Abstract
A set of coordinated interactions between gut microbiota and the immune cells surveilling the intestine play a key role in shaping local immune responses and intestinal health. Gpr109a is a G-protein coupled receptor expressed at a very high level on innate immune cells and previously shown to play a key role in the induction of colonic Tregs. Here we show that Gpr109a−/−Rag1−/− mice exhibit spontaneous rectal prolapse and colonic inflammation, characterized by the presence of an elevated number of IL-17-producing Rorγt+ innate lymphoid cells (ILC3). Genetic deletion of Rorγt ameliorated the spontaneous colonic inflammation in Gpr109a−/−Rag1−/− mice. Gpr109a-deficient colonic dendritic cells produce higher amounts of IL-23, and thereby promote ILC3. Moreover, the depletion of gut microbiota by antibiotics treatment decreased IL-23 production, ILC3, and colonic inflammation in Gpr109a−/−Rag1−/− mice. The caecums of Gpr109a−/−Rag1−/− mice showed significantly increased colonization by members of Bacteroidaceae, Porphyromonadaceae, Prevotellacea, Streptococcaceae, Christensenellaceae, and Mogibacteriaceae as well as IBD-associated microbiota such as Enterobacteriaceae and Mycoplasmataceae than Rag1−/− mice, housed in a facility positive for Helicobacter and murine norovirus. Niacin, a Gpr109a agonist, suppressed both IL-23 production by colonic DCs and ILC3 number in a Gpr109a-dependent manner. Collectively, our data presents a model suggesting that targeting Gpr109a will be potentially beneficial in the suppression of IL-23 mediated immunopathologies.
Introduction
Ulcerative colitis (UC) and Crohn’s disease (CD), together termed inflammatory bowel diseases (IBD), are chronic and relapsing inflammatory diseases of the gut with unknown etiology. Recent observations indicate that a complex interaction between an individual’s genetics, immune system, diet, and gut microbiota plays a critical role in the development of IBD (1, 2). Genetic studies have identified more than 100 susceptibility loci linked to IBD (3). Dysregulation of adaptive immune responses is thought to be a dominant reason for the induction of IBD (4–8). However, recent findings indicate that an anomaly in innate responses also plays a critical role in the initiation and progression of IBD. Single nucleotide polymorphisms in nucleotide-binding oligomerization domain 2 (NOD2) are associated with susceptibility to Crohn’s disease (9, 10). In addition, Crohn’s disease is also associated with mutations in the ATG16L1 and IRGM genes (11, 12). These findings have generated considerable interest in the activation and regulation of the innate immune system at the gut mucosal surface in the pursuit of understanding the mechanisms regulating the pathogenesis of IBD.
Innate lymphoid cells or ILCs are a recently identified family of immune cells that are found at a much lower frequency than adaptive immune cells. They are mostly present at barrier surfaces such as the gut, lungs, and skin. They play an important role in the induction, regulation, and resolution of inflammatory responses. ILCs are of lymphoid origin but lack antigenic receptor expressed by B and T cells, as well as the markers associated with myeloid cells. ILCs are stimulated by microbes and cytokines present in the microenvironment to rapidly produce proinflammatory and regulatory cytokines. Based on the expression of transcription factors and related cytokines, ILCs are classified into three groups. ILC1 express T-bet and produce IFN-γ and TNF-α. ILC2 express high levels of GATA3 and produce IL-5 and IL-13. ILC3 express transcription factor Rorγt and produce IL-17 and IL-22. ILC3 play a critical role in regulating and promoting inflammation in the intestine. IL-22 producing ILC3 protect the intestine by inducing production of anti-bacterial peptides by the gut epithelium (13, 14), promoting protein fucosylation, which modifies bacterial metabolism and attenuates their virulence (15), containing intestinal bacteria (16), inducing T cell tolerance to gut bacteria antigens (17, 18), maintaining the tolerogenic potential of intestinal DCs (19), and protecting intestinal stem cells during graft versus host disease (20). In contrast, in Helicobacter hepaticus- or anti-CD40-driven colitis in Rag1−/− mice and in spontaneous colonic inflammation in TRUC (Tbx21−/−Rag2−/−) mice, ILC3 induce colonic inflammation (21–25). Similarly, in IBD, IL-22 producing ILC3 are decreased (26, 27). IL-22 produced by ILC3 promotes colon carcinogenesis (28). In the azoxymethane (AOM)/ dextran sulfate sodium (DSS) model, IL-22 production at the peak of inflammation leads to the resolution of inflammation and the inhibition of carcinogenesis. In contrast, uncontrolled IL-22 production in the same model supports tumorigenesis (29).
Gpr109a is a G protein coupled receptor that is highly expressed on innate immune cells and adipose tissue. Gpr109a is activated by niacin and butyrate, a short chain fatty acid and fermentation product of dietary fiber by gut microbiota. Selected gut microbiota such as Lactobacillus acidophilus produce niacin. Niacin deficiency results in intestinal inflammation and diarrhea (30, 31). We have previously shown that Gpr109a signaling plays a critical role in the homeostasis of Treg cells in the colon and in the suppression of colonic inflammation and carcinogenesis (32). However, whether Gpr109a regulates innate lymphoid cells and its relevance to colonic inflammation and carcinogenesis remain unknown. Our study identifies a critical role for Gpr109a in the inhibition of IL-23 production by colonic DCs leading to suppression of ILC3 and colonic inflammation.
Materials and Methods
Mice
Rag1−/− (C57BL/6 background), Rorγt−/− (C57BL/6 background) and C57BL/6 mice were originally from Jackson Laboratory (Bar Harbor, ME) and bred on-site. Gpr109a−/− mice (C57BL/6 background) have been described. Gpr109a−/− and Rag1−/− mice were interbred to generate Gpr109a−/−Rag1−/− mice. Murine norovirus and Helicobacter were detected in our mouse colony. The Institutional Animal Care and Use Committee (IACUC), Augusta University approved all animal procedures.
Histopathology
Colons were excised and fixed in neutral buffered formalin (Thermo Fisher, Waltham, MA). Fixed colon tissues were embedded in paraffin and 5 µm thick sections were sliced and placed on glass microscope slides. Hematoxylin and eosin (H&E) staining on sections were visualized using Olympus BX43 microscope. H&E stained sections were scored for colitis based on 4 histologic parameters and leukocyte infiltration, as follows: 0 = normal histology, 1 = mild hyperplasia of epithelium, 2 = moderate hyperplasia with marked leukocyte infiltration, 3 = severe hyperplasia with leukocyte infiltration and significant decrease in goblet cells, 4 = severe hyperplasia with inflammatory cells, ulceration, crypt abscesses and severe depletion of goblet cells.
Cell Isolation and Analysis
For the isolation of intestinal lamina propria cells, colons and small intestines were opened and luminal contents were removed. Intestines were cut into ~ 1 centimeter pieces and shaken in Hanks Balanced salt solution (HBSS, Corning, Corning NY) containing 5% fetal bovine serum (GE Healthcare, Logan, Utah), 5mM EDTA, 10 mM HEPES pH 7.4 (Sigma, St. Louis, MO) twice for 20 minutes each to remove the epithelial layer. The left over intestinal pieces were digested with 0.5 mg/ml of collagenase D and DNAse 1 (Roche Diagnostics, Germany) twice for 30 minutes each. The released cells were collected by centrifugation and contaminating epithelial cells were removed by centrifugation over 40% percoll. The cell pellet was washed and used as lamina propria immune cells. Cells were stained for CD45, Rorγt, Ly6G, GATA3, CD11b, CD11c, CD103, NKP46, Sca, CD127, IL-17, IL-22 (Thermo-Fisher, Waltham, MA), Thy1.2, T-bet, Ly6G (Biolegend, San Diego, CA) and analyzed using a LSRII flow cytometer (BD Biosciences, San Jose, CA). Colonic DCs were sorted as CD45+Thy1.2−Ly6G−CD11c+ population by FACS Aria Flow cytometer (BD Biosciences). In some experiments colonic DCs were sorted into CD103− and CD103+ subsets.
Cell Culture
Isolated mesenteric lymph node and colonic or small intestinal lamina propria cells were cultured in 0.2 ml of RPMI 1640 (Corning, Corning NY) fortified with 10% fetal bovine serum (GE Healthcare, Logan, Utah), 10 mM HEPES pH 7.4 (Sigma, St. Louis, MO) and 50µM of 2-mercaptoethanol (Thermo-Fisher, Waltham, MA), 100 ng/ml of PMA (Sigma, St. Louis, MO) and 1 µM of ionomycin (EMD Millipore Billerica, MA), monensin and brefeldin A (Thermo-Fisher, Waltham, MA). Four hours later cells were fixed and intracellular staining for transcription factors and cytokines was performed. In some experiments, mesenteric lymph node Thy1.2+ cells were cultured with colonic DCs in the presence of IL-2 (10 ng/ml) and IL-7 (10 ng/ml, both from Peprotech, Rocky Hill, NJ). Three days later cells were analyzed for Rorγt and IL-17 as above.
Antibiotics and niacin treatment
Mice were given a cocktail of ciprofloxacin.HCl (TCI, Portland, Oregon, 0.15 g/liter), gentamycin sulfate (0.2 g/liter), bacitracin (1g/liter) and streptomycin (2 g/liter) (Thermo-Fisher, Waltham, MA) in drinking water ad libitum. In some experiments, niacin (10 mg/ml) was given in drinking water ad libitum for the indicated period of time.
Quantitative real time PCR
Total RNA was extracted from cells using the RNeasy micro kit (Qiagen, Hilden, Germany). RNA was converted into cDNA using the Superscript III reverse transcription system (Thermo-Fisher, Waltham, MA). Quantitative polymerase chain reaction (qPCR) was performed using Sybr green PCR mix (Bio-rad, Hercules, CA) and StepOnePlus machine (Applied Biosystems). PCR primers were Gpr109a; forward 5’-ATGGCGAGGCATATCTGTGTAGCA-3’, reverse 5’-TCCTGCCTGAGCAGAACAAGATGA-3’ Gapdh; forward 5’AGGTCGGTGAACGGATTTG-3’, reverse 5’-TGTAGACCATGTAGTTGAGGTCA-3’ Hprt; forward 5’-GCGTCGTGATTAGCGATGAAC-3’ reverse 5’CCTCCCATCTCCTTCATGACATCT-3’, Il23P19; forward 5’-GACCCACAAGGACTCAAGGA-3’, reverse 5’-CATGGGGCTATCAGGGAGTA-3’.
Antibody treatment
Animals were intraperitoneally injected with anti-Thy1.2 (Clone 30H12) or isotype control (Clone LTF2, both from BioXcell, West Lebanon, NH) antibodies (100 µg/mouse/week). Two months later animals were sacrificed and used in experiments.
Microbiome sequencing
DNA from caecal content was isolated as described previously. Briefly ~100–200 mg of caecal content was suspended with 710 µL disruption buffer and 500 µL phenol/chloroform/isoamyl alcohol, pH 8.0, inside tubes containing Zirconium beads (0.1mm diameter, Benchmark Scientific). Following centrifugation, DNA was precipitated from the aqueous phase using isopropanol. Sequencing (300-bp paired end) of V3–V4 regions of bacterial 16S rDNA was performed on the Illumina MiSeq platform and data was analyzed using Quantitative Insights into Microbial Ecology (QIIME).
Statistical analysis
Statistical significance was calculated using Student’s t-test with two-tailed analysis.
Results
Spontaneous Colitis in Gpr109a−/−Rag1−/− mice
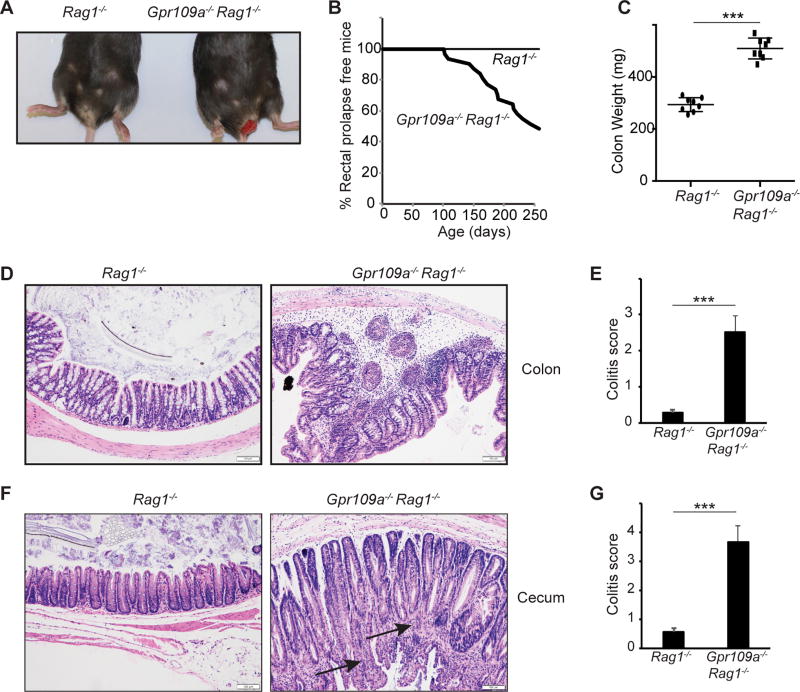
To test the role of Gpr109a in the absence of the adaptive immune system, we crossed Gpr109a−/− animals onto the Rag1−/− background. Gpr109a−/−Rag1−/− mice spontaneously develop rectal prolapse. Rectal prolapse starts appearing as early as 3 months of age and approximately 50% of the animals developed rectal prolapses by 9 months of age (Fig. 1A, 1B). In contrast, Rag1−/− mice housed in the same colony and on the same rack did not show any sign of rectal prolapse (Fig. 1A, 1B). Colons of Gpr109a−/−Rag1−/− mice showed more mass than Rag1−/− mice (Fig. 1C). Moreover, colons and caecum of Gpr109a−/−Rag1−/− mice show signs of inflammation with infiltration of immune cells within the epithelial and sub-epithelial spaces. In addition, highly proliferative, crowded, and hyper cellular colonic crypts with elongated, hyperchromatic, and pseudostratified nuclei were present in the colons of Gpr109a−/−Rag1−/− mice. (Fig. 1D, 1F). The numbers of infiltrating neutrophils were also significantly increased in the colons of Gpr109a−/−Rag1−/− mice compared to Rag1−/− mice (Supplemental Fig. 1A). This resulted in significantly higher colitis scores for colons and caecum of Gpr109a−/−Rag1−/− mice than Rag1−/− mice (Fig. 1E, 1G). Chronic colitis is one risk factor for the development of colon cancers. Signs of ongoing adenomatous transformation were present in the caecum of Gpr109a−/−Rag1−/− mice. (arrows in Fig. 1F). Spleens of Gpr109a−/−Rag1−/− mice were significantly enlarged and contained significantly more cells than Rag1−/− mice (Supplemental Fig. 1B–C).
FIGURE 1.
Spontaneous colitis in Gpr109a−/−Rag1−/− mice. (A) Photograph showing rectal prolapse in Gpr109a−/−Rag1−/− mice. (B) Incidence of rectal prolapse in Gpr109a−/−Rag1−/− mice and Rag1−/− mice (n = 22 mice of each genotype). (C) Colon mass of Rag1−/− and Gpr109a−/−Rag1−/− mice. Each circle represents an individual mouse, lines represent the mean (n = 9 mice of each genotype). A representative photomicrograph of H&E stained cross sections of colons (D) and caecum (F) of indicated mice at 6 months of age. (E, G) Histopathological score of colons and caecum of mice indicated in D and F, respectively. Error bars represent standard deviation of the mean (n = 4–5 mice/group). *** P<0.0005
Increased numbers of ILC3 in Gpr109a−/−Rag1−/− mice
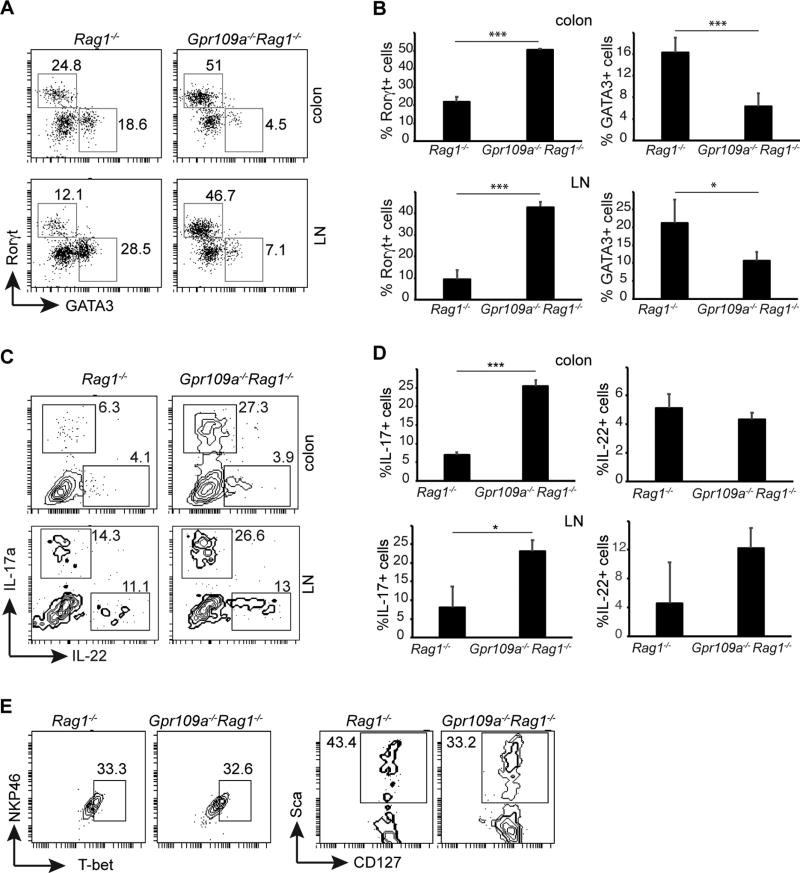
To characterize the mechanisms underlying the enhanced colonic inflammation in Gpr109a−/−Rag1−/− mice, we analyzed the presence of innate lymphoid cell subsets in intestinal lamina propria and mesenteric lymph node cells. CD45+Thy1.2+ cells were examined for expression of Rorγt and GATA3, markers for ILC3 and ILC2 respectively. Significantly higher numbers of Rorγt+ cells (ILC3) were present among CD45+Thy1.2+ cells in the colonic lamina propria of Gpr109a−/−Rag1−/− mice than Rag1−/− mice (Fig. 2A, 2B). Similarly, compared to Rag1−/− mice, Gpr109a−/−Rag1−/− mice exhibited increased numbers of ILC3 in their mesenteric lymph node and small intestines (Fig. 2A, 2B and Supplemental Fig. 2A, 2B). In sharp contrast, the frequency of CD45+Thy1.2+ GATA3+ cells (ILC2) was significantly decreased in colons, mesenteric lymph node, and small intestines of Gpr109a−/−Rag1−/− mice compared to Rag1−/− mice (Fig. 2A, 2B and Supplemental Fig. 2A, 2B). Next, cytokine production by ILC3 was analyzed. IL-17-producing ILC3 were present at a significantly higher percentage in the colonic lamina propria of Gpr109a−/−Rag1−/− mice (Fig. 2C, 2D). A similar trend was observed in mesenteric lymph nodes. On the other hand, IL-22 production by ILC3 was present at a comparable frequency in colons and lymph nodes of both Gpr109a−/−Rag1−/− mice and Rag1−/− mice (Fig. 2C, 2D). GM-CSF produced by ILC3 plays a critical role in innate immune cell driven colitis (33). Proportions of ILC3 producing GM-CSF were similar in Gpr109a−/−Rag1−/− and Rag1−/− mice (Supplemental Fig 2C).
FIGURE 2.
Increased number of ILC3 in Gpr109a−/−Rag1−/− mice. (A) Frequency of Rorγt+ and GATA3+ cells among CD45+Thy1.2+ cells in colon and mesenteric lymph node cells of indicated mice at 6 months of age. (B) Number of Rorγt+ and GATA3+ cells in colons and mesenteric lymph node of Rag1−/− and Gpr109a−/−Rag1−/− mice. Error bars represent standard deviation of mean (n = 5 mice/group). (C) Frequency of IL-17 and IL-22 producing cells following PMA and ionomycin stimulation among CD45+Thy1.2+Rorγt+ cells in colons and mesenteric lymph node cells of indicated mice. (D). Summary of IL-17 and IL-22 producing cells in indicated organs of Rag1−/− and Gpr109a−/−Rag1−/− mice. Error bars represent standard deviation of mean (n = 3 mice/group). * P<0.05, *** P<0.0005. (E) Phenotype of colonic CD45+Thy1.2+Rorγt+ cells in Rag1−/− and Gpr109a−/−Rag1−/− mice. A representative or pooled data of at least two experiments is shown.
ILC3 consists of 2 major subsets, lymphoid tissue-inducer (LTi)-like ILC3 and NKP46+ ILC3 (34–36). Both subsets are dependent on Rorγt and aryl hydrocarbon receptor (AHR). LTi-like ILC3 are CCR6+ and negative for natural killer (NK) cell markers such as NKP46. In contrast to LTi-like ILC3, NKP46+ ILC3 express NK cell markers and require T-bet, Notch, and the presence of gut microbiota for their development (37–41). Similar fractions of ILC3 in Gpr109a−/−Rag1−/− and Rag1−/− mice were positive for NKP46 and T-bet (Fig 2E). SCA-1+ and CD127 (IL-7R)+ ILCs mediate Helicobacter-induced colonic inflammation on the 129SvEvRag2−/− background (21, 22). The frequency of SCA-1+ and CD127+ ILC3 was comparable between ILC3 from Gpr109a−/−Rag1−/− and Rag1−/− mice (Fig 2E). ILC2 produce IL-13 and regulate homeostasis at mucosal surfaces (42, 43). IL-13 production by ILC2 cells was unchanged in the colons of Gpr109a−/−Rag1−/− mice when compared to Rag1−/− mice (Supplemental Fig. 2D, 2E). Taken together, these data demonstrate that colons of Gpr109a−/−Rag1−/− mice possess an increased number of IL-17 producing ILC3.
Colonic inflammation in Gpr109a−/−Rag1−/− mice is ILC3 dependent
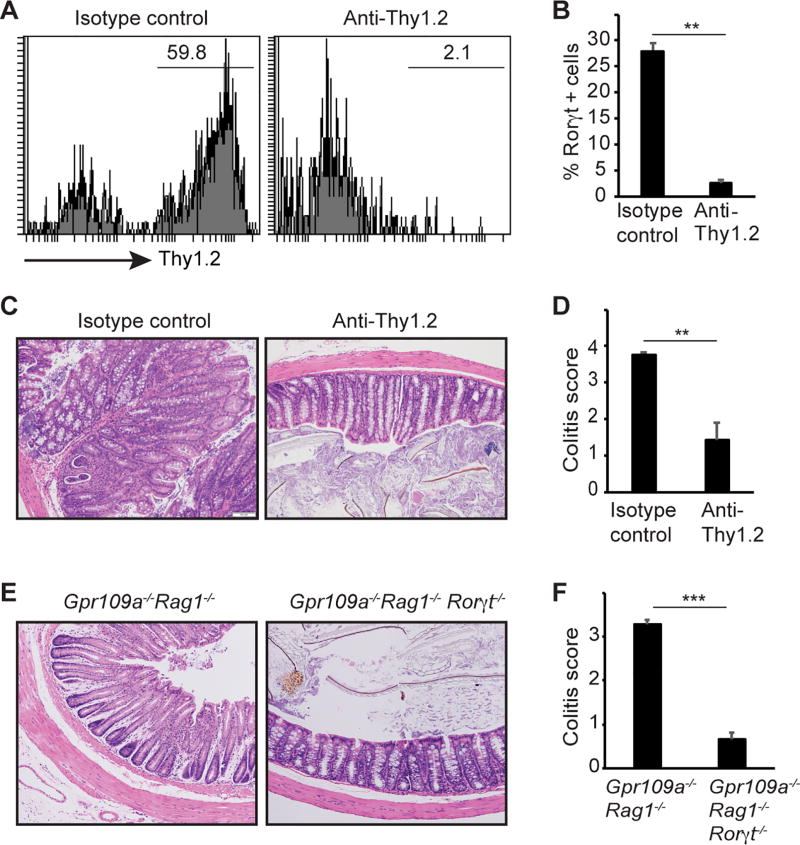
To test the functional relevance of ILC3’s role in inducing colitis in Gpr109a−/−Rag1−/− mice, we first treated these animals with depleting antibody against Thy1.2. Anti-Thy1.2 treatment reduced the frequency of Thy1.2+ cells among CD45+ cells by >90% (Fig. 3A). Compared to ~ 27.9% ILC3 of CD45+ cells in the isotype control-treated animals, ~ 1.5% of the CD45+ cells were ILC3 in anti-Thy1.2 treated animals (Fig. 3B). Anti-Thy1.2 treatment also reduced the colon mass and spleen cellularity (Supplemental Fig. 3). Most importantly, colons of anti-Thy1.2 treated animals showed a significant reduction in colitis histopathological than control antibody treated animals (Fig. 3C, 3D). Anti-Thy1.2 antibody depletes all the ILC on Rag1−/− background. To test a direct role of ILC3 in colonic inflammation in Gpr109a−/−Rag1−/− mice, we crossed these animals on Rorγt−/− background. No sign of colonic inflammation or hyperplasia was present in Gpr109a−/−Rag1−/−Rorγt−/− mice (Fig. 3E). Accordingly, histopathological scores for colitis were significantly reduced in Gpr109a−/−Rag1−/−Rorγt−/− mice than Gpr109a−/−Rag1−/− mice (Fig. 3F). Collectively, these findings demonstrate that ILC3 mediates colonic inflammation in Gpr109a−/−Rag1−/− mice.
FIGURE 3.
ILC3 mediate spontaneous colitis in Gpr109a−/−Rag1−/− mice. (A). Thy1.2 staining on CD45+ cells in Gpr109a−/−Rag1−/− mice treated with isotype control or anti-Thy1.2 antibody. (B) Frequency of Rorγt+ cells among CD45+ cells in colonic lamina propria of mice treated as in A (n = 3 mice/group). (C) Photomicrographs of H&E stained cross sections of Gpr109a−/−Rag1−/− mice indicated (original magnification X200). (D) Histopathological colitis score of Gpr109a−/−Rag1−/− mice treated as in C (n = 5 mice/group). Photomicrographs of H&E stained cross sections (E) and histopathological colitis score (F) of Gpr109a−/−Rag1−/− and Gpr109a−/−Rag1−/−Rorγt−/− mice. Error bars represent standard deviation of mean (n = 5 mice/group). ** P<0.005. A representative or pooled data of at least two experiments is shown.
A role for Gpr109a in regulating ILC3 homeostasis
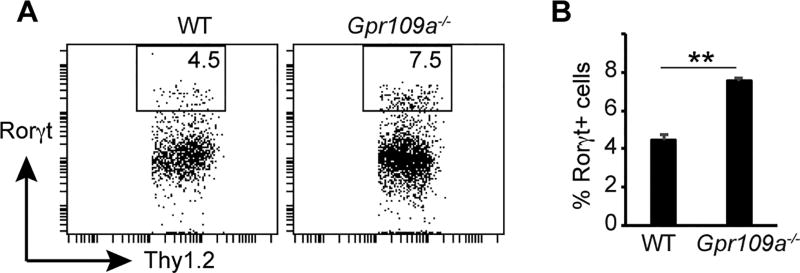
Data presented above show that in the absence of Gpr109a, ILC3 numbers are increased on a Rag1−/− background. A recent report shows that the number and distribution of ILC3 subsets are altered in Rag1−/− mice (44). Therefore, to directly assess the role of Gpr109a in influencing ILC3 population in the gut, ILC3 numbers were enumerated in WT and Gpr109a−/− animals. Fig. 4 shows that the colonic lamina propria of Gpr109a−/− mice contained a significantly higher number of Rorγt+ cells among Lin−CD45+Thy1.2+ cells than WT mice. Collectively, these data show that Gpr109a regulates ILC3 number in intestine.
FIGURE 4.
A critical role for Gpr109a in ILC3 homeostasis. (A) A representative flow cytometric staining showing Rorγt and GATA3 expression by colonic CD45+Lin−Thy1.2+ cells from WT and Gpr109a−/− mice. (B) Quantification of Rorγt and GATA3 cells among colonic CD45+Lin−Thy1.2+ cells from indicated mice. Error bars represent standard deviation of mean (n = 3 mice of each genotype). A representative of two experiments is shown.
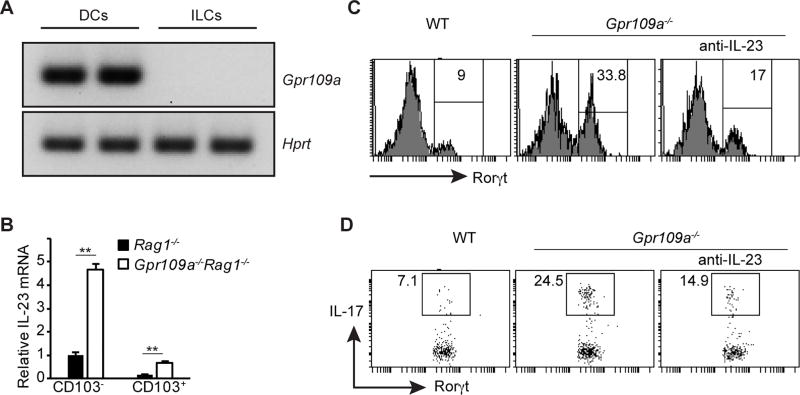
Gpr109a signaling suppresses IL-23 production
To characterize the mechanism underlying the Gpr109a mediated suppression of the ILC3 population in the gut, the expression of Gpr109a on various cells was examined. We have shown that in the gut, innate immune cells such as macrophages and dendritic cells express very high levels of Gpr109a. On the other hand, Gpr109a is expressed at very low levels by the colonic epithelium and it is undetectable in T and B cells (32). As expected, dendritic cells (DCs) expressed Gpr109a. In contrast, Gpr109a expression was undetectable on ILCs (Fig. 5A). To understand how the deficiency of Gpr109a in DCs can influence ILC3 numbers in the gut, we focused on IL-23 production by DCs. IL-23 is a key cytokine that promotes differentiation of ILC3 and regulates colonic inflammation (21, 45–47). Two main subsets of intestinal DCs, CD103− and CD103+, differ in their ability to produce IL-23. Therefore, the expression of IL-23 by colonic CD103− and CD103+ DCs was analyzed. Both subsets of colonic DCs from Gpr109a−/−Rag1−/− mice produced higher amounts of IL-23 than their counterparts from Rag1−/− mice (Fig. 5B). To test the functional relevance of overproduction of IL-23 by gut DCs from Gpr109a−/−Rag1−/− mice, we cultured sorted colonic DCs from Gpr109a−/−Rag1−/− and Rag1−/− mice with Thy1.2+ cells from Rag1−/− mice. Three days later, the development of ILC3 in these cultures was analyzed. Significantly higher numbers of ILC3 developed in cultures containing DCs from colons of Gpr109a−/−Rag1−/− mice than Rag1−/− mice (Fig. 5C). Moreover, higher proportions of ILC3 from cultures containing Gpr109a−/−Rag1−/− colonic DCs produced IL-17 than Rag1−/− cultures (Fig. 5D). The ability of colonic DCs from Gpr109a−/−Rag1−/− mice to promote the development of ILC3 was shown to be dependent on IL-23 production because the addition of neutralizing antibody against IL-23 significantly reduced the development of ILC3 in the cultures (Fig. 5C, 5D). Collectively, these data demonstrate that Gpr109a suppresses ILC3 by diminishing IL-23 production by intestinal DCs.
FIGURE 5.
Role of IL-23 in Gpr109a mediated promotion of ILC3. (A) Expression of Gpr109a and Hprt by CD11c+ and ILC (CD45+Thy1.2+) was analyzed by reverse transcriptase polymerase chain reaction (RT-PCR). (B) IL-23 expression by colonic CD103− and CD103+ DCs from Rag1−/− and Gpr109a−/−Rag1−/− mice was analyzed by quantitative RT-PCR (qRT-PCR) Error bars represent standard deviation of triplicates. (C) Rorγt expression by Thy1.2+ cells cultured with colonic DCs in the presence or absence of anti-IL-23 from indicated mice. (D) IL-17 production by Rorγt cells from C following PMA and ionomycin. Error bars represent standard deviation of triplicates (n = 3 mice/group). A representative of two experiments is shown.
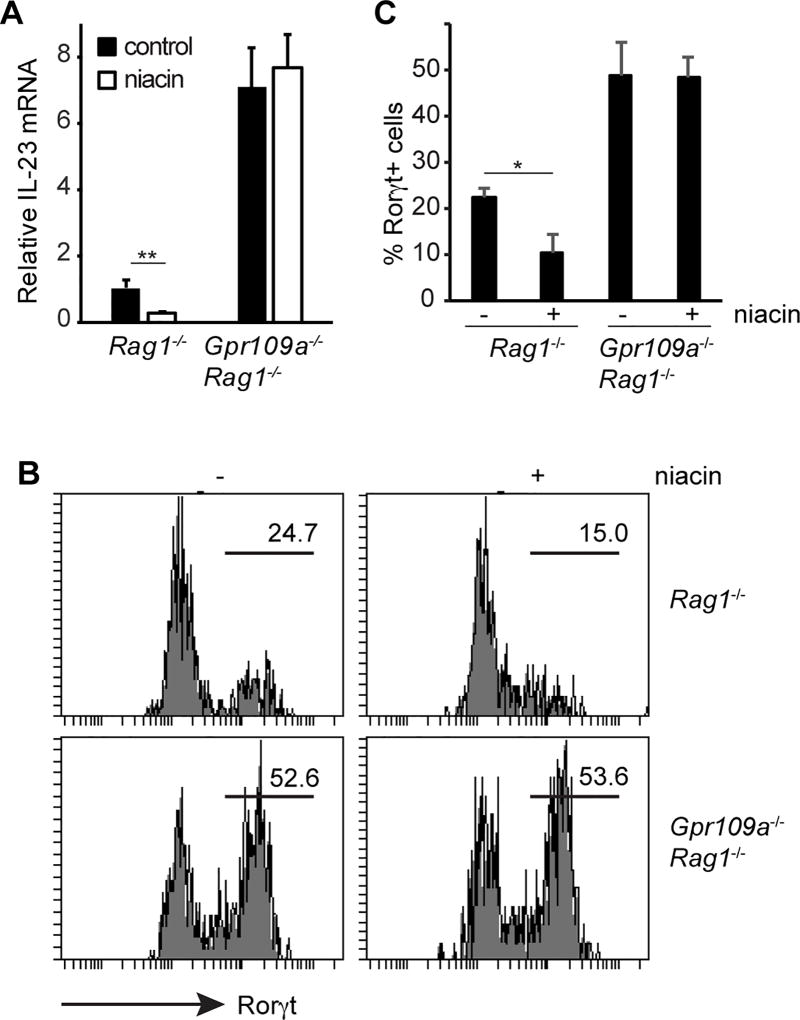
Niacin, a Gpr109a ligand, inhibits IL-23 and ILC3
Based on the data presented above, we tested whether niacin, a Gpr109a agonist, suppresses IL-23 production and ILC3 generation. Rag1−/− and Gpr109a−/−Rag1−/− mice were administered niacin in their drinking water. One month later, the number of ILC3 was analyzed from their colonic lamina propria. Niacin significantly diminished IL-23 production by colonic DCs from Rag1−/− mice, whereas IL-23 production remained unaffected in Gpr109a−/−Rag1−/− mice by niacin treatment (Fig. 6A). In accordance with this, niacin treatment significantly reduced the numbers of ILC3 in Rag1−/− mice (Fig. 6B, 6C). On the other hand, under the same treatment conditions, niacin failed to affect the ILC3 in Gpr109a−/−Rag1−/− mice. Collectively, these data suggest that Gpr109a activation decreased IL-23 production by DCs, leading to the suppression of ILC3.
FIGURE 6.
Gpr109a agonist suppresses IL-23 and ILC3. (A) Rag1−/− and Gpr109a−/−Rag1−/− mice were treated with niacin in drinking water. One month later, IL-23 expression by colonic DCs was analyzed. Error bars represent standard deviation of triplicates. (B) Rorγt expression among colonic CD45+Thy1.2+ cells from mice treated as in (A). (C) Rorγt+ cell frequency among colonic CD45+Thy1.2+ cells from mice treated as in A. (n = 3 mice/group). A representative of two experiments is shown.
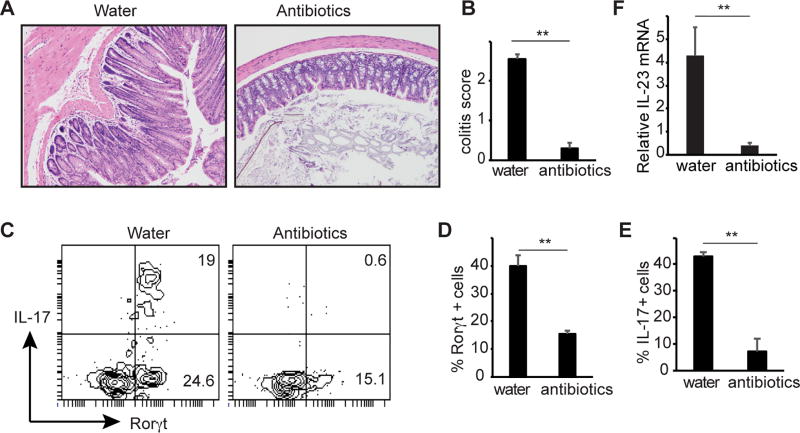
Colonic inflammation in Gpr109a−/−Rag1−/− mice is dependent on gut microbiota
Trillions of bacteria, collectively called gut microbiota, reside in the gut and impact inflammation. To test the role of gut microbiota in the colonic inflammation of Gpr109a−/−Rag1−/− mice, the animals were given a cocktail of antibiotics at 3 months of age. Colonic inflammation and hyperplasia of the epithelial layer were significantly reduced in antibody treated animals and lead to a significant decrease in histopathological score for colitis (Fig. 7A, 7B). Consistent with this, overall ILC3 number and the frequency of IL-17 producing ILC3 were significantly decreased in the colons of antibiotics treated mice compared to control untreated mice (Fig. 7C, 7D, 7E). Moreover, antibiotics treatment significantly inhibited IL-23 production by colonic DCs (Fig. 7F). Taken together, these data suggest that Gpr109a limits microbiota induced IL-23 production and ILC3 to regulate colonic homeostasis.
FIGURE 7.
Role of microbiota in spontaneous colitis of Gpr109a−/−Rag1−/− mice. (A) Gpr109a−/−Rag1−/− mice were given plain water or water containing a cocktail of antibiotics. Two months later colons of mice were analyzed for colitis. A representative photomicrograph of H&E stained cross section of colon. (B) Histopathological score of colitis. n = 4 (water) and 5 (antibiotics) mice. (C) A representative flow cytometric plot showing expression of Rorγt and IL-17 by CD45+Thy1.2+ cells. (D) and (E) Enumeration of frequency of Rorγt and IL-17 positive cells among CD45+Thy1.2+ cells (n = 3 mice/group). (F) IL-23 expression by colonic DCs from mice treated as in A. Error bars represent standard deviation of triplicates. A representative or pooled data of two experiments is shown.
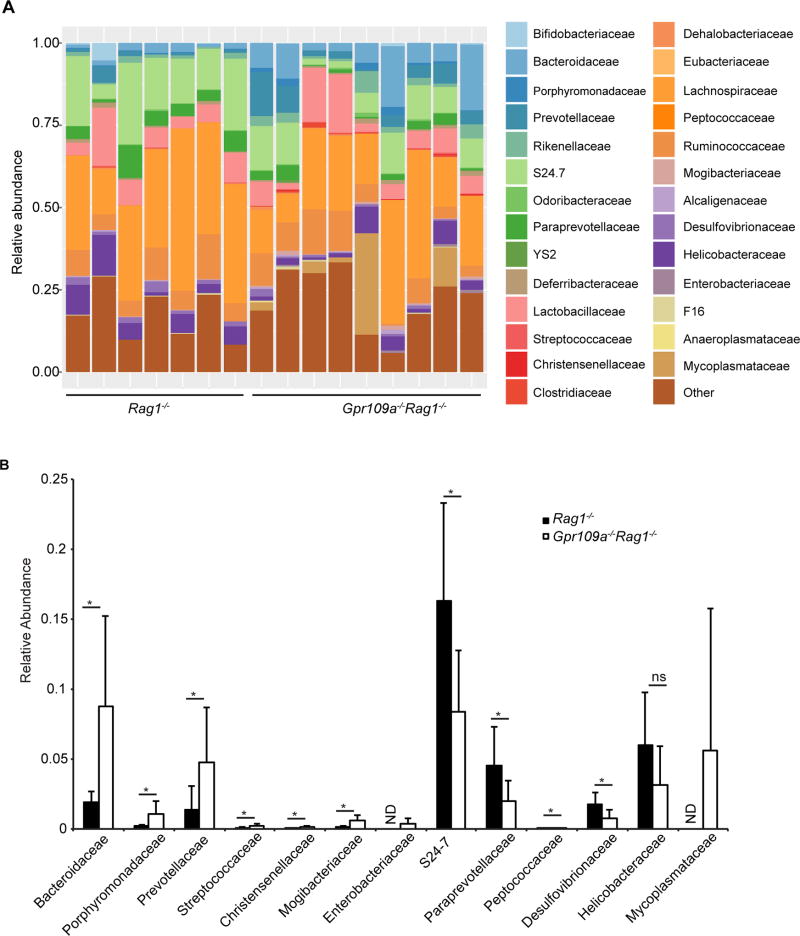
Altered gut microbiota in Gpr109a−/−Rag1−/− mice
Since antibiotics treatment ameliorated spontaneous colonic inflammation in Gpr109a−/−Rag1−/− mice, the composition of the gut microbiota between Rag1−/− and Gpr109a−/−Rag1−/− mice was compared. An average of 158160 ± 29409 sequences per mouse were analyzed. The relative abundance of Bacteroidetes belonging to family Bacteroidaceae, Porphyromonadaceae, Prevotellaceae, and Firmicutes belonging to family Streptococcaceae, Christensenellaceae, and Mogibacteriaceae among all bacteria was significantly higher in Gpr109a−/−Rag1−/− mice than Rag1−/− mice (Fig. 8A, 8B). On the other hand, Bacteroidetes belonging to family S24-7, Paraprevotellaceae, Firmicutes belonging to family Peptococcaceae, and Proteobacteria belonging to family Desulfovibrionaceae were less abundant in Gpr109a−/−Rag1−/− mice compared to Rag1−/− mice (Fig. 8A, 8B). Microbiota of the family Helicobacteraceae have been shown to cause colitis in mouse models, while in humans the results have been inconclusive and even indicated a potential protective effect against IBD (21, 46, 48–50). Furthermore, Enterobacteriaceae and Mycoplasmataceae in the gut have been associated with IBD (51–54). Bacteria of the family Enterobacteriaceae were present in the Gpr109a−/−Rag1−/− mice (7 out of 9 mice), whereas they were undetectable in Rag1−/− mice (Fig. 8B). Similarly, Mycoplasmataceae were found to be present in the gut of Gpr109a−/−Rag1−/− mice, while not in the gut of Rag1−/− mice (Fig. 8B). Helicobacteraceae, on the other hand, were present in comparable numbers between Gpr109a−/−Rag1−/− and Rag1−/− mice (Fig. 8B). Collectively, these data indicate that the gut of Gpr109a−/−Rag1−/− mice show an increased colonization by some microbiota associated with IBD than Rag1−/− mice.
FIGURE 8.
Microbial communities in the gut of Rag1−/− and Gpr109a−/−Rag1−/− mice. Sequencing of V3–V4 regions of bacterial 16S rDNA was performed on DNA isolated from caecum content. Shown is the (A) relative abundance of family-level phylotypes present within total bacterial taxonomic units sequenced in the indicated mice. Each bar in A represents an individual mouse. (B) Data from A is replotted for bacterial families showing significant differences between Rag1−/− and Gpr109a−/−Rag1−/− mice, as well as Helicobacteraceae. n = 7 and 9 mice of Rag1−/− and Gpr109a−/−Rag1−/− genotype respectively. * P<0.05, ND = not detected.
Discussion
In the current study, we showed that Gpr109a signaling suppresses IL-23 production by dendritic cells. Gpr109a-deficient colonic DCs promote ILC3 via a mechanism dependent on IL-23. Opposing roles of ILC3 in intestinal inflammation have been reported. IL-23 and IL-1β stimulate ILC3 to produce IL-22 and/or IL-17. IL-22 acts on gut epithelial cells and promotes their proliferation, wound healing, and barrier function (55, 56). Moreover, IL-22 also induces expression of anti-microbial peptides by the gut epithelia and thus protects against pathogenic bacteria (13). Thus, the actions of IL-22 improve intestinal health. IL-17 favors the release of chemoattractant factors and chemokines, which recruit pro-inflammatory neutrophils and leads to intestinal inflammation. Our data shows that in contrast to IL-22, IL-17 production is significantly increased by ILC3 in Gpr109a−/−Rag1−/− mice than Rag1−/− mice. These findings are consistent with an inflammatory role for ILC3 in colons of Gpr109a−/−Rag1−/− mice. Although IL-23 stimulates production of both IL-17 and IL-22 by ILC3, the conditions favoring preferential IL-17 production remain poorly defined and identification of such mechanisms will be key in understanding the regulation of ILC3 homeostasis by Gpr109a.
Dendritic cells play a key role in the homeostasis of ILC1 and ILC3 through the production of IL-12 and IL-23 (45). IL-23 favors the differentiation of ILCs to ILC3, whereas IL-12 promotes differentiation to ILC1. Data obtained in animals as well as humans unequivocally demonstrate a pathogenic role for IL-23 in the induction of inflammatory bowel disease (IBD). Whole genome association and transmission disequilibrium studies have identified that the point mutations Arg381Gln and R381Q in the IL-23 receptor (IL-23R) confer protection against Crohn’s disease (57, 58). Elevated levels of IL-23 mRNA are present in the mucosa of individuals with IBD and its expression level is correlated with disease severity (59, 60). Myeloid DCs from the mesenteric lymph nodes of Crohn’s disease subjects produce higher amounts of IL-23 and lower levels of IL-10 than the healthy controls. However, what factors regulate increased IL-23 and decreased IL-10 expression in IBD individuals remains unknown. In the current study, we have identified Gpr109a as one the determinants that suppresses IL-23 production. We have previously published that Gpr109a-deficient colonic DCs produce lower amounts of IL-10 and higher amounts of IL-6 (32). These findings establish a critical role for Gpr109a in the regulation of multiple cytokines relevant to the etiology of IBD. Future studies aimed at understanding the molecular mechanisms underlying Gpr109a-mediated regulation of IL-23 and IL-10 will be important in understanding the dysregulation of these cytokines in IBD.
Gut microbiota play a critical role in intestinal homeostasis. The engagement of pathogen-associated molecular patterns, or PAMPs, derived from gut microbiota to toll-like receptors (TLRs) or NOD expressed by innate immune cells induces production of inflammatory molecules. Therefore, the presence of trillions of bacteria in the gut poses a threat of inflammatory responses. However, the presence of overlapping immunoregulatory mechanisms arising from both the host and microbiota prevent the undesirable induction of inflammatory responses in the gut (2). Several studies show that a breakdown in these immunoregulatory mechanisms results in IBD. In mice lacking IL-10 or TGF-β and IL-10, the presence of commensals like Helicobacter hepaticus or Bacteroides is enough to induce colonic inflammation (50). Similarly, the Helicobacter hepaticus strain 51449 induces colitis in 129SvEvRag2−/− mice and Helicobacter infection is associated with rectal prolapses in mice (21, 61). Murine norovirus infection has previously been linked to the induction of colon inflammation and Crohn’s Disease-like symptoms in mice expressing hypomorphic Atg16L1 and also accelerates Helicobacter bilis-induced colitis in Mdr1a−/− mice (62, 63). In this study, Rag1−/− and Gpr109a−/−Rag1−/− mice (both on a C57BL/6 background) were housed in a colony that is positive for Helicobacter and murine norovirus. Gut microbiota analysis using 16S rDNA sequencing of caecum bacteria confirmed the presence of a comparable number of Helicobacter in Rag1−/− mice and Gpr109a−/−Rag1−/− mice. Further investigation is required to determine the impact Helicobacter and murine norovirus may have on the observed differences in bacterial colonization of the gut between Gpr109a−/−Rag1−/− and Rag1−/− mice as well as the spontaneous colitis of the former. The preferential presence of Enterobacteriaceae, which is associated with IBD, in Gpr109a−/−Rag1−/− mice is interesting and future studies should be aimed at investigating its role in inducing colonic inflammation, IL-23 production, and the promotion of ILC3 in Gpr109a−/−Rag1−/− mice. Niacin suppresses LPS mediated production of IL-6, TNF-α, IL-12p40, and IL-1β in a Gpr109a dependent manner (64). Therefore, it is possible that Gpr109a signaling inhibits microbiota-induced production of several inflammatory cytokines, including IL-23 in the colon, leading to suppression of inflammation. Identification of the signaling mechanisms underlying Gpr109a mediated suppression of the production of inflammatory cytokines will be very important in understanding the beneficial role of this receptor in the intestine and will facilitate the design of new therapeutics aimed at ameliorating colonic inflammation.
Supplementary Material
Acknowledgments
We thank the Georgia Cancer Center Flow Cytometry Core of Augusta University for technical help.
This research was supported in part by National Institutes of Health grant R01DK103576 to N.S.
Abbreviations used in this article
- DC
dendritic cell
- Gpr109a
G protein coupled receptor 109a
- H&E
hematoxylin and eosin
- IBD
inflammatory bowel disease
- ILC
innate lymphoid cell
- PMA
phorbol 12-myristate-13-acetate
- Treg
regulatory T cell
Footnotes
Disclosures
The authors declare no competing financial interests.
References
- 1.Wu GD, Bushmanc FD, Lewis JD. Diet, the human gut microbiota, and IBD. Anaerobe. 2013;24:117–120. doi: 10.1016/j.anaerobe.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 2.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, Essers J, Mitrovic M, Ning K, Cleynen I, Theatre E, Spain SL, Raychaudhuri S, Goyette P, Wei Z, Abraham C, Achkar JP, Ahmad T, Amininejad L, Ananthakrishnan AN, Andersen V, Andrews JM, Baidoo L, Balschun T, Bampton PA, Bitton A, Boucher G, Brand S, Buning C, Cohain A, Cichon S, D'Amato M, De Jong D, Devaney KL, Dubinsky M, Edwards C, Ellinghaus D, Ferguson LR, Franchimont D, Fransen K, Gearry R, Georges M, Gieger C, Glas J, Haritunians T, Hart A, Hawkey C, Hedl M, Hu X, Karlsen TH, Kupcinskas L, Kugathasan S, Latiano A, Laukens D, Lawrance IC, Lees CW, Louis E, Mahy G, Mansfield J, Morgan AR, Mowat C, Newman W, Palmieri O, Ponsioen CY, Potocnik U, Prescott NJ, Regueiro M, Rotter JI, Russell RK, Sanderson JD, Sans M, Satsangi J, Schreiber S, Simms LA, Sventoraityte J, Targan SR, Taylor KD, Tremelling M, Verspaget HW, De Vos M, Wijmenga C, Wilson DC, Winkelmann J, Xavier RJ, Zeissig S, Zhang B, Zhang CK, Zhao H, Silverberg MS, Annese V, Hakonarson H, Brant SR, Radford-Smith G, Mathew CG, Rioux JD, Schadt EE, Daly MJ, Franke A, Parkes M, Vermeire S, Barrett JC, Cho JH. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monteleone G, Biancone L, Marasco R, Morrone G, Marasco O, Luzza F, Pallone F. Interleukin 12 is expressed and actively released by Crohn's disease intestinal lamina propria mononuclear cells. Gastroenterology. 1997;112:1169–1178. doi: 10.1016/s0016-5085(97)70128-8. [DOI] [PubMed] [Google Scholar]
- 5.Heller F, Florian P, Bojarski C, Richter J, Christ M, Hillenbrand B, Mankertz J, Gitter AH, Burgel N, Fromm M, Zeitz M, Fuss I, Strober W, Schulzke JD. Interleukin-13 is the key effector Th2 cytokine in ulcerative colitis that affects epithelial tight junctions, apoptosis, and cell restitution. Gastroenterology. 2005;129:550–564. doi: 10.1016/j.gastro.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 6.Monteleone G, Kumberova A, Croft NM, McKenzie C, Steer HW, MacDonald TT. Blocking Smad7 restores TGF-beta1 signaling in chronic inflammatory bowel disease. J Clin Invest. 2001;108:601–609. doi: 10.1172/JCI12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fantini MC, Rizzo A, Fina D, Caruso R, Sarra M, Stolfi C, Becker C, Macdonald TT, Pallone F, Neurath MF, Monteleone G. Smad7 controls resistance of colitogenic T cells to regulatory T cell-mediated suppression. Gastroenterology. 2009;136:1308–1316. e1301–1303. doi: 10.1053/j.gastro.2008.12.053. [DOI] [PubMed] [Google Scholar]
- 8.Geremia A, Biancheri P, Allan P, Corazza GR, Di Sabatino A. Innate and adaptive immunity in inflammatory bowel disease. Autoimmun Rev. 2014;13:3–10. doi: 10.1016/j.autrev.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M, Binder V, Finkel Y, Cortot A, Modigliani R, Laurent-Puig P, Gower-Rousseau C, Macry J, Colombel JF, Sahbatou M, Thomas G. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 10.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, Achkar JP, Brant SR, Bayless TM, Kirschner BS, Hanauer SB, Nunez G, Cho JH. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 11.Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J, Gunther S, Prescott NJ, Onnie CM, Hasler R, Sipos B, Folsch UR, Lengauer T, Platzer M, Mathew CG, Krawczak M, Schreiber S. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 12.Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, Fisher SA, Roberts RG, Nimmo ER, Cummings FR, Soars D, Drummond H, Lees CW, Khawaja SA, Bagnall R, Burke DA, Todhunter CE, Ahmad T, Onnie CM, McArdle W, Strachan D, Bethel G, Bryan C, Lewis CM, Deloukas P, Forbes A, Sanderson J, Jewell DP, Satsangi J, Mansfield JC, Cardon L, Mathew CG. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, Ouyang W. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 14.Tumanov AV, Koroleva EP, Guo X, Wang Y, Kruglov A, Nedospasov S, Fu YX. Lymphotoxin controls the IL-22 protection pathway in gut innate lymphoid cells during mucosal pathogen challenge. Cell Host Microbe. 2011;10:44–53. doi: 10.1016/j.chom.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pickard JM, Maurice CF, Kinnebrew MA, Abt MC, Schenten D, Golovkina TV, Bogatyrev SR, Ismagilov RF, Pamer EG, Turnbaugh PJ, Chervonsky AV. Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature. 2014;514:638–641. doi: 10.1038/nature13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sonnenberg GF, Monticelli LA, Alenghat T, Fung TC, Hutnick NA, Kunisawa J, Shibata N, Grunberg S, Sinha R, Zahm AM, Tardif MR, Sathaliyawala T, Kubota M, Farber DL, Collman RG, Shaked A, Fouser LA, Weiner DB, Tessier PA, Friedman JR, Kiyono H, Bushman FD, Chang KM, Artis D. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336:1321–1325. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hepworth MR, Monticelli LA, Fung TC, Ziegler CG, Grunberg S, Sinha R, Mantegazza AR, Ma HL, Crawford A, Angelosanto JM, Wherry EJ, Koni PA, Bushman FD, Elson CO, Eberl G, Artis D, Sonnenberg GF. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 2013;498:113–117. doi: 10.1038/nature12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hepworth MR, Fung TC, Masur SH, Kelsen JR, McConnell FM, Dubrot J, Withers DR, Hugues S, Farrar MA, Reith W, Eberl G, Baldassano RN, Laufer TM, Elson CO, Sonnenberg GF. Group 3 innate lymphoid cells mediate intestinal selection of commensal bacteria-specific CD4(+) T cells. Science. 2015;348:1031–1035. doi: 10.1126/science.aaa4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mortha A, Chudnovskiy A, Hashimoto D, Bogunovic M, Spencer SP, Belkaid Y, Merad M. Microbiota-dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science. 2014;343:1249288. doi: 10.1126/science.1249288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dudakov JA, Hanash AM, van den Brink MR. Interleukin-22: immunobiology and pathology. Annu Rev Immunol. 2015;33:747–785. doi: 10.1146/annurev-immunol-032414-112123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buonocore S, Ahern PP, Uhlig HH, Ivanov, Littman DR, Maloy KJ, Powrie F. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 2010;464:1371–1375. doi: 10.1038/nature08949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Powell N, Walker AW, Stolarczyk E, Canavan JB, Gokmen MR, Marks E, Jackson I, Hashim A, Curtis MA, Jenner RG, Howard JK, Parkhill J, MacDonald TT, Lord GM. The transcription factor T-bet regulates intestinal inflammation mediated by interleukin-7 receptor+ innate lymphoid cells. Immunity. 2012;37:674–684. doi: 10.1016/j.immuni.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian Sarkis K, Ito S, Glickman JN, Glimcher LH. Communicable Ulcerative Colitis Induced by T-bet Deficiency in the Innate Immune System. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ermann J, Staton T, Glickman JN, de Waal Malefyt R, Glimcher LH. Nod/Ripk2 signaling in dendritic cells activates IL-17A–secreting innate lymphoid cells and drives colitis in T-bet−/−.Rag2−/− (TRUC) mice. Proceedings of the National Academy of Sciences. 2014;111:E2559–E2566. doi: 10.1073/pnas.1408540111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song C, Lee JS, Gilfillan S, Robinette ML, Newberry RD, Stappenbeck TS, Mack M, Cella M, Colonna M. Unique and redundant functions of NKp46+ILC3s in models of intestinal inflammation. The Journal of Experimental Medicine. 2015;212:1869–1882. doi: 10.1084/jem.20151403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernink JH, Peters CP, Munneke M, te Velde AA, Meijer SL, Weijer K, Hreggvidsdottir HS, Heinsbroek SE, Legrand N, Buskens CJ, Bemelman WA, Mjosberg JM, Spits H. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat Immunol. 2013;14:221–229. doi: 10.1038/ni.2534. [DOI] [PubMed] [Google Scholar]
- 27.Takayama T, Kamada N, Chinen H, Okamoto S, Kitazume MT, Chang J, Matuzaki Y, Suzuki S, Sugita A, Koganei K, Hisamatsu T, Kanai T, Hibi T. Imbalance of NKp44(+)NKp46(−) and NKp44(−)NKp46(+) natural killer cells in the intestinal mucosa of patients with Crohn's disease. Gastroenterology. 2010;139:882–892. 892.e881–883. doi: 10.1053/j.gastro.2010.05.040. [DOI] [PubMed] [Google Scholar]
- 28.Kirchberger S, Royston DJ, Boulard O, Thornton E, Franchini F, Szabady RL, Harrison O, Powrie F. Innate lymphoid cells sustain colon cancer through production of interleukin-22 in a mouse model. J Exp Med. 2013;210:917–931. doi: 10.1084/jem.20122308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huber S, Gagliani N, Zenewicz LA, Huber FJ, Bosurgi L, Hu B, Hedl M, Zhang W, O'Connor W, Jr, Murphy AJ, Valenzuela DM, Yancopoulos GD, Booth CJ, Cho JH, Ouyang W, Abraham C, Flavell RA. IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature. 2012;491:259–263. doi: 10.1038/nature11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hegyi J, Schwartz RA, Hegyi V. Pellagra: dermatitis, dementia, and diarrhea. Int J Dermatol. 2004;43:1–5. doi: 10.1111/j.1365-4632.2004.01959.x. [DOI] [PubMed] [Google Scholar]
- 31.Segal I, Ou Tim L, Demetriou A, Paterson A, Hale M, Lerios M. Rectal manifestations of pellagra. Int J Colorectal Dis. 1986;1:238–243. doi: 10.1007/BF01648345. [DOI] [PubMed] [Google Scholar]
- 32.Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, Thangaraju M, Prasad PD, Manicassamy S, Munn DH, Lee JR, Offermanns S, Ganapathy V. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40:128–139. doi: 10.1016/j.immuni.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pearson C, Thornton EE, McKenzie B, Schaupp AL, Huskens N, Griseri T, West N, Tung S, Seddon BP, Uhlig HH, Powrie F. ILC3 GM-CSF production and mobilisation orchestrate acute intestinal inflammation. Elife. 2016;5:e10066. doi: 10.7554/eLife.10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diefenbach A, Colonna M, Koyasu S. Development, differentiation, and diversity of innate lymphoid cells. Immunity. 2014;41:354–365. doi: 10.1016/j.immuni.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eberl G, Colonna M, Di Santo JP, McKenzie AN. Innate lymphoid cells: a new paradigm in immunology. Science. 2015;348:aaa6566. doi: 10.1126/science.aaa6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sonnenberg GF, Artis D. Innate lymphoid cells in the initiation, regulation and resolution of inflammation. Nat Med. 2015;21:698–708. doi: 10.1038/nm.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klose CS, Kiss EA, Schwierzeck V, Ebert K, Hoyler T, d'Hargues Y, Goppert N, Croxford AL, Waisman A, Tanriver Y, Diefenbach A. A T-bet gradient controls the fate and function of CCR6-RORgammat+ innate lymphoid cells. Nature. 2013;494:261–265. doi: 10.1038/nature11813. [DOI] [PubMed] [Google Scholar]
- 38.Kiss EA, Vonarbourg C, Kopfmann S, Hobeika E, Finke D, Esser C, Diefenbach A. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science. 2011;334:1561–1565. doi: 10.1126/science.1214914. [DOI] [PubMed] [Google Scholar]
- 39.Lee JS, Cella M, McDonald KG, Garlanda C, Kennedy GD, Nukaya M, Mantovani A, Kopan R, Bradfield CA, Newberry RD, Colonna M. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat Immunol. 2011;13:144–151. doi: 10.1038/ni.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qiu J, Heller JJ, Guo X, Chen ZM, Fish K, Fu YX, Zhou L. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity. 2012;36:92–104. doi: 10.1016/j.immuni.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vonarbourg C, Mortha A, Bui VL, Hernandez PP, Kiss EA, Hoyler T, Flach M, Bengsch B, Thimme R, Holscher C, Honig M, Pannicke U, Schwarz K, Ware CF, Finke D, Diefenbach A. Regulated expression of nuclear receptor RORgammat confers distinct functional fates to NK cell receptor-expressing RORgammat(+) innate lymphocytes. Immunity. 2010;33:736–751. doi: 10.1016/j.immuni.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nussbaum JC, Van Dyken SJ, von Moltke J, Cheng LE, Mohapatra A, Molofsky AB, Thornton EE, Krummel MF, Chawla A, Liang H-E, Locksley RM. Type 2 innate lymphoid cells control eosinophil homeostasis. Nature. 2013;502:245–248. doi: 10.1038/nature12526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spencer SP, Wilhelm C, Yang Q, Hall JA, Bouladoux N, Boyd A, Nutman TB, Urban JF, Jr, Wang J, Ramalingam TR, Bhandoola A, Wynn TA, Belkaid Y. Adaptation of innate lymphoid cells to a micronutrient deficiency promotes type 2 barrier immunity. Science. 2014;343:432–437. doi: 10.1126/science.1247606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mackley EC, Houston S, Marriott CL, Halford EE, Lucas B, Cerovic V, Filbey KJ, Maizels RM, Hepworth MR, Sonnenberg GF, Milling S, Withers DR. CCR7-dependent trafficking of RORgamma(+) ILCs creates a unique microenvironment within mucosal draining lymph nodes. Nat Commun. 2015;6:5862. doi: 10.1038/ncomms6862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bernink JH, Krabbendam L, Germar K, de Jong E, Gronke K, Kofoed-Nielsen M, Munneke JM, Hazenberg MD, Villaudy J, Buskens CJ, Bemelman WA, Diefenbach A, Blom B, Spits H. Interleukin-12 and -23 Control Plasticity of CD127(+) Group 1 and Group 3 Innate Lymphoid Cells in the Intestinal Lamina Propria. Immunity. 2015;43:146–160. doi: 10.1016/j.immuni.2015.06.019. [DOI] [PubMed] [Google Scholar]
- 46.Arnold IC, Mathisen S, Schulthess J, Danne C, Hegazy AN, Powrie F. CD11c+ monocyte/macrophages promote chronic Helicobacter hepaticus-induced intestinal inflammation through the production of IL-23. Mucosal Immunology. 2015;9:352. doi: 10.1038/mi.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen L, He Z, Slinger E, Bongers G, Lapenda TL, Pacer ME, Jiao J, Beltrao MF, Soto AJ, Harpaz N, Gordon RE, Ochando JC, Oukka M, Iuga AC, Chensue SW, Blander JM, Furtado GC, Lira SA. IL-23 activates innate lymphoid cells to promote neonatal intestinal pathology. Mucosal Immunology. 2014;8:390. doi: 10.1038/mi.2014.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luther J, Dave M, Higgins PDR, Kao JY. Association Between Helicobacter pylori Infection and Inflammatory Bowel Disease: A Meta-analysis and Systematic Review of the Literature. Inflammatory bowel diseases. 2010;16:1077–1084. doi: 10.1002/ibd.21116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Papamichael K, Konstantopoulos P, Mantzaris GJ. Helicobacter pylori infection and inflammatory bowel disease: Is there a link? World Journal of Gastroenterology : WJG. 2014;20:6374–6385. doi: 10.3748/wjg.v20.i21.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bloom Seth M, Bijanki Vinieth N, Nava Gerardo M, Sun L, Malvin Nicole P, Donermeyer David L, Dunne WM, Allen Paul M, Stappenbeck Thaddeus S. Commensal Bacteroides Species Induce Colitis in Host-Genotype-Specific Fashion in a Mouse Model of Inflammatory Bowel Disease. Cell Host & Microbe. 2011;9:390–403. doi: 10.1016/j.chom.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rajilic-Stojanovic M, Shanahan F, Guarner F, de Vos WM. Phylogenetic analysis of dysbiosis in ulcerative colitis during remission. Inflamm Bowel Dis. 2013;19:481–488. doi: 10.1097/MIB.0b013e31827fec6d. [DOI] [PubMed] [Google Scholar]
- 52.Knights D, Silverberg MS, Weersma RK, Gevers D, Dijkstra G, Huang H, Tyler AD, van Sommeren S, Imhann F, Stempak JM, Huang H, Vangay P, Al-Ghalith GA, Russell C, Sauk J, Knight J, Daly MJ, Huttenhower C, Xavier RJ. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Medicine. 2014;6:107. doi: 10.1186/s13073-014-0107-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seksik P, Rigottier-Gois L, Gramet G, Sutren M, Pochart P, Marteau P, Jian R, Doré J. Alterations of the dominant faecal bacterial groups in patients with Crohn's disease of the colon. Gut. 2003;52:237–242. doi: 10.1136/gut.52.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen W, Li D, Paulus B, Wilson I, Chadwick VS. High prevalence of Mycoplasma pneumoniae in intestinal mucosal biopsies from patients with inflammatory bowel disease and controls. Digestive diseases and sciences. 2001;46:2529–2535. doi: 10.1023/a:1012352626117. [DOI] [PubMed] [Google Scholar]
- 55.Pickert G, Neufert C, Leppkes M, Zheng Y, Wittkopf N, Warntjen M, Lehr HA, Hirth S, Weigmann B, Wirtz S, Ouyang W, Neurath MF, Becker C. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J Exp Med. 2009;206:1465–1472. doi: 10.1084/jem.20082683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sugimoto K, Ogawa A, Mizoguchi E, Shimomura Y, Andoh A, Bhan AK, Blumberg RS, Xavier RJ, Mizoguchi A. IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J Clin Invest. 2008;118:534–544. doi: 10.1172/JCI33194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, Dassopoulos T, Bitton A, Yang H, Targan S, Datta LW, Kistner EO, Schumm LP, Lee AT, Gregersen PK, Barmada MM, Rotter JI, Nicolae DL, Cho JH. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dubinsky MC, Wang D, Picornell Y, Wrobel I, Katzir L, Quiros A, Dutridge D, Wahbeh G, Silber G, Bahar R, Mengesha E, Targan SR, Taylor KD, Rotter JI. IL-23 receptor (IL-23R) gene protects against pediatric Crohn's disease. Inflamm Bowel Dis. 2007;13:511–515. doi: 10.1002/ibd.20126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmidt C, Stallmach A. Etiology and pathogenesis of inflammatory bowel disease. Minerva gastroenterologica e dietologica. 2005;51:127–145. [PubMed] [Google Scholar]
- 60.Punkenburg E, Vogler T, Buttner M, Amann K, Waldner M, Atreya R, Abendroth B, Mudter J, Merkel S, Gallmeier E, Rose-John S, Neurath MF, Hildner K. Batf-dependent Th17 cells critically regulate IL-23 driven colitis-associated colon cancer. Gut. 2016;65:1139–1150. doi: 10.1136/gutjnl-2014-308227. [DOI] [PubMed] [Google Scholar]
- 61.Miller CL, Muthupalani S, Shen Z, Fox JG. Isolation of Helicobacter spp. from mice with rectal prolapses. Comparative medicine. 2014;64:171–178. [PMC free article] [PubMed] [Google Scholar]
- 62.Cadwell K, Patel KK, Maloney NS, Liu T-C, Ng ACY, Storer CE, Head RD, Xavier R, Stappenbeck TS, Virgin HW. Virus-Plus-Susceptibility Gene Interaction Determines Crohn's Disease Gene Atg16L1 Phenotypes in Intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lencioni KC, Seamons A, Treuting PM, Maggio-Price L, Brabb T. Murine Norovirus: An Intercurrent Variable in a Mouse Model of Bacteria-Induced Inflammatory Bowel Disease. Comparative medicine. 2008;58:522–533. [PMC free article] [PubMed] [Google Scholar]
- 64.Zandi-Nejad K, Takakura A, Jurewicz M, Chandraker AK, Offermanns S, Mount D, Abdi R. The role of HCA2 (GPR109A) in regulating macrophage function. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2013;27:4366–4374. doi: 10.1096/fj.12-223933. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.