Abstract
Mammalian Toll-like receptors (TLR) recognize microbial infection or cell death associated danger signals and trigger appropriate cellular response. These responses determine the strength and the outcome of the host-microbe interaction. The TLRs are transmembrane proteins located on the plasma or the endosomal membrane. Their ectodomains recognize specific microbial or endogenous ligands and the cytoplasmic domains interact with specific proteins to activate intracellular signaling pathways. TLR9, an endosomal TLR, is activated by endocytosed DNA. The activated TLR9 recruits the cytoplasmic adaptor MyD88 and other signaling proteins to induce the synthesis of inflammatory cytokines and interferon. Uncontrolled activation of TLR9 leads to undesired overproduction of inflammatory cytokines and consequent pathogenesis. Therefore, appropriate activation and the regulation of TLR9 signaling are critical. Tyrosine phosphorylation of TLR9 is essential for its activation; however, the role of specific tyrosine kinase is not clear. Here, we report that epidermal growth factor receptor (EGFR), a membrane-bound protein tyrosine kinase, is essential for TLR9 signaling. Genetic ablation of EGFR or pharmacological inhibition of its kinase activity attenuates TLR9-mediated induction of genes in both myeloid and non-myeloid cell types. EGFR is constitutively bound to TLR9; upon ligand stimulation, it mediates TLR9 tyrosine phosphorylation, which leads to the recruitment of MyD88, activation of the signaling kinases and transcription factors, and gene induction. In mice, TLR9-mediated liver injury and death are blocked by an EGFR inhibitor or deletion of the EGFR gene from myeloid cells, which are the major producers of the inflammatory cytokines.
INTRODUCTION
Production of inflammatory cytokines and interferons by innate immune cells is triggered by Toll-like receptors (TLR) which are pattern recognition receptors for pathogen-associated molecular patterns (PAMP) and damage-associated molecular patterns (DAMP) (1–4). Upon activation, TLRs recruit specific cytoplasmic adaptors which nucleate the assembly of signaling proteins, protein kinases and transcription factors; the activated transcription factors translocate to the nucleus and drive induced transcription of the target genes. TLRs are transmembrane proteins; their ectodomains interact with the ligands whereas their short carboxyl-terminal regions are in the cytoplasm where they signal. Some TLRs are located on the plasma membrane and others are on the endosomal membrane (5). TLR9 is an endosomal DNA-recognizing receptor, which is primarily expressed in myeloid cells and activated by both microbial PAMPs and mammalian DAMPs; it can provide protection against pathogenesis caused by selected bacteria and viruses (6, 7). In human cancers, TLR9 can be beneficial or detrimental. In lung cancer, TLR9 expression in tumor-infiltrating mononuclear cells is associated with a worse outcome (8, 9); on the other hand, high TLR9 expression in triple negative breast cancer is associated with better survival of the patients (10).
Experimentally, unmethylated CpG-rich oligodeoxynucleotide (ODN) is used to stimulate TLR9; it is endocytosed and binds to the ectodomain of TLR9 in the endosomal lumen. TLR9 dimerizes in endoplasmic reticulum and translocates to endolysomes, a process regulated by UNC93B1, a membrane protein. In endolysomes of mouse cells, TLR9 is proteolytically cleaved and undergoes ligand-induced conformational changes leading to post-translational modifications of amino acid residues in its cytoplasmic domain to generate a functional receptor (11–15). TLR9-mediated induction of proinflammatory cytokines and interferon (IFN) may originate from two different membrane compartments (16). To signal, TLR9 uses MyD88 as the adaptor protein and IKKs and IRAKs as the protein kinases to activate transcription factors. Genome-wide RNAi screening identified many proteins that affect TLR9 trafficking or signaling (17). Recent observations indicate that, to signal, many TLRs require ligand-dependent phosphorylation of specific tyrosine residues in their cytoplasmic domains. The phosphotyrosine residues of the TLR cytoplasmic domains can potentially serve as docking sites of signaling proteins that contain SH2 domains and thereby, expand the repertoire of signaling pathways. Ligand-dependent Tyr phosphorylation of TLR9 has been reported; however, the specific phosphorylated Tyr residue and the relevant protein tyrosine kinase (PTK) have not been identified (4, 18). A Tyr-containing motif, in the cytoplasmic domain of TLR9, is important for its appropriate intracellular localization and ability to signal; Tyr888, a part of this motif, is required for TLR9 Tyr-phosphorylation, although this residue itself is not phosphorylated (19). CpG ODN-treatment of cells causes rapid activation of Src family PTKs, such as Hck and Lyn, without any involvement of TLR9; this leads to the activation of another PTK, Syk, which binds to TLR9 (20).
Our investigation of the biochemistry of TLR3 signaling led to the identification of the epidermal growth factor receptor (EGFR) as the critical PTK required for TLR3 signaling. EGFR binds TLR3 only after ligand stimulation, recruits another PTK, Src, and the two kinases phosphorylates two specific Tyr residues in the cytoplasmic domain of TLR3. This chain of event is required for the recruitment of the adaptor, TRIF, and subsequent signaling (21). For TLR4 signaling in mouse myeloid cells, only the IRF3-mediated branch of endosomal signaling requires EGFR; however, no physical interaction between EGFR and TLR4 has been noted (22). Here, we report that EGFR kinase activity is required for all TLR9 signaling in vitro and in vivo. However, unlike TLR3, endosomal TLR9 binds EGFR constitutively.
MATERIALS AND METHODS
Reagents and antibodies
CpG oligonucleotides (CpG-A and CpG-B) were purchased from Integrated DNA Technologies (IDT). EGFR inhibitors, gefitinib, erlotinib and AG1478 were obtained from Selleckchem, Santa Cruz and Calbiochem respectively. Phospho-tyrosine antibody was obtained from Millipore, antibodies against EGFR, MyD88, pAKT, pIkBα, pERK, pJNK, pTBK1, AKT, IkBα, TBK1 and actin were from Cell Signaling, HA antibody was purchased from Abcam, antibody against murine Ifit proteins were raised in our laboratory (23). TLR4 antibody was obtained from Santa Cruz Biotechnology. TLR9 antibody (Clone: ABM1C51), which recognizes the N-terminal regions of both murine and human TLR9 was from Abeomics. Cetuximab (Erbitux, Bristol-Myers Squibb) was obtained from Cleveland Clinic pharmacy. D-galactosamine (GalN) was purchased from Sigma. Poly(I:C) was obtained from GE Healthcare and Lipofectamine 2000 was obtained from Invitrogen.
Cell lines
Primary bone marrow derived dendritic cells (BMDCs), macrophages (BMDMs) or plasmacytoid dendritic cells (pDCs) were isolated and differentiated as described before (24). RAW264.7 cells (ATCC) were maintained in DMEM containing 10% FBS and penicillin-streptomycin; 293XL, 293XL-human TLR9-HA cells and 293-CD14.MD2.TLR4 cells were from Invivogen and maintained in DMEM containing 10% FBS, penicillin-streptomycin, normocin and blasticidin. EGFR-knockdown 293XL-human TLR9-HA cells were generated by lentiviral transduction of human EGFR-specific shRNA (Sigma, #TRCN0000121202), followed by selection under puromycin; a non-targeting shRNA (#SHC002) was used as control. HT1080-human TLR9 (with double epitope tags, YFP and Flag) cells were generated by lentiviral transduction.
RLR stimulation
4μg of poly(I:C) with 10μl of lipofectamine (LF2000) was incubated in 250 μl of OptiMEM (10μl of lipofectamine (LF2000) was incubated in 250 μl of OptiMEM as control) for 30 min and then added to the cell culture medium containing 10% of FBS.
Mouse experiments
WT, EGFRfl/− and EGFRfl/− LysMCre+/+ mice (all in C57Bl/6 genetic background) were used for the experiments. WT mice were obtained from The Jackson Laboratory. EGFRfl/fl mice, originally generated by Dr. David Threadgill’s lab at University of North Carolina (25), were a gift from Dr. Xiaoxia Li’s lab at the Cleveland Clinic. EGFRfl/− were generated in the laboratory by crossing EGFRfl/fl mice with CMVCre+ mice purchased from The Jackson Laboratory. EGFRfl/− LysMCre+/+ mice were generated by crossing EGFRfl/− mice with LysMCre +/+ mice purchased from The Jackson Laboratory. For inducing septic shock, 8–10 week old WT, EGFRfl/− or EGFRfl/− LysCre+/+ mice were intraperitoneally injected with CpG (30 μg/mouse) along with D-galactosamine (20 mg/mouse) and their survival was monitored for 5 days. To investigate the effect of EGFR inhibitor on the CpG-induced septic shock, the mice were orally gavage with gefitinib (2.4 mg/mouse) or vehicle (DMSO) followed by intraperitoneal injection of CpG and galactosamine (26, 27). Blood samples were obtained by cardiac puncture after one hour of CpG/GalN injection and serum was isolated. The livers were harvested from the animals at 8 hours post CpG/GalN treatment for histopathological analyses. All mice procedures were approved by IACUC.
Quantitative real-time PCR
RNA was isolated using Roche RNA isolation kit (Roche). cDNA was prepared using ImprompII Reverse Transcription Kit (Promega) and 0.5 ng of cDNA was applied to 384-well plate for realtime PCR using Applied Biosystem’s Power SYBR Green PCR mix in Roche LightCycler 480 II. The expression levels of the induced mRNAs were normalized to 18S rRNA or RPL32 mRNA.
Microarray analyses
RAW264.7 cells (in biological triplicate) were pretreated with DMSO or gefitinib (10 μM) for 1h, and stimulated with CpG (10μg/ml) for 6 hours along with DMSO or Gefitinib. Total RNA was isolated, treated with DNase I and the RNA were further purified by using the RNAeasy kit (Qiagen). The purified RNA was then analyzed in an Illumina Mouse Ref-8 gene array and the data analysis was carried out by using Illumina Genome Studio V2011.1. We selected the mRNAs, which were induced at least two-fold by CpG, and quantified the inhibition index (E/F) (as presented in Supplemental Table 1). All datasets have been deposited at National Center for Biotechnology Information/Gene Expression Omnibus under accession number GSE97366 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE97366).
Enzyme-linked immunosorbent assay
Culture supernatants from CpG-treated cells were used for quantification of the secreted TNF-α and IFN-β by ELISA using manufacturers’ instructions. Blood was collected by cardiac puncture from anesthetized mice into serum separator tubes (BD Biosciences) and sera were isolated following manufacturer’s protocol for measuring the secreted cytokines. TNF-α levels were then assessed in serum samples using mouse TNF-α ELISA kit from eBioscience.
In situ proximity ligation assay (PLA)
Duolink In situ Detection kit (DUO92008, Sigma Aldrich) was used to perform PLA following the manufacturer’s instructions. In brief, cells were grown on glass coverslips. Cells were incubated with wheat germ agglutinin (WGA) 633 (Thermo Fisher Scientific) for 10 min at room temperature to label the plasma membrane. They were then fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100 followed by blocking with 2.5% goat serum. Cells were then incubated overnight with primary antibodies, rabbit antibody targeting Flag (Cell Signaling Technology) and mouse antibody targeting HA (Sigma), in 2.5% goat serum. PLA probes corresponding to the primary antibodies, Duolink In situ PLA Probe Anti-Mouse MINUS (DUO92004, Sigma Aldrich) and Duolink In situ PLA Probe Anti-Rabbit MINUS (DUO92005, Sigma-Aldrich), in 2.5% goat serum were used for further incubation for 1 h at 37°C followed by incubation with a DNA ligase diluted in ligation buffer for 30 min at 37°C. Finally, cells were incubated with a DNA polymerase diluted in amplification buffer for 100 min at 37°C and then mounted in VectaShield/DAPI. Confocal images were acquired using a Leica laser scanning confocal microscope.
Confocal microscopy
Human HT1080-TLR9 cells were grown on glass coverslips. The cells were fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100 for 20 min each. The cells were blocked with 5% normal goat serum for 1 hr and then labeled overnight with anti-EGFR (C74B9, Cell Signaling) and anti-EEA1 (BD Transduction Labs) for staining early endosomes. Goat anti-rabbit Alexa Fluor 594 (Invitrogen) and goat anti-mouse Alexa Fluor 647 (Invitrogen) (1 hr) were used respectively as secondary antibodies. Objects were mounted using VectaShield/DAPI and images were taken by confocal laser scanning microscopy (Leica TCS SP8). Images were processed with Leica LCS software. ImageJ software Co-localization plugin were used for determining co-localization of two proteins where the white dots represents co-localization.
Liver histology
Livers were harvested, fixed in formalin (Sigma) and then paraffin embedded. The 5μm sections were cut and then stained with hematoxylin and eosin to view liver pathology. Immunohistochemistry staining for infiltrated macrophages on 5μm sections was performed using MAC-2 antibody. In brief, antigen retrieval was performed using a tris/borate/EDTA buffer (Discovery CC1, Ventana), pH 8.0 to 8.5, for 32 minutes at 95°C. Slides were incubated with MAC-2 at a 1:1800 dilution (Cedarlane Labs), for 40 minutes at room temperature. The antibody was visualized using a biotinylated rabbit anti-rat secondary at 1:200 dilution (Vector Laboratories) and the DABMap detection kit (Ventana). Lastly, the slides were counterstained with hematoxylin and bluing agent.
Statistical analysis
All statistical analyses were performed using GraphPad Prism 5.02 software. P values were calculated using two-tailed, un-paired Student’s t Tests. P values for survival curves were calculated using Log-rank test.
RESULTS
Gene induction by TLR9 signaling requires EGFR kinase activity
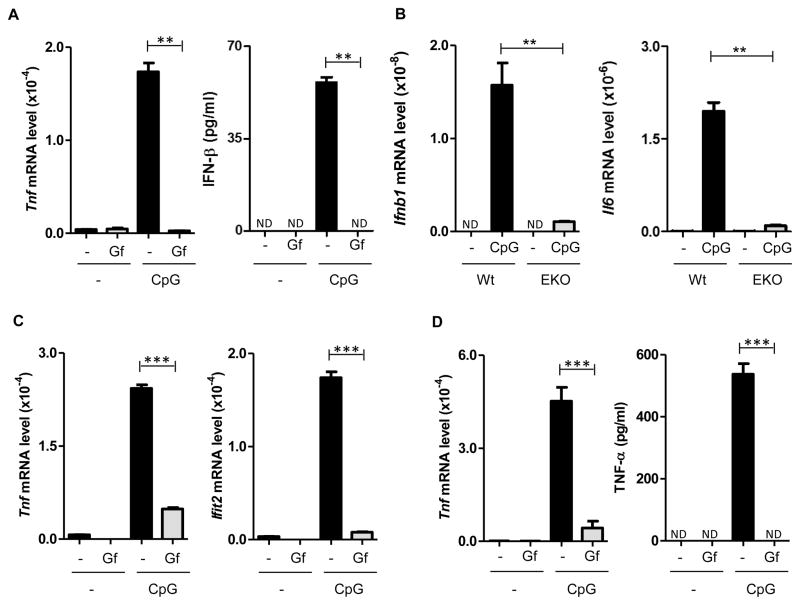
In view of our observation that TLR3 and TLR4 signaling requires EGFR activity, we inquired whether the same is true for TLR9 signaling. TLR9 activation by CpG ODN treatment of myeloid cells induced transcription of many cytokine mRNAs, including Tnfα and IFNβ secretion; such induction in primary mouse macrophages was completely blocked by gefitinib (Gf), an inhibitor of EGFR kinase (Fig. 1A). Testing of Gf dose response indicated that 50% inhibition was achieved in the presence of 1–5 μM concentration of the inhibitor (Supplemental Fig. 1A); the high dose was needed possibly because the inhibitor had to reach intracellular EGFR which activated endosomal TLR9. To exclude any off-target effect of Gf, we pursued a genetic approach; TLR9 signaling was totally blocked in primary macrophages obtained from mice in which the EGFR gene had been selectively knocked-out in myeloid cells (Fig. 1B). Similar to macrophages, in bone marrow-derived dendritic cells, Gf inhibited the induction of Tnf mRNA and Ifit2 (an IFN-induced gene) mRNA (Fig. 1C). In contrast, in the same cells, as reported before (22), TLR4-mediated Tnf mRNA induction by LPS was not at all inhibited by Gf (Supplemental Fig. 1B). A different inhibitor of EGFR, AG1478, had similar inhibitory effects on Il6 and Ifit2 induction by TLR9 (Supplemental Fig. 1B). In plasmacytoid dendritic cells, Gf inhibited Tnf induction, measured at both mRNA and protein levels (Fig. 1D); inhibition was also observed for three other mRNAs (Supplemental Fig. 1C). Similarly, in the monocytic RAW264.7 cell line, induction of Tnf, Il6 and Ifnβ mRNAs by CpG ODN was strongly inhibited by Gf (Fig 2A, Supplemental Fig. 2A); two other inhibitors of EGFR kinase, erlotinib and AG1478, had similar effects (Supplemental Fig. 2B). To ascertain whether the inhibitory effects of Gf on gene induction by TLR9 was global, we compared gene expression profiles, by microarray analyses, using RNAs isolated from untreated, CpG ODN-treated, Gf-treated and CpG ODN plus Gf-treated cells. It was clear that CpG ODN could not induce any gene in Gf-treated cells (Supplemental Table 1). None of the 170 mRNAs, which were induced by CpG ODN by more than two fold, was induced if the cells were treated with Gf as well. The microarray results were verified by qRT-PCR analyses of four induced mRNAs that had not been examined before (Fig. 2B). The above results demonstrated that EGFR kinase activity was globally needed for gene induction by TLR9 signaling.
Fig. 1. EGFR activity is required for TLR9-mediated gene induction in primary myeloid cells.
Cells, as indicated, were pre-treated with DMSO or gefitinib (Gf, 10 μM) for 1 h and stimulated with CpG (10 μg/ml) for 6 h in the presence of DMSO or Gf. mRNA induction was analyzed by qRT–PCR. TNF-α and IFN-β secretion in the supernatants was quantified by ELISA. (A) BMDM were used to analyze Tnf mRNA and IFN-β secretion. (B) BMDM from wild type and myeloid specific EGFR knock out mice (EKO). (C) BMDC. (D) pDC. ND: not detectable. Error bars were calculated as mean ± SEM from three biological replicates, and the data are representative of at least three independent experiments. P-values were calculated using two-tailed unpaired Student’s t-test; **P < 0.01, ***P < 0.001.
Fig. 2. EGFR activity is required for TLR9-mediated gene induction in a mouse monocytic cell line.
RAW264.7 cells were pre-treated with DMSO or gefitinib (Gf, 10 μM) for 1 h and stimulated with CpG (10 μg/ml) for 6 h in the presence of DMSO or Gf. mRNA induction (as indicated in each panel) was analyzed by qRT–PCR. Error bars were calculated as mean ± SEM from three biological replicates, and the data are representative of at least three independent experiments. P-values were calculated using two-tailed unpaired Student’s t-test; **P < 0.01, ***P < 0.001.
Activation of signaling protein kinases by TLR9 requires EGFR kinase activity
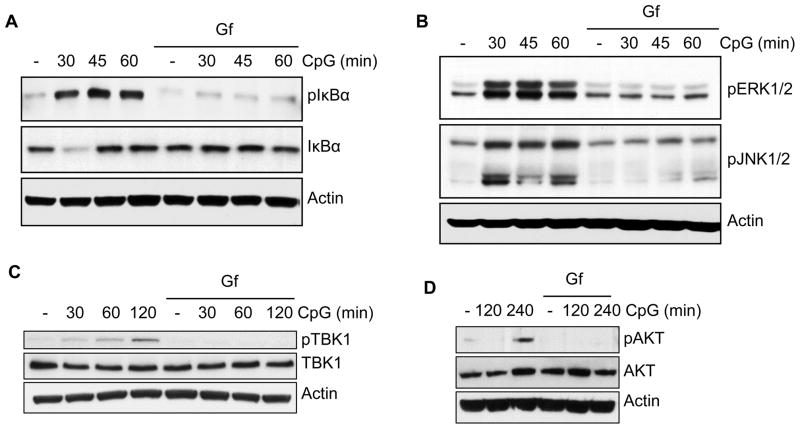
Gene induction by TLR9 requires the activation of transcription factors, such as, NFκB and IRF, which, in turn, requires the activation of specific signaling kinases; we inquired whether TLR9 stimulation could activate these kinases if EGFR activity was blocked. NFκB is activated by its release from IκB, which is degraded upon its phosphorylation. As expected, TLR9 stimulation caused IκBα phosphorylation and degradation but both processes were blocked by Gf (Fig. 3A). Similarly, activation of ERK1, ERK2, JNK1 and JNK2, as measured by their phosphorylation, was impaired by Gf-treatment (Fig. 3B). TBK1, which phosphorylates and activates IRF, was also not activated in the presence of Gf (Fig. 3C). The same was true for AKT, which is activated by PI3 kinase and required for the full transcriptional activity of NFκB and IRF (Fig. 3D).
Fig. 3. TLR9-mediated activation of signaling kinases needs EGFR activity.
(A–D) RAW264.7 cells were pre-treated with DMSO or gefitinib (Gf, 10 μM) for 1 h and were stimulated with CpG (10 μg/ml) for the indicated times in the presence of DMSO or Gf. The cell lysates were analyzed for (A) pIkBα (on Ser32) and total IkBα; (B) pERK1/2 (on Thr202/Tyr204), pJNK1/2 (phosphor- p54/p46); (C) pTBK1 (on Ser172) and total TBK1 and (D) pAKT (on Ser473) and total AKT. Actin was used as a loading control. The data are representative of at least three independent experiments.
Mechanism of EGFR action on TLR9 signaling
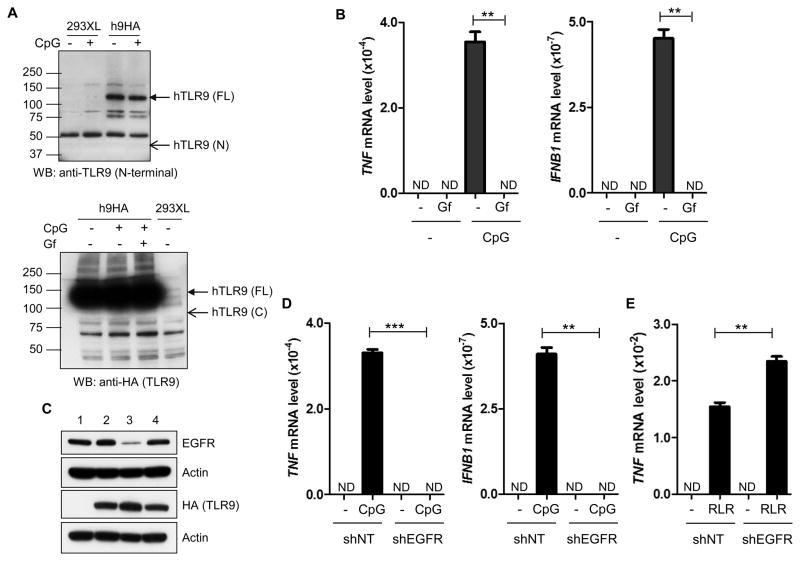
To facilitate our investigation of the biochemical basis of the observed need of EGFR for TLR9 signaling, we utilized 293XL cells expressing human TLR9 with a HA-epitope tag at its C-terminus. Unlike murine TLR9, no cleavage of human TLR9 could be detected in this cell line even after ligand stimulation; an antibody, that recognizes the N-terminal regions of both murine and human TLR9s, readily detected the full length hTLR9 (FL, upper arrow), but not a putative N-terminal fragment (N, lower arrow) (Fig. 4A, upper panel). Similarly, HA antibody detected the FL protein (upper arrow, Fig. 4A, lower panel), but not a putative C-terminal fragment (lower arrow, Fig. 4A, lower panel). The absence of N-terminal and C-terminal fragments having molecular weights similar to their murine counterparts, prompted us to ensure that the signaling properties of the TLR9-expressing 293XL cells were similar to those of murine myeloid cells. CpG ODN robustly induced TNF and IFNβ mRNAs in these cells and the induction was totally abolished by Gf treatment (Fig. 4B).
Fig. 4. TLR9-signaling in a human experimental cell line requires EGFR.
(A) Upper panel: 293XL-TLR9-HA and 293XL cells were stimulated with CpG for 30 min. The cells were lysed and analyzed by western blot using a TLR9 antibody which can detect the N-terminal region of TLR9. Lower panel: 293XL-TLR9-HA and 293XL cells were pre-treated with DMSO or Gf for 1h and then stimulated with CpG for 30 min in the presence of DMSO or Gf. The cells were lysed and analyzed for TLR9 (anti-HA) by WB. (B) 293XL cells expressing human TLR9-HA were pre-treated with DMSO or gefitinib (Gf, 10 μM) for 1 h and stimulated with CpG (10 μg/ml) in the presence of DMSO or Gf for 6 h; the induction of TNF and IFNB1 mRNA were analyzed by qRT–PCR. (C) Western analyses of EGFR expression; 1: 293XL cells; 2: 293XL-TLR9-HA cells; 3: 293XL-TLR9-HA-shEGFR expressing a shRNA against EGFR; 4: 293XL-TLR9-HA-shNT expressing a non-targeting control shRNA. (D) 293XL-TLR9-HA-shEGFR and 293XL-TLR9-HA-shNT cells were stimulated with or without CpG for 6 h and analyzed as in (B). (E) 293XL-TLR9-HA-shNT and 293XL-TLR9-HA-shEGFR cells were transfected with lipofectamine 2000 or poly(I:C) (RLR). The cells were harvested after 6 h of post transfection and TNF mRNA induction was analyzed by qRT-PCR. ND: not detectable. Error bars were calculated as mean ± SEM from three biological replicates. P-values were calculated using two-tailed unpaired Student’s t-test; **P < 0.01, ***P < 0.001. The data are representative of at least three independent experiments.
Because EGFR has several isoforms with kinase activity, we determined which isoform is functionally required for TLR9 signaling in our system. For this purpose, expression of different EGFR isoforms was knocked down by respective shRNAs, clonal cell lines were established and TLR9 signaling was assessed. In one such clone (clone 3), the expression of a shRNA for EGFR1 (erbB1) strongly impaired the expression of the EGFR1 protein without affecting the expression of TLR9 (Fig. 4C); neither TNF mRNA nor IFNβ mRNA was induced by CpG ODN treatment of these cells (Fig. 4D). In contrast, as expected, TNF mRNA induction in the same cells, by poly(I:C) transfection (RLR signaling), was unimpaired (Fig. 4E). The above results demonstrated that our experimental cell line, 293XL-TLR9, shares the properties of primary myeloid cells; moreover, EGFR1 is the relevant isoform that is required for TLR9 signaling.
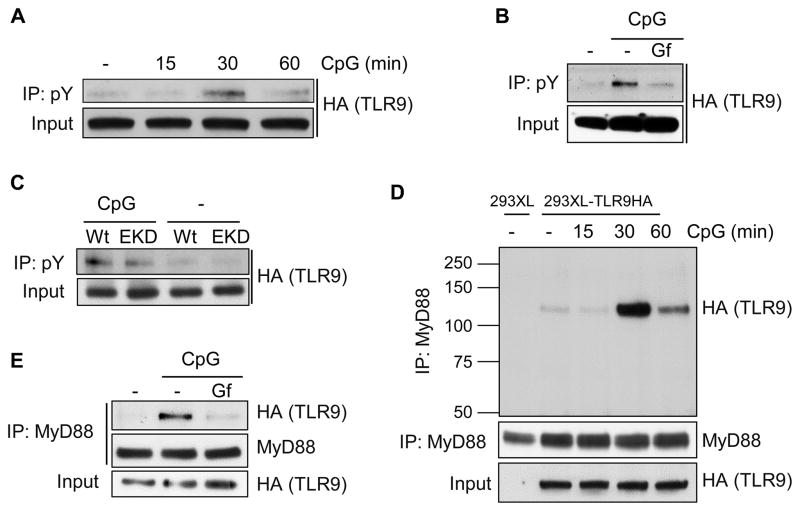
To understand the basis of the requirement of the EGFR tyrosine kinase activity for TLR9 signaling, we examined whether TLR9 itself is Tyr-phosphorylated; we observed ligand-dependent and transient Tyr-phosphorylation of TLR9 (Fig. 5A) which required EGFR kinase activity (Fig. 5B). Similar results were obtained with EKD cells, in which EGFR expression had been knocked down (Fig. 5C). The absence of total elimination of TLR9 phosphorylation (Figs. 5B, 5C) could be due to the residual EGFR in EKD cells (see Fig. 4C, lane 3) or the action of another protein tyrosine kinase. Assembly of the TLR9 signaling complex requires interaction of TLR9 with the adaptor protein, MyD88. We detected such an interaction which, as expected, was ligand-dependent and transient (Fig. 5D); note that MyD88 interacted with FL TLR9 to form the signaling complex. More importantly, this interaction was EGFR kinase-dependent as well (Fig. 5E). The above results suggested that ligand-induced EGFR-mediated Tyr-phosphorylation of TLR9 leads to MyD88 recruitment by TLR9 and the triggering of the consequent signaling pathways.
Fig. 5. TLR9 tyrosine phosphorylation and MyD88 recruitment require both ligand-stimulation and EGFR kinase activity.
(A) TLR9 tyrosine phosphorylation requires ligand stimulation. 293XL-TLR9-HA cells were stimulated with CpG for the indicated time, cell lysates were immunoprecipitated with anti-phosphotyrosine antibody and the immunoprecipitates were analyzed for TLR9 (anti-HA) by Western blot. (B) TLR9 tyrosine phosphorylation requires EGFR kinase activity. Tyrosine phosphorylation of TLR9 was analyzed after 30 min of CpG treatment in the presence of DMSO or Gf; cells were pre-treated with DMSO or gefitinib (Gf, 10 μM) for 1 h. (C) TLR9 tyrosine phosphorylation requires EGFR. Tyrosine phosphorylation of TLR9 was analyzed as in (A) after 30 min of CpG treatment. Wt is 293XL-TLR9-HA-shNT cells expressing a non-targeting control shRNA and EKD is 293XL-TLR9-HA-shEGFR expressing a shRNA against EGFR. (D) MyD88 recruitment by TLR9 requires ligand stimulation. 293XL and 293XL-TLR9-HA cells were stimulated with CpG for the indicated time, cell lysates were immunoprecipitated with anti-MyD88 antibody and the immunoprecipitates were analyzed for TLR9 (anti-HA) by Western blot. (E) MyD88 recruitment by TLR9 requires EGFR kinase activity. TLR9 and MyD88 co-immunoprecipitation was analyzed after 30 min of CpG treatment in the presence of DMSO or Gf; cells were pre-treated with DMSO or gefitinib (Gf, 10 μM) for 1 h. The data are representative of at least three independent experiments.
Characteristics of EGFR-TLR9 interaction
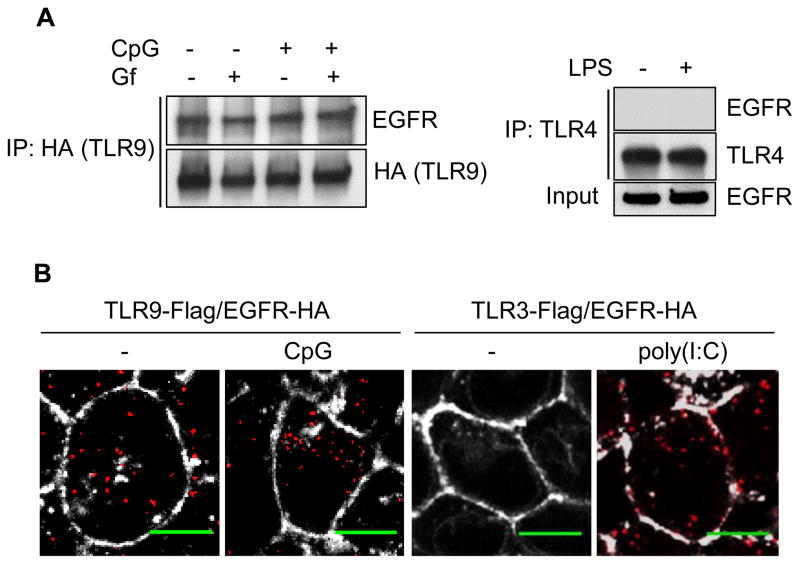
Because EGFR was required for ligand-induced Tyr phosphorylation of TLR9, we wondered whether the two proteins physically interact. We used two experimental approaches to measure EGFR-TLR9 interaction. We could readily demonstrate their interaction by co-immunoprecipitation assays (Fig. 6A, left panel); their interaction was not affected by Gf and more surprisingly, it did not require ligand stimulation. In contrast, we detected no interaction between EGFR and TLR4, even after LPS stimulation (Fig 6A, right panel). The above conclusion was confirmed by a cell-based imaging assay; in the proximal ligation Duolink assay, a strong signal is generated if two proteins are in close proximity in a cell (28). As shown in Fig. 6B, cells scored positive for TLR9/EGFR interaction both before and after CpG ODN treatment. In contrast, as reported before, TLR3/EGFR interaction required ligand stimulation (21). The above results demonstrated that EGFR was bound to TLR9 constitutively, but ligand stimulation was required for TLR9 phosphorylation and signaling.
Fig. 6. TLR9-EGFR interaction does not need CpG stimulation or EGFR kinase activity.
(A) TLR9-EGFR co-immunoprecipitation does not require ligand stimulation or EGFR kinase activity. Left panel: 293XL-TLR9-HA cells were pre-treated DMSO or Gf (10 μM) for 1 h and then untreated or stimulated with CpG for 30 min in presence of DMSO or Gf. The cell lysates were immunoprecipitated with anti-HA (TLR9) and the immunoprecipitates were analyzed for EGFR by Western blot. Right panel: 293-CD14-MD2-TLR4 cells were stimulated with LPS for 1 h. The cell lysates were immunoprecipitated with TLR4 antibody and immunoprecipitates were analyzed for EGFR by Western blot. (B) TLR9-EGFR interaction in cells does not require ligand stimulation. TLR9-EGFR interaction was analyzed by Duolink assay, with or without CpG treatment, of 293XL cells co-expressing TLR9-Flag and EGFR-HA. TLR3-Flag and EGFR-HA interaction, with or without poly(I:C) treatment, was used as a positive control. Duolink signals were visualized as red dots. Wheat germ agglutinin (WGA) 633 was used for staining the plasma membrane (white). Scale bars: 5 μm. The data are representative of at least three independent experiments.
Subcellular location of TLR9-EGFR interaction
Because cytoplasmic signaling is elicited by endosomal membrane-bound TLR9, we postulated that TLR9-EGFR interaction occurs, not on the plasma membrane, but on the internal membranes of the cell. In cells exposed to EGFR ligands, EGFR is found not only on the plasma membrane but also on the endosomal membrane (21). However, in cells cultured in serum-free medium, which does not contain EGF or other EGFR ligands, EGFR is localized almost exclusively on the plasma membrane. We observed that in serum-starved (SS) cells, there was no interaction between TLR9 and EGFR even after CpG ODN stimulation (Fig. 7A). Consequently there was no TLR9 phosphorylation (Fig. 7B) and gene induction (Fig. 7C) in serum-starved cells indicating that TLR9 interaction with intracellular EGFR was required for its ability to signal. This conclusion was supported by the observation that cetuximab, an EGFR antibody, that blocked cell surface EGFR signaling (Fig. 7D), could not block gene induction by CpG ODN (Fig. 7E). Unlike Gf, cetuximab, could not enter the cell and hence could not inhibit the function of endosomal EGFR. Confocal microscopy confirmed co-localization of TLR9 and EGFR on the early endosomal membrane. Co-localization of an early endosomal marker (EEA1), EGFR and TLR9-YFP, visualized by three different colors, (Fig. 7F, three left panels) was confirmed by image-analysis, using a software that produces white dots if two colors are co-localized (Fig. 7F, three right panels).
Fig. 7. EGFR on internal membranes, but not plasma membrane, is required for TLR9 signaling.
(A) No interaction of EGFR with TLR9 in 293XL-TLR9-HA cells cultured in serum-free (starved) medium, as opposed to serum containing (normal) medium, with and without 30 min of CpG stimulation (serum starved before and during CpG treatment). (B) Lack of CpG-induced tyrosine phosphorylation of TLR9 in 293XL-TLR9-HA cells cultured in starved medium. Tyrosine phosphorylation of TLR9 was analyzed after 30 min of CpG treatment (serum starved before and during CpG treatment). (C) Serum-free (starved) condition inhibits IFNB1 and IL6 production. 293XL-TLR9-HA cells were treated with CpG under normal or serum starved (SS) conditions (serum starved before and during CpG treatment). The induction of IFNB1 mRNA and IL6 mRNA were analyzed by qRT–PCR. (D) EGFR activity is inhibited by cetuximab (Cet). Inhibition of EGF (100 ng/ml) induced EGFR tyrosine 1068 phosphorylation with different concentrations of Cet. HEK293 cells were pretreated with different concentrations of Cet for 1 h, the cells were then stimulated with EGF for 10 min in the absence or presence of Cet. The cell lysates were analyzed for EGFR pY1068, EGFR and actin. (E) No effect on gene induction by CpG by inhibiting EGFR on the cell surface. 293XL-TLR9-HA cells were pretreated with Cet (100 μg/ml) for 1h, the cells were stimulated with CpG for 6h in the absence or the presence of Cet and the induction of TNF and IFNB1 mRNA were analyzed by qRT–PCR. (F) Confocal microscopy to demonstrate co-localization of TLR9 and EGFR on early endosomal membrane. HT1080 cells expressing TLR9-YFP were used. The left three panels show the sub-cellular locations of EGFR (red), TLR9 (green) and early endosomal marker (magenta); the right three panels show their co-localization using ImageJ software Co-localization plugin where the white dots represents co-localization. Scale bars: 10 μm. Error bars were calculated as mean ± SEM from three biological replicates. P-values were calculated using two-tailed unpaired Student’s t-test; ***P < 0.001. The data are representative of at least three independent experiments.
Demonstration of the need of EGFR for TLR9 signaling in mice
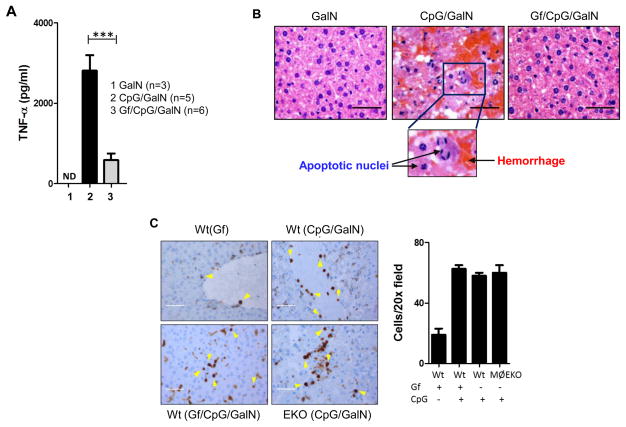
To establish the need of EGFR for TLR9 signaling in vivo, we used a liver injury model in mice. When CpG ODN is injected to galactosoamine-sensitized mice, TLR9-induced TNF-α causes rapid liver damage and death (26). To test the need of EGFR in this process, mice were pre-treated with Gf before CpG ODN challenge. TNF-α was strongly induced in the serum of CpG ODN-injected mice, but not if they were pre-treated with Gf (Fig. 8A). The liver architectures of the untreated and the CpG ODN plus Gf-treated mice were very similar; in contrast, there were massive hemorrhage and cell apoptosis in the livers of mice injected with CpG ODN (Fig. 8B). There was pronounced macrophage-infiltration in the livers of CpG ODN-treated mice but the absence of EGFR or its kinase activity had no effect on the magnitude of the infiltration (Fig. 8C).
Fig. 8. Gefitinib inhibits serum TNF production and liver toxicity in CpG-treated mice.
(A) Gf treatment inhibits TNF production. C57Bl/6 mice were pre-treated with vehicle or Gf (oral gavaged) for 7 days and injected (i.p.) with galactosamine (GalN) or CpG in combination with GalN, along with vehicle or Gf (oral gavage). 1 hour later serum TNF production was determined by ELISA. (B) Gf treatment inhibits changes in liver structures. Mice were treated as above and 8h later liver histology was analyzed. Scale bars: 10 μm. (C) Immunohistochemical (IHC) staining for infiltrated macrophages using MAC2 antibody of liver tissues from Wt treated with Gf, CpG and Gf+CpG and of myeloid specific EGFR knock out (EKO) treated with CpG as described in (A). Positive stained macrophages are indicated with yellow arrowheads. Cells/20X field is plotted as bar graph. Scale bars: 10 μm. Error bars were calculated as mean ± SEM from biological replicates. P-values were calculated using two-tailed unpaired Student’s t-test; ***P < 0.001. The data are representative of at least three independent experiments.
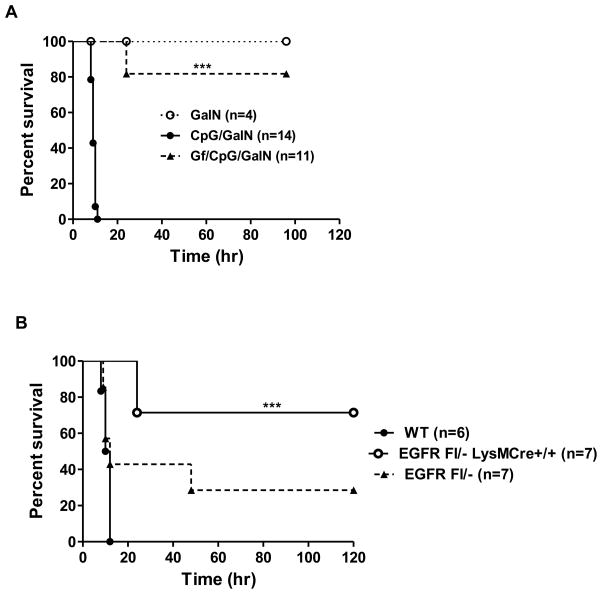
The observed liver damage caused the death of all CpG-injected mice within 12h whereas 80% of the Gf-treated mice survived (Fig. 9A). We used this mouse survival assay to ascertain the shortest time period needed for Gf administration, prior to CpG ODN treatment, to protect the mice. 16h of pretreatment (Supplemental Fig. 3A) and 1h of pretreatment (Supplemental Fig. 3B) were effective; surprisingly, even co-administration of CpG ODN and Gf was protective (Supplemental Fig. 3C). These results clearly demonstrated that TLR9 signaling in vivo required EGFR kinase activity. To strengthen the conclusion drawn from pharmacological intervention EGFR activity in vivo, we tested mice in which the EGFR gene has been manipulated. The need of EGFR for pathogenesis was supported by the observation that mice harboring only one allele of EGFR gene were less susceptible to TLR9-induced pathogenesis (Fig. 9B). To ascertain the cell type in which EGFR expression was required to elicit the phenotype, we selectively deleted both alleles of the EGFR gene in myeloid cells; the corresponding mice were quite resistant to pathogenesis (Fig. 9B). These results indicated that EGFR-dependent TLR9 signaling in myeloid cells was essential for the deleterious effects of CpG ODN administration to mice.
Fig. 9. Lack of EGFR activity in myeloid cells protects mice from CpG induced lethality.
(A) Gf-treatment protects mice from CpG ODN induced lethality. C57Bl/6 mice were treated as in Fig. 8 and survival was monitored. (B) EGFR expression in myeloid cells is required for lethality. CpG ODN induced death was measured in C57Bl/6 mice with three genotypes: Wt mice, EGFR Fl/− mice carrying only one allele of the EGFR gene and EGFR Fl/− LysM Cre mice, in which both alleles of the EGFR gene had been selectively deleted only from the myeloid cells. All mice were treated with CpG/GalN. ND: not detectable. P values for survival curves were calculated using Log-rank test, ***P < 0.001.
DISCUSSION
This study provides support for the paradigm that endosomal TLRs require ligand-induced EGFR-mediated Tyr phosphorylation to trigger signaling; our results demonstrated an absolute need of EGFR kinase activity for TLR9 to induce gene transcription. This is reminiscent of a similar need for TLR3 signaling, but not for TLR4 signaling, in which only the IRF3 branch requires EGFR (21, 22). Among the different ligands of TLR9, CpG A ODN is a better inducer of IFN, as compared to CpG B ODN, which was used for all the experiments reported here. EGFR kinase activity was required for IFN induction by CpG A ODN as well (data not shown). Consistent with the need of EGFR for all gene induction by TLR9, its PTK activity was also needed for activating the signaling kinases.
Several highly specific pharmacological inhibitors of EGFR, gefitinib, erlotinib and AG1478, inhibited gene induction by TLR9; however, for effective inhibition, the required dose of Gf was high, compared to what is needed to inhibit biochemically purified EGFR in vitro (29). Because our assays were cell-based, inefficient cellular uptake and intracellular degradation might have contributed to inefficient inhibition. Moreover, in contrast to inhibiting cell surface EGFR, to inhibit TLR9, Gf had to reach endosomal EGFR, which binds to TLR9. Nonetheless, to rule out off-target effects of high doses of Gf, we sought genetic evidence for the need of EGFR in TLR9 signaling; knocking down the expression of EGFR (Fig. 4D) or knocking out the EGFR gene itself (Fig. 1B) confirmed the conclusion that EGFR was necessary for TLR9 to signal.
To facilitate the analysis of EGFR’s involvement in TLR9 signaling, we took advantage of the cell line, 293XL-TLR9, which expresses epitope-tagged human TLR9. Unlike murine myeloid cells, in human 293 cells, TLR9 was not cleaved and neither its N-terminal fragment nor its C-terminal fragment was detected (Fig. 4A); similar observations had been made before by Latz et al (15). We confirmed that full length TLR9 was the signaling receptor by demonstrating its ligand-depended Tyr-phosphorylation and consequent recruitment of MyD88. TLR9 signaling in 293XL-TLR9 cells had characteristics similar to those in mouse myeloid cells; most importantly, these cells also required EGFR kinase activity for CpG ODN-induced gene induction and like TLR3, TLR9 used the erbB1 isoform of EGFR for this purpose. In these cells, we observed transient Tyr-phosphorylation of TLR9 in response to CpG ODN stimulation, a process that required EGFR kinase activity, which was also required for the recruitment of MyD88 to TLR9. From these observations we concluded that EGFR directly, or indirectly through the acquisition of another PTK, phosphorylates TLR9, which leads to the binding of MyD88, triggering the complete signaling pathway. It is worth noting that a basal low level of Tyr-phosphorylation of TLR9 was detectable in unstimulated cells (Figs. 5A, 5B, 5C), even in the presence of gefitinib (Fig. 5B). The role and the nature of the basal phosphorylation of TLR9 remain to be investigated. There are six Tyr residues in the cytoplasmic region of TLR9 and it is possible that both basal and CpG ODN-induced phosphorylation of multiple Tyr residues is required for the receptor to signal. A specific Tyr, Tyr888, has been shown to be a part of a structural motif required for correct intracellular localization of TLR9; although this Tyr is not phosphorylated, it is required for TLR9 Tyr phosphorylation and signaling (19).
The functional requirement of EGFR for ligand-induced activation of TLR9-signaling was consistent with the observed physical interaction between the two proteins. Their interaction was constitutive; it did not require ligand-stimulation or EGFR kinase activity. This is in contrast to the nature of the EGFR interaction with TLR3, which depends on ligand-induced conformational change of the receptor allowing accessibility of EGFR to its binding site on TLR3 (21). In the case of TLR9, most probably, ligand-induced conformational change allows pre-bound EGFR to access its target Tyr moiety on TLR9 and phosphorylate it. It is likely that the two proteins encounter each other on the endosomal membrane. In serum-starved cells, without exogenous ligands, EGFR resides almost exclusively on the plasma membrane; we observed no interaction with TLR9 and no Tyr phosphorylation of TLR9 in those cells (Fig. 7). But, in cells cultured in media containing growth factors, EGFR is abundant in internal membranes, including endosomal membrane where TLR9 binds to it. Consequently, inhibiting EGFR kinase activity on the cell surface by a neutralizing antibody had no effect on TLR9 signaling; whereas cell-permeable small chemical inhibitors, such as Gf, could inhibit the action of endosomal EGFR and block TLR9 signaling. It is surprising that EGFR was not identified as a TLR9 partner in a previous study (17); however, because genome-wide RNAi screening was used in this study, it is possible that the target cells were not viable in the absence of EGFR.
To verify the need of EGFR for TLR9 signaling in vivo, we resorted to a convenient model of CpG ODN mediated liver injury in mice (26). In this model, administration of CpG stimulates TLR9 to induce TNF production, which promotes massive apoptotic death of hepatocytes in D-galactosamine-sensitized mice causing fulminant liver failure and consequent death. We tested whether pre-treatment of the mice with gefitinib would block TLR9 signaling and its deleterious effects in this model. Indeed, pre-treatment, or even co-treatment (Supplemental Fig. 3), with gefitinib was very effective in preventing death caused by CpG ODN administration. As expected, the protected mice suffered lesser liver damage and the level of TNF in their circulation was much lower. These pharmacological data were supported by results obtained from genetic manipulation of EGFR expression. Mice harboring only one functional allele of the EGFR gene were less susceptible than Wt mice. Because TNF is primarily produced by the macrophages of CpG-treated mice (26), when we deleted the remaining allele of EGFR gene selectively only in those cells, the mice were highly protected from CpG-induced death. The pharmacological and genetic data suggest that TLR9 expressed in myeloid cells allows is activated by CpG ODN to produce TNF, a process that requires physical and functional presence of EGFR in those cells. Thus, the in vivo results are in complete agreement with the mechanistic conclusions drawn from our in vitro experiments.
The requirement for EGFR kinase activity for signaling and gene induction by endosomal TLRs can be potentially exploited for clinical benefits. Especially in situations, such as autoimmunity, in which TLR signaling is harmful, blocking EGFR therapeutically may be beneficial. Many EGFR chemical inhibitors and antibodies are widely used clinically to treat specific cohorts of cancer patients; thus, their safety and efficacies are well documented. These drugs can be re-purposed for treating patients suffering from hyper-inflammation due to potent TLR signaling. On the flip side, in cancer patients receiving EGFR blockers on a long term basis, signaling by the endosomal TLRs may be impaired. These TLRs are often activated by endocytosis of cellular DNA or RNA produced by apoptotic cancer cells; the consequent cytokine-mediated inflammation may promote disease progression, depending on the specific type of cancer. In such cases, the beneficial effects of EGFR inhibitors may be mediated by not only impairing cancer cell proliferation but also blocking inflammatory cytokine production.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health grants CA062220 and CA068782. M.Y. received funding from UCLA CTSI Grant (UL1TR001881) and American Heart Association Grant (17SDG33660947). Leica SP8 confocal microscope was purchased with funding from the National Institutes of Health SIG grant 1S10OD019972-01.
We thank Xiaoxia Li for providing the EGFR fl/fl mice and Volker Fensterl for helpful suggestions. We thank Judy Drazba for help with confocal microscopy using the Leica SP8 confocal microscope.
Footnotes
The datasets presented in this article have been submitted to the National Center for Biotechnology Information/Gene Expression Omnibus under accession number GSE97366.
References
- 1.Pandey S, Kawai T, Akira S. Microbial sensing by Toll-like receptors and intracellular nucleic acid sensors. Cold Spring Harb Perspect Biol. 2014;7:a016246. doi: 10.1101/cshperspect.a016246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 3.Severa M, Fitzgerald KA. TLR-mediated activation of type I IFN during antiviral immune responses: fighting the battle to win the war. Curr Top Microbiol Immunol. 2007;316:167–192. doi: 10.1007/978-3-540-71329-6_9. [DOI] [PubMed] [Google Scholar]
- 4.Chattopadhyay S, Sen GC. Tyrosine phosphorylation in Toll-like receptor signaling. Cytokine Growth Factor Rev. 2014;25:533–541. doi: 10.1016/j.cytogfr.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gay NJ, Symmons MF, Gangloff M, Bryant CE. Assembly and localization of Toll-like receptor signalling complexes. Nat Rev Immunol. 2014;14:546–558. doi: 10.1038/nri3713. [DOI] [PubMed] [Google Scholar]
- 6.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 7.Yasuda K, Richez C, Uccellini MB, Richards RJ, Bonegio RG, Akira S, Monestier M, Corley RB, Viglianti GA, Marshak-Rothstein A, Rifkin IR. Requirement for DNA CpG content in TLR9-dependent dendritic cell activation induced by DNA-containing immune complexes. J Immunol. 2009;183:3109–3117. doi: 10.4049/jimmunol.0900399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krieg AM. Development of TLR9 agonists for cancer therapy. J Clin Invest. 2007;117:1184–1194. doi: 10.1172/JCI31414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sorrentino R, Morello S, Luciano A, Crother TR, Maiolino P, Bonavita E, Arra C, Adcock IM, Arditi M, Pinto A. Plasmacytoid dendritic cells alter the antitumor activity of CpG-oligodeoxynucleotides in a mouse model of lung carcinoma. J Immunol. 2010;185:4641–4650. doi: 10.4049/jimmunol.1000881. [DOI] [PubMed] [Google Scholar]
- 10.Krieg AM. Toll-like receptor 9 (TLR9) agonists in the treatment of cancer. Oncogene. 2008;27:161–167. doi: 10.1038/sj.onc.1210911. [DOI] [PubMed] [Google Scholar]
- 11.Kim YM, Brinkmann MM, Paquet ME, Ploegh HL. UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature. 2008;452:234–238. doi: 10.1038/nature06726. [DOI] [PubMed] [Google Scholar]
- 12.Pelka K, Phulphagar K, Zimmermann J, Stahl R, Schmid-Burgk JL, Schmidt T, Spille JH, Labzin LI, Agrawal S, Kandimalla ER, Casanova JL, Hornung V, Marshak-Rothstein A, Honing S, Latz E. Cutting edge: the UNC93B1 tyrosine-based motif regulates trafficking and TLR responses via separate mechanisms. J Immunol. 2014;193:3257–3261. doi: 10.4049/jimmunol.1301886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee BL, Moon JE, Shu JH, Yuan L, Newman ZR, Schekman R, Barton GM. UNC93B1 mediates differential trafficking of endosomal TLRs. Elife. 2013;2:e00291. doi: 10.7554/eLife.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ewald SE, Lee BL, Lau L, Wickliffe KE, Shi GP, Chapman HA, Barton GM. The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature. 2008;456:658–U688. doi: 10.1038/nature07405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Latz E, Verma A, Visintin A, Gong M, Sirois CM, Klein DCG, Monks BG, McKnight CJ, Lamphier MS, Duprex WP, Espevik T, Golenbock DT. Ligand-induced conformational changes allosterically activate Toll-like receptor 9 (vol 8, pg 772, 2007) Nat Immunol. 2007;8:1266–1266. doi: 10.1038/ni1479. [DOI] [PubMed] [Google Scholar]
- 16.Lee BL, Barton GM. Trafficking of endosomal Toll-like receptors. Trends Cell Biol. 2014;24:360–369. doi: 10.1016/j.tcb.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiang CY, Engel A, Opaluch AM, Ramos I, Maestre AM, Secundino I, De Jesus PD, Nguyen QT, Welch G, Bonamy GM, Miraglia LJ, Orth AP, Nizet V, Fernandez-Sesma A, Zhou Y, Barton GM, Chanda SK. Cofactors required for TLR7- and TLR9-dependent innate immune responses. Cell Host Microbe. 2012;11:306–318. doi: 10.1016/j.chom.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasan M, Gruber E, Cameron J, Leifer CA. TLR9 stability and signaling are regulated by phosphorylation and cell stress. J Leukoc Biol. 2016;100:525–533. doi: 10.1189/jlb.2A0815-337R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chockalingam A, Rose WA, 2nd, Hasan M, Ju CH, Leifer CA. Cutting edge: a TLR9 cytoplasmic tyrosine motif is selectively required for proinflammatory cytokine production. J Immunol. 2012;188:527–530. doi: 10.4049/jimmunol.1102713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanjuan MA, Rao N, Lai KT, Gu Y, Sun S, Fuchs A, Fung-Leung WP, Colonna M, Karlsson L. CpG-induced tyrosine phosphorylation occurs via a TLR9-independent mechanism and is required for cytokine secretion. J Cell Biol. 2006;172:1057–1068. doi: 10.1083/jcb.200508058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamashita M, Chattopadhyay S, Fensterl V, Saikia P, Wetzel JL, Sen GC. Epidermal growth factor receptor is essential for Toll-like receptor 3 signaling. Sci Signal. 2012;5:ra50. doi: 10.1126/scisignal.2002581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chattopadhyay S, Veleeparambil M, Poddar D, Abdulkhalek S, Bandyopadhyay SK, Fensterl V, Sen GC. EGFR kinase activity is required for TLR4 signaling and the septic shock response. EMBO Rep. 2015;16:1535–1547. doi: 10.15252/embr.201540337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terenzi F, White C, Pal S, Williams BR, Sen GC. Tissue-specific and inducer-specific differential induction of ISG56 and ISG54 in mice. J Virol. 2007;81:8656–8665. doi: 10.1128/JVI.00322-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uchijima M, Nagata T, Aoshi T, Koide Y. IFN-gamma overcomes low responsiveness of myeloid dendritic cells to CpG DNA. Immunol Cell Biol. 2005;83:92–95. doi: 10.1111/j.1440-1711.2005.01307.x. [DOI] [PubMed] [Google Scholar]
- 25.Lee TC, Threadgill DW. Generation and validation of mice carrying a conditional allele of the epidermal growth factor receptor. Genesis. 2009;47:85–92. doi: 10.1002/dvg.20464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yi AK, Yoon H, Park JE, Kim BS, Kim HJ, Martinez-Hernandez A. CpG DNA-mediated induction of acute liver injury in D-galactosamine-sensitized mice: the mitochondrial apoptotic pathway-dependent death of hepatocytes. J Biol Chem. 2006;281:15001–15012. doi: 10.1074/jbc.M601337200. [DOI] [PubMed] [Google Scholar]
- 27.Lee JY, Lee YM, Chang GC, Yu SL, Hsieh WY, Chen JJ, Chen HW, Yang PC. Curcumin induces EGFR degradation in lung adenocarcinoma and modulates p38 activation in intestine: the versatile adjuvant for gefitinib therapy. PLoS One. 2011;6:e23756. doi: 10.1371/journal.pone.0023756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nilsson I, Bahram F, Li X, Gualandi L, Koch S, Jarvius M, Soderberg O, Anisimov A, Kholova I, Pytowski B, Baldwin M, Yla-Herttuala S, Alitalo K, Kreuger J, Claesson-Welsh L. VEGF receptor 2/−3 heterodimers detected in situ by proximity ligation on angiogenic sprouts. Embo J. 2010;29:1377–1388. doi: 10.1038/emboj.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wakeling AE, Guy SP, Woodburn JR, Ashton SE, Curry BJ, Barker AJ, Gibson KH. ZD1839 (Iressa): an orally active inhibitor of epidermal growth factor signaling with potential for cancer therapy. Cancer Res. 2002;62:5749–5754. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.