of CONCR, we conducted loss-of-function studies using RNAi-mediated depletion of the lncRNA. Cells were transfected with two independent siRNAs targeting CONCR, alone or in combination, a scrambled oligonucleotide as a control, or left untransfected (Figure 3A). Cells depleted of CONCR were assayed for their proliferation ability, apoptosis, and cell-cycle progression. A significant reduction in the number of proliferating cells was observed following depletion of CONCR (Figure 3B), concurrent with an increase in the number of apoptotic cells (Figure 3C) and with the number of cells blocked at the G0/G1 phases of the cell cycle (Figure 3D), suggesting a potential role for CONCR in cell division and survival.
Figure 3. CONCR Expression Is Periodic in the Cell Cycle and Necessary for Cell Division and Proliferation.
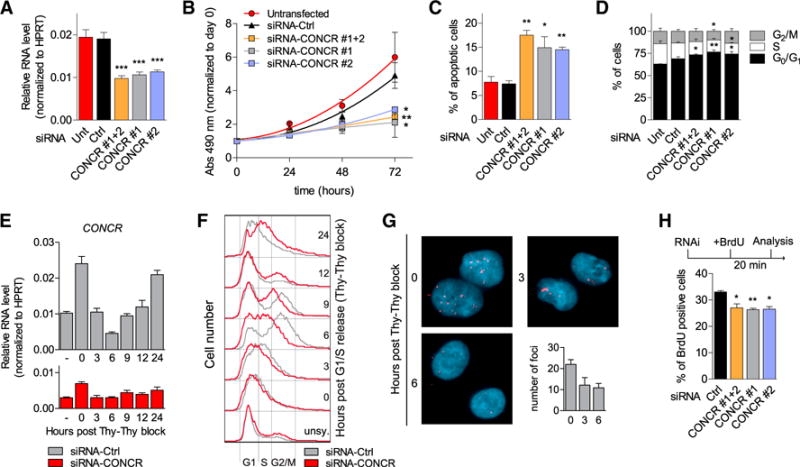
(A–D) A549 cells left untransfected (Unt), transfected with a control siRNA (Ctrl), or with two siRNAs targeting CONCR, separately (#1 and #2) or in combination (#1+2). Graphs show mean ± SEM of at least three independent experiments. Significance was determined comparing to siRNA-Ctrl.
(A) RNA knockdown efficiencies determined by qRT-PCR.
(B) Cell proliferation measured by MTS assay.
(C) Percentage of apoptotic cells determined by flow cytometry of annexin V and 7-AAD stained cells.
(D) Analysis of cell-cycle phase distribution by flow cytometry of propidium iodide-stained cells.
(E and F) A549 cells either transfected with a control siRNA (Ctrl) or with two siRNAs targeting CONCR in combination. G1/S synchronized cells obtained by double thymidine block procedure. Normal medium was then used for the release, and cells collected at the different time points indicated for CONCR expression analysis by qRT-PCR (E) and cell-cycle analysis (F). (E) Graph shows mean ± SD of two independent experiments, while in (F) cell-cycle profiles of one representative experiment are shown.
(G) CONCR RNA-FISH performed on A549 cells G1/S synchronized and released as before using LNA #2 probe. Fluorescent foci were quantified on 100 cells per condition. Graph shows mean ± SD of two independent experiments.
(H) Percentage of A549 BrdU-positive cells (20 min BrdU pulse) transfected with the indicated siRNAs. Graph shows mean ± SD of three independent experiments.
To gain insight into the function of CONCR, we used microarray technology to analyze gene expression changes in cells depleted of CONCR compared to cells transfected with a control siRNA. We identified approximately 500 genes affected by CONCR inhibition (B > 0), both coding and noncoding (Table S3 and Figure S2A). Moreover, gene ontology analysis identified a significant enrichment in pathways related to apoptosis and cell cycle progression, including downregulation of numerous genes involved in the G1 to S-phase transition of the cell cycle (Figure S2B) consistent with a functional role of CONCR in these processes and in agreement with the phenotype observed.
The observation that silencing of CONCR affected cell cycle progression (Figure 3D) prompted us to investigate the expression of the lncRNA across the different phases of the cell cycle. Cells were therefore synchronized in G1/S by double thymidine block and synchrony of cells monitored by flow cytometry of propidium iodide-stained cells. Analysis of the RNA content at the different time points showed that CONCR expression was periodic, with peaks of expression matching with the mid-late G1 phase of the cell cycle (Figures 3E and 3F). Expression levels of well-known periodic genes, such as CCNE1 (peaks in G1), CCNA2 (peaks in G2), and CCNB1 (peaks in M), were used as controls (Figures S2C–S2E). Similar results were obtained by RNA FISH (Figure 3G). Moreover, silencing of CONCR followed by double thymidine block and release as before showed a clear impairment in the ability of cells to S-phase re-start, with a large proportion of cells found to be delayed when compared to siRNA-control transfected cells (Figure 3F).
To further investigate the nature of the delay observed for CONCR-depleted cells in cell-cycle progression, we performed an experiment of bromodeoxyuridine (BrdU) pulse labeling. CONCR was silenced and replicating DNA labeled with a 20-min pulse of BrdU. Cells were then collected and analyzed for BrdU incorporation by flow cytometry (Figure 3H). BrdU incorporation into newly synthesized DNA was significantly reduced in CONCR-depleted cells compared to control cells (Figure 3H), suggesting an involvement of CONCR in DNA replication.
Altogether, the results obtained suggest that the expression of CONCR is tightly regulated across the cell cycle and its presence is required for efficient G1/S transition and DNA replication.
Cells Depleted of CONCR Show Severe Defects in Sister Chromatid Cohesion
In order to gain more insight into CONCR function, we performed a correlation analysis using RNA-seq data from 495 samples, including normal and tumor tissues (Cancer Genome Atlas Research Network, 2014). This analysis indicated that most of the genes co-expressed with CONCR (correlation value > 0.45) encode for proteins known to be involved in DNA replication and chromosome maintenance (Figures S2F and S2G and Table S4), including DDX11 and several of its interacting partners, such as TIMELESS (Calì et al., 2016), ESPL1, and components of the minichromosome maintenance complex (MCM) (Figure S2H). This, together with the known function of DDX11 in sister chromatid cohesion, prompted us to investigate the functional role of CONCR in this process.
Interestingly, CONCR depletion caused a dramatic increase in the percentage of cells with sister chromatid cohesion loss consistently observed with two independent siRNAs (Figures 4A–4C). Cohesion loss was found to affect entire metaphases rather than sparse sister chromatids within different meta-phases, while the degree of the cohesion defect varied from loosely paired to more widely separated chromatids, referred to as “loss of cohesion” (Figure 4A, ii–v), in contrast with the canonical X-shaped conformation observed in the control cells (Figure 4A, i). Similarly, loss in sister chromatid cohesion was also observed when the protein-coding gene DDX11 was silenced by RNAi as expected and previously described (Farina et al., 2008; Parish et al., 2006) (Figures 4B and 4D). In both cases, i.e., CONCR and DDX11 knockdown (Figures 4C, 4D, and S3A), cell death was observed following RNAi-mediated silencing, reaching 30% of apoptotic cells for DDX11 silencing (Figures 3C and S3B), supporting the notion that Ddx11 knockout in mouse was found to be lethal (Inoue et al., 2007) and suggesting that a ~50% reduction in the levels of CONCR or DDX11 (Figures 4C and 4D) were sufficient to cause substantial cohesion defects that ultimately result in cell death. To better appreciate the extent of the cohesion defects, we subclassified the “loss of cohesion” phenotype into either “loosely paired” or “completely separated” chromatids and compared the effect of CONCR and DDX11 knockdowns with that obtained by silencing the cohesion complex component RAD21 (Figures S3C–S3I). The same was done in HCT116 (Figures S3F and S3G) and HeLa cells (Figures S3H and S3I) to evaluate possible cell-type variability. Results showed that the “loosely paired” phenotype appeared predominant in respect to “completely separated” and that both the aspect of the chromatids and the percentages of the phenotype classifications appeared comparable across cell lines (Figure S3). RAD21 depletion showed a higher number of metaphases with cohesion defects compared to CONCR or DDX11 knockdowns, although it may reflect the differences observed in terms of knockdown efficiencies (Figure S3). On the other hand, the knockdown of WAPL, a regulator of sister chromatid resolution (Gandhi et al., 2006), which is known to restore the cohesion defect caused by DDX11 depletion (de Lange et al., 2015), was also able to restore cohesion defects in CONCR-deficient cells (Figures S3J–S3L), suggesting that DDX11 and CONCR affect chromosomal cohesion at the same level.
Figure 4. Depletion of CONCR Causes Sister Chromatid Cohesion Defects.
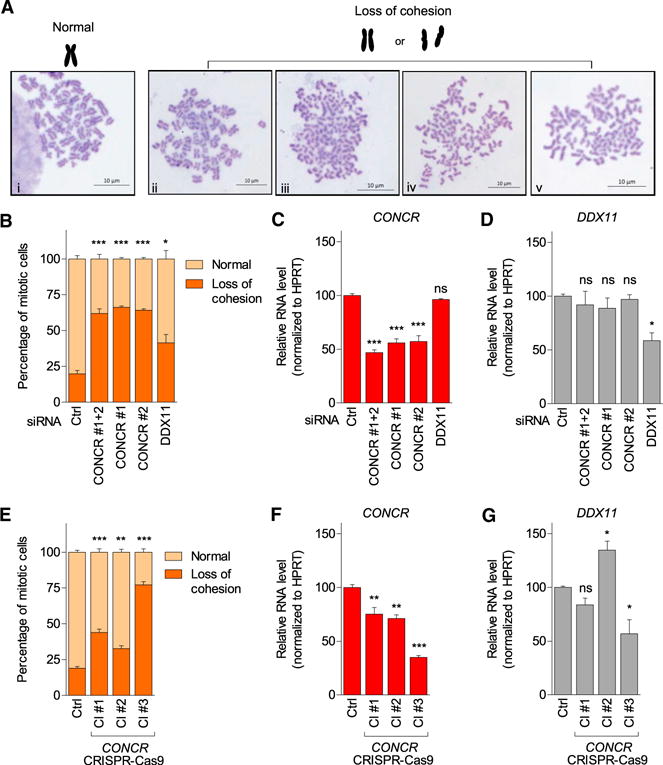
(A) Representative images of chromosome spreads showing normal X-shaped conformation observed in control cells (i) or representative images of cohesion defects observed in cells depleted of CONCR or DDX11 (ii–v).
(B) Percentage of mitotic cells showing normal sister chromatid cohesion or loss of cohesion. At least 50 metaphases per condition were scored and each experiment blindly scored twice. Graph shows mean ± SD of three independent experiments.
(C and D) CONCR and DDX11 knockdown effi-ciencies determined by qRT-PCR. Graphs show mean ± SD of three independent experiments.
(E) Percentage of mitotic cells showing normal sister chromatid cohesion or loss of cohesion measured as in (B) in A549 control cells (Ctrl) and in different clones (Cl #1, Cl #2, and Cl #3) of A549 cells showing silencing of CONCR by insertion of a selection marker gene with a polyadenylation signal. Graph shows mean ± SD of three (Cl #1 and #2) or two (Cl #3) independent experiments. (F and G) CONCR and DDX11 levels determined by qRT-PCR.
See also Figures S3 and S4.
To further confirm these results, we silenced CONCR in A549 cells by inserting a polyadenylation signal immediately downstream of CONCR promoter using the CRISPR-Cas9 system. We obtained several clones with CONCR expression reduced to different levels, probably corresponding to different levels of heterozygosity of A549 polyploid cells (Figure S4). Notably, the two clones that presented lower expression of CONCR, marked as clones #3 and #4, did not survive cell culture passaging, allowing us to perform only two independent chromosomes spread preparations, or none in the case of clone #3 and clone #4. Nevertheless, the results obtained using the CONCR CRISPR-Cas9-edited cells showed a consistent increase in the number of metaphases with cohesion defects (Figure 4E), supporting the results previously obtained by RNAi (Figure 4B). Moreover, both in the case of RNAi- and CRISPR-Cas9-mediated depletion of CONCR, the percentage of cells with cohesion loss clearly correlated with the level of knockdown achieved for CONCR (Figures 4B, 4C, 4E, and 4F). Remarkably, changes in the levels of DDX11 did not appear to contribute to the phenotype observed (Figures 4B, 4D, 4E, and 4G).
Together, these results suggest that CONCR has a biological function in sister chromatid cohesion.
CONCR Modulates the Activity of the Helicase DDX11
The common sister chromatid cohesion phenotype suggested a functional relationship between CONCR and DDX11, which are co-regulated divergent genes (Figure S2F and Figure 1A). To date, several lncRNAs have been shown to modulate the expression levels of neighboring protein-coding genes in a mechanism known as regulation in cis (Guil and Esteller, 2012; Villegas and Zaphiropoulos, 2015). However, as shown above, CONCR depletion caused cohesion defects without affecting DDX11 RNA levels (Figure 4). Indeed, DDX11 RNA levels remained unchanged when silencing the lncRNA and increased when silencing p53 (Figures S5A–S5C and S2A). In agreement with this observation, histone H3K9 acetylation at DDX11 promoter region was not affected by knockdown of CONCR (Figure S5D). Similarly, western blot analysis did not show changes in DDX11 protein levels upon CONCR knockdown (Figure S5E). We then concluded that CONCR does not regulate DDX11 RNA or protein levels.
Having excluded a possible regulation in cis of DDX11 by the lncRNA, we investigated the possibility of a physical interaction between them. To that end, we incubated cell extracts with biotinylated oligonucleotides with sequence complementarity to CONCR, and bound material was then pulled down using streptavidin beads. Analysis of a fraction of the pull-down eluates confirmed specific CONCR enrichment using two independent oligos compared to the lacZ control or other control RNAs (Figure 5A). Interestingly, the protein DDX11 was found to associate with the pulled down CONCR (Figure 5B). In contrast, a control nuclear protein, WDR5 (subunit of the MLL1/MLL complex), was not detected bound to CONCR (Figure 5B). Moreover, the interaction between CONCR and DDX11 was confirmed using RNA immunoprecipitation (RIP) from cross-linked nuclear extracts. A significant enrichment of CONCR, but not several control RNAs in DDX11 immunoprecipitates, was observed, whereas no CONCR was detected when using the IgG control (Figures 5C and 5D).
Figure 5. CONCR Interacts with DDX11 Protein and Regulates Its Function.
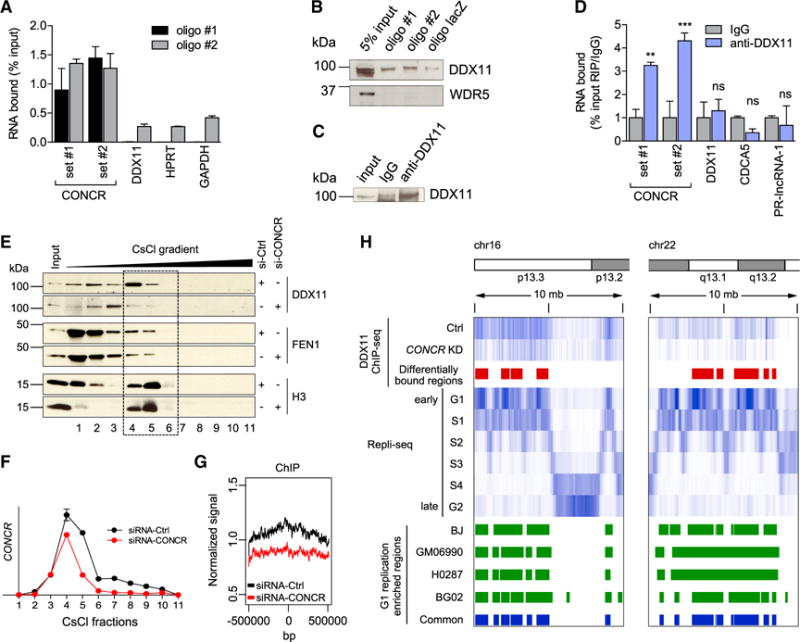
(A) Percentage of CONCR and control RNAs (DDX11, HPRT, and GAPDH) pulled down with two independent biotinylated oligonucleotides (oligo #1 and oligo #2) in A549 cells.
(B) DDX11 and WDR5 (control) western blots of the CONCR associated proteins pulled down as in (A).
(C) Western blot of DDX11 immunoprecipitation as in (D).
(D) Percentage of CONCR and control RNAs (DDX11, CDCA5, and PR-lncRNA-1) immunoprecipitated with an anti-DDX11 antibody or IgG control. Graph shows mean ± SD of three independent experiments. Significance was determined comparing to IgG.
(E) DDX11, FEN1, and histone 3 (H3) distribution in the fractions obtained from CsCl density-gradient centrifugation of A549 cells transfected with control or CONCR siRNAs as indicated.
(F) Quantification by qRT-PCR of CONCR in the gradient fractions shown in (E).
(G) Mean DDX11 ChIP-seq signal around the center of all the genomic regions identified as differentially bound by DDX11 in CONCR-depleted cells compared to siRNA-Ctrl-transfected cells.
(H) DDX11 ChIP-seq. Top to bottom: chromosome schematic of two representative regions of chr16 and chr22; DDX11 ChIP-seq signal in control (Ctrl) and CONCR-depleted cells (KD); regions with differential binding of DDX11 (Ctrl versus KD); signals of DNA replicating regions of BJ cells in G1 to G2 phases of the cell cycle as reported in Hansen et al. (2010); G1-replication-enriched regions common to BJ, GM06990, H0287, and BG02 cell types as reported in Hansen et al. (2010).
See also Figures S5 and S6, and Table S5.
DDX11 is known to function at the replication fork, coordinating lagging strand synthesis and sister chromatid cohesion (Bharti et al., 2014). Therefore, we hypothesized that CONCR association with DDX11 may affect its function in DNA replication. To test this, we performed chromatin fractionation by CsCl density-gradient centrifugation. This protocol allows separating the chromatin fractions that are enriched in DNA replication factors (Dellino et al., 2013), as the gradient can separate the proteins that are bound to the chromatin from those in the soluble fraction of the nucleus, as well as from the protein-free DNA (Figures 5E, S5F, and S5G). We observed that in control conditions DDX11 peaks at the chromatin fraction, co-localizing with CONCR, while knockdown of CONCR induces a shift in DDX11 from the chromatin-bound to the soluble fractions (Figures 5E and 5F). Importantly, this shift seems specific, as FEN1, a protein known to function in coordination with DDX11 (Farina et al., 2008), remains in the same fractions upon CONCR knockdown (Figure 5E). These data show that CONCR and DDX11 co-localize on the chromatin, and the association of DDX11 with the chromatin is highly dependent on CONCR. We further investigated this relationship between DDX11 and CONCR by performing DDX11 ChIP-seq in cells depleted of CONCR by RNAi or transfected with a siRNA-control and synchronized in G1/S (Figures S5H and S5I). The analysis of the ChIP-seq data revealed broad binding of DDX11 to the chromatin, covering genomic regions in the order of mega bases (Figures 5G, 5H, and S6). As expected, DDX11 ChIP-seq signal preferentially matched to early DNA replicating regions previously identified by Repli-seq analysis of four different cell types (Hansen et al., 2010) (Supplemental Experimental Procedures, Figure 5H, and Figure S6). Interestingly, silencing of CONCR caused a significant decrease in the association of DDX11 to 43 of these 867 replicating regions, while no significant differences were observed in the remaining regions (Figures 5G, 5H, and S6 and Table S5), supporting the notion that the function of DDX11 is dependent on CONCR.
To further investigate the functional relationship between CONCR and DDX11 protein, we overexpressed DDX11 in cells depleted or not of CONCR, and chromosome spreads were prepared and analyzed for cohesion defects. As observed before, the number of metaphases with sister chromatid cohesion loss significantly increased in cells depleted of CONCR (Figures 6A, S5J, and S5K). Furthermore, the cohesion defect was rescued by the overexpression of DDX11, confirming that both factors function in the same pathway (Figures 6A, S5J, and S5K). On the other hand, when instead of the wild-type form, the DDX11 ATPase defective mutant K50R (Farina et al., 2008) was overexpressed, no rescue of the cohesion defect was observed (Figures 6A, S5J, and S5K).
Figure 6. CONCR Enhances the ATPase Activity of DDX11.
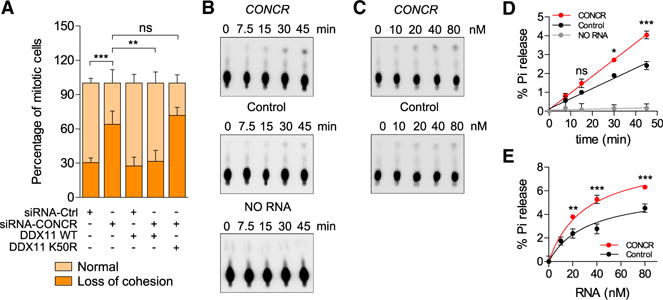
(A) Percentage of mitotic cells showing normal sister chromatid cohesion or loss of cohesion transfected with the indicated combination of siRNAs and DDX11 expression plasmids. Graph shows mean ± SD of two independent experiments, each blindly scored twice.
(B–E) Representative images (B and C) and quantification (D and E) of three independent ATPase assays (mean ± SD) using recombinant purified human DDX11 and in vitro transcribed RNAs, CONCR, or a control RNA (antisense sequence of CONCR). The standard reaction mixture contained 80 nM (B and
D) or the indicated concentration of RNA (C and E) and was incubated for the indicated time (B and D) or 45 min (C and E).
See also Figure S5.
Since we observed a physical and functional interaction between the lncRNA and DDX11, but not the catalytic mutant, we hypothesized that CONCR could have an effect on the enzymatic activity of DDX11. To test this hypothesis, we assayed the ATPase activity of DDX11 either in the presence of CONCR or a control RNA of the same length. Both time course and RNA titration experiments showed a significant increase in the ability of DDX11 to hydrolyze ATP when incubated in the presence of the lncRNA, significantly higher than that observed when the enzyme was incubated with the control RNA (Figures 6B–6E). The result of these in vitro assays suggests a potential function for CONCR as a modulator of DDX11 enzymatic activity, which is required for the proper function of the protein in DNA replication and sister chromatid cohesion.
Collectively, our results show that CONCR is an MYC-regulated lncRNA upregulated in cancer, which modulates DNA replication and sister chromatid cohesion by enhancing the catalytic activity of DDX11 and binding to DNA replicating regions.
DISCUSSION
Long noncoding RNAs represent more than half of the total of human transcripts. Although it still remains to be shown how many of these lncRNAs are functional, it is suspected that they intervene in most cellular processes. Here we report, for the first time to our knowledge, an lncRNA involved in sister chromatid cohesion.
The establishment of sister chromatid cohesion is tightly linked to DNA replication. The role of CONCR at this specific stage of cell division is consistent with its regulated expression by MYC and also in agreement with its repression by p53 and up-regulation in highly proliferative cells, such as cancer cells. CONCR, similar to other previously described lncRNAs, is a transcriptional target of MYC (Hart et al., 2014; Kim et al., 2015). This highlights how the oncogenic MYC orchestrates a cell-cycle-regulated transcriptional response that includes lncRNAs to promote cell proliferation by diverse mechanisms (Hart et al., 2014; Kim et al., 2015), and opens up the possibility to novel cancer treatments that involve targeting lncRNAs such as CONCR.
CONCR is transcribed divergently from DDX11 promoter region, sharing with the protein-coding gene a common pattern of expression and regulation. Although CONCR does not affect the levels of DDX11 mRNA or protein, we show that both genes are coordinately regulated to participate in the same biological process. This may represent a common feature shared with other lncRNA-protein-coding gene pairs that are transcribed from bidirectional promoters in the genome.
DDX11 is a DNA helicase that links the replication of the lagging DNA strand with the establishment of chromosomal cohesion (Bharti et al., 2014), and it is especially required in order to solve stalled replication forks (Calì et al., 2016). Although we cannot exclude that CONCR has additional activities, our results show that CONCR mainly functions at this level. We show that the interaction between DDX11 and CONCR is required for the proper function of DDX11 in vivo, reflected by its efficient binding to DNA at replicating regions. Interestingly, in vitro assays show that CONCR enhances the ATPase activity of DDX11, acting as an RNA effector for the enzyme. As observed for RNA helicases, RNA seems to stimulate ATPase activity of DDX11; however, the degree of stimulation varies depending on the RNA species and is greater in the presence of CONCR. It remains to be elucidated whether CONCR has a direct effect on DDX11 catalytic activity in vivo, or whether it regulates its function by other means. Like CONCR, two other lncRNAs have been recently shown to act by modulating the activity of RNA and DNA helicases (Han et al., 2014; Huang et al., 2015). We speculate that the mode of action of CONCR may represent a more widely spread mechanism in which lncRNAs interact with DNA/RNA helicases to modulate their activity.
In summary, we have uncovered a so far unknown function for an lncRNA in a critical step of cell division, which impacts genome instability, a major hallmark of cancer cells.
EXPERIMENTAL PROCEDURES
Cell Culture, RNAi, and Transfection
A549 and HCT116 (gift from Dr. Vogelstein) cells were cultured in RPMI 1640 medium (GIBCO) supplemented with 10% fetal calf serum (GIBCO). HeLa cells were cultured in DMEM medium (GIBCO) supplemented with 10% fetal calf serum (GIBCO). Cells were maintained at 37 C in the presence of 5% CO2. For DNA damage, cells were treated with 350 μM 5-fluorouracil (F6627; Sigma) or 500 nM doxorubicin hydrochloride (Sigma D1515). For RNAi, cells were transfected twice, 24 hr apart, using 40 nM siRNA (final concentration) and Lipofectamine 2000 (Invitrogen). All siRNAs used in this study were obtained from Sigma and are listed in Table S6. pcDNA3-His6-DDX11-3xFLAG wild-type and K50R constructs were previously described (Wu et al., 2012) and transfected using Lipofectamine 2000.
RNA Extraction and qRT-PCR
Total RNA was isolated using TRIzol (Sigma). cDNA was generated following DNase I (Invitrogen) treatment using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystem) with random primers. This cDNA was analyzed by qPCR using SYBR Green reagent (Applied Biosystem). Relative quantitation with standard curves was used for RNA quantitation using HPRT as normalizer. All primers used in the study are listed in Table S6.
Nuclear-Cytoplasmic Fractionation
A total of 107 cells were collected by trypsinization and divided into two tubes. One cell pellet represented the whole-cell extract, while the other one was processed for the remaining subcellular fractions. Both pellets were resuspended in 500 μL of buffer A (10 mM Tris-HCl [pH 7.5], 1.5 mM MgCl2, 140 mM NaCl, 0.05% Igepal supplemented with 1× cOmplete Protease Inhibitor Cocktail [Roche] and SUPERaseIN [Ambion] 10 U/mL), incubated for 10 min on ice and kept for subsequent RNA extraction. A total of 500 μL of buffer A containing 50% sucrose was settled at the bottom of a clean tube and the whole-cell extract in buffer A was gently added on top, preventing mixture of the two phases and centrifuged at 12,000 × g for 10 min at 4 °C to obtain nuclear and cytoplasmic fractions. RNA was then extracted from fractions using TRIzol.
RNA FISH
Cells were washed in 1× PBS and fixed with freshly prepared 3.7% formaldehyde for 15 min. For RNA-FISH, fluorescein-labeled Locked Nucleic Acid (LNA) DNA probes were synthesized by Exiqon and hybridized according to manufacturer’s protocol with some modifications. LNA sequences are listed in Table S6. Fixed cells were first incubated with 70% ethanol for 1 hr and then with acetylation buffer (0.1 M triethanol amine, 0.5% [v/v]acetic anhydrid) for 30 min. To avoid unspecific probe binding, warm hybridization buffer (10% dextran sulfate, 50% formamide, 2× SSC) was added and cells incubated for 1 hr at 55 °C. Meanwhile, LNA probes were denatured at 92 °C for 4 min and then mixed with hybridization buffer to a final concentration of 25 nM. The specific probe-target RNA hybridization was performed overnight at 55 °C. The following day, the probes’ residues were eliminated through extensive washes with 2× SSC buffer and fixed cells were incubated with 3% hydrogen peroxide for 30 min. For fluorescein (FAM) detection, cells were first incubated with blocking buffer (10% heat-inactivated goat serum, 0.5% Blocking Reagent [Roche, 11096176001] in PBS-0.5% Tween-20) and then with 1.5 U/ml of specific anti-FAM-POD antibody (Roche, 11426346910) diluted in blocking buffer. After washing three times with 2× SSC solution, the signal was developed through incubation with TSA-Cy3 solution (Perkin Elmer). Antibody residues were eliminated through extensive washing with 4× SSC solution and the slides were prepared for microscope imaging using mounting solution with DAPI.
Mouse Xenograft
Ten million HCT116 cells transfected with CONCR-targeting siRNAs or with a control siRNA were collected and subcutaneously injected in the flanks of 6- to 7-week-old female BALB/c-Rag2/-IL2cc/immunodeficient mice (n = 6 per experimental condition). Tumor size was measured using a precision caliper and tumor volume calculated using the following equation: V = π/6 × width × height × length. Tumor growth was measured every 3 days for 1 month.
Tumor Analysis
Gene expression was determined in colon adenocarcinoma and lung adenocarcinoma RNA-seq data available through the TCGA database (https://gdc-portal.nci.nih.gov). The aligned reads were assigned and quantified using Cufflinks v2.2.1 (Trapnell et al., 2012). CONCR expression was compared in each cancer type between: (1) normal tissue samples and primary tumor samples, and (2) from those tumor samples, individuals bearing mutations in TP53 and those with the wild-type gene. Statistical significance was determined by unpaired Student’s t test. See also Table S2 and Table S4.
Cell Proliferation, Apoptosis, and Cell-Cycle Analyses
Cell proliferation was measured using the CellTiter96 Aqueous Non-Radioactive Cell Proliferation Assay (MTS) kit (Promega) following the manufacturer’s instructions. Apoptosis was measured by Annexin V and 7-AAD staining using the Apoptosis Detection Kit I (BD Biosciences) and a fluorescence-activated cell sorting (FACS) flow cytometer (FACSCalibur, BD Biosciences). For cell-cycle analysis, cells were labeled with propidium iodide and sorted in the FACS-Calibur flow cytometer (BD Biosciences). Data were acquired and analyzed by BD CellQuest and Flow Jo software. G1/S synchronized cells were obtained by double thymidine block procedure; cells were cultured for 16 hr in the presence of 2 mM thymidine, for 9 hr in normal medium, and then again for 16 hr in the presence of 2 mM thymidine. Normal medium was then used for the release and cells were collected at different time points for cell-cycle analysis.
Chromosome Spreads
Chromosome spreads were performed as follows: cells were grown at 37 °C in the presence of 30 ug/ml KaryoMax Colcemid Solution (GIBCO) for 12 hr to enrich mitotic cells. Cells were then harvested by trypsinization and incubated in a hypotonic solution of KCl 0.075M for 30 min at 37 °C. Cells were subsequently fixed with freshly made Carnoy’s buffer (1:3 acetic acid:methanol) for 10 min at room temperature (RT) and pelleted. This fixation step was repeated two times. The suspension of cells was dropped onto a clean slide and stained with Giemsa. Chromosome spreads from individual cells were imaged and scored with regards to the status of sister chromatid. At least 50 metaphases per slide were scored and each experiment was blindly scored twice.
CRISPR-Cas9 Editing
The Streptococcus pyogenes Cas9 (SpCas9) target site in CONCR exon 1 to design the single guide RNA (sgRNA) was found using the CRISPR Design Tool from the Zhang Lab (http://crispr.mit.edu/). Oligonucleotides to clone the guide RNA (Table S6) were then annealed and cloned into the CAS9-containing plasmid pX330 (Cong et al., 2013). The repair template used to insert the Neomycin (Neo)-SV40pA sequence at the site of cleavage was cloned as follows (see also Figure S4 for a schematic). The Neo-SV40pA sequence was amplified by PCR from a pcDNA3 backbone using the primers listed in Table S6. Right and left flanking regions to the cleavage site were amplified by PCR from genomic DNA (A549 cells) using the primers listed in Table S6. The three parts, i.e., right, Neo-SV40pA, and left, were then joined and cloned into pcDNA3 in the following order (KpnI-right-BamHI-Neo-SV40pA-NotI-left-XhoI). The repair template was therefore obtained by KpnI-XhoI digestion of the construct and gel extraction. All constructs were verified by sequencing. pX330-sgRNA and repair template were then co-transfected into A549 cells using Lipofectamine 2000. Cells were grown for 48 hr and then G418 (GIBCO) was added to the culture medium to select for cells with recombined Neomycin sequence. Single cell derived-clones were isolated and expanded. Genomic DNA and total RNA were therefore extracted and used for screening analysis by PCR, sequencing, and qRT-PCR (Figure S4).
RNA Pull-down
RNA pull-down was performed as previously described (Marín-Béjar and Huarte, 2015), except that cell extracts were incubated with biotinylated oligonucleotides with sequence complementarity to CONCR and then with streptavidin magnetic beads. Oligonucleotide sequences are listed in Table S6.
Microarray Hybridization and Data Analysis
Total RNA was isolated using TRIzol. As a last step of the extraction procedure, the RNA was purified with the RNeasy Mini-kit (QIAGEN). Before cDNA synthesis, RNA integrity from each sample was confirmed on Agilent RNA Nano Lab-Chips (Agilent Technologies). The sense cDNA was prepared from 300 ng of total RNA using the AmbionWT Expression Kit. The sense strand cDNA was then fragmented and biotinylated with the Affymetrix GeneChip WT Terminal Labeling Kit (PN900671). Labeled sense cDNA was hybridized to the Affymetrix Human Transcriptome Array 2.0 according to the manufacturer protocols and using GeneChip Hybridization, Wash, and Stain Kit. GeneChips were scanned with the Affymetrix GeneChip Scanner 3000. Both background correction and normalization were done using RMA (Robust Multichip Average) algorithm (Irizarry et al., 2003) using Affymetrix Power Tools. After quality assessment, a filtering process was performed to eliminate low expression probe sets. R and Bioconductor were used for preprocessing and statistical analysis. LIMMA (Smyth, 2004) was used to find the probe sets that showed significant differential expression between experimental conditions. Functional and pathway analyses were performed using GREAT (McLean et al., 2010). See also Table S3.
Supplementary Material
Highlights.
CONCR is a human lncRNA activated by MYC and upregulated in multiple cancer types
CONCR expression is periodic and required for cell division and survival
Inactivation of CONCR causes severe defects in sister chromatid cohesion
CONCR modulates the activity of the helicase DDX11
Acknowledgments
We are most grateful to Dr. Victor Segura and Carmen Ferreira-Espinar for technical assistance. We thank the ENCODE project and The Cancer Genome Atlas database for their valuable datasets. Our research was supported by the European Research Council Starting Grant 281877 and the Spanish Ministry of Science Grants BFU2014-58027-R and SRYC1100I008347XV0. This work was supported in part by the Intramural Research Program of the NIH, National Institute on Aging.
Footnotes
ACCESSION NUMBERS
The accession number for the RNA-seq, ChIP-seq, and microarray data reported in this paper is GEO: GSE81602.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, six figures, and six tables and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2016.06.031.
AUTHOR CONTRIBUTIONS
Conceptualization, F.P.M. and M.H.; Investigation, F.P.M., E.G., O.M.-B., S.K.B., I.R., and J.G.; Formal Analysis, D.J.M.-H., A. Athie, and A. Amadoz; Writing, F.P.M. and M.H.; Resources, R.M.B.; Funding Acquisition, R.M.B. and M.H.; Supervision, M.H.
References
- Bharti SK, Khan I, Banerjee T, Sommers JA, Wu Y, Brosh RM., Jr Molecular functions and cellular roles of the ChlR1 (DDX11) helicase defective in the rare cohesinopathy Warsaw breakage syndrome. Cell Mol Life Sci. 2014;71:2625–2639. doi: 10.1007/s00018-014-1569-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas AK, Johnson DG. Transcriptional and nontranscriptional functions of E2F1 in response to DNA damage. Cancer Res. 2012;72:13–17. doi: 10.1158/0008-5472.CAN-11-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R, Shiekhattar R. Regulation of transcription by long noncoding RNAs. Annu Rev Genet. 2014;48:433–455. doi: 10.1146/annurev-genet-120213-092323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calì F, Bharti SK, Di Perna R, Brosh RM, Jr, Pisani FM. Tim/Timeless, a member of the replication fork protection complex, operates with the Warsaw breakage syndrome DNA helicase DDX11 in the same fork recovery pathway. Nucleic Acids Res. 2016;44:705–717. doi: 10.1093/nar/gkv1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capo-Chichi JM, Bharti SK, Sommers JA, Yammine T, Chouery E, Patry L, Rouleau GA, Samuels ME, Hamdan FF, Michaud JL, et al. Identification and biochemical characterization of a novel mutation in DDX11 causing Warsaw breakage syndrome. Hum Mutat. 2013;34:103–107. doi: 10.1002/humu.22226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange J, Faramarz A, Oostra AB, de Menezes RX, van der Meulen IH, Rooimans MA, Rockx DA, Brakenhoff RH, van Beusechem VW, King RW, et al. Defective sister chromatid cohesion is synthetically lethal with impaired APC/C function. Nat Commun. 2015;6:8399. doi: 10.1038/ncomms9399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellino GI, Cittaro D, Piccioni R, Luzi L, Banfi S, Segalla S, Cesaroni M, Mendoza-Maldonado R, Giacca M, Pelicci PG. Genome-wide mapping of human DNA-replication origins: levels of transcription at ORC1 sites regulate origin selection and replication timing. Genome Res. 2013;23:1–11. doi: 10.1101/gr.142331.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina A, Shin JH, Kim DH, Bermudez VP, Kelman Z, Seo YS, Hurwitz J. Studies with the human cohesin establishment factor, ChlR1. Association of ChlR1 with Ctf18-RFC and Fen1. J Biol Chem. 2008;283:20925–20936. doi: 10.1074/jbc.M802696200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- Fischer M, Steiner L, Engeland K. The transcription factor p53: not a repressor, solely an activator. Cell Cycle. 2014;13:3037–3058. doi: 10.4161/15384101.2014.949083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard H, García-Muse T, Aguilera A. Replication stress and cancer. Nat Rev Cancer. 2015;15:276–289. doi: 10.1038/nrc3916. [DOI] [PubMed] [Google Scholar]
- Gandhi R, Gillespie PJ, Hirano T. Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr Biol. 2006;16:2406–2417. doi: 10.1016/j.cub.2006.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guil S, Esteller M. Cis-acting noncoding RNAs: friends and foes. Nat Struct Mol Biol. 2012;19:1068–1075. doi: 10.1038/nsmb.2428. [DOI] [PubMed] [Google Scholar]
- Han P, Li W, Lin CH, Yang J, Shang C, Nurnberg ST, Jin KK, Xu W, Lin CY, Lin CJ, et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature. 2014;514:102–106. doi: 10.1038/nature13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen RS, Thomas S, Sandstrom R, Canfield TK, Thurman RE, Weaver M, Dorschner MO, Gartler SM, Stamatoyannopoulos JA. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc Natl Acad Sci USA. 2010;107:139–144. doi: 10.1073/pnas.0912402107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart JR, Roberts TC, Weinberg MS, Morris KV, Vogt PK. MYC regulates the non-coding transcriptome. Oncotarget. 2014;5:12543–12554. doi: 10.18632/oncotarget.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JS, Ma W, Mao DY, Benchimol S. p53-Dependent transcriptional repression of c-myc is required for G1 cell cycle arrest. Mol Cell Biol. 2005;25:7423–7431. doi: 10.1128/MCB.25.17.7423-7431.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Thomas B, Flynn RA, Gavzy SJ, Wu L, Kim SV, Hall JA, Miraldi ER, Ng CP, Rigo F, et al. DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions. Nature. 2015;528:517–522. doi: 10.1038/nature16193. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- Inoue A, Li T, Roby SK, Valentine MB, Inoue M, Boyd K, Kidd VJ, Lahti JM. Loss of ChlR1 helicase in mouse causes lethality due to the accumulation of aneuploid cells generated by cohesion defects and placental malformation. Cell Cycle. 2007;6:1646–1654. doi: 10.4161/cc.6.13.4411. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeser MD, Pebernard S, Iggo RD. Regulation of p53 stability and function in HCT116 colon cancer cells. J Biol Chem. 2004;279:7598–7605. doi: 10.1074/jbc.M311732200. [DOI] [PubMed] [Google Scholar]
- Kim T, Jeon YJ, Cui R, Lee JH, Peng Y, Kim SH, Tili E, Alder H, Croce CM. Role of MYC-regulated long noncoding RNAs in cell cycle regulation and tumorigenesis. J Natl Cancer Inst. 2015;107:107. doi: 10.1093/jnci/dju505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A. Cohesin in cancer: chromosome segregation and beyond. Nat Rev Cancer. 2014;14:389–393. doi: 10.1038/nrc3743. [DOI] [PubMed] [Google Scholar]
- Marín-Béjar O, Huarte M. RNA pulldown protocol for in vitro detection and identification of RNA-associated proteins. Methods Mol Biol. 2015;1206:87–95. doi: 10.1007/978-1-4939-1369-5_8. [DOI] [PubMed] [Google Scholar]
- Masai H, Matsumoto S, You Z, Yoshizawa-Sugata N, Oda M. Eukaryotic chromosome DNA replication: where, when, and how? Annu Rev Biochem. 2010;79:89–130. doi: 10.1146/annurev.biochem.052308.103205. [DOI] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parish JL, Rosa J, Wang X, Lahti JM, Doxsey SJ, Androphy EJ. The DNA helicase ChlR1 is required for sister chromatid cohesion in mammalian cells. J Cell Sci. 2006;119:4857–4865. doi: 10.1242/jcs.03262. [DOI] [PubMed] [Google Scholar]
- Peters JM, Nishiyama T. Sister chromatid cohesion. Cold Spring Harb Perspect Biol. 2012;4:4. doi: 10.1101/cshperspect.a011130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Huarte M. To repress or not to repress: this is the guardian’s question. Trends Cell Biol. 2011;21:344–353. doi: 10.1016/j.tcb.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Sabò A, Amati B. Genome recognition by MYC. Cold Spring Harb Perspect Med. 2014;4:4. doi: 10.1101/cshperspect.a014191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, Elble R, Watabe K, Mo YY. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci USA. 2009;106:3207–3212. doi: 10.1073/pnas.0808042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez Y, Segura V, Marín-Béjar O, Athie A, Marchese FP, González J, Bujanda L, Guo S, Matheu A, Huarte M. Genome-wide analysis of the human p53 transcriptional network unveils a lncRNA tumour suppressor signature. Nat Commun. 2014;5:5812. doi: 10.1038/ncomms6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens RV, Colquhoun JM, Green MJ, Molnar CA, Sin DN, Sullivan BJ, Tanzosh EE. Cohesinopathies of a feather flock together. PLoS Genet. 2013;9:e1004036. doi: 10.1371/journal.pgen.1004036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004 doi: 10.2202/1544-6115.1027. Published online February 12 2004 http://dx.doi.org/10.2202/1544-6115.1027. [DOI] [PubMed]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lelij P, Chrzanowska KH, Godthelp BC, Rooimans MA, Oostra AB, Stumm M, Zdzienicka MZ, Joenje H, de Winter JP. Warsaw breakage syndrome, a cohesinopathy associated with mutations in the XPD helicase family member DDX11/ChlR1. Am J Hum Genet. 2010;86:262–266. doi: 10.1016/j.ajhg.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villegas VE, Zaphiropoulos PG. Neighboring gene regulation by antisense long non-coding RNAs. Int J Mol Sci. 2015;16:3251–3266. doi: 10.3390/ijms16023251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Sommers JA, Khan I, de Winter JP, Brosh RM., Jr Biochemical characterization of Warsaw breakage syndrome helicase. J Biol Chem. 2012;287:1007–1021. doi: 10.1074/jbc.M111.276022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


