SUMMARY
Changes in social preference of amphibian larvae result from sustained exposure to kinship odorants. To understand the molecular and cellular mechanisms of this neuroplasticity, we investigated the effects of olfactory system activation on neurotransmitter (NT) expression in accessory olfactory bulb (AOB) interneurons during development. We show that protracted exposure to kin or non-kin odorants changes the number of dopamine (DA)- or gamma aminobutyric acid (GABA)-expressing neurons, with corresponding changes in attraction/aversion behavior. Changing the relative number of dopaminergic and GABAergic AOB interneurons or locally introducing DA or GABA receptor antagonists alters kinship preference. We then isolate AOB microRNAs (miRs) differentially regulated across these conditions. Inhibition of miR-375 and miR-200b reveals that they target Pax6 and Bcl11b to regulate the dopaminergic and GABAergic phenotypes. The results illuminate the role of NT switching governing experience-dependent social preference.
In Brief
Acquisition of social preference for siblings versus non-siblings is dynamic, but the developmental regulatory mechanisms have been unclear. Dulcis et al. found that sustained exposure to olfactory cues converts non-kin aversion to attraction via neurotransmitter switching coordinated by microRNAs.
INTRODUCTION
Many animals use olfactory-mediated sibling and non-sibling recognition via kinship odorants to distinguish between siblings and non-siblings. 4-day-old (stage 44) X. laevis larvae display attraction and aversion behavior to water-borne kin and non-kin odorants, respectively (Waldman and Adler, 1979). Kin and non-kin refer to odorants while sibling and non-sibling refer to raising conditions. The olfactory basis of kinship recognition was demonstrated by showing that larvae with external nares blocked by a gelatinous paste did not discriminate behaviorally between siblings and non-siblings (Waldman, 1985).
This social preference of Xenopus tadpoles can be altered by their exposure to kinship-signaling molecules from non-siblings versus siblings during embryonic development (Waldman, 1981) and at later stages (Yamazaki et al., 1988) as well as by major histocompatibility complex (MHC) genotype differences (Villinger and Waldman, 2008). Self- and non-self-recognition, which lie at the core of social behavior, are mediated by activation of vomeronasal organ (VNO) neurons in mice and frogs (Ben-Shaul et al., 2010; Chamero et al., 2012; Hagino-Yamagishi et al., 2004). These olfactory neurons express receptors that are activated by peptides cleaved from major urinary proteins (MUPs) or MHC proteins (Kaur et al., 2014; Manning et al., 1992; Sturm et al., 2013). The larval frog olfactory system has been the object of extensive studies (Jungblut et al., 2012; Manzini and Schild, 2010; Sansone et al., 2014). VNO receptor cells project to the accessory olfactory bulb (AOB). These findings suggest that sibling recognition may result from activation of VNO neurons by kinship-signaling odorants, generating activity patterns that lead to circuit plasticity.
Electrical activity and calcium signaling have significant roles in directing several aspects of neuronal differentiation, including neurotransmitter (NT) respecification. Sustained changes in electrical activity by experimental manipulations or by natural sensory stimuli cause the expression of NTs in CNS neurons that otherwise produce different ones, both during development and in the mature nervous system (Borodinsky et al., 2004; Dulcis et al., 2013; Dulcis and Spitzer, 2008; Gu and Spitzer, 1995; Gutiérrez et al., 2003). Transmitter respecification during development occurs in reserve pool neurons characterized by distinct patterns of calcium signaling that combine with transcription factors to create responsiveness to activity manipulations (Demarque and Spitzer, 2010; Dulcis and Spitzer, 2012; Guemez-Gamboa et al., 2014; Marek et al., 2010; Velázquez-Ulloa et al., 2011).
Here we examine the mechanisms underlying plasticity of social preference in the developing accessory olfactory system of Xenopus larvae and the behaviors that ensue. We discover social experience-dependent changes in dopamine (DA) and gamma aminobutyric acid (GABA) expression in granule and periglomerular interneurons of the AOB affecting kinship recognition and preference. These findings may shed light on the olfactory imprinting of fish (Gerlach et al., 2008; Harden et al., 2006; Hino et al., 2009; Johnstone et al., 2011) and neonatal attachment in mammals (Janus, 1993; Kendrick et al., 1997a; Porter and Etscorn, 1974), potentially including humans.
The molecular players coordinating the transcriptional changes at the core of this experience-dependent NT plasticity are largely unexplored. MicroRNAs (miRs) are strong candidates because they mediate post-transcriptional regulation of expression of multiple genes (Bartel, 2004; Izaurralde, 2015). miRs have crucial roles at many steps in neurogenesis and brain development (Li and Jin, 2010; Yoo et al., 2009), including neuronal fate determination (Chang et al., 2004; de Chevigny et al., 2012; Kim et al., 2007). miRs execute and refine these complex developmental transitions by targeting the mRNAs encoding key regulatory proteins such as transcription factors and repressors (Visvanathan et al., 2007; Xu et al., 2013). miRs also help brain adaptation to environmental perturbations through activity-dependent forms of plasticity (Krol et al., 2010; McNeill and Van Vactor, 2012). Testing the hypothesis that miRs are involved in kinship-induced DA and GABA plasticity observed in the AOB, we show that miR-200b and miR-375 control Pax6 and Bcl11b expression and produce NT plasticity that determines kinship recognition behavior.
RESULTS
Kinship Odorants Drive an Activity-Dependent DA/GABA Switch in AOB Neurons
Amphibian embryos are laid in clutches, and larvae are normally exposed to kinship odorants after hatching. To investigate whether exposure to these odorants affects NT expression in the AOB, we raised Xenopus larvae with several hundred siblings until they hatched (Nieuwkoop and Faber, 1967; stage 39) at 2 days after fertilization. Larvae were then single housed (Figure S1A) and exposed for 2 days (stage 39 to 44) to kin or non-kin odorants or to no odorants (Figure 1A). Brains were fixed, sectioned, and analyzed immunohistochemically (IHC) for the presence of tyrosine hydroxylase (TH, a marker for DA expression) and GABA in AOB neurons (Figure 1B). Genetic programs had likely set the baseline NT identity (Spitzer, 2017). We found that the numbers of dopaminergic and GABAergic periglomerular and granule neurons (Figure S1B, arrows and arrowheads) depend on exposure to water-borne kin odorants, non-kin odorants, or the absence of odorants (Figures 1B and 1C; Figure S1C). There was no effect on the total number (mean ± SD) of AOB granule (297 ± 5 non-sibling; 338 ± 26 orphan; 308 ± 27 sibling) and periglomerular (198 ± 30 non-sibling; 191 ± 9 orphan; 197 ± 12 sibling) neurons (Figure 1C) and total cells labeled with the nuclear marker DRAQ5 (Figure 2A). Exposure to kin odorants led to the largest numbers of interneurons expressing only TH and the smallest numbers of interneurons expressing GABA in both granule and periglomerular cells. Exposure to non-kin odorants, in contrast, led to the smallest numbers of neurons expressing only TH and the largest numbers of neurons expressing only GABA. The orphan condition generated the highest number of TH/GABA co-expressing neurons. This result suggests that co-expression is the default NT expression in the absence of odorants, potentially generated in response to remaining spontaneous activity.
Figure 1. Neurotransmitter Phenotype Switching between Sibling and Non-sibling Conditions Is Activity Dependent.
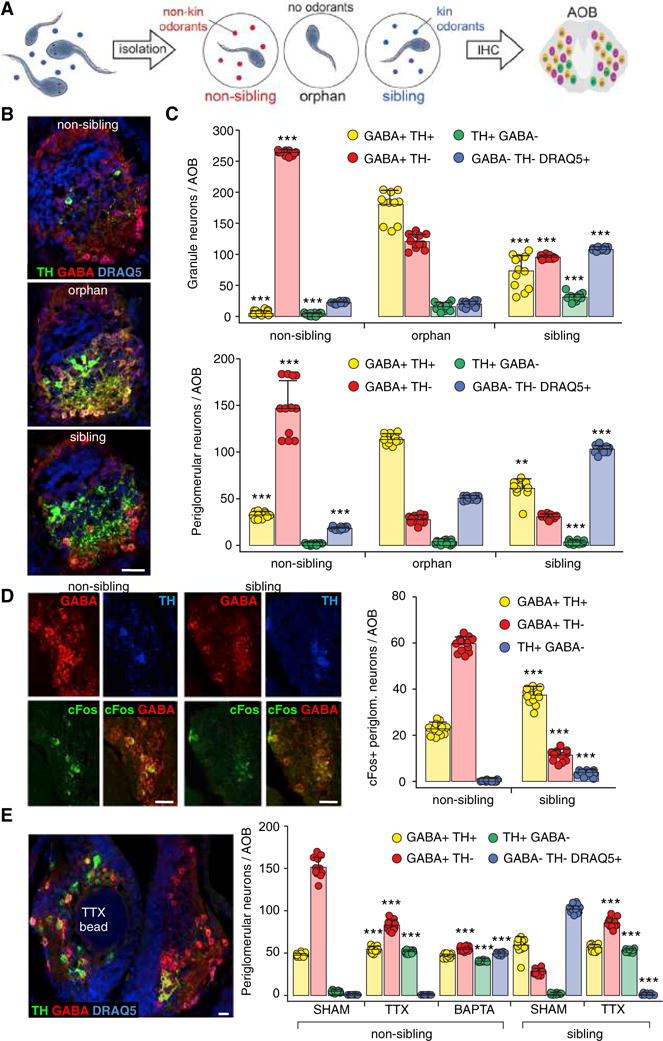
(A) Exposure of larvae to different odorants. Larvae cohabit until stage 39 (left). They are then individually housed (middle) during 48 hr exposure to either non-kin (red, non-sibling raising condition) or kin (blue, sibling raising condition) odorants or no odorants (orphan raising condition). AOBs were processed for immunohistochemistry (IHC) at stage 44.
(B) IHC identifies neurotransmitter expression. Triple labeling of TH, GABA, and DRAQ5 in single AOB sections of animals raised in non-sibling, orphan, and sibling conditions. Scale bar, 60 μm.
(C) Raising conditions determine the number of granule neurons/AOB (above) and periglomerular neurons/AOB (below) co-expressing both GABA and TH (yellow) or expressing GABA only (red), TH only (green), or neither (nuclear marker DRAQ5+, blue). Graphs show data points (circles) and the mean ± SD. n = 12 larvae from 3 independent experiments. One-way ANOVA (granule neurons: GABA+TH+: F(2,33) = 233.6, p < 0.0001; GABA+ TH−: F(2,33) = 2,137, p < 0.0001; TH+GABA−: F(2,33) = 154.3, p < 0.0001; GABA−TH−DRAQ5+: F(2,33) = 4,019, p < 0.0001) (periglomerular neurons: GABA+TH−: F(2,33) = 196.3, p < 0.0001; TH+ GABA−: F(2,33) = 59.11, p < 0.0001; GABA−TH−DRAQ5+: F(2,33) = 2,276, p < 0.0001) with Bonferroni’s multiple comparison test, except GABA+TH+ (periglomerular neurons), where Kruskal-Wallis with Dunn’s multiple comparison test was used. Comparisons of each subgroup are between the mean of the orphan condition and the means of non-sibling and sibling conditions. **p < 0.01, ***p ≤ 0.001.
(D) The pattern of cFos expression in AOB neurons expressing GABA or TH (stage 44) changes following exposure (48 hr) of larvae to non-kin or kin odorants (non-sibling or sibling condition). Images of sections immunostained for GABA, TH, and cFos show cFos localization in periglomerular neurons. Quantification shows the reduction of cFos expression in TH+ cells and the increase in GABA+ cells induced by non-kin exposure, compared to the sibling condition. Scale bars, 40 μm. Graphs show data points (circles) and the mean ± SD. n = 12 larvae from 3 independent experiments. Unpaired Student’s t test, except TH+GABA−, where Mann-Whitney U test was used. ***p ≤ 0.001.
(E) Activity dependence of transmitter respecification. Horizontal section through the AOB (stage 44) triple labeled for TH, GABA, and DRAQ5 shows that the effect of a TTX-loaded agarose bead on the GABA and TH phenotypes in non-sibling raising conditions is restricted to the AOB surrounding the bead (left; compare with contralateral internal control). Quantification of periglomerular neurons/AOB (right) shows that both TTX and BAPTA reduced the number of GABA neurons and increased the number of TH neurons compared to sham in the non-kin condition. In the kin condition, TTX increased the number of both GABA neurons and TH neurons compared to sham. Cells that do not express GABA or TH are marked by DRAQ5 (blue bar). Scale bar, 30 μm. Graphs show data points (circles) and the mean ± SD. n = 12 larvae from 3 independent experiments. Non-sibling: one-way ANOVA (GABA+TH+: F(2,33) = 33.57, p < 0.0001; GABA+TH−: F(2,33) = 520.1, p < 0.0001; TH+GABA−: F(2,33) = 2,367, p < 0.0001; GABA−TH−DRAQ5+: F(2,33) = 7,200, p < 0.0001) with Bonferroni’s multiple comparison test. Comparisons of each subgroup are between the mean of the sham condition and the means of TTX and BAPTA conditions. Sibling: Mann-Whitney U test (GABA+TH+), unpaired Student’s t test (GABA+TH−), Welch’s t test (TH+GABA−, GABA−TH−DRAQ5+). ***p ≤ 0.001.
Figure 2. Neurotransmitter Switching between Sibling and Non-sibling Conditions Occurs within a PAX6+ Reserve Pool.
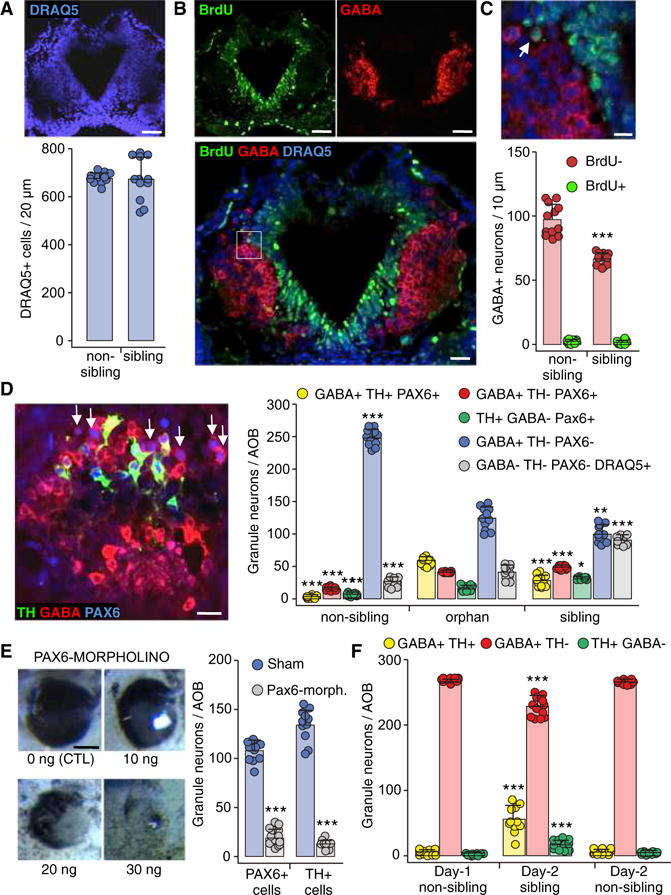
(A) The total number of AOB cells does not change across raising conditions. Horizontal section through the middle of the AOB (stage 44) labeled for DRAQ5. Quantification of DRAQ5+ cells/20 μm section. Scale bar, 60 μm. Graph shows data points (circles) and the mean ± SD. n = 12 larvae from 3 independent experiments.
(B) BrdU labeling to test for neurogenesis. Images of a horizontal section through the AOB (stage 44) labeled for BrdU (left) and GABA (right); superimposed image of the same BrdU and GABA channels above, plus DRAQ5, are shown at higher magnification (below). Scale bars, 60 μm (top) and 30 μm (bottom).
(C) Inset in (B) at higher magnification shows a BrdU+GABA+ cell (left, arrow). Quantification of the number of BrdU+GABA+ neurons/10 μm section across non-sibling and sibling raising conditions (below). Scale bar, 20 μm. Graphs show data points (circles) and the mean ± SD. n = 12 larvae from 3 independent experiments. Welch’s t test. ***p ≤ 0.001.
(D) GABAergic Pax6+ reserve pool neurons (arrows) shown in an AOB section triple labeled with antibodies for TH, GABA, and PAX6 (stage 44). Scale bar, 45 μm. Quantification of PAX6+ granule neurons/AOB co-expressing both GABA and TH (yellow) or expressing GABA only (red) or TH only (green) for non-sibling, orphan, and sibling raising conditions. The number of GABAergic neurons that are PAX6− and TH−is also quantified (blue). Granule cells that do not express TH, GABA, or Pax6 were identified by the nuclear marker DRAQ5 (gray, compare with Figure 1C). Graphs show data points (circles) and the mean ± SD. n = 11 larvae from 3 independent experiments. One-way ANOVA (GABA+TH+PAX6+: F(2,30) = 247.5, p < 0.0001; GABA+TH−PAX6+: F(2,30) = 779.1, p < 0.0001; GABA+TH−PAX6−: F(2,30) = 308.4, p < 0.0001; GABA−TH−PAX6−DRAQ5+: F(2,33) = 190, p < 0.0001) with Bonferroni’s multiple comparison test, except TH+GABA−PAX6+, where Kruskal-Wallis with Dunn’s multiple comparison test was used. Comparisons of each subgroup are between the mean of the orphan condition and the means of non-sibling and sibling conditions. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
(E) PAX6-morpholino-injected larvae. Bright field images of the eye phenotype of larvae injected with different morpholino concentrations (left). Scale bar, 100 μm. Quantification of PAX6+ (left) and TH+ (right) granule neurons/AOB of larvae raised in the sibling condition and injected with PAX6-morpholino (blue) or sham (gray). Graphs show data points (circles) and the mean ± SD. n = 11 larvae from 3 independent experiments. Unpaired Student’s t test (PAX6+ cells), Welch’s t test (TH+ cells). ***p ≤ 0.001.
(F) Partial reversibility of dopaminergic plasticity in larvae exposed to the non-sibling raising condition for 24 hr (from stage 39 to stage 42) and then exposed to either sibling or non-sibling raising condition for another 24 hr (stage 42 to stage 44). Quantification of the number of AOB granule neurons that are co-expressing both GABA and TH, GABA only, or TH only. Graphs show data points (circles) and the mean ± SD. n = 11 from 3 independent experiments. One-way ANOVA (GABA+TH+: F(2,30) = 61.91, p < 0.0001; GABA+TH−: F(2,30) = 56.91, p < 0.0001; TH+GABA−: F(2,30) = 28.57, p < 0.0001) followed by Bonferroni’s multiple comparison test. Comparisons of each subgroup are between the mean of the day 1 non-sibling condition and the means of day 2 sibling and non-sibling conditions. ***p ≤ 0.001.
See also Figures S1–S4.
To test the activity dependence of kinship-induced NT plasticity, we first quantified AOB interneurons expressing cFos along with TH and/or GABA in response to kin or non-kin odorants. Only a subset of AOB interneurons was cFos+ in response to odorants. In non-sibling conditions, cFos was expressed in many GABA-only cells and GABA/TH co-expressing cells but not detected in TH-only cells. In sibling conditions, cFos was expressed in a small number of GABA-only cells, in a large number of GABA/TH co-expressing cells, and in a significant number of TH-only cells (Figure 1D).
We then implanted drug-loaded agarose beads unilaterally in the AOB at the time of single housing to deliver tetrodotoxin (TTX) to block action potentials or a calcium chelator (BAPTA-AM) to selectively suppress calcium-mediated activity during non-kin or kin exposure. Bead delivery of TTX and BAPTA suppresses sodium channel-dependent action potentials and calcium-mediated activity in Xenopus larvae (Borodinsky et al., 2004; Dulcis and Spitzer, 2008). Because BAPTA-AM and calcein-AM are similar in molecular weight, fluorescent calcein-AM-loaded beads were used to estimate the extent of drug diffusion from the site of implantation (Figure S2C). Layers of cells that were 70 μm from the bead had a level of fluorescence (F1) equal to background (F0) after 24 hr, suggesting that drug application was local. Both TTX and BAPTA reduced the number of GABA neurons and increased the number of TH neurons compared to sham in the non-sibling condition. In the sibling condition, TTX increased the number of both GABA neurons and TH neurons compared to sham (Figure 1E). These results imply that neurons that appear to be respecifying their NT possess activity necessary for responsiveness to sensory circuit activation (Velázquez-Ulloa et al., 2011). Exposure to non-kin odorants (Figure 1E, left) caused the expected large number of GABA+ and small number of TH-only neurons in the AOB contralateral to the bead-implanted side (compare sham with Figure 1C), confirming local application of the drugs.
Kinship Odorants Do Not Change Cell Number and Instead Alter the DA/GABA Phenotype by Acting on a Pax6+ Reserve Pool
The constant number of AOB cells quantified with the nuclear marker DRAQ5 across conditions (Figure 2A) suggests that NT switching is not paralleled by changes in the number of neurons. Experiments testing the presence of cell proliferation (Figures 2B and 2C) revealed that 3 ± 2 GABA+ neurons/AOB section incorporated BrdU following non-kin odorant exposure for 24 hr (stages 39–42). In contrast, non-kin exposure led to 30 ± 12 newly GABA+ neurons/section (the difference between the non-sibling and sibling red GABA+ bars in Figure 2C). Thus, the GABAergic phenotype is expressed in neurons that underwent their last DNA synthesis before the last 24 hr of non-kin exposure, and the increased number of GABA neurons is not due to cellular proliferation. To test whether cell differentiation is affected more generally during NT respecification, we co-labeled AOB TH+ and GABA+ interneurons with doublecortin (DCX), a marker expressed by neuronal precursor cells and immature neurons. Following non-kin-induced GABA respecification (Figure S3A), only 6 ± 4 GABA+ cells/AOB section were DCX+ at stage 42 in contrast with 30 ± 12 newly GABA+ neurons/section (the difference between the non-sibling and sibling red GABA+ bars in Figure S3B). These GABA+DCX+ neurons were not periglomerular (green arrows) or granule GABA+ neurons (yellow arrows) of the AOB (Figure S3A) but were located dorsally and ventrally to developed glomeruli (Figures S3A and S3B, blue arrows). They are likely to be future interneurons serving developing glomeruli that will become active after stage 49 when class II olfactory receptors recognizing air-borne odorants are expressed (Mezler et al., 1999). The lack of DCX expression in AOB TH+ and GABA+ neurons provides further evidence that this experience-dependent NT respecification is not occurring in newborn neurons.
Since neurons expressing a newly specified NT are not recruited from undifferentiated neurons, we immunostained the AOB for transcription factors Pax6 and Lim1,2, which are associated with dopaminergic differentiation (Hack et al., 2005), to identify the switching reserve pool of neurons (Figure 2D; Figures S1D, S1E, S3C, and S3D). Pax6 expression begins in mouse AOB progenitor cells and is maintained in mature TH+ neurons, while it is downregulated in cells that differentiate as GABAergic granule neurons (Hack et al., 2005). GABA+ cells were rarely Lim1,2+ (Figure S3D), and its role was not considered further. To test the hypothesis that Pax6+ neurons represent the reserve pool able to respecify NT following exposure to different kinship odorants, we first determined whether Pax6 expression changes in response to different kinship odorant exposure during development. We found that the number of Pax6+ neurons varies across the three kinship-raising conditions (Figure 2D). Pax6 is expressed in all TH+ cells across conditions; however, it is also expressed in 8% of GABA-only cells in the non-sibling condition, 35% of GABA-only cells in the orphan condition, and 50% of GABA-only cells in the sibling condition. These results indicate that the reserve pool of Pax6+ cells is dynamic.
To test whether Pax6 expression is necessary for recruitment to the dopaminergic phenotype during kinship odorant exposure, we electroporated fluorescein-tagged oligo morpholinos selectively targeting Pax6 transcripts (Rungger-Brändle et al., 2010) into the two-cell stage embryo. This electroporation down-regulates Pax6 expression during induction of NT respecification in sibling conditions. A dose-response analysis identified the minimal concentration of Pax6 morpholino (10 ng/embryo) required to achieve a marginal eye disruption phenotype (Figure 2E). Knocking down 83% of Pax6 expression blocked 90% of the kin-induced increase in TH-expression. Pax6+ neurons are unlikely to be immature precursors waiting to acquire a NT identity since quantification of the Pax6+ pool double stained for TH and GABA showed that all Pax6+ cells already express either TH or GABA. Additionally, Pax6+ neurons do not express DCX in both kin and non-kin conditions (data not shown). However, we cannot exclude that early Pax6 morpholino treatment may have affected NT switching by a different mechanism other than a purely kin odorant effect.
Because neurogenesis is absent, the sibling condition is likely to recruit a number of GABAergic neurons expressing the Pax6 transcription factor (Figure S1E, arrows) to become dopaminergic neurons expressing Pax6 but no longer expressing GABA. Consistent with the association of Pax6 expression with dopaminergic differentiation, the Pax6 morpholino reduced the number of TH+ cells. This result suggests that neurons switch transmitters in response to kin odorants, losing expression of one (GABA) and gaining expression of another (DA).
Transmitter Switching Is Partially Reversible and Specific to AOB Neurons
To test the reversibility of transmitter switching, we first exposed larvae to non-kin odorants for 24 hr (from stage 39 to stage 42) and then exposed these larvae to either kin or non-kin odorants for another 24 hr (stage 42 to stage 44). Granule and periglomerular interneurons were identified by their positions in the AOB. The induction of GABA expression observed after 1 day of exposure to non-kin odorants was not fully reversed by a second day of exposure to kin odorants. However, this second day exposure induced a significant increase in the number of TH+ cells co-expressing GABA (Figure 2F; Figure S4A). These results indicate partial reversibility that may become greater with longer periods of reversed odorant exposure.
In contrast to NT switching in the AOB granule and periglomerular neurons, there was no change in the number of neuropeptide-Y+ (NPY+) neurons located adjacent to AOB GABAergic neurons (Figure S4B) or in the number of TH+ neurons of another dopaminergic nucleus, the suprachiasmatic nucleus (Figure S4C), between sibling and non-sibling raising conditions. To investigate other neuronal components of the AOB circuit, we immunostained the vesicular glutamate transporter (VGLUT1,2) and neuronal nitric oxide synthase (nNOS). This identified the glutamatergic olfactory receptor neurons (ORNs) that provide VNO sensory input to the AOB and the NOS+ mitral cells that are the principal AOB output. We observed a conserved glutamatergic ORN projection pattern and glomerular neuropil and detected no changes in the number of NOS+ mitral cells across conditions (Figures S4D and S4E).
Neurotransmitter Switching Is Paralleled by Changes in Social Preference
We next tested kin recognition behavior in groups of 50 free-swimming larvae using a novel larval distribution assay (Figure 3A). We found that different numbers of TH-only and GABA-only neurons are correlated with the way in which AOB circuitry processes kin/non-kin odorants and generates an attractive or aversive response to these odorants. Shortly after odorant-conditioned saline is placed in one of the inserts at each end of the test tank, larvae initiate active swimming that lasts 5–10 min (sampling phase). Each larva can sample the entire length of the container in as little as 5 s (Figure 3A, dashed line). Sibling-raised larvae cluster at one end of the container by swimming either toward the kin odorant-releasing insert (Figure 3B) or away from the non-kin odorant-diffusing insert until they stop swimming after about 10 min (kinship recognition phase). Swimming gradually resumes as the odorant gradient in the solution between the two inserts dissipates, and larvae then distribute equally across the two sides of the tank (rebound phase). Since the differences in larval distribution are most significant after 10 min and 15 min from insert loading, these data points were averaged. A percent larval distribution ratio (LDR) was calculated by dividing the average number of larvae distributed on the side containing the kinship odorant by the total number of larvae for each experiment and condition. The initial LDR value before odorant introduction was always 50%.
Figure 3. Kinship Recognition Is Odorant Dependent.
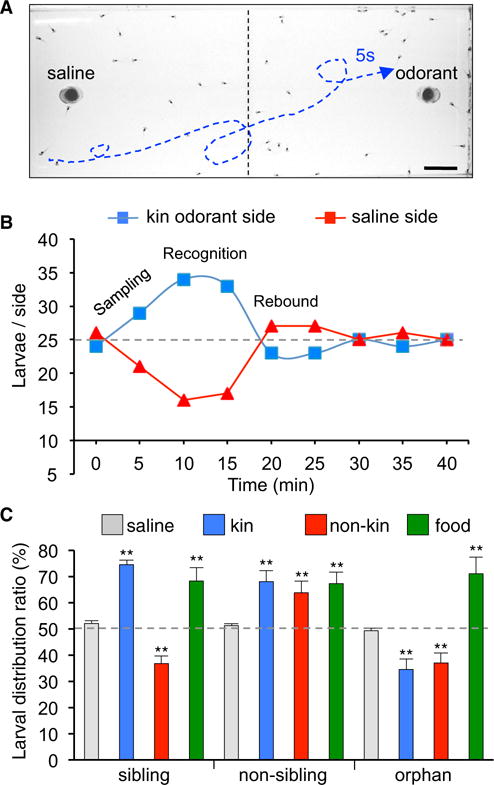
(A) Photograph of larvae swimming in a water-filled tank prior to loading of odorant in the insert at right. Representative larval trajectory during a 5 s interval in the presence of odorant is indicated by the blue dashed line. Scale bar, 1.5 cm.
(B) Results of a larval distribution assay with 50 larvae raised in sibling conditions and tested with kin odorants. The greatest difference of larval distribution across the left and right sides of the tank occurs between 10 and 15 min. The average at these times is expressed as a percent of the total number of larvae to define the larval distribution ratio (LDR, %).
(C) The LDR of larvae raised in sibling, non-sibling, or orphan conditions changes systematically when tested with kin or non-kin odorants or food, compared to saline. Graph shows the mean ± SD. n = 4 independent experiments with 50 larvae/experiment. One-way ANOVA (sibling: F(3,12) = 32.01, p < 0.0001; non-sibling: F(3,12) = 10.36, p = 0.0012; orphan: F(3,12) = 27.69, p < 0.0001) with Bonferroni’s multiple comparison test. Comparisons of each subgroup are between the mean of the saline condition and the means of all other conditions. **p ≤ 0.01.
See also Figure S5 and Movie S1.
Larvae raised in the sibling condition, which is typical in nature, display attraction to kin odorants (60% LDR) and aversion to non-kin odorants (37% LDR) (Figures S5A and S5B; Movie S1 illustrates behavior during 20 s of free swimming). Changes in the numbers of granule and periglomerular interneurons expressing TH and/or GABA obtained by raising larvae in the non-sibling and orphan conditions are correlated with altered kinship recognition behavior. Larvae raised in the non-sibling condition, with an AOB configuration of a high number of GABA-only neurons and reduced number of TH-only neurons, broaden their social preference. These larvae now display an attractive response to non-kin odorants (65% LDR) in addition to kin odorants (Figure 3C; Figures S5C and S5D). Retention of attraction to kin odorants suggests that this is innate or learned during the first 2 days of development and is preserved after experience-dependent learning of attraction to non-kin odorants. Larvae raised in the orphan condition, with an AOB configuration characterized by the highest TH/GABA co-expression, display a general reduction of social preference.
These larvae exhibit aversion to non-kin odorants (37% LDR) as well as to kin odorants (34% LDR; Figure 3C; Figures S5E and S5F). Loss of the attraction response to kin odorants displayed by orphan larvae suggests that the lack of kinship cue experience during development from stage 39 to stage 44 puts the AOB in a default state that overrides the innate or learned attraction response to kin odorants. We also measured the LDR of sibling-, non-sibling-, and orphan-raised larvae in response to powdered pellets of Xenopus food (Zeigler), a mix containing 12 different amino acids. This is a non-social cue and was consistently attractive during changes in social preference across raising conditions, demonstrating specificity of the effects of social cue-induced AOB processing of social preference (Figure 3C; Figures S5G and S5H).
Newly Dopaminergic Neurons Appear Sufficient to Drive Sibling Recognition
We then investigated the role of newly dopaminergic neurons in altered kinship recognition behavior, since kin and non-kin odorants may change other features of olfactory circuitry in addition to TH and GABA expression in granule and periglomerular neurons. To detect a causal connection between neurons with newly respecified NTs and altered kinship recognition behavior, we first ablated pre-existing dopaminergic neurons in the AOB by local exposure to the dopaminergic neurotoxin MPTP (Dulcis and Spitzer, 2008) delivered via agarose bead implantation (Figures 4A and 4B; Figure S6B). Dose/response quantification of MPTP efficacy showed that 800 μM MPTP ablates 100% of TH+ interneurons while sparing TH neurons in the AOB as well as TH+ neurons of the neighboring ventral main olfactory bulb (Figure 4C; Figure S6B, arrows). This treatment prevented attraction to kin odorants (50% LDR; Figure 4D), suggesting that dopaminergic neurons in the AOB are necessary for normal processing of kinship odorants to elicit attraction to siblings. We next exposed MPTP-treated larvae to kin odorants for 24 hr to induce DA respecification of spared GABA+TH− reserve pool neurons in the presence of the MPTP blocker deprenyl (Tatton and Greenwood, 1991) (Figure 4A). This treatment led to 88% restoration of attraction to kin odorants (70% LDR; Figure 4D). The functional rescue suggests that newly specified DA-expressing neurons are sufficient to fulfill the dopaminergic requirement mediating kinship recognition behavior toward siblings.
Figure 4. Contribution of Dopaminergic and GABAergic AOB Interneurons to Kinship Recognition Behavior.
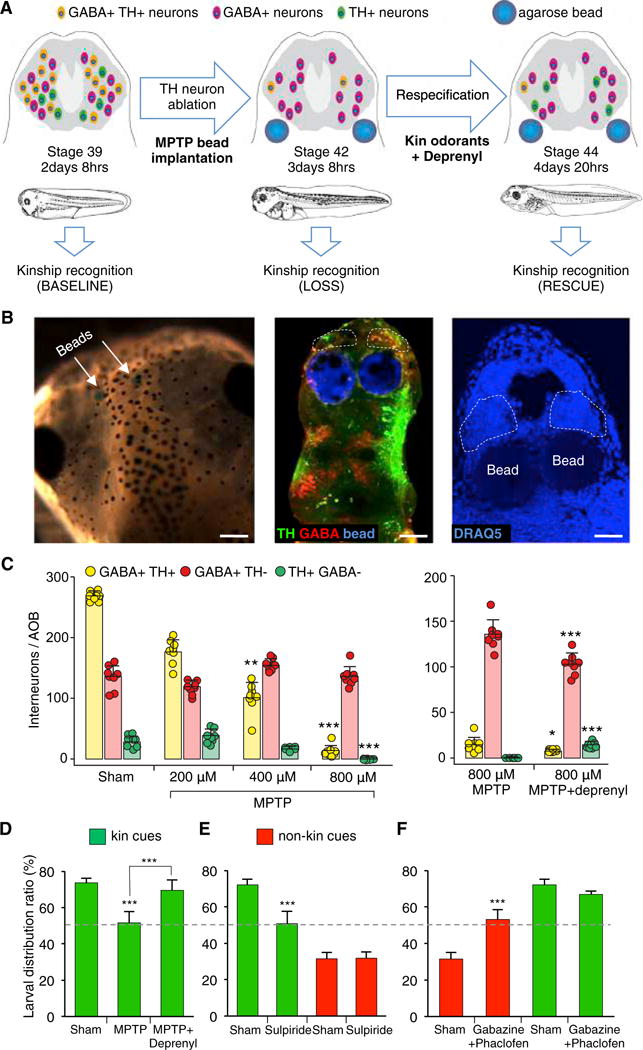
(A) Experimental design to identify the causal connection between neurons with newly respecified NTs and altered kinship recognition behavior. Kinship recognition was measured before (baseline, stage 39) and after (loss of function, stage 42) MPTP-mediated ablation of native dopaminergic neurons in the AOB (green cells) of sibling-raised larvae. Larvae were then exposed to kin odorants for 24 hr in the presence of the MPTP blocker deprenyl to induce spared GABA+TH−reserve pool neurons (purple cells) to newly express DA (yellow cells) and re-tested for kinship recognition (rescue of function, stage 44).
(B) Delivery of drugs via bilateral agarose bead implantations at stage 39. Beads (blue) are visible through the skin of stage 44 larvae (left, arrows). Beads are adjacent to the AOBs (dashed lines) in whole-mount preparations of the CNS double labeled for TH (green) and GABA (red) (middle). They are ventral to the AOBs (dashed lines) in horizontal sections counterstained with DRAQ5 (right). Scale bars, 200 μm (left), 80 μm (middle), and 50 μm (right).
(C) Quantification of the number of interneurons/AOB (stage 44, sibling raised) co-expressing both GABA and TH or expressing GABA only or TH only following dopaminergic ablation with different MPTP concentrations (left). Deprenyl induces partial rescue of dopaminergic ablation (right). Graphs show data points (circles) and the mean ± SD. n = 8 larvae from 3 independent experiments. Kruskal-Wallis with Dunn’s multiple comparison test (left). Comparisons of each subgroup are between the mean of the sham condition and the means of different MPTP conditions. Mann-Whitney U test (right). *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
(D) Larval distribution assay (stage 44 sibling raised) shows MPTP-induced loss of attraction to kin odorants and functional restoration by newly induced dopaminergic neurons (deprenyl) compared to sham. Graph shows the mean ± SD. n = 10 independent experiments with 50 larvae/experiment, except MPTP + Deprenyl: n = 6. One-way ANOVA (F(2,23) = 55.15, p < 0.0001) with Bonferroni’s multiple comparison test. Comparisons are between the mean of each column against the mean of every other column. ***p ≤ 0.001.
(E) Larval distribution assay (stage 44 sibling raised) shows selective loss of attraction to kin odorants and intact aversion to non-kin odorants following sulpiride-mediated DA receptor blockade compared to sham. Graph shows the mean ± SD. n = 6 independent experiments with 50 larvae/experiment. Unpaired Student’s t test. ***p ≤ 0.001.
(F) Larval distribution assay (stage 44 sibling raised) shows selective loss of aversion to non-kin odorants and intact attraction to kin odorants following gabazine/phaclofen-mediated GABA receptor blockade compared to sham. Graph shows the mean ± SD. n = 6 independent experiments with 50 larvae/experiment, except gabazine/phaclofen: n = 9. Unpaired Student’s t test. ***p ≤ 0.001.
See also Figure S6.
Selective Blockade of AOB DA or GABA Receptors Suppresses Attraction or Aversion Behaviors
Because we found that the numbers of GABA- and TH-expressing neurons in the AOB are experience dependent (Figures 1B and 1C), we investigated how GABA and DA contribute to AOB processing of attraction versus aversion behaviors in response to kin and non-kin odorants. To achieve local receptor blockade in the AOB of larvae raised in the sibling condition, we implanted agarose beads impregnated with NT receptor antagonists. Given the involvement of D2 receptors in frog olfactory processing (Davison et al., 2004), we blocked D2 receptors with sulpiride and found that this abolished attraction (50% LDR) to kin odorants without affecting aversion (30% LDR) to non-kin signals (Figure 4E). In contrast, blockade of GABAAB receptors by application of gabazine and phaclofen in the AOB suppressed aversion toward non-kin odorants (50% LDR) while leaving the ability to recognize siblings intact (65% LDR; Figure 4F).
These experiments identify reciprocal roles for GABA- and DA-expressing AOB interneurons in regulation of attractive and aversive kinship recognition behaviors when larvae are raised in the sibling condition. The profiles of GABA-only, TH-only, and GABA/TH neurons in the AOBs of non-sibling and orphan larvae are different from the DA and GABA AOB profiles of sibling larvae, suggesting that each raising condition generates a distinct circuit configuration. Thus, NT switching affecting the locations of synaptic release of GABA and DA within the AOB may lead to the atypical recognition behaviors observed in non-sibling and orphan larvae.
Small RNA Profiling Identifies miRs Differentially Regulated by Kinship Exposure
miRs have frequently emerged as regulators of coordinated multicomponent signaling pathways. To test the involvement of miRs in regulating transmitter switching, we sequenced small RNAs from AOB tissue harvested from animals exposed to sibling and non-sibling conditions and obtained close to 400 miRs in each case. To select miR candidates that could mediate NT respecification, we applied three criteria. First, we focused on medium-to-highly expressed miRs (1%–20% of the total reads) that more likely mediate significant regulation of gene expression (Mullokandov et al., 2012). Second, we chose miRs that had the largest fold changes between sibling and non-sibling conditions. Third, we identified miRs predicted to regulate genes in the DA or GABA pathways. We included regulatory genes (transcription factors and genes determining neuronal NT identity) and genes involved in NT metabolism (synthetic enzymes, transporters, degradative enzymes, and receptors) (Table S1). Bioinformatics tools were used to identify predicted miR responsive elements (MREs) in each transcript; excluding the miRs that were not predicted to target the transcripts reduced the number for qPCR validation. This strategy produced a list of 18 miR candidates potentially involved in NT switching between dopaminergic and GABAergic phenotypes in sibling and non-sibling conditions (Figures 5A and 5B; Figure S7C).
Figure 5. Small RNA Profiling Reveals miRs Differentially Regulated by Sustained Kinship Exposure.
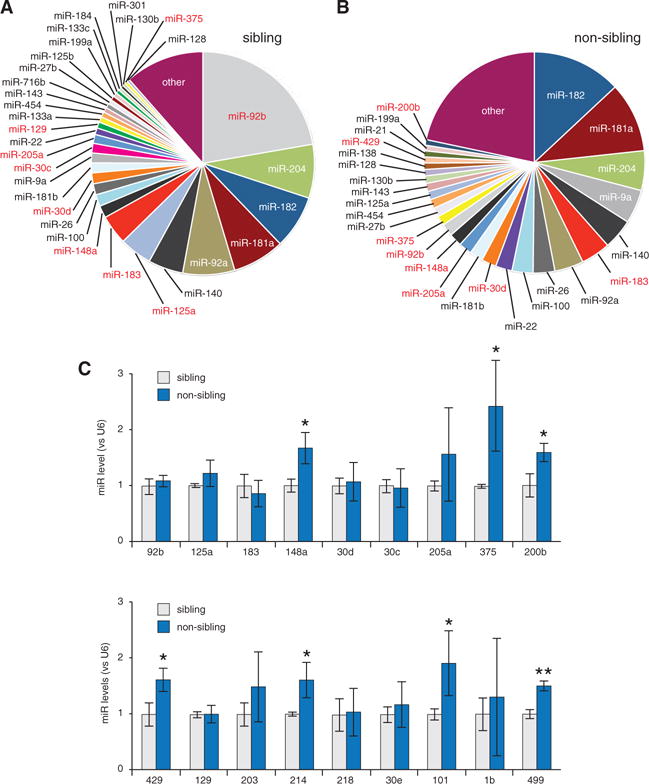
(A) Fractions of total sequenced reads for the indicated miRs in AOB tissue harvested from animals exposed to sibling condition.
(B) Equivalent fractions for exposure to non-sibling condition. Red entries in (A) and (B) were validated by qPCR.
(C) qPCR validation of the top 18 miR candidates. miR-148a, miR-375, miR-200b, miR-429, miR-214, miR-101, and miR-499 are significantly different in non-sibling versus sibling conditions. Graphs show the mean ± SD. n = 3 independent experiments with 15 AOB samples per condition, except miR-375 and miR-200b: n = 5. Unpaired Student’s t test. *p ≤ 0.05; **p ≤ 0.01.
See also Figure S7.
miR-375/miR-200b Regulate DA/GABA Phenotype Switching in the AOB
To validate the top miR candidates, we performed qPCR from three independent clutches of larvae raised in the presence of kin or non-kin odorants. This analysis revealed significantly elevated expression of miR-148a, miR-375, miR-200b, miR-429, miR-214, miR-101, and miR-499 in non-sibling versus sibling conditions (Figure 5C; miR-200b and miR-429 are similar in sequence, and their seeds are identical). We performed a secondary analysis among these kinship-dependent miRs to identify those directly targeting two genes known to be determinants of the dopaminergic and GABAergic phenotypes in the olfactory bulb: Pax6 (Hack et al., 2005) and Bcl11b (Enomoto et al., 2011). The expected interactions of these miR candidates with Pax6 or Bcl11b mRNAs were suggested by the presence of their highly conserved MREs on the target genes (Figures 6A and 6B). Interestingly, both miR-375 and miR-200b are predicted to target MREs on Pax6 mRNA; miR-200b, miR-214, miR-101, and miR-499 are predicted to recognize MREs on Bcl11b mRNA (Figures S7A and S7B). MiR-148a targeted neither Pax6 nor Bcl11b transcripts and was not analyzed further.
Figure 6. miR-375/miR-200b Regulate Pax6 and Bcl11b Levels in the AOB.
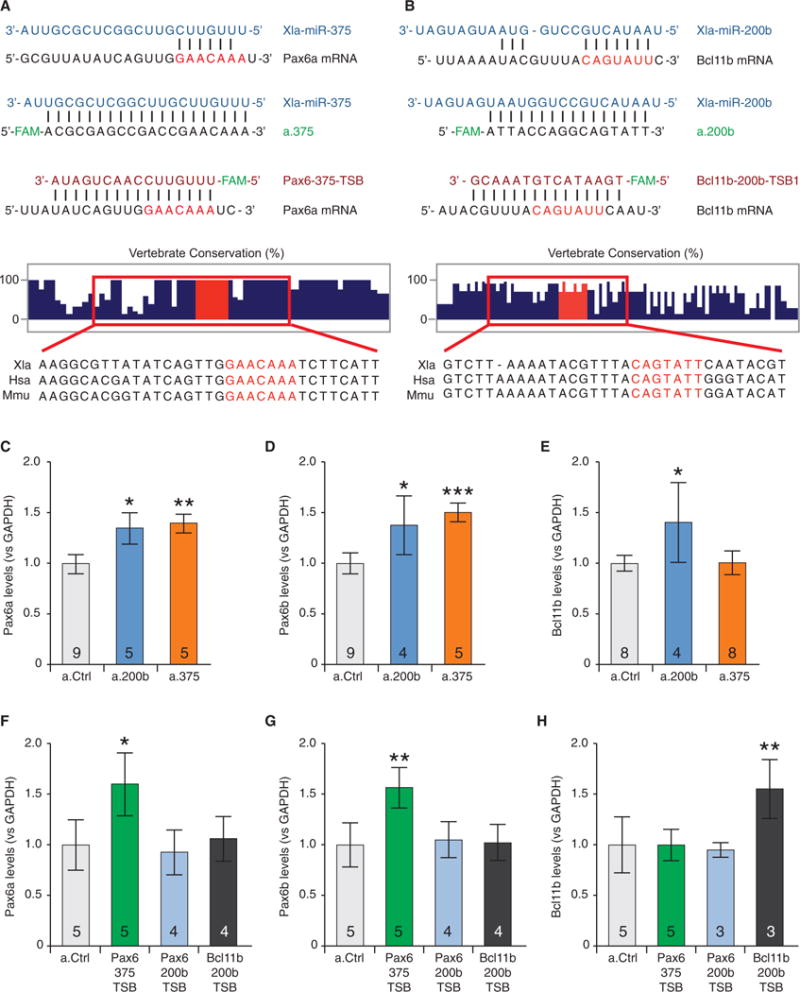
(A) The interactions between miR-375 and the MRE on Pax6a mRNA (top), between miR-375 and the antagonist a.375 (second from top), and between the target-site blocker Pax6-375-TSB and the MRE on the Pax6a mRNA (third from top). Vertical lines indicate base pair matches. Vertebrate conservation of the region flanking the MRE on Pax6a mRNA (second from bottom), adapted from basewise conservation by PhyloP (UCSC Genome Browser), with Multiz alignments for Xenopus laevis (Xla), Homo sapiens (Hsa), and Mus musculus (Mmu) (bottom).
(B) The interactions between miR-200b and the first MRE on Bcl11b mRNA (top), between miR-200b and the antagonist a.200b (second from top), and between the target-site blocker Bcl11b-200b-TSB1 and the first MRE on Bcl11b mRNA (third from top). Vertebrate conservation of the region flanking the MRE on Bcl11b mRNA (second from bottom), with Multiz alignments (bottom).
(C and D) Injection of a.200b and a.375 in the AOB increased levels of the Pax6a (C) and Pax6b (D) isoforms compared to a.Ctrl, measured via qPCR.
(E) a.200b, but not a.375, increased levels of Bcl11b compared to a.Ctrl.
(F and G) Pax6-375-TSB, but not Pax6-200b-TSB or Bcl11b-200b-TSB, increased Pax6a (F) and Pax6b (G) levels compared to a.Ctrl.
(H) Only Bcl11b-200b-TSB increased Bcl11b levels compared to a.Ctrl.
(C–H) Graphs show the mean ± SD. n for each condition is reported in the bar graph and represent independent experiments with 15 AOB samples per condition. One-way ANOVA (C: F(2,16) = 9.143, p = 0.0022; D: F(2,15) = 12.65, p = 0.006; E: F(2,17) = 3.286, p = 0.0622; F: F(3,14) = 3.671, p = 0.0386; G: F(3,14) = 5.274, p = 0.0121; H: F(3,12) = 9.900, p = 0.0014;) followed by Bonferroni’s multiple comparison test. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001.
See also Figure S6.
To specifically inhibit each miR individually in vivo, we designed (Exiqon) locked nucleic acid (LNA) miR antagonists and delivered them locally in the AOB. The levels of the mRNA for Pax6 and Bcl11b were then determined by qPCR to evaluate their potential roles as regulators of NT respecification. MiR-375 and miR-200b emerged as the most promising candidates (Figures 6C–6E; Figures S7D–S7H). Consistent with previous findings (de Chevigny et al., 2012), the inhibitor for miR-375 (a.375) increased Pax6 levels (Figures 6C and 6D) but had no effect on Bcl11b levels (Figure 6E). MiR-200b, however, appeared to play a double role as a coherent regulator targeting both Pax6 and Bcl11b transcripts (Figures 6C–6E). While the miR-200b MREs on Bcl11b are well conserved, the MREs on Pax6 are nearly exclusive to Xenopus, suggesting species-specific regulation (Figures S6A and S6B). These data indicate that increased miR-375 and miR-200b levels in non-sibling conditions could contribute both to the reduction of TH-only neurons by suppressing expression of the stimulatory Pax6 transcription factor and to the increase in GABA-only neurons by suppressing production of the inhibitory Bcl11b transcription factor.
To determine whether miR-375 and miR-200b regulate mRNA levels of Pax6 and Bcl11b by direct interaction with their transcripts, we utilized LNA target-site blockers (TSBs, Exiqon). A TSB selectively protects the MRE on a specific mRNA by stopping a given miR from binding and repressing its expression, leaving all other mRNA targets unaffected (Lippi et al., 2016) (Figures 6A and 6B, third oligonucleotide pair). Local delivery to the AOB of the TSB protecting the miR-375 MRE on Pax6 (Pax6-375-TSB) increased Pax6 mRNA levels comparable to those achieved with a.375 (Figures 6F and 6G). Similarly, the TSB protecting the miR-200b MRE on Bcl11b (Bcl11b-200b-TSB) upregulated Bcl11b transcripts (Figure 6H). No effect on Pax6 levels in the AOB was observed with TSBs protecting the two miR-200b sites on Pax6 (Pax6-200b-TSB) (Figures 6F and 6G), suggesting that regulation of Pax6 by miR-200b is through a non-canonical MRE or indirect.
To ascertain whether changes in the levels of miR-375 and/or miR-200b transcripts were sufficient to drive changes in the number of dopaminergic and/or GABAergic neurons in vivo, we performed TH/GABA immunostaining on AOBs from antagonist- or TSB-treated larvae raised in the non-sibling condition (Figure 7). Larvae received a.375 or a.200b either by electroporation at an early stage (Figure S6A) or by infusion of in vivo ready LNAs/TSBs at a later stage. Electroporation of a.375 blocked the typical reduction of the dopaminergic phenotype induced by non-kin exposure (Figure 7A). In contrast, electroporation of a control antagonist (a.Ctrl) not targeting any miR under the same conditions did not block the reduction. The ability of miR-375 to repress the dopaminergic phenotype was confirmed when the AOB was infused with an in vivo ready Pax6-375-TSB (Figures 7B and 7E). This experiment demonstrates that miR-375 exerts its inhibitory function by direct interaction with the Pax6 target transcript. Notably, the Pax6-375-TSB had no effect on the number of GABAergic neurons (Figure 7E).
Figure 7. miR-375 and miR-200b Inhibitors Regulate Neurotransmitter Expression.
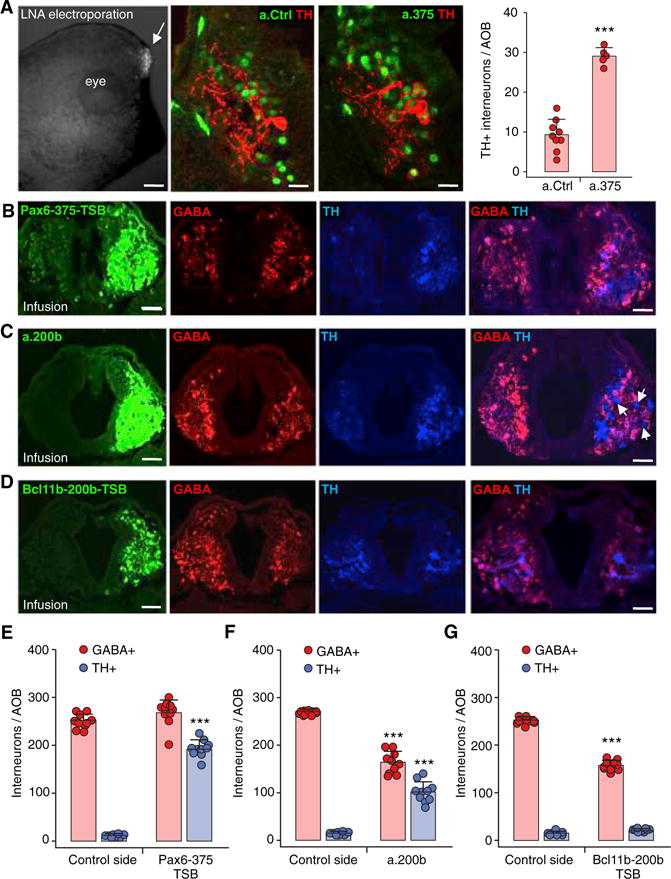
(A) Electroporation of 6-carboxyfluroscein (FAM)-tagged antagonists (LNA) in the AOB performed on stage 21 larvae raised in non-sibling conditions and scored at stage 33 (left, arrow). Images demonstrate a.375 blockade, compared to a.Ctrl, of the reduction in dopaminergic phenotype (TH) normally induced by non-kin exposure (middle). Quantification of the number of TH+ interneurons/AOB (right) following electroporation of miR antagonists (a.375 and a.200b) and control LNA (a.Ctrl). Scale bars, 300 μm (left), 30 μm (middle). Graph shows the mean ± SD. n = 5 larvae from 3 independent experiments. ***p < 0.001.
(B) Infusions of in vivo ready Pax6-375-TSB in the AOB performed on stage 39 larvae raised in non-sibling conditions and analyzed in stage 42 larvae (left). AOB sections immunostained for GABA and TH show the Pax6-375-TSB-mediated block on the infused side of the reduction in dopaminergic phenotype normally induced by non-kin exposure, which is still observed on the control side. Scale bars, 40 μm (3 left panels), 50 μm (right).
(C) Infusions of in vivo ready miR antagonist a.200b in the AOB performed on stage 39 larvae raised in non-sibling conditions and analyzed at stage 42 (left). AOB sections immunostained for GABA and TH show the simultaneous blocking by a.200b of both the reduction in dopaminergic phenotype and the induction of the GABAergic phenotype normally caused by non-kin exposure; these effects are confined to the infused side and do not extend to control side. Scale bars, 40 μm (3 left panels), 50 μm (right).
(D) Infusions of in vivo ready Bcl11b-200b-TSB in the AOB performed on stage 39 larvae raised in non-sibling conditions and analyzed in stage 42 larvae (left). AOB sections immunostained for GABA and TH show Bcl11b-200b-TSB-mediated blockade of the increased GABAergic phenotype normally induced by non-kin exposure. There is no effect on TH expression, and the GABAergic block does not extend to the control side. Scale bars, 40 μm (3 left panels), 50 μm (right).
(E) The number of TH+ and GABA+ interneurons/AOB following infusion of in vivo ready Pax6-375-TSB compared to the control side.
(F) The number of TH+ and GABA+ interneurons/AOB following infusion of in vivo ready miR antagonist a.200b compared to the control side.
(G) The number of TH+ and GABA+ interneurons/AOB following infusion of in vivo ready Bcl11b-200b-TSB compared to the control side.
(E–G) Graphs show data points (circles) and the mean ± SD. n = 10 larvae from 3 independent experiments. Unpaired Student’s t test, except for (E), (F), and (G) GABA+, where Mann-Whitney U test was used. ***p ≤ 0.001.
See also Figure S7A.
When we infused in vivo ready a.200b into the AOB of larvae raised in non-sibling conditions to selectively block miR-200b function, we observed two distinct effects: a reduction in the number of GABAergic neurons with a simultaneous increase in the number of dopaminergic neurons (arrows, Figures 7C and 7F). The ability of miR-200b to reduce the GABAergic phenotype was confirmed by infusing the AOB with an in vivo ready Bcl11b-200b-TSB (Figures 7D and 7G). This test demonstrates that miR-200b exerts its function on the GABA phenotype by direct interaction with the Bcl11b target transcript. The infusion of Bcl11b-200b-TSB did not, however, replicate the effect of a.200b on the dopaminergic neurons. Since a.200b increases both Pax6 and Bcl11b transcript levels, these data suggest an important role for miR-200b as a regulator of the genetic switch between DA and GABA (Ebert and Sharp, 2012).
DISCUSSION
Here we identify cellular mechanisms of activity-dependent neuroplasticity that drive activity-dependent acquisition of social preference. We show that prolonged exposure of sibling larvae to kin odorants drives changes in NT expression from GABA to DA in Xenopus AOB neurons recruited from a GABA+PAX6+ pool, accompanied by behavioral preference for kin odorants. In contrast, sustained exposure of sibling larvae to non-kin odorants induces a DA-to-GABA shift in AOB neurons and a parallel aversion-to-attraction shift in social preference toward the same non-kin odorants. These findings are reminiscent of the mechanism described for acquired imprinting that occurs in early neonatal life in sheep (Kendrick et al., 1997b), resulting in an increase in the ratio of excitatory to inhibitory tone in granule cell synapses onto mitral cells. Orphan-raised larvae have a large number of neurons co-expressing DA and GABA and avoid both kin and non-kin odorants. The number of AOB interneurons expressing only DA and only GABA changes in response to developmental exposure to kinship odorants in the absence of neurogenesis. NT respecification has been shown to occur through activation of the visual pathway both in the developing amphibian nervous system (Dulcis and Spitzer, 2008) and in the adult rat brain (Dulcis et al., 2013). NT plasticity in the mouse AOB and the main olfactory bulb (Cave et al., 2010) indicates that this form of transmitter reconfiguration can be elicited by activation of other sensory modalities. Induction of NT switching has been described in adult mice (Aumann et al., 2013; Kesby et al., 2017) and may occur in humans (Aumann et al., 2016).
NT switching in a given circuit has powerful consequences for circuit function, since neuromodulation of a single target neuron can dramatically alter the performance of an entire network (Gutierrez and Marder, 2014). Selective blockade of DA or GABA transmission in the AOB indicated the specific roles of these two transmitters in kinship recognition displayed by larvae raised in sibling conditions, as described for zebrafish (Scerbina et al., 2012; Souza et al., 2011). Ablation of endogenously dopaminergic neurons in sibling-raised larvae followed by kin odorant induction of newly dopaminergic neurons recruited from the reserve pool affirmed their contribution to attraction to kin. However, experience-dependent NT respecification leading to increased DA and GABA co-expression (orphan condition) led to aversion to both kin and non-kin odorants. Respecification to an increased number of GABA-only neurons (non-sibling condition) led to attraction to both non-kin and kin odorants. We conclude that the different configuration of NT expression and release in the AOB, and not the class of NTs alone, determines the control of aversion versus attraction behaviors toward kinship cues. Reversal of social preference in frogs and zebrafish with sexual maturation (Gerlach and Lysiak, 2006) suggests that experience-dependent DA/GABA transmitter switching in the AOB induced by atypical kinship exposure during development may also be activated in adults.
Cellular changes in NT expression are achieved by genetic regulation mediated by miRs. RNA profiling of the AOB detected miRs that are regulated across kinship conditions and identified miR-375 and miR-200b as key regulators mediating DA and GABA switching (Figure 8). We find that miR-375 inhibits Pax6 through a previously described MRE (Bhinge et al., 2016; de Chevigny et al., 2012). Although the role of Pax6 as a determinant of the dopaminergic phenotype in the olfactory bulb is well established (Brill et al., 2008; Hack et al., 2005; Kohwi et al., 2005; Ninkovic et al., 2010), its regulation by miRs as a developmental switch appeared only recently. miRs regulate transcription factor levels during development to modulate many aspects of neuronal identity (Amin et al., 2015; Bhinge et al., 2016; Chen et al., 2011; de Chevigny et al., 2012; Thiebes et al., 2015).
Figure 8. Model of the Mechanisms Involved in Neurotransmitter Respecification Affecting Social Preference.
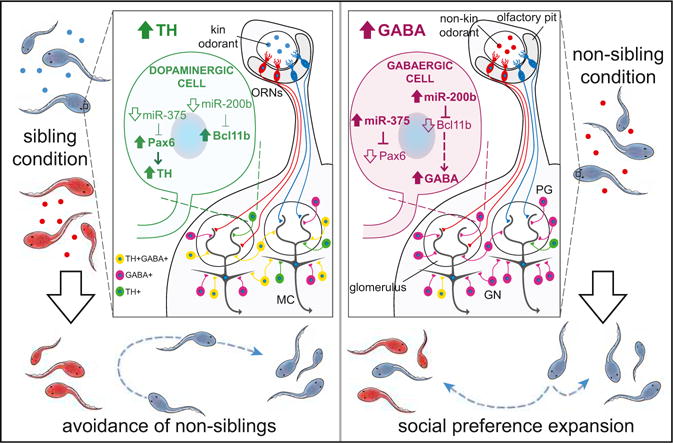
Kinship raising conditions provide olfactory cues that determine behavioral attraction or aversion toward siblings and non-siblings. These cues can also affect social preference. Sustained kin (sibling condition, left) or non-kin (non-sibling condition, right) odorant exposure for 48 hr during development drives activity-dependent DA/GABA switching in neurons of the developing AOB. Signaling is regulated by miRs acting on Pax6 and Bcl11b transcripts to control DA and GABA expression that determines social preference.
Because kinship odorants also modulate GABA expression in AOB interneurons, we investigated the functional role of miRs that are upregulated in non-sibling conditions and are predicted to recognize a conserved MRE on transcription factor Bcl11b transcripts. We show that miR-200b, controlling olfactory neurogenesis in mice (Choi et al., 2008) and eye development in X. laevis (Gessert et al., 2010), directly regulates Bcl11b expression levels. Bcl11b inhibits brain-derived neurotrophic factor (BDNF) signaling (Tang et al., 2011), potentially providing a link between miR-200b-mediated Bcl11b regulation and GABA expression since BDNF stimulates the GABA phenotype of developing spinal neurons (Guemez-Gamboa et al., 2014). Accordingly, we suggest that non-kin odorants detected by ORN receptors increase neural activity in odor-specific pathways, inducing c-Fos expression and driving the activity-dependent increase of miR-200b in VNO interneurons that inhibits translation of its target Bcl11b. A reduction of Bcl11b expression may increase BDNF and upregulate GABA in AOB interneurons.
MiR-200b has a pivotal role in reciprocal regulation of DA/GABA phenotype switching. Infusion of a.200b to selectively block miR-200b function in the AOB elicited both a reduction in the number of GABAergic neurons and a coordinated increase in the number of dopaminergic neurons. Since a.200b increases both Pax6 and Bcl11b mRNA levels, it appears to regulate a genetic switch for DA and GABA, sharpening the transition to GABA phenotype by inhibiting both the activator, Pax6, and the repressor, Bcl11b. Future work will determine the molecular basis of NT switching induced by kin odorants. miR-dependent coordination of gene expression regulates behavior in a variety of settings (Gascon et al., 2014; Tan et al., 2013) that can depend on social context (Shi et al., 2013).
How are odor responses linked to miR expression? There are two classes of odor response. The first, in response to a brief exposure to an odor, is behavioral. The second, in response to a sustained exposure to an odor, is metabolic, changing the behavioral response when the larvae are later briefly exposed to a particular odor. Different miRs lead to expression of different NTs, which in turn activate circuits, causing different behaviors. We speculate that the ecological functions of attraction to siblings and avoidance of non-siblings are to promote the survival of the respective gene pools in the face of predation. Sibling acquisition of attraction to non-siblings, which constitutes a form of adoption, may promote survival of the species. Whether or not there are other longer-term benefits to these behaviors remains to be determined.
STAR+METHODS
Detailed methods are provided in the online version of this paper and include the following:
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-Tyrosine Hydroxylase (clone LNC1) | Chemicon/Millipore | Cat#MAB318; RRID: AB_2313764 |
| Guinea pig polyclonal anti-GABA | Millipore | Cat#AB175; RRID: AB_91011 |
| Rabbit polyclonal anti-Doublecortin | Abcam | Cat#Ab18723; RRID: AB_732011 |
| Rabbit polyclonal anti-Pax6 | Covance | Cat#PRB-278P; RRID: AB_2313780 |
| Rat monoclonal anti-BrdU | Bio-Rad | Cat#OBT0030S; RRID: AB_609570 |
| Rabbit polyclonal anti-VGLUT1,2 | Synaptic Systems | Cat#135503; RRID: AB_2285905 |
| Rabbit polyclonal anti-nNOS | Millipore | Cat#07-571; RRID: AB_310722 |
| Rabbit anti-Neuropeptide Y | ImmunoStar | Cat#22940; RRID: AB_10720817 |
| Sheep polyclonal anti-Tyrosine Hydroxylase | Imgenex | Cat#IMG-5070; RRID: AB_317561 |
| Goat polyclonal anti-cFos | Santa Cruz Biotechnology | Cat#Sc-52-G; RRID: AB_2629503 |
| Biological Samples | ||
| Xenopus laevis Accessory Olfactory Bulb (AOB) | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Calcein-AM | Invitrogen | Cat#C3099; CAS: 148504-34-1 |
| BAPTA-AM | Invitrogen | Cat#B6769; CAS: 126150-97-8 |
| TTX | Sigma | Cat#T8024; CAS: 4368-28-9 |
| Sulpiride | Sigma | Cat#S8010; CAS: 15676-16-1 |
| Gabazine | Tocris | Cat#1262; CAS: 104104-50-9 |
| Phaclofen | Tocris | Cat#0178; CAS: 114012-12-3 |
| Deprenyl | Sigma | Cat#M003; CAS: 14611-52-0 |
| DRAQ5 | BioStatus | Cat#BOS-889-001 |
| Tricaine | Pentair | Cat#TRS4 |
| Human chorionic gonadotropin | Merck | Cat#057176 |
| MPTP | Sigma | Cat#M0896; CAS: 23007-85-4 |
| Xenopusfood | Zeigler | http://www.zeiglerfeed.com/Literature/Adult%20Xenopus%20Diet.pdf |
| Critical Commercial Assays | ||
| Recover All Total Nucleic Acid | Ambion | Cat#AM1975 |
| TaqMan microRNA assays | Applied Biosystems | Cat#4366596 |
| Universal Probe Library | Roche | 04683633001 |
| Agarose beads (Affi-gel blue) | Bio-Rad | Cat#153-7302 |
| Deposited Data | ||
| Small RNA-seq (GEO: GSE78854) | GEO | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=gdcrgkkelrchfmv&acc=GSE78854 |
| Experimental Models: Organisms/Strains | ||
| Xenopus laevis pigmented (Lab bred WT adult, 9+ cm) | Xenopus Express | Cat# LB OP XL FM |
| Xenopus laevis pigmented (Lab bred adult, 9+ cm) | Nasco | Cat# LM00531M |
| Oligonucleotides | ||
| Morpholinos | Gene Tools | http://www.gene-tools.com |
| Custom LNA miR inhibitors | Exiqon | Cat#500150 |
| Custom LNA Target Site Blockers | Exiqon | Cat#500150 |
| Software and Algorithms | ||
| Targetscan | N/A | http://www.targetscan.org/vert_71/ |
| iHOP | N/A | http://www.ihop-net.org/UniPub/iHOP/ |
| BioGRID | N/A | https://thebiogrid.org/ |
| KEGG Pathway | N/A | http://www.genome.jp/kegg/pathway.html |
| Assay Design Center (UPL) | N/A | https://lifescience.roche.com/en_us/brands/universal-probe-library.html |
| Prism7 | GraphPad | http://www.graphpad.com |
| Excel | Microsoft | https://products.office.com/en/excel |
CONTACT FOR REAGENTS AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Davide Dulcis (ddulcis@ucsd.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Establishment of Unrelated Xenopus Lines
To ensure that we used two groups of Xenopus laevis embryos that shared no relation to each other, we performed the following crosses: Colony1 ♀ × Colony1 ♂, yielding “population 1” and Colony2 ♀ × Colony2 ♂, yielding “population 2.” Colony1 frogs were purchased from Nasko; Colony2 frogs were bought from Xenopus Express. These two groups of animals were kept separate throughout the experiments. Ovulation was induced by injection of human chorionic gonadotropin (Merck). Female frogs were primed 12 hr prior to collection and fertilization of eggs. Embryos were allowed to develop for 2 days (to stage 39) in closely packed clusters and then exposed to odorants. The correct developmental stage was determined according to developmental tables (Nieuwkoop and Faber, 1967; http://www.xenbase.org/anatomy/alldev.do); following odorant exposure, stage 44 was confirmed by quantification of the total number of DRAQ5+ cells per AOB performed in histological sections.
METHOD DETAILS
Collection of Kinship Odorants
Two days post-fertilization, when the majority of the embryos had hatched, odorants were collected by first washing the embryos with 10% MMR (composition: NaCl (100 mM), KCl (2 mM), MgSO4 (1 mM), HEPES (5 mM), EDTA (0.1 mM) and CaCl2 (2 mM) in Fisher water). Concentrated odorant solution was then generated by 125 embryos placed in bottles containing 50 mL 10% MMR for 2 days.
Exposure of Embryos to Odorants
Fifty stage 39 embryos were individually placed in 200 μl of solution conditioned by sibling or non-sibling embryos (Figure S1A). To minimize the accumulation of self-generated kin cues, the solution of individual wells containing single-housed larvae was replaced every 12 hr with fresh, concentrated odorant solution conditioned by the same class of odorants until the larvae had developed to stage 44 (2 days later) and were ready for immunocytochemical examination or behavioral testing.
For reversibility experiments, larvae were first exposed to non-kin odorants for 24 hr (from stage 39 to stage 42), then exposed to either kin or non-kin odorants for another 24 hr (stage 42 to stage 44) followed by processing for immunocytochemistry for TH and GABA.
Local Delivery of Pharmacological Agents
Spatial and temporal control of delivery of pharmacological agents from 80 μm agarose beads loaded with solutions of these agents has proved highly effective (Borodinsky et al., 2004; Dulcis and Spitzer, 2008). Beads were washed in 2 mM calcium saline for 2–4 hr at 22°C and loaded for 1 hr in a bi-directional rotator (Barnstead International) with a Marc’s Modified Ringers (MMR) solution containing 10 μM calcein-AM (Invitrogen), 10 μM BAPTA-AM (Invitrogen), 10 μM TTX (Sigma), 10 nM sulpiride (Sigma) for D2 DA receptor blockade, 500 μM gabazine/phaclofen (Tocris) for GABAA,B receptor blockade, or 800 μM MPTP (Sigma) for DA neuron ablation. Deprenyl (100 μM, Sigma) was used as MPTP-blocker during kin-induced respecification. Stage 39 larvae were anesthetized with 0.05% tricaine (Pentair) before the lateral margin of the AOB was incised and the bead was inserted (Figures S2A and S2B), then allowed to develop for 24–48 hr before behavioral testing and fixation.
Immunocytochemistry
Larvae were fixed in 4% paraformaldehyde and 0.025% glutaraldehyde in PBS at pH 7.4 for 1–2 h at 4° C, rinsed twice in Dulbecco’s calcium–magnesium-free PBS, cryoprotected in 30% sucrose and embedded in optimal cutting temperature medium (Tissue-Tek) for cryostat sections. 10 μm sections were taken following an anterior to posterior progression through the forebrain. One or 2 night incubation at 4°C with primary antibodies to TH (Imgenex), GABA (Millipore), Doublecortin (Abcam), Pax6 (Covance); BrdU (AbD Serotec); VGLUT 1,2 (Synaptic Systems), nNOS (Millipore), and c-Fos (Santa Cruz Biotechnology, Inc.) was followed by incubation with fluorescently tagged secondary antibodies for 1 hr 30 min at 20–22°C. Immunoreactivity of stained sections was examined on a SP5 Leica confocal system; z-stacks were acquired to confirm colocalization and to generate through-series projections. TH, the rate-limiting enzyme for DA synthesis, was used as a marker for DA and confirmed by DA immunostaining.
BrdU Staining
To understand whether AOB neurons recruited for transmitter respecification represent newly born cells or pre-existing neurons, cells proliferating during kin/non-kin conditioning were identified by BrdU (Sigma) labeling. Stage 39 larvae were exposed to BrdU by immersion in 4 mg/ml in 10% MMR for 24 hr. Specimens (stage 42 and 45) were fixed and sucrose-cryoprotected. 10 μm cryostat sections were treated for 20 min in 2 M hydrochloric acid (HCl; Fisher Scientific) for antigen retrieval and then washed and incubated with rat anti-BrdU antibody (AbD Serotec) overnight.
Morpholino Knockdown
Morpholino oligomers (Gene Tools) were used to knock down Pax6 expression. The Pax6 morpholino sequence (5′ GCTGTGAC TGTTCTGCATGTCGAG 3′) and concentration (15 ng/embryo) have been shown to be effective in Xenopus embryos (Rungger-Brändle et al., 2010). The manufacturer’s unspecific standard oligo (5′ CCTCTTACCTCAGTTACAATTTATA 3′) was used as control. The morpholino was fluorescein-labeled to follow its distribution during embryonic development. Effective protein knockdown was quantified by Pax6 immunolocalization.
Larval Distribution Behavioral Assay
A glass tank 9.0 × 21.× 3 cm was washed with deionized water and placed on top of a white light source that provided uniform illumination for photography. The tank was leveled and filled with 100 mL of 10% MMR. Two Millipore Millicell culture plate inserts that had been soaked in 10% MMR for an hr were placed on opposite ends of this container, 1.5 cm from the edge. An opaque, white paper barrier was placed around the container to eliminate external visual cues that might distract larvae and affect their swimming. Fifty stage 44 larvae were washed twice in 10% MMR and transferred to the container as a single group, allowed to distribute equally to the two sides of the dish and photographed from above the container to mark the time zero distribution. Liquid was removed from the inserts, one was refilled with 10% MMR and the other at the opposite end was refilled with odorant; the end at which the odorants were placed was switched for each experiment. To assay the behavioral response to food, powdered pellets of Xenopus diet (Zeigler) were crushed and dissolved in 10% MMR to obtain a mix containing 12 different amino acids (Arginine, Lysine, Methionine, Cysteine, Tryptophan, Histidine, Leucine, Isoleucine, Phenylalanine, Tyrosine, Threonine, Valine). Tests were carried out in the morning when swimming is most robust; photographs were taken every 5 min for 30 min to record changes in the distribution of larvae in the container (Movie S1). The numbers of larvae on each side were photographed, scored and plotted as a function of time (Figures 3A and 3B; Figure S5). A percent larval distribution ratio (LDR) was calculated by dividing the average number of larvae distributed on the side containing the kinship odorant (mean of 15 min and 20 min data points) by the total number of larvae for each experiment and condition.
MicroRNA Analysis
The AOBs of 15 larvae (stage 44) raised in sibling or non-sibling conditions were frozen, cryosectioned at 50 μm and neuronal tissue was harvested with scalpels under the scope for selective AOB isolation. Total RNA was extracted from tissue with the “Recover All Total Nucleic Acid” isolation kit (Ambion). RNA quality was assessed on a Bioanalyzer (Agilent) and degraded samples excluded from RNA sequencing. Library preparation and RNA sequencing (HiSeq, Illumina) were performed at the UCSD Biogem core following manufacturer’s recommendations. To identify miRs potentially involved in NT respecification we compiled a list of proteins (including transcription factors and proteins involved in NT metabolism (Table S1) involved in the production and release of DA and GABA, searching databases available at iHOP, BioGRID, and KEGG Pathway websites. We then used Targetscan (Lewis et al., 2005) to identify predicted miR Responsive elements (MRE) in their 3′-UTRs and cross-referenced this list with the miRs altered by sibling versus non-sibling conditions. Top candidates targeted key genes that were differentially expressed in the three conditions.
Semiquantitative PCR
qPCR was used to measure levels of both miRs and their mRNA targets. Total RNA was extracted as described above. Levels of miRs were assessed using TaqMan microRNA assays (Applied Biosystems) following manufacturer’s recommendations. For reverse transcription, sequence-specific primers were added for each miR being tested and for the housekeeping gene U6. The assays used (assay number in brackets) were: miR-92b (007028_mat); miR-125a (007071_mat); miR-183 (000484); miR-148a (000470); miR-30d (000420); miR-30c (000419); miR-205a (000509); miR-375 (007201_mat); miR-200b (006005); miR-429 (001077); miR-129 (000590); miR-203 (004590_mat); miR-214 (000517); miR-218 (000521); miR-30e (002223); miR-101 (000438); miR-1b (008022); miR-499 (005772);
Levels of miR-target mRNAs were assessed with Universal Probe Library (UPL) Taqman Assays (Roche), following manufacturer’s recommendations. Reverse transcription was performed as previously described (Lippi et al., 2011). Each qPCR experiment used 3–6 clutches of larvae per condition. Primers for each assay were designed using the Roche online tool (Assay Design Center). The list of the primers used includes their respective UPL probe:
Pax6a. Forward: catctggcaccacctctaca; Reverse: ggttcactgccgggtactt; Probe: 95.
Pax6b. Forward: tgggcaacaatctacctatgc; Reverse: ggttcactgccgggtactt; Probe: 95.
Bcl11b. Forward: cgcttaaaccccatgacaa; Reverse: ttctcttagccttcgggaaa; Probe: 67.
GAPDH. Forward: agggaatcctgggatacacac; Reverse: attccagcatcagcatcaaa; Probe: 25
Real-Time PCR analysis was conducted on a Light Cycler 480 system (Roche), and data were processed and analyzed using the comparative DCT method (Livak and Schmittgen, 2001).
MiR Antagonists and TSBs
Fluorescein-tagged LNA antagonists (Exiqon) were designed to inhibit miR candidates:
miR-375 (a.375: 5′/56-FAM/ACGCGAGCCGAACGAACAAA/3′);
miR-200b (a.200b: 5′/56-FAM/ATTACCAGGCAGTATT/3′);
miR-214 (a.214: 5′/56-FAM/CTGTCTGTGCCTGCTG/3′);
miR-148a (a.148a: 5′/56-FAM/AAGTTCTGTAGTGCAC/3′);
miR-101 (a.101: 5′/56-FAM/TCAGCTATCACAGTACT/3′);
miR-499 (a.499: 5′/56-FAM/ATCACTGCAAGTCTTA/3′);
control LNA antagonist (a.Ctrl: 5′/56-FAM/GTGTAACACGTCTATACGCCCA/3′).
Fluorescein-tagged LNA TSBs (Exiqon) were designed to protect mRNA targets from miR regulation:
Pax6-375-TSB (5′/56-FAM/TTTGTTCCAACTGATA/3′);
Pax6-200b-TSB (5′/56-FAM/AAATACTGCTGAACAT/3′);
Bcl11b-200b-TSB1 (5′/56-FAM/TGAATACTGTAAACG/3′);
Bcl11b-200b-TSB2 (5′/56-FAM/TACAAATACTGGTCCA/3′);
Electroporation of miR LNA Antagonists
Antagonists were electroporated (50 nM) at stage 21–23 (Figure S6A). Successful electroporations exhibited fluorescence localization limited to the AOB at stage 31. Embryos were allowed to develop until stage 39, exposed for 48 hr to non-kin odorants and processed for TH immunocytochemistry at stage 44.
Infusions of In Vivo Ready Antagonists and TSBs
Injection needles were prepared from glass capillaries (1 mm OD × 0.5 mm ID; FHC) using a Brown-Flaming electrode puller and loaded with 2 mL of 10 μM 6-carboxyfluorescein (FAM)-tagged antagonists or TSB aliquots using Eppendorf microloader tips. Electrode tips were then manually broken under a dissecting microscope using forceps to achieve a size that ejected 5 nL with a 200–250 msec pulse at 30–60 psi. 20–30 stage 39 larvae were transferred to a mesh-lined Petri dish containing 0.05% tricaine in 10% MMR. Larvae were injected unilaterally (one pulse per side) in the AOB just dorsally to the olfactory pit. LNA-injected larvae were transferred to a Petri dish with fresh 10% MMR saline for recovery. Development was monitored until stage 42 when larvae were fixed and sectioned for immunoprocessing.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data acquisition and quantification were performed blind. Data analysis was performed with Microsoft Excel and Statistical analysis with GraphPad Prism 7. Statistical details (definition of test, exact value of N, means ± standard deviations, p values) are listed in each Figure Legend.
All datasets were tested for normal distribution and equal variances. Departure from normality was assessed with D’Agostino-Pearson omnibus normality test. The variance was calculated with F-test. Grubbs’ test was used to exclude outliers from normally distributed datasets.
Two-groups comparison: Unpaired Student’s t test was used for all experiments with normal distribution and equal variances. Welch’s t test was used when the variances were not equal. Mann-Whitney U test was used for datasets that were not normally distributed.
Three- or more groups comparison: One-way analysis of variance (ANOVA), followed by a Bonferroni’s multiple comparison test, was used for experiments with normal distribution. Kruskal-Wallis’ test followed by a Dunn’s multiple comparisons test was used for datasets that were not normally distributed.
For all tests, p < 0.05 was used as threshold for significant difference. For all tests P values are two-tailed.
DATA AND SOFTWARE AVAILABILITY
For alignment and statistical analysis, RNA sequencing data were first analyzed with Bowtie (version 0.12.8) and mapped reads were aligned with their mature miR (miRNA) sequences using using miRDeep2 with miRBase version-19 (Friedländer et al., 2008; Kozomara and Griffiths-Jones, 2011). miR read counts were quantile normalized using preprocesscore library (Bioconductor 3.1, R 3.2) to determine relative fold changes of miRs between experiments; miR read counts were normalized within each sample (standard score from normal distribution). Due to the limited coverage of the Xenopus laevis genome we aligned the small RNA sequencing reads to all known miR databases (miRBase 20), giving priority to closely related species (e.g., Xenopus tropicalis). Redundant miRs (for example hsa-miR-1 and mmu-miR-1) were manually excluded based on sequence similarity. Sequence and processed data are available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=gdcrgkkelrchfmv&acc=GSE78854 through GEO: GSE78854.
Supplementary Material
Highlights.
Developmental odorant exposure reversibly changes attraction and aversion behavior
Kinship odorants drive dopamine-GABA switching in accessory olfactory bulb neurons
MicroRNAs act on Pax6/Bcl11b to control DA/GABA levels and social preference
Results may be relevant to olfactory imprinting and neonatal attachment in mammals
Acknowledgments
We thank Sean Enright, Melissa Lau, Geetika Arora, Nishant Munugala, Murtaza Bharmal, Zheng Zeng, and Melanie Kucharski for their excellent technical assistance. We are grateful to James Sprague for his assistance on the RNA sequencing at the Biogen core facility. Special thanks go to lab managers Armando De La Torre and James Lee for their assistance with the morpholino electroporations and LNA injections, respectively. Dr. Byungkook Lim and Dr. Brenda Bloodgood provided critical reviews of the manuscript. This work was supported by NIH-NINDS (R01NS015918 to N.C.S. and R01NS057690 to D.D. and N.C.S.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures, one table, and one movie and can be found with this article online at http://dx.doi.org/10.1016/j.neuron.2017.08.023.
AUTHOR CONTRIBUTIONS
D.D. and N.C.S. planned the project; D.D. designed and carried out the immunohistochemical, pharmacological, and behavioral experiments, the imaging, and the data analysis in this study; G.L. designed the miR antagonists and TSB experiments and performed the statistical and genetic analyses and qPCR validation; C.J.S. performed the infusions of LNAs and TSBs; L.H.D. and G.L. performed the small RNA profiling; D.D., G.L., D.K.B., and N.C.S. wrote the manuscript.
References
- Amin ND, Bai G, Klug JR, Bonanomi D, Pankratz MT, Gifford WD, Hinckley CA, Sternfeld MJ, Driscoll SP, Dominguez B, et al. Loss of motoneuron-specific microRNA-218 causes systemic neuromuscular failure. Science. 2015;350:1525–1529. doi: 10.1126/science.aad2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumann TD, Tomas D, Horne MK. Environmental and behavioral modulation of the number of substantia nigra dopamine neurons in adult mice. Brain Behav. 2013;3:617–625. doi: 10.1002/brb3.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumann TD, Raabus M, Tomas D, Prijanto A, Churilov L, Spitzer NC, Horne MK. Differences in number of midbrain dopamine neurons associated with summer and winter photoperiods in humans. PLoS ONE. 2016;11:e0158847. doi: 10.1371/journal.pone.0158847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Ben-Shaul Y, Katz LC, Mooney R, Dulac C. In vivo vomeronasal stimulation reveals sensory encoding of conspecific and allospecific cues by the mouse accessory olfactory bulb. Proc Natl Acad Sci USA. 2010;107:5172–5177. doi: 10.1073/pnas.0915147107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhinge A, Namboori SC, Bithell A, Soldati C, Buckley NJ, Stanton LW. MiR-375 is essential for human spinal motor neuron development and may be involved in motor neuron degeneration. Stem Cells. 2016;34:124–134. doi: 10.1002/stem.2233. [DOI] [PubMed] [Google Scholar]
- Borodinsky LN, Root CM, Cronin JA, Sann SB, Gu X, Spitzer NC. Activity-dependent homeostatic specification of transmitter expression in embryonic neurons. Nature. 2004;429:523–530. doi: 10.1038/nature02518. [DOI] [PubMed] [Google Scholar]
- Brill MS, Snapyan M, Wohlfrom H, Ninkovic J, Jawerka M, Mastick GS, Ashery-Padan R, Saghatelyan A, Berninger B, Götz M. A dlx2- and pax6-dependent transcriptional code for periglomerular neuron specification in the adult olfactory bulb. J Neurosci. 2008;28:6439–6452. doi: 10.1523/JNEUROSCI.0700-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cave JW, Akiba Y, Banerjee K, Bhosle S, Berlin R, Baker H. Differential regulation of dopaminergic gene expression by Er81. J Neurosci. 2010;30:4717–4724. doi: 10.1523/JNEUROSCI.0419-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamero P, Leinders-Zufall T, Zufall F. From genes to social communication: molecular sensing by the vomeronasal organ. Trends Neurosci. 2012;35:597–606. doi: 10.1016/j.tins.2012.04.011. [DOI] [PubMed] [Google Scholar]
- Chang S, Johnston RJ, Jr, Frøkjaer-Jensen C, Lockery S, Hobert O. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature. 2004;430:785–789. doi: 10.1038/nature02752. [DOI] [PubMed] [Google Scholar]
- Chen JA, Huang YP, Mazzoni EO, Tan GC, Zavadil J, Wichterle H. Mir-17-3p controls spinal neural progenitor patterning by regulating Olig2/Irx3 cross-repressive loop. Neuron. 2011;69:721–735. doi: 10.1016/j.neuron.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi PS, Zakhary L, Choi WY, Caron S, Alvarez-Saavedra E, Miska EA, McManus M, Harfe B, Giraldez AJ, Horvitz HR, et al. Members of the miRNA-200 family regulate olfactory neurogenesis. Neuron. 2008;57:41–55. doi: 10.1016/j.neuron.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison IG, Boyd JD, Delaney KR. Dopamine inhibits mitral/tufted/granule cell synapses in the frog olfactory bulb. J Neurosci. 2004;24:8057–8067. doi: 10.1523/JNEUROSCI.2138-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Chevigny A, Coré N, Follert P, Gaudin M, Barbry P, Béclin C, Cremer H. miR-7a regulation of Pax6 controls spatial origin of forebrain dopaminergic neurons. Nat Neurosci. 2012;15:1120–1126. doi: 10.1038/nn.3142. [DOI] [PubMed] [Google Scholar]
- Demarque M, Spitzer NC. Activity-dependent expression of Lmx1b regulates specification of serotonergic neurons modulating swimming behavior. Neuron. 2010;67:321–334. doi: 10.1016/j.neuron.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulcis D, Spitzer NC. Illumination controls differentiation of dopamine neurons regulating behaviour. Nature. 2008;456:195–201. doi: 10.1038/nature07569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulcis D, Spitzer NC. Reserve pool neuron transmitter respecification: novel neuroplasticity. Dev Neurobiol. 2012;72:465–474. doi: 10.1002/dneu.20920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulcis D, Jamshidi P, Leutgeb S, Spitzer NC. Neurotransmitter switching in the adult brain regulates behavior. Science. 2013;340:449–453. doi: 10.1126/science.1234152. [DOI] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto T, Ohmoto M, Iwata T, Uno A, Saitou M, Yamaguchi T, Kominami R, Matsumoto I, Hirota J. Bcl11b/Ctip2 controls the differentiation of vomeronasal sensory neurons in mice. J Neurosci. 2011;31:10159–10173. doi: 10.1523/JNEUROSCI.1245-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedländer MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- Gascon E, Lynch K, Ruan H, Almeida S, Verheyden JM, Seeley WW, Dickson DW, Petrucelli L, Sun D, Jiao J, et al. Alterations in microRNA-124 and AMPA receptors contribute to social behavioral deficits in frontotemporal dementia. Nat Med. 2014;20:1444–1451. doi: 10.1038/nm.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach G, Lysiak N. Kin recognition and inbreeding avoidance in zebrafish, Danio rerio, is based on phenotype matching. Anim Behav. 2006;71:1371–1377. [Google Scholar]
- Gerlach G, Hodgins-Davis A, Avolio C, Schunter C. Kin recognition in zebrafish: a 24-hour window for olfactory imprinting. Proc Biol Sci. 2008;275:2165–2170. doi: 10.1098/rspb.2008.0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessert S, Bugner V, Tecza A, Pinker M, Kühl M. FMR1/FXR1 and the miRNA pathway are required for eye and neural crest development. Dev Biol. 2010;341:222–235. doi: 10.1016/j.ydbio.2010.02.031. [DOI] [PubMed] [Google Scholar]
- Gu X, Spitzer NC. Distinct aspects of neuronal differentiation encoded by frequency of spontaneous Ca2+ transients. Nature. 1995;375:784–787. doi: 10.1038/375784a0. [DOI] [PubMed] [Google Scholar]
- Guemez-Gamboa A, Xu L, Meng D, Spitzer NC. Non-cell-autonomous mechanism of activity-dependent neurotransmitter switching. Neuron. 2014;82:1004–1016. doi: 10.1016/j.neuron.2014.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez GJ, Marder E. Modulation of a single neuron has state-dependent actions on circuit dynamics. eNeuro 1 Published online November. 2014;12:2014. doi: 10.1523/ENEURO.0009-14.2014. http://dx.doi.org/10.1523/ENEURO.0009-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez R, Romo-Parra H, Maqueda J, Vivar C, Ramìrez M, Morales MA, Lamas M. Plasticity of the GABAergic phenotype of the “glutamatergic” granule cells of the rat dentate gyrus. J Neurosci. 2003;23:5594–5598. doi: 10.1523/JNEUROSCI.23-13-05594.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hack MA, Saghatelyan A, de Chevigny A, Pfeifer A, Ashery-Padan R, Lledo PM, Götz M. Neuronal fate determinants of adult olfactory bulb neurogenesis. Nat Neurosci. 2005;8:865–872. doi: 10.1038/nn1479. [DOI] [PubMed] [Google Scholar]
- Hagino-Yamagishi K, Moriya K, Kubo H, Wakabayashi Y, Isobe N, Saito S, Ichikawa M, Yazaki K. Expression of vomeronasal receptor genes in Xenopus laevis. J Comp Neurol. 2004;472:246–256. doi: 10.1002/cne.20073. [DOI] [PubMed] [Google Scholar]
- Harden MV, Newton LA, Lloyd RC, Whitlock KE. Olfactory imprinting is correlated with changes in gene expression in the olfactory epithelia of the zebrafish. J Neurobiol. 2006;66:1452–1466. doi: 10.1002/neu.20328. [DOI] [PubMed] [Google Scholar]
- Hino H, Miles NG, Bandoh H, Ueda H. Molecular biological research on olfactory chemoreception in fishes. J Fish Biol. 2009;75:945–959. doi: 10.1111/j.1095-8649.2009.02341.x. [DOI] [PubMed] [Google Scholar]
- Izaurralde E. GENE REGULATION. Breakers and blockers—miRNAs at work. Science. 2015;349:380–382. doi: 10.1126/science.1260969. [DOI] [PubMed] [Google Scholar]
- Janus C. Stability of preference for odors after short-term exposure in young spiny mice. Dev Psychobiol. 1993;26:65–79. doi: 10.1002/dev.420260106. [DOI] [PubMed] [Google Scholar]
- Johnstone KA, Lubieniecki KP, Koop BF, Davidson WS. Expression of olfactory receptors in different life stages and life histories of wild Atlantic salmon (Salmo salar) Mol Ecol. 2011;20:4059–4069. doi: 10.1111/j.1365-294X.2011.05251.x. [DOI] [PubMed] [Google Scholar]
- Jungblut LD, Pozzi AG, Paz DA. A putative functional vomeronasal system in anuran tadpoles. J Anat. 2012;221:364–372. doi: 10.1111/j.1469-7580.2012.01543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur AW, Ackels T, Kuo TH, Cichy A, Dey S, Hays C, Kateri M, Logan DW, Marton TF, Spehr M, Stowers L. Murine pheromone proteins constitute a context-dependent combinatorial code governing multiple social behaviors. Cell. 2014;157:676–688. doi: 10.1016/j.cell.2014.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick KM, Da Costa APC, Broad KD, Ohkura S, Guevara R, Lévy F, Keverne EB. Neural control of maternal behaviour and olfactory recognition of offspring. Brain Res Bull. 1997a;44:383–395. doi: 10.1016/s0361-9230(97)00218-9. [DOI] [PubMed] [Google Scholar]
- Kendrick KM, Guevara-Guzman R, Zorrilla J, Hinton MR, Broad KD, Mimmack M, Ohkura S. Formation of olfactory memories mediated by nitric oxide. Nature. 1997b;388:670–674. doi: 10.1038/41765. [DOI] [PubMed] [Google Scholar]
- Kesby JP, Najera JA, Romoli B, Fang Y, Basova L, Birmingham A, Marcondes MCG, Dulcis D, Semenova S. HIV-1 TAT protein enhances sensitization to methamphetamine by affecting dopaminergic function. Brain Behav Immun. 2017;65:210–221. doi: 10.1016/j.bbi.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi M, Osumi N, Rubenstein JLR, Alvarez-Buylla A. Pax6 is required for making specific subpopulations of granule and periglomerular neurons in the olfactory bulb. J Neurosci. 2005;25:6997–7003. doi: 10.1523/JNEUROSCI.1435-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Busskamp V, Markiewicz I, Stadler MB, Ribi S, Richter J, Duebel J, Bicker S, Fehling HJ, Schübeler D, et al. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell. 2010;141:618–631. doi: 10.1016/j.cell.2010.03.039. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li X, Jin P. Roles of small regulatory RNAs in determining neuronal identity. Nat Rev Neurosci. 2010;11:329–338. doi: 10.1038/nrn2739. [DOI] [PubMed] [Google Scholar]
- Lippi G, Steinert JR, Marczylo EL, D’Oro S, Fiore R, Forsythe ID, Schratt G, Zoli M, Nicotera P, Young KW. Targeting of the Arpc3 actin nucleation factor by miR-29a/b regulates dendritic spine morphology. J Cell Biol. 2011;194:889–904. doi: 10.1083/jcb.201103006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippi G, Fernandes CC, Ewell LA, John D, Romoli B, Curia G, Taylor SR, Frady EP, Jensen AB, Liu JC, et al. MicroRNA-101 regulates multiple developmental programs to constrain excitation in adult neural networks. Neuron. 2016;92:1337–1351. doi: 10.1016/j.neuron.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Manning CJ, Wakeland EK, Potts WK. Communal nesting patterns in mice implicate MHC genes in kin recognition. Nature. 1992;360:581–583. doi: 10.1038/360581a0. [DOI] [PubMed] [Google Scholar]
- Manzini I, Schild D. Olfactory coding in larvae of the African clawed frog Xenopus laevis. In: Menini A, editor. The Neurobiology of Olfaction. CRC Press/Taylor & Francis Llc; 2010. pp. 114–129. [PubMed] [Google Scholar]
- Marek KW, Kurtz LM, Spitzer NC. cJun integrates calcium activity and tlx3 expression to regulate neurotransmitter specification. Nat Neurosci. 2010;13:944–950. doi: 10.1038/nn.2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill E, Van Vactor D. MicroRNAs shape the neuronal landscape. Neuron. 2012;75:363–379. doi: 10.1016/j.neuron.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezler M, Konzelmann S, Freitag J, Rössler P, Breer H. Expression of olfactory receptors during development in Xenopus laevis. J Exp Biol. 1999;202:365–376. doi: 10.1242/jeb.202.4.365. [DOI] [PubMed] [Google Scholar]
- Mullokandov G, Baccarini A, Ruzo A, Jayaprakash AD, Tung N, Israelow B, Evans MJ, Sachidanandam R, Brown BD. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat Methods. 2012;9:840–846. doi: 10.1038/nmeth.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal Table of Xenopus laevis (Daudin) North-Holland Publishing Company; 1967. [Google Scholar]
- Ninkovic J, Pinto L, Petricca S, Lepier A, Sun J, Rieger MA, Schroeder T, Cvekl A, Favor J, Götz M. The transcription factor Pax6 regulates survival of dopaminergic olfactory bulb neurons via crystallin αA. Neuron. 2010;68:682–694. doi: 10.1016/j.neuron.2010.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter RH, Etscorn F. Olfactory imprinting resulting from brief exposure in Acomys cahirinus. Nature. 1974;250:732–733. doi: 10.1038/250732a0. [DOI] [PubMed] [Google Scholar]
- Rungger-Brändle E, Ripperger JA, Steiner K, Conti A, Stieger A, Soltanieh S, Rungger D. Retinal patterning by Pax6-dependent cell adhesion molecules. Dev Neurobiol. 2010;70:764–780. doi: 10.1002/dneu.20816. [DOI] [PubMed] [Google Scholar]
- Sansone A, Syed AS, Tantalaki E, Korsching SI, Manzini I. Trpc2 is expressed in two olfactory subsystems, the main and the vomeronasal system of larval Xenopus laevis. J Exp Biol. 2014;217:2235–2238. doi: 10.1242/jeb.103465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scerbina T, Chatterjee D, Gerlai R. Dopamine receptor antagonism disrupts social preference in zebrafish: a strain comparison study. Amino Acids. 2012;43:2059–2072. doi: 10.1007/s00726-012-1284-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Luo G, Fu L, Fang Z, Wang X, Li X. miR-9 and miR-140-5p target FoxP2 and are regulated as a function of the social context of singing behavior in zebra finches. J Neurosci. 2013;33:16510–16521. doi: 10.1523/JNEUROSCI.0838-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza BR, Romano-Silva MA, Tropepe V. Dopamine D2 receptor activity modulates Akt signaling and alters GABAergic neuron development and motor behavior in zebrafish larvae. J Neurosci. 2011;31:5512–5525. doi: 10.1523/JNEUROSCI.5548-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer NC. Neurotransmitter switching in the developing and adult brain. Annu Rev Neurosci. 2017;40:1–19. doi: 10.1146/annurev-neuro-072116-031204. [DOI] [PubMed] [Google Scholar]
- Sturm T, Leinders-Zufall T, Maček B, Walzer M, Jung S, Pömmerl B, Stevanović S, Zufall F, Overath P, Rammensee HG. Mouse urinary peptides provide a molecular basis for genotype discrimination by nasal sensory neurons. Nat Commun. 2013;4:1616. doi: 10.1038/ncomms2610. [DOI] [PubMed] [Google Scholar]
- Tan CL, Plotkin JL, Venø MT, von Schimmelmann M, Feinberg P, Mann S, Handler A, Kjems J, Surmeier DJ, O’Carroll D, et al. MicroRNA-128 governs neuronal excitability and motor behavior in mice. Science. 2013;342:1254–1258. doi: 10.1126/science.1244193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B, Di Lena P, Schaffer L, Head SR, Baldi P, Thomas EA. Genome-wide identification of Bcl11b gene targets reveals role in brain-derived neurotrophic factor signaling. PLoS ONE. 2011;6:e23691. doi: 10.1371/journal.pone.0023691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatton WG, Greenwood CE. Rescue of dying neurons: a new action for deprenyl in MPTP parkinsonism. J Neurosci Res. 1991;30:666–672. doi: 10.1002/jnr.490300410. [DOI] [PubMed] [Google Scholar]
- Thiebes KP, Nam H, Cambronne XA, Shen R, Glasgow SM, Cho HH, Kwon JS, Goodman RH, Lee JW, Lee S, Lee SK. miR-218 is essential to establish motor neuron fate as a downstream effector of Isl1-Lhx3. Nat Commun. 2015;6:7718. doi: 10.1038/ncomms8718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velázquez-Ulloa NA, Spitzer NC, Dulcis D. Contexts for dopamine specification by calcium spike activity in the CNS. J Neurosci. 2011;31:78–88. doi: 10.1523/JNEUROSCI.3542-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villinger J, Waldman B. Self-referent MHC type matching in frog tadpoles. Proc Biol Sci. 2008;275:1225–1230. doi: 10.1098/rspb.2008.0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman B. Sibling recognition in toad tadpoles—the role of experience. Zeitschrift Fur Tierpsychologie-Journal of Comparative Ethology. 1981;56:341–358. [Google Scholar]
- Waldman B. Olfactory basis of kin recognition in toad tadpoles. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1985;156:565–577. [Google Scholar]
- Waldman B, Adler K. Toad tadpoles associate preferentially with siblings. Nature. 1979;282:611–613. [Google Scholar]
- Xu B, Hsu PK, Stark KL, Karayiorgou M, Gogos JA. Derepression of a neuronal inhibitor due to miRNA dysregulation in a schizophrenia-related microdeletion. Cell. 2013;152:262–275. doi: 10.1016/j.cell.2012.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki K, Beauchamp GK, Kupniewski D, Bard J, Thomas L, Boyse EA. Familial imprinting determines H-2 selective mating preferences. Science. 1988;240:1331–1332. doi: 10.1126/science.3375818. [DOI] [PubMed] [Google Scholar]
- Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


