Abstract
Development of a highly effective vaccine or antibodies for prevention and ultimately elimination of malaria is urgently needed. Here, we report the isolation of a number of human monoclonal antibodies (mAbs) directed against the Plasmodium falciparum (Pf) circumsporozoite protein (CSP) from several subjects immunized with an attenuated whole sporozoite (SPZ) vaccine (Sanaria® PfSPZ Vaccine). Passive transfer of one of these antibodies, mAb CIS43, conferred high-level, sterile protection in two different mouse models of malaria infection. Stoichiometry and affinity of mAb CIS43 for PfCSP indicate two sequential multivalent binding events to six sites: the first 7-fold higher affinity binding event is to a unique “junctional” epitope positioned between the N-terminus and the central repeat domain of PfCSP. Moreover, mAb CIS43 prevented proteolytic cleavage of PfCSP on PfSPZ. Crystal structures of the CIS43 fragment antigen binding (Fab) in complex with the junctional epitope determined the molecular interactions of binding, revealed the epitope’s conformational flexibility, and defined NPN as the structural repeat motif. The demonstration that mAb CIS43 is highly effective for passive prevention of malaria has potential application for use in travelers, military personnel and elimination campaigns and identifies a new and conserved site of vulnerability on PfCSP for next generation rational vaccine design.
Keywords: Malaria, circumsporozoite protein, neutralizing antibody, cleavage, crystal structure, vaccine
Malaria is a mosquito-borne parasitic disease causing high morbidity and mortality, primarily in infants and young children in sub-Saharan Africa. Infection with Plasmodium falciparum (Pf) is responsible for the majority of deaths due to malaria. The Pf circumsporozoite protein (PfCSP) covers the surface of the infecting sporozoites and has a critical role in sporozoite development in the mosquito, and invasion of hepatocytes necessary for initiation of malaria infection1–4. PfCSP is comprised of an N terminal domain, which contains a highly conserved pentapeptide sequence, termed region I (RI), followed by an immunodominant central repeat region, consisting of ~40 NANP and up to 4 NVDP motifs, and a C-terminal domain5,6 (Fig. 1a). PfCSP is a major target for neutralizing antibodies and provided the rationale for using it as a vaccine 7. High titer antibodies against the NANP repeats have been associated with some clinical protection following vaccination with RTS,S/ASO1, a truncated formulation of PfCSP containing only NANP repeats and the C-terminal region8–10. Moreover, human monoclonal antibodies (mAbs) against the NANP repeats isolated from subjects vaccinated with RTS,S/ASO1 protect mice following malaria challenge11,12. Of note, antibody levels and protection in humans wane over several years following RTS,S/ASO1 immunization13,14. These data highlight the need to induce more durable and higher potency antibodies against NANP repeats or new regions of PfCSP. Heretofore, there are limited data characterizing human mAbs to sites other than the repetitive NANP motifs that mediate sterile protection in mice11,12,15. Thus, isolation of antibodies from humans exposed to whole sporozoites by vaccination or infection provides a powerful approach to identify such mAbs since full length PfCSP is presented in its native conformation. Here we isolated several human mAbs specific for PfCSP from subjects immunized with an attenuated whole sporozoite vaccine (Sanaria® PfSPZ Vaccine). We then determined their capacity to mediate protection in vivo, and used structural and biophysical analyses to define their specificity and provide insights into the mechanisms of protection.
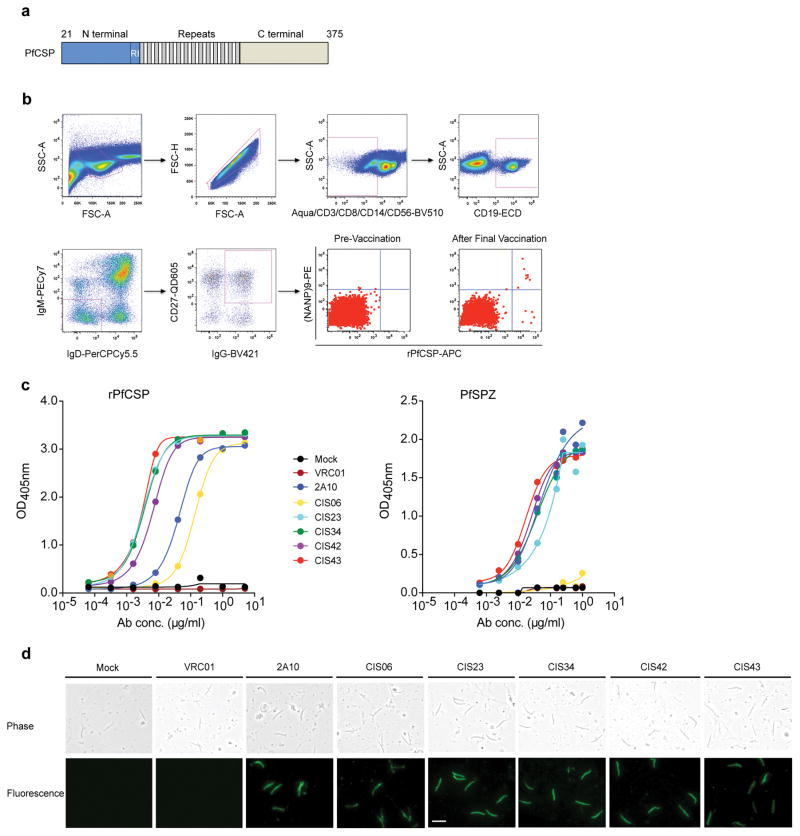
Figure 1. Isolation and binding specificity of mAbs from rPfCSP specific-memory B cells.
a, Schematic representation of rPfCSP (residues 21–375). Signal (1–21) and anchor (375–397) residues are excluded. The N-, C-terminal, and repeat domains are shown. The conserved region I (RI) is indicated. b, Gating strategy for sorting rPfCSP and (NANP)9 memory B cells. rPfCSP-specific, CD19+ IgG+ CD27+ memory B cells from pre-vaccination or after the 5th (last) vaccination. c, Binding of varying concentrations of mAbs to rPfCSP by ELISA. OD405nm, optical density at 405 nm. d, Binding of mAbs to PfSPZ by ELISA. e, Binding of mAbs to PfSPZ by immunofluorescence assay (IFA). Phase contrast and fluorescence channels are shown. Scale bar, 10 μm. In c–e, Negative controls: Mock, transfection filtrate; VRC01, a human anti-HIV-1 IgG1 isotype control mAb. Positive control: 2A10, mouse anti-PfCSP repeat mAb. Data are representative of two independent experiments (c–e).
RESULTS
Isolation of PfCSP mAbs from memory B cells and plasmablasts
To isolate PfCSP mAbs, two different experimental approaches were used. The first approach used recombinant PfCSP (rPfCSP) and (NANP)9 repeat peptide probes to directly identify and sort PfCSP-reactive IgG+ memory B cells from peripheral blood two weeks after the final vaccination (Fig. 1b) of a malaria naïve subject who received PfSPZ Vaccine. The subject had high serum PfCSP antibody titers with inhibition of sporozoite invasion of hepatocytes in vitro and was protected after controlled human malaria infection. Four of the five mAbs (CIS23, CIS34, CIS42, CIS43) showed dose- dependent binding to rPfCSP and PfSPZ by ELISA, with EC50s ranging from 0.003–0.134 μg/ml and 0.017–0.08 μg/ml, respectively (Fig. 1c–d). These antibodies also bound PfSPZ by immunofluorescent assay (IFA) (Fig. 1e). Additional PfCSP mAbs were generated by a second approach using plasmablasts derived from two subjects immunized with the PfSPZ Vaccine. This method provides a direct and unbiased approach for isolating mAbs against PfCSP and other PfSPZ proteins. Plasmablasts were sorted 7 days after the third immunization without any antigen-specific probe, and Ig heavy and light chain sequences recovered from mRNA were expressed as recombinant antibodies. Of the 141 mAbs expressed, 68 were rPfCSP-specific as determined by ELISA and twelve Abs were selected for additional analysis (Supplementary Fig. 1a).
Each PfCSP mAb isolated utilized distinct VH and VL gene families with 95 to 100% and 89 to 100% nucleotide identity to their germline for heavy and light chains, respectively. All PfCSP mAbs had canonical third heavy complimentarity-determining region (CDRH3) lengths ranging from 12–17 amino acid residues (Supplementary Table 1)16. For the PfCSP mAbs isolated from plasmablasts, the IGVH3 gene family predominated, particularly VH3~33 consistent with recent reports by others 12,15 (Supplementary Table 1).
PfCSP mAbs inhibit sporozoite infection in vitro
The functional inhibitory capacity of the PfCSP mAbs was first assessed using an in vitro model of liver stage development with primary human hepatocytes17,18. This analysis provides a high throughput initial screening assay to assess hundreds of antibodies to downselect those showing inhibition for in vivo protection studies. Four of the PfCSP mAbs (CIS23, CIS34, CIS42, and CIS43) isolated by the rPfCSP probe sorted memory B cells mAbs and three (mAb04, mAb09, and mAb10) of the PfCSP-specific antibodies isolated from plasmablasts showed dose-dependent inhibition (Supplementary Fig. 1b).
mAb CIS43 confers sterile protection in two mouse models of malaria infection
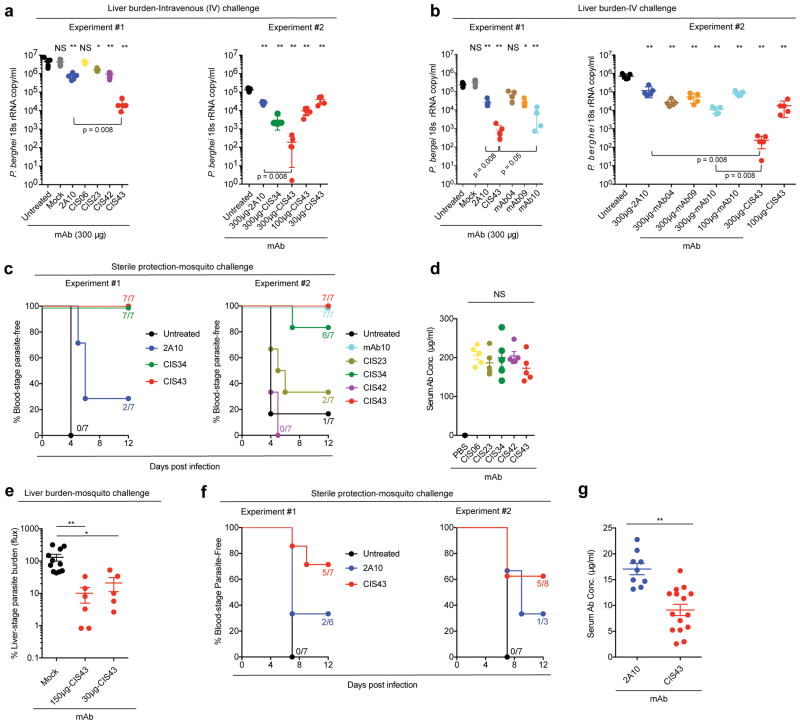
Functional inhibition of the PfCSP mAbs was then assessed in vivo using two different and complementary mouse models of malaria infection. The first model uses a transgenic strain of P. berghei (Pb) in which the endogenous PbCSP has been replaced with full-length PfCSP (Pb-PfCSP)19. As rodents are permissive for Pb infection, this allows assessment of the PfCSP mAbs in the context of a natural infection in a normal mouse. In this model, all antibodies that showed in vitro inhibition were tested. Passive transfer of mAb CIS43 led to the highest reduction (~2–4 logs) in a dose-dependent manner in parasite liver-stage burden (p < 0.008) (Fig. 2a, b), followed by mAb CIS34 which showed ~2 log reduction compared to untreated mice (p < 0.008) following intravenous infection (Fig. 2a). We then assessed the inhibitory capacity of the three mAbs (mAb04, mAb09, and mAb10) isolated from plasmablasts that showed in vitro inhibition (Supplementary Fig. 1b). Of these antibodies, mAb10 showed the most significant reduction (~1–2 log) in parasite liver-stage burden compared to untreated mice (p < 0.008) (Fig. 2b). In all 4 experiments shown, mAb CIS43 showed significantly better protection than mAb2A10 or mAb10 (p < 0.008). Of note, mAbs CIS42, mAb04 and mAb09 which all had potent inhibitory activity in vitro, did not significantly reduce liver burden in vivo. These data are consistent with prior reports showing similar differences20,21 and highlight the importance of the in vivo studies for demonstrating protective efficacy. Since malaria infection is naturally transmitted by mosquito bites and antibodies to PfCSP have inhibitory effects in the skin22, the protective capacity of the PfCSP mAbs was further assessed in mice challenged with mosquitoes. In two independent experiments, all 14 mice that received mAb CIS43, 13 of 14 mice that received mAb CIS34, and 7 of 7 mice that received mAb10 were parasite-free in blood (p = 0.0001). In contrast, only 2 of 7 mice that received mAb2A10 were parasite-free (Fig. 2c). Last, serum antibody levels following passive transfer were approximately 150–200 μg/ml for all PfCSP mAbs (Fig. 2d). These data show that differences in protective efficacy of the mAbs in vivo are likely related to binding specificity and affinity rather than differing amounts of antibody in vivo.
Figure 2. Protection against malaria infection by PfCSP mAbs.
Protective effect of PfCSP mAbs isolated from PfCSP-specific memory B cells (a) and plasmablasts (b) on liver burden. Following passive transfer of the indicated mAbs, C57BL/6 mice (n=5/group) were challenged intravenously (IV) with chimeric PbSPZ expressing PfCSP (Pb-PfCSP). Liver burden is expressed as Pb 18s ribosomal RNA (rRNA) copies/mL and differences between each antibody and the untreated group was determined using the two-tailed Mann–Whitney test. * p = 0.016, and ** p = 0.008, and NS = not significant. Brackets reflect comparisons between mAb CIS43 and mAb 2A10 or mAb10. c, Sterile protection by PfCSP mAbs following Pb-PfCSP SPZ infection by mosquito bite. C57BL/6 mice challenged with 5 infected mosquitoes following passive transfer (300 ug) of the indicated mAbs. Kaplan Meier curves, analyzed by the log rank test, show frequencies of mice free of parasites as determined by Giemsa staining of blood. Differences between mAbs CIS43, CIS34 and mAb10 as compared to untreated mice were significant (p = 0.0001). d, Serum PfCSP mAb levels one hour after passive transfer of 300 μg the indicated mAbs as determined by ELISA in a separate group of C57BL/6 mice (n = 5/group). Differences in mAb levels were compared for significance using one-way ANOVA, with Dunnett’s correction for multiple comparisons. e, Protective effect of mAb CIS43 on parasite liver burden following Pf infection in FRG-huHep mice with 50 mosquitoes infected with Pf expressing GFP-luciferase. Parasite burden determined 6 days later by bioluminescent imaging (flux). Results were normalized to mice receiving a non-specific IgG (Mock). Mock, n = 12 mice; mAb CIS43 150 and 30 μg, n = 7 and 6 mice, respectively. The Kruskal-Wallis test, with Dunn’s correction was used for multiple comparisons (* p = 0.041 and ** p = 0.001). f, Sterile protection by PfCSP mAbs following PfSPZ infection. Following passive transfer (50 μg) of the indicated mAbs, FRG-huHep mice challenged with 5 infected mosquitoes. Kaplan Meier curves, analyzed by the log rank test, show frequencies of mice free of parasites as determined by Pf 18s rRNA on day 7 and 9. In both experiments mAb CIS43 was significantly more protective than untreated mice (p = 0.0002). g, Serum PfCSP mAb levels in FRG-huHep mice used in f were assessed at the time of challenge. Data were compared for significance using the two-tailed Mann–Whitney test (** p = 0.001). Data are representative of two independent experiments (d and e). Bar denotes geometric mean (panel d, e, f) and median (panel a, b, g). Each dot represents one mouse.
To substantiate the in vivo protective capacity of PfCSP mAbs in the second mouse model, human liver-chimeric mice (FRG-huHep) permissive to infection with Pf were used23. Given the limited availability of these mice, studies focused on mAb CIS43 which was consistently the most protective antibody in vivo (Fig. 2a–b). In the first experiment following passive transfer of mAb CIS43, mice were challenged with 50 infected mosquitoes. This high dose challenge model provides a dynamic range to assess reduction in parasite load in the liver as a first demonstrstion of protection. Compared to the mock control mAb-treated mice, there was a dose-dependent reduction (p = 0.001 and p = 0.041, respectively) of Pf in liver burden by 80%–90% by mAb CIS43 (Fig. 2e). We then performed challenge using 5 infected mosquitoes and transferred human red blood cells to assess sterile protection in blood. Of note, this challenge of 5 infected mosquitoes is similar to what is used in controlled human malaria infection following vaccination24,25. In two independent experiments, 10 of 15 mice were parasite-free (p = 0.0002) in blood by passive transfer of only 50 μg of mAb CIS43 (Fig. 2f) with serum levels of ~10 μg/ml at the time of mosquito challenge (Fig. 2g). Collectively, these data show that mAb CIS43 conferred high-level, sterile protection in two mouse models of malaria infection.
mAb CIS43 preferentially binds to a unique “junctional” epitope between the N-terminus and central repeat domains of PfCSP
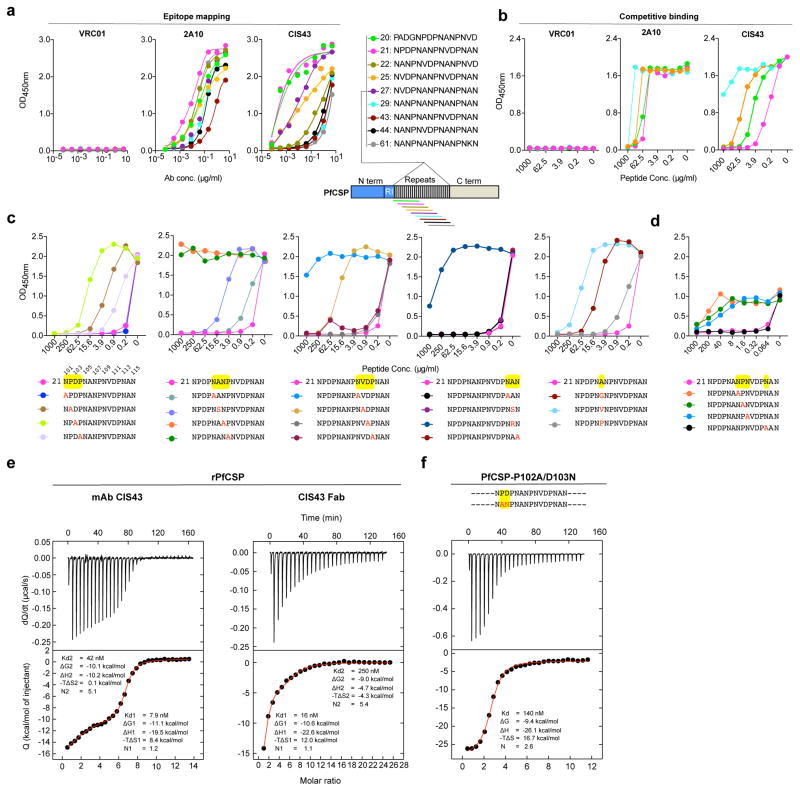
To understand the mechanisms for the differential in vivo potency of the PfCSP mAbs, binding analysis by mapping to PfCSP and structural studies were performed. Binding specificity was first assessed against polypeptides representing the N-terminal, central repeat and C-terminal regions. All PfCSP mAbs tested for in vivo function bound only within the central repeats highlighting the immunodominance of this region by the PfSPZ Vaccine (Supplementary Fig. 1c). More detailed mapping was performed using a series of 15-mer linear peptides overlapping by 11 amino acids encompassing the central repeat region (peptides 20–61, residues 97–276) (Fig. 3a, Supplementary Fig. 1d, e) showed that PfCSP mAbs isolated from plasmablasts bound only NVDP and NANP repeats (Supplementary Fig. 1e). In striking contrast, mAb CIS43 showed the highest binding to peptide 20 (P97ADGNPDPNANPNVD111) and peptide 21 (N101PDPNANPNVDPNAN115) with an EC50 < 0.0001 μg/ml, and reduced binding to the representative NANP-repeat peptide 29 (N132ANPNANPNANPNAN147) with an EC50 >5.0 μg/ml (Fig. 3a, Supplementary Fig. 1d). Furthermore, preincubation of mAb CIS43 with peptides 20 or 21 significantly inhibited binding to rPfCSP in a dose-dependent manner (Fig. 3b) with IC50 values of 4.4 μg/ml or 0.29 μg/ml, respectively. This striking inhibitory capacity of peptide 21 for mAb CIS43 binding to rPfCSP was not observed with the other PfCSP mAbs (Fig. 3b and Supplementary Fig. 1e, g). Peptide 21 is a unique region at the junction of the N-terminus and the central repeat domains that we define here as the “junctional” epitope.
Figure 3. Epitope mapping and ITC analysis of mAb CIS43.
a, Binding of mAb CIS43 to overlapping peptides of PfCSP, with specified sequences numbered and color coded 20 – 61 (representing amino acid residues 97–276) by ELISA. Peptides 28–41 and 46–60, consist only of NANP repeats, and are represented by peptide 29. b, Binding of mAb CIS43 to rPfCSP in the presence of varying concentrations of peptides. Peptide color code as in a. c, Binding of mAb CIS43 to rPfCSP in the presence of peptide 21 sequence variants. Wild type peptide 21 sequence with numeric position listed above and mutated residues highlighted in yellow. d, Binding of mAb CIS43 to PfSPZ in the presence of peptide 21 and its sequence variants. e, ITC of mAb CIS43 and CIS43 Fab binding to rPfCSP. Data are representative of two independent experiments (a–e). f, ITC of mAb CIS43 binding to PfCSP mutant, PfCSP(P102A,D103N), with changes in the junctional epitope sequence depicted in red and highlighted in yellow. Upper panels show the output signal, dQ/dt, as a function of time. Lower panels show the integrated heats as a function of the antibody-site/PfCSP molar ratio in the cell. The solid line represents the result from best non-linear leastsquares fit of the data to a binding model that takes into account two sets of sites with different affinities for rPfCSP. Data shown are representative of three independent experiments. Dissociation constant (Kd), changes in Gibbs energy (ΔG) of binding, enthalpy (ΔH) and entropy (−TΔS) and stoichiometry (N) are shown.
Alanine scanning mutagenesis of peptide 21 was done to define the critical residues for mAb CIS43 binding to rPfCSP. If an alanine occurred in the original sequence, it was substituted with either an arginine, glycine, proline, serine, or valine (Fig. 3c). Alanine substitutions at Asn107, Pro108, or Asn109, abrogated the capacity of peptide 21 to compete for mAb CIS43 binding to rPfCSP as well as to PfSPZ (Fig 3c, d). Mutating Pro102, Asp103, Ala106, Val110 also affected binding. Moreover, peptide 25, which is identical to peptide 21, except for the substitution of a single residue at position 102 (from Pro to Val), limited the inhibitory capacity of peptide 21, suggesting a key role for Pro102 interacting with mAb CIS43 (Fig. 3b, c).
mAb CIS43 shows sequential, multivalent, high affinity binding to PfCSP
In assessing affinity first by biolayer inferometry, mAb CIS43 had the highest apparent affinity (<0.001 nM) to peptide 21 compared to the other PfCSP mAbs (range <0.001–6.06 nM) (Supplementary Fig. 2). These results are consistent with the epitope mapping data (Fig. 3a,b). Thermodynamic parameters and stoichiometry of binding of mAbs CIS43, CIS42, CIS23, CIS34 and mAb10 to rPfCSP were then determined by isothermal titration calorimetry (ITC). Remarkably, the IgG and Fab of mAb CIS43 showed two sequential binding events, the first with a single binding site per antibody with an affinity of 7.9 nM and 16 nM, respectively, and the second with 5 binding sites with 42 nM and 250 nM, respectively (Fig. 3e)26. Similarly, mAb CIS34 also had two binding events with one site bound at 31 nM, and 8 other sites bound at 80 nM affinity (Supplementary Fig. 3). In contrast, mAbs CIS23, CIS42 and mAb10, had only one binding event with 6, 7 or ~9 binding sites at 25 nM, 51 nM or 27 nM respectively (Supplementary Fig. 3), consistent with recent reports using other mouse and human mAbs against the NANP repeat epitopes12,27. To further understand the sequential binding event observed for mAb CIS43, we performed ITC analysis of mAb CIS43 with peptide 21, peptide 29 and a mutant of rPfCSP. mAb CIS43 binding to peptides 21 and 29 had affinities of 7.5 nM and 53 nM, respectively (Supplementary Fig. 3b) which were similar to values obtained with rPfCSP suggesting that the first binding event is to peptide 21. Indeed, ITC analysis with mAb CIS43 and a mutant PfCSP in which two unique residues specific for the “junctional” epitope were mutated (Pro102Ala and Asp103Asn), indicated that only one multivalent binding event occurred with a reduced affinity of 140 nM and stoichiometry of 3 binding sites (Fig. 3f). As a control, the binding affinity and stoichiometry of mAb10, which does not bind peptide 21 was not affected with the mutant PfCSP compared to rPfCSP (Supplementary Fig. 3c). These data strongly suggest that the first high affinity binding event of mAb CIS43 to rPfCSP is to the junctional epitope and that such binding may induce conformational changes of rPfCSP, which can influence the stoichiometry of the remaining binding events of mAb CIS43.
Crystal structures of CIS43 Fab in complex with multiple PfCSP peptides
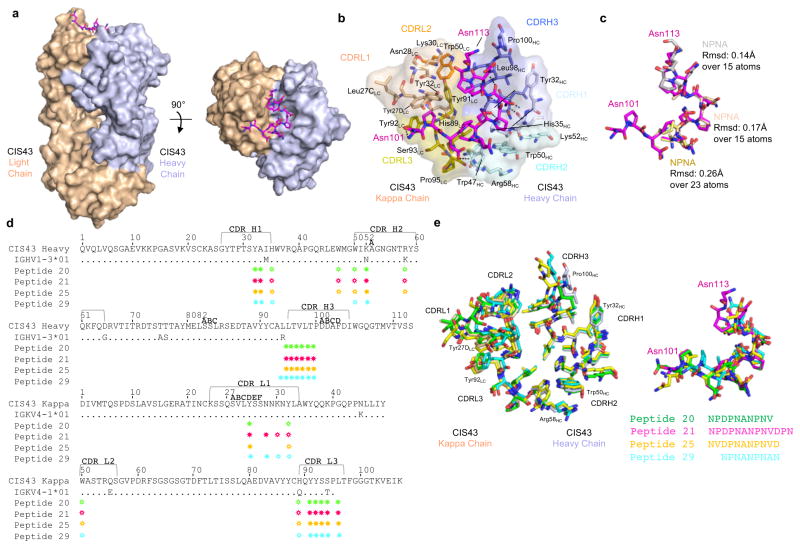
To determine the molecular interactions of epitope binding by mAb CIS43, we co-crystallized CIS43 Fab with peptides 20, 21, 25 and 29 since these peptides differentially competed with binding to rPfCSP (Fig. 3b). Attempts to co-crytallize rPfCSP with mAb CIS43 were unsuccessful likely due to the multivalent stoichiometry of antibody binding to rPfCSP and the flexibility of this protein. Crystals were obtained that diffracted X-rays to 2.4 Å, 1.8 Å, 2.0 Å and 2.2 Å, respectively (Supplementary Table 3). These peptides inserted into a hydrophobic groove formed at the interface of the heavy and light chains, which is positively charged, tyrosine-rich and involved all the CDRs (Fig. 4a, b, d, Supplementary Fig. 4 and Supplementary Table 4). Some conformational changes were observed: residues Trp47HC, Arg58HC and Pro100HC of the heavy chain and residues Tyr27LC, Asn28LC, Trp50LC, Tyr91LC and Tyr92LC of the light chain showed movements when bound to different peptides (Fig. 4e). While peptides 20, 21 and 25 adopted a similar overall conformation (except at the N-terminus of peptide 25 and at all the C-termini) (Fig. 4e), peptide 29 adopted a clearly different conformation. mAb CIS43 has low affinity maturation (Supplementary Table 1). Two of the seven residues that are affinity matured in the V-gene heavy chain interact with peptide 21 (Fig. 4d, Supplementary Table 4c). Similarly, two of the four somatically mutated residues in the V-gene kappa chain interact with the epitope (Fig. 4d, Supplementary Table 4d). The β-1 turn conformation defined previously for the ANPNA peptide28,29 was observed in the peptide-antibody bound structures and three β-1 turns could be aligned consecutively to peptide 21 (Fig. 4c). We did not observe the stem-like structure reported previously30. Peptide 21 showed the most contacts with the CIS43 Fab compared to other peptides (Fig. 4d, Supplementary Table 4). To confirm the role of mAb CIS43 residues that mediate binding to peptide 21, variants of this antibody were generated. One variant (Asn56Gly, Thr57Gly, Arg58Gly, Glu61Gly in the heavy chain) (Supplementary Fig. 5a) abrogated binding to peptide 21 (Supplementary Fig. 5b). Of note, Arg58HC on mAb CIS43 had been shown to be a key contact residue by structural analysis for binding to peptide 21 (Fig. 4). Moreover, computational free energy calculations revealed Arg58Gly to be the most critical for binding of all four mutations (Supplementary Fig. 5c). Importantly, this antibody variant had minimal binding to rPfCSP and no functional inhibition of PfSPZ invasion in vitro (Supplementary Fig. 5b,d).
Figure 4. Crystal structures of CIS43 Fab in complex with PfCSP peptides.
a, Surface representation of CIS43 Fab (light chain in wheat and heavy chain in light blue) with peptide 21 N101PDPNANPNVDPN113 from the repeat region of PfCSP shown in sticks (magenta) and 90° rotation with view down towards the combining sites. b, Details of the interactions between peptide 21 and CIS43 Fab. The peptide 21 is shown in magenta as sticks representation, the CIS43 epitope is shown as sticks and semi-transparent surface and residues are colored based on the CDR regions, with light chain in shades of wheat and heavy chain in shades of light blue. c, Stick representation of peptide 21 shown in magenta in the conformation bound to CIS43 with superposition of three type-I β-turn NPNA repeat structures of CSP as described by Ghasparian et al.28 Each NPNA repeat is labeled for clarity and shown in different colors. Root mean square deviation (Angstroms, Å) is indicated over the total number of atoms used in the alignment. d, Sequence of CIS43 Fab following Kabbat numbering and alignment with germline gene. Residues that contact each peptide (green, magenta, yellow and cyan for peptide 20, 21, 25 and 29, respectively) are shown as closed circle for main chain only, open star for side chains only and closed star for both main and side chains. e, Detailed of the interactions of the peptides with CIS43 Fabs. Residues within 5 Å of the peptides are shown as sticks in wheat for the light chain residues and light blue for the heavy chain residues when bound to peptide 21 and as green, yellow and cyan for peptide 20, peptide 25 and peptide 29, respectively. (Right) Superposition of the peptides shown as sticks and colored as in (d) with sequences observed in electron density.
Crystal structures of CIS42 Fab in complex with multiple PfCSP peptides
To further understand the relative functional potency of mAb CIS43, we co-crystallized the Fab of mAb CIS42, which has limited in vivo function (Fig. 2a, c), with the same peptides (20, 21, 25 and 29) and obtained crystals that diffracted X-rays to 2.4 Å, 1.8 Å, 2.0 Å and 2.2 Å, respectively (Supplementary Fig. 6, Supplementary Table 5). The structures show that, unlike with CIS43 Fab, all four peptides adopted an almost identical conformation when bound to CIS42 Fab, with peptide 29 exhibiting the most contacts with CIS42 Fab (Supplementary Fig. 6). These data are consistent with the peptide competition analysis of mAb CIS42 (Supplementary Fig. 1c). Additionally, the conformation of peptide 21 differed when bound to CIS42 and CIS43 Fabs as well as the angles of approach of the antibodies (Supplementary Fig. 7). Molecular dynamics (MD) simulations were then used to further analyze the physical movements and stability of mAbs CIS43 and CIS42 bound to peptide 21 (Supplementary Fig. 8a–c). After 500ns, peptide 21 remained buried in the hydrophobic groove of the mAb CIS43 CDR while it detached from its initial binding pocket of mAb CIS42, losing most of its electrostatic interactions and confirming the clear preference of mAb CIS43 for this epitope (Supplementary Fig. 8d–e, Supplementary Videos 1–3). Principal component analysis (PCA) of peptide 21 MD simulation showed that mAb CIS43 binds to a rare conformation of peptide 21 (Supplementary Fig. 8f). Lastly, since this is the first time structures of multiple PfCSP motifs are reported, we sought to define the PfCSP repeat structure. The analysis showed Ramachandran angles with antibody-bound repeat peptides had similar phi-psi angles every four residues with a few notable outliers (Supplementary Fig. 9). These data substantiated that NPN is the structural repeat that defines PfCSP sequences that contain NANP12,28,31.
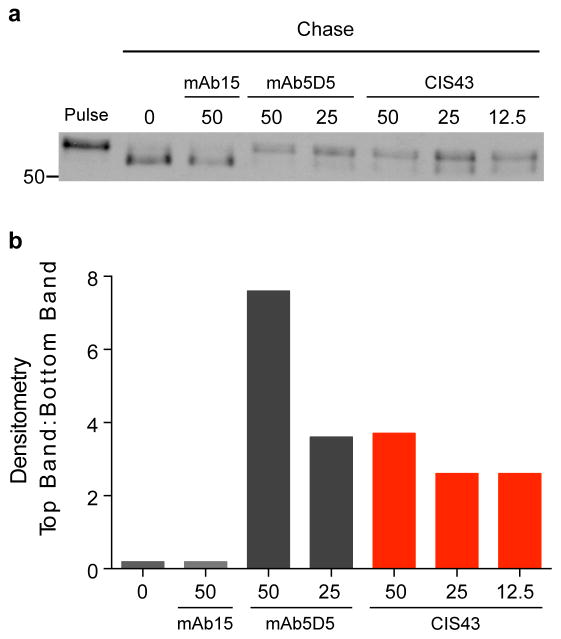
mAb CIS43 inhibits cleavage of PfCSP
A key requirement for sporozoite infection of hepatocytes is the proteolytic cleavage of PfCSP at the putative processing site, RI32,33, which is only 3 amino acids upstream of the junctional epitope. Thus, to gain additional insight into the mechanism for how mAb CIS43 could mediate protection, we assessed whether mAb CIS43 could affect processing or cleavage of PfCSP. Metabolically labeled PfSPZ were chased in the presence of mAb CIS43. As a positive control mAb5D5, a mouse mAb specific to an epitope upstream of R1 which has been shown to block cleavage, was used33. Additionally, mAb15 which is specific for the C-terminus (Supplementary Fig. 1) was used as a negative control. Indeed, mAb CIS43, does limit cleavage of PfCSP in a dose-responsive manner, albeit to a lesser extent than 5D5 (Fig. 5).
Figure 5. Effect of mAb CIS43 on cleavage of PfCSP.
a, Metabolically-labeled Pf sporozoites were kept on ice (Pulse) or chased for 90 min in the absence, or presence of the indicated mAb concentrations (μg/ml). PfCSP was immunoprecipitated from sporozoite lysates and analyzed by SDS-PAGE and autoradiography. Negative control: mAb15 (anti C-terminus PfCSP mAb, Supplementary Fig. 1c). Positive control: mAb5D5 (mouse anti-N terminus PfCSP mAb). Molecular mass is indicated in kilodaltons on the left side of the autoradiograph. b, Densitometry analysis of scanned autoradiograph shown in a. The density ratio of top to bottom PfCSP bands is shown for each chased sample with mAb concentrations shown below in (μg/ml). A ratio of 1 indicates the density of the top and bottom bands is equal. Data are representative of 3 independent experiments (a–b).
DISCUSSION
Here, we show that the human mAb CIS43 binding to a structurally defined junctional epitope on PfCSP confers high-level, sterile protection in vivo. Biophysical and structural analyses showing sequential and multivalent, high affinity binding of mAb CIS43 to rPfCSP demonstrate a unique mechanism for neutralization. Moreover, binding of mAb CIS43 at a specific angle and rare conformation of the junctional epitope (Supplementary Fig. 7 and Supplementary Fig. 8) highlights how the immunodominant NANP sequence, now structurally defined as NPN, may potentially shield the “NPDP” junctional site from recognition and thus may divert the immune response away from this or other sites of vulnerability (Fig. 3c–f). Indeed, this unique NPDP sequence is critical for the high affinity binding of mAb CIS43 to the junctional epitope and may play a role in the overall conformation of rPfCSP for the sequential, multivalent binding of mAb CIS43 (Fig. 3e–f). We speculate mAb CIS43 has multiple mechanisms for mediating protection in vivo. Multivalent binding of mAb CIS43 to PfCSP could inhibit sporozoite motility in the skin22. In addition, by interfering with cleavage of PfCSP, this mAb could limit invasion of hepatocytes by sporozoites32.
There are two potential clinical applications of the findings reported here. One is related to using mAbs as passive prevention and the other is using the junctional epitope as part of a next generation vaccine. As the junctional epitope sequence of mAb CIS43 occurs only once in PfCSP and is highly conserved (>99.8%) amongst thousands of Pf strains recovered globally (Supplementary Fig. 10)34,35, it provides a conserved site for antibody neutralization by passive transfer and vaccine design to induce such antibodies. The most critical features for conferring protection by passive transfer of mAbs or vaccine induced immunity is potency and durability. The findings that mAb CIS43 leads to a 10–100 fold reduction in liver burden compared to mAb10, an antibody against the NANP repeat region only (Fig 2b) provides evidence for greater potency. Additional studies comparing mAb CIS43 and other newly identified human mAbs against PfCSP to NANP12,15 will need to be performed to broaden these findings. The ability of mAb CIS43 to protect against Pf infection in the majority of mice at a concentration of ~10 μg/ml in vivo is notable since this is an achievable concentration for up to 6 months by subcutaneous immunization with a human mAb specific to an HIV envelope protein36. These data suggest that mAb CIS43 could be used alone or in combination with other antibodies for passive prevention in humans in which high–level protection would be required for up to 6 months. Such indications would be used by travelers, military personnel and elimination campaigns in combination with mass drug administration.
Finally, this junctional epitope provides a new and conserved site of vulnerability, allowing the design of next-generation subunit vaccines that may increase the breadth and potency of humoral immunity to improve protective efficacy against malaria.
Methods
Study subjects and clinical specimens
Sera from protected volunteers in the previously described VRC 312 clinical trial (https://clinicaltrials.gov; NCT01441167) were screened for Plasmodium falciparum (Pf) circumsporozoite protein (CSP) antibodies and functional capacity for inhibition of sporozoite invasion of hepatocytes in vitro 37–40. Briefly, this was a phase 1, open-label, dose-escalation study investigating safety, immunogenicity, and protective efficacy of the radiation-attenuated aseptic, purified, cryopreserved Pf (NF54 strain) sporozoites (PfSPZ) vaccine, Sanaria® PfSPZ Vaccine. Memory B cell PfCSP monoclonal antibodies (mAbs) were isolated from peripheral blood mononuclear cells (PBMCs) taken two weeks after the last dose in a protected donor (VRC312-402) who received 5 doses of 1.35×105 PfSPZ. Plasmablast PfCSP mAbs were isolated from 2 volunteers in the VRC 314 clinical trial (https://clinicaltrials.gov; NCT02015091). These subjects received a total of 4 doses of PfSPZ vaccine intravenously at 0, 4, 12 and 20 weeks (first 2 doses at 2.7 × 105 and 2 doses at 4.5 × 105). Plasmablasts were isolated from PBMCs, 7 days after the 3rd immunization.
Ethics Statement for animal studies
All animal research rigorously complied with the Institutional Animal Care and Use Committee of Johns Hopkins University (Approved protocol permit #: MO16H35) and in accordance with and approved by the Center for Infectious Disease Research Institutional Animal Care and Use Committee (IACUC) under protocol SK-16. The Seattle Biomed IACUC adheres to the NIH Office of Laboratory Animal Welfare standards (OLAW welfare assurance # A3640-01).
PfCSP probes generation
The amino acid sequence of PfCSP of 3D7 (Plasmodb ID PF3D7_0304600.1), a clone of the NF54 strain, was used to generate a codon optimized synthetic gene for expression in mammalian cells (GenScript). The DNA construct corresponding to the full length PfCSP, replacing the leader residues 1–20 with a mammalian secretion signal sequence derived from the modified bovine prolactin (MDSKGSSQKGSRLLLLLVVSNLLLPQGVLA) and excluding the GPI anchor residues 376–397, was cloned into a CMV/R-expression vector along with the C-terminal Avi-tag, HRV3C –processing tag, and the His-purification tag 41–43. The resulting construct encodes for the native N-terminal domain, the central domain consisting of (NANP)38 tandem repeats with 4 interspersed NVDP, as well as the C-terminal domain. PfCSP mutants were generated by site-directed mutagenesis (GenScript). Recombinant wild type PfCSP (rPfCSP) or PfCSP mutants were expressed by transient transfection in Expi293 cells (Life Technologies), according to the manufacturer’s instructions, and purified from culture supernatants by polyhistidine-tag affinity chromatography (GE Healthcare), followed by gel-filtration. Fractions containing monomers were pooled, concentrated and frozen at −80°C. For tetramer probe generation, peptides/proteins were first biotinylated and then conjugated to fluorophore. rPfCSP was biotinylated using ligase Bir A (Avidity) at 30°C for 4h and buffer exchanged with PBS (pH 7.4) over a 30-kDa Centricon plus filter (Millipore) to remove excess free biotin. The repeat peptide (NANP)9 was synthetically made and N-terminally biotinylated through the lysine analog Ahx linker (GenScript). Biotinylated rPfCSP and (NANP)9 peptide were fluorescently labeled by sequential addition of streptavidin conjugated to allophycocyanin (SA-APC) or phycoerythrin (SA-PE) (Life Technologies) in a 4:1 molar ratio 41.
Isolation of PfCSP-specific memory B cells
PBMCs (106) were stained for viability using the amine-reactive dye Aqua LIVE/DEAD (Invitrogen) followed by staining for surface markers. In addition to the tetramer probes rPfCSP-APC, (NANP)9-PE generated above, the staining panels included: anti-CD3-BV510 (BioLegend), CD8-BV510 (BioLegend), CD14-BV510 (BioLegend), CD56-BV510 (BioLegend), CD19-ECD (Beckman), CD27-QD605 (Invitrogen), CD21-Cy5PE (Becton Dickenson), CD38-Alexa Fluor 680 (Becton Dickenson), IgD-Cy7PE (Becton Dickenson), IgM-Cy5.5-PerCP (Becton Dickenson), and IgG- Cy7PE (Becton Dickenson). Cells were acquired and sorted using a BD FACS Aria II instrument (BD Immunocytometry Systems), and fluorescence-activated cell sorting (FACS) data was analyzed using FlowJo software (Tree Star). Gating strategy is shown in Fig. 1a. PfCSP-reactive (rPfCSP+ and/or (NANP)9+) CD19+CD27+IgG+ IgD−IgM− memory B cells were single cell, dry-sorted into 96-well PCR plates, rapidly frozen on dry ice and stored at −80°C until processing41,42.
Production of recombinant immunoglobulins (IgG)
Following single rPfCSP-specific memory B cell sort, lysis buffer was added to the plate, and the variable region of the heavy and light chains of the IgG genes were amplified by RT–PCR and re-expressed as described previously44. Briefly, amplification of rearranged IgG VH and Vκ or Vλ genes was performed using a cocktail of primers followed by sequencing and cloning into expression vectors containing the relevant constant region. Matched heavy and light chain constructs derived from each sorted cell were coexpressed using Expi293 cells, and 28 full-length IgG were purified using a recombinant protein-A column (GE Healthcare). Sequence analysis was performed using IMGT (http://www.igmt.org)45.
Isolation of plasmablasts
PBMCs were freshly isolated from blood samples collected 7 days after PfSPZ immunization and stained for viability with Aqua Live/Dead dye(Invitrogen) followed by surface-staining for the following markers: CD20-Cy7APC (BD Bioscience), CD19-FITC (BD Bioscience), CD3-Cy7PE (BD Bioscience), CD38-PE (BD Bioscience), and CD27-APC (ThermoFisher). Plasmablasts were gated as live, CD3-CD20-CD19+CD27+CD38+ and were sorted as single cells into 96-well PCR plates containing 20 μl/well of RT reaction buffer that included 5 μl of 5× First strand cDNA buffer, 0.5 μl of RNAseOut (Invitrogen), 1.25 μl of DTT, 0.0625 μl of Igepal and 13.25 μl of dH2O (Invitrogen) as previously described46.
Antibody production from plasmablasts
Immunoglobulin genes were amplified by RT and nested PCR without cloning from RNA of single sorted cells as previously described46,47. Isolated Ig V(D)J gene pairs were assembled by PCR into linear full-length Ig heavy- and light-chain gene expression cassettes as previously described46,47. Heavy and light chain linear cassettes were co-transfected in 293T cells using Effectene with enhancer (Qiagen)46,47. Transfected cultures were incubated at 37°C 5% CO2 for 3 days. Supernatants were harvested, concentrated and purified using HiTrap Protein A pre-packed high performance plates (GE Healthcare) for 20 min at RT on a shaker. Following PBS/NaCl wash, eluates were neutralized with trizma hydrochloride and buffer exchanged with PBS before determining antibody concentration by Nanodrop.
ELISA
MaxiSorp ELISA plates (Thermo Scientific Nunc) were coated with 100 μL of rPfCSP (1μg/ml) per well for one hour at room temperature (RT) according to the manufacturer instructions (KPL). Coated plates were blocked with 100 μl of 1× blocking solution for one hour at RT, followed by incubation with 100 μl of PfCSP mAbs or controls at varying concentrations (0.00006–5.0 μg/ml). After one hour, plates were incubated with 100 μl/well of 1.0 μg/ml peroxidase-labeled goat anti-human IgG antibody. Plates were washed six times with PBS-Tween between each step. After a final wash, samples were incubated for about 15 min with the ABTS peroxidase substrate. The optical density was read at 450 nm after addition of stopping solution (100 μl/well).
For the PfSPZ ELISA, serially diluted (0.06–1.0 μg/ml) PfCSP mAbs or controls were added to the PfSPZ-coated plates and incubated for one hour and binding was assessed in a similar manner as for the rPfCSP ELISA described above. The mouse mAb2A1048,49, which is specific for the NANP repeat region of PfCSP was used as a positive control and VRC-0141, a human anti-HIV-1 IgG1, as an isotype matched negative control. To determine the inhibitory effect of PfCSP peptides on binding of mAb CIS43 to PfSPZ, MaxiSorp ELISA plates (Thermo Scientific Nunc-Immuno™) pre-coated with 5×103 PfSPZ were blocked with PBS-BSA (2% BSA –SIGMA) for one hour at RT and washed with PBS prior to adding mAb CIS43 (10 ng/ml), preincubated overnight with varying concentrations (0–1000 μg/ml) of the PfCSP peptides. ELISA was performed as described above.
Epitope mapping of PfCSP mAbs
Mapping of the PfCSP mAb epitopes was performed as previously described50 using linear PfCSP 15-mer peptides (GenScript) overlapping by 11 residues spanning the full length of PfCSP. MaxiSorp ELISA plates (Thermo Scientific Nunc) were coated with 100 μl of peptides (10 μg/ml). Following overnight incubation, plates were blocked with PBS + 0.05% Tween-20 and 1% BSA (Sigma-Aldrich) for one hour. Plates were incubated with 100 μL of PfCSP mAbs or controls at varying concentrations (0.00006–5.0 μg/mL). After one hour, plates were incubated with 100 μl/well of peroxidase-labeled goat anti-human or goat anti-mouse (IgG H + L) secondary antibody (Thermo Fisher Scientific) at 1:20,000 dilution in PBST-1% BSA. All the incubation steps were done at RT. Plates were washed six times with PBS-Tween between each step. After a final wash, samples were incubated for about15 min with the TMB Plus Reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. The optical density was read at 450 nm after addition of stopping solution (100 μl/well).
Competitive ELISA was performed as previously reported33 using selected peptides. Briefly, ELISA plates were coated with 100μL of rPfCSP (200 ng/ml) overnight. After blocking plates with PBS-1% BSA for one hour, PfCSP mAbs (10 ng/ml), preincubated overnight with varying concentrations (0–1000 μg/ml) of selected PfCSP peptides in PBS-1% BSA, were added to the rPfCSP-coated plates and ELISA proceeded as described above. For the alanine scanning mutagenesis experiments, competitive ELISA was performed as described above using peptide 21 variants (GenScript).
Indirect fluorescence assay (IFA)
Freshly isolated PfSPZ (3×103) were air-dried on poly-L-lysine-coverslips (Tekdon Inc.) and incubated for half an hour at RT with varying concentrations (0.0002–1.0 μg/ml) of PfCSP mAbs or controls. After washing with PBS-1% BSA, slides were incubated with FITC-labeled goat anti-Human IgG (Fcγ) and green-fluorescent sporozoites were visualized using an upright Nikon Eclipse 90i fluorescence microscope33.
Kinetic binding assay by biolayer interferometry
Antibody binding kinetics were measured using biolayer interferometry on an Octet Red384 instrument (fortéBio) using streptavidin capture biosensors (fortéBio). PfCSP mAb solutions were plated in solid black tilt-well 96-well plates (Geiger Bio-One); assays were performed with agitation at 30°C. Loading of biotinylated rPfCSP or peptides 21 and 29 was performed for 300s, followed by a 60s baseline in buffer (PBS + 1% BSA). Association with whole IgG (serially diluted from 267 to 33 μM) was done for 300s, followed by a dissociation step in buffer for 600s. Background subtraction of non-specific binding was performed by measuring association in buffer alone. Data analysis and curve fitting were performed using Octet software, version 7.0. Experimental data were fitted with the binding equations describing a 1:1 heterologous ligand interaction. Global analyses of the complete data sets, assuming binding was reversible (full dissociation), were carried out using nonlinear least-squares fitting allowing a single set of binding parameters to be obtained simultaneously for all concentrations of a given mAb dilution series.
In vitro functional inhibition assay of PfSPZ invasion of hepatocytes
Micropatterned co-culture (MPCC) preparation and Pf infection were carried out as described previously17,18. Briefly, glass-bottom 96-well plates were coated homogenously with rat tail type I collagen (50 μg/ml) and subjected to soft-lithographic techniques to pattern the collagen into microdomains of 500 μm islands that mediate selective hepatocyte adhesion. To create MPCCs, cryopreserved primary human hepatocytes (BioreclamationIVT) were pelleted by centrifugation at 100 g for 6 min at 4°C, assessed for viability using trypan blue exclusion (typically, 70 to 90% excluded the dye), and seeded on collagen-micropatterned plates. Each well contained approximately 10,000 hepatocytes organized in colonies of 500 μm in serum-free DMEM with 1% PenStrep. Two to 3 h later, cells were washed with serum-free DMEM–1% PenStrep, and the medium was switched to human hepatocyte culture medium. One day after seeding, PfCSP mAbs were added, in triplicate, at four concentrations (10, 1.0, 0.1 and 0.01 μg/ml) to the MPCCs, 30–45 minutes prior to infection with 7.5×104 fresh PfSPZ per well. After 3 hours, cultures were washed with hepatocyte culture medium with 3% PenStrep and 0.1% Fungizone, and 7,000 3T3-J2 fibroblasts were added to create the co-culture. Medium was replaced daily. Samples were fixed on day 3.5 post infection. The impact of the antibodies on hepatocyte infection was assessed by calculating the number of liver-stage parasites or exoerythrocytic forms (EEFs) present at day 3.5 of infection. EEFs were determined by staining for PfHSP70 and visualized with a Nikon Eclipse Ti fluorescence microscope.
In vivo protection in C57BL/6 mice with chimeric Pb-PfCSP SPZ
For intravenous (IV) challenge, 6–8 week old C57BL/6 mice (5/group) were IV injected with controls or varying concentrations of PfCSP mAbs diluted in PBS (pH 7.4) in a total volume of 200 μL per mouse, and immediately challenged with 2000 P. berghei chimeric SPZ expressing PfCSP (Pb-PfCSP SPZ). Forty hours later, livers were harvested and RNA was isolated to quantify the Pb-specific 18s rRNA levels by quantitative real-time PCR (RT-qPCR)51,52.
For mosquito bite challenge, Anopheles stephensi female mosquitoes were allowed to feed on 8 weeks old Swiss webster infected mice with blood stages Pb-PfCSP. Eighteen days after blood feeding on infected mice, the proportion of infected mosquitoes was 9 out of 12 (75%) as determined by salivary gland dissection. Based on this calculation, 6–7 mosquitoes were needed to challenge mice with the equivalent of ~5 infected mosquitoes. Mice (up to 7 per group) were intravenously injected with PfCSP mAbs (300 μg/mAb) as described above. Ten minutes later, mice were anesthetized with 2% Avertin (Alfa Aesar) and mosquitoes were allowed to feed on mice for ~10 minutes, after which mosquito abdomens were visually inspected for positive blood meal. Mouse parasitemia was assessed daily by Giemsa staining of blood smears from day 4 through day 12 post-infection.
In vivo protection in FRG-huHep mice
Passive transfer of controls and PfCSP mAbs, and infectious challenge in human hepatocyte donor-matched (FRG-huHep) mice was done as previously described23. Briefly, FRG-huHep mice, purchased from Yecuris, Inc, were given 30–150 μg of PfCSP mAbs or controls intraperitoneally (IP) 16–24 hours prior to infection. Fifty Anopheles mosquitos infected with Pf expressing GFP-luciferase53 were allowed to feed for ten minutes on FRG-huHep mice. Six days post challenge, mice were imaged following IP-administration of 100 μl of Rediject D-luciferin (Perkin Elmer) using bioluminescence and IVIS imaging to determine the parasite liver burden as previously described23,54. Total flux (pixels/second) was measured after choosing an equivalent region of interest around each mouse liver. Parasite liver burden was normalized using the average of the negative control group that received the isotype species-matched IgG.
For sterile protection, FRG-huHep mice were IP-injected with controls or PfCSP mAb (50 μg/mouse) as described above. The following day, mice were challenged by the bite of 5 Pf-infected mosquitoes/mouse by aliquoting 5 infected mosquitos per mouse into a single cage based on mosquito midgut oocyst infection quantification. At approximately day 6.5, all mice were injected with 400 μl of packed human red blood cells at 70% hematocrit in RPMI. On day 7 and day 9, mice were bled via the retroorbital plexus and 50 μl was mixed into 1 ml of NucliSens EasyMag buffer (BioMerieux) and placed at −80°C until extraction. Total RNA extraction and qPCR for number of Pf 18s rRNA copies was performed as previously published55. Animals were considered positive if any parasite RNA above background was detected in blood.
ELISA quantitation of mAb serum concentration
To assess circulating levels of passively transferred PfCSP mAbs, FRG-huHep mice were bled via the retroorbital plexus immediately prior to infectious mosquito bite challenge. ELISA was performed from serum as previously described using of rPfCSP-coated plates (2 μg/ml). A standard curve for each mAb was generated by applying 8 two-fold dilutions of mAb in dilution/blocking buffer starting at 625 ng/ml. Serum samples were applied at a 1:160 and 1:320 dilution in dilution/blocking buffer. The average of the 1:160 and 1:320 calculated concentration values was used for each individual sample.
Fab expression and purification
Antibodies heavy and light chains were cotransfected into GnTI- cells (ATCC). Six days following transfection, supernatant was harvested and antibodies were purified over protein A resin column, washed with 5 column volumes of PBS, and eluted with IgG Elution buffer (Pierce). Tris (1M) was added immediately to neutralize the pH to ~7.4. IgGs were then digested with Lys C overnight at 37 °C (1 μg for 10 mg of IgG) and the antigen-binding fragments (Fab) was purified further by collecting the flow through of Protein A column and by size exclusion chromatography in 5 mM Hepes, 150 mM NaCl. For Fab-peptide preparations, peptides were dissolved in DMSO and added to the Fabs at a 50% molar excess.
Isothermal titration calorimetry (ITC)
ITC was carried out using VP-ITC microcalorimeters from MicroCal/Malvern Instruments. In all titration experiments, rPfCSP, PfCSP mutants and PfCSP mAbs (whole IgG or Fab) were prepared in PBS, pH 7.4. Each antibody solution, prepared at a concentration of ~100 μM (expressed per site or Fab fragment), was injected in 10 μl aliquots into the calorimetric cell containing rPfCSP at a concentration of ~1 μM. Peptides 21 or 29 and mAb CIS43 were prepared in PBS, pH 7.4, with 2 % DMSO. mAb CIS43 solution, prepared at a concentration of 50 μM (expressed per Fab site), was injected in 10 μl aliquots into the calorimetric cell containing the peptide at a concentration of 2 μM. All titrations were performed at 25 °C. The exact concentrations of the reactants in each experiment were determined from the absorbance at 280nm. The heat evolved upon each injection of antibody was obtained from the integral of the calorimetric signal. The heat associated with binding to rPfCSP or the peptide in the cell was obtained by subtracting the heat of dilution from the heat of reaction. The individual heats were plotted against the molar ratio, and the enthalpy change, ΔH, the association constant, Ka (the dissocation constant, Kd =1/Ka) and the stoichiometry, N, were obtained by nonlinear regression of the data to a model that takes into account the binding to two sets of sites with different binding energetics for rPfCSP or to a single-binding-site model for the peptides. Gibbs energy, ΔG, was calculated from the relation ΔG = −RTlnKa, where R is the is the universal gas constant, (1.987 cal/(K × mol)) and T the absolute temperature in kelvin. The entropy contribution to Gibbs energy, −TΔS, was calculated from the known relation ΔG = ΔH −TΔS.
Crystallization and data collection
Crystals of CIS43 Fab with peptide 20, 21, 25 and 29, respectively, were obtained using a mosquito and NT8 dispensing robots and screening was done with Rigaku Wizard Precipitant Synergy block#2, Molecular Dimensions Proplex screen HT-96, Hampton Research Crystal Screen HT by the vapor diffusion method. Crystals used for diffraction data were grown in the following conditions; CIS43 Fab with peptide 20: 0.2M Ammonium Sulfate, 0.1M Sodium Acetate Trihydrate pH 4.6, 30% PEG MME 2000; CIS43 Fab with peptide 21: 0.1M Sodium Acetate Trihydrate pH 4.6, 2M Ammonium Sulfate; CIS43 Fab with peptide 25: 0.1M Sodium Hepes pH 7.5, 1.4M Sodium Citrate Tribasic Dihydate; CIS43 Fab with peptide 29: 1.6M Sodium Citrate Tribasic Dihydrate pH 6.5, using the vapor diffusion method. Crystals were cryo-protected in solutions containing 30% molar excess of their original reagents and 20% Ethylene Glycol. Crystal diffracted to 2.4 angstroms (Å), 1.8 Å, 2.0 Å and 2.2 Å, respectively (Supplementary Table 3).
Crystals of CIS42 Fab with peptide 20, 21, 25 and 29 were obtained following the same procedure described above. Crystals used for diffraction data were all grown in 15% PEG3350, 9% Isopropanol, 0.12M Ammonium Citrate pH 8.5. Crystals were cryo-protected in solutions containing 30% molar excess of their original reagents and 20% Ethylene Glycol. Crystals of CIS42 Fab with peptide 20, peptide 21, peptide 25 and peptide 29 diffracted to 2.3 Å, 1.8 Å, 2.0 Å and 2.2 Å, respectively (Supplementary Table 5). Data were collected at ALS 5.1 and 5.2 on the Fred Hutchinson Cancer Research Center home source (Rigaku) and processed using HKL2000 56.
Structure solution and refinement
The structures of CIS43 Fab and CIS42 Fab with peptides were solved by molecular replacement using Phaser in CCP457. The peptides were manually fitted using COOT58 and refinement of the structures was done in Phenix59. The refinement statistics are summarized in Supplementary Table 3 and 5. Structural figures were made with Pymol60.
Molecular dynamic simulations
Four protein systems were created based on the crystal structures: CIS43 Fab in complex with peptide 21, CIS42 Fab in complex with peptide 21, free peptide 21 in CIS43 Fab-bound conformation, and free peptide 21 in CIS42 Fab-bound conformation. All protein models were immersed in a water box neutralized to 0.15M with sodium chloride ions. The water box was built using VMD’s Solvate plugin and the ions were added with VMD’s Autoionize plugin (Visual Molecular Dynamics)61. Protein structure files were created using VMD’s psfgen function.
The four protein systems were then configured to run with NAMD2.12 (Scalable Molecular Dynamics)62 with the CHARMM36 force field63. TIP3P water parameterization was used to describe the water molecules. All systems were fully minimized with 20,000 conjugate gradient steps before proceeding with equilibration runs. Equilibration began with a slow heating starting at 100 K until reaching a final temperature of 310 K. Simulations were performed with a 1 fs timestep for all equilibration and production runs. The periodic electrostatic interactions were computed using particle-mesh Ewald (PME) summation with a grid spacing smaller than 1 Å. Constant temperature was imposed by using Langevin dynamics with a damping coefficient of 1.0 ps. Constant pressure of 1 atm was maintained with Langevin piston dynamics, a 200 fs decay period and a 50 fs time constant. 500 ns of molecular dynamics simulation was achieved for all four protein systems on the NIH High-Performance Computing Cluster (https://hpc.nih.gov).
Peptide 21 was assessed with root mean square deviation (RMSD) and root mean square fluctuation (RMSF). RMSD was calculated using VMD’s Trajectory Tool plugin, while RMSF was calculated with Gromacs64. Both were calculated in angstroms over the full 500 ns trajectory and were compared side-by-side. For RMSD and RMSF, the antibodies were aligned to frame 1 of the respective trajectories in order to compare conformation changes and movement of peptide 21. Additionally, principal component analysis (PCA) was performed using mdtraj65, scikit66, and graphed with matplotlib67 to visualize the distribution of structural conformations of free peptide 21. Peptide 21 in its conformations bound to CIS43 Fab and CIS42 Fab were plotted on the PCA density heatmap. Eigenvalues are listed to show principal component 1 and principal component 2 as the dominant components. Hydrogen bond analysis was performed with UCSF Chimera68 on peptide 21 for both the CIS43 Fab and CIS42 Fab simulations and number of hydrogen bonds are shown over the 500 ns trajectory. Lastly, the crystal structures of CIS43 Fab and CIS42 Fab were aligned to their 500 ns frames and RMSD was calculated to compare movement and flexibility of the antibody CDR loops. Free energy calculations of the CIS43 variant mutations were performed computationally using FoldX. CIS43 Fab bound to peptide 21 was used as input. The protein structure was first repaired, then mutated, then analyzed for interaction binding energy between the mutants and wild-type69.
Pulse-Chase Metabolic Labeling Assay
Pulse-chase metabolic labeling of Plasmodium sporozoites has been previously described32,33. Briefly, freshly dissected Pfsporozoites were incubated in Dulbecco’s modified Eagle’s medium (DMEM) without Cys/Met, 1% BSA and 400 μCi/mL L-[35S]-Cys/Met for 45 min at 28°C and then kept on ice or chased at 28°C for 1.5 hours in DMEM with Cys/Met and 1% BSA in the absence or presence of the indicated concentrations of antibodies: mAb15 (isotype control human mAb specific for PfCSP C terminus), mAb5D5 (positive control mouse mAb specific for PfCSP N terminus), and mAb CIS43. Following the lysis of chased sporozoites, labeled PfCSP was immunoprecipitated with mAb2A10 (mouse mAb specific for PfCSP repeats) conjugated to sepharose. Labeled PfCSP was eluted from the beads and analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography. Densitometry of the high and low molecular weight bands was performed using Image J software. The ratio of the density, or area under the curve, of the top band to the bottom band was calculated.
Statistics
For liver parasite burden in C57BL/6 mice, data were compared for significance using a Mann–Whitney test, whereas in FRG-huHep mice the Kruskal-Wallis test, with Dunn’s correction for multiple comparisons, was used. Kaplan-Meier parasitemia curves were analyzed by the log rank test. For measurement of PfCSP mAbs in mouse serum, standard curves were fitted with a hyperbolic parameter curve and concentration values were interpolated. For the stoichiometry data, errors with 95% confidence were estimated from the fits of the data. Unless otherwise indicated, all data were plotted and graphed using GraphPad Prism 7.0. p value <0.05 indicated statistically significant differences.
Data availability
The authors declare that all the data supporting the findings of this study are available within the paper and supplementary information files. The structure factors and coordinates of CIS43 and CIS42 bound to peptide 20, 21, 25 and 29 have been deposited in the Protein Data Bank (PDB) with accession codes 6B5L, 6B5M, 6B5N, 6B5O, 6B5P, 6B5R, 6B5S and 6B5T, respectively.
Supplementary Material
Acknowledgments
We thank the study volunteers VRC312-402, VRC314-705 and 709. We thank R. Bailer for providing PBMC samples. We thank R. Lynch, S. Narpala, M. Prabhakaran, R. Nguyen, and Xuejun Chen for technical help and advice on the experiments. We thank I. Cockburn (Australian National University College of Health and Medicine) for providing some of the biotinylated (NANP)9 probe used here. We thank T. Zhou (Vaccine Research Center, NIH) for providing mAb VRC01. We thank C. Peckels, A. Foulger, A. Holland and M. Wang (Duke Human Vaccine Institute, DHVI) for technical assistance. H. Bouton-Verville (DHVI) for project management. We are grateful to C. A. Schramm’s expertise and assistance in analyzing the malaria Pf3K database. We thank the Sanaria Manufacturing Team for the production of fresh PfSPZ. We thank M. Nason for technical help with the statistcs. We thank B. Graham for insightful discussion on the project. We are particularly grateful to L. Stamatatos for use of laboratory space and equipment. We thank the J. B. Pendleton Charitable Trust for its generous support of Formulatrix robotic instruments. This work was supported by US N.I.H. grant GM56550 and N.S.F. grant MCB-1157506 to E.F. M.P. was supported by a Vaccine and Infectious Disease Division Faculty Initiative Grant through the Fred Hutchinson Cancer Research Center. X-ray diffraction data was collected at the Berkeley Center for Structural Biology beamlines 5.0.1 and 5.0.2, which are supported in part by the National Institutes of Health, National Institute of General Medical Sciences. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Footnotes
Author Contributions
N.K.K., A.H.I., B.K.S., M.P., F.Z., and R.A.S. planned the studies. N.K.K., A.H.I., B.K.S., S.Mu., C.W., Y.F.-G, B.J.F, A.S., J.R.F., S.Ma., A.B.M., M.K., A.K.W, S.K.F., G.-Y.C., S.C., N.K.C., B.Z., J.G., P.S. and M.P. conducted experiments. F.Z. provided mAb2A10 and mAb5D5. K.W., K.O.S., A.M.T., M.A.G. and A.M.: contributed new methodologies, analytic tools and performed research; M.B., H.-X.L., B.F.H.: isolated, cloned and produced plasmablast antibodies and analyzed data. N.K.K., A.H.I., B.K.S, M.P., F.Z., S.H.I.K., S.C., A.S., E.F., S.Ma., A.B.M., P.D.K., P.S., S.N.B., B.K.L.S., S.L.H. and R.A.S. interpreted the studies. N.K.K., A.H.I., M.P. and R.A.S. wrote the paper. All authors reviewed, edited and approved the paper.
Competing financial interests
N.K.C., S.C., B.K.L.S., and S.L.H. are salaried employees of Sanaria Inc., the developer and owner of PfSPZ Vaccine and the sponsor of the clinical trials. In addition, S.L.H. and B.K.L.S. have a financial interest in Sanaria Inc. All other authors declare no competing financial interests.
References
- 1.Menard R, et al. Circumsporozoite protein is required for development of malaria sporozoites in mosquitoes. Nature. 1997;385:336–340. doi: 10.1038/385336a0. [DOI] [PubMed] [Google Scholar]
- 2.Coppi A, et al. Heparan sulfate proteoglycans provide a signal to Plasmodium sporozoites to stop migrating and productively invade host cells. Cell Host Microbe. 2007;2:316–327. doi: 10.1016/j.chom.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ancsin JB, Kisilevsky R. A binding site for highly sulfated heparan sulfate is identified in the N terminus of the circumsporozoite protein: significance for malarial sporozoite attachment to hepatocytes. J Biol Chem. 2004;279:21824–21832. doi: 10.1074/jbc.M401979200. [DOI] [PubMed] [Google Scholar]
- 4.Rathore D, Sacci JB, de la Vega P, McCutchan TF. Binding and invasion of liver cells by Plasmodium falciparum sporozoites. Essential involvement of the amino terminus of circumsporozoite protein. J Biol Chem. 2002;277:7092–7098. doi: 10.1074/jbc.M106862200. [DOI] [PubMed] [Google Scholar]
- 5.Dame JB, et al. Structure of the gene encoding the immunodominant surface antigen on the sporozoite of the human malaria parasite Plasmodium falciparum. Science. 1984;225:593–599. doi: 10.1126/science.6204383. [DOI] [PubMed] [Google Scholar]
- 6.Enea V, et al. DNA cloning of Plasmodium falciparum circumsporozoite gene: amino acid sequence of repetitive epitope. Science. 1984;225:628–630. doi: 10.1126/science.6204384. [DOI] [PubMed] [Google Scholar]
- 7.Nussenzweig RS, Nussenzweig V. Antisporozoite vaccine for malaria: experimental basis and current status. Rev Infect Dis. 1989;11(Suppl 3):S579–585. doi: 10.1093/clinids/11.supplement_3.s579. [DOI] [PubMed] [Google Scholar]
- 8.Casares S, Brumeanu TD, Richie TL. The RTS,S malaria vaccine. Vaccine. 2010;28:4880–4894. doi: 10.1016/j.vaccine.2010.05.033. [DOI] [PubMed] [Google Scholar]
- 9.White MT, et al. The relationship between RTS,S vaccine-induced antibodies, CD4(+) T cell responses and protection against Plasmodium falciparum infection. PLoS One. 2013;8:e61395. doi: 10.1371/journal.pone.0061395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stoute JA, et al. A preliminary evaluation of a recombinant circumsporozoite protein vaccine against Plasmodium falciparum malaria. RTS,S Malaria Vaccine Evaluation Group. N Engl J Med. 1997;336:86–91. doi: 10.1056/NEJM199701093360202. [DOI] [PubMed] [Google Scholar]
- 11.Foquet L, et al. Vaccine-induced monoclonal antibodies targeting circumsporozoite protein prevent Plasmodium falciparum infection. J Clin Invest. 2014;124:140–144. doi: 10.1172/JCI70349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oyen D, et al. Structural basis for antibody recognition of the NANP repeats in Plasmodium falciparum circumsporozoite protein. Proc Natl Acad Sci U S A. 2017;114:E10438–E10445. doi: 10.1073/pnas.1715812114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olotu A, et al. Four-year efficacy of RTS,S/AS01E and its interaction with malaria exposure. N Engl J Med. 2013;368:1111–1120. doi: 10.1056/NEJMoa1207564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olotu A, et al. Seven-Year Efficacy of RTS,S/AS01 Malaria Vaccine among Young African Children. N Engl J Med. 2016;374:2519–2529. doi: 10.1056/NEJMoa1515257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Triller G, et al. Natural Parasite Exposure Induces Protective Human Anti-Malarial Antibodies. Immunity. 2017 doi: 10.1016/j.immuni.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Briney BS, Willis JR, Hicar MD, Thomas JW, Crowe JE. Frequency and genetic characterization of V(DD)J recombinants in the human peripheral blood antibody repertoire. Immunology. 2012;137:56–64. doi: 10.1111/j.1365-2567.2012.03605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.March S, et al. A microscale human liver platform that supports the hepatic stages of Plasmodium falciparum and vivax. Cell Host Microbe. 2013;14:104–115. doi: 10.1016/j.chom.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.March S, et al. Micropatterned coculture of primary human hepatocytes and supportive cells for the study of hepatotropic pathogens. Nat Protoc. 2015;10:2027–2053. doi: 10.1038/nprot.2015.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Espinosa DA, et al. Robust antibody and CD8+ T-cell responses induced by P. falciparum CSP adsorbed to cationic liposomal adjuvant CAF09 confer sterilizing immunity against experimental rodent malaria infection. npj Vaccines. 2017;2 doi: 10.1038/s41541-017-0011-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sack BK, et al. Humoral protection against mosquito bite-transmitted Plasmodium falciparum infection in humanized mice. npj Vaccines. 2017;2 doi: 10.1038/s41541-017-0028-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kublin JG, et al. Complete attenuation of genetically engineered Plasmodium falciparum sporozoites in human subjects. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aad9099. [DOI] [PubMed] [Google Scholar]
- 22.Vanderberg JP, Frevert U. Intravital microscopy demonstrating antibody-mediated immobilisation of Plasmodium berghei sporozoites injected into skin by mosquitoes. Int J Parasitol. 2004;34:991–996. doi: 10.1016/j.ijpara.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 23.Sack BK, et al. Model for in vivo assessment of humoral protection against malaria sporozoite challenge by passive transfer of monoclonal antibodies and immune serum. Infect Immun. 2014;82:808–817. doi: 10.1128/IAI.01249-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Epstein JE, et al. Safety and clinical outcome of experimental challenge of human volunteers with Plasmodium falciparum-infected mosquitoes: an update. J Infect Dis. 2007;196:145–154. doi: 10.1086/518510. [DOI] [PubMed] [Google Scholar]
- 25.Rickman LS, et al. Plasmodium falciparum-infected Anopheles stephensi inconsistently transmit malaria to humans. Am J Trop Med Hyg. 1990;43:441–445. doi: 10.4269/ajtmh.1990.43.441. [DOI] [PubMed] [Google Scholar]
- 26.Freire E, Schon A, Velazquez-Campoy A. Isothermal titration calorimetry: general formalism using binding polynomials. Methods Enzymol. 2009;455:127–155. doi: 10.1016/S0076-6879(08)04205-5. [DOI] [PubMed] [Google Scholar]
- 27.Fisher CR, et al. T-dependent B cell responses to Plasmodium induce antibodies that form a high-avidity multivalent complex with the circumsporozoite protein. PLoS Pathog. 2017;13:e1006469. doi: 10.1371/journal.ppat.1006469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghasparian A, Moehle K, Linden A, Robinson JA. Crystal structure of an NPNA-repeat motif from the circumsporozoite protein of the malaria parasite Plasmodium falciparum. Chem Commun (Camb) 2006:174–176. doi: 10.1039/b510812h. [DOI] [PubMed] [Google Scholar]
- 29.Dyson HJ, Satterthwait AC, Lerner RA, Wright PE. Conformational preferences of synthetic peptides derived from the immunodominant site of the circumsporozoite protein of Plasmodium falciparum by 1H NMR. Biochemistry. 1990;29:7828–7837. doi: 10.1021/bi00486a008. [DOI] [PubMed] [Google Scholar]
- 30.Plassmeyer ML, et al. Structure of the Plasmodium falciparum circumsporozoite protein, a leading malaria vaccine candidate. J Biol Chem. 2009;284:26951–26963. doi: 10.1074/jbc.M109.013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Topchiy E, et al. T1BT* structural study of an anti-plasmodial peptide through NMR and molecular dynamics. Malar J. 2013;12:104. doi: 10.1186/1475-2875-12-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coppi A, Pinzon-Ortiz C, Hutter C, Sinnis P. The Plasmodium circumsporozoite protein is proteolytically processed during cell invasion. J Exp Med. 2005;201:27–33. doi: 10.1084/jem.20040989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Espinosa DA, et al. Proteolytic Cleavage of the Plasmodium falciparum Circumsporozoite Protein Is a Target of Protective Antibodies. J Infect Dis. 2015;212:1111–1119. doi: 10.1093/infdis/jiv154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aurrecoechea C, et al. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37:D539–543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.The Pf3K Project: pilot data release 5. 2016 www.malariagen.net/data/pf3k-5.
- 36.Gaudinski MR, et al. Safety and pharmacokinetics of the Fc-modified HIV-1 human monoclonal antibody VRC01LS: A Phase 1 open-label clinical trial in healthy adults. PLoS Med. 2018;15:e1002493. doi: 10.1371/journal.pmed.1002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoffman SL, et al. Development of a metabolically active, non-replicating sporozoite vaccine to prevent Plasmodium falciparum malaria. Hum Vaccin. 2010;6:97–106. doi: 10.4161/hv.6.1.10396. [DOI] [PubMed] [Google Scholar]
- 38.Epstein JE, et al. Live attenuated malaria vaccine designed to protect through hepatic CD8(+) T cell immunity. Science. 2011;334:475–480. doi: 10.1126/science.1211548. [DOI] [PubMed] [Google Scholar]
- 39.Seder RA, et al. Protection against malaria by intravenous immunization with a nonreplicating sporozoite vaccine. Science. 2013;341:1359–1365. doi: 10.1126/science.1241800. [DOI] [PubMed] [Google Scholar]
- 40.Ishizuka AS, et al. Protection against malaria at 1 year and immune correlates following PfSPZ vaccination. Nat Med. 2016;22:614–623. doi: 10.1038/nm.4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu X, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wheatley AK, et al. H5N1 Vaccine-Elicited Memory B Cells Are Genetically Constrained by the IGHV Locus in the Recognition of a Neutralizing Epitope in the Hemagglutinin Stem. J Immunol. 2015;195:602–610. doi: 10.4049/jimmunol.1402835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanekiyo M, et al. Rational Design of an Epstein-Barr Virus Vaccine Targeting the Receptor-Binding Site. Cell. 2015;162:1090–1100. doi: 10.1016/j.cell.2015.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tiller T, et al. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lefranc MP, et al. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37:D1006–1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liao HX, et al. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. J Virol Methods. 2009;158:171–179. doi: 10.1016/j.jviromet.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bonsignori M, et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J Virol. 2011;85:9998–10009. doi: 10.1128/JVI.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nardin EH, et al. Circumsporozoite proteins of human malaria parasites Plasmodium falciparum and Plasmodium vivax. J Exp Med. 1982;156:20–30. doi: 10.1084/jem.156.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zavala F, Hollingdale MR, Schwartz AL, Nussenzweig RS, Nussenzweig V. Immunoradiometric assay to measure the in vitro penetration of sporozoites of malaria parasites into hepatoma cells. J Immunol. 1985;134:1202–1205. [PubMed] [Google Scholar]
- 50.Douglas AD, et al. Neutralization of Plasmodium falciparum merozoites by antibodies against PfRH5. J Immunol. 2014;192:245–258. doi: 10.4049/jimmunol.1302045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 52.Bruña-Romero O, et al. Detection of malaria liver-stages in mice infected through the bite of a single Anopheles mosquito using a highly sensitive real-time PCR. Int J Parasitol. 2001;31:1499–1502. doi: 10.1016/s0020-7519(01)00265-x. [DOI] [PubMed] [Google Scholar]
- 53.Vaughan AM, et al. A transgenic Plasmodium falciparum NF54 strain that expresses GFP-luciferase throughout the parasite life cycle. Mol Biochem Parasitol. 2012;186:143–147. doi: 10.1016/j.molbiopara.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 54.Miller JL, et al. Quantitative bioluminescent imaging of pre-erythrocytic malaria parasite infection using luciferase-expressing Plasmodium yoelii. PLoS One. 2013;8:e60820. doi: 10.1371/journal.pone.0060820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murphy SC, et al. Real-time quantitative reverse transcription PCR for monitoring of blood-stage Plasmodium falciparum infections in malaria human challenge trials. Am J Trop Med Hyg. 2012;86:383–394. doi: 10.4269/ajtmh.2012.10-0658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Otwinowski Z, Minor W. [20] Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 57.Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 58.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 59.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Delano WL. The PyMOL Molecular Graphics System. DeLano Scientific; 2002. [Google Scholar]
- 61.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 62.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang J, et al. Isolation of human monoclonal antibodies from peripheral blood B cells. Nat Protoc. 2013;8:1907–1915. doi: 10.1038/nprot.2013.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Páll S, Abraham MJ, Kutzner C, Hess B, Lindahl E. Tackling Exascale Software Challenges in Molecular Dynamics Simulations with GROMACS. 2015;8759:3–27. doi: 10.1007/978-3-319-15976-8_1. [DOI] [Google Scholar]
- 65.McGibbon RT, et al. MDTraj: A Modern Open Library for the Analysis of Molecular Dynamics Trajectories. Biophys J. 2015;109:1528–1532. doi: 10.1016/j.bpj.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pedregosa F, et al. Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research. 2011 [Google Scholar]
- 67.Hunter JD. Matplotlib: A 2D Graphics Environment. Computing in Science & Engineering. 2007;9:90–95. doi: 10.1109/mcse.2007.55. [DOI] [Google Scholar]
- 68.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 69.Guerois R, Nielsen JE, Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J Mol Biol. 2002;320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- 70.Rich SM, Hudson RR, Ayala FJ. Plasmodium falciparum antigenic diversity: evidence of clonal population structure. Proc Natl Acad Sci U S A. 1997;94:13040–13045. doi: 10.1073/pnas.94.24.13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zeeshan M, et al. Genetic variation in the Plasmodium falciparum circumsporozoite protein in India and its relevance to RTS,S malaria vaccine. PLoS One. 2012;7:e43430. doi: 10.1371/journal.pone.0043430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zakeri S, Avazalipoor M, Mehrizi AA, Djadid ND, Snounou G. Restricted T-cell epitope diversity in the circumsporozoite protein from Plasmodium falciparum populations prevalent in Iran. Am J Trop Med Hyg. 2007;76:1046–1051. [PubMed] [Google Scholar]
- 73.Tanabe K, et al. Within-population genetic diversity of Plasmodium falciparum vaccine candidate antigens reveals geographic distance from a Central sub-Saharan African origin. Vaccine. 2013;31:1334–1339. doi: 10.1016/j.vaccine.2012.12.039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all the data supporting the findings of this study are available within the paper and supplementary information files. The structure factors and coordinates of CIS43 and CIS42 bound to peptide 20, 21, 25 and 29 have been deposited in the Protein Data Bank (PDB) with accession codes 6B5L, 6B5M, 6B5N, 6B5O, 6B5P, 6B5R, 6B5S and 6B5T, respectively.