Abstract
Early studies indicate that rats may have a repertoire of MHC class Ib-reactive Ly49 stimulatory receptors capable of mounting memory-like NK cell alloresponses, and here we provide molecular and functional evidence for this assumption. Pairs of Ly49 receptors with sequence similarities in the lectin-like domains, but with opposing signalling functions, showed specificity for ligands with class Ia-like structural features encoded from the first telomeric MHC class Ib gene cluster, RT1-CE, which is syntenic with the H2-D/-L/-Q cluster in mice. The activating Ly49s4 receptor and its inhibitory counterparts, Ly49i4 and Ly49i3, reacted with all allelic variants of RT1–U, while Ly49s5 and Ly49i5 were specific for RT1-Eu. NK cell cytolytic responses were predictably activated and inhibited, and potent in vivo NK alloresponses were induced by repeated MHC class Ib alloimmunizations. Additional Ly49-class Ib interactions, including RT1-Cl with the Ly49s4/-i4/-i3 group of receptors, were characterized using overexpressed receptor/ligand pairs, in vitro functional assays, and limited mutational analyses. Obvious as well as subtle Ly49-class Ib interactions led to ligand-induced receptor calibration and NK subset expansions in vivo. Together these studies suggest that in vivo NK alloresponses are controlled by pleomorphic Ly49-class Ib interactions, some of which may not be easily detectable in vitro.
Introduction
NK cells are innate immune lymphocytes that respond to infection and malignancy, and they regulate placentation and reproductive fitness. NK cell responses are controlled by arrays of activating and inhibitory receptors. In accordance with the “missing-self” hypothesis, NK cells discriminate normal “self” cells from foreign or abnormal cells predominantly through inhibitory NK cell receptors that interrupt cellular activation in the presence of self MHC class I ligands (1). Whereas inhibitory receptors for MHC are a fundamental feature of NK cells, activating receptors for MHC ligands have also been identified. The NK cell receptors that recognize polymorphic MHC class I molecules are encoded by polymorphic and rapidly evolving gene families that contribute to the diversity and target cell repertoire of NK cells. Among different mammalian species, analogous MHC recognition functions are often performed by structurally unrelated glycoproteins. For instance, NK cell recognition of polymorphic MHC class I ligands is performed by Killer cell Immunoglobulin-like Receptors (KIR) in humans, higher primates, and cattle, but by lectin-like members of the Ly49 family in rodents and horses (2,3). These species-specific differences in the evolution of KIR and Ly49 emphasize the evolutionary plasticity and versatility of NK receptor families, and they suggest that the differential evolution of KIR and Ly49 gene families may be determined, in part, by pressures unique to the environmental niche of each species.
Among MHC binding NK receptors, inhibitory function is conferred by an ITIM motif in the cytoplasmic tail, and activating function is affected by a positively charged transmembrane residue that recruits stimulatory signalling adaptors such as DAP-12 (4,5). In contrast to humans and mice, where inhibitory NK alloresponses predominate, early genetic and functional studies pointed to the existence of powerful stimulatory NK cell responses directed against MHC encoded ligands in rats (6–9). A phylogenetic analysis of KIR and Ly49 genes suggests that activating variants evolve from inhibitory receptors by species-specific evolutionary events (10). In the rat, several clusters of Ly49 genes containing both activating and inhibitory members, as well as “transitional” variants containing both activating and inhibitory features, appear to have expanded in a species-specific fashion (11).
In order to establish the physiologic role of activating NK cell alloreceptors, we have undertaken an extensive functional analysis of select rat Ly49 receptors and ligands encoded within the rat MHC locus, RT1. We show that the recognition of nonclassical MHC class Ib molecules is a generalizable feature of activating rat Ly49 receptors. These ligands are encoded within the first class I gene cluster in the telomeric part of the rat MHC, termed RT1-CE and situated in a position syntenic to the H2-D/L/Q cluster in the mouse. The RT1-CE cluster contains an expanded array of nonclassical class Ib genes, which show extensive genomic plasticity based on the “presence or absence” of genes rather than nucleotide substitutions at the allele level. Phylogenetic analyses have shown that the RT1-CE genes are more related to each other and to the classical RT1-A genes, than to the H-2K, -D, -L, and –Q genes in mice. Sharing of several unique rat-specific genomic features among RT1-CE and RT1-A genes, suggest independent evolution following speciation of rat and mouse as a reflection of the unique challenges encountered in their respective environmental niches (12). Unlike classical RT1-A molecules, which are expressed on nearly all nucleated cells and restrict T cell functions, RT1-CE proteins typically do not present antigenic peptides to T cells (13,14). Moreover, RT1-CE molecules are normally expressed at low levels, but their cell surface expression may be upregulated during inflammation or infections with specific pathogens such as Listeria monocytogenes (15,16). In this manner, the upregulated expression of class Ib molecules might serve as a danger signal promoting NK cell mediated immune surveillance during pathologic conditions. These findings point to the species-specific evolutionary expansion of activating Ly49 receptors and their polymorphic nonclassical RT1-CE ligands in rats. These paired NK cell receptor-ligand systems likely constitute a novel rat innate immune surveillance mechanism involving an expanded repertoire of activating receptors that have evolved in parallel with a unique array of polymorphic nonclassical MHC class Ib ligands.
Materials and Methods
Rats
The MHC congenic and intra-MHC recombinant rat strains PVG-RT7b (PVG.7B) [RT1c or c], PVG.1N (n), PVG.1LLEW (l), PVG.1LF344 (lv1), PVG.1U (u), and PVG.R23 (RT1-Au -B/Da -CE/N/M av1 [class Ia-class II-class Ib] abbreviated u-a-av1) were bred in Oslo. BN (n), F344 (lv1) and congenitally athymic nude rats (c) with the Han/rnu mutation (HsdHan™: RNU-Foxn1rnu/Foxn1+) were from Harlan UK (Bicester, UK). Rats were used at 8–12 weeks of age. Animal experiments were conducted in compliance with “The Norwegian Regulations on Animal Experimentation” and approved by the institutional veterinarian with delegated authority from the Norwegian Animal Research Authority (NARA). Intraperitoneal injections were performed under neuroleptanalgesia with fentanyl citrate and fluanisone (Hypnorm®; VetaPharma, UK).
In vivo alloimmunization
A modification of a previously described immunization protocol was used (15–17). Rats (congenitally athymic nude or F344) were injected i.p. once weekly for a total of four weeks with 107 mononuclear splenocytes from PVG.1U or PVG.1N rats, or with irradiated YB2/0 cells transfected with RT1-Un (YB.Un) or control wild type YB2/0 cells. Peritoneal cells were retrieved with cold PBS three days after the last injection. Mononuclear cells were obtained by centrifugation on Lymphoprep. DAR13+ and DAR13− IL-2 activated NK cells were generated as described (18). Results are presented as proportions of indicated cell subpopulations in individual rats. Statistical differences between groups were evaluated with Student’s t-test based on the individual values.
Cytotoxicity assay
Peritoneal cells or Ly49 transfected RNK-16 cells were used as effector cells against the indicated target cells in a 4-hr 51Cr release assay as previously described (6). Results are presented as mean values of triplicates with bars representing one SD. Statistical difference was evaluated with Student’s t-test using the triplicate values.
Reporter assay
For the reporter assay, 5 × 104 BWZ reporter cells were incubated with 2.5 × 104 stimulator cells in a total volume of 200 μl in U-bottomed 96-well plates. 1 μg of mAb DAR13 or FLY5 together the FcR+ P815 cells or 3 μM ionomycin was added for positive control. Cells were incubated for 24 h at 37˚C in RPMI-1640, 1% FCS, and 10 ng/ml of PMA, washed, lysed with 0.125% NP-40 in PBS containing MgCl, 2-mercaptoethanol and chlorophenol red red-β-D galactopyranoside reagent, and incubated for 1–3 hr at 37°C or for up to 24 hrs at room temperature before determination of OD. Results are presented as mean values of triplicates (A595–A650) and bars representing one SD. Statistical differences were evaluated in individual experiments with Student’s t-test using the triplicates values.
Antibodies and flow cytometry
1–10 × 105 cells were labelled with different combinations of the following conjugated mAbs: Biotinylated DAR13 (anti-Ly49s3/-i3/-s4/-i4 (18,19); FLY5 (anti-Ly49i5/-s5 (20)), STOK2 (anti-Ly49i2; (21)), STOK9 (anti-KLRH1; (22), or WEN23 (anti-NKp46; (23)), Alexa-conjugated 3.2.3 (anti-NKR-P1A/B) or STOK27 (anti-NKR-P1B; (24)); PE or FITC-conjugated G4.18 (anti-CD3) or 10/78 (anti-NKR-P1A/B) (both from Pharmingen). Biotinylated mAbs were revealed by PE-Cy5-conjugated streptavidin and cells were analysed on a FACSCalibur.
Cloning and stable transfection
Total RNA was generated from Con A lymphoblasts from the different MHC congenic strains on PVG background or from the R2 macrophage cell line (d haplotype) and reverse transcribed to cDNA. Gene-specific primers used for the amplification by pwo DNA polymerase (Roche) are specified in Supplemental Table I. The identities of receptor and ligand genes were confirmed by the sequencing of more than 10 clones/isolates on an ABI3730 sequencer. cDNA for RT1-EU, RT1-Au, and rat β2m was kindly provided by Dr. Etienne Joly. MHC class I genes containing an N-terminal FLAG tag were subcloned into the EMCV.SRα expression vector and used for stable transfection of the rat YB2/0 cell line. For MHC class I transfection of mouse NSO cells we used the pMXpuro vector (puromycin resistance), with or without rat β2m using a picornavirus-derived co-expression strategy (from the foot-and-mouth-disease virus; F2A) inducing a balanced expression of two partner chains (25). This is based on transfection of a single open reading frame with cleavage of the polypeptide chain at the translational stage by ribosomal “skipping” (the core recognition site of a “2A peptide skipping motif” is underlined and the cleavage site marked: QTLNFDLLKLAGDVESNPG^P). RNK-16 cells were transfected with full length Ly49 receptor constructs using the pMXpuro vector using a Neon® electroporation system for RNK-16 (26). 293T cells were transfected with full-length Ly49 receptors with or without rat DAP12 using pMXpuro vector and FuGene (Roche) and the F2A co-expression strategy as above. BW5147 cells expressing the LacZ gene under the control of 3×NFAT-1 promoter (BWZ) (from Nilabh Shastri, Univ. California Berkeley), were transfected with a construct encoding parts of a reversed ζ-chain together with most of the Ly49 molecule, using the pMXpuro vector. MHC reporter cells were generated using a fusion construct containing the extracellular part of RT1-Eu or RT1-Un, transmembrane part of rat CD8 and intracellular part of rat ζ chain, together with rat β2m using the F2A co-expression strategy.
Real-time RT-PCR
NK cells from the peritoneal cavity of alloimmunized F344 rats were enriched with WEN23-coated Microbeads, stained with appropriate antibodies, and subjected to sorting separating DAR13+ and DAR13− cells using a FACStar (BD Biosciences). cDNA was generated from total RNA. Allele-specific FAM-TAMRA conjugated primers were used for Ly49i3, s3, i4 and s4 expression, with CD45 used as an endogenous control. Real-time PCR was performed using an ABI7900H analyser. Relative expressions were calibrated based on 1/216 of CD45 expression level. Results are presented as mean values of triplicates and bars representing one SD. Statistical differences were evaluated with Student’s t-test using the triplicates values.
Results
Ly49s4, Ly49i4, and Ly49i3 reporter cells react with a ligand in the n, av1, lv1, and c RT1 haplotypes, while Ly49s5 is specific for u
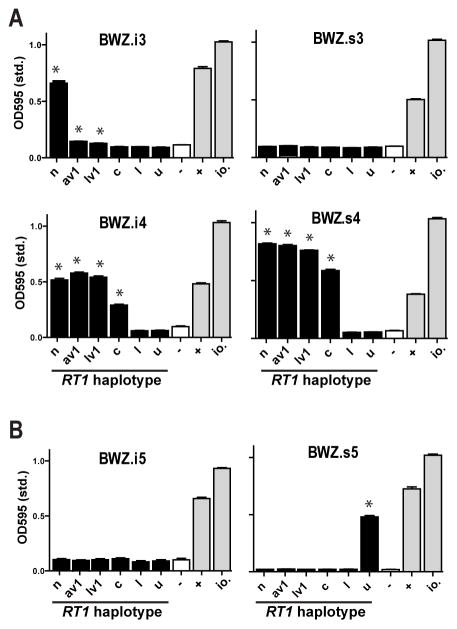
A map of the rat NK gene complex and the MHC in the reference n haplotype is depicted in Fig. 1A and B. The receptors investigated in the present study are encoded by a chromosomal cluster of rat Ly49 genes (block II) that are closely related phylogenetically. This species-specific clustering suggests relatively recent expansion and homogenization events following speciation of rat and mouse (11). Previous studies using subsets of IL-2 activated NK cells and Con A lymphoblast target cells have shown that the Ly49s3 and/or Ly49s4 receptor reacts with a class Ib allodeterminant expressed by most RT1 haplotypes from many rat strains, while the Ly49s5 receptor displays a narrower class Ib specificity (18–20). Ly49s4 has close sequence homology with Ly49s3, and with the inhibitory receptors Ly49i4 and Ly49i3, whereas Ly49s5 and Ly49i5 constitute a separate group. Select receptor-ligand interactions were investigated in more detail with a panel of Ly49 transfectants of BWZ reporter cells (Suppl. Fig. 1A). In agreement with previous work, the Ly49s4 reporter reacted with lymphoblast stimulator cells from a panel of MHC congenic PVG rats expressing the n, av1, lv1, and c, but not the u or l RT1 haplotypes. The same was true for Ly49i4, while Ly49i3 showed weak or negative responses to av1, lv1, and c. The Ly49s5 reporter cells, on the other hand, specifically reacted with u stimulator cells (Fig. 2). The Ly49s3 and Ly49i5 reporters failed to react with any of the stimulator cells, but as discussed below, the BWZ reporter may lack the sensitivity necessary to detect all relevant receptor-ligand interactions.
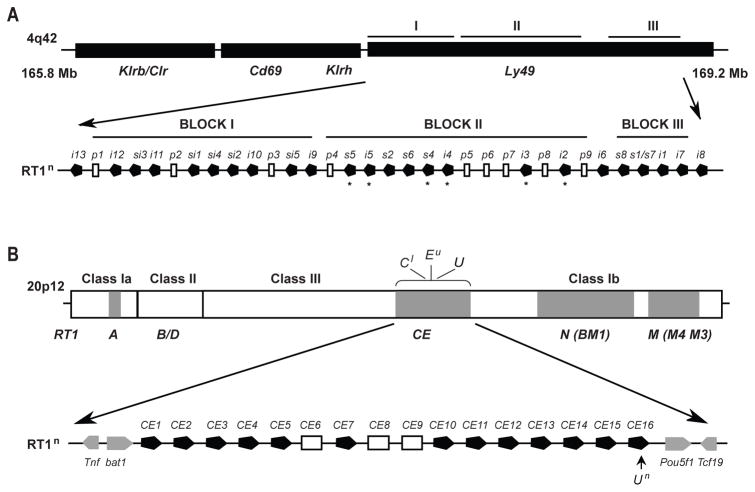
FIGURE 1.
Schematic maps of the rat NKC and MHC (RT1) gene complexes. (A) The Ly49 region is located in the telomeric part of the rat NKC and contains three major blocks of Ly49 genes in the reference n haplotype. Block II and III encode both inhibitory (i) and activating (s) receptors, while block I contains uncharacterized inhibitory and bifunctional (si) receptors. The Ly49 receptors characterized in the present study are mainly encoded from block II, but members from block III were also included. Ly49 receptors with defined MHC ligands are marked with an asterisk. The Ly49s3 gene is missing in the n haplotype and its location is currently not known. (B) RT1 class I gene clusters are shaded in grey. Classical class Ia molecules are encoded by the RT1-A cluster which is located centromeric to the class II/III regions. The RT1-CE cluster is in the same genomic location as the H2-D/L/Q cluster in the mouse and contains 16 class I genes (CE1–CE16) in the reference n haplotype, functional genes are shown in black.
FIGURE 2.
MHC specificity of Ly49 reporter cells.(A and B) Ly49 reporter cells were generated by stable Ly49 transfection of the BWZ reporter line expressing the LacZ gene under the control of NFAT promoters (Ly49i3, BWZ.i3; Ly49s3, BWZ.s3 etc). Reporter cells were co-incubated with Con A-activated lymphoblasts from a panel of MHC congenic PVG strains expressing the n, av1, lv1, c, l, and u rat MHC (RT1) haplotypes. As positive controls are shown reporter cells stimulated by Ionomycin (io.) or by a combination of a crosslinking mAb (DAR13 in panel A and FLY5 in panel B) and FcR-expressing P815 cells (+). As negative controls (-) are shown reporter cells incubated with culture medium only. Data are representative of at least 3 independent experiments, *p<0.01.
Ly49s4, Ly49i4, and Ly49i3 react with the MHC class Ib molecule RT1-Un (RT1-CE16) in the reference n haplotype
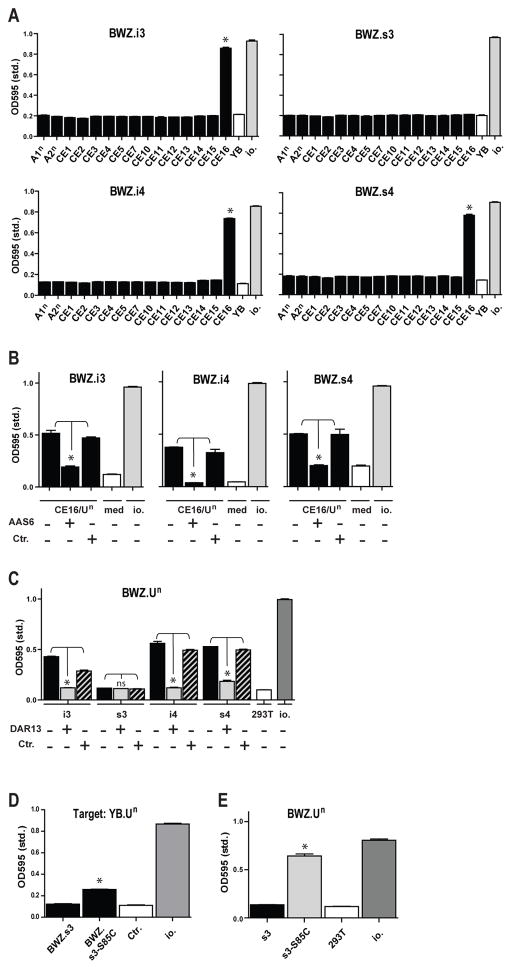
We searched for ligand(s) for the Ly49s4/-i4/-s3/-i3 group of receptors within the n RT1 haplotype, in which the full genomic sequence is available. We focused on the first telomeric class Ib cluster RT1-CE based on earlier data which mapped an activating NK allodeterminant to a class Ib deletion within the LEW.1LM1mutant rat strain (6). All 13 functional RT1-CE members together with the classical class Ia molecules RT1-A1n and RT1-A2n were stably transfected into the YB2/0 rat lymphoma line (Table I and Suppl. Fig. 1B). The most distal RT1-CE member, RT1-CE16, elicited a brisk response by the Ly49s4, Ly49i4, and Ly49i3 reporters, but no response by Ly49s3 (Fig. 3A). The protein encoded by RT1-CE16 corresponds to a previously identified molecule, known alternatively as RT1-Un (27), and addition of the RT1-Un reactive mAb AAS6 specifically blocked these responses (Fig. 3B).
Table I.
Rat MHC molecules, expression levels and mAb staining profiles1
| Annotations | MHC-cluster | Accession numbers | Expression in YB 2/0 (anti-FLAG) | OX18 mAb | AAS1 mAb | AAS5 mAb | AAS6 mAb | 8G10 mAb |
|---|---|---|---|---|---|---|---|---|
| A1n | RT1-A | MG963095 | NA | +++ | +++ | − | +++ | − |
| A2n | RT1-A | MG963096 | +++ | ++ | − | − | − | − |
| A3n | RT1-A | MG963097 | ND | + | − | − | − | − |
| CE1 | RT1-CE | MG963098 | ++ | + | − | − | − | − |
| CE2 | RT1-CE | MG963099 | ++ | ++ | − | − | +/− | − |
| CE3 | RT1-CE | MG963100 | + | − | − | − | − | − |
| CE4 | RT1-CE | MG963101 | + | − | − | − | − | − |
| CE5 | RT1-CE | MG963102 | +/− | − | − | − | − | − |
| CE7 | RT1-CE | MG963103 | + | − | − | − | − | − |
| CE10 | RT1-CE | MG963104 | +/− | +/− | − | − | − | − |
| CE11 | RT1-CE | MG963105 | ND | − | − | − | − | − |
| CE12 | RT1-CE | MG963106 | ++ | ++ | − | − | − | − |
| CE13 | RT1-CE | MG963107 | + | − | − | − | − | − |
| CE14 | RT1-CE | MG963108 | ++ | ++ | − | − | − | − |
| CE15 | RT1-CE | MG963109 | + | − | − | − | − | − |
| CE16/Un | RT1-CE | MG963110 | ++ | ++ | ++ | ++ | ++ | − |
| Uc/d | RT1-CE | MG963112 | + | + | + | + | + | − |
| Ulv1 | RT1-CE | MG963113 | + | + | + | + | + | − |
| Uav1 | RT1-CE | MG963111 | + | + | + | + | + | − |
| Au | RT1-A | X82106 | +++ | +++ | − | − | − | − |
| Eu | RT1-CE | AJ306619 | + | − | − | − | − | + |
| Cl | RT1-CE | X70066.1 | + | ND | ND | ND | ND | − |
FLAG-tagged MHC class Ia and Ib molecules (A1n was not FLAG-tagged) were transfected into the rat YB2/0 cell line (expressing rat β2m) and tested for expression levels with anti-FLAG or different MHC reactive mAbs. Functional RT1-CE genes were cloned from BN strain rats expressing the reference n haplotype and are numbered according to their positioning in this class Ib gene cluster (RT1-CE6, -CE8, and -CE9 are pseudogenes and were not studied). The presumed alleles of RT1-CE16/Un were cloned from the c, d, lv1, and av1 haplotypes, and are designated Uc/d, Ulv1, and Uav1 (Uc and Ud were identical at DNA level). cDNA clones of Au, Eu and Cl (kindly provided by Etienne Joly, Toulouse, France) were subcloned into relevant expression constructs. Note that the broadly reactive mouse anti-rat mAb OX18 failed to stain several of the RT1-CE molecules in BN and the Eu molecule. The AAS1, AAS5, and AAS6 allo-mAbs reacted with all alleles of RT1-U, but not with the other RT1-CE molecules tested (CE1–15) or with RT1-Eu. The sequences can be accessed from https://www.ncbi.nlm.nih.gov/nuccore/. ND, not done; NA, not applicable.
FIGURE 3.
RT1-Un (RT1-CE16) is a ligand for the Ly49s4/-i4/-s3/-i3 group of receptors. (A) Ly49s4/-i4/-s3/-i3 reporter cells were co-incubated with YB2/0 stimulator cells stably transfected with different MHC class Ia (A1n, A2n) or class Ib (CE1–CE16) molecules from the n haplotype. As negative and positive controls are shown stimulation with YB2/0 wild-type cells (YB) and Ionomycin (io.), respectively. (B) Blocking of the specific response induced by the RT1-CE16/Un transfectant by addition of 1 μg of mAb AAS6 (anti-CE16/RT1-U). As isotype control (Ctr.) was added purified mAb LOV8 (anti- CDw93). (C) Testing of a reciprocal RT1-Un reporter cell against 293T stimulator cells stably transfected with Ly49i3,-s3,-i4 or-s4. The response could be blocked with mAb DAR13 binding these four Ly49 receptors. As isotype control (Ctr.) was used mAb FLY5. (D) Comparison of wild-type (BWZ.s3) and point-mutated Ly49s3 reporter (Ser85Cys in the stem region; BWZ.s3-S85C) against YB2/0.RT1-Un (YB.Un) stimulator cells. (E) Comparison of a reciprocal RT1-Un reporter cell against 293T stimulator cells transfected with wild-type Ly49s3 (s3) and the point-mutated variant (Ser85Cys in the stem region; s3-S85C). Data are representative of at least 3 independent experiments, *p<0.01.
The failure of the Ly49s3 reporter to react with the RT1-Un stimulator cell was unexpected since we previously showed that the surface expression of this receptor is strongly down-modulated in n haplotype rats, suggesting ligand-induced Ly49s3 receptor downregulation in this haplotype (19). We repeated these experiments with a separate GFP reporter cell line yielding similar results (data not shown), so this negative Ly49s3 triggering result was not due to a signalling defect in BWZ reporter cells. We also tested a reciprocal reporter expressing a chimeric RT1-Un/CD3zeta receptor together with rat β2m against Ly49 transfected 293T cells as stimulator cells (Suppl. Fig. 1A and C). Reciprocal reporters can be informative about successful receptor-ligand interactions per se, but not of functional consequences of such interactions in NK cells. Consistent with the Ly49 reporter experiments, the BWZ.RT1-Un (BWZ-Un) ligand reporter specifically reacted with Ly49s4, Ly49i4, and Ly49i3, but not Ly49s3 stimulator cells (Fig. 3C).
We also performed limited site-directed mutagenesis of Ly49s3 by converting Ser in the stem region to the consensus Cys presumably involved in homodimerization of Ly49 molecules. Interestingly, this Ser85Cys Ly49s3 reporter displayed a weak response to RT1-Un stimulator cells, but not against other MHC transfectants tested (Fig. 3D and data not shown). This interaction was confirmed with the reverse BWZ.Un reporter (Fig 3E). These results suggest that Ly49s3 has an inherent capacity for MHC binding, consistent with the in vivo calibration effect mentioned above, but that the serine in the stem region of Ly49s3 likely lowers the MHC binding affinity out of the range of detection in our reporter assays.
Ly49s4, Ly49i4, and Ly49i3 react with all allelic variants of RT1-U tested; specificity is consistent with “gain and loss” variation in different haplotypes
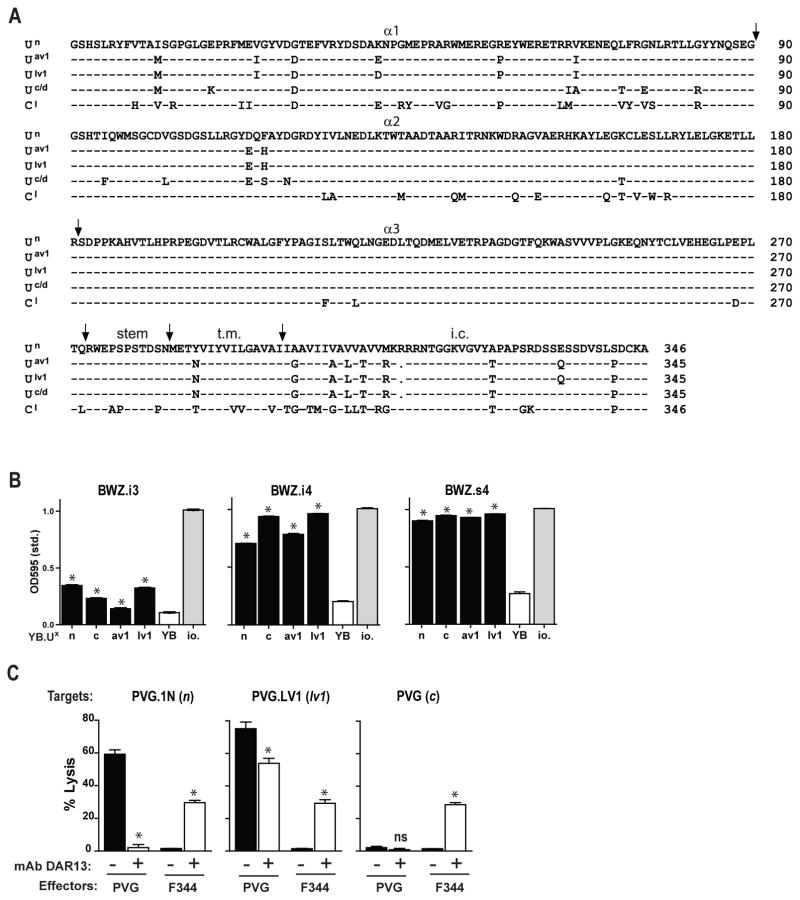
The RT1-U molecule is present in a relatively broad range of haplotypes and displays considerable allelic polymorphisms that can be discriminated by alloantibodies and alloreactive T cells (27). To test whether the Ly49s4/-i4/-i3 receptors could distinguish between allelic variants of RT1-U, we isolated and stably expressed RT1-U molecules from four additional haplotypes encoding three different variants at the protein level (Fig. 4A and Suppl. Fig. 1B). We were unable to isolate RT1-U transcripts from the two NK ligand-negative MHC haplotypes u or l (refer to Fig. 2A), suggesting that the RT1-U locus is either non-functional or deleted in these two haplotypes. This conclusion is in line with the previous finding that u or l cells do not stain with a panel of RT1-U specific alloantibodies (27) and supports a “gain and loss” paradigm of genomic plasticity within this part of the rat MHC (28).
FIGURE 4.
All cloned variants of RT1-U functions as ligands for Ly49s4/-i4/-i3. (A) Amino acid sequence of the different allelic variants of RT1-U used in this study, i.e. Un (corresponds with CE16 in the n haplotype), Uav1, Ulv1, and Uc/d and RT1-Cl. (B) Testing of the response of Ly49s4/-i4/-i3 reporter cells against YB2/0 stimulator cells stably transfected with the different RT1-U alleles. As negative and positive controls are shown reporters incubated with wild-type YB2/0 cells (YB) or with Ionomycin (io.) (C) Cytolytic activity of DAR13+ IL-2 activated NK cells generated from spleens of normal PVG and F344 rats was tested against Con A lymphoblast targets from the MHC congenic strains PVG.1N (n), PVG.LV1 (lv1), and PVG (c), in the presence of blocking quantities of mAb DAR13 (+) or the isotype control mAb FLY5 (-). Results are only shown for the effector:target cell ratio of 50:1. Data are representative of at least 3 independent experiments, *p<0.01.
The Ly49s4 and Ly49i4 reporters displayed brisk responses against YB2/0 cells expressing either RT1-U variant, i.e. Un, Uav1, Ulv1, and Uc. Ly49i3 reporter cells showed weaker responses (Fig. 4B). It is unknown whether these ligands are encoded by a single locus or by two duplicated loci (U1 and U2) (27), but the former is more likely since we have not been able to isolate more than one variant from any single haplotype (data not shown).
We then investigated the functions of the stimulatory Ly49s3/-s4 and the inhibitory Ly49i3/-i4 receptors in IL-2 activated splenic NK cells from two rat strains with divergent NK gene complexes. The function of the inhibitory variants was assessed in the F344 strain, in which the activating variants are either missing (Ly49s3) or mutated to non-function (Ly49s4) (11,19). As shown in Fig. 4C, Ly49i3/i4+ NK cells from F344 (lv1) rats failed to react with Con A blast target cells of the n, lv1, and c haplotypes, but cytolysis was induced by addition of the anti-Ly49s3 mAb DAR13, which also binds the Ly49s4/-i4/-i3 receptors (19). This is consistent with the “missing self” hypothesis and blocking of an inhibitory signal mediated by Ly49i3/i4 in F344 NK cells. By contrast, PVG (c) rat NK cells, which also express the activating Ly49s3 and Ly49s4 receptors, showed brisk lysis of allogeneic n and lv1 targets, while sparing syngeneic c targets. Cytolysis was partially or completely blocked by the addition of mAb DAR13 in line with previous data (18). Ligand-negative u targets were effectively lysed by both the F344 and PVG effector cells, and the addition of mAb DAR13 had no significant effect (data not shown).
Ly49s5 and Ly49i5 react with the class Ib molecule RT1-Eu
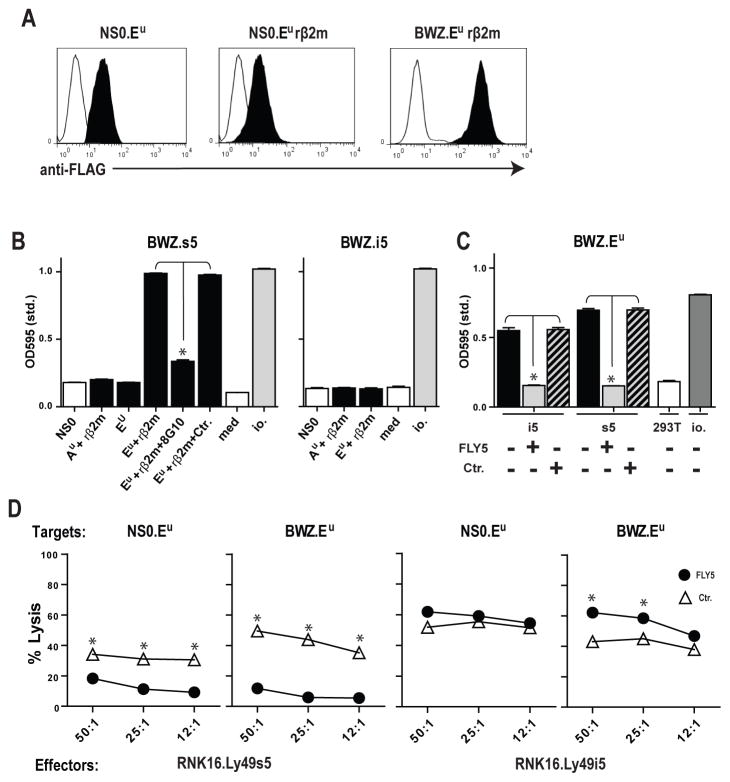
The RT1-Eu class Ib molecule resembles RT1-U in that it is class Ia-like and is encoded within the first telomeric class Ib cluster, RT1-CE. It has been identified as an activating MHC-ligand for rat NK cells, but its cognate receptor still awaits identification (17,20). Previous genomic data have shown that the u haplotype is divergent from the reference n haplotype in the proximal part of the RT1-CE cluster, with no class I genes detected within 20-kb of the Bat1 gene where CE1 is located in the n haplotype (28). The rest of the RT1-CE cluster is most likely very different, as we have not been able to clone RT1-U or other functional RT1-CEn genes from u strain cells (data not shown). We stably expressed RT1-Eu in NSO mouse myeloma cells in the presence or absence of rat β2m (Fig. 5A). Ly49s5 reporter cells reacted specifically with RT1-Eu stimulator cells in a rat β2m-dependent manner, while the class Ia molecule RT1-Au was not recognized. The Ly49s5-RT1-Eu reaction was blocked by the addition of the anti-RT1-Eu mAb 8G10 (Fig. 5B). The Ly49i5 reporter failed to respond to RT1-Eu stimulator cells. This was not because Ly49i5 fails to recognize RT1-Eu, but rather the activation threshold of the reporter was not reached. A reciprocal BWZ.Eu reporter, in which the ζ-chain coupled RT1-Eu molecule is expressed at a high level on the cell surface (Fig. 5A), showed brisk responses to both Ly49s5 and Ly49i5 transfected 293T stimulator cells, and these responses could be blocked by addition of the anti-Ly49s5/-i5 mAb FLY5 (Fig. 5C).
FIGURE 5.
RT1-Eu is a ligand for the Ly49s5/-i5 pair of structurally related receptors. (A) Single-color flow staining with anti-FLAG mAb M2 of stable RT1-Eu transfectants in NSO with and without rat β2m (rβ2m) and BWZ with rat β2m. (B) Testing of the response of the Ly49s5/-i5 reporter cells against mouse myeloma NSO cells stably transfected with the class Ia molecule RT1-Au (Au) and class Ib molecule RT1-Eu (Eu) in the presence (+ β2m) or absence of rat β2m. The response of the Ly49s5 reporter against Eu together with rat β2m was blocked by addition of the anti-Eu mAb 8G10, but not a control mAb (AAS5). (C) Testing of a reciprocal RT1-Eu reporter cell (BWZ.Eu) against 293T stimulator cells stably transfected with Ly49s5 or -i5. The response could be blocked with mAb FLY5 reacting with these two Ly49 receptors. As isotype control (Ctr.) was used mAb DAR13. (D) Cytolytic activity of RNK cells stably transfected with Ly49i5 (RNK.Ly49i5) and Ly49s5 (RNK.Ly49s5) against Eu transfected NSO or BWZ target cells in a 51Chromium release assay, in the presence of blocking quantities of anti-Ly49s5/-i5 mAb FLY5 or the isotype control (Ctr.) mAb DAR13. Data are representative of at least 3 independent experiments, *p<0.01.
The function of the Ly49s5/-i5 receptors in NK cells was assessed by stable transfection of the RNK-16 NK cell line (Suppl. Fig. 1C). In line with the reporter data, RNK-16.Ly49s5 cells efficiently killed both NSO and BWZ cells expressing RT1-Eu together with rat β2m, and lysis could be blocked by addition of mAb FLY5. By contrast, RT1-Eu mediated weak inhibition of RNK-16.Ly49i5 cells, but only when it was expressed at high levels in the BWZ line (Fig. 5D). It should be noted that this inhibition was considerably weaker than that previously observed using IL-2 activated splenic Ly49i5+ PVG NK cells and u haplotype Con A blasts as target cells (20), again indicating that the reporter system and the RNK-16 assay have limitations in their sensitivities.
Induction of highly alloreactive Ly49+ NK cell subsets in vivo by immunization with normal allogeneic cells
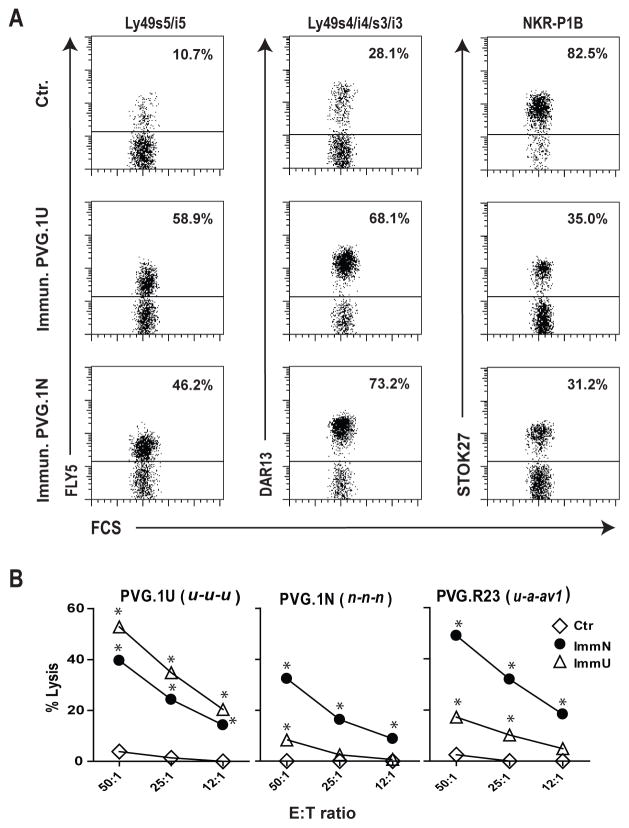
The activation potential of Ly49s receptors in vivo was evaluated by their ability to recruit and/or expand alloreactive NK cell subsets in the peritoneal cavity upon repeated alloimmunizations. Inbred athymic nude rats (c) were immunized once weekly (four in total) with mononuclear splenocytes from MHC allogeneic strains expressing either the n or u haplotypes. Peritoneal exudate cells were retrieved one week after the last injection. Following alloimmunization we observed a massive recruitment of leukocytes including NK cells to the peritoneum (data not shown). Immunization with u and n cells led to a preferential expansion of FLY5+ (anti-Ly49s5/-i5) and DAR13+ (anti-Ly49s4/-i4/-s3/-i3) NK cells, respectively, although both subsets were heavily expanded by either stimulus compared to control rats receiving PBS (Fig. 6A). Ly49i2, which is an inhibitory receptor for a class Ia molecule (RT1-A1c) in the strain used for immunization (29,30), was also markedly expanded (data not shown). The marked co-clustering of different Ly49 receptors following alloimmunization was expected since rat NK cells express more than one Ly49 receptor and most Ly49 receptors are expressed within a common subpopulation of NK cells lacking the NKR-P1B receptor (31,32). Concordantly, the NKR-P1B+ subset (relative to the Ly49+ subset) contracted following alloimmunization (Fig. 6A). The in vivo skewing of NK subsets appeared to be limited to the peritoneal cavity, as no skewing effects were observed among splenic NK cells (data not shown).
FIGURE 6.
Specific adaptation of the NK cell repertoire to repeated alloimmunizations in vivo. (A) Congenitally athymic nude rats (c haplotype) were immunized once weekly for a total of four weeks i.p. with mononuclear splenocytes from PVG.1U (u) or PVG.1N (n) rats. Nude rats receiving PBS only were used as negative controls (Ctr.). Peritoneal cells were retrieved one week after the last injection and analysed by flow cytometry, gating on CD3−NKR-P1A+ NK cells. Data are representative of 4 experiments, the percentage of positive cells are given. (B) Cytolytic activity of ex vivo peritoneal cells from nude rats immunized with PVG.1U (ImmU) or PVG.1N (ImmN) or non-immunized nude controls (Ctr.) against lymphoblast target cells from PVG.1U (u), PVG.1N (n) and PVG.R23 (u-a-av1) rats. Data are representative of at least 3 independent experiments, *p<0.01.
The in vivo induced NK cells were also tested for cytotoxicity against normal lymphoblast targets from informative donor strains. Cytolytic activity against u target cells was strongly enhanced following immunization with u cells, and much more so than against third party n targets or the intra-MHC recombinant u-a-av1 (RT1-Au-B/Da-CEav1) which expresses u in the RT1-A region. This suggests that this response was not directed to a classical class Ia molecule (compare the three panels of Fig. 6B). Conversely, NK alloreactivities against targets from haplotypes n, u, and intra-MHC congenic u-a-av1 lymphoblasts were all strongly enhanced upon immunization with n cells (Fig. 6B). Taken together, these results suggest that the observed NK alloresponses were directed against RT1-CE class Ib ligands, and that the key components of these responses may vary considerably based upon the Ly49 receptor repertoire of the induced NK cells.
Induction of highly alloreactive Ly49s4/-i4/-s3/-i3+ NK cells in vivo by repeated allostimulation in vivo with RT1-Un expressing cells
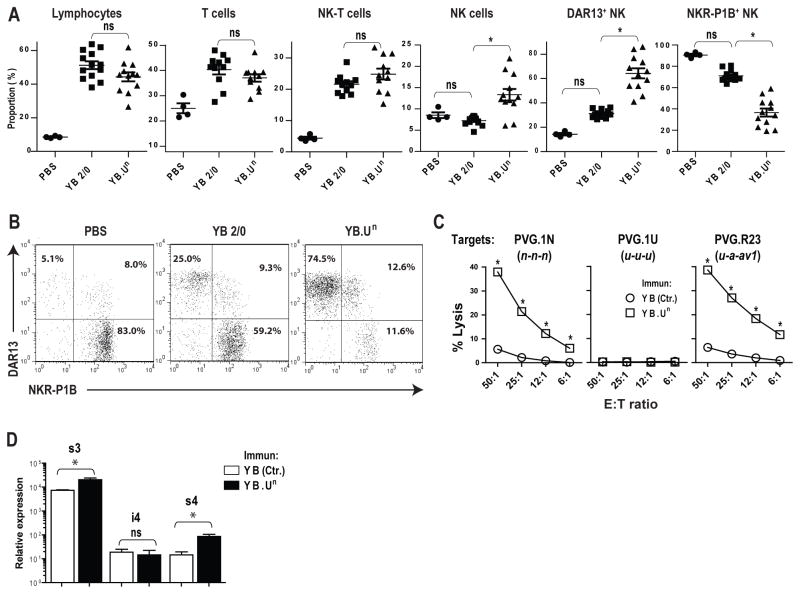
To investigate in detail single receptor-ligand interaction(s) in the induction of alloreactive NK cells in vivo, we immunized the RT1-U negative strain PVG.1U with an RT1-Un transfected cell line YB2/0 (YB.Un). This line is identical with the recipient strain PVG.1U in the MHC (both u haplotype), thus avoiding T cell alloresponses against major class Ia/ class II mismatches which might potentially override the stimulatory effects on NK cells. Repeated i.p. immunizations led to a marked accumulation of NK cells compared to immunization with YB2.0 wild-type cells or PBS only. There was a marked expansion of DAR13+ (Ly49s4/-i4/-s3/-i3) NK cells and a comparable contraction of NKR-P1B+ NK cells (Fig. 7A and B). T cells and NK-T cells were similarly expanded by both the YB.Un and the YB2/0 control cells (Fig. 7A).
FIGURE 7.
Allostimulation with an RT1-Un transfectant induces highly alloreactive Ly49s4/-i4/-s3/-i3+ NK cells in vivo. (A) PVG.1U rats were immunized once weekly for a total of four weeks i.p. with RT1-Un transfected YB2/0 cells (YB.Un), wild-type YB2/0 cells (YB2/0) or PBS only. Proportions of lymphocytes, T cells (CD3+), NK-T cells (CD3+NKR-P1+), NK cells (CD3−NKR-P1A+), DAR13+ NK cells and NKR-P1B+ NK cells were evaluated by flow cytometry, data for individual rats (n=4–13) are given for each group, *p<0.01. (B) Two-color plots showing the two dominant DAR13+ and NKR-P1B+ NK subsets in the three experimental groups. (C) Cytolytic activity of ex vivo peritoneal cells from immunized (YB.Un) versus control (Ctr.) PVG.1U rats against Con A-activated lymphoblast target cells from PVG.1N (n), PVG.R23 (u-a-av1), or syngeneic PVG.1U (u) rats. Data are representative of at least 3 independent experiments, *p<0.01 (D) qPCR of Ly49s4, -i4 and -s3 expression among DAR13+ sorted NK cells from PVG.1U rats immunized with transfected YB.Un cells compared to untransfected YB2/0 cells as control (YB Ctr.), *p<0.05.
The YB.Un stimulator cells induced a strong NK cell cytolytic alloresponse as shown in Fig. 7C. The peritoneal NK cells efficiently killed RT1-Un expressing PVG.1N (n) lymphoblasts, while sparing syngeneic PVG.1U (u) targets. Target cells from the intra-MHC recombinant strain PVG.R23 (u-a-av1) were also effectively lysed, consistent with recognition of both the n and av1 alleles of RT1-U (refer to Fig. 4B).
We predicted that RT1-Un allostimulation would induce a preferential expansion of Ly49s4/-s3 and not Ly49i4/-i3 expressing cells. This would parallel our previous observation that the inhibitory receptor Ly49i5 leads to contraction of the Ly49i5+ NK subset upon allostimulation of BN strain rats with cells expressing its cognate Eu ligand (20). It is not possible to distinguish between the Ly49s4/-i4/-s3/-i3 receptors with our current mAbs, so we performed a quantitative PCR to measure their relative contribution to the receptor pool among DAR13+ cells from immunized rats. Interestingly, transcripts for the two stimulatory variants (Ly49s3 and Ly49s4) increased following stimulation with YB.Un as compared to YB2/0 control cells, and transcripts for Ly49i4 diminished slightly (Fig. 7D). We were unable to assess Ly49i3, as we have never been able to amplify Ly49i3 from PVG strain rats (data not shown), suggesting that this gene is either non-functional or missing in the PVG NKC genome. These data show that there is a specific increase in Ly49s4 and Ly49s3 transcripts as a result of immunization with RT1-Un.
The class Ib molecule RT1-Cl is a ligand for the Ly49s4/-i4/-s3/-i3 group of receptors
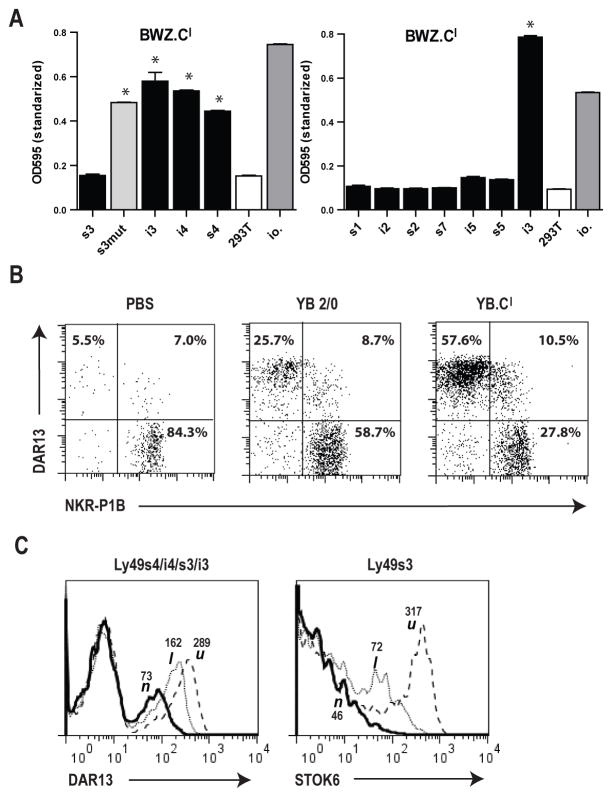
Early studies of the LEW.1LM1 mutant strain with a genomic deletion in the RT1-CE cluster (including the polymorphic RT1-Cl molecule) (6), prompted us to investigate RT1-Cl as an alloactivating ligand. Our panel of Ly49 reporter cells failed to trigger against l haplotype cells (Fig. 1), from which the LEW.1LM1 mutant is derived. To increase the sensitivity, we generated an RT1-Cl reverse reporter in the BWZ line overexpressing ζ-chain coupled RT1-Cl and tested it against a panel of Ly49 transfected 293T stimulator cells (the activating Ly49 variants co-transfected with DAP12 in order to facilitate receptor expression). As shown in Fig. 8A, the BWZ.Cl reporter showed brisk responses against Ly49i3, -i4, -s4, and the –s3 Ser85Cys mutant, whereas several other Ly49 receptors including native Ly49s3 were negative. To verify that this result is not due to reporter artifact, we investigated whether RT1-Cl transfected target cells could selectively expand relevant NK subsets in vivo. This was indeed the case, as RT1-Cl negative PVG1.U rats repeatedly immunized i.p. with YB.Cl cells showed a marked expansion and contraction, respectively, of DAR13+ and NKR-P1B+ subsets compared with rats injected with YB2/0 control cells (Fig. 8B). Furthermore, the Ly49s4/-i4/-s3/-i3 group of receptors were down-modulated in NK cells from l compared with u haplotype rats, as detected by reduced staining with the anti-s4/-i4/-s3/-i3 mAb DAR13 (Fig. 8C), a clear indication of the presence of cognate ligand(s) in the l haplotype (18,19). Using the Ly49s3 specific mAb STOK6, we were able to show that even native Ly49s3 was downmodulated in l haplotype strains (Fig. 8C). These experiments are a further indication that in vitro assays may not always be sufficiently sensitive to detect some relevant in vivo receptor-ligand interactions. Based on these data and the aforementioned LEW.1LM1 class Ib deletion data, it is likely that the haplotype 1 encoded alloactivating ligand is indeed RT1-Cl. It should also be noted that Ly49s4/-i4/-s3/-i3 down-modulation was particularly pronounced in n haplotype NK cells (Fig. 8C), in agreement with previous studies (18,19). As can be deduced from Supplemental Fig. 1B, its cognate ligand RT1-Un is expressed at particularly high levels, leading to predictably stronger receptor down-modulation.
FIGURE 8.
Recognition of RT1-Cl by the Ly49s3/-s4/-i3/-i4 group of receptors. (A) Testing of a reciprocal BWZ.RT1-Cl reporter cell against 293T stimulator cells stably transfected with various Ly49 receptors (the activating variants cotransfected with DAP12). Data are representative of at least 3 independent experiments, *p<0.01 (B) PVG.1U rats were immunized once weekly for a total of four weeks i.p. with RT1-Cl transfected YB2/0 cells (YB.Cl), wild-type YB2/0 cells or PBS only. Two-color plots show the distribution of the two dominant Ly49s3+ vs. the NKR-P1B+ NK subsets. (C) Down-modulation of the Ly49s3/-s4/-i3/-i4 receptors on ex vivo splenic NK cells in n and l, but not the u haplotype rats as detected with the anti-s3/-s4/-i3/-i4 mAb DAR13 and the Ly49s3-specific alloantibody STOK6, MFI values are given.
Discussion
Unlike inhibitory mouse Ly49 receptors, which primarily bind classical MHC Ia molecules, we show here that activating rat Ly49 receptors react with three different polymorphic nonclassical class Ib RT1-CE proteins, RT1-U (also termed CE16 in the reference n haplotype), RT1-Eu, and RT1-Cl. These ligands are encoded from the first telomeric class Ib gene cluster, RT1-CE, which shares structural features with classical class Ia genes, RT1-A, and are located in the same position as H2-D/-L/-Q in the mouse. Expansion of activating receptors for class Ib encoded ligands may have driven the parallel evolution of inhibitory receptors for similar ligands such as the inhibitory receptors Ly49i3, -i4, and –i5 which bind the same RT1-CE ligands as their activating counterparts. The Ly49s4/-i4/-s3/-i3 receptors react with RT1-Cl as well as with all known allelic variants of RT1–U present in a broad range of haplotypes (n, lv1, av1, c, and d). Ly49s5/-i5 are specific for RT1-Eu in the u haplotype. We also show that potent NK alloresponses can be induced in vivo by repeated alloimmunizations with either allogeneic cells or class Ib transfectants.
Early studies in the rat have suggested that alloreactive NK cells may have adaptive features. Alloimmunization of BN rats with spleen cells from WF (u) rats led to expansion of allospecific NK cells (33,34) that selectively killed target cells derived from the immunizing rat strain. An increased percentage of activated NK cells was observed both in the peritoneum and spleen of i.p. immunized BN rats. This was caused by an increased proliferation of NK cells at both sites (35). The heightened NK alloresponse in BN rats was directed against the nonclassical MHC I molecule RT1-Eu (17) as shown by in vitro cytotoxic assays. This finding was in accordance with our early data showing that rat NK cell alloreactivity is controlled mainly by molecules encoded within the nonclassical MHC class Ib region (6). In this study, we show that the activating Ly49s5 receptor recognizes RT1-Eu and may potently induce in vivo alloresponses in PVG rats. This receptor, however, is nonfunctional in BN rats, suggesting that another activating Ly49 receptor, yet to be defined, is responsible for previously described adaptive NK alloresponse in the BN strain (20). We also show that Ly49s4, and possibly Ly49s3, recognize the MHC class Ib molecule RT1-U and evoke potent in vivo responses in PVG rats. Alloimmunization of PVG rats with cells expressing RT1-Eu or RT1-U led to expansion and enhanced cytotoxicity of the Ly49s5 and Ly49s3/s4 positive NK cell subsets, respectively. In contrast to the aforementioned experiments in BN rats, we only observed expansion of peritoneal, not splenic alloreactive NK cells in PVG rats, suggesting that the immunization-induced expansion of alloreactive NK cells may be more localized in PVG rats. In a separate set of experiments, we have been able to confirm marked skewing of the NK subset distribution in the spleen upon i.p. alloimmunization of BN strain rats (L. Kveberg and J.T. Vaage unpublished data), in line with the original data referred to above (17,34,35). Together, these findings show that activating Ly49 receptors can play an important role in shaping the NK cell repertoire and may induce strong in vivo responses with some adaptive features. The mouse activating Ly49H receptor has been shown to evoke adaptive NK cell responses during mouse CMV infection in vivo (36). After an expansion and contraction phase, memory Ly49H positive NK cells reside in the animals for months and can mediate specific protective immunity (36). Expansion and differentiation of Ly49D+ mouse NK cells have also been studied upon alloantigen stimulation of mice (37). These studies showed that activating Ly49 receptors have the capacity to induce adaptive immune-responses in mouse NK cells. The adaptive features of alloreactive rat NK cells resemble those of mouse Ly49H and Ly49D responses after CMV or alloantigen challenge. We do not know how long these enhanced responses persist, however, and at present we cannot assume that activating NK alloresponses exhibit strict memory features in the rat.
Other evolutionary pressures are evident from an analysis of ligand specificities of MHC-binding receptors in rats. Not all activating rat Ly49 receptors have identifiable RT1-CE-encoded ligands. Despite its structural similarity to Ly49s4/-i4/-i3, a single amino acid difference in the extracellular domain of Ly49s3 (compared with Ly49s4) appears to significantly reduce or abrogate the binding of this receptor to RT1-encoded ligands in vitro. This might, however, be an in vitro artifact as the expression of Ly49s3 is clearly down-modulated in the presence of its ligand in vivo (19) suggesting that Ly49s3 is functional in vivo. The functional attenuation of stimulatory innate immune receptors against self MHC might be evolutionarily advantageous, as some high affinity activating self-receptors may promote excessive inflammation, autoimmunity, or anergy due to immune exhaustion.
A careful analysis or our data and previously published studies suggests that the binding of rat and mouse Ly49 receptors to their respective MHC class I ligands is controlled by orthologous structural constraints. The first co-crystallization studies of mouse Ly49A and its ligand H-2Dd showed that Ly49 molecules had two potential interaction sites with MHC class I molecules. Site 1 was found at one end of the peptide-binding groove; while site 2 was below the peptide-binding groove were the Ly49 molecule makes contact with the α1/α2, α3 and β2m-domains. Later studies have shown site 2 to be most important. Studies in the rat with Ly49i2 and its ligand RT1-A1c have supported this view. It has been shown that the P2 anchor amino acid (Pro/Val) and its associated B pocket are especially important. It has been proposed that changes here alter the conformation of solvent exposed residues at site 2 important for Ly49 binding (38,39). Interestingly, when we generated RT1-Eu transfectants in mouse NS0 cells expressing the mouse β2m, target cells were not recognized by BWZ Ly49s5 reporter cells. But when NS0.Eu cells co-expressed rat β2m, they were recognized by Ly49s5 (Fig. 5B), despite the fact that both transfectants expressed similar levels of RT1-Eu. The requirement for rat β2m was confirmed in the RT1-U interactions, when NS0-RT1-Un transfectants without rat β2m either failed or were recognized to a much lower degree by reporter cells (Ly49s4/i4/i3, data not shown). These findings support the notion that site 2 is important for optimal binding of rat Ly49 receptors, but these interactions are likely sensitive to minimal differences in ligand structure
All rat Ly49 receptors that have been functionally characterized to date belong to the same phylogenetic clade, which is significantly expanded within the second cluster of rat Ly49 genes (11). This cluster includes the Ly49s4/-i4 /-i3 and Ly49s5/-i5 molecules, as well as the inhibitory Ly49i2 receptor. Ly49s3 likely also maps to this cluster, but this gene is missing in the n haplotype. Two additional activating receptors in this cluster Ly49s2 and –s6, as well as the activating receptors Ly49s1, -s7 and –s8 in another phylogenetic clade remain functionally uncharacterized, and their ligands are not known. In contrast to activating KIR in humans and Ly49 in mice, our data suggest that the recognition of nonclassical RT1-CE-encoded ligands by activating Ly49 receptors may be a generalizable feature of rat NK cells. Moreover, our reporter data demonstrate that reporter activation by RT1-CE proteins is exquisitely sensitive to changes in ligand density on target cells and to subtle structural changes in specific receptors or their ligands. The telomeric part of the rat MHC contains several clusters of class Ib genes in addition to RT1-CE, with RT1-N being homologous to H2-T and RT1-M with H2-M in mice. In this context, it is interesting that the mouse class Ib molecules H2-Q10 and H2-M3 function as ligands for the Ly49C and Ly49A inhibitory receptors, respectively (40,41). The mouse MHC does not contain formal orthologs of the RT1-CE genes located in the first class Ib gene cluster (refer to Fig. 1B), and this cluster does not contain direct H2-D/L orthologs. The number of RT1-CE genes varies remarkably between different rat MHC haplotypes. The NK ligands defined here, RT1-U, -Eu, and -Cl, however, display some class Ia like structural features and contribute significantly to haplotype diversity (28). Whereas polymorphic rat RT1-A molecules and mouse H-2K/D/L molecules are expressed on all nucleated cells, nonclassical class Ib proteins are normally expressed at low levels and do not typically serve as restriction elements for cytotoxic T cells (14,27). Therefore, the rat RT1-CE cluster contains an array of polymorphic nonclassical MHC molecules with unique features. Although they are normally expressed at low levels, their expression may be upregulated by inflammatory cytokines or by specific pathologic conditions. We propose that rats have selectively co-evolved an expanded array of activating Ly49 receptors and class Ib RT1-CE ligands, which together have evolved to serve as a uniquely calibrated immune sensing system for inflamed, infected, or otherwise abnormal target cells.
Supplementary Material
Abbreviations
- NKC
NK gene complex
- RT1
rat MHC complex
Footnotes
This work was supported by the Norwegian Cancer Society and the South-Eastern Norway Regional Health Authority. KZD received a Fellowship from the Norwegian Cancer Society during the early phase of this project. JCR received support from NIH RO1 AI 083113, NIH P30 DK 02673 (UCSF Liver Center), from the US Deparment of Veterans Affairs, and the United States Veterans Administration.
Bent Rolstad passed away in 2015 but he was involved and very enthusiastic about this project, which addressed receptor-ligand interactions explaining early NK findings of allogeneic lymphocyte cytotoxicity dating back to the seventies. He was an inspirational leader and is deeply missed.
Reference List
- 1.Ljunggren HG, Kärre K. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol Today. 1990;11:237–244. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 2.Rahim MM, Makrigiannis AP. Ly49 receptors: evolution, genetic diversity, and impact on immunity. Immunol Rev. 2015;267:137–147. doi: 10.1111/imr.12318. [DOI] [PubMed] [Google Scholar]
- 3.Guethlein LA, Norman PJ, Hilton HG, Parham P. Co-evolution of MHC class I and variable NK cell receptors in placental mammals. Immunol Rev. 2015;267:259–282. doi: 10.1111/imr.12326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomasello E, Olcese L, Vely F, Geourgeon C, Blery M, Moqrich A, Gautheret D, Djabali M, Mattei MG, Vivier E. Gene structure, expression pattern, and biological activity of mouse killer cell activating receptor-associated protein (KARAP)/DAP-12. J Biol Chem. 1998;273:34115–34119. doi: 10.1074/jbc.273.51.34115. [DOI] [PubMed] [Google Scholar]
- 5.Lanier LL, Corliss BC, Wu J, Leong C, Phillips JH. Immunoreceptor DAP12 bearing a tyrosine-based activation motif is involved in activating NK cells [see comments] Nature. 1998;391:703–707. doi: 10.1038/35642. [DOI] [PubMed] [Google Scholar]
- 6.Vaage JT, Naper C, Løvik G, Lambracht D, Rehm A, Hedrich HJ, Wonigeit K, Rolstad B. Control of rat natural killer cell-mediated allorecognition by a major histocompatibility complex region encoding nonclassical class I antigens. J Exp Med. 1994;180:641–651. doi: 10.1084/jem.180.2.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rolstad B. The early days of NK cells: an example of how a phenomenon led to detection of a novel immune receptor system - lessons from a rat model. Front Immunol. 2014;5:283. doi: 10.3389/fimmu.2014.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rolstad B, Ford WL. The rapid elimination of allogeneic lymphocytes: relationship to established mechanisms of immunity and to lymphocyte traffic. Immunol Rev. 1983;73:87–113. doi: 10.1111/j.1600-065x.1983.tb01080.x. [DOI] [PubMed] [Google Scholar]
- 9.Dissen E, Ryan JC, Seaman WE, Fossum S. An autosomal dominant locus, Nka, mapping to the Ly-49 region of a rat natural killer (NK) gene complex, controls NK cell lysis of allogeneic lymphocytes. J Exp Med. 1996;183:2197–2207. doi: 10.1084/jem.183.5.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abi-Rached L, Parham P. Natural selection drives recurrent formation of activating killer cell immunoglobulin-like receptor and Ly49 from inhibitory homologues. J Exp Med. 2005;201:1319–1332. doi: 10.1084/jem.20042558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nylenna Ø, Naper C, Vaage JT, Woon PY, Gauguier D, Dissen E, Ryan JC, Fossum S. The genes and gene organization of the Ly49 region of the rat natural killer cell gene complex. Eur J Immunol. 2005;35:261–272. doi: 10.1002/eji.200425429. [DOI] [PubMed] [Google Scholar]
- 12.Hurt P, Walter L, Sudbrak R, Klages S, Müller I, Shiina T, Inoko H, Lehrach H, Günther E, Reinhardt R, Himmelbauer H. The genomic sequence and comparative analysis of the rat major histocompatibility complex. Genome Res. 2004;14:631–639. doi: 10.1101/gr.1987704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang C-R, Livingstone A, Butcher GW, Hermel E, Howard JC, Lindahl KF. Antigen presentation by neoclassical MHC class I gene products in murine rodents. In: Klein J, Klein D, editors. NATO ASI Series, Vol. H 59. Molecular evolution of the major histocompatibility complex. Springer-Verlag; Berlin: 1991. pp. 441–462. [Google Scholar]
- 14.Günther E, Wurst W. Cytotoxic T lymphocytes of the rat are predominantly restricted by RT1.A and not RT1.C-determined major histocompatibility class I antigens. Immunogenetics. 1984;20:1. doi: 10.1007/BF00373442. [DOI] [PubMed] [Google Scholar]
- 15.Shegarfi H, Dai KZ, Daws MR, Ryan JC, Vaage JT, Rolstad B, Naper C. The rat NK cell receptors Ly49s4 and Ly49i4 recognize nonclassical MHC-I molecules on Listeria monocytogenes-infected macrophages. J Leukoc Biol. 2011;89:617–623. doi: 10.1189/jlb.1010593. [DOI] [PubMed] [Google Scholar]
- 16.Shegarfi H, Dai KZ, Inngjerdingen M, Ryan JC, Vaage JT, Rolstad B, Naper C. The activating rat Ly49s5 receptor responds to increased levels of MHC class Ib molecules on Listeria monocytogenes-infected enteric epithelial cells. Eur J Immunol. 2010;40:3535–3543. doi: 10.1002/eji.201040651. [DOI] [PubMed] [Google Scholar]
- 17.Petersson E, Holmdahl R, Butcher GW, Hedlund G. Activation and selection of NK cells via recognition of an allogeneic, non-classical MHC class I molecule, RT1-E. Eur J Immunol. 1999;29:3663–3673. doi: 10.1002/(SICI)1521-4141(199911)29:11<3663::AID-IMMU3663>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 18.Naper C, Hayashi S, Kveberg L, Niemi EC, Lanier LL, Vaage JT, Ryan JC. Ly-49s3 is a promiscuous activating rat NK cell receptor for nonclassical MHC class I-encoded target ligands. J Immunol. 2002;169:22–30. doi: 10.4049/jimmunol.169.1.22. [DOI] [PubMed] [Google Scholar]
- 19.Kveberg L, Dai KZ, Dissen E, Ryan JC, Rolstad B, Vaage JT, Naper C. Strain-dependent expression of four structurally related rat Ly49 receptors; correlation with NK gene complex haplotype and NK alloreactivity. Immunogenetics. 2006;58:905–916. doi: 10.1007/s00251-006-0154-x. [DOI] [PubMed] [Google Scholar]
- 20.Naper C, Dai KZ, Kveberg L, Rolstad B, Niemi EC, Vaage JT, Ryan JC. Two structurally related rat Ly49 receptors with opposing functions (Ly49 stimulatory receptor 5 and Ly49 inhibitory receptor 5) recognize nonclassical MHC class Ib-encoded target ligands. J Immunol. 2005;174:2702–2711. doi: 10.4049/jimmunol.174.5.2702. [DOI] [PubMed] [Google Scholar]
- 21.Naper C, Hayashi S, Joly E, Butcher GW, Rolstad B, Vaage JT, Ryan JC. Ly49i2 is an inhibitory rat natural killer cell receptor for an MHC class Ia molecule (RT1-A1c) Eur J Immunol. 2002;32:2031–2036. doi: 10.1002/1521-4141(200207)32:7<2031::AID-IMMU2031>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 22.Naper C, Hayashi S, Løvik G, Kveberg L, Niemi EC, Rolstad B, Dissen E, Ryan JC, Vaage JT. Characterization of a novel killer cell lectin-like receptor (KLRH1) expressed by alloreactive rat NK cells. J Immunol. 2002;168:5147–5154. doi: 10.4049/jimmunol.168.10.5147. [DOI] [PubMed] [Google Scholar]
- 23.Westgaard IH, Berg SF, Vaage JT, Wang LL, Yokoyama WM, Dissen E, Fossum S. Rat NKp46 activates natural killer cell cytotoxicity and is associated with FcepsilonRIgamma and CD3zeta. J Leukoc Biol. 2004;76:1200–1206. doi: 10.1189/jlb.0903428. [DOI] [PubMed] [Google Scholar]
- 24.Kveberg L, Back CJ, Dai KZ, Inngjerdingen M, Rolstad B, Ryan JC, Vaage JT, Naper C. The novel inhibitory NKR-P1C receptor and Ly49s3 identify two complementary, functionally distinct NK cell subsets in rats. J Immunol. 2006;176:4133–4140. doi: 10.4049/jimmunol.176.7.4133. [DOI] [PubMed] [Google Scholar]
- 25.de FP, Ryan MD. Targeting of proteins derived from self-processing polyproteins containing multiple signal sequences. Traffic. 2004;5:616–626. doi: 10.1111/j.1398-9219.2004.00205.x. [DOI] [PubMed] [Google Scholar]
- 26.Ryan JC, Niemi EC, Nakamura MC. Functional analysis of natural killer cell receptors in the RNK-16 rat leukemic cell line. Methods Mol Biol. 2000;121:283–295. doi: 10.1385/1-59259-044-6:283. [DOI] [PubMed] [Google Scholar]
- 27.Leong LYW, Le Rolle AF, Deverson EV, Powis SJ, Larkins AP, Vaage JT, Stokland A, Lambracht-Washington D, Rolstad B, Joly E, Butcher GW. RT1-U: Identification of a novel, active, class Ib alloantigen of the rat MHC. J Immunol. 1999;162:743–752. [PubMed] [Google Scholar]
- 28.Roos C, Walter L. Considerable haplotypic diversity in the RT1-CE class I gene region of the rat major histocompatibility complex. Immunogenetics. 2005;56:773. doi: 10.1007/s00251-004-0744-4. [DOI] [PubMed] [Google Scholar]
- 29.Naper C, Ryan JC, Kirsch R, Butcher GW, Rolstad B, Vaage JT. Genes in two major histocompatibility complex class I regions control selection, phenotype, and function of a rat Ly-49 natural killer cell subset. Eur J Immunol. 1999;29:2046–2053. doi: 10.1002/(SICI)1521-4141(199906)29:06<2046::AID-IMMU2046>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 30.Naper C, Hayashi S, Joly E, Butcher GW, Rolstad B, Vaage JT, Ryan JC. Ly49i2 is an inhibitory rat natural killer cell receptor for an MHC class Ia molecule (RT1-A1c) Eur J Immunol. 2002;32:2031–2036. doi: 10.1002/1521-4141(200207)32:7<2031::AID-IMMU2031>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 31.Kveberg L, Bäck CJ, Dai KZ, Inngjerdingen M, Rolstad B, Ryan JC, Vaage JT, Naper C. The novel inhibitory NKR-P1C receptor and Ly49s3 identify two complementary, functionally distinct NK cell subsets in rats. J Immunol. 2006;176:4133–4140. doi: 10.4049/jimmunol.176.7.4133. [DOI] [PubMed] [Google Scholar]
- 32.Kveberg L, Jiménez-Royo P, Naper C, Rolstad B, Butcher GW, Vaage JT, Inngjerdingen M. Two complementary rat NK cell subsets, Ly49s3+ and NKR-P1B+, differ in phenotypic characteristics and responsiveness to cytokines. J Leukoc Biol. 2010;88:87–93. doi: 10.1189/jlb.0110039. [DOI] [PubMed] [Google Scholar]
- 33.Hedlund G, Brodin T, Sjögren HO. Selective induction of OX19+ (CD5+) or OX19− (CD5−) alloreactive cytolytic lymphocytes in the rat. Cell Immunol. 1987;105:366–373. doi: 10.1016/0008-8749(87)90084-0. [DOI] [PubMed] [Google Scholar]
- 34.Ericsson PO, Hansson J, Dohlsten M, Sjögren HO, Hiserodt JC, Hedlund G. In vivo induced allo-reactive natural killer cells. J Immunol. 1992;149:1504–1509. [PubMed] [Google Scholar]
- 35.Petersson E, Hedlund G. Proliferation and differentiation of alloselective NK cells after alloimmunization - evidence for an adaptive NK response. Cell Immunol. 1999;197:10–18. doi: 10.1006/cimm.1999.1560. [DOI] [PubMed] [Google Scholar]
- 36.Sun JC, Beilke JN, Lanier LL. Adaptive immune features of natural killer cells. Nature. 2009;457:557–561. doi: 10.1038/nature07665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nabekura T, Lanier LL. Antigen-specific expansion and differentiation of natural killer cells by alloantigen stimulation. J Exp Med. 2014;211:2455–2465. doi: 10.1084/jem.20140798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lavender KJ, Kane KP. Cross-species dependence of Ly49 recognition on the supertype defining B-pocket of a class I MHC molecule. J Immunol. 2006;177:8578–8586. doi: 10.4049/jimmunol.177.12.8578. [DOI] [PubMed] [Google Scholar]
- 39.Ma BJ, Kane KP. Recognition of class I MHC by a rat Ly49 NK cell receptor is dependent on the identity of the P2 anchor amino acid of bound peptide. J Immunol. 2011;187:3267–3276. doi: 10.4049/jimmunol.1002809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sullivan LC, Berry R, Sosnin N, Widjaja JM, Deuss FA, Balaji GR, LaGruta NL, Mirams M, Trapani JA, Rossjohn J, Brooks AG, Andrews DM. Recognition of the Major Histocompatibility Complex (MHC) Class Ib Molecule H2-Q10 by the Natural Killer Cell Receptor Ly49C. J Biol Chem. 2016;291:18740–18752. doi: 10.1074/jbc.M116.737130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andrews DM, Sullivan LC, Baschuk N, Chan CJ, Berry R, Cotterell CL, Lin J, Halse H, Watt SV, Poursine-Laurent J, Wang CR, Scalzo AA, Yokoyama WM, Rossjohn J, Brooks AG, Smyth MJ. Recognition of the nonclassical MHC class I molecule H2-M3 by the receptor Ly49A regulates the licensing and activation of NK cells. Nat Immunol. 2012;13:1171–1177. doi: 10.1038/ni.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.