Abstract
Eukaryotic elongation factor 1α (eEF1A) can be post-translationally modified by the addition of phosphorylglycerylethanolamine (PGE). [14C]Ethanolamine was incorporated into the PGE modification, and with carrot (Daucus carota L.) suspension culture cells, eEF1A was the only protein that incorporated detectable quantities of [14C]ethanolamine (Ransom et al., 1998). When 1 mm CaCl2 was added to microsomes containing [14C]ethanolamine-labeled eEF1A ([14C]et-eEF1A), there was a 60% decrease in the amount of [14C]et-eEF1A recovered after 10 min. The loss of endogenous [14C]et-eEF1A was prevented by adding EGTA. Recombinant eEF1A, which did not contain the PGE modification, also was degraded by microsomes in a Ca2+-regulated manner, indicating that PGE modification was not necessary for proteolysis; however, it enabled us to quantify enodgenous eEF1A. By monitoring [14C]et-eEF1A, we found that treatment with phospholipase D or C, but not phospholipase A2, resulted in a decrease in [14C]et-eEF1A from carrot microsomes. The fact that there was no loss of [14C]et-eEF1A with phospholipase A2 treatment even in the presence of 1 mm Ca2+ suggested that the loss of membrane lipids was not essential for eEF1A proteolysis and that lysolipids or fatty acids decreased proteolysis. At micromolar Ca2+ concentrations, proteolysis of eEF1A was pH sensitive. When 1 μm CaCl2 was added at pH 7.2, 35% of [14C]et-eEF1A was lost; while at pH 6.8, 10 μm CaCl2 was required to give a similar loss of protein. These data suggest that eEF1A may be an important downstream target for Ca2+ and lipid-mediated signal transduction cascades.
As a critical protein for cell survival, elongation factor 1 alpha (eEF1A) has generated a lot of interest in recent years. eEF1A is a multifunctional protein that is essential for protein translation (Browning, 1996; Merrick and Hershey, 1996). In addition, it binds and bundles actin (Demma et al., 1990; Edmonds et al., 1995), activates phosphatidylinositol (PI) 4-kinase (PI4K) (Yang et al., 1993), binds (Durso and Cyr, 1994; Durso et al., 1996) and severs microtubules (Shiina et al., 1994), and binds calmodulin (Durso and Cyr, 1994; Kaur and Ruben, 1994; Moore et al., 1998) and Ca2+/calmodulin-dependent protein kinases (Wang and Poovaiah, 1999). Putative roles in the ubiquitin-dependent protein degradation pathway (Gonen et al., 1994) and apoptosis (Duttaroy et al., 1998) have also been described.
eEF1A is a soluble protein that is found associated with the cytoskeleton (Demma et al., 1990; Dharmawardhane et al., 1991; Collings et al., 1994; Clore et al., 1996), protein bodies (Clore et al., 1996), and microtubules in situ (Ohta et al., 1990; Hasezawa and Nagata, 1993; Durso et al., 1996; Hasezawa et al., 1997). eEF1A fractionates with the endoplasmic reticulum (Hayashi et al., 1989), cytoskeleton (Yang et al., 1990; Tan and Boss, 1992; Durso and Cyr, 1994; Shiina et al., 1994; Ransom et al., 1998), and plasma membranes (Ransom et al., 1998). There are several lines of evidence indicating that the function and distribution of eEF1A are sensitive to changes in cytosolic pH (Condeelis, 1995; Liu et al., 1996a). At a cellular pH above 7.0, eEF1A loses its ability to bundle F-actin (Edmonds et al., 1995). The decrease in actin bundling is consistent with observed changes in eEF1A distribution in vivo (Aerts et al., 1987; van Duijn and Inouye, 1991). Liu et al. (1996b) also discovered that the bundling of F-actin by eEF1A precludes it from interaction with aminoacyl-tRNA for protein translation. Thus, in response to an increase in cytosolic pH, a change in the binding of eEF1A to F-actin would decrease actin bundling while at the same time permitting eEF1A to bind aminoacyl-tRNA and facilitate protein synthesis.
Changes in cytosolic pH and Ca2+ have also been implicated as critical factors affecting cytoskeletal structure during cell elongation in plants. For example, in alfalfa root hairs, exposure to nod factor causes changes in pH (Ehrhardt et al., 1992; Felle et al., 1996), Ca2+ (Ehrhardt et al., 1996), and actin depolymerization (Cárdenas et al., 1998). In pollen tubes, Ca2+ gradients correlate positively with tip growth (Pierson et al., 1994, 1996; Holdaway-Clarke et al., 1997). Holdaway-Clarke et al. (1997) suggest that there are coordinated changes in Ca2+ and cytoskeleton in the growing pollen tube. Subsequent work indicates that in the region of the pollen tube where actin filaments are being depolymerized as the tip is extended, the pH increases to 7.2 (Feijó et al., 1999).
In previous work we characterized the phosphorylglycerylethanolamine (PGE) modification of carrot (Daucus carota L.) cell eEF1A (Ransom et al., 1998). PGE-modified eEF1A in carrot suspension culture cells was present in soluble, microsomal, and plasma membrane fractions. It co-purified with an F-actin-enriched fraction and bound F-actin in vitro. Although the PGE modification provides a possible mechanism for eEF1A to interact with lipids or membranes in the cell, the function of the PGE modification in vivo is not known.
We wanted to determine if PGE-modified eEF1A is sensitive to Ca2+, pH, and lipases. We reasoned that lipases would hydrolyze the lipid moiety and release eEF1A from a hydrophobic (lipid) environment. We found that both Ca2+ phospholipase C and D decrease the amount of eEF1A associated with a carrot microsomal fraction; however, the loss does not result from the release of a soluble [14C]et-eEF1A, but, rather, from proteolysis. PGE modification provided a means for quantifying the loss of eEF1A, but was not essential for proteolysis. Proteolysis of eEF1A is sensitive to physiological changes in pH and could be inhibited by treatment with EGTA or benzamidine.
We propose that this essential, multifunctional protein is a sensitive monitor of intracellular pH and Ca2+. By analogy to electronic circuitry, eEF1A serves as a gate that can process an input signal (e.g. a change in pH) to generate multiple output signals (e.g. changes in cytoskeletal structure and protein synthesis). Furthermore, because eEF1A is so abundant, we propose that proteolysis may provide an effective means of rapidly decreasing functional activity.
MATERIALS AND METHODS
Wild carrot (Daucus carota L.) cells were grown in suspension culture and transferred weekly as previously described (Chen and Boss, 1990). Trichloroacetic acid (TCA), phospholipase D (Streptomyces species), phospholipase A2 (Crotalus durissus), PI-specific phospholipase C (Bacillus cereus), aprotinin, leupeptin, and benzamidine were purchased from Sigma-Aldrich (St. Louis). [14C]Ethanolamine (55 mCi/mmol) was purchased from American Radiolabeled Chemicals (St. Louis). Phospholipase C (Bacillus cereus) was purchased from Calbiochem-Novabiochem (San Diego). Lysed, transformed Escherichia coli expressing recombinant castor bean phospholipase Dα, Arabidopsis phospholipase Dβ, phospholipase Dγ, and vector alone were prepared as previously described (Pappan et al., 1998). Caspase inhibitors were a gift from Dr. Gary Smith (Glaxo Wellcome Inc., Research Triangle Park, NC). Antibodies to ABP-50 (Dictyostelium discoideum eEF1A) were a gift from Dr. John S. Condeelis (Albert Einstein College of Medicine, New York). Micro-BCA protein assay reagent was purchased from Pierce Chemical (Rockford, IL). Bradford protein assay reagent was purchased from Bio-Rad Laboratories (Hercules, CA). The phosphor imager (Storm, Molecular Dynamics, Sunnyvale, CA) and a phosphor imager tritium screen were used for the radioisotopic analysis.
In Vivo Labeling and Isolation of Membranes
Two days after transfer, carrot cells were incubated with [14C]ethanolamine (4 μCi/0.3–0.8 g fresh weight) for 48 h. The carrot cells (2 g/2 mL buffer/0.5 g polyvinylpolypyrrolidone) were homogenized using a Dounce homogenizer and the following buffer: 30 mm Tris, pH 7.2, at 25°C, 2 mm EGTA, 1 mm EDTA, 1 mm sodium molybdate, 25% [v/v] glycerol, 1 mm phenylmethylsulfonyl fluoride (PMSF), and 10 mm β-mercaptoethanol at 4°C. The homogenate was centrifuged at 700g for 5 min. The 700g supernatant was separated by centrifugation for 1 h at 40,000g, and the resulting pellet was used as the microsomal fraction. The microsomal fraction was kept on ice and either used immediately or frozen at −80°C until use. Protein concentration was determined using the Micro-BCA assay with bovine serum albumin as a standard.
TCA Precipitation of [14C]et-eEF1A
TCA precipitation of [14C]et-eEF1A was performed and the filters were washed to remove non-covalently bound lipids, as previously described (Ransom et al., 1998). The washed filters were air-dried, placed in scintillation vials with Biosafe II cocktail, and counted in a scintillation counter (Minaxi Tri-CARB 4000 series, United Technologies-Packard, Meriden, CT). Data are reported as disintegrations per minute recovered minus a control of the same size filter paper that was incubated in the same beaker throughout the entire procedure to determine non-specific binding to the filter paper.
Phospholipase Treatment of the Microsomal Fraction
Prior to treatment of the microsomal fraction with phospholipases, microsomes were resuspended in 10 mL of ice-cold buffer consisting of 20 mm Tris-HCl, 3 mm MgCl2, pH 7.2, and concentrated by centrifugation at 40,000g for 30 min at 4°C. The washed pellet was resuspended in 0.1 mL of 20 mm Tris-HCl, pH 6.8 or 7.2 (25°C). The phospholipase treatments were carried out using 112 μg of membrane protein in a total volume of 120 μL. The membranes were added to the reaction buffer consisting of 20 mm Tris-HCl, 1 mm EGTA, 0.1% (v/v) Triton X-100, with and without 2 mm CaCl2 (unless otherwise indicated). Phospholipases were added immediately after addition of the membranes (phospholipase A2, 20 units; phospholipase D, 20 units; phospholipase C, 20 units; PI-specific phospholipase C, 3 units unless otherwise indicated), and the reaction mixture was incubated at 25°C on a rotary shaker (200 rpm) for 10 min. The reaction was stopped by adding 3 mm EGTA and placing the reactions on ice.
Aliquots were removed from each reaction for analysis of [14C]et-eEF1A by TCA precipitation, by SDS-PAGE through 10% (w/v) polyacrylamide, and for analysis of residual [14C]phosphatidylethanolamine by lipid extraction and thin layer chromatography. The lipids were separated in a CHCl3:MeOH:NH4OH (65:25:4) solvent system and quantitated with a scanner (System 500, Bioscan, Inc., Washington, DC). After these initial experiments characterizing the different phospholipases, we optimized the phospholipase assays to determine the least amount of phospholipase D needed to observe a reproducible loss in [14C]et-eEF1A from microsomal membranes by TCA precipitation. Based on the concentration curve performed (data not shown), the rest of the phospholipase D assays were carried out using 5 units. When we used 5 units of phospholipase D/112 μg of membrane protein, there was no discernible loss of membrane protein on a Coomassie Blue-stained SDS-polyacrylamide gel; however, we still were able to quantitate a loss of [14C]et-eEF1A by TCA precipitation.
Experiments using protease inhibitors were performed as follows: benzamidine (100 mm in water), aprotinin (5 mm in water), and leupeptin (3.5 mm in water) were added 1 min before the addition of phospholipase D. The reactions were incubated at 25°C on a rotary shaker (200 rpm) for 10 min. The reactions were stopped by adding 15 μL of ice-cold 25 mm EGTA and placing the test tubes on ice. Aliquots were removed from each reaction for analysis of [14C]et-eEF1A by TCA precipitation.
For experiments using extracts from E. coli expressing recombinant phospholipase Dα, phospholipase Dβ, phospholipase Dγ, or vector alone, carrot microsomes were resuspended in 20 mm Tris-HCl at pH 6.5 for phospholipase Dα and at pH 7.0 for phospholipase Dβ and phospholipase Dγ. Phospholipase Dα activity was tested using the conditions described by Pappan et al. (1997, 1998). Carrot microsomes (30 μg) were added to the reaction buffer consisting of 100 mm 2-(N-morpholino)-ethanesulfonic acid (MES), pH 6.5, 5 mm CaCl2, 0.5 mm SDS, and 1% (v/v) ethanol. Phospholipase Dα or vector alone (30 μg of E. coli extracts) was added immediately after addition of the carrot microsomes, and the reaction was incubated at 30°C in a shaking water bath for 30 min. The reaction was stopped by placing the test tubes on ice. Aliquots were removed from each reaction for analysis as described above. For phospholipase Dβ and phospholipase Dγ, the carrot microsomes (30 μg) were added to the reaction buffer consisting of 100 mm MES (pH 7.0), 80 mm KCl, and 1% (v/v) ethanol. An extract of phospholipase Dβ, phospholipase Dγ, or vector alone (30 μg of lysed E. coli extracts) was added immediately after addition of the carrot microsomes and the reaction was incubated at 30°C in a shaking water bath. After 30 min the tubes were placed on ice to stop the reaction and aliquots were removed from each reaction for analysis as described above.
His-Tagged Recombinant eEF1A
Tomato eEF1A cDNA fused to a six-His tag was expressed in E. coli and purified using resin (Probond, Invitrogen, Carlsbad, CA) according to the manufacturer's protocol (Invitrogen). The recombinant eEF1A (1 μg unless indicated otherwise) was added to (112 μg) microsomes and incubated for 10 min in the presence or absence of 1 mm Ca2+, as indicated above. The proteins were separated by SDS-PAGE and analyzed by western blotting using chemiluminescence to detect the His-tagged antibody according to the manufacturer's protocol (Pierce Chemical). As a control, the catalytic domain of PI4Kα fused to a six-His tag, a 62-kD polypeptide (Stevenson et al., 1998), was expressed in E. coli and purified in the same manner.
RESULTS
Treatment with Phospholipase D or C Results in a Loss of [14C]et-eEF1A from Microsomal Membranes
If the PGE post-translational modification of eEF1A facilitates the association of eEF1A with membranes, then we hypothesized that treating the membranes with phospholipases D, C, or A2 might hydrolyze the lipid-anchored [14C]ethanolamine eEF1A to produce phosphatidic acid, diacylglycerol, or fatty acids, respectively, and the remaining 14C-labeled peptide should be recovered in the soluble fraction. In addition, treatment with PI-specific phospholipase C should release [14C]phosphoethanolamine eEF1A into the supernatant if the PGE attachment is part of a GPI anchor. We tested these hypotheses by incubating different phospholipases with microsomal membranes. The lipase treatments were carried out with and without 1 mm Ca2+, because Ca2+ is generally required for maximum phospholipase activity (Wang, 1993; Wang et al., 1993).
After the reactions were completed, the reaction mixture was separated into microsomal and supernatant fractions by centrifugation. Each fraction was analyzed by SDS-PAGE, followed by transfer to polyvinylidene difluoride membrane for western-blot analysis using an antibody to D. discoideum eEF1A (Demma et al., 1990). Western-blot analysis indicated a loss of protein from both the supernatant and pellet when microsomal membranes were treated with Ca2+ alone, with phospholipase C, or with phospholipase D, suggesting that eEF1A was being proteolytically degraded and not just released from the microsomes to a soluble fraction (data not shown). To determine whether eEF1A was being proteolytically degraded, we added recombinant eEF1A to a microsomal fraction and analyzed total protein in the reaction mixture. As shown in Figure 1A, in the presence of 1 mm Ca2+, recombinant eEF1A was degraded within 20 min, as determined by western-blot analysis using a His-tagged antibody.
Figure 1.
eEF1A is sensitive to Ca2+-regulated, membrane-associated proteases. Recombinant eEF1A or PI4Kα catalytic domain was added to 112 μg of microsomal protein in the absence (−) or presence (+) of CaCl2 in a 120-μL reaction mixture, as described in “Materials and Methods.” Thirty-five-microliter aliquots were removed at the times indicated, and proteins were separated by SDS-PAGE and analyzed by western blots using an antibody to the six-His tag. The Ca2+ concentration was 1 mm in A and 100 μm in B. A, The amount of recombinant eEF1A (reEF1A) added to the microsomes was varied. Lane 1, 1.2 μg; lane 2, 0.8 μg; lane 3, 0.4 μg; lane 4, 1.2 μg; lane 5, 1.2 μg; lane 6, 0.8 μg; lane 7, 0.4 μg. Lanes 1 through 4 are −Ca2+ and lanes 5 through 7 are +1 mm Ca2+. Lanes 1 through 3 and 5 through 7 were incubated 20 min; lane 4 was placed immediately into sample buffer. B, Recombinant proteins (1 μg) were incubated with the microsomes for 10 min (lanes 2–9) or added immediately to sample buffer, lane 1. Lanes 1 through 5, recombinant eEF1A; lane 1, 2, and 3 are −Ca2+. Lanes 4 and 5 are +100 μm Ca2+. Lanes 6 through 9, recombinant PI4Kα (rPI4Kα) catalytic domain; lanes 6 and 8, are +100 μm Ca2+ and lanes 7 and 9 are −Ca2+.
To show a clear effect of the protease(s), eEF1A was added at three different concentrations (1.2, 0.8, and 0.4 μg) to the same amount of microsomal protein. The protease activity was inhibited by adding EGTA (compare lanes 1–3 and 5–7, Fig. 1A). Lane 4 is the time zero control with 1.2 μg of eEF1A added in the presence of EGTA. A similar loss of eEF1A could be seen after only 10 min in the presence of 100 μm Ca2+ (Fig. 1B, lanes 1–5). To determine whether the microsomal proteases would degrade recombinant proteins in general, the recombinant PI4Kα catalytic domain, a 62-kD recombinant polypeptide expressed and purified with the same protocols used for eEF1A, was added. Using the same microsomal fraction, we found no detectable loss of the PIKα polypeptide after 10 min in the presence or absence of 100 μm Ca2+ (Fig. 1B, lanes 6–9).
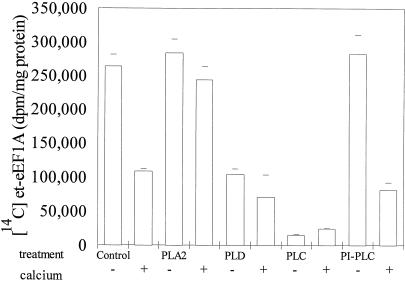
Although we could detect a loss of eEF1A on western blots, in order to quantify the loss and to study the fate of the endogenous protein, we used [14C]ethanolamine to label the PGE post-translational modification and monitored the recovery of TCA-precipitable 14C-labeled protein. From previous work we knew that eEF1A was the only protein that incorporated [14C]ethanolamine in carrot microsomal membranes and that [14C]ethanolamine was incorporated into a PGE post-translational modification (Ransom et al., 1998). The TCA precipitation data were consistent with the results of the western blot. TCA precipitation showed that Ca2+ alone, phospholipase C with and without Ca2+, phospholipase D with and without Ca2+, and PI-specific phospholipase C plus Ca2+ caused a loss of [14C]et-eEF1A from microsomal membranes and phospholipase A2 did not (Fig. 2). These data suggested that a Ca2+- and/or lipase-activated protease was present.
Figure 2.
[14C]et-eEF1A is lost from microsomes treated with 1 mm Ca2+, phospholipase D (PLD) or phospholipase C (PLC), but not phospholipase A2 (PLA2). Carrot cells were incubated with [14C]ethanolamine (10 μCi/g fresh weight) for 2 d, and harvested as described in “Materials and Methods.” The 40,000g microsomal pellet was washed, concentrated by centrifugation, and resuspended in reaction buffer. Phospholipases were added to 112 μg of membrane protein, and the reaction was incubated at 25°C with shaking (200 rpm) for 10 min. The reaction was stopped by adding ice-cold EGTA. Aliquots were removed from each reaction, and the recovery of [14C]et-eEF1A was monitored by TCA precipitation analysis. The means of four values from two experiments are reported. PI-PLC, PI-phospholipase C.
The loss of eEF1A was not the result of a general loss of membrane lipid because when phospholipase A2 was added, there was no significant loss of eEF1A plus or minus Ca2+. In fact, more [14C]et-eEF1A was recovered from microsomal membranes treated with phospholipase A2 plus Ca2+ than from those treated with Ca2+ alone, and under these conditions, phospholipase A2 degraded more than 98% of the lipids, as determined by TLC (data not shown). One explanation is that the lysolipids and fatty acids produced by phospholipase A2 protected [14C]et-eEF1A from Ca2+-induced proteolysis. A 1 mm Ca2+ concentration tended to enhance the loss of [14C]et-eEF1A when phospholipase D but not phospholipase C was added. With PI-specific phospholipase C, there was no loss unless Ca2+ was added, and the loss of [14C]et-eEF1A from microsomal membranes was equal to that of Ca2+ alone.
To study the specificity of the response to phospholipase D and because commercial lipases could have proteases present, we used three different isoforms of plant phospholipase D expressed in E. coli. Only castor bean phospholipase Dα caused a significant loss of [14C]et-eEF1A (Table I). An equivalent E. coli extract containing the vector alone was used as a control. The fact that under the same conditions, the E. coli extract with vector alone did not enhance the loss of [14C]et-eEF1A compared with the buffer control suggested that the castor bean phospholipase Dα was the active agent and that phospholipase Dα enhanced Ca2+-induced proteolysis of eEF1A. In similar experiments, the vector control, Arabidopsis phospholipase Dβ, and Arabidopsis phospholipase Dγ all failed to show a loss of [14C]et-eEF1A (data not shown).
Table I.
Phospholipase Dα causes a decrease of [14C]et-eEF1A recovered from microsomes
| Sample | [14C]et-EF1A |
|---|---|
| dpm/mg | |
| MES buffer | 32,250 ± 10,000 |
| Vector alone | 47,375 ± 13,812 |
| Phospholipase Dα | 16,875 ± 9,870 |
Carrot microsomes (30 μg of protein) resuspended in 20 mm Tris-HCl (pH 6.5) were added to reaction buffer consisting of 100 mm MES (pH 6.5), 5 mm CaCl2, 0.5 mm SDS, and 1% (v/v) ethanol. The reaction was incubated at 30°C in a shaking water bath for 30 min. Aliquots were removed from each reaction for recovery of [14C]et-eEF1A by TCA precipitation analysis. The means of four values from two experiments are reported.
EGTA and Benzamidine Inhibit the Loss of [14C]et-eEF1A from Microsomes Treated with Ca2+ or Phospholipase D
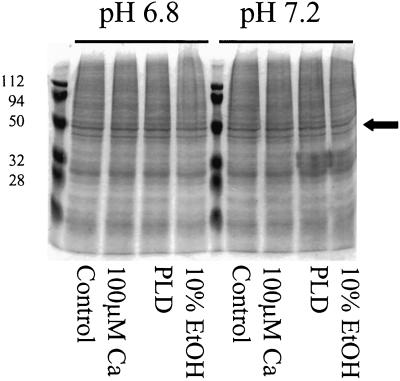
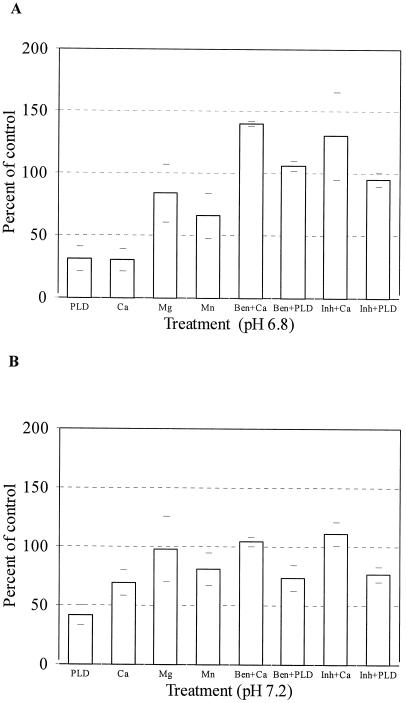
To characterize the types of proteases involved in the loss of [14C]et-eEF1A from microsomal membranes, we pretreated the membranes with different protease inhibitors. For these experiments we reduced the amount of phospholipase D added from 20 units to 5 units/112 μg of membrane protein. As can be seen in Figure 3, under these conditions, there was not a visible loss of membrane proteins based on Coomassie Blue staining. The common protease inhibitors PMSF and pepstatin did not prevent eEF1A proteolysis (data not shown); however, this may be because PMSF and pepstatin are not water soluble and were therefore dissolved in ethanol, and the high percentage of ethanol required in the reaction (>16%) would have activated phospholipase D (Munnik et al., 1998). To avoid activation of phospholipase D, we used water-soluble protease inhibitors. Pretreatment of the [14C]et-eEF1A microsomal membranes with a cocktail mix of leupeptin (3.5 mm), aprotinin (5 mm), and benzamidine (100 mm) prevented the loss of [14C]et-eEF1A when either phospholipase D or Ca2+ was added (Fig. 4, A and B).
Figure 3.
Phospholipase D (PLD) treatment did not result in a general loss of membrane protein. Either Ca2+ (100 μm), 10% (v/v) ethanol, or phospholipase D (5 units) were added to microsomes prepared as described in Figure 2. The reaction containing 112 μg of membrane protein was incubated at 25°C with shaking (200 rpm) for 10 min. The reaction was stopped by adding ice-cold EGTA. An aliquot from each reaction (20 μL) was analyzed by SDS-PAGE (10%, [w/v]). The SDS-polyacrylamide gel was stained with Coomassie Blue dye and dried. The arrow denotes the migration of eEF1A. Molecular mass markers are shown on the left (in kD).
Figure 4.
Protease inhibitors prevent the Ca2+- or phospholipase D (PLD)- induced proteolysis of [14C]et-eEF1A at pH 6.8 and pH 7.2. A, Either benzamidine (Ben) alone or a cocktail (Inh) of aprotinin (5 mm), leupeptin (3.5 mm), and benzamidine (100 mm) were added to microsomes (112 μg of membrane protein) for 1 min, followed by the addition of phospholipase D (5 units) or Ca2+ (100 μm) as indicated, or microsomes were treated with CaCl2 (100 μm), MgCl2 (100 μm), or MnCl2 (100 μm). The reactions were incubated at 25°C with shaking (200 rpm) for 10 min, as described in “Materials and Methods” and analyzed by TCA precipitation. B, The reaction conditions are the same as in A except the pH of the reactions was 7.2. The recovery of [14C]et-eEF1A was analyzed by TCA precipitation. The means of four values from two experiments are reported.
To determine if one inhibitor alone would prevent proteolysis of [14C]et-eEF1A, we pretreated the membranes with either benzamidine (a Ser protease inhibitor, 100 mm), aprotinin (a Ser protease inhibitor, 5 mm), or leupeptin (a thiol protease inhibitor, 3.5 mm) for 1 min before the addition of Ca2+ or phospholipase D. Only benzamidine prevented the loss of [14C]et-eEF1A (Fig. 4, A and B). At these concentrations, benzamidine also inhibited phospholipase D activity (data not shown) so that we cannot determine whether phospholipase D actvity was necessary for proteolysis. In addition, because Ca2+ is known to increase phospholipase Dα activity (Wang, 1993, 1997; Wang et al., 1993), we could not distinguish between a direct activation of a protease by Ca2+ or indirect activation resulting from Ca2+ activation of phospholipase D.
Calpeptin, an inhibitor of calpain (100 μm) and cyclosporin A, an inhibitor of calcineurin inhibitor, were also tried but had no effect on [14C]et-eEF1A proteolysis using isolated membranes (data not shown). Ca2+ and calmodulin can destabilize microtubules in vitro (Durso and Cyr, 1994; Moore et al., 1998), so we reasoned that calmodulin might enhance the proteolysis of [14C]et-eEF1A. However, when excess calmodulin (100 μm) was added to microsomes in the presence or absence of Ca2+, the loss of [14C]et-eEF1A caused by treatment with phospholipase D was not affected (data not shown). These data do not rule out a role for calmodulin, because there may have been sufficient calmodulin already present in the microsomal fraction; however, they do not support a role for a Ca2+-calmodulin-mediated proteolysis of eEF1A.
Because Ca2+ and eEF1A can affect the viability of cells, we asked if inhibitors of programmed cell death would prevent the loss of [14C]et-eEF1A from microsomes treated with phospholipase D with and without Ca2+. The caspase inhibitors ZVAD-FMK and ACDEVD-CHO (50 μm dissolved in DMSO and water, respectively) did not prevent proteolysis of [14C]et-eEF1A (data not shown). We also tried to use these inhibitors in vivo by adding them to carrot cells prior to the addition of Mas 7. Mas 7 increases intracellular Ca2+ (Tucker and Boss, 1996) and results in a 45% loss of eEF1A recovered from whole-cell microsomes; however, neither caspase inhibitor prevented the loss of [14C]et-eEF1A (data not shown). The lack of effect of caspase inhibitors in vivo is equivocal, because the inhibitors may not have entered the cells.
The effect of other divalent cations was compared. Mn2+ and Mg2+ at equal molar concentrations could not substitute for 1 mm CaCl2 at pH 6.8 (Fig. 4, A and B). At pH 7.2, 1 mm Ca2+ tended to be more effective, but there was no statistically significant difference in the [14C]et-eEF1A recovered. In all the experiments, EGTA was the only treatment that routinely decreased the loss of eEF1A during the 10-min incubation, suggesting that the protease was Ca2+ regulated.
Effects of Ca2+ and pH on the Loss of [14C]et-eEF1A
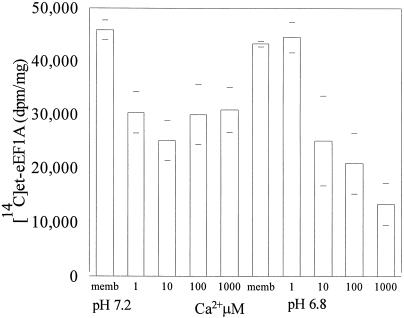
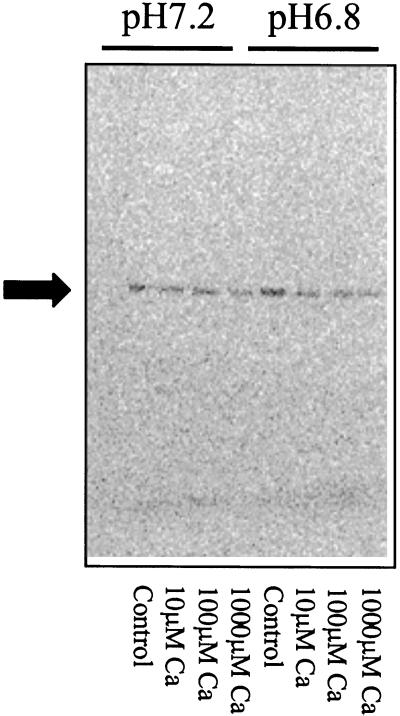
Edmonds et al. (1995) observed that eEF1A loses its ability to bundle F-actin at pH 7.2 compared with pH 6.8. Therefore, we asked what effect pH would have on the amount of [14C]et-eEF1A we recovered from microsomal membranes treated with various concentrations of Ca2+. Carrot microsomal membranes containing [14C]et-eEF1A were treated with four concentrations of CaCl2 (1, 10, 100, and 1,000 μm) at pH 6.8 and 7.2. At pH 7.2, 1 μm CaCl2 resulted in a 35% loss of [14C]et-eEF1A (Fig. 5). However, the amount lost did not increase with increasing CaCl2. At pH 6.8, 10 μm CaCl2 was needed to cause a significant loss of [14C]et-eEF1A. At pH 6.8, increasing concentrations of CaCl2 resulted in decreased recovery of [14C]et-eEF1A. Radiographic analysis of microsomal proteins blotted onto polyvinylidene difluoride indicated that the decrease in [14C] resulted from a loss of [14C]et-eEF1A (Fig. 6). These data suggest that in a living cell at physiological levels of Ca2+, eEF1A will be most sensitive to Ca2+-mediated proteases at pH 7.2.
Figure 5.
The effect of Ca2+ on [14C]et-eEF1A is pH dependent. Ca2+ (1, 10, 100, and 1,000 μm) was added to microsomes (112 μg of membrane protein) prepared as described in Figure 2. The reaction was incubated at 25°C with shaking (200 rpm) for 10 min and stopped by adding ice-cold EGTA. Recovery of [14C]et-eEF1A was analyzed by TCA precipitation. The means of four values from two experiments are reported.
Figure 6.
Treatment with Ca2+ causes a decrease of [14C]et-eEF1A recovered from microsomes. Ca2+ (1, 10, 100, and 1,000 μm) was added to microsomes (112 μg of membrane protein) prepared as described in Figure 2. The reaction was incubated at 25°C with shaking (200 rpm) for 10 min. The reaction was stopped by adding ice-cold EGTA. An aliquot of each reaction (20 μL) was separated by 10% (w/v) SDS-PAGE. The gel was dried and exposed to a phosphor imager tritium screen for 1 week.
DISCUSSION
We have demonstrated a loss of [14C]et-eEF1A from carrot microsomal fractions treated with Ca2+ alone, phospholipase D, or phospholipase C. The loss of eEF1A was pH and Ca2+ sensitive and could be prevented by adding EGTA or benzamidine. Proteolysis may seem like a wasteful mechanism for altering cellular metabolism; however, eEF1A is a very abundant protein within the cell and may act as a cellular buffer by binding soluble proteins and aminoacyl-tRNA. Proteolysis would provide a rapid means of decreasing functional activity and provide a ready source of amino acids for protein synthesis in response to stress. It is conceivable that eEF1A is synthesized and degraded in a manner similar to key proteins involved in cell cycle regulation, whose appearance and disappearance signal each stage of the cell cycle (Wilkinson, 1995; Hershko and Ciechanover, 1998).
The observation that no [14C]et-eEF1A is lost after treatment with phospholipase A2 with or without Ca2+ indicates that proteolysis does not result from a general hydrolysis of membrane lipid. Furthermore, the fact that there is no apparent proteolysis when phospholipase A2 is added in the presence of Ca2+ points to a key role for phospholipase D, because lysolipids formed by phospholipase A2 treatment would inhibit endogenous phospholipase D activity even in the presence of Ca2+ (Ryu et al., 1997; Kawabe et al., 1998). The proteolysis of [14C]et-eEF1A in the presence of the plant recombinant phospholipase Dα but not phospholipase Dβ or phospholipase Dγ also provides evidence for a Ca2+/phospholipase D sensitive protease(s).
Proteolysis is Ca2+ regulated. Benzamidine, a Ser protease inhibitor, prevented proteolysis but is also known to inhibit phospholipase D activity (X. Wang, unpublished results), so it is not clear if a Ser-protease is involved. EGTA, however, consistently prevented eEF1A proteolysis whether we used recombinant eEF1A or the endogenous protein. Ca2+ could directly activate a membrane-associated protease(s) or, by activating a phospholipase D or phospholipase C, the increased Ca2+ could release a protease(s) or eEF1A and thereby enhance enzyme-substrate interaction. In either event, the sensitivity of eEF1A to μm Ca2+ provides a mechanism whereby eEF1A could serve as a very important target in signal transduction pathways.
We used [14C]et-eEF1A to quantify the loss of eEF1A. Although measurement reflects the amount of PGE-modified eEF1A in the microsomes and not the total pool of available eEF1A, we found no indication of a selective loss of the [14C]et-eEF1A isoform of eEF1A based on comparative analysis of western blots using both immunodetection and radiography (with a phosphor imager). Under conditions where there was a quantifiable loss of [14C]et-eEF1A, there was a visible decrease in eEF1A on the western blots, and, most importantly, recombinant eEF1A, which did not exhibit post-translational modification, was also sensitive to proteolysis.
While our experimental treatments were only for 10 min, prolonged increases in phospholipase D and Ca2+ can lead to cell death (Wang, 1997; Berridge et al., 1998). Sustained increases in phospholipase D are associated with senescence (Paliyath et al., 1987; Paliyath and Droillard, 1992) or hypersensitive response (Young et al., 1996), and elevated intracellular Ca2+ levels are associated with the onset of apoptosis (McConkey and Orrenius, 1997). It is reasonable that changes in the amount of eEF1A, which would affect both protein synthesis and cytoskeletal structure, would be an important factor in regulating the programming of cell death (Duttaroy et al., 1998) or senescence.
In summary, we have shown that proteolysis of eEF1A is sensitive to physiologically relevant changes in Ca2+ and pH. The sensitivity of eEF1A to Ca2+ provides a mechanism whereby eEF1A could serve as a very important target in signal transduction pathways. For example, turnover of eEF1A would contribute to the restructuring of the actin cytoskeleton in growing tip cells in response to an oscillation in cytosolic pH and Ca2+ (Pierson et al., 1994, 1996; Holdaway-Clarke et al., 1997; Feijó et al., 1999). In addition, proteolysis of eEF1A could contribute to the decrease in bundling of the F-actin cytoskeleton with a change in pH from 6.8 to 7.2 (Edmonds et al., 1995). The decrease in F-actin bundling in turn could increase protein synthesis mediated by the eEF1A that remains bound to the actin (Liu et al., 1996b). Thus, this essential, multifunctional protein, eEF1A, provides a sensitive monitor of intracellular pH and Ca2+ and can impact multiple downstream responses. By analogy to an electronic circuit, eEF1A could serve as a gate, which would sense an input signal such as a change in pH or Ca2+ and generate multiple output signals such as changes in cytoskeletal structure and protein synthesis.
ACKNOWLEDGMENTS
The authors would like to thank Dr. Gary Smith for the gift of the Caspase inhibitors, John S. Condeelis for the antibody to Dictyostelium eEF1A (ABP-50), and Dr. Christine K. Shewmaker (Calgene, Davis, CA) for the tomato eEF1A clone.
Footnotes
This work was supported by grants from the National Science Foundation (grant no. MCB–9604285 to W.F.B. and IBN–9808729 to X.W.) and by a Patricia Robert Harris fellowship to W.D.R.-H.
LITERATURE CITED
- Aerts RJ, DeWit RJW, Van Lokeren Campagne MM. Cyclic AMP induces alkalinization in Dictyostelium. FEBS Lett. 1987;220:366–370. doi: 10.1016/0014-5793(87)80848-7. [DOI] [PubMed] [Google Scholar]
- Berridge MJ, Bootman MD, Lipp P. Calcium-a life and death signal. Nature. 1998;395:645–648. doi: 10.1038/27094. [DOI] [PubMed] [Google Scholar]
- Browning K. The plant translational apparatus. Plant Mol Biol. 1996;32:107–144. doi: 10.1007/BF00039380. [DOI] [PubMed] [Google Scholar]
- Cárdenas L, Vidali L, Dominguez J, Perez H, Sanchez F, Hepler PK, Quinto C. Rearrangement of actin microfilaments in plant root hairs responding to Rhizobium etlinodulation signals. Plant Physiol. 1998;116:871–877. doi: 10.1104/pp.116.3.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Boss WF. Short-term treatment with cell wall degradation enzymes increases the activity of the inositol phospholipid kinases and the vanadate-sensitive ATPase of carrot cells. Plant Physiol. 1990;94:1820–1829. doi: 10.1104/pp.94.4.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore AM, Dannenhoffer JM, Larkins BA. EF-1α is associated with a cytoskeletal network surrounding protein bodies in maize endosperm cells. Plant Cell. 1996;8:2003–2014. doi: 10.1105/tpc.8.11.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collings DA, Wasteneys GO, Miyazaki M, Williamson RE. Elongation factor 1α is a component of the subcortical actin bundles of characean algae. Cell Biol Int. 1994;18:1019–1024. doi: 10.1006/cbir.1994.1025. [DOI] [PubMed] [Google Scholar]
- Condeelis J. Elongation factor 1α, translation and the cytoskeleton. Trends Biol Sci. 1995;20:169–170. doi: 10.1016/s0968-0004(00)88998-7. [DOI] [PubMed] [Google Scholar]
- Demma M, Warren V, Hick R, Dharmawardhane S, Condeelis J. Isolation of an abundant 50,000-dalton actin filament bundling protein from Dictyosteliumamoebae. J Biol Chem. 1990;265:2286–2291. [PubMed] [Google Scholar]
- Dharmawardhane S, Demma N, Yang F, Condeelis J. Compartmentalization and actin binding properties of ABP-50: the elongation factor-1 alpha of Dictyostelium. Cell Motil Cytoskelet. 1991;20:279–288. doi: 10.1002/cm.970200404. [DOI] [PubMed] [Google Scholar]
- Durso NA, Cyr RJ. A calmodulin-sensitive interaction between microtubules and a higher plant homolog of elongation factor-1α. Plant Cell. 1994;6:893–905. doi: 10.1105/tpc.6.6.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durso NA, Leslie JD, Cyr RJ. In situ immunocytochemical evidence that a homolog of protein translation elongation factor EF-1α is associated with microtubules in carrot cells. Protoplasma. 1996;190:141–150. [Google Scholar]
- Duttaroy A, Bourbeau D, Wang XL, Wang E. Apoptosis rate can be accelerated or decelerated by overexpression or reduction of the level of elongation factor-1α. Exp Cell Res. 1998;238:168–176. doi: 10.1006/excr.1997.3819. [DOI] [PubMed] [Google Scholar]
- Edmonds BT, Murray J, Condeelis J. pH regulation of the F-actin binding properties of Dictyosteliumelongation factor 1α. J Biol Chem. 1995;270:15222–15230. doi: 10.1074/jbc.270.25.15222. [DOI] [PubMed] [Google Scholar]
- Ehrhardt DW, Atkinson EM, Long SR. Depolarization of alfalfa root hair membrane potential by Rhizobium melilotiNod factors. Science. 1992;256:998–1000. doi: 10.1126/science.10744524. [DOI] [PubMed] [Google Scholar]
- Ehrhardt DW, Wais R, Long SR. Calcium spiking in plant root hairs responding to rhizobium nodulation signals. Cell. 1996;85:673–681. doi: 10.1016/s0092-8674(00)81234-9. [DOI] [PubMed] [Google Scholar]
- Feijó JA, Sainhas J, Hacket GR, Kunkel JG, Hepler PK. Growing pollen tudes possess a constiutive alkaline band in the clear zone and a growth-dependent acidic tip. J Cell Biol. 1999;144:483–496. doi: 10.1083/jcb.144.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felle HE, Kondorosi E, Kondorosi A, Schultze M. Rapid alkalinization in alfalfa root hairs in response to rizobial lipochitooligosacharide signals. Plant J. 1996;10:295–301. [Google Scholar]
- Gonen H, Smith CE, Siegel NR, Kahana C, Merrick WC, Chakraburtty K, Schwartz AL, Ciechanover A. Protein synthesis elongation factor EF-1α is essential for ubiquitin-dependent degradation of certain Nα-acetylated proteins and may be substituted by the bacterial elongation factor EF-TU. Proc Natl Acad Sci USA. 1994;91:7648–7652. doi: 10.1073/pnas.91.16.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasezawa S, Kumagai F, Nagata T. Sites of microtubule reorganization in tobacco BY-2 cells during cell-cycle progression. Protoplasma. 1997;198:202–209. [Google Scholar]
- Hasezawa S, Nagata T. Microtubule organizing centers in plant cells: localization of a 49 kDa protein from sea urchin centrosomes in synchronized tobacco BY-2 cells. Protoplasma. 1993;176:64–74. [Google Scholar]
- Hayashi Y, Urade R, Utsumi S, Kito M. Anchoring of peptide elongation factor EF-1α by phosphatidylinositol at the endoplasmic reticulum membrane. J Biochem. 1989;106:560–563. doi: 10.1093/oxfordjournals.jbchem.a122895. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. In: Richardson CC, Abelson JN, Raetz CRH, Thorner JW, editors. Annual Review of Biochemistry 67. Palo Alto, CA: Annual Reviews Inc.; 1998. pp. 425–479. [DOI] [PubMed] [Google Scholar]
- Holdaway-Clarke TL, Feijó JA, Hackett GR, Kunkel JF, Hepler PK. Pollen tube growth and the intracellular cytosolic calcium gradient oscillate in phase while extracellular calcium influx is delayed. Plant Cell. 1997;9:1999–2010. doi: 10.1105/tpc.9.11.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur KJ, Ruben L. Protein translation elongation factor-1α from Trypanosoma bruceibinds calmodulin. J Biol Chem. 1994;269:23045–23050. [PubMed] [Google Scholar]
- Kawabe K, Kodaki T, Katayama K, Okamura Si, Mori M, Yamashita S. Identification of lipid inhibitors of mammalian phospholipase D. J Biochem. 1998;123:870–875. doi: 10.1093/oxfordjournals.jbchem.a022018. [DOI] [PubMed] [Google Scholar]
- Liu G, Edmonds BT, Condeelis J. pH, EF-1α and the cytoskeleton. Trends Cell Biol. 1996a;6:168–171. doi: 10.1016/0962-8924(96)20013-3. [DOI] [PubMed] [Google Scholar]
- Liu G, Tang J, Edmonds BT, Murray J, Levin S, Condeelis J. F-actin sequesters elongation factor 1α from interaction with aminoacyl-tRNA in a pH-dependent reaction. J Cell Biol. 1996b;135:953–963. doi: 10.1083/jcb.135.4.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConkey DJ, Orrenius S. The role of calcium in the regulation of apoptosis. Biochem Biophys Res Commun. 1997;239:357–366. doi: 10.1006/bbrc.1997.7409. [DOI] [PubMed] [Google Scholar]
- Merrick WC, Hershey JWB. The pathway and mechanism of eukaryotic protein synthesis. In: Hershey JWB, Mathews MB, Sonenberg N, editors. Translational Control. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1996. pp. 31–69. [Google Scholar]
- Moore RC, Durso NA, Cyr RC. Elongation factor-1α stabilizes microtubules in a calcium/calmodulin-dependent manner. Cell Motil Cytoskelet. 1998;41:168–180. doi: 10.1002/(SICI)1097-0169(1998)41:2<168::AID-CM7>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Munnik T, Irvine RF, Musgrave A. Phospholipid signaling in plants. Biochim Biophys Acta. 1998;1389:222–272. doi: 10.1016/s0005-2760(97)00158-6. [DOI] [PubMed] [Google Scholar]
- Ohta k, Toriyama M, Miyazaki M, Murofushi H, Hosoda S, Endo S, Sakai H. The mitotic apparatus-associated 51-kDa protein from sea urchin eggs is a GTP-binding protein and is immunologically related to yeast polypeptide elongation factor 1α. J Biol Chem. 1990;265:3240–3247. [PubMed] [Google Scholar]
- Paliyath G, Droillard MJ. The mechanisms of membrane deterioration and disassembly during senescence. Plant Biochem Physiol. 1992;30:789–812. [Google Scholar]
- Paliyath G, Lunch DV, Thompson JE. Regulation of membrane phospholipid catabolism in senescing carnation flowers. Physiol Plant. 1987;71:503–511. [Google Scholar]
- Pappan K, Austin-Brown S, Chapman KD, Wang X. Substrate selectivities and the lipid modulation of plant phospholipase Dα, -β, and -γ. Arch Biochem Biophys. 1998;353:131–140. doi: 10.1006/abbi.1998.0640. [DOI] [PubMed] [Google Scholar]
- Pappan K, Zheng S, Wang X. Identification and characterization of a novel plant phospholipase D that requires polyphosphoinositides and submicromolar calcium for activity in Arabidopsis. J Biol Chem. 1997;272:7048–7054. doi: 10.1074/jbc.272.11.7048. [DOI] [PubMed] [Google Scholar]
- Pierson ES, Miller DD, Callaham DA, Shipley AM, Rivers BA, Cresti M, Hepler PK. Pollen tube growth is coupled to the extracellular calcium ion flux and the intracellular calcium gradient: effect of BAPTA-type buffers and hypertonic media. Plant Cell. 1994;6:1815–1828. doi: 10.1105/tpc.6.12.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson ES, Miller DD, Callaham DA, van Aken J, Hackett G, Hepler PK. Tip-localized entry fluctuates during pollen tube growth. Dev Biol. 1996;174:160–173. doi: 10.1006/dbio.1996.0060. [DOI] [PubMed] [Google Scholar]
- Ransom WD, Lao PC, Gage DA, Boss WF. Phosphoglycerylethanolamine posttranslational modification of plant eukaryotic elongation factor 1α. Plant Physiol. 1998;117:949–960. doi: 10.1104/pp.117.3.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu SB, Karlesson BH, Ozgem M, Palta JP. Inhibition of phospholipase D by lysophosphatidylethanolamine, a lipid-derived senescence retardant. Proc Natl Acad Sci USA. 1997;94:12717–12721. doi: 10.1073/pnas.94.23.12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina N, Gotoh Y, Kubomura N, Iwamatsu A, Nishida E. Microtubule severing by elongation factor 1α. Science. 1994;266:282–285. doi: 10.1126/science.7939665. [DOI] [PubMed] [Google Scholar]
- Stevenson JM, Perera IY, Boss WF. A phosphatidylinositol 4-kinase pleckstrin homology (PH) domain that binds phosphatidylinositol 4-monophosphate. J Biol Chem. 1998;273:22761–22767. doi: 10.1074/jbc.273.35.22761. [DOI] [PubMed] [Google Scholar]
- Tan Z, Boss WF. Association of phosphatidylinositol kinase, phosphatidylinositol monophosphate kinase, and diacylglycerol kinase with the cytoskeleton and F-actin fractions of carrot (Daucus carotaL.) cells grown in suspension culture. Plant Physiol. 1992;100:2116–2120. doi: 10.1104/pp.100.4.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker E, Boss WF. Mastoparan-induced intracellular Ca2+fluxes may regulate cell-to-cell communication in plants. Plant Physiol. 1996;111:459–467. doi: 10.1104/pp.111.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duijn V, Inouye K. Regulation of movement speed by intracellular pH during Dictyostelium discoideumchemotaxis. Proc Natl Acad Sci USA. 1991;88:4951–4955. doi: 10.1073/pnas.88.11.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. Phospholipases. In: Moore TS, editor. Lipid Metabolism in Plants. Boca Raton, FL: CRC Press; 1993. pp. 505–525. [Google Scholar]
- Wang X. Molecular analysis of phospholipase D. Trends Plant Sci. 1997;2:261–266. [Google Scholar]
- Wang X, Dyer JH, Zheng L. Purification and immunological analysis of phospholipase D from germinating castor bean endosperm. Arch Biochem Biophys. 1993;306:486–494. doi: 10.1006/abbi.1993.1541. [DOI] [PubMed] [Google Scholar]
- Wang W, Poovaiah BW. Interaction of plant chimeric calcium/calmodulin-dependent protein kinase with a homolog of eukaryotic elongation factor-1α. J Biol Chem. 1999;274:12001–12008. doi: 10.1074/jbc.274.17.12001. [DOI] [PubMed] [Google Scholar]
- Wilkinson KD. Roles of ubiquitinylation in proteolysis and cellular regulation. In: McCormick DB, Bier DM, Goodridge AG, editors. Annual Review of Nutrition 15. Palo Alto, CA: Annual Reviews, Inc.; 1995. pp. 161–189. [DOI] [PubMed] [Google Scholar]
- Yang F, Demma M, Warren V, Dharmawardhane S, Condeelis J. Identification of an actin-binding protein from Dictyosteliumas elongation factor 1α. Nature. 1990;347:494–496. doi: 10.1038/347494a0. [DOI] [PubMed] [Google Scholar]
- Yang W, Burkhart W, Cavallius J, Merrick WC, Boss WF. Purification and characterization of a phosphatidylinositol 4-kinase activator in carrot cells. J Biol Chem. 1993;268:392–398. [PubMed] [Google Scholar]
- Young SA, Wang X, Leach JE. Changes in the plasma membrane distribution of rice phospholipase D during resistant interaction with Xanthomonas oryzae pv oryzae. Plant Cell. 1996;8:1079–1090. doi: 10.1105/tpc.8.6.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]