ABSTRACT
Live attenuated influenza vaccines (LAIV) induce CD8+ T lymphocyte responses that play an important role in killing virus-infected cells. Despite the relative conservation of internal influenza A proteins, the epitopes recognized by T cells can undergo drift under immune pressure. The internal proteins of Russian LAIVs are derived from the master donor virus A/Leningrad/134/17/57 (Len/17) isolated 60 years ago and as such, some CD8+ T cell epitopes may vary between the vaccine and circulating wild-type strains. To partially overcome this issue, the nucleoprotein (NP) gene of wild-type virus can be incorporated into LAIV reassortant virus, along with the HA and NA genes. The present study compares the human CD8+ T cell memory responses to H3N2 LAIVs with the Len/17 or the wild-type NP using an in vitro model.
KEYWORDS: Antigenic escape, influenza, LAIV, nucleoprotein, T cell epitope
Introduction
Immune escape is a major mechanism of influenza virus evolution. The emergence of viruses with random mutations in the antigenic epitopes under the selective pressure of neutralizing antibody, a process termed antigenic drift, has been intensively studied and methods to quantify and visualize these antigenic changes have been devised.1 Less well understood is evolution in the regions of viral proteins acting as epitopes for CD8+ T cells. These T cells provide an important role in clearance of established infections by lysis of infected cells2 and can provide protection against severe disease.3 CD8+ T cells recognize infected cells through the expression of processed viral peptides in the context of class I HLA molecules on the infected cell surface. As these peptide epitopes can be derived not only from the variable surface glycoproteins that are the targets for neutralizing antibody, but also on the conserved internal proteins of the virus, pre-existing CD8+ T cell immunity can be effective against serologically distant influenza A viruses, including zoonotic influenza viruses.4 In addition, human HLA-heterogeneity implies that different peptide epitopes may be available for presentation to CD8+ T cells in different hosts.5
Recently, evidence for positive pressure of host CD8+ T cell immunity on influenza A virus has been observed.6 Epidemiological data suggests that in a prolonged infectious process, especially in immunocompromised individuals,7 viruses arise with mutations in CD8+ T cell epitopes. In addition, mutations in dominant epitopes have been shown to diminish cross-recognition of heterologous viruses by human CD8+ T cells in vitro as shown for evolutionary distant influenza A viruses and, moreover, within an influenza A virus subtype.8
Of the available influenza prophylactic vaccines, live attenuated influenza vaccines (LAIV) have been shown to boost cross-reactive CD8+ T cell responses in children.9,10 LAIV is administered locally by intranasal spray and replication is restricted to the nasal mucosa cells due to the temperature sensitive and cold-adapted phenotype of the virus. In this way the infection mimics natural upper-respiratory influenza infection without or with mild clinical symptoms11 but does not progress to the lower respiratory tract. Depending on preexisted immunity, low-level virus shedding in upper-respiratory tract can be observed for several days after LAIV administration,12 sufficient to trigger pronounced immunity.
The Russian type A LAIVs are produced by reassortment of the seasonal vaccine strain and the attenuated A/Leningrad/134/17/57 (Len/17) master-donor virus (MDV), with the internal proteins being derived from the MDV. As Len/17 is an H2N2 strain isolated in 1957,13 there is a risk of reduced efficiency of LAIV-induced CD8+ T cells against recent influenza A virus infection. Previously we predicted that the cross-reactive MHC-I-restricted nucleoprotein (NP) epitopes of MDV Len/17 had likely evolved in recent human influenza A viruses.14 Accordingly, less than 40% of potential Len/17 NP-specific MHC-I-restricted epitopes were conserved in human influenza A viruses isolated in 2009–2014 (H3N2 and H1N1 subtypes). One strategy to partially overcome this issue, is to incorporate the NP gene of wild-type virus into the LAIV reassortant virus, along with the HA and NA genes.15 This is possible because none of the temperature-sensitive or cold-adaptive mutations in the Len/17 MDV are present in the NP gene and so substitution of this gene will not compromise attenuation.16
In recent murine studies we demonstrated diminished CD8+ T cell responses of LAIVs with classical Len/17 NP compared to LAIVs containing the wild-type H1N1 and H7N9 nucleoproteins.14,17 We also showed impaired in vitro cytotoxicity of cells from mice immunized with H7N9 LAIV Len/17 NP against wild-type infected target cells14 or pdmH1N1 wild-type NP366–374 peptide pulsed target cell.17 In the LAIV H1N1 study, there was diminished clearance of the H1N1 challenge virus in mice immunized by LAIV with classical Len/17 NP in contrast to mice immunized with LAIV expressing wild-type NP. In both H1N1 and H7N9 LAIVs, the immunodominant CD8+ T cell epitope for C57BL mice (NP366–374) differed between Len/17 and H1N1 or H7N9 nucleoproteins which may have impacted on CD8+ T cell efficiency.
Despite evidence for the impact of CD8+ T cell epitope variation on viral clearance in the murine model of LAIV vaccination, whether these changes will alter human CD8+ T cell anti-influenza immune responses is unknown. In this study, we compared the variation of experimentally-determined human class I HLA-restricted epitopes in NPs of Len/17 MDV and recent H3N2 isolate A/Hong Kong/4801/2014 (HK). Using reverse genetics, H3N2 LAIV candidates bearing Len/17 NP (LAIV 6:2) or wild-type NP (LAIV 5:3) were generated. By using human T cell cultures the reactivity of CD8+ T cells against several NP epitopes that differed between LAIV 6:2 and LAIV 5:3 was estimated.
Results and discussion
Influenza H3N2 viruses are characterized by more rapid antigenic evolution compared to the H1N1 influenza A viruses, and as a result the H3N2 vaccine component needs to be updated on a regular basis.18 We explored the strategy of inducing enhanced T cell immune responses by incorporating the NP gene of recent H3N2 A/Hong Kong/4801/2014 (HK) virus into the LAIV genome, along with HA and NA genes (i.e. LAIV 5:3 genome composition)
To predict the impact of nucleoprotein diversity on the epitope-specific human CD8+ T cell response, the class I-restricted epitopes published in IEDB (on December 2016) were mapped on the LAIVs nucleoprotein sequences (SUP.FIG 3.pdf). There were 209 experimentally estimated class I HLA-restricted influenza A NP epitopes that were recognized by human hosts. The number of epitopes was restricted further to 96 by exclusion of non-confirmed queries (refer to Materials and Methods: CD8+ T cell epitope database analysis). LAIV 5:3 and 6:2 nucleoprotein sequence diversity is presented in Table 1. From a total of 37 substitutions, there were 28 (more than 75%) located in the selected class I-restricted epitopes. Still, from the list of relevant experimentally-estimated HLA-I-restricted epitopes, more 70% of epitopes were conserved between H2N2 MDV Len/17 and modern wild-type H3N2 nucleoproteins.
Table 1.
Diversity of the experimentally estimated CTL epitopes between H2N2 A/Leningrad/134/17/57 (Len/17) and H3N2 A/Hong Kong/4801/2014 (HK) nucleoproteins.
| Substitution position, aa | Len17 letter | Position within epitope | HK letter | Epitope sequence in Len/17 | Epitope sequence in HK | Predicted cleavage site involved, (position in epitope) | EpitopeID | HLA restriction |
|---|---|---|---|---|---|---|---|---|
| 18 | E | 2 | D | GERQNATEI | GDRQNATEI | 19421 | B*18:01 B*40:02 B*45:01 | |
| 52 | Y | 9 | H | CTELKLSDY | CTELKLSDH | HK-/Len+(9) | 7136 | A*01:01 A1 |
| 52 | Y | 5 | H | KLSDYEGRL | KLSDHEGRL | 32157 | A*02:01 | |
| 65 | K | 1 | R | RMVLSAFDER | KMVLSAFDER | 97614 | A3 | |
| 98 | K | 8 | R | KTGGPIYRR | KTGGPIYKR | 33682 | A68 | |
| 101 | D | G | ||||||
| 127 | D | E | ||||||
| 131 | A | S | ||||||
| 136 | M | I | ||||||
| 146 | T | 7 | A | HSNLNDTTY | HSNLNDATY | 24819 | A*01:01 A*26:01 A*30:02 | |
| 146 | T | 2 | A | DTTYQRTRALVR | DATYQRTRALVR | 181194 | A*68:01 | |
| 186 | V | 17 | I | STLPRRSGAAGAAVKGV | STLPRRSGAAGAAVKGI | 97664 | class I | |
| 197 | I | 9 | V | MVMELIRMI | MVMELIRMV | HK+/Len-(9) | 42974 | A*02:01 |
| 217 | I | 7 | S | NGRKTRIAYERMCNILKG | NGRKTRSAYERMCNILKG | HK-/Len+(7) | 145915 | B*15:01 |
| 217 | I | 1 | S | IAYERMCNILKGKFQTAA | SAYERMCNILKGKFQTAA | HK-/Lan+(1) | 145824 | B*15:01 |
| 239 | M | 11 | V | KFQTAAQRAMMDQVRESR | KFQTAAQRAMVDQVRESR | HK+/Len-(14,15) | 97417 | class I |
| 280 | V | 8 | A | KSCLPACVY | KSCLPACAY | HK+/Len-(4,6) | 33285 | A*01:01 A*30:02 |
| 280 | V | 6 | A | CLPACVYGP | CLPACAYGP | HK+/Len-(2,4) | 6615 | A*02:01 A*02:02 A*02:03 A*02:06 A*68:02 |
| 286 | A | S | ||||||
| 290 | E | D | ||||||
| 312 | V | I | ||||||
| 334 | N | 8 | H | QLVWMACNSAA | QLVWMACHSAA | 97583 | A2 | |
| 343 | V | 6 | L | FEDLRVSSF | FEDLRLLSF | 97298 | B44 | |
| 344 | S | 7 | L | FEDLRVSSF | FEDLRLLSF | 97298 | B44 | |
| 353 | I | 17 | S | AFEDLRVSSFIRGTKVI | AFEDLRLLSFIRGTKVS | Hk-/Len+(17) | 97180 | class I |
| 373 | T | 1 | N | TMESSTLEL | NMGSSTLEL | Hk-/Len+(2) | 3078 | A*02:01 |
| 375 | E | 3 | G | TMESSTLEL | NMGSSTLEL | Hk-/Len+(2) | 3078 | A*02:01 |
| 384 | R | 5 | G | ELRSRYWAI | ELRSGYWAI | 13263 | B*08:01 B8 | |
| 384 | R | 2 | G | SRYWAIRTR | SGYWAIRTR | 60867 | A*02:01 B*27:05 B*27:09 B27 B8 | |
| 406 | I | 3 | T | GQISVQPTFS | GQTSVQPTFS | 145805 | B*15:01 | |
| 421 | D | 4 | E | LPFDKPTIM | LPFEKSTIM | 38466 | B*07:02 B*35:01 | |
| 423 | P | 6 | S | LPFDKPTIM | LPFEKSTIM | 38466 | B*07:02 B*35:01 | |
| 421 | D | 3 | E | PFDKPTIMAAF | PFEKSTIMAAF | HK-/Len+(10,11) | 176373 E3D S5P | |
| 423 | P | 5 | S | PFDKPTIMAAF | PFEKSTIMAAF | HK-/Len+(10,11) | 176373 E3D S5P | |
| 433 | A | T | ||||||
| 454 | E | D | ||||||
| 459 | Q | 2 | R | FQGRGVFEL | FRGRGVFEL | 144292 | A*02:01 A*02:02 A*02:03 A*02:06 |
Cleavage cites were predicted by IEDB netChop algorithm: presence (+) or absence (-) of epitope cleavage site at position (brackets) was shown. EpitopeID is an epitope identification number in Immune Epitope Data Base (www.iedb.org)
Improper processing of antigenic peptides can occur if the proteasome cleavage site is mutated, resulting in CTL immune escape.19 We estimated the risks of cleavage sites mutations within diversified epitopes by computational cleavage site prediction modeling. From the proteasomal cleavage prediction model, 5 of the 28 substitutions were associated with the C-terminal cleavage site needed for proper epitope generation and 6 of the 28 substitutions were involved in generating a cleavage site within the epitope sequence (Table 1).
For the assessment of cross-reactivity of epitope-specific CD8+ T cells, virus-specific T-cells were expanded from PBMCs of HLA-A*01:01 and HLA-B*35:01 positive donors by in vitro stimulation with 6:2 and 5:3 H3N2 LAIVs for 10 days. Both 5:3 and 6:2 LAIVs successfully re-stimulated virus-specific CD8+ T cell proliferation from donor PBMCs (SUP.FIG 4.pdf). Of note, there was less virus-specific T cell expansion in samples of HLA B*35:01 donors, compared to HLA A*01:01 donors.
Pairs of immunodominant HLA-A*01:01-restricted NP44–52 (Len/17: CTELKLSDY, HK: CTELKLSDH) and HLA-B*35:01-restricted NP418–426 (Len/17: PFDKPTIMAAF, HK: PFEKSTIMAAF) epitopes were selected to study in vitro for human CD8+ T cell cross-reactivity due to their high immunogenicity in our previous experiments,20-22 and also because they differed between Len/17 and HK viruses. HK nucleoprotein (LAIV 5:3) contains Y9H substitution in NP44–52, and two D3E and P5S substitutions in NP418–426. The cross-reactivity of HLA-A*01:01-restricted CD8+ T cells to NP44–52 variants is shown in Fig. 1. Only in LAIV 6:2-stimulated PBMC samples were NP44–52 specific CD8+ T cells detected, particularly those reactive against Len/17-NP44–52. However, these LAIV 6:2-stimulated T cells did not cross-react with the HK-NP44–52 peptide variant. The failure to detect NP44–52-specific CD8+ T cells in LAIV 5:3-stimulated PBMC cultures, suggests that the substitution in the class I-anchoring position 9 in HK NP44–52 epitope completely abrogated the capability to stimulate NP44–52-specific CD8+ T cells. Interestingly, these results were complemented by the low probability of proteasomal cleavage site occurrence in position 52 of HK nucleoprotein (C-terminal for NP44–52 epitope) (Table 1). These data suggest that Len/17-NP44–52-specific CD8+ T cells might be ineffective against circulating wild-type H3N2 influenza viruses. Moreover, Len/17-NP44–52-specific CD8+ T cells generation by 6:2 LAIV vaccination might be antigenically redundant for H3N2 influenza prophylaxis.
Figure 1.
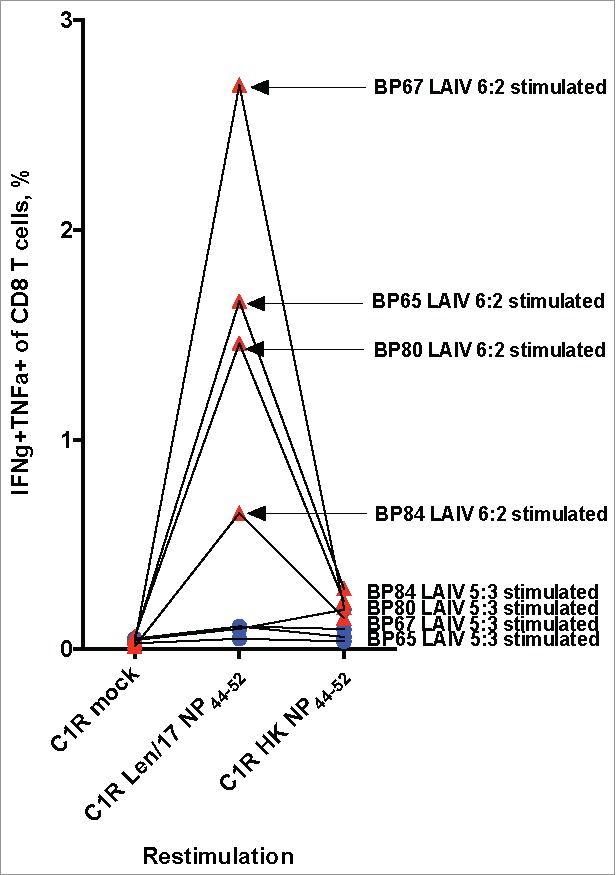
Cross-reactivity of NP44–52 specific CD8 T cells. PMBC samples of HLA-A*01:01-positive donors were stimulated with 10 MOI LAIV 5:3 (Blue circle) or 10 MOI LAIV 6:2 (Red triangle) for 10 days. The expression of cytokines were estimated by ICS assay after 6 hours of in vitro cultures re-stimulation with Len/17 or HK NP44–52 peptide loaded C1R cells.
Of the two HLA-B*35:01 positive donors, the BP75 donor samples showed no expansion of NP418–426-specific CD8+ T cells upon LAIVs stimulation (Fig. 2). However, for the BP25 donor samples, there was robust homologous reactivity for their cognate peptides. Both Len/17-NP418–426 and HK-NP418–426-specific CD8+ T cells expanded well after LAIV 6:2 and 5:3 virus stimulations respectively. However, there was diminished cross-reactivity of these CD8+ T cells against the heterologous NP418–426-peptide, both for LAIV 6:2- and LAIV 5:3-stimulated PBMC cultures. Thus, HK-NP418–426-specific CD8+ T cells that were generated from the LAIV 5:3 stimulated cultures failed to recognize Len/17-NP418–426 peptide-loaded C1Rs and Len/17-NP418–426-specific CD8+ T cells from LAIV 6:2 stimulated cultures had diminished reactivity to HK-NP418–426 epitope. The two substitutions in the NP418–426 epitope in non-anchoring positions 3 and 5 might have changed the MHC-I-peptide complex recognition by TCR dramatically. These data suggest that immunization with classical LAIVs 6:2 might be less effective in generating NP418–426 epitope-specific CD8+ T cells targeted to recent H3N2 viruses.
Figure 2.
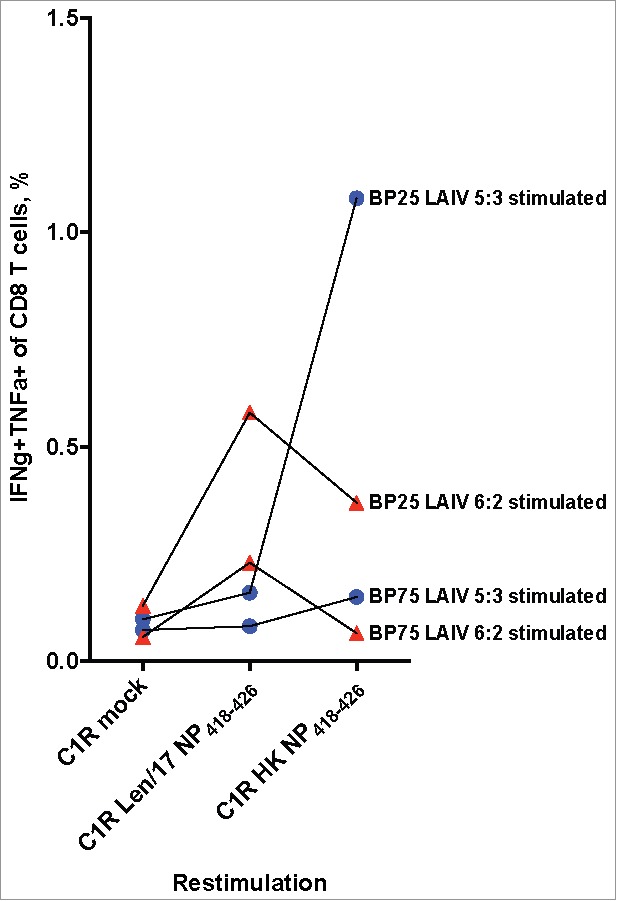
Cross-reactivity of NP418–426 specific CD8 T cells. PMBC samples of HLA-B*35:01-positive donors were stimulated with 10 MOI LAIV 5:3 (Blue circle) or 10 MOI LAIV 6:2 (Red triangle) for 10 days. The expression of cytokines were estimated by ICS assay after 6 hours of in vitro cultures re-stimulation with Len/17 or HK NP44–52 peptide loaded C1R cells.
Materials and methods
Viruses. A pair of H3N2 LAIV reassortant viruses with 6:2 and 5:3 genome compositions were generated by the means of reverse genetics from Len/17 MDV and A/Hong Kong/4801/2014 (H3N2) wild-type virus genes. Six genes coding for internal and non-structural proteins of A/Leningrad/134/17/57 (H2N2) were previously cloned into RG vectors.16 HA, NA and NP genes of H3N2 virus A/Hong Kong/4801/2014 were cloned into dual-promoter plasmids and rg-viruses were rescued in MDCK/293T cells, as previously described.16 Both viruses were fully sequenced and found to be identical apart from the NP gene by using ABI BigDye® Terminator v1.1 Cycle Sequencing kits and capillary based 3500 Series Genetic Analyzer (ThermoFisher scientific).23 Viruses were grown in 10 day-old embryonated chicken eggs at 33°C. Allantoic fluid was harvested, clarified by centrifugation and stored at −70°C. Viral infectivity was quantified as spots/ml by ViroSpot assay.24
Donors. Buffy packs from healthy donors were obtained from the Australian Red Cross Blood Service (ARCBS) (Victoria, Australia). HLA class I molecular typing was performed by the Victorian Transplantation and Immunogenetics Service (ARCBS, West Melbourne, Victoria, Australia). Four HLA-A*01:01-positive and two HLA-B*35:01 donors were used in the study (SUP.TABLE1.pdf). Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll-Paque (GE Healthcare, Uppsala, Sweden) density gradient centrifugation and cryopreserved at 196°C until required.
CD8+ T cell epitope database analysis. The Immune Epitope Data Base (IEDB, www.iedb.org) was utilized as a source of consolidated data about experimentally estimated Influenza NP epitopes.25 The local dataset was generated by IEDB search query: Organism: Influenza A virus (ID:11320, influenza A) AND Antigen: Nucleoprotein [P03466] (Nucleoprotein Influenza A virus) AND MHC Restriction Type: Class I AND Host: Homo sapiens (human). The data was further processed by Microsoft Access database. The IEDB netChop algorithm was utilized for proteasomal c-terminal cleavage sites prediction with default settings.26 For each epitope entry, IEDB generates statistics of frequency of experimental data with positive (assay positive) and negative (assay negative) immune responses. To achieve stricter epitope inclusion criteria, the local dataset was restricted to entries with «assay positive» – «assay negative» > 1. The resulting HLA class-I-restricted epitopes were mapped on the NP protein sequence by Geneious 6.0 software (Biomatters Ltd).
Virus-specific T cell expansion in vitro. PBMCs were thawed and seeded in complete RPMI-1640 medium containing 2 mM L-glutamine, 1 mM MEM sodium pyruvate, 100µM MEM non-essential amino acids, 5 mM HEPES buffer solution, 55 µM 2-mercaptoethanol and 100 U/ml penicillin/100 µg/ml streptomycin, with 10% heat-inactivated fetal bovine serum (cR-10) at 3–5*106 cells/ml in 24-well plates. Media reagents were from Gibco (Thermo Fisher Scientific, Scoresby, Victoria, Australia). One tenth of the cells was washed in complete RPMI-1640 medium without serum (cR-0) and infected with LAIVs at 10 MOI for 1 hour at 37°C/5% CO2. These infected “stimulator” cells were washed and added to uninfected “responder” cells at 1 to 9 ratio. Cultures were maintained for 10 days at 37°C/5% CO2 with daily observation. After every 3 days of culturing, 10 U/ml of human recombinant IL-2 (Roche Diagnostics, Mannheim, Germany) were added. If necessary, cultures were split at a 1:2 ratio. Harvested cells were counted and washed in cR-10 prior to intracellular staining (ICS) analysis. Percent of recovery was estimated as a live cell count ratio of day 10 to day 0 values.
ICS assay. C1R.A*01:01 and C1R.B*35:01 B-lymphoblastoid cell lines (B-LCLs) derived from the class 1 reduced (C1R) cell line27 were kindly provided by Dr. Nicole Mifsud (Monash University, Clayton, Victoria, Australia). These cells were used as antigen presenting cells for epitope-specific T cell activation. C1R cell lines were maintained in cR-10 at cell density 0.5–1.0 × 106 cells/ml at 35°C with selection reagents 0.3 mg/ml Hygromycin B (Gibco) for C1R.A*01:01 cells and 0.5 mg/ml Geneticin (G418, Gibco) for C1R.B*35:01 cells. Preliminary infection of C1R cells showed similar infectious activity at 33°C and 35°C for both 6:2 and 5:3 H3N2 LAIV strains, hence for all the experiments C1R cells were infected with 10 MOI of LAIVs at 35°C/5% CO2 for 1 hour in cR-0 followed by 16h in cR-10. Intracellular nucleoprotein antigen staining was performed using anti-influenza A NP antibodies conjugated with FITC (GeneTex, Irvine, CA, USA) to study the infection of C1R cells with LAIV viruses (SUP.FIG 1.pdf). In peptide-pulsing experiments, C1R cells were pulsed with 10 uM of the selected peptides in cR-0 for 1 h at 37°C with occasional mixing. Next, C1R stimulator cells were washed twice and resuspended in cR-10.
Day 10 PBMC cultures and antigen-loaded C1R cells were mixed at a responder to stimulator ratio of 2:1 for 6h in cR-10 with GolgiPlug (BD Biosciences) at 37°C. Samples were processed for ICS staining according to BD Cytofix/Cytoperm (BD Biosciences) protocol with Live/Dead Fixable Aqua blue stain (Invitrogen), CD4-PE, CD8-PerCP5.5, CD3-PC7, CD14-APC-H7, CD19-APC-H7, IFN-gamma-V450 (BD Biosciences) and NP-FITC antibodies. Flow cytometric analyses were performed on a BD FACS Canto II flow cytometer (BD Biosciences). The gating strategy for virus-specific CD8+ T cells (IFNγ and TNFα positive) is presented in supplementary materials (SUP.FIG 2.pdf).
Statistics. Descriptive statistical analysis and graphical data representation were performed with Prism 6 software (GraphPad Software).
Conclusions
It is generally accepted that influenza T cell epitopes are more conserved in comparison to B cell epitopes. Therefore, little attention is being paid to the evolution of known MHC-I-restricted epitopes that may lead to differences between these sequences in vaccine and wild-type viruses. In this study we found that at least 24 human HLA class I-restricted epitopes were different between the Len/17 MDV and recent H3N2 influenza viruses. These are about 30% of experimentally–estimated HLA-I-restricted nucleoprotein epitopes. The results of our in vitro study confirmed that classical 6:2 LAIV (with MDV nucleoprotein) might be inefficient in inducing appropriate epitope-specific CD8+ T cell immunity targeted to circulating wild-type viruses, at least for two immunodominant HLA-A1-restricted NP44–52 and HLA-B35-restricted NP418–426 epitopes. These results illustrate the necessity of an LAIV nucleoprotein “actualization” strategy to improve the efficiency of the epitope-specific CD8+ T cell response against recent influenza viruses. Further studies are necessary to assess how particular epitope structure changes might influence the overall T cell response and the outcome of influenza infection.
Supplementary Material
Funding Statement
The study was supported by Russian Science Foundation Grant №14–15–00034.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We sincerely appreciate Dr. Kanta Subbarao, Dr. Ian Barr, Dr. Karen Laurie, Dr. Kok Fei Chan, Louise Caroline and all WHO CCRR Influenza staff for the consultations and the virus strains management at The Peter Doherty Institute for Infection & Immunity, Melbourne, Australia.
References
- 1.Smith DJ, Lapedes AS, de Jong JC, Bestebroer TM, Rimmelzwaan GF, Osterhaus ADME, Fouchier RAM. Mapping the Antigenic and Genetic Evolution of Influenza Virus. Science. 2004;305:371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- 2.Moskophidis D, Kioussis D. Contribution of Virus-specific CD8(+) Cytotoxic T Cells to Virus Clearance or Pathologic Manifestations of Influenza Virus Infection in a T Cell Receptor Transgenic Mouse Model. J Exp Med. 1998. 188:223–232. doi: 10.1084/jem.188.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rimmelzwaan GF, Fouchier RA, Osterhaus AD. Influenza virus-specific cytotoxic T lymphocytes: a correlate of protection and a basis for vaccine development. Curr Opin Biotechnol. 2007;18:529–536. doi: 10.1016/j.copbio.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Wang M, Lamberth K, Harndahl M, Roder G, Stryhn A, Larsen MV, Nielsen M, Lundegaard C, Tang ST, Dziegiel MH, et al;. CTL epitopes for influenza A including the H5N1 bird flu; genome-, pathogen-, and HLA-wide screening. Vaccine. 2007;25:2823–2831. doi: 10.1016/j.vaccine.2006.12.038. [DOI] [PubMed] [Google Scholar]
- 5.Alexander J, Bilsel P, del Guercio M-F, Marinkovic-Petrovic A, Southwood S, Stewart S, Ishioka G, Kotturi MF, Botten J, Sidney J, et al;. Identification of broad binding class I HLA supertype epitopes to provide universal coverage of influenza A virus. Human Immunology. 2010;71:468–474. doi: 10.1016/j.humimm.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Machkovech HM, Bedford T, Suchard MA, Bloom JD. Positive Selection in CD8+ T-Cell Epitopes of Influenza Virus Nucleoprotein Revealed by a Comparative Analysis of Human and Swine Viral Lineages. J Virol. 2015;89:11275–11283. doi: 10.1128/JVI.01571-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valkenburg SA, Quiñones-Parra S, Gras S, Komadina N, McVernon J, Wang Z, Halim H, Iannello P, Cole C, Laurie K, et al;. Acute emergence and reversion of influenza A virus quasispecies within CD8+ T cell antigenic peptides. Nat Commun. 2013;4. doi: 10.1038/ncomms3663. [DOI] [PubMed] [Google Scholar]
- 8.Rimmelzwaan GF, Kreijtz JH, Bodewes R, Fouchier RA, Osterhaus AD. Influenza virus CTL epitopes, remarkably conserved and remarkably variable. Vaccine. 2009;27:6363–6365. doi: 10.1016/j.vaccine.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 9.Hoft DF, Babusis E, Worku S, Spencer CT, Lottenbach K, Truscott SM, Abate G, Sakala IG, Edwards KM, Creech CB, et al;. Live and inactivated influenza vaccines induce similar humoral responses, but only live vaccines induce diverse T-cell responses in young children. J Infect Dis. 2011;204:845–853. doi: 10.1093/infdis/jir436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohn KGI, Zhou F, Brokstad KA, Sridhar S, Cox RJ. Boosting of Cross-Reactive and Protection-Associated T Cells in Children After Live Attenuated Influenza Vaccination. J Infect Dis. 2017;215:1527–1535. doi: 10.1093/infdis/jix165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brooks WA, Zaman K, Lewis KDC, Ortiz JR, Goswami D, Feser J, Sharmeen AT, Nahar K, Rahman M, Rahman MZ, Barin B, Yunus M, Fry AM, Bresee J, Azim T, Neuzil KM. Efficacy of a Russian-backbone live attenuated influenza vaccine among young children in Bangladesh: a randomised, double-blind, placebo-controlled trial. Lancet Glob Health. 2016;4:e946−954. doi: 10.1016/S2214-109X(16)30200-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Block SL, Yogev R, Hayden FG, Ambrose CS, Zeng W, Walker RE. Shedding and immunogenicity of live attenuated influenza vaccine virus in subjects 5-49 years of age. Vaccine. 2008;26:4940–4946. doi: 10.1016/j.vaccine.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 13.Alexandrova GI, Smorodintsev AA. Obtaining of an additionally attenuated vaccinating cryophilic influenza strain. Revue Roumaine d'Inframicrobiologie. 1965;2:179–186. [Google Scholar]
- 14.Isakova-Sivak I, Korenkov D, Smolonogina T, Tretiak T, Donina S, Rekstin A, Naykhin A, Shcherbik S, Pearce N, et al;. Comparative studies of infectivity, immunogenicity and cross-protective efficacy of live attenuated influenza vaccines containing nucleoprotein from cold-adapted or wild-type influenza virus in a mouse model. Virology. 2017;500:209–217. doi: 10.1016/j.virol.2016.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Isakova-Sivak I, Korenkov D, Rudenko L. Reassortant viruses for influenza vaccines: is it time to reconsider their genome structures? Expert Rev Vaccines. 2016;15:565–567. doi: 10.1586/14760584.2016.1158109. [DOI] [PubMed] [Google Scholar]
- 16.Isakova-Sivak I, Chen LM, Matsuoka Y, Voeten JT, Kiseleva I, Heldens JG, den Bosch H, Klimov A, Rudenko L, Cox NJ, et al;. Genetic bases of the temperature-sensitive phenotype of a master donor virus used in live attenuated influenza vaccines: A/Leningrad/134/17/57 (H2N2). Virology. 2011;412:297–305. doi: 10.1016/j.virol.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 17.Rekstin A, Isakova-Sivak I, Petukhova G, Korenkov D, Losev I, Smolonogina T, Tretiak T, Donina S, Shcherbik S, Bousse T, et al;. Immunogenicity and Cross Protection in Mice Afforded by Pandemic H1N1 Live Attenuated Influenza Vaccine Containing Wild-Type Nucleoprotein. BioMed Research International. 2017;2017:11. doi: 10.1155/2017/9359276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holmes EC. Evolution in health and medicine Sackler colloquium: The comparative genomics of viral emergence. Proc Natl Acad Sci U S A. 2010;107 Suppl 1:1742–1746. doi: 10.1073/pnas.0906193106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beekman NJ, van Veelen PA, van Hall T, Neisig A, Sijts A, Camps M, Kloetzel P-M, Neefjes JJ, Melief CJ, Ossendorp F. Abrogation of CTL Epitope Processing by Single Amino Acid Substitution Flanking the C-Terminal Proteasome Cleavage Site. The Journal of Immunology. 2000;164:1898–1905. doi: 10.4049/jimmunol.164.4.1898. [DOI] [PubMed] [Google Scholar]
- 20.Quinones-Parra S, Grant E, Loh L, Nguyen TH, Campbell KA, Tong SY, Miller A, Doherty PC, Vijaykrishna D, Rossjohn J, et al;. Preexisting CD8+ T-cell immunity to the H7N9 influenza A virus varies across ethnicities. Proc Natl Acad Sci U S A. 2014;111:1049–1054. doi: 10.1073/pnas.1322229111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gras S, Kedzierski L, Valkenburg SA, Laurie K, Liu YC, Denholm JT, Richards MJ, Rimmelzwaan GF, Kelso A, Doherty PC, et al;. Cross-reactive CD8+ T-cell immunity between the pandemic H1N1–2009 and H1N1–1918 influenza A viruses. Proc Natl Acad Sci U S A. 2010;107:12599–12604. doi: 10.1073/pnas.1007270107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valkenburg SA, Josephs TM, Clemens EB, Grant EJ, Nguyen TH, Wang GC, Price DA, Miller A, Tong SY, Thomas PG, et al;. Molecular basis for universal HLA-A*0201-restricted CD8+ T-cell immunity against influenza viruses. Proc Natl Acad Sci U S A. 2016;113:4440–4445. doi: 10.1073/pnas.1603106113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.WHO information for molecular diagnosis of influenza virus - update, on WHO. http://www.who.int/influenza/gisrs_laboratory/molecular_diagnosis/en/. Accessed July2017
- 24.van Baalen CA, Jeeninga RE, Penders GH, van Gent B, van Beek R, Koopmans MP, Rimmelzwaan GF. ViroSpot microneutralization assay for antigenic characterization of human influenza viruses. Vaccine. 2017;35:46–52. doi: 10.1016/j.vaccine.2016.11.060. [DOI] [PubMed] [Google Scholar]
- 25.Vita R, Overton JA, Greenbaum JA, Ponomarenko J, Clark JD, Cantrell JR, Wheeler DK, Gabbard JL, Hix D, Sette A, et al;. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015;43:D405−412. doi: 10.1093/nar/gku938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nielsen M, Lundegaard C, Lund O, Kesmir C. The role of the proteasome in generating cytotoxic T-cell epitopes: insights obtained from improved predictions of proteasomal cleavage. Immunogenetics. 2005;57:33–41. doi: 10.1007/s00251-005-0781-7. [DOI] [PubMed] [Google Scholar]
- 27.Zemmour J, Little AM, Schendel DJ, Parham P. The HLA-A,B “negative” mutant cell line C1R expresses a novel HLA-B35 allele, which also has a point mutation in the translation initiation codon. J Immunol. 1992;148:1941–1948. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


