Abstract
Eligibility criteria are important for clinical research protocols or clinical practice guidelines for determining who qualify for studies and to whom clinical evidence is applicable, but the free-text format is not amenable for computational processing. In this paper, we described a practical method for transforming free-text clinical research eligibility criteria of Alzheimer’s clinical trials into a structured relational database compliant with standards for medical terminologies and clinical data models. We utilized a hybrid natural language processing system and a concept normalization tool to extract medical terms in clinical research eligibility criteria and represent them using the OMOP Common Data Model (CDM) v5. We created a database schema design to store syntactic relations to facilitate efficient cohort queries. We further discussed the potential of applying this method to trials on other diseases and the promise of using it to accelerate clinical research with electronic health records.
Keywords: Clinical Research Informatics, Relational Data Management, Electronic Health Record
Introduction
Randomized controlled trials are the gold standard for medical evidence generation. Eligibility criteria (EC) are the essential elements of clinical study protocols for specifying qualification of participants but often exist as free text, which are not amendable for computer processing. They are also found to have poor comprehensibility [1]. Given the wide adoption of electronic health records (EHRs), there is a great need for improving the interoperability of EC with EHRs to better integrate clinical research and patient care towards the development of a learning health system.
Multiple methods, such as ERGO [2], for structuring EC were developed before the widespread adoption of EHRs. As a result, such representations do not interoperate well with EHRs. Levi-Fix et al. developed EliXR-CDM [3] to structure criteria using the OMOP Common Data Model v4. This system was the first of its kind to transform free-text EC into a structured format using a standardized common data model. However, with a rule-based natural language processing system, it could not deal with the complex preprocessing and the scarcity of evaluation, which limited its generalization.
In this study, we extended this method and adopted the latest OMOP data standard, OMOP CDM version 5 [4], a model that is more comprehensive and better integrated than OMOP CDM version 4 for facilitating the interoperability among disparate observational databases. To the best of our knowledge, this study is amongst the first to build a relational database of clinical trial eligibility criteria using a widely adopted EHR data standard, OMOP CDM v5. Our method helps bridge the gap between clinical trials and EHRs by enabling fast and accurate patient cohort searching for trial recruiters, protocol designers, and healthcare providers.
Method
Our method consists of the following steps: (1) criteria relational database design; (2) criteria parsing; (3) concept normalization using terminologies; (4) relation extraction; and (5) ETL (extract, transform and load) for criteria using the OMOP CDM v5. We used a hybrid machine learning-based natural language processing toolkit, CLAMP, for name entity recognition to extract medical terms in EC. We matched the extracted terms to the standardized concept identifiers in the OMOP CDM v5. Aside from the entity recognition, we also used the SVM classifier to obtain relations between entities and attributes. Finally, we built a relational database for fast querying via Django. We also provided a RESTful API for retrieving information.
Step 1: Database schema design
The EHR data standard of OMOP CDM v5 was described by the Observational Health Data Sciences and Informatics (OHDSI) community [5]. In this data model, medical terms were categorized into seven types including four entities (Condition, Observation, Drug, Procedure) and three attributes (Qualifier, Measurements, Temporal_constraints). Each attribute has a close relationship with a corresponding entity. For instance, a relation of has_value shows a quantitative measurement value of one entity. The four entities consist of medical terms with similar characteristics, while the three attributes differ from each other. Due to this, we decided to build an efficient schema in which the four entities could be stored into one table while the three attributes could be saved in three separate tables. The benefit of categorizing attributes in individual tables is to handle measurement and temporal constraints independently. This will prevent disarrangement with other terms in the criteria database as these two attributes are lab values or time phrases that need to be split in future work. We also used three types of relations to build the connections between entities and attributes. Given the fact that one entity has several attributes and one attribute corresponds to many entities, the relationship between entities and attributes were considered as many-to-many in the database. With this design, the relations could be saved in the database and the pattern of entities and attributes could be queried.
Step 2: Name entity recognition (NER)
To achieve precise name entity recognition, we implemented a comprehensive clinical natural language processing software, CLAMP [6], designed by Hua et al in 2015. We used annotated criteria corpus of 230 Alzheimer’s disease clinical trial provided by previous lab members [7] to train the name entity recognition model. We implemented brown-clustering, n-gram, prefix-suffix, random-indexing, sentence-pattern, word-embedding, word-shape and word regular expression as name entity recognition features with a five-fold cross validation. Then we applied a NLP pipeline consisting of NegEx assertion, sentence detector, tokenizer, POS tagger, CRF-based NER and UMLS encoder. An example NER output is shown in Table 1.
Table 1.
Structured Output of Entity and Attribute in EC
|
NCT00000171 Exclusion Criteria: Sleep disturbance is acute (within the last 2 weeks). | |||
|---|---|---|---|
| Condition | present | Sleep disturbance | T0 |
| Qualifier | present | acute | T1 |
| Temporal constraints | present | within the last 2 weeks | T2 |
Step 3: Concept normalization
Once we finished the name entity recognition, we mapped the extracted clinical terms into the concept standardization identifiers (CONCEPT_ID) using the open-source software, Usagi [8]. Each concept has a distinctive CONCEPT_ID, which is mapped to multiple CONCEPT_CODES across domains such as ICD9CM, SNOMED_CT, etc. With the matching, we were able to map the concepts in clinical research eligibility criteria into terminology standards. Usagi provided an algorithm to evaluate the effect of the matching by giving a score; a higher score represents better match, and a score of 1.00 is a 100% match. We manually reviewed 100 randomly chosen terms of each domain and analyzed a statistical performance of the matching score. After an assessment of the matching score, we set the matching threshold at 0.80.
Step 4: Relation extraction
We applied our previously developed open-source criteria parser [7] to extract relations between entities and attributes using the Support Vector Machine (SVM) classifier. The direction of each relation was defined from each entity to its corresponding attributes. This method used the basic function of LibSVM [9] with features including the class of head entity, the class of attribute, the shortest path between two terms in the dependency tree and whether or not the entity is the only one in its class in the corpus. The classifier inspected each entity-attribute pair and projected them into four classes: no_relation, has_value, modified by, has_temp. An example of relation extraction output is shown in Table 2. The relation between entities T4 (“liver or kidney disease”) and T3 (“clinically significant”) is “modified by”. The relation between entities T16 (“alcohol abuse and dependence”) and T15 (“current”) is “has temporal relation” or “has-temp” in short form.
Table 2.
Structured Output of Relation in EC
|
NCT00007189 Exclusion Criteria: Clinically significant liver or kidney disease. Current alcohol abuse or dependence. | ||
|---|---|---|
|
| ||
| T4 | T3 | |
| liver or kidney disease | Clinically significant | Modified by |
|
| ||
| T16 | T15 | |
| alcohol abuse or dependence | Current | Has_temp |
Step 5: Data storage
In the last step, we created an efficient schema using Django [10] and loaded all the extracted entities, attributes and relations into respective tables. The most economical method of storing relations is through many-to-many relationships. In addition, we used REST architecture [11] to build an API to provide a convenient interface for users to retrieve information.
Results
Database Infrastructure Description
The database is comprised of five major tables: (1) clinical trial metadata information (2) entity table (3) qualifier table (4) measurement table (5) temporal constraints table. The detailed schema and formulation of the database provided as an appendix is available at https://github.com/Yuqi92/DBMS_EC.
Descriptive Statistical Analysis
To understand how well the name entity recognition and relation extraction performs at each step, we designed an evaluation framework by using classical classification metrics: precision, recall and F-score, which are defined below: (TP: true positive; FP: false positive; FN: false negative; TN: true negative).
To ensure that we had an ample amount of training set for good performance on name entity recognition, we analyzed the performance of varying sizes of annotated files. Based on the learning curve shown in Figure 2, we confirmed that the training set of 230 annotated trials is sufficient to achieve good performance of name entity recognition.
Figure 2.
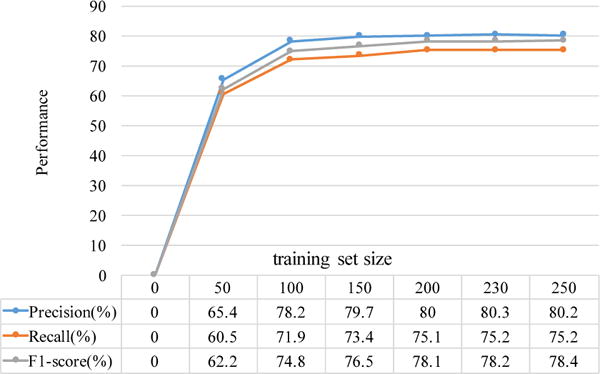
Learning curve for NER tasks
The performances of the different domains are variable due to the size of the training data. Of the annotated trials, the Condition domain is the largest (4136 terms) while the Procedure domain is the smallest (652 terms). According to the evaluation result of NER (Table 4), the Condition domain happened to achieve the best performance while the Procedure domain happened to have the poorest performance. Comparing these results, a larger amount correlates with a better performance and vice versa.
Table 4.
Evaluation of Name Entity Recognition
| Domain | Precision | Recall | F1-score |
|---|---|---|---|
| Condition | 0.835 | 0.836 | 0.831 |
| Observation | 0.748 | 0.745 | 0.793 |
| Drug | 0.852 | 0.790 | 0.820 |
| Procedure | 0.721 | 0.583 | 0.645 |
| Qualifier | 0.820 | 0.756 | 0.786 |
| Measurement | 0.820 | 0.770 | 0.794 |
| Temporal_constraints | 0.826 | 0.788 | 0.807 |
We matched the four entities (Condition, Observation, Drug, Procedure) and the qualifier attribute to the CONCEPT_ID in OMOP CDM v5. We used the matching score to evaluate the mapping results for different domains on decreasing thresholds. Figure 3 is the descriptive statistical analysis curve of the matching score for the different domains. It’s apparent and reasonable that when the matching score threshold was decreased from 0.9 to 0.7, the false positive rate (CONCEPT_ID incorrectly matched the term) decreased, while the false negative rate (CONCEPT_ID lost the term) increased. Therefore, we set the matching score threshold to 0.80 to trade off the balance between the error and the missing.
Figure 3.
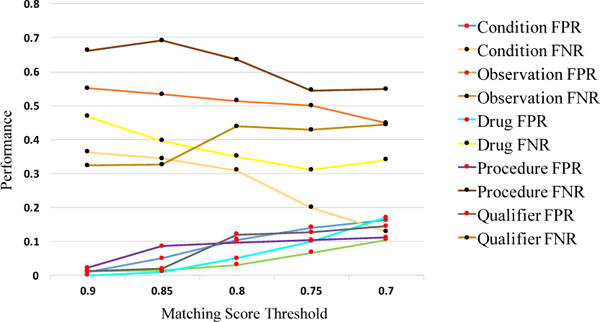
Mapping evaluation statistical analysis result
When the threshold reached 0.80, 68.80% of terms could be matched to CONCEPT_ID. Among the different domains, Qualifier terms reached the highest matching proportion (86.53%), Condition terms reached 74.88% and Observation terms reached only 44.41%. To evaluate the compression efficiency using CONCEPT_ID, we calculated the unique terms amount (in exacted terms), the compression ratio between number of unique terms and number of extracted terms, the unique CONCEPT_ID amount (in matched terms) and the compression ratio between number of unique CONCEPT_ID and number of matched terms. The ratio of the unique CONCEPT_ID is much lower than that of the unique terms in all domains. In total, the unique CONCEPT_ID ratio was 0.08, while the unique terms ratio was 0.19. Therefore, the representation ability of CONCETP_ID was well proved. The details are shown in Table 5.
Table 5.
Statistical Matching Result of Extracted Terms
| Extra cted term | Num. of match | Perc. of match (%) | Unique term (compression ratio) | Unique CONCEPT_ID (compressionn ratio | |
|---|---|---|---|---|---|
| Condition | 23336 | 17474 | 74.88 | 4453 (0.19) | 1336 (0.08) |
| Observation | 8824 | 3919 | 44.41 | 2360 (0.27) | 391 (0.10) |
| Drug | 6775 | 3694 | 54.52 | 1930 (0.28) | 624 (0.17) |
| Procedure | 3195 | 2136 | 66.85 | 626 (0.20) | 193 (0.09) |
| Qualifier | 9354 | 8094 | 86.53 | 449 (0.05) | 188 (0.02) |
| Total | 51484 | 35317 | 68.60 | 9819 (0.19) | 2660 (0.08) |
Relation extraction was evaluated separately by using the gold standard relations marked in annotated texts. The performance of SVM relation classifier is shown in Table 6. We counted the number of extracted relations and the number of attributes covered by the relations. Ideally, the attributes should not exist independently, and the cover percentage should be 100%. By dividing the number of corresponding attributes (has_value & Measurement; has_temp & Temporal Constraints; modified by & Qualifier), we calculated the percentage of extracted relations from the existing relations (Perc. of Extracting in Table 6). Our method extracted 54.81% of relations in general, 79.93% of relations between qualifier and entity, and 38.24% measurement.
Table 6.
Statistical Matching Result of Extracted Relations
| Number | Unique number | Attribute number | Perc. of Extracting (%) | |
|---|---|---|---|---|
| Has_value | 3005 | 2224 | 5816 | 38.24 |
| Has_temp | 4632 | 3051 | 4507 | 67.69 |
| Modified by | 10400 | 7477 | 9354 | 79.93 |
|
| ||||
| Total | 18037 | 12752 | 19677 | 54.81 |
We also evaluated the relation extraction performance by manually reviewing 100 randomly-selected trials and counting statistical measurements including true positive, false positive and false negative. Then, we calculated the precision and recall of the three types of relation, “modified by”, “has temporal constraints” and “has value” as shown in Figure 4. The performance of relation “modified by” was the best among these three relations, while “has value” was the poorest. The performance evaluation result corresponds to the descriptive statistic matching result of extracted relations.
Figure 4.
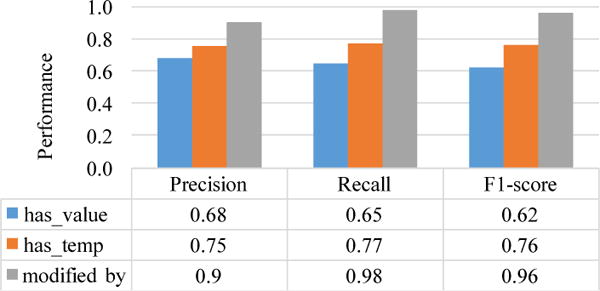
Evaluation of Relation Extraction
Sample Use Cases
This database of structured, standard-based eligibility criteria enables several use cases for integrating clinical research studies and electronic health records.
We have designed a RESTful API for users to input terms and search studies with certain criteria. Our http request accepted several parameters including entity key word (entity), criteria type (c-type), temporal constraint key word (t-constrain), qualifier (qua), etc. The sample request is in following format: GET {domain}/?entity=#&c_type=#&t_constrain=#&qua=#. The response to this request is a list of clinical trials NCT identifiers that match the request.
Here we take several pair querying examples. For instance, if we are concerned about which trial has exclusion criteria involving participants with severe psychotic features within the previous three months, we are going to fetch all the available information for those specific parameters in the format {“entity”: psychotic, “qualifier”: severe, “criteria type”: exclusion, “temporal constraints”: three + months}. The complete URL for this request as it appears on the GET{domain}/?entity=psychotic&qualifier=severe&c_type=e x&t-constrain=three+month. The response to this request comes with two trials NCT identifiers: NCT01822951 and NCT00911807.
Another pairing example is to look for participants who have had stable AD therapy, which often occurs as essential inclusion criteria in Alzheimer’s disease clinical studies. In this case, the parameters should consist of entity (AD) and qualifier (stable). The following list of NCT identifiers is the response to this request: NCT00495417,NCT02051608,NCT01122329,NCT02670083, NCT02386306,NCT01954550,NCT02423122,NCT00299988. Therefore, by using the RESTful API, healthcare providers or clinical research investigators can request relevant study criteria information from our database.
Discussion
We fetched 1587 trials of Alzheimer’s disease as of September 2016 from ClinicalTrial.gov [12] and captured 4453, 2360, 1930, 626, 449 unique terms of Condition, Observation, Drug, Procedure, and Qualifier respectively. We also matched extracted terms into CONCEPT_ID in OMOP CDM v5. The compression ratio of Condition, Observation, Drug, Procedure, and Qualifier were respectively 0.08, 0.10, 0.17, 0.09 and 0.02. Then we associated attributes with entities via relations including “has_value”, “has_temp” and “modified by” into a relational database. The relation “modified by” can be found and extracted from 79.99% of Qualifier. We justified the benefit of this method by descriptive statistical analysis and detailed user cases. We then further discussed the great potential and future application of this method in bridging the gap between EHR and EC.
Error Analysis
Errors of NER and relation extraction mainly resulted from wrong classified predictions. As for NER, the performance of the Procedure domain was poorer than that of other domains because the Procedure had the smallest number of instances in the training set. Since the output of NER is the input of the relation extraction, the errors in NER task will be multiplied in the relation extraction step.
Another cause of errors is the incomplete coverage of entities in the OMOP CDM v5. In other words, not all the terms existing in the criteria text have already been modeled in the OMOP CDM v5. Scarcity in the OMOP terminology dictionary is the reason why the matching score of some terms are lower than 0.50. Terms consisting of capital letters such as AChEI, NIA-AA criteria, MI are not identified correctly. An entire list of recommended terminology that could be added to the Concept table of OMOP CDM v5 will be provided.
Primary Contributions
This study has made four primary novel contributions.
First, we enabled semantic search of criteria by normalizing clinical terms using standard terminologies and by mapping them to CONCEPT_IDs in OMOP CDM. In this way, terms that share one meaning were regarded as the same. For instance, in the previous search methods, the term “AV block” was not returned by the query using the term “atrioventricular block”. In our database, these two terms are referred to one CONCEPT_ID, 316135. Users will no longer be inconvenienced by incomplete search results revolving around heterogeneous semantic representations for the same concept. Furthermore, each CONCEPT_ID has an associated clinical code such as ICD9CM, SNOMED_CT in the CONCEPT table of OMOP CDM. Users will be able to search for a specific disease by inputting its ICD9CM code.
The second primary contribution is the transformation of free-text criteria into a computable relational database compliant with an EHR common data model. The way relations were stored is a highlight of our work. For example, the many-to-many relationships in the database schema can retrieve relations between an entity and its respective attribute. Also, the clear definition and completeness of the attribute category will become a strong tool for handling pair querying, that the advanced search function provided by ClinicalTrial.gov could not achieve. For instance, if we input a combined search of several different domains such as “severe” + “Alzheimer’s Disease” + “for three years” + “inclusion”, then the search result will include all the trials with participants who have had severe Alzheimer’s disease for three years. Therefore, users can query and search the database for sophisticated logical queries, which can essentially improve the efficiency of clinical trial EC reuse.
Thirdly, the database of Alzheimer’s disease provides different audiences with an effective computer-based knowledge representation of EC. Study investigators could query in both the hospital data warehouse and the database of EC to target eligible participants. Another use case for a trial designer is that the computable format of the criteria could help them define future study guidelines by comparing differences and commonalities of EC and study contents.
Finally, as an evidence-based clinical support method, the combination of searching the databases of EHR and EC allows healthcare providers to determine if a patient’s treatment will benefit from a particular study or decide whether the patient is eligible for a study. Essentially, EHRs can be automatically matched to computably formatted clinical trial EC in our database. We could design a pipeline for patient screening with a combination of EHRs and the database of EC.
Limitations and Future Work
Based on the work we have done, researchers could build a database with more comprehensive information from clinical trial studies. Our future work will concentrate on two areas: performance and completeness. To improve the performance of our transformation pipeline for the free-text criteria, we will need to explore methods to extract complex expressions of Temporal Constraints [13] and Measurement [14]. We plan to extract specific numerical and temporal expressions from complex attributes. For example, temporal information such as “for three months” should be extracted and stored as “three” + “month” into different columns. Measurement information like “Hemoglobin ≥ 9.0g/dL” should be extracted and stored by number and unit separately and the unit for the same test should be unified. Further collaborative research on natural language processing of free-text information is desired.
To improve the completeness, the outcomes and other sections of the trial will need to be transformed into structured output and stored into the database. We would also like to expand the method to cover the entire disease spectrum from ClincalTrial.gov. More studies are warranted to test how this method would work for other eligibility features of other diseases. We may need to expand the database to better cover the eligibility features and elements. Additional tables may need to be added such as the Anatomic Location or Genetic Name when it comes to cancer. Furthermore, we will design a user-friendly interface to retrieve the necessary features from our database. The implementation of Django, a high-level web framework, also encourages rapid development and design that significantly reduces the workload of the back-end development. Therefore, we successfully transformed free-text EC into a computable, relational database following OMOP CDM v5. We hope that this computable format of EC can support the need of predictive analysis of targeted participants of clinical trials in the near future.
Conclusions
We contributed a practical method for transforming free-text eligibility criteria into a computable, relational database following OMOP Common Data Model (CDM) version 5. This method promises to be applicable to all disease trials in ClinicalTrial.gov and to accelerate EHR-based clinical research.
Figure 1.
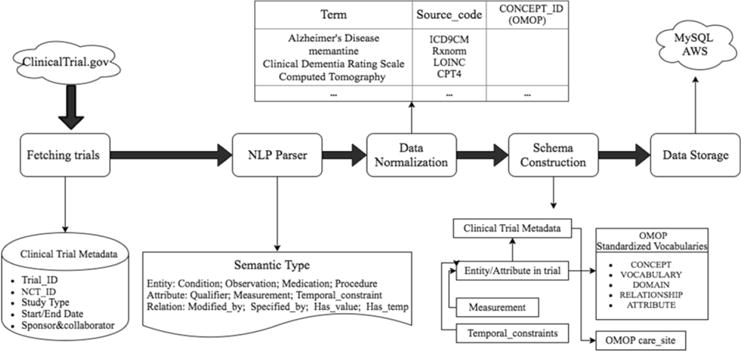
Workflow of Transformation and Reconstruction
Table 3.
Definition of TP, FP, FN, TN of a NER System
| True positive | System extracts a concept that matches the label |
| False positive | System extracts a concept but there is no label or doesn’t match the correct label |
| False negative | System doesn’t extract a concept but there is a label |
| True negative | System doesn’t extract a concept and there is no label |
Acknowledgments
This research was supported by R01 LM009886 from The National Library of Medicine (Bridging the semantic gap between research eligibility criteria and clinical data; PI:Weng).
References
- 1.Wu DT, Hanauer DA, Mei Q, Clark PM, An LC, Proulx J, Zeng QT, Vydiswaran VV, Collins-Thompson K, Zheng K. Assessing the readability of ClinicalTrials.gov. Journal of the American Medical Informatics Association. 2016 Mar 1;23(2):269–75. doi: 10.1093/jamia/ocv062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tu SW, Peleg M, Carini S, Bobak M, Ross J, Rubin D, Sim I. A practical method for transforming free-text eligibility criteria into computable criteria. Journal of biomedical informatics. 2011 Apr 30;44(2):239–50. doi: 10.1016/j.jbi.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levy-Fix G, Yaman A, Weng C. Proceedings of 2015 AMIA Joint Summits for Translational Science. San Francisco: 2015. Structuring Clinical Trial Eligibility Criteria with Common Data Model; pp. 194–198. [Google Scholar]
- 4.Observational Health Data Sciences and Informatics (OHDSI) OMOP Common Data Model V5.0. Available at https://www.ohdsi.org. Accessed Nov 2016
- 5.Hripcsaka G, Dukeb JD, Shahc NH, Reichd CG, Husere V, Schuemief MJ, Suchardh MA, Parki RW, Wongf IC, Rijnbeekj PR. Observational Health Data Sciences and Informatics (OHDSI): Opportunities for Observational Researchers. Studies in health technology and informatics. 2015;216:574. [PMC free article] [PubMed] [Google Scholar]
- 6.CLAMP: Clinical Language Annotation, Modeling and Processing Toolkit. Available at http://clamp.uth.edu. Accessd Nov 2016.
- 7.Kang T, Zhang S, Tang Y, Hruby GW, Rusanov A, Elhadad N, Weng C. EliIE: An open-source information extraction system for clinical trial eligibility criteria. Journal of the American Medical Informatics Association. 2017 Apr 1; doi: 10.1093/jamia/ocx019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Usagi, an application to help create mappings between coding systems and the Vocabulary standard concepts. Available at https://github.com/OHDSI/usagi. Accessed Nov 2016.
- 9.Chang CC, Lin CJ. LIBSVM: a library for support vector machines. ACM Transactions on Intelligent Systems and Technology (TIST) 2011 Apr 1;2(3):27. [Google Scholar]
- 10.Django. Available at https://www.djangoproject.com/. Accessed Nov 2016.
- 11.RESTful Web services. Available at http://www.restapitutorial.com/. Accessed Dec 2016.
- 12.ClinicalTrial.gov. http://www.clinicaltrials.gov/. Accessed Dec 2016.
- 13.Boland MR, Tu SW, Carini S, Sim I, Weng C. EliXR-TIME: a temporal knowledge representation for clinical research eligibility criteria. AMIA Summits Transl Sci Proc. 2012;2012:71–80. [PMC free article] [PubMed] [Google Scholar]
- 14.Hao T, Liu H, Weng C. Valx: a system for extracting and structuring numeric lab test comparison statements from text. Methods of information in medicine. 2016;55(3):266–75. doi: 10.3414/ME15-01-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]


