ABSTRACT
The aim of this study was to produce a humanized single chain antibody (scFv) as a potential improved product design to target EGFR (Epidermal Growth Factor Receptor) overexpressing cancer cells. To this end, CDR loops of cetuximab (an FDA-approved anti-EGFR antibody) were grafted on framework regions derived from type 3 (VH3 and VL3 kappa) human germline sequences to obtain recombinant VH and VL domainslinked together with a flexible linker [(Gly4Ser)3] to form a scFv. Codon optimized synthetic gene encoding the scFv (with NH2-VH-linker-VL-COOH orientation) was expressed in E. coli Origami™ 2(DE3) cells and the resultant scFv purified by using Ni-NTA affinity chromatography. The scFv, called cet.Hum scFv, was evaluated in ELISA and immunoblot to determine whether it can recognize EGFR. The scFv was able to recognize EGFR over-expressing cancer cells (A-431) but failed to detect cancer cells with low levels of EGFR (MCF-7 cells). Although the affinity of the scFv forA-431 cells was 9 fold lower than that of cetuximab, it was strong enough to recognize these cells. Considering its ability to bind EGFR molecules, the scFv may exhibit a potential application for the detection of EGFR-overexpressing cancer cells.
KEYWORDS: cetuximab, germline humanization, scFv, VH3, VL3 kappa
Introduction
Epidermal growth factor receptor (EGFR) is a member of receptor tyrosine kinase (RTK) family involving in a variety of human cancers, including breast cancer, colorectal cancer, non-small cell lung cancer (NSCLC), pancreatic cancer, and head and neck squamous cell carcinoma (HNSCC).1-4 So far, several antibodies have been developed against EGFR, among which are nimotuzumab [(squamous cell carcinoma in head and neck-(SCCHN))], matuzumab (for metastatic colorectal cancer), panitumumab (for metastatic colorectal cancer) and cetuximab (for SCCHN and metastatic colorectal cancer).5-7 The latest, cetuximab, has been widely used for the treatment and detection of EGFR-overexpressing cancers and presented as a promising anti-EGFR antibody8-11 since 2008, the Fab fragment of cetuximab has also been produced and characterized (PDP ID: 1YY8) 12 It exhibits rapid tumor penetration and excellent tumor-to-normal tissue ratio in animal studies.13 In addition to Fab fragment, numerous single chain formats of cetuximab have been constructed and evaluated, most of which are humanized scFvs. Jalalypour and colleagues (2016) produced a murine scFv by linking VH (heavy chain variable domain) and VL (light chain variable domain) domains of cetuximab and found it to be able to recognize A-431 cells.14 Chi and colleagues produced a minibody composed of cetuximab VH, linker, cetuximab VL, a hinge region (with amino acid sequences of EPKSPKSADKTHTAP), and an IgG1 CH3 domain. The minibody was found to be able to inhibit EGFR signaling in HT29 colon cancer cells and suppress tumor growth in mice bearing HT29 tumor xenografts.15 CH3 domain of IgG1 molecules promotes dimerization of scFv molecules, leading to improved targeting.16
Among the anti-EGFR antibodies mentioned above, cetuximab is a chimeric antibody composed of human constant (C) domains and mouse variable domains. Hence, joining of cetuximab VH and VL domains will result in the production of a murine scFv. Murine antibodies elicit HAMA (human anti-mouse antibody), which restricts their applications for human use.17 Therefore, we decided to humanize variable domains of cetuximab to construct a humanized single chain antibody. Antibody humanization reduces potential immunogenicity of non-human antibodies.18,19 One of the most common humanization techniques is CDR grafting, in which non-human CDR loops are grafted on human framework regions extracted from either IgG or germline sequences. Humanized antibodies with germline framework regions are expected to show lower degrees of immunogenicity than those with non-germline framework regions; they carry fewer somatic hypermutations and therefore are less immunogenic.20 Using CDR grafting technique, Veisi and colleagues produced a cetuximab-based humanized scFv. They grafted the CDR loops of cetuximab on framework regions driven from human germline sequences and then mutated a number of special residues within these regions (referred to as vernier zone residues) back to their counterparts in cetuximab to retain the antibody affinity.21 Despite its good affinity for EGFR, the scFv (called scFv) has been reported to be expressed in bacterial cells largely as inclusion body, which needs to be denatured and then refolded to be active.22 hscFv carries framework regions from type 3 variable domain [IGHV3-3304 (a heavy chain sequence) and IGKV3-15 * 01 (a kappa light chain sequence)] and, thus is expected to be a soluble scFv. We analyzed the amino acid sequence of hscFv VH and found that the composition of its hydrophobic core residues more looks like to that of VH4 subclasses (IGHV4), one of the most unstable sequences.23 In fact, the retention of cetuximab vernier zone residues in hscFv has changed the amino acid composition of hydrophobic cores. Hydrophobic cores (upper and lower hydrophobic cores) play a critical role in scFv stability.23 CDR homology approach makes it possible to retain antibody affinity without back-mutation of vernier zone residues24 and therefore, makes it possible to use intact framework regions containing hydrophobic core residues of interest. We used this method in our previous work and obtained an active scFv for targeting EGFRvIII.25 In the current study, we report the production of a novel humanized anti-EGFR scFv using IgG3 variable domains (VH3 and VL3 kappa) based on CDR homology approach, and demonstrate its ability to recognize EGFR expressing cancer cells. Subtype 3 sequences have been proven to be more stable than other subtypes.23
Results
3-D structure of Cet.Hum scFv
3-D modeling was performed using I-TASSER online server to predict tertiary structure of cet.Hum scFv. The server predicted five potential 3-D models (with different C-score values) for cet.Hum scFv, two of which (models 1 and 2) were found by superposition to cetuximab Fab fragment (PDB ID: 1YY8) to be more likely to reflect the 3-D structure of cet.Hum scFv. Despite a huge difference between their C-scores (0.94 and −3.17, respectively), the two models differed only in the spatial position of their linker region (Fig. 1). The maximum free energy of Cet.Hum scFv/EGFR interaction was calculated by Hex 8.0.0 to be −537.44 kCal/mol, while that of cetuximab/EGFR interaction was −796.68. These data indicate that cetuximab has higher affinity for EGFR as compared to cet.Hum scFv.
Figure 1.
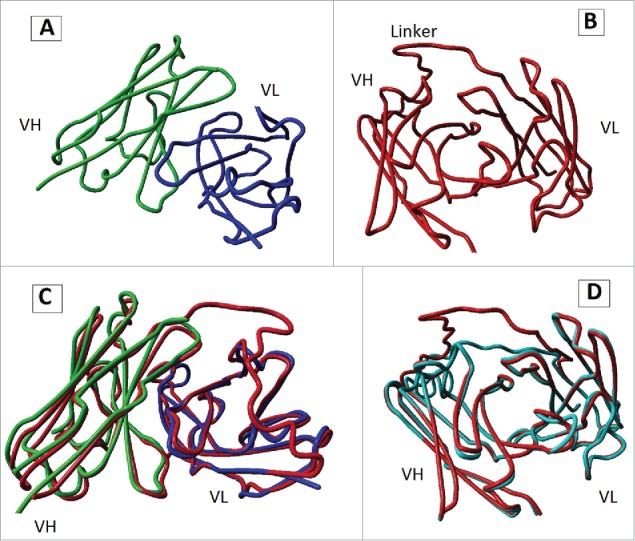
3-D structures predicted for cet.Hum scFv. (A): 3-D structure of cetuximab Fab fragment (PDB ID: 1YYB). Constant domains are not shown. (B) 3-D structure of model 1, the best model (in terms of C-score value) predicted for 3-D structure of cet.Hum scFv. (C): superposition of cetuximab Fab fragment and model 1. (D) Superposition of model 1 and model 2 (a predicted model for 3-D structure cet.Hum scFv).
Expression and purification of Cet.HumscFv
Bacterial expression of cet.Hum scFv gene resulted in production of a 27 kDa protein. SDS-PAGE analysis of bacterial samples (before IPTG induction, after IPTG induction, supernatant fraction of lysed bacteria, and bacterial pellet) revealed that majority of the expressed cet.Hum scFv (≥70%) was present in the supernatant fraction (Fig. 2).
Figure 2.
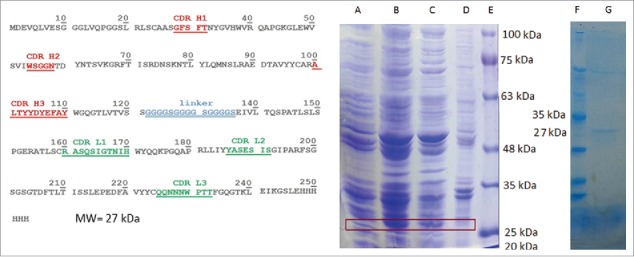
cet.Hum scFv amino acid sequence (in VH-linker-VL format) and expression analysis. (The left panel): The underlined red-colored letters indicate the CDR regions of VH domain and green-colored letters display the CDR regions of VL domain. 6-His tag appears in the C-terminal end of the protein. (The right panel): (A) bacterial cell lysate before IPTG addition, (B) bacterial cell lysate after IPTG addition, (C) supernatant faction after sonication and centrifugation, (D) bacterial pellet, (E) protein ladder, (F) protein ladder, and (G) Hid-tag purified cet. Hum scFv. Addition of IPTG to bacterial culture medium results in expression of a 27 KD protein that appears in supernatant fraction.
Results of ELISA with EGFR protein
Cet.Hum scFv could recognize EGFR protein in ELISA. The graphs of OD values versus the logarithms of antibody concentrations were sigmoidal, indicating that Cet.Hum scFv molecules react to EGFR molecules in a dose dependent manner and finally saturate them. OD100% value (upper plateau) increased by increase of EGFR concentration; the higher the peptide concentration, the higher the OD 100 value (Fig. 3). The affinity of cet.Hum scFv for EGFR protein was calculated to be 253nM.
Figure 3.
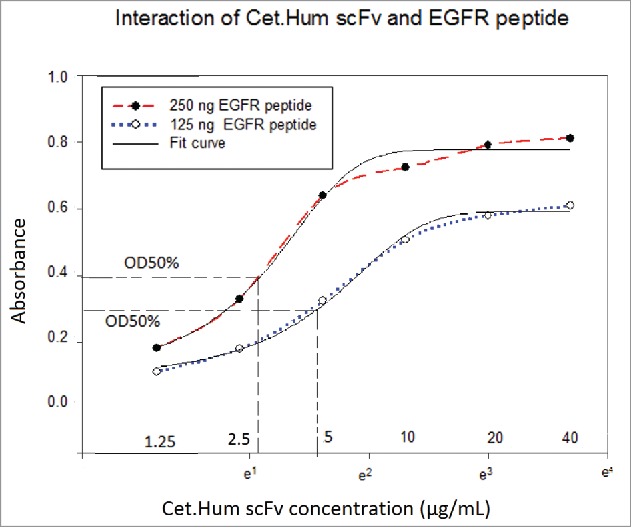
Interaction of EGFR protein and cet.Hum scFv in ELISA. Relatively higher OD values are obtained when EGFR concentration increased from 125 to 250 ng. At both EGFR protein concentrations (250 and 125ng), OD values increase upon increase of Cet.Hum scFv concentration, finally forming a plateau.
Results of ELISA with A-431 and MCF-7
Both cet.Hum scFv and cetuximab reacted to A-431 cells in a dose-dependent manner. For both antibodies, the curves of OD values versus the logarithms of antibody concentrations were sigmoidal (Fig. 4-A and -B). Mean comparison (the mean of three repetitions) using T-test revealed significant differences between OD100% values of cetuximab and cet.Hum scFv. The OD100% values of cetuximab/A-431 cell lysate interaction were relatively higher than those of cet.Hum scFv/A-431 cell lysate interaction (Fig 4-B). In the case of both antibodies, OD100% values increased significantly when antigen concentration (cell density) increased from 10000 cells/well to 20000 (Table 1). At cell density of 20000 cells/well, OD50% points for Cet.Hum scFv and cetuximab were calculated to be 6.14 µg/mL and 5.52 µg/mL, respectively. Using the mathematical equation optimized for single chain antibodies [8], the affinity of cet.Hum scFv for A-431 cell was calculated to be 153.8 nM. Affinity of cetuximab for A-431 cells was calculated [using the mathematical equation developed for full-length antibodies26 to be 17.2 nM (Table 2). As expected, cet.Hum scFv was unable to recognize MCF-7 cells in an efficient manner. At its lowest concentration (2.5 µg/mL), cet.Hum scFv produced OD values of ≤ 0.3 with the lysate of 10000 MCF-7 cells. No significant increase was observed in OD values when higher concentration of cet.Hum (5, 10, 20, and 40 µg/mL) were used. In addition, no significant increase was observed in OD values when the higher level of MCF-7 cells (lysate of 20000 cells) was used, indicating that cet.Hum scFv does not recognize MCF-7 cells in an efficient manner (Fig. 4-C). The maximum OD value registered for cet.Hum scFv/A-431 cell interaction (OD = 1.134, mean of three repetitions) was 3.06 fold greater than that registered for cet.Hum scFv/MCF-7 (OD = 0.37, mean of three repetitions). At MCF-7 cell density of 40000 cells/well, cetuximab produced OD values ranging from 0.268 to 0.918. The OD values increased in a linear fashion; the higher the concentration of cetuximab, the higher the OD values. Surprisingly, cetuximab (at all concentrations, except for 40 µg/mL) produced relatively higher OD values with the lower level of MCF-7 cells (density of 20000 cells/well). At cell density of 20000 cells/well, OD values for cetuximab concentrations of 3.25, 6.25, 12.5, 25 and 50 µg/mL were 0.649, 0.729, 0.729, 0.769, and 0.824 respectively. At MCF-7 cell density of 10000 cells/well, OD values were even higher than those observed at the densities of 20000 and 40000 cell/well (Fig. 4-D).
Figure 4.
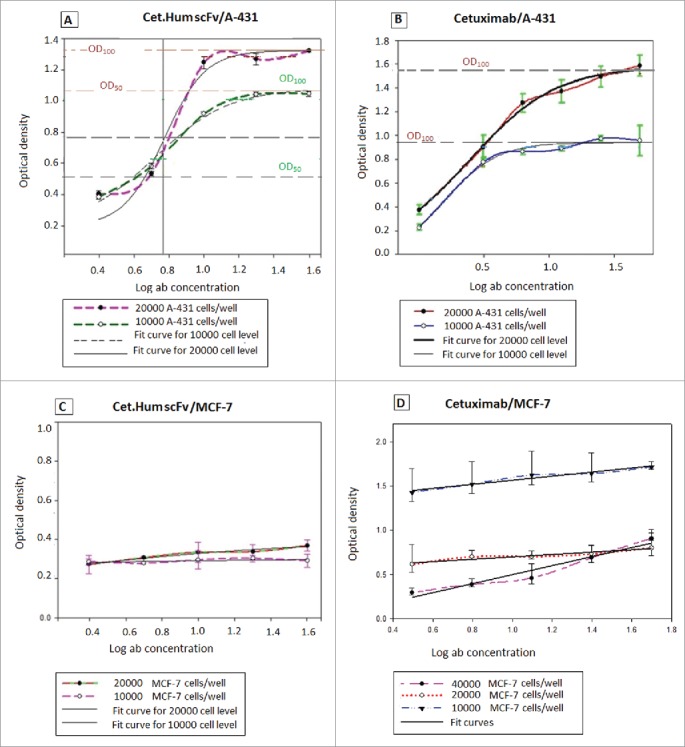
Interaction of scFv and cetuximab with A-431 and MCF-7 cells. (A): cet.Hum scFv can recognize A-431 cells in a dose dependent manner. The plots of logarithms of antibody concentrations versus OD values are sigmoidal. (B): Cetuximab can also recognize A-431 cells in a dose dependent manner. (C) cet.Hum scFv is unable to significantly recognize MCF-cells. (D): At lower concentration of MCF-7 cells, cetuximab produces higher OD values. None of the graphs form upper plateau.
Table 1.
Mean comparison of OD100 values at different A-431cell concentrations using t-test.
| Antibody | The number of lysed cells/well | OD100% (3 repetitions) | SD | SEM | t- test result |
|---|---|---|---|---|---|
| Cet.Hum scFv | 20000 | 1.13 | 0.012 | 0.007 | There is a statistically significant difference between the input groups (P = <0.001). |
| 1.13 | |||||
| 1.11 | |||||
| Cetuximab | 20000 | 1.60 | 0.087 | 0.05 | |
| 1.66 | |||||
| 1.49 | |||||
| Cet.Hum scFv | 10000 | 0.87 | 0.02 | 0.012 | There is a statistically significant difference between the input groups (P = 0.002). |
| 0.85 | |||||
| 0.8300 | |||||
| Cetuximab | 10000 | 0.98 | 0.021 | 0.012 | |
| 0.99 | |||||
| 0.95 | |||||
| cetuximab | 10000 | 0.95 | 0.14 | 0.081 | There is a statistically significant difference between the input groups (P = 0.003). |
| 1.09 | |||||
| 0.81 | |||||
| 20000 | 1.60 | 0.086 | 0.05 | ||
| 1.66 | |||||
| 1.49 | |||||
| cet.Hum scFv | 10000 | 0.87 | 0.02 | 0.012 | There is a statistically significant difference between the input groups (P = <0.001). |
| 0.85 | |||||
| 0.8300 | |||||
| 20000 | 1.13 | 0.012 | 0.007 | ||
| 1.13 | |||||
| 1.11 |
Notes: OD100%, OD value at upper plateau of sigmoidal curve; SD, Standard deviation; SEM, standard error of the mean.
Table 2.
Affinity of Cet.Hum scFv and cetuximab for A-431 cells.
| Antibody | The number of lysed cells/well) | OD100% | OD50% | Antibody concentration at OD50% | Antibody affinity (nM) |
|---|---|---|---|---|---|
| Cet.Hum-scFv (27.02 kDa) | 20000 | 1.12 | 0.56 | 6.14 µg/mL | 153.8 |
| (227.24 nM) | |||||
| 10000 | 0.86 | 0.43 | 5.13 µg/mL | ||
| (189.86 nM) | |||||
| Cetuximab (145.78kDa) | 20000 | 1.6 | 0.76 | 5.52 µg/mL | 17.2 |
| (37.87 nM) | |||||
| 10000 | 0.96 | 0.47 | 6.15 µg/mL | ||
| (42.19 nM) |
Notes: Ab, antibody concentration; OD100, OD value at upper plateau of sigmoid curve; OD50%, OD value at midpoint of sigmoid curve.
Immunoblotting
Cetuximab recognizes a conformational epitope on the surface of EGFR molecule. Therefore, instead of SDS-PAGE, which destroys 3-D structure of proteins, we used Native-PAGE) to separate cell lysate proteins. Crystal structure of the extracellular domain of the EGFR in complex with the Fab fragment of cetuximab is also freely available at PDB (ID: 1YY9). A relatively strong band with the size of approximately 145 kDa appeared in the lane of A-431 cell lysate on the PVDF membrane incubated with cet.Hum scFv (concentration of 50 µg/mL). A band with the same size appeared in the lane of A-431 cell lysate on the PVDF membrane incubated with cetuximab (25 µg/mL). 50 µg of cet.Hum scFv (MW = 27.02 kDa) makes 111.43 × 1013 molecules and therefore the same number of antigen binding sites, while 25 µg of cetuximab (MW = 145.78 kDa) makes 10.33 × 1013 molecules and 2 × (10.33 × 1013) antigen binding sites. 25 µg of cetuximab in 1 mL PBS makes a 171.49 nanomolar (nM) solution while 50 µg of cet.Hum scFv in the same volume makes a 925.24 nM solution. In fact, concentration of cet.Hum scFv has been 5.4 times higher than that of cetuximab. Neither cetuximab nor cet.Hum scFv recognized the MCF-7 cell lysate (Fig. 5), perhaps due to either the absence or at least an undetectable number of EGFR molecules. The results of immunoblot assay indicate that both cet.Hum scFv and cetuximab can recognize EGFR molecules in a specific manner.
Figure 5.
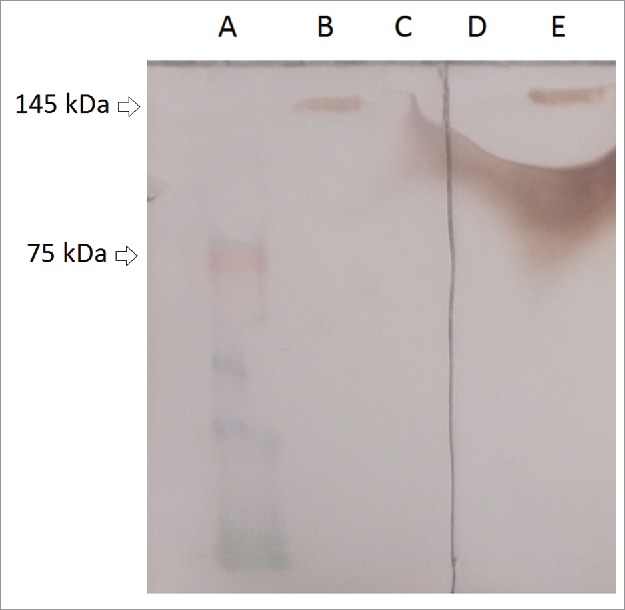
Activity assay of cetuximab and cet.Hum scFv by Immunoblotting. (A) Pre-stained protein ladder. (B) Interaction of cetuximab and A-431 cell lysate. (C) Interaction of cetuximab and MCF-7 cell lysate. (D) Interaction of cet.Hum scFv and MCF-7 cell lysate. (E) Interaction of cet.Hum scFv and A-431 cell lysate. (F) Interaction of cet.Hum scFv and EGFR protein.
Discussion
Cet.Hum scFv, which contains cetuximab CDR loops and human germline framework regions, is active and able to recognize EGFR. This indicates that recombinant VH and VL domains of this scFv fold and assemble correctly. I-TASSER online server predicted 5 potential models for cet.Hum scFv, two of which (models 1 and 2) were found by superposition to cetuximab Fab fragment (PDB ID: 1YY8) to be more likely to reflect 3-D structure of cet.Hum scFv. Spatial position of linker was the main difference of these two models. Linker region is not involved in antigen binding activity; therefore the two models present the same information on spatial position and 3-D structure of variable domains. 3-D modeling indicates that glycine-serine linker [(Gly4Ser)3] is flexible enough to allow the VH and VL domains to assemble and form a scFv with the3-D structure of interest.
Kappa and lambda light chains have indicated to display different biophysical properties; the latter are assumed to be less stable.23,27,28 Single chain antibodies with kappa-3 light chains have been shown to be more thermodynamically stable than those containing lambda-1 or lambda-3 light chains.27 Cet.Hum scFv has a VH3 heavy chain and a kappa-3 light chain; therefore, it should be thermodynamically stable. In SDS-PAGE, ELISA and immunoblot, the cet.Hum scFv was found to be soluble and was able to recognize EGFR molecules.
Both cetuximab and cet.Hum scFv were found in ELISA to be able to interact with A-431 cells in a dose dependent manner. In the case of both antibodies, the curves of OD values versus the logarithms of antibody concentrations were sigmoidal. Sigmoidal curves have an upper plateau. Formation of upper plateau for antigen-antibody interaction in ELISA indicates that antigen molecules are saturated with antibody molecules. Saturation occurs when a receptor is a specific target of a ligand.29,30 Since the curves of OD versus antibody concentrations were sigmoidal for both cetuximab and cet.Hum scFv, it can be deduced that they interact with A-431 cells in a specific manner. The OD values of cet.Hum scFv/ A-431 cell interaction in ELISA were relatively lower than those of cetuximab/ A-431 cell interaction, indicating that cetuximab recognizes A-431 cells with a higher affinity than the cet.Hum scFv does. As expected, cet.Hum scFv did not recognize MCF-7 cells in ELISA, whereas cetuximab produced unexpected OD values when interacted with these cells. Surprisingly, cetuximab produced higher OD values with lower concentrations of MCF-7 cells. At cell densities of 10000 and 40000 cells/well, OD values increased in a linear manner without forming upper plateaus. Using immunoblot assay, we found that both cetuximab and cet.Hum scFv are able to recognize A-431 cells but unable to recognize MCF-7 cells. These findings are consistent with the findings of Mamot and colleagues (2003). They demonstrated that EGFR-targeted immunoliposomes were able to bind and enter A-431 cells but failed to bind the MCF-7 cells.31 Strong reaction of cetuximab with MCF-cells seems to be an unusual phenomenon. Cetuximab has been proven to be unable to significantly recognize and/or influence MCF-7 cells. For example, Song and colleagues (2013) demonstrated by flow cytometry that cetuximab was unable to recognize MCF-7 cells. They also found that radiolabeled cetuximab (213Bi-Cetuximab) was incapable of affecting the viability of MCF-7 cells in an efficient manner.32 Cetuximab has been reported to be able to inhibit the proliferation of MCF-10 A and MDA-MB-468 breast cancer cells (EGFR-expressing cells lines) but not MCF-7 cells.33
In the current work we demonstrated that cet.Hum scFv is active and able to recognize A-431. However, this study has several limitations; we did not test cet.Hum scFv on more EGFR- expressing cell lines or cell lines expressing other HER family members. We did not test the scFv on triple-negative cancer cells (e.g. MDA-MB-231 and MDA-MB-468) to be certain that it cannot recognize these cells. We also did not evaluate anti-proliferative capacity or immunological effects of cet.Hum svFv. Hence, cet.Hum scFv needs to undergo further in vitro and in vivo (tumor xenograft) studies to be introduced as a promising anti-EGFR scFv.
ScFv molecules lack constant domains and hence are unable to evoke the activation of complement system and eradication of cancer cells. However, they can be useful for several purposes. For example, they can block cancer cell surface receptors and inhibit their downstream signaling.34 In addition, they can detect human cancers by binding to overexpressed receptors on the cancer cell surface. We proved that cet.Hum scFv is able to bind EGFR molecules expressed on the surface of EGFR-overexpressing cancer cells; therefore, it can be radiolabeled or conjugated to a fluorescent dye (e.g. IRDye800CW) and used to detect EGFR- expressing tumors. It may be used for blocking EGF (epidermal growth factor) receptors on the surface of cancer cells and inhibiting their downstream signaling. The scFv can also be used as a targeting ligand on the surface of drug carriers for targeted drug delivery to EGFR-overexpressing cancer cells.
Materials and methods
Cetuximab variable regions and CDR loops
Crystal structure of cetuximab Fab fragment is freely available at Protein Data Base, PDB (PDB ID: 1YY8). Each chain of the Fab fragment consists of two domains: a constant domain (C) and a variable domain (VH or VL). Variable domain of each chain was determined using AbNum tool (http://www.bioinf.org.uk/abs/abnum/). This tool is able to discriminate variable regions from constant domains and number amino acids within these regions based on Kabat or Chothia numbering schemes. CDR regions of variable domains were determined using the guideline described by Hwang and colleagues.24
Framework-donor human germline sequences
The amino acid sequences of cetuximab VH and VL domains were separately inserted in the search box of IMGT Domain Gap Align tool (www.imgt.org/IMGTindex/IMGTDomainGapAlign.php) to find the most homologous sequences. The tool arranged a large number of human germline sequences, among which two sequences (one VH3 and one VL3 kappa, both carrying CDR loops with canonical classes of interest) were selected for use in CDR grafting.
Cet.Hum scFv gene sequence
Cetuximab CDR Loops were grafted on the framework regions of the selected human germline sequences to obtain recombinant VH and VL sequences. The variable sequences were linked together with a flexible linker (Gly4Ser)3 to form a scFv. The Resultant protein sequence was reverse-translated into a single nucleotide sequence (using jCAT online tool, http://www.jcat.de/) encoding the scFv with NH2-VH-linker-VL-COOH orientation. The nucleotide sequence was flanked by two restriction sites (NcoI recognition site at 5′ end and XhoI recognition site at 3′ end) for insertion in pET22b(+) expression vector. This vector contains a His-Tag encoding sequence after XhoI recognition site, resulting in the addition of six histidine residues at carboxyl end of recombinant protein. The scFv-encoding sequence was synthesized By Bioneer™ Corporation (South Korea) in pGH8 cloning vector.
Three-dimensional (3-D) modeling and molecular docking
3-D modeling was carried out using I-TASSER (Iterative Threading Assembly Refinement) online server (http://zhanglab.ccmb.med.umich.edu/I-TASSER). The predicted models were visualized and analyzed by YASARA (Yet Another Scientific Artificial Reality Application) viewer (http://yasara.org).
Cell lines and reagents
A-431 and MCF-7 cell lines were purchased from cell bank of Pasteur Institute of Iran. HRP-Protein L (GenScript, Cat. Number: M00098) and DAB (Diaminobenzidine) tablets (Cat. Number: D4293) were purchased from GenScript (NJ, USA) and Sigma Aldrich, respectively. Cetuximab (Erbitux) (concentration of 5mg/mL, total volume of 20 mL) was obtained from Red Cross Pharmacy of Gorgan city (Golestan Province, Northern Iran). EGFR protein was purchased from Abcam (Cat. Number: ab155726).
Expression and purification of cet.Hum scFv
The nucleotide sequence encoding cet.Hum scFv was excised from pGH8 vector by NcoI/XhoI double-digestion and then inserted into pET22b(+) expression vector. The recombinant plasmid was transformed into E. coli Origami™ 2(DE3) competent cells using heat-shock method. A single colony of transformed bacteria was transferred into a conical flask containing 50 mL LB medium and allowed to grow until OD600 reached 0.6. At this stage, 50 µL IPTG (concentration of 100 mM) was added to the LB medium and the flask incubated overnight at 21°C. The bacterial cells were then precipitated by centrifuging the flask content at 8000 rpm for 10 min at 4°C. Bacterial pellet was suspended in 10 mL of lysis buffer [50 mM NaH2PO4, 50 mM NaCl, 1 mM phenylmethylsulfonyl fluoride (PMSF)] and then sonicated (80 amp) for 30 cycles (30 s on/30 s off) while cooling on ice. After confirming its expression by SDS-PAGE analysis, the scFvwas purified using Ni-NTA affinity chromatography (Qiagen, Cat. No: 30210).
ELISA with cancer cell lines
A-431 and MCF-7 cells were grown in RPMI culture medium (1 × 106 cells/10mL) containing 10% FBS and then precipitated by centrifuging at 3000 rpm for 5 min in 5°C. The cells were separately suspended in PBS buffer (at two densities of 200000 and 100000 cells/mL) and then lysed by sonication (15 cycles: 10 s on/10 s off) at 80 Amp. For each cellular density, 30 determinations (6 rows, each consisted of 5 wells) were considered for ELISA; 15 wells for A-431 and 15 wells for MCF-7 cells. Each well on ELISA plate was filled with 100 µl of cell lysate (lysate of 20000 or 10000 cells). After overnight incubation at 4°C, the wells were washed three times with wash buffer (PBS buffer containing tween 20) and then blocked with 300 µl of blocking buffer [(PBS buffer containing 5% BSA (Sigma, Cat. No: 05482)]. After 2 h, the wells were washed again and each was filled with100 µl of antibody (cetuximab or cet.Hum scFv) for 2h. For each antibody, five concentrations were considered (serial dilutions of 40, 20, 10, 5, 2.5 µg for cet.Hum scFv and 50, 25, 12.5, 6.25, 3.125 µg for cetuximab). (Total concentration of the scFv was determined using Bradford assay). After another round of washing, 100 µL of HRP-Protein L (concentration of 0.5 µg/mL) was added to each well. After 1 h incubation, the wells were washed again (five times, each 5 min) and each was filled with100 µL TMB [(3, 3′,5,5′-Tetramethylbenzidine), Sigma, Cat. Number T0440]. OD values were registered using spectrophotometer at wavelength of 450nm. Serial dilutions of antibody in ELISA make it possible to determine the linear range and OD50% points of antibody-antigen interaction curves. For each antigen level, a graph of OD versus antibody concentration was plotted using statistical software SigmaPlot 11. After determining OD50% points of each curve (by moving the computer mouse over the curves), the affinities of antibodies were calculated using mathematical equations, as described previously.26,35
ELISA with EGFR protein
EGFR protein solution (with dilutions of 250 and 125ng/ml) was prepared in 50 mM carbonate-bicarbonate buffer (1.59g/L Na2CO3 and 2.93g/L NaHCO3, pH = 9.6). Each well of ELISA plate was filled with 100 µL of the solution and the plate incubated overnight at 4°C. After being blocked with blocking buffer (PBS buffer containing 5% BSA) for 2 h, the wells were washed with wash buffer (PBS buffer containing tween 20). Then, each was filled with 100 µL cet.Hum scFv (serial dilutions of 40, 20, 10, 5, 2.5 µg). The other stages of ELISA, determination of OD50% points of antigen-antibody interaction curves, and affinity calculation were carried out in the same way as described for ELISA with cell lysates.
Immunoblot assay
The lysates of A-431 and MCF-7 cells (density of 400000 cells/mL) were run on a Native-PAGE gel (30μL cell lysate per well)and then transferred to a PVDF membrane. The PVDF membrane was immersed in blocking buffer (PBS buffer containing 5% skim milk) overnight to block non-specific sites. After being washed three times with wash buffer, the membrane was divided into two moieties to be treated with two different antibodies, for cet.Hum scFv and cetuximab (as a control). Each moiety contained lines of MCF-7 and A-431 cells. One of the moieties was treated with cetuximab (25 µg/mL) and the other treated with cet.Hum scFv (50 µg/mL). After 1 h incubation, the membranes were washed again and treated with HRP- Protein L (concentration of 0.5 µg/mL) for 1 h. The membranes were washed three times and then soaked in freshly prepared DAB substrate.
Statistical analysis
The results of ELISA were analyzed using statistical software SigmaPlot 11.
Disclosure of potential conflicts of interest
There are no conflicts of interests to declare.
Funding
This project was financially supported by the Deputy for Research of Golestan University of Medical Sciences (Gorgan, Iran) under the approval code of 950124008 and ethical approval code of IR.goums.1394.339.
Authorship contributions
Participated in research design: Yaghoub Safdari, Anvarsadat Kianmehr
Conducted experiments: Arsham Banisadr, Mahdieh Pourafshar
References
- 1.Masuda H, Zhang D, Bartholomeusz C, Doihara H, Hortobagyi GN, Ueno NT. Role of epidermal growth factor receptor in breast cancer. Breast Cancer Res Treat. 2012;136:331–345. doi: 10.1007/s10549-012-2289-9. PMID:23073759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miyamoto Y, Suyama K, Baba H. Recent advances in targeting the EGFR signaling pathway for the treatment of metastatic colorectal cancer. Int J Mol Sci. 2017;18:E75. doi: 10.3390/ijms18040752. PMID:28045433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prabhakar CN. Epidermal growth factor receptor in non-small cell lung cancer. Transl Lung Cancer Res. 2015;4:110–118. PMID:25870793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martinelli E, De PR, Orditura M, De VF, Ciardiello F. Anti-epidermal growth factor receptor monoclonal antibodies in cancer therapy. Clin Exp Immunol. 2009;158:1–9 doi: 10.1111/j.1365-2249.2009.03992.x. PMID:19737224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gridelli C, Maione P, Ferrara ML, Rossi A. Cetuximab and other anti-epidermal growth factor receptor monoclonal antibodies in the treatment of non-small cell lung cancer. Oncologist. 2009;14:601–611. doi: 10.1634/theoncologist.2008-0153. PMID:19482958 [DOI] [PubMed] [Google Scholar]
- 6.Capdevila J, Elez E, Macarulla T, Ramos FJ, Ruiz-Echarri M, Tabernero J. Anti-epidermal growth factor receptor monoclonal antibodies in cancer treatment. Cancer Treat Rev. 2009;35:354–363. doi: 10.1016/j.ctrv.2009.02.001. PMID:19269105 [DOI] [PubMed] [Google Scholar]
- 7.Ramakrishnan MS, Eswaraiah A, Crombet T, Piedra P, Saurez G, Iyer H, Arvind AS. Nimotuzumab, a promising therapeutic monoclonal for treatment of tumors of epithelial origin. MAbs. 2009;1:41–48. doi: 10.4161/mabs.1.1.7509. PMID:20046573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Day KE, Sweeny L, Kulbersh B, Zinn KR, Rosenthal EL. Preclinical comparison of near-infrared-labeled cetuximab and panitumumab for optical imaging of head and neck squamous cell carcinoma. Mol Imaging Biol. 2013;15:722–729. doi: 10.1007/s11307-013-0652-9. PMID:23715932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenthal EL, Moore LS, Tipirneni K, de BE, Stevens TM, Hartman YE, Carroll WR, Zinn KR, Warram JM. Sensitivity and specificity of Cetuximab-IRDye800CW to identify regional metastatic disease in head and neck cancer. Clin Cancer Res. 2017;23(16):4744–4752 doi: 10.1158/1078-0432.CCR-16-2968. PMID:28446503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenthal EL, Warram JM, de BE, Chung TK, Korb ML, Brandwein-Gensler M, Strong TV, Schmalbach CE, Morlandt AB, Agarwal G, et al.. Safety and tumor specificity of Cetuximab-IRDye800 for surgical navigation in head and neck cancer. Clin Cancer Res. 2015;21:3658–3666. doi: 10.1158/1078-0432.CCR-14-3284. PMID:25904751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sobrero AF, Maurel J, Fehrenbacher L, Scheithauer W, Abubakr YA, Lutz MP, Vega-Villegas ME, Eng C, Steinhauer EU, Prausova J, et al.. EPIC: phase III trial of cetuximab plus irinotecan after fluoropyrimidine and oxaliplatin failure in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:2311–2319 doi: 10.1200/JCO.2007.13.1193. PMID:18390971 [DOI] [PubMed] [Google Scholar]
- 12.Li S, Schmitz KR, Jeffrey PD, Wiltzius JJ, Kussie P, Ferguson KM. Structural basis for inhibition of the epidermal growth factor receptor by cetuximab. Cancer Cell. 2005;7:301–311 doi: 10.1016/j.ccr.2005.03.003. PMID:15837620 [DOI] [PubMed] [Google Scholar]
- 13.Chakravarty R, Goel S, Valdovinos HF, Hernandez R, Hong H, Nickles RJ, Cai W. Matching the decay half-life with the biological half-life: ImmunoPET imaging with (44)Sc-labeled cetuximab Fab fragment. Bioconjug Chem. 2014;25:2197–2204. doi: 10.1021/bc500415x. PMID:25389697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jalalypour F, Farajnia S, Mahmoudi F, Baradaran B, Farajzadeh D, Rahbarnia L, Majidi J. Cloning and expression of the variable regions of Anti-EGFR monoclonal antibody in E. coli for production of a single chain antibody. Iranian J Biotech. 2016;12:9–14 [Google Scholar]
- 15.Chi W, Kim H, Yoo H, Kim Y, Hong S. Periplasmic expression, purification, and characterization of an anti-epidermal growth factor receptor antibody fragment in escherichia coli. Biotechnol Bioprocess Engineering. 2016;21:321–330. doi: 10.1007/s12257-015-0817-2 [DOI] [Google Scholar]
- 16.Hu S, Shively L, Raubitschek A, Sherman M, Williams LE, Wong JY, Shively JE, Wu AM. Minibody: A novel engineered anti-carcinoembryonic antigen antibody fragment (single-chain Fv-CH3) which exhibits rapid, high-level targeting of xenografts. Cancer Res. 1996;56:3055–3061. PMID:8674062 [PubMed] [Google Scholar]
- 17.Klee GG. Human anti-mouse antibodies. Arch Pathol Lab Med. 2000;124:921–923. PMID:10835540 [DOI] [PubMed] [Google Scholar]
- 18.Almagro JC, Fransson J. Humanization of antibodies. Front Biosci. 2008;13:1619–1633. PMID:17981654 [DOI] [PubMed] [Google Scholar]
- 19.Safdari Y, Farajnia S, Asgharzadeh M, Khalili M. Antibody humanization methods – a review and update. Biotechnol Genet Eng Rev. 2013;29:175–186 doi: 10.1080/02648725.2013.801235. PMID:24568279 [DOI] [PubMed] [Google Scholar]
- 20.Pelat T, Bedouelle H, Rees AR, Crennell SJ, Lefranc MP, Thullier P. Germline humanization of a non-human primate antibody that neutralizes the anthrax toxin, by in vitro and in silico engineering. J Mol Biol. 2008;384:1400–1407. doi: 10.1016/j.jmb.2008.10.033. PMID:18976662 [DOI] [PubMed] [Google Scholar]
- 21.Veisi K, Farajnia S, Zarghami N, Khorshid HR, Samadi N, Safdari Y, Ahmadzadeh V. Development and evaluation of a Cetuximab-based humanized single chain antibody against EGFR-overexpressing tumors. Drug Res (Stuttg). 2015;65:624–628. PMID:25333654 [DOI] [PubMed] [Google Scholar]
- 22.Veisi K, Farajnia S, Zarghami N, Khoram Khorshid HR, Samadi N, Ahdi KS, Zarei JH. Chaperone-assisted soluble expression of a humanized Anti-EGFR ScFv antibody in E. Coli. Adv Pharm Bull. 2015;5:621–627 doi: 10.15171/apb.2015.084. PMID:26793607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ewert S, Huber T, Honegger A, Pluckthun A. Biophysical properties of human antibody variable domains. J Mol Biol. 2003;325:531–553. doi: 10.1016/S0022-2836(02)01237-8. PMID:12498801 [DOI] [PubMed] [Google Scholar]
- 24.Hwang WY, Almagro JC, Buss TN, Tan P, Foote J. Use of human germline genes in a CDR homology-based approach to antibody humanization. Methods. 2005;36:35–42. doi: 10.1016/j.ymeth.2005.01.004. PMID:15848073 [DOI] [PubMed] [Google Scholar]
- 25.Safdari Y, Farajnia S, Asgharzadeh M, Omidfar K, Khalili M. humMR1, a highly specific humanized single chain antibody for targeting EGFRvIII. Int Immunopharmacol. 2014;18:304–310. doi: 10.1016/j.intimp.2013.12.006. PMID:24370392 [DOI] [PubMed] [Google Scholar]
- 26.Beatty JD, Beatty BG, Vlahos WG. Measurement of monoclonal antibody affinity by non-competitive enzyme immunoassay. J Immunol Methods. 1987;100:173–179. doi: 10.1016/0022-1759(87)90187-6. PMID:2439600 [DOI] [PubMed] [Google Scholar]
- 27.Lehmann A, Wixted JH, Shapovalov MV, Roder H, Dunbrack RL Jr, Robinson MK. Stability engineering of anti-EGFR scFv antibodies by rational design of a lambda-to-kappa swap of the VL framework using a structure-guided approach. MAbs. 2015;7:1058–1071 doi: 10.1080/19420862.2015.1088618. PMID:26337947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ewert S, Cambillau C, Conrath K, Pluckthun A. Biophysical properties of camelid V(HH) domains compared to those of human V(H)3 domains. Biochemistry. 2002;41:3628–3636. doi: 10.1021/bi011239a. PMID:11888279 [DOI] [PubMed] [Google Scholar]
- 29.Krebs LT, Hanesworth JM, Sardinia MF, Speth RC, Wright JW, Harding JW. A novel angiotensin analog with subnanomolar affinity for angiotensin-converting enzyme. J Pharmacol Exp Ther. 2000;293:260–267. PMID:10734177 [PubMed] [Google Scholar]
- 30.Taylor HP, Cooper NR. Human cytomegalovirus binding to fibroblasts is receptor mediated. J Virol. 1989;63:3991–3998. PMID:2548011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mamot C, Drummond DC, Greiser U, Hong K, Kirpotin DB, Marks JD, Park JW. Epidermal growth factor receptor (EGFR)-targeted immunoliposomes mediate specific and efficient drug delivery to. Cancer Res. 2003;63:3154–3161. PMID:12810643 [PubMed] [Google Scholar]
- 32.Song H, Hedayati M, Hobbs RF, Shao C, Bruchertseifer F, Morgenstern A, Deweese TL, Sgouros G. Targeting aberrant DNA double-strand break repair in triple-negative breast cancer with alpha-particle emitter radiolabeled anti-EGFR antibody. Mol Cancer Ther. 2013;12:2043–2054. doi: 10.1158/1535-7163.MCT-13-0108. PMID:23873849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oliveras-Ferraros C, Vazquez-Martin A, Lopez-Bonet E, Martin-Castillo B, Del BS, Brunet J, Menendez JA. Growth and molecular interactions of the anti-EGFR antibody cetuximab and the DNA cross-linking agent cisplatin in gefitinib-resistant MDA-MB-468 cells: new prospects in the treatment of triple-negative/basal-like breast cancer. Int J Oncol. 2008;33:1165–1176. PMID:19020749 [PubMed] [Google Scholar]
- 34.Beerli RR, Wels W, Hynes NE. Inhibition of signaling from Type 1 receptor tyrosine kinases via intracellular expression of single-chain antibodies. Breast Cancer Res Treat. 1996;38:11–17. doi: 10.1007/BF01803779. PMID:8825118 [DOI] [PubMed] [Google Scholar]
- 35.Safdari Y, Farajnia S, Asgharzadeh M, Khalili M, Jaliani HZ. Affinity measurement of single chain antibodies: a mathematical method facilitated by statistical software SigmaPlot. Monoclon Antib Immunodiagn Immunother. 2014;33:13–19. PMID:24555931 [DOI] [PubMed] [Google Scholar]


