Abstract
Our aim was to investigate whether molecular classification can be used to refine prognosis in grade 3 endometrioid endometrial carcinomas (EECs). Grade 3 EECs were classified into four subgroups: p53-abnormal, based on mutant-like immunostaining (p53abn); MMR-deficient, based on loss of mismatch repair protein expression (MMRd); presence of POLE exonuclease domain hotspot mutation (POLE); no specific molecular profile (NSMP), in which none of these aberrations were present. Overall (OS), and recurrence-free survival (RFS) rates were compared using the Kaplan-Meier method (Log-Rank test) and univariable and multivariable Cox proportional hazard models. In total, 381 patients were included. The median age was 66 years (range 33–96). FIGO stages (2009) were as follows: IA, 171 (44.9%), IB, 120 (31.5%), II, 24 (6.3%), III, 50 (13.1%), IV, 11 (2.9%). There were 49 (12.9%) POLE, 79 (20.7%) p53abn, 115 (30.2%) NSMP, and 138 (36.2%) MMRd tumors. Median follow-up of patients was 6.1 years (range 0.2–17.0). Compared to patients with NSMP, patients with POLE mutant grade 3 EEC (OS: Hazard Ratio [HR] 0.36 [95%CI: 0.18–0.70], p=0.003; RFS: HR 0.17 [0.05–0.54], p=0.003) had a significantly better prognosis; patients with p53abn tumors had a significantly worse RFS (HR 1.73 [1.09–2.74], p0.021); patients with MMRd tumors showed a trend towards better RFS. Estimated 5-year OS rates were as follows: POLE 89%, MMRd 75%, NSMP 69%, p53abn 55% (Log Rank p=0.001). Five-year RFS rates were as follows: POLE 96%, MMRd 77%, NSMP 64%, p53abn 47% (p=0.000001), respectively. In a multivariable Cox model that included age and FIGO stage, POLE and MMRd status remained independent prognostic factors for better RFS; p53 status was an independent prognostic factor for worse RFS. Molecular classification of grade 3 EECs reveals that these tumors are a mixture of molecular subtypes of endometrial carcinoma, rather than a homogeneous group. The addition of molecular markers identifies prognostic subgroups, with potential therapeutic implications.
Keywords: POLE exonuclease domain mutations, Endometrial cancer, Molecular classification
Introduction
Endometrial cancer is a heterogenous disease, with an overall 5-year survival rate of approximately 80%.1 There is a critical need to improve the identification of early-stage patients who may benefit from adjuvant therapy, and to identify patients with metastatic disease for whom a specific targeted therapy may be advantageous. To facilitate this, clinicopathologic variables are incorporated into the current risk assessment. For example, Stage I/grade 3 endometrioid-type endometrial cancers (EECs) are considered to pose a higher risk of recurrence; therefore, adjuvant radiotherapy is administered in many centers to treat these tumors. Recent molecular profiling has provided novel insight into the molecular heterogeneity of endometrial cancer.
Studies documenting molecular heterogeneity in endometrial cancer using genome-wide analyses, particularly those from the Cancer Genome Atlas (TCGA), have identified four distinct molecular subclasses based on genomic architecture/mutational burden.2 These include the ultramutated subtype, defined by mutation in the exonuclease domain of DNA Polymerase epsilon (POLE); the hypermutated, microsatellite-unstable subclass, with deficiency of one or more mismatch repair proteins (MMRd); the copy number-high subtype, which is characterized by mutations in p53; and the copy number-low subtype that does not have a surrogate marker. More recently, clinically applicable methodologies have been applied to identify these groups. These methodologies may also impact prognosis and identify potentially targetable underlying molecular abnormalities.3–8 With the use of immunohistochemistry (IHC) and targeted sequencing of POLE, this approach becomes cost-effective and may be rapidly implemented in current practice. Recent studies have independently proven this concept using clinical trials materials4,5 and unselected cases drawn from multiple institutions.7 The Institute of Medicine guidelines for development of “omics-based” biomarkers were followed in the latter study.9
In the current study, we focused our efforts on grade 3 EECs. These tumors comprise a subset of endometrial carcinoma that has engendered significant controversy with respect to pathogenesis (type I or type II?), prognosis (comparable to that of serous carcinoma, or more favorable?), and treatment. Diagnosis is associated with significant interobserver variability and suboptimal reproducibility. Furthermore, this is the only tumor type represented in each of the four genomic subgroups of the TCGA cohort. We tested the hypothesis that the molecular subtypes of grade 3 EECs are associated with significant differences in prognosis, using a clinically applicable molecular classification tool in a large, well-characterized cohort of grade 3 EECs from 6 centers in North America and Europe.
Methods
Institutional approval for this study was obtained from each of the participating centers.
FIGO grade 3 EECs with clinical follow-up data were collected from 6 institutions in Europe and North America (Table 1). Strict diagnostic criteria were applied, as follows: 1) tumors demonstrated endometrioid lineage evidenced by a component of low-grade endometrioid adenocarcinoma with low-intermediate nuclear grade and/or metaplasias typical of endometrioid differentiation, with exclusion of histological mimics (i.e. “confirmatory endometrioid features”); 2) tumors were characterized by predominantly solid architecture exclusive of squamous differentiation, or mixtures of glandular and solid architecture with diffusely distributed high-grade nuclei (Figure 1).
Table 1.
Case distribution by center
| Centers | |||||||
|---|---|---|---|---|---|---|---|
| NL | BC | NY | ES | MA | UK | Total | |
| N | 113 | 116 | 65 | 47 | 25 | 15 | 381 |
| % | 29.7% | 30.4% | 17.1% | 12.3% | 6.6% | 3.9% | 100.0% |
NL=Netherlands; BC=British Columbia; NY=New York (USA); ES=Spain; MA=Massachusetts (USA); UK=United Kingdom
Figure 1.

Use of immunohistochemistry for molecular subgroup assignment (A) FIGO grade 3 endometrioid carcinoma with p53 overexpression (p53abn subgroup). (B) FIGO grade 3 endometrioid carcinoma with loss of MLH1 expression (MMRd subgroup). (C) FIGO grade 3 endometrioid carcinoma with normal expression p53 staining, no DNA MMR abnormalities (not shown) and no POLE hotspot mutation (NSMP subgroup). (D) FIGO grade 3 endometrioid carcinoma lacking immunohistochemical abnormalities. POLE hotspot mutation identified (POLE subgroup).
Molecular subgroup assignment
POLE sequencing for hotspots in the exonuclease domain (exons 9–14) was performed using either Sanger or next-generation approaches, as described previously.4,7 Either 2 (PMS2 and MSH6) or 4 (MLH1, PMS2, MSH2, MSH6) DNA mismatch repair IHC markers were performed on representative sections at the referring institution, as well as p53 IHC staining (Figure 1). DNA mismatch repair results were scored as abnormal if no tumor nuclear staining was found in the presence of an intact internal control. Mutation-type p53 staining was recognized either by overexpression (intense staining of >75% tumor cell nuclei) or no expression in tumor cell nuclei, in the presence of an intact internal control. In a subset of cases, sequencing was performed on tumors with ambiguous p53 IHC results. Participating institutions used slightly different approaches, the details of which are described in the Supplemental Methodology.
Assignation of carcinomas to TCGA-like subgroups (genomic subgroup classification) resulted in the following categories:
POLE mutation (POLE)—pathogenic variants in the exonuclease domain of DNA Polymerase epsilon, which were characterized by the TCGA as “ultramutated” (in the absence of MMRd or abnormal p53 expression).
Mismatch repair deficient (MMRd)—abnormal expression of one or more mismatch repair proteins by IHC, which is highly concordant with MSI-H status and assignment to the TCGA “hypermutated” group (in the absence of POLE mutation or abnormal p53 expression).10,11
p53 abnormal (p53abn)—exhibiting aberrant p53 immunostaining (IHC score 0 for complete absence of protein expression, as in null mutations or overexpression, defined as intense staining of >75% of tumor cell nuclei) in the absence of POLE mutations or MMRd. This group largely corresponds to the “copy number-high/serous-like” TCGA group.
No specific molecular profile (NSMP)—exhibiting normal p53 and MMR expression by IHC and no mutations in the exonuclease domain of POLE, analogous to the “copy number-low” subgroup in the TCGA.
Cases that could not be assigned to any of the above categories—either because one or more assays failed (incomplete profile) or because more than one classifier was present (e.g. POLE and p53abn)—were excluded from this study.
Clinical data were obtained by review of medical records, and appropriate statistical tests (Log-rank for OS and RFS [Kaplan-Meier] and Cox proportional hazards modeling for uni- and multivariable analyses) were performed to investigate the clinical significance of the molecular classification. FIGO stages (2009) were grouped for statistical analysis, providing alignment with most clinical algorithms, as follows: Stage IA, Stage IB, and Stages II–IV.
As we anticipated that a significant percentage of the FIGO grade 3 EECs would map to the p53abn category, we collected staging data from the Memorial Sloan Kettering Cancer Center database on endometrial serous carcinoma patients who were surgically staged (n=409). The aim was to detect differences in stage distribution by comparing the p53abn endometrioid category with serous carcinomas, tumors that were clustered together in the TCGA study.2
Results
Three hundred and eighty-one patients met the inclusion criteria. Median follow-up was 6.1 years (range 0.2–17.0). Median age was 66 years (range 33–96). The FIGO 2009 stage distribution was as follows: IA, 44.9%; IB, 31.5%; II, 6.3%; III, 13.1%; IV, 2.9%.
Genomic subgroup classification and demographics
The classification tool yielded the following distribution of subgoups: POLE—12.9%; MMRd—36.2%; p53abn—20.7%; NSMP—30.2%. Table 2 shows the distribution of genomic subgroups stratified by stage. An overwhelming majority of POLE mutant carcinomas were Stage I (47/48, 98%), in contrast to other types (p=0.0003). MMRd and NSMP carcinomas were relatively evenly distributed in Stages I–IV. The majority (44/79, 56%) of p53abn carcinomas were Stage IA, with only 22% (17/79) in Stages II–IV. Of note, MMRd tumors comprised a relatively large proportion of Stages II–IV (41/85, 48%). Median age also varied in relation to genomic subgroup, with POLE mutant carcinoma patients being youngest (median age 60 years, p=0.00006). Patients with tumors in the three other categories presented at nearly identical median ages (65–69 years).
Table 2.
Stage distribution and age stratified by molecular subgroup
| POLE | MMRd | NSMP | TP53 | Total | p-value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| FIGO (2009) | N | % | N | % | N | % | N | % | N | ||
| Stage IA | 35 | 20.5 | 48 | 28.1 | 44 | 25.7 | 44 | 25.7 | 171 | 0.0003 | |
| Stage IB | 12 | 10.0 | 47 | 39.2 | 43 | 35.8 | 18 | 15.0 | 120 | ||
| Stage II–IV | 1 | 1.2 | 41 | 48.2 | 26 | 30.6 | 17 | 20.0 | 85 | ||
| Total | 48 | 12.8 | 136 | 36.2 | 113 | 30.1 | 79 | 21.0 | 376 | ||
| Age, years | mean (range) | 60 | 38–93 | 65 | 33–92 | 67 | 36–96 | 69 | 47–93 | 0.00006 | |
Stage comparison: p53abn FIGO grade 3 endometrioid versus serous carcinoma
As stated above, 22% of patients with p53abn grade 3 EECs presented at Stages II–IV. This stands in contrast to the institutional series of surgically staged serous carcinoma patients, of whom 58.36% presented at Stages II–IV (p<0.0001). Presentation at Stages IIIC (17.76%) and IVB (22.65%) was notable.
Outcomes analyses
Table 3 shows the relationships between stage and clinical outcomes, and age and clinical outcomes. Stage was independently associated with OS and RFS, whereas age was independently associated only with OS. Compared to outcomes in Stage IA tumors, Stage IB tumors had a hazard ratio (HR) of 1.64 (confidence interval [CI]: 1.00–2.69) for RFS (p=0.059); Stage II–IV tumors had HRs of 3.73 (CI: 2.54–5.49) and 4.55 (CI: 2.83–7.33) for OS and RFS, respectively (p<0.001 for both) (Figure 2).
Table 3.
Survival and associations with FIGO stage and age
| Overall Survival | Recurrence-Free Survival | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| univariable | multivariable | univariable | multivariable | |||||||||
| HR | 95%CI | p-value | HR | 95%CI | p-value | HR | 95%CI | p-value | HR | 95%CI | p-value | |
| FIGO IA | 1 | - | 1 | - | 1 | - | - | - | ||||
| IB | 1.19 | 0.80–1.78 | 0.395 | 1.06 | 0.71–1.58 | 0.792 | 1.64 | 1.00–2.69 | 0.049 | 1.61 | 0.98–2.65 | 0.059 |
| II–IV | 3.55 | 2.42–5.10 | <0.001 | 3.73 | 2.54–5.49 | <0.001 | 4.49 | 2.79–7.21 | <0.001 | 4.55 | 2.83–7.33 | <0.001 |
| age | 1.04 | 1.03–1.06 | <0.001 | 1.04 | 1.03–1.06 | <0.001 | 1.00 | 0.98–1.02 | 0.889 | 1.01 | 0.99–1.03 | 0.456 |
Figure 2.
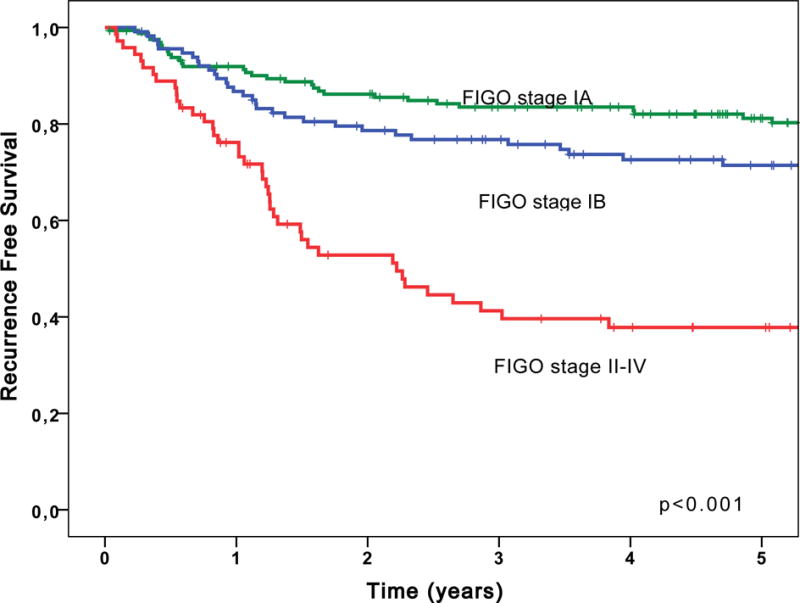
FIGO stage is associated with recurrence-free survival.
Genomic subgroup classification was independently associated with RFS (Table 4). Patients with POLE mutant carcinomas had significantly better RFS, compared to patients with NSMP tumors (HR=0.23, 0.07–0.77) p=0.017. The MMRd group showed a trend towards better RFS, compared to the NSMP group (HR=0.61, 0.37–1.00 CI, p=0.052). RFS in patients with p53abn cancers was unfavorable compared to RFS in patients with NSMP tumors (HR=1.92, 1.20–3.07 CI, p=0.007) (Figure 3). Multivariable analysis was performed for Stage I only, demonstrating that patients with POLE mutant tumors had significantly longer RFS (p=0.05) (Supplemental Table 1A–B) (Figure 4). Following this multivariable analysis, p53abn remained prognostically unfavorable compared to NSMP, with respect to RFS (p=0.004).
Table 4.
Survival and associations with stage and molecular subgroup
| Overall Survival | Recurrence-Free Survival | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| univariable | multivariable | univariable | multivariable | |||||||||
| HR | 95%CI | p-value | HR | 95%CI | p-value | HR | 95%CI | p-value | HR | 95%CI | p-value | |
| POLE | 0.36 | 0.18–0.70 | 0.003 | 0.56 | 0.27–1.15 | 0.112 | 0.17 | 0.05–0.54 | 0.003 | 0.23 | 0.07–0.77 | 0.017 |
| MMRd | 0.80 | 0.55–1.18 | 0.261 | 0.84 | 0.57–1.25 | 0.394 | 0.66 | 0.41–1.08 | 0.098 | 0.61 | 0.37–1.00 | 0.052 |
| NMSP | 1 | - | 1 | - | 1 | - | 1 | - | ||||
| TP53 | 1.30 | 0.86–1.97 | 0.215 | 1.37 | 0.90–2.09 | 0.143 | 1.73 | 1.09–2.74 | 0.021 | 1.92 | 1.20–3.07 | 0.007 |
| FIGO IA | 1 | - | 1 | - | 1 | - | - | - | ||||
| IB | 1.19 | 0.80–1.78 | 0.395 | 1.09 | 0.73–1.64 | 0.679 | 1.64 | 1.00–2.69 | 0.049 | 1.78 | 1.07–2.96 | 0.027 |
| II–IV | 3.55 | 2.42–5.10 | <0.001 | 3.60 | 2.43–5.32 | <0.001 | 4.49 | 2.79–7.21 | <0.001 | 4.28 | 2.64–6.95 | <0.001 |
| age | 1.04 | 1.03–1.06 | <0.001 | 1.04 | 1.02–1.06 | <0.001 | 1.00 | 0.98–1.02 | 0.889 | 1.00 | 0.98–1.02 | 0.611 |
Figure 3.

Molecular subgroup is associated with overall (A) and recurrence-free survival (B) in FIGO grade 3 endometrioid carcinoma.
Figure 4.

Molecular subgroup is associated with overall (A) and recurrence-free survival (B) in FIGO stage I, grade 3 endometrioid carcinoma.
Discussion
Bokhman’s subclassification of endometrial carcinomas into estrogen-driven type I and estrogen-independent type II tumors12 was a landmark achievement, and provided a framework for understanding most endometrial carcinomas. However, some of these neoplasms defied classification as type I or II, and the TCGA-based molecular classification offers both an improved system for subclassifcation and an explanation for the limitations of the type I versus II categorization. Particularly problematic are the 30% of endometrial carcinomas that are neither type I/low-grade endometrioid nor type II/serous, but are characterized by very high mutation burdens (with resulting neoantigen formation and prominent host immune response, as well as intratumoral morphological heterogeneity), i.e. MMRd and POLE subtypes.2 With regard to histotype, almost all copy number-low (NSMP), POLE mutant and MMRd tumors are endometrioid; although most of the copy number-high/p53abn tumors are serous, a significant minority are grade 3 EECs. Thus, grade 3 EECs are uniquely distributed amongst the four molecular subtypes, and our study confirms this. The outstanding question addressed herein was whether the molecular subtypes are of prognostic significance within the group of grade 3 EECs. In this group, stage remains an important prognostic factor with respect to both OS and RFS. POLE mutations are frequent, are associated with early stage disease, and arise in patients younger than those in other genomic subgroups. POLE mutant group assignment is independently associated with favorable OS and RFS, whereas p53abn is associated with a significantly worse prognosis.
In addition to the fact that histotype assignment is suboptimally reproducible in high-grade EECs without molecular classification,13–16 the features reported as characteristic of each TCGA group are not sufficiently specific or reproducible to supplant the molecular classification itself. For example, dense tumor infiltrating lymphocytes and peritumoral lymphocytes are encountered in both the POLE mutant and MMRd groups.17–20 Intratumoral heterogeneity may be seen in those groups as well as in the p53abn group, which contains an appreciable number of “mixed endometrioid and serous carcinomas”.2 Additional reasons for pursuing molecular classification of FIGO grade 3 EECs include its potential usefulness in 1) Lynch Syndrome screening, 2) therapeutic prediction, and 3) differential diagnosis.
Classification as MMRd endometrioid carcinoma should trigger application of an algorithm that stratifies patients into two groups, each with a different likelihood of harboring germline abnormalities associated with Lynch Syndrome. The lowest-risk patients with MMRd carcinomas have tumors demonstrating abnormal MLH1 expression in the presence of MLH1 promoter methylation, or those with tumors harboring somatic MMR gene mutations, in the absence of germline mutations, in the same genes. Nearly all of the highest-risk patients have germline mutations in one of the DNA MMR genes or EPCAM; rarely, patients have “constitutive epimutation”, in which there is widespread methylation of the genome, including the MMR genes. The prognostic significance of a germline MMR mutation is currently unclear.
Ongoing interest in and success with immune checkpoint inhibitor therapy has encouraged the study of molecular phenotypes that are predictive of response to these agents. Although PD-1 blockade, for example, is not currently a first-line therapy for endometrial carcinoma patients, it has recently been FDA-approved for MMRd endometrial carcinomas in the setting of recurrence. The approval was based, in part, on the recognition that microsatellite unstable carcinomas, the vast majority of which are MMRd, are sensitive to immune checkpoint inhibition.21 MMRd FIGO grade 3 EECs are more likely than other genomic subgroups to present at an advanced stage, in which such therapy may be most applicable. This is based on the work of McMeekin,22 who reported heightened chemosensitivity and favorable prognosis for MMRd endometrial carcinomas treated with platinum-based agents. There are theoretical reasons to account for these responses in both MMRd and POLE mutant EECs. Deficient DNA repair results in at least a 100-fold increase in somatic mutations in tumor cell lines,23 resulting in the creation of “neo-antigens,” some of which may drive apoptosis and increase sensitivity to chemo- and radiotherapy. An immune-mediated role is also suggested by the presence of rich tumor-infiltrating and peritumoral lymphocytes in these tumors.19 More recently, researchers have investigated the role of tumor-infiltrating lymphocytes in POLE-ultramutated tumors.24–26 Howitt et al showed that POLE-ultramutated endometrial carcinomas are associated with a high level of neoantigens and elevated CD8+ tumor-infiltrating lymphocytes, counterbalanced by overexpression of PDL-1 (an immune checkpoint); this suggests that such tumors might be excellent candidates for anti-PDL1 target therapies.24
Given that ~18% of grade 3 EECS and all serous carcinomas fall into the p53abn subgroup, we must ask whether it is necessary to rigorously distinguish these two subtypes of endometrial cancer. A definitive answer is not obvious, although review of the demographic data reported herein reveals some potentially important differences, most notably the prevalence of Stage II–IV disease in serous carcinoma. In our cohort, only 20% of p53abn grade 3 EECs presented at a high stage. This stands in contrast to the 58% prevalence of surgically staged, high-stage endometrial serous carcinomas reported in the Memorial Sloan Kettering Cancer Center institutional database. It is possible that histotype and molecular profile provide different and potentially complementary information (which would support the practice of reporting both).
Although the NSMP (copy number-low) category of grade 3 EEC appeared homogeneous at first, review of the TCGA data disclosed heterogeneity, with probable clinical relevance. For example, amplification of chromosome 1q2,27 and mutations in exon3 of CTNNB1 (ß-catenin)5,28,29 are prognostically unfavorable. Unfortunately, ß-catenin IHC is not a useful surrogate method for detecting prognostically unfavorable CTNNB1 exon 3 mutations. Going forward, the identification of these and other abnormalities may prove important, facilitating the stratification of tumors within this largest molecular subclass.
The proposed molecular integrated risk stratification will require sequencing of the exonuclease domain of POLE. Although there are commercially available POLE IHC antibodies, unpublished observations from several centers indicate that these cannot be applied to clinical practice at this time. It is possible that POLE antibodies with good performance characteristics, or surrogates thereof, could be developed. There may still be a role for assessing histological features that are likely to be encountered in POLE mutant tumors, such as obvious peritumoral and tumor-infiltrating lymphocytes.30 When endometrial cancers with pronounced TILS are found to be DNA MMR-proficient, the likelihood of POLE mutant subgroup assignment increases. There may also be circumstances in which more costly sequencing could be avoided; for example, in advanced stage tumors, in which the likelihood of POLE mutations is extremely small. Additional study is needed to determine how POLE status impacts triage for surgical staging and treatment. If POLE mutation is identified in a biopsy, can lymph node dissection be avoided? Is adjuvant chemo- or radiotherapy beneficial in the setting of a grade 3 deeply invasive POLE-mutant tumor? Would the excellent prognosis, observed in this and prior studies, be possible without additional treatment? Only with this information can the role and timing of POLE mutational analysis be defined. The increasing use of next-generation sequencing platforms that recognize hundreds of somatic mutations simultaneously, including the exonuclease domains of POLE, is likely to supplant Sanger sequencing or IHC in detecting such mutations. It is encouraging that next-generation sequencing platforms including the DNA mismatch repair genes, and assessment of microsatellite instability, are now FDA-approved, and should [theoretically] be reimbursable by insurance companies.
The current study included patients with grade 3 EECs of all stages, irrespective of adjuvant treatment given. This resulted in a large but clinically heterogeneous retrospective study cohort. Therefore, we cannot make any definitive conclusions with respect to the putative effect of adjuvant treatment on the TCGA subclasses. Molecular analysis of a study cohort from a randomized controlled trial, such as PORTEC-3, will be of interest in exploring the effects of adding chemotherapy to the treatment regimen. Another potential limitation of our study is the fact that there were some minor differences in methodology between the participating centers, and not all cases were centrally reviewed. POLE testing was performed by NGS covering exon 9–14 by some centers; others limited their analysis to exon 9 and 13 by Sanger sequencing. It remains to be seen which methods will emerge as the standard in clinical practice. As molecular subclassification enters clinical practice, it will be necessary to develop appropriate proficiency testing programs to ensure that the quality of biomarker testing meets the expected standards (e.g. the ASCO-CAP guidelines for breast cancer biomarker assessment). However, the technical differences between participating laboratories are unlikely to have had any major impact on our results and conclusions. Unlike the TCGA study, the current study utilized techniques that are widely available and commonly used in clinical practice. Our study population included a well-annotated cohort of 381 patients with FIGO grade 3 EECs, with a median follow-up of almost 7 years.
In summary, we have shown that the pathologic entity typically recognized as “FIGO grade 3 endometrioid carcinoma” is, in reality, a collection of at least four distinct disease types. In this era of increasingly individualized patient care, the elucidation and recognition of these subgroups will contribute significantly to prognostication and the selection of novel therapeutics.
Supplementary Material
Acknowledgments
Funding: This study was funded in part by the Dutch Cancer Society (KWF-UL2012-5719) (Dr. Bosse, Dr. Nout).
This study was funded in part through the NIH/NCI Support Grant P30 CA008748 (Dr. Abu-Rustum, Dr. Levine, Dr. Soslow).
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to disclose.
References
- 1.NIH National Cancer Institute. Surveillance Epidemiology, and End Results Program. (Cancer Statistics).Cancer Stat Facts: Endometrial Cancer. 2017 Available at: http://seer.cancer.gov/statfacts/html/corp.html. Accessed 10/12/2017.
- 2.Kandoth C, Schultz N, Cherniack AD, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Proctor L, Pradhan M, Leung S, et al. Assessment of DNA Ploidy in the ProMisE molecular subgroups of endometrial cancer. Gynecol Oncol. 2017;146:596–602. doi: 10.1016/j.ygyno.2017.06.020. [DOI] [PubMed] [Google Scholar]
- 4.Stelloo E, Bosse T, Nout RA, et al. Refining prognosis and identifying targetable pathways for high-risk endometrial cancer; a TransPORTEC initiative. Mod Pathol. 2015;28:836–844. doi: 10.1038/modpathol.2015.43. [DOI] [PubMed] [Google Scholar]
- 5.Stelloo E, Nout RA, Osse EM, et al. Improved Risk Assessment by Integrating Molecular and Clinicopathological Factors in Early-stage Endometrial Cancer-Combined Analysis of the PORTEC Cohorts. Clin Cancer Res. 2016;22:4215–4224. doi: 10.1158/1078-0432.CCR-15-2878. [DOI] [PubMed] [Google Scholar]
- 6.Talhouk A, Hoang LN, McConechy MK, et al. Molecular classification of endometrial carcinoma on diagnostic specimens is highly concordant with final hysterectomy: Earlier prognostic information to guide treatment. Gynecol Oncol. 2016;143:46–53. doi: 10.1016/j.ygyno.2016.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Talhouk A, McConechy MK, Leung S, et al. A clinically applicable molecular-based classification for endometrial cancers. Br J Cancer. 2015;113:299–310. doi: 10.1038/bjc.2015.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Talhouk A, McConechy MK, Leung S, et al. Confirmation of ProMisE: A simple, genomics-based clinical classifier for endometrial cancer. Cancer. 2017;123:802–813. doi: 10.1002/cncr.30496. [DOI] [PubMed] [Google Scholar]
- 9.Micheel CM, Nass SJ, Omenn GS, editors. Committee on the Review of Omics-Based Tests for Predicting Patient Outcomes in Clinical Trials; Board on Health Care Services; Board on Health Sciences Policy; Institute of Medicine. Evolution of Translational Omics: Lessons Learned and the Path Forward. Washington (DC): National Academies Press (US). National Academy of Sciences; 2012. 2012. [PubMed] [Google Scholar]
- 10.McConechy MK, Talhouk A, Li-Chang HH, et al. Detection of DNA mismatch repair (MMR) deficiencies by immunohistochemistry can effectively diagnose the microsatellite instability (MSI) phenotype in endometrial carcinomas. Gynecol Oncol. 2015;137:306–310. doi: 10.1016/j.ygyno.2015.01.541. [DOI] [PubMed] [Google Scholar]
- 11.Stelloo E, Jansen AML, Osse EM, et al. Practical guidance for mismatch repair-deficiency testing in endometrial cancer. Ann Oncol. 2017;28:96–102. doi: 10.1093/annonc/mdw542. [DOI] [PubMed] [Google Scholar]
- 12.Bokhman JV. Two pathogenetic types of endometrial carcinoma. Gynecol Oncol. 1983;15:10–17. doi: 10.1016/0090-8258(83)90111-7. [DOI] [PubMed] [Google Scholar]
- 13.Gilks CB, Oliva E, Soslow RA. Poor interobserver reproducibility in the diagnosis of high-grade endometrial carcinoma. Am J Surg Pathol. 2013;37:874–881. doi: 10.1097/PAS.0b013e31827f576a. [DOI] [PubMed] [Google Scholar]
- 14.Hoang LN, Kinloch MA, Leo JM, et al. Interobserver Agreement in Endometrial Carcinoma Histotype Diagnosis Varies Depending on The Cancer Genome Atlas (TCGA)-based Molecular Subgroup. Am J Surg Pathol. 2017;41:245–252. doi: 10.1097/PAS.0000000000000764. [DOI] [PubMed] [Google Scholar]
- 15.Hoang LN, McConechy MK, Kobel M, et al. Histotype-genotype correlation in 36 high-grade endometrial carcinomas. Am J Surg Pathol. 2013;37:1421–1432. doi: 10.1097/PAS.0b013e31828c63ed. [DOI] [PubMed] [Google Scholar]
- 16.Hussein YR, Broaddus R, Weigelt B, et al. The Genomic Heterogeneity of FIGO Grade 3 Endometrioid Carcinoma Impacts Diagnostic Accuracy and Reproducibility. Int J Gynecol Pathol. 2016;35:16–24. doi: 10.1097/PGP.0000000000000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bakhsh S, Kinloch M, Hoang LN, et al. Histopathological features of endometrial carcinomas associated with POLE mutations: implications for decisions about adjuvant therapy. Histopathology. 2016;68:916–924. doi: 10.1111/his.12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garg K, Leitao MM, Jr, Kauff ND, et al. Selection of endometrial carcinomas for DNA mismatch repair protein immunohistochemistry using patient age and tumor morphology enhances detection of mismatch repair abnormalities. Am J Surg Pathol. 2009;33:925–933. doi: 10.1097/PAS.0b013e318197a046. [DOI] [PubMed] [Google Scholar]
- 19.Hussein YR, Weigelt B, Levine DA, et al. Clinicopathological analysis of endometrial carcinomas harboring somatic POLE exonuclease domain mutations. Mod Pathol. 2015;28:505–514. doi: 10.1038/modpathol.2014.143. [DOI] [PubMed] [Google Scholar]
- 20.Shia J, Black D, Hummer AJ, et al. Routinely assessed morphological features correlate with microsatellite instability status in endometrial cancer. Hum Pathol. 2008;39:116–125. doi: 10.1016/j.humpath.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 21.Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McMeekin DS, Tritchler DL, Cohn DE, et al. Clinicopathologic Significance of Mismatch Repair Defects in Endometrial Cancer: An NRG Oncology/Gynecologic Oncology Group Study. J Clin Oncol. 2016;34:3062–3068. doi: 10.1200/JCO.2016.67.8722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parsons R, Li GM, Longley MJ, et al. Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell. 1993;75:1227–1236. doi: 10.1016/0092-8674(93)90331-j. [DOI] [PubMed] [Google Scholar]
- 24.Howitt BE, Shukla SA, Sholl LM, et al. Association of Polymerase e-Mutated and Microsatellite-Instable Endometrial Cancers With Neoantigen Load, Number of Tumor-Infiltrating Lymphocytes, and Expression of PD-1 and PD-L1. JAMA Oncol. 2015;1:1319–1323. doi: 10.1001/jamaoncol.2015.2151. [DOI] [PubMed] [Google Scholar]
- 25.Piulats JM, Guerra E, Gil-Martin M, et al. Molecular approaches for classifying endometrial carcinoma. Gynecol Oncol. 2017;145:200–207. doi: 10.1016/j.ygyno.2016.12.015. [DOI] [PubMed] [Google Scholar]
- 26.van Gool IC, Eggink FA, Freeman-Mills L, et al. POLE Proofreading Mutations Elicit an Antitumor Immune Response in Endometrial Cancer. Clin Cancer Res. 2015;21:3347–3355. doi: 10.1158/1078-0432.CCR-15-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Depreeuw J, Stelloo E, Osse EM, et al. Amplification of 1q32.1 refines the molecular classification of endometrial carcinoma. Clin Cancer Res. 2017 Sep 22; doi: 10.1158/1078-0432.CCR-17-0566. pii: clincanres.0566.2017. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 28.Kurnit KC, Kim GN, Fellman BM, et al. CTNNB1 (beta-catenin) mutation identifies low grade, early stage endometrial cancer patients at increased risk of recurrence. Mod Pathol. 2017;30:1032–1041. doi: 10.1038/modpathol.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, Patel L, Mills GB, et al. Clinical significance of CTNNB1 mutation and Wnt pathway activation in endometrioid endometrial carcinoma. J Natl Cancer Inst. 2014 Sep 10;106106(9) doi: 10.1093/jnci/dju245. pii: dju245. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4200060. Accessed 10/12/2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Gool IC, Ubachs JEH, Stelloo E, et al. Blinded histopathological characterisation of POLE exonuclease domain-mutant endometrial cancers: sheep in wolf’s clothing. Histopathology. 2017 Aug 10; doi: 10.1111/his.13338. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


