Abstract
Charentais melons (Cucumis melo cv Reticulatus) are climacteric and undergo extremely rapid ripening. Sixteen cDNAs corresponding to mRNAs whose abundance is ripening regulated were isolated to characterize the changes in gene expression that accompany this very rapid ripening process. Sequence comparisons indicated that eight of these cDNA clones encoded proteins that have been previously characterized, with one corresponding to ACC (1-aminocyclopropane-1-carboxylic acid) oxidase, three to proteins associated with pathogen responses, two to proteins involved in sulfur amino acid biosynthesis, and two having significant homology to a seed storage protein or a yeast secretory protein. The remaining eight cDNA sequences did not reveal significant sequence similarities to previously characterized proteins. The majority of the 16 ripening-regulated cDNAs corresponded to mRNAs that were fruit specific, although three were expressed at low levels in vegetative tissues. When examined in transgenic antisense ACC oxidase melon fruit, three distinct patterns of mRNA accumulation were observed. One group of cDNAs corresponded to mRNAs whose abundance was reduced in transgenic fruit but inducible by ethylene treatment, indicating that these genes are directly regulated by ethylene. A second group of mRNAs was not significantly altered in the transgenic fruit and was unaffected by treatment with ethylene, indicating that these genes are regulated by ethylene-independent developmental cues. The third and largest group of cDNAs showed an unexpected pattern of expression, with levels of mRNA reduced in transgenic fruit and remaining low after exposure to ethylene. Regulation of this third group of genes thus appears to ethylene independent, but may be regulated by developmental cues that require ethylene at a certain stage in fruit development. The results confirm that both ethylene-dependent and ethylene-independent pathways of gene regulation coexist in climacteric fruit.
Fruit ripening is characterized by a number of biochemical and developmental processes that result in changes in color, texture, flavor, and aroma. Like other types of programmed organ senescence, fruit ripening is genetically determined, and ripening-regulated genes have been identified in a large number of fruit (Hadfield and Bennett, 1997). The function of many ripening-regulated genes has been critically tested by altering their expression in transgenic plants, resulting in the identification of components of biochemical pathways involved in pigmentation, soluble carbohydrate metabolism, cell wall metabolism, and ethylene biosynthesis (Giovannoni et al., 1989; Hamilton et al., 1990; Oeller et al., 1991; Fray and Grierson, 1993; Ayub et al., 1996; Klann et al., 1996).
In addition to elucidating the biochemical pathways that determine fruit ripening, the modification of gene expression offers the potential to improve fruit quality by altering biochemical pathways that contribute to flavor, color, and aroma or by increasing shelf-life and/or regulating the initiation and rate of the ripening process. Differential screening of cDNA libraries has proven to be a useful approach to identify developmentally regulated genes that encode proteins critical to developmentally regulated processes (Slater et al., 1985; Lincoln et al., 1987; Dopico et al., 1993; Ledger and Gardner, 1994; Aggelis et al., 1997; Clendennen and May, 1997; Medina-Suarez et al., 1997). Some of these genes encode proteins such as 1-aminocyclopropane-1-carboxylic acid (ACC) oxidase that had been previously difficult to characterize biochemically, and others have proven to be valuable as sources of developmentally regulated promoters (Lincoln et al., 1987; Holdsworth et al., 1988; Pear et al., 1989; Spanu et al., 1991; Lasserre et al., 1997). Examination of ripening-regulated gene expression has also provided insight into the hormonal and developmental signals that regulate fruit ripening. While ethylene is a dominant hormonal trigger for ripening of climacteric fruit, it has been suggested that both ethylene-dependent and ethylene-independent regulatory pathways coexist to coordinate the ripening process in both climacteric and non-climacteric fruit (Lelièvre et al., 1997).
Tomato is the best-studied model system used to characterize ripening-regulated gene expression because of its economic importance, ease of genetic manipulation, and relatively small genome, and because of the availability of developmental mutants that are ripening impaired. However, we felt that the use of alternative systems that may differ from tomato in quantitative and/or qualitative aspects of ripening could provide additional insight into general ripening processes. To this end, we studied the ripening of Charentais melon (Cucumis melo cv Reticulatus F1 Alpha; Hadfield et al., 1995). The usefulness of melon as an alternative model system to study fruit ripening was recently demonstrated by experiments examining the temporal sequence of cell wall disassembly during ripening (Rose et al., 1998). We showed that xyloglucan disassembly occurs during the early stages of softening and is initiated before the extensive pectin disassembly that occurs late in ripening, when tissue breakdown is greatest (Rose et al., 1998). Because the two phases of softening are more distinct in Charentais melon relative to tomato, these melons provided new information on the molecular events associated with the disassembly of each cell wall polymer network.
In the present study, we identified mRNAs that increase in expression during the ripening of Charentais melons. The pattern of gene expression of each of 16 unique cDNAs was examined during the ripening of wild-type fruit and transgenic melons suppressed >99% for ethylene production by the presence of an ACC oxidase antisense transgene (Ayub et al., 1996). The results indicated that ripening-regulated genes in Charentais melon fall into three classes that appear to be regulated by distinct ripening-associated developmental signals.
MATERIALS AND METHODS
Plant Material
Vegetative tissues and fruit tissue used as a source for the cDNA libraries and differential screen probes were obtained from Charentais melon (Cucumis melo cv Reticulatus F1 Alpha) plants grown in Davis, CA. Fruit, stem, and expanding leaf tissues were collected from plants grown under standard field conditions, and root tissue was collected from seedlings grown in a growth chamber (16 h/8 h day/night, 25°C) for 9 d. Wild-type and ACC oxidase antisense Charentais melon (C. melo cv Reticulatus Vedrantais) plants were grown in Toulouse, France under standard field conditions. Flowers were tagged on the day of hand-pollination, and fruit harvested at 32, 36, 40, 43, and 46 d after pollination. The fruit harvested at these time points corresponded to the immature green 1 (IG1), mature green (MG), ripening 1 (R1), ripening 3 (R3), and ripening 4 (R4) stages, respectively, as described in Rose et al. (1998). ACC oxidase antisense fruit were harvested at an additional time point, 50 d after pollination., a subset of which was exposed to 50 μL L−1 ethylene for 24 or 96 h.
Differential Screening of cDNAs
The RNA used for probes in the differential screen was extracted from melon fruit tissues using the hot borate method, and selected for poly(A+) RNA as described in Hadfield et al. (1998). The RNA used for the preripe probe was extracted from IG1 stage fruit, and the RNA for the ripe probe was co-extracted from R2 and R3 fruit. Low-density (5,000 recombinants/plate) duplicate filter lifts of 5 × 105 recombinants from a ripe melon fruit cDNA library (Hadfield et al., 1998) were made according to the manufacturer's instructions (Optitran, Schleicher & Schuell, Keene, NH) and prehybridized in 1× 1,4-piperazinediethanesulfonic acid (PIPES) buffer, 50% (v/v) formamide, 0.5% (w/v) SDS, and 100 μg/mL heat-denatured sheared salmon sperm DNA, overnight at 42°C. The filters were then hybridized overnight at 42°C in fresh buffer (same composition as the prehybridization buffer) with 32P-labeled cDNA probes synthesized from 1 μg of poly(A+) RNA. Poly(A+) RNA in a volume of 20 μL was isotopically labeled by adding 1.5 μL of 1 μg/μL oligo dT(12–18) (Pharmacia Biotech, Piscataway, NJ) and heating to 65°C for 3 min.
After allowing the mixture to come to room temperature, 1 μL of RNasin (40 IU/μL, Promega, Madison, WI), 2.5 μL of 10 mm each dC, dG, and dT, 10 μL of 5× single-strand reverse transcriptase buffer (GIBCO-BRL, Gaithersburg, MD), and 6 μL of [α-32P]dATP (6,000 Ci/mmol) were added. After incubating at 42°C for 3 min, 3 μL of single-strand reverse transcriptase buffer (200 units/μL, GIBCO-BRL) was added and incubated at 42°C for 1 h; then, 2.5 μL of 10 mm cold dA and 1 μL of single-strand reverse transcriptase buffer were added and incubated at 42°C for an additional 30 min. Five microliters of 5 n NaOH and 1 μL of 0.5 m EDTA (pH 8.0) were added and incubated at 70°C for 30 min, at which time 100 μL of 3 m Na acetate (pH 5.2) was added and the unincorporated nucleotides were removed by running the entire reaction mixture through an STE-(20 mm TrisCl, pH 7.5, 100 mm NaCl, and 10 mm EDTA) equilibrated Sephadex G-50 spin column (Pharmacia).
After hybridization, the filters were washed twice in 1× SSC, 0.1% (w/v) SDS at 60°C, and twice in 0.2× SSC, 0.1% (w/v) SDS at 60°C, and exposed to film with one intensifying screen (Reflection, DuPont-NEN, Boston) for 48 h at −80°C. Forty-nine clones that hybridized to the ripe probe but not the preripe probe, were carried through one round of purification by hybridizing duplicate filter lifts of each positive clone to labeled cDNA as described above. In addition, a duplicate filter lift of a phage containing the ripening-regulated MPG1 cDNA (Hadfield et al., 1998) was used as a positive control. After the secondary screen, 29 clones remained positive and the phagemid from each was in vivo excised from the λ-ZAP vector according to the manufacturer's instructions (Stratagene, La Jolla, CA).
Partial Sequencing of Positive Differential cDNA Clones
A partial-length sequence of each of the positive cDNA clones was obtained using the dideoxynucleotide method (Sanger et al., 1977) or by automated sequencing in the Plant Genetics Facility (University of California, Davis, CA) using an automated DNA sequencer (ABI 377, Perkin Elmer/ABI, Foster City, CA) and the vector-specific primers T3 and T7. Sequencing resulted in the identification of 16 non-redundant families, and a representative of each family was used for further analysis. The partial-length sequences of the representative cDNAs were analyzed using the MacDNASIS Pro 3.5 software package (Hitachi, San Bruno, CA). Predicted open reading frames compared with all known translated DNA sequences using the National Center for Biotechnology Information TBLASTN (1.4.11) algorithm and default settings (Altschul et al., 1990). Results of homology searches that had P(N) scores <1e−5 were considered significant, and sequences that did not show significant homologies to proteins in the database were re-analyzed by comparing the DNA sequence translated in six reading frames with all known translated DNA sequences using the BLASTX (1.4.11) algorithm (Gish and States, 1993).
RNA Gel-Blot Hybridization
Total RNA was isolated from the tissues described above and 15 μg of each of the samples separated on 1% (w/v) agarose formaldehyde gels and transferred to Hybond-N nylon membranes (Amersham, Piscataway, NJ) as previously described (Hadfield et al., 1998). A total of eight blots/set of RNA samples were made and each blot was hybridized twice to distinct cDNAs. In between probing, the blots were stripped two times by adding boiling 0.1% (w/v) SDS and incubating at 65°C for 1 h. The blots were checked with a Geiger counter to verify that the probe had been completely stripped before rehybridizing the blot with a second probe. Inserts from each of the representative plasmid cDNAs were isolated and labeled and the blots hybridized and washed, as previously described (Hadfield et al., 1998).
RESULTS AND DISCUSSION
Isolation and Sequence Analysis of Ripening-Regulated cDNA Clones
A differential screen of 50,000 cDNAs from a ripe melon fruit cDNA library, using radiolabeled reverse transcribed RNA from preripe and ripe fruit as probes, resulted in the identification of 29 putative positive-ripening up-regulated clones. Each of the in vivo-excised plasmids were partially sequenced, resulting in the identification of 16 unique cDNAs, each represented by one to six independently isolated cDNA clones. Database searches performed using a partial-length sequence from each unique cDNA revealed high levels of homology between eight of the cDNAs and sequences in the database. Table I summarizes the results of the differential screen. The largest group of ripening-regulated cDNAs, with six independent isolates, RM1, encodes ACC oxidase (MEL1), which has been previously shown to be abundantly expressed in ripening Charentais melons (Balagué et al., 1993). ACC oxidase catalyzes the final step in ethylene biosynthesis, and down-regulation of ACC oxidase gene expression in transgenic melon expressing an ACC oxidase transgene suppresses ethylene production and reversibly inhibits ripening (Ayub et al., 1996).
Table I.
Melon ripening-regulated cDNAs
| cDNA Clone | Homology | P (N)a | Accession No. | Transcript Size | Related Sequence Accession Nos. |
|---|---|---|---|---|---|
| kb | |||||
| RM1 (MEL1) | ACC oxidase | 3.0e−147 | 1.4 | X58885, U07953 | |
| RM2 (MEL7) | Major latex protein | 1.9e−31 | Z70522 | 1.9 | AJ001449, X91914, Z70522, X54305, X79230 |
| RM3 | Chitinase | 6.9e−05 | AF206621 | ND | D11335, X88802 |
| RM4 | SAHH | 1.2e−28 | AF206620 | 1.2 | L11872, Z26881, U79766 |
| RM5 (MEL2) | Hypersensitivity-related gene | 3.4e−49 | Z70521 | 1.3 | X95343, A20628 |
| RM6 | Noneb | NA | 2.0 | ||
| RM7 | Noneb | NA | 2.6 | ||
| RM8 | None | AF206622 | 2.6 | ||
| RM9 | None | AF206623 | 1.4 | ||
| RM10 | None | AF206624 | 1.5 | ||
| RM11 | None | AF206625 | 1.6 | ||
| RM12 | CGS | 2.8e−09 | AF206626 | 3.3 | AF007785, U62147 |
| RM13 | Seed storage protein | 5.4e−16 | AF206627 | 1.8 | Z93107, X75426, Z50778 |
| RM14 | Secretory Protein | 8.1e−26 | AF206628 | 1.0 | L09260, L05929 |
| RM15 | None | AF206629 | 1.6 | ||
| RM16 | Noneb | NA | 1.7 |
Smallest sum probability of homology occurring by chance.
In addition to RM1, two other ripening-regulated cDNAs, RM2 (MEL7) and RM5 (MEL2), have been previously identified in melon and shown to be expressed abundantly during melon fruit ripening (Aggelis et al., 1997). The RM2 sequence was isolated three times and the RM5 four times. Results of our database search for sequences homologous to RM2 agreed with those of Aggelis et al. (1997) showing similarity between RM2 and the major latex protein from poppy (Nessler and Burnett, 1992). In addition, RM2 showed lower (yet significant) homology to a family of genes that encode a fruit-specific, wound-stimulated protein, Sn, of unknown function in nonclimacteric bell pepper fruit. The Sn-1 gene is expressed at low levels in flowers and developing fruit, but its mRNA increases dramatically during ripening and in response to wounding (Pozueta-Romero et al., 1995).
The MEL2 (RM5) sequence revealed similarity with the hypersensitivity-related gene hsr201 from tobacco and to a ripening-regulated gene from tomato, pTOM36. The function of the protein encoded by pTOM36 is not known but its expression is induced during tomato fruit ripening and leaf senescence (Davies and Grierson, 1989).
The RM3 sequence was isolated once and shares high sequence homology to acidic chitinases. A ripening up-regulated gene encoding a putative chitinase has been identified in avocado (Dopico et al., 1993), and both ripening up- and down-regulated genes have been identified in banana (Clendennen and May, 1997; Medina-Suarez et al., 1997). The abundance of RM3 transcript in melon was very low and its pattern of ripening-related expression could not be determined.
The RM2, RM3, and RM5 predicted proteins are all homologous to proteins associated with defense responses. The expression of pathogen-response proteins has been observed to increase during normal ripening of other fruit as well (Dopico et al., 1993; Atkinson et al., 1996; Meyer et al., 1996; Clendennen and May, 1997), suggesting that the processes of fruit ripening and defense responses may share common elements. Ethylene is a key regulator in the ripening of climacteric fruit and in the response to stresses such as pathogens and wounding. Pathogen-response proteins such as chitinase and thaumatin-like proteins, however, accumulate to very high levels in cherry and grape (Fils-Lycaon et al., 1996; Waters et al., 1996; Tattersall et al., 1997), both non-climacteric fruit, indicating that the commonalities between ripening and pathogen responses extend beyond their regulation by ethylene.
Two of the ripening-regulated cDNAs, RM4 and RM12, encode predicted proteins that are homologous to enzymes involved in sulfur-containing amino acid metabolism. RM4 is highly homologous to S-adenosyl-l-homocysteine hydrolase (SAHH) (Schroder et al., 1994), and RM12 is homologous to cystathionine γ-synthase (CGS) (Kim and Leustek, 1996). SAHH catalyzes one of the steps involved in the regeneration of S-adenosylmethionine (SAM) during methyl transfer reactions. SAHH could be involved in ethylene biosynthesis by prohibiting the accumulation of adenosylhomocysteine, a known competitive inhibitor of SAM-dependent methyl transferase reactions and a potential inhibitor of the hydrolysis of SAM to methylthioadenosine and ACC, the precursor of ethylene. A ripening- regulated cDNA clone with high homology to SAHH was recently identified in banana (Medina-Suarez et al., 1997), suggesting that the expression of SAHH during ripening may be a common feature in climacteric fruit. Plant CGS catalyzes the first committed step in Met biosynthesis, the precursor to SAM. The possible functions of CGS and SAHH in ripening fruit aren't readily apparent, but their increased expression may indicate that methylation of one or more acceptor molecules using SAM as the methyl donor may play an important role in the process of fruit ripening.
The predicted protein encoded by RM13 is highly homologous to pBAN UU80, a ripening-regulated gene from banana that was not assigned a putative identity (Medina-Suarez et al., 1997). When the RM13-predicted amino acid sequence was used in database searches, significant homology was revealed between RM13 and seed storage proteins from ginkgo (Arahira and Fukazawa, 1994). In seeds, storage proteins function as nitrogen reserves to be used during seedling growth, but it is unlikely that seed-storage-like proteins function as nitrogen reserves in ripening fruit. The RM14-predicted amino acid sequence has high sequence homology to the human homolog of the yeast secretory pathway gene SEC13 (Swaroop et al., 1994). In fruit, the RM14 protein may be involved in transporting proteins through the secretory system to deliver cell wall hydrolases known to accumulate to high levels during this period. Seven additional ripening-regulated cDNAs, RM6, RM7, RM8, RM9, RM10, RM11, and RM15 did not have significant sequence homology to any protein or nucleotide sequence in the database.
Expression Patterns of Melon Ripening-Regulated mRNAs
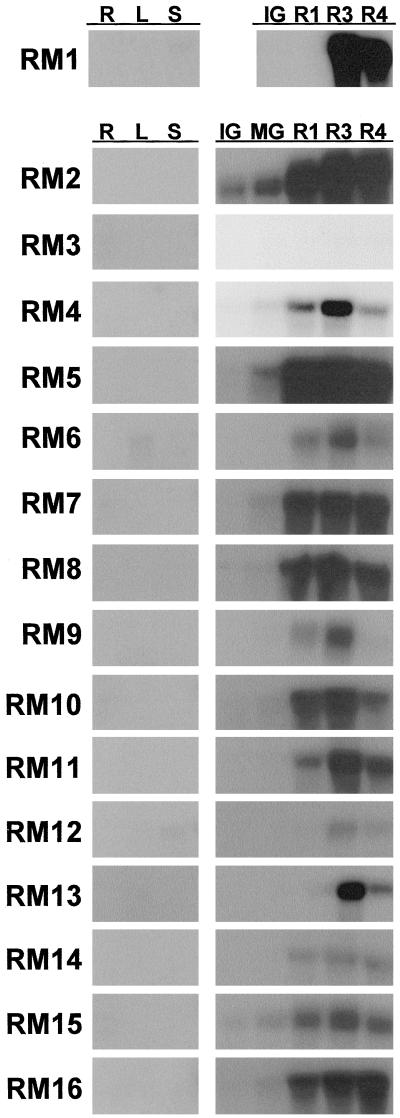
The mRNA abundance corresponding to the 16 unique ripening-regulated cDNA clones was examined in roots, leaves, and stems as well as during the ripening of normal and ACC oxidase suppressed melon fruit. The majority of the cDNAs corresponded to mRNAs that were fruit specific (Fig. 1). However, RM12, the CGS homolog, was expressed at low levels in stems. During ripening, mRNA abundance corresponding to most of the cDNAs was high at the onset of ripening (R1) and remained at similar levels throughout ripening. However, RM12 (CGS) and RM13 (seed-storage-like protein) were both induced later in ripening, between the R1 and R3 stages, and mRNA accumulation was greatest at the R3 stage. The abundance of RM4 (SAHH), RM6, RM9, and RM11 mRNA was easily detectable at the R1 stage but accumulated to higher levels at the R3 stage. This late regulation of gene expression is similar to the pattern of melon fruit PG gene expression, where mRNA accumulation is detectable after the onset of ripening and accumulates to higher levels during the late stage of ripening (Hadfield et al., 1998). The mRNA that hybridized to the putative acidic chitinase clone RM3 was very low and barely detectable after a multiday exposure to film, making it difficult to assess its pattern of expression.
Figure 1.
RNA-blot analysis of RM1 to RM16 RNA in developing melon fruit and non-fruit tissues. Each lane was loaded with 15 μg of total RNA isolated from fruit tissues at the five stages of development (see text) and from roots (R), young leaves (L), and stems (S). The blot hybridized with RM1 did not have RNA from MG fruit. The blots were probed with gel-purified labeled inserts, washed in 0.2× SSC at 65°C, and exposed to film for approximately 2 h.
To assess the regulation of mRNA accumulation corresponding to the ripening-regulated cDNA clones by ethylene, their expression was examined in melons that are suppressed for ethylene production by the presence of an antisense ACC oxidase transgene (Ayub et al., 1996). These melons produce very low levels of ethylene and do not undergo the normal processes associated with ripening, including softening and tissue deterioration. When transgenic melons were exposed to 50 μL L−1 ethylene for 24 to 96 h, the ripening phenotype was restored and the fruit softened and entered the overripe stage (Guis et al., 1997).
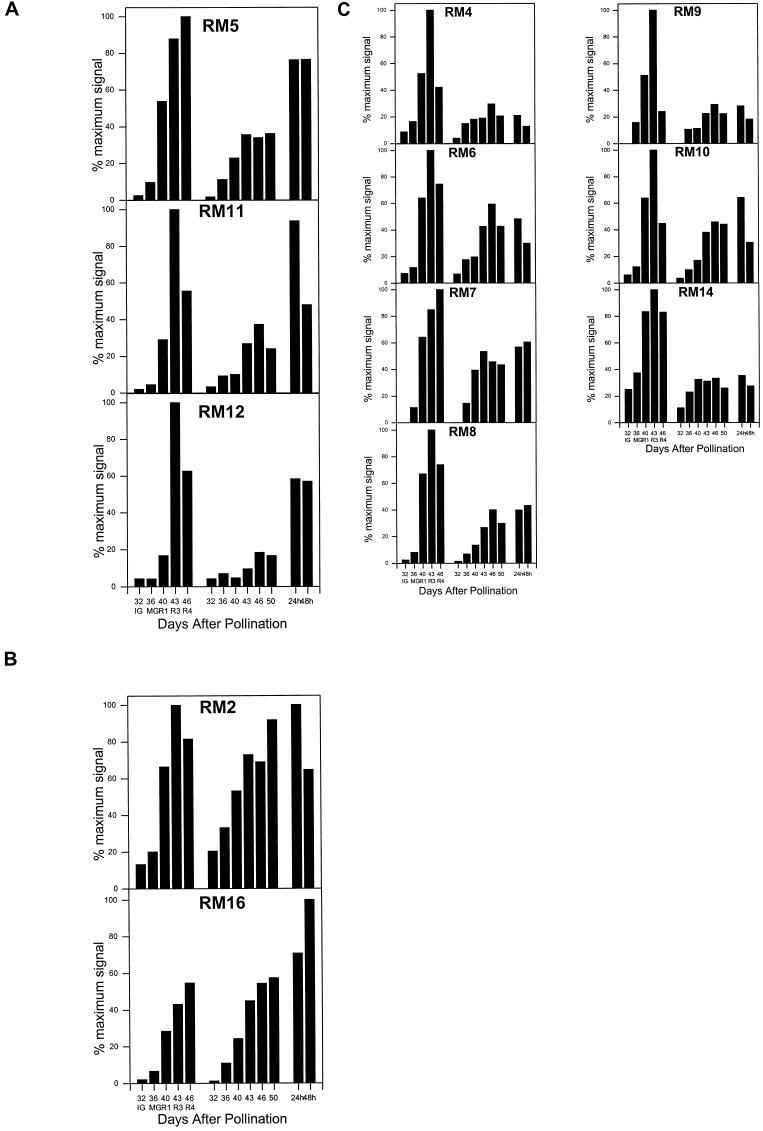
The mRNA accumulation patterns of the ripening-regulated cDNA clones fell into three general categories. The first group showed a typical pattern of expression for ethylene-regulated genes. That is, the expression was greatly reduced in transgenic antisense ACC oxidase fruit during the period of ripening of wild-type fruit, but was induced by subsequent exposure of these fruit to ethylene (Fig. 2A). This group was comprised of RM1 (ACC oxidase) (data previously shown in Ayub et al., 1996), RM5 (HSR gene), RM11, RM12 (CGS), and RM13 (seed storage protein). The second group (Fig. 2B) was comprised of two clones, RM2 (major latex protein) and RM16, and showed a pattern of expression that was not significantly reduced in the transgenic antisense ACC oxidase fruit (and the levels were not affected by subsequent exposure to ethylene). This result may indicate that expression of group 2 genes is ethylene independent, or that expression is very sensitive to ethylene and that the low levels of ethylene produced by these transgenic fruit exceed the threshold required for their expression. The third pattern of expression (Fig. 2C) was characterized by significantly reduced levels of mRNA accumulation in the transgenic antisense ACC oxidase fruit relative to wild-type fruit, but, interestingly, expression was not induced in fruit that were exposed to ethylene. This pattern of expression was unexpected and may reflect a combination of independent developmental- and ethylene-dependent pathways of regulation of gene expression. The expression of group 2 genes in transgenic antisense ACC oxidase fruit and the lack of induction of gene expression of the group 3 genes by ethylene suggests that the proteins encoded by these genes do not play a role in the ripening processes associated with the presence of ethylene. This analysis of gene expression provides critical insight into the role of specific genes and their encoded proteins in ethylene-regulated ripening processes.
Figure 2.
Phosphor imager data of RNA gel-blot analysis of RM2 to RM7 and RM9 to RM16 RNA in developing wild-type and ACC oxidase antisense melon fruit. Each lane was loaded with 15 μg of total RNA isolated from fruit tissues at five stages of development (see text), from ACC oxidase antisense fruit harvested at 32, 36, 40, 43, 46, and 50 d after pollination, and antisense fruit 50 d after pollination treated with 50 μL L−1 ethylene for 24 h or 96 h. The fruit harvested at these time points corresponded to the IG1, MG, R1, R3, and R4 stages, respectively. The phosphor imager data are expressed for each individual blot in terms of percentage maximum signal detected on that blot. A, Group 1 cDNAs showed a decrease in gene expression in antisense fruit relative to wild type and were induced in antisense fruit treated with ethylene. B, Group 2 cDNA expression was not greatly reduced in antisense fruit relative to wild type, and gene expression was relatively unchanged when antisense fruit were exposed to ethylene. C, Group 3 cDNA expression was reduced in antisense fruit relative to wild type but remained low in antisense fruit exposed to ethylene.
CONCLUSIONS
Differential screening of a ripe melon fruit cDNA library resulted in the identification of 16 unique cDNAs corresponding to mRNAs whose accumulation was stimulated by ripening. Database searches for homologies of the predicted amino acid sequences to previously identified genes resulted in the putative identification of seven of the differentially expressed cDNA clones. Three of the sequences were homologous to proteins associated with pathogen responses in other, non-fruit systems. The ripening-regulated expression of genes encoding pathogen-response-like proteins has been shown in a wide range of climacteric and non-climacteric fruit, suggesting that the processes of pathogen response and fruit ripening may share common elements. Two of the sequences were homologous to proteins involved in sulfur amino acid biosynthesis, and the remaining two showed significant amino acid homology to seed storage proteins or a yeast secretory protein. The remaining eight cDNA sequences did not reveal significant homologies to any proteins in the database.
Expression of 15 of the 16 cDNAs was shown to be ripening regulated and expression in 12 was fruit specific. The abundance of the 12 fruit-specific and ripening-regulated mRNAs was very high, and these cDNAs may be useful in identifying strong promoters that are fruit specific and ripening regulated.
Three patterns of gene expression were apparent when the expression of cDNA clones was examined in ethylene-suppressed transgenic antisense ACC oxidase melon fruit. One group of cDNAs corresponded to mRNAs whose abundance was reduced in transgenic fruit but inducible by ethylene treatment, indicating that these genes are directly regulated by ethylene. A second group of mRNAs was not significantly altered in the transgenic fruit and was unaffected by treatment with ethylene, indicating that these genes are regulated by ethylene-independent developmental cues. The third and largest group of cDNAs showed an unexpected pattern of expression, with levels of mRNA reduced in transgenic fruit and remaining low after exposure to ethylene. Regulation of this third group of genes thus appears to be ethylene independent, but may be regulated by developmental cues that require ethylene at a certain stage in fruit development. The results confirm that both ethylene-dependent and ethylene-independent pathways of gene regulation coexist in climacteric fruit (Lelièvre et al., 1997). The ethylene-independent genes identified here and transgenic melon fruit with severely suppressed ethylene levels provide a means to identify ethylene-independent signals that contribute to ripening-regulated gene expression.
Footnotes
This research was supported by grants to A.B.B. from Zeneca Plant Science and by the U.S. Department of Agriculture-National Research Initiative (grant no. 97–35304–4627).
LITERATURE CITED
- Aggelis A, John I, Karvouni Z, Grierson D. Characterization of two cDNA clones for mRNAs expressed during ripening of melon (Cucumis meloL.) fruits. Plant Mol Biol. 1997;33:313–322. doi: 10.1023/a:1005701730598. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arahira M, Fukazawa C. Ginkgo 11S seed storage protein family mRNA: unusual Asn-Asn linkage as post-translational cleavage site. Plant Mol Biol. 1994;25:597–605. doi: 10.1007/BF00029599. [DOI] [PubMed] [Google Scholar]
- Atkinson RG, Perry J, Matsui T, Ross GS, Macrae EA. A stress-, pathogenesis-, and allergen-related cDNA in apple fruit is also ripening-related. N Z J Crop Hortic Sci. 1996;24:103–107. [Google Scholar]
- Ayub R, Guis M, Ben-Amor M, Gillot L, Roustan J-P, Latché A, Bouzayen M, Pech J-C. Expression of ACC oxidase antisense gene inhibits ripening of cantaloupe melon fruits. Nat Biotech. 1996;14:862–866. doi: 10.1038/nbt0796-862. [DOI] [PubMed] [Google Scholar]
- Balagué C, Watson CF, Turner AJ, Rouge P, Picton S, Pech JC, Grierson D. Isolation of a ripening and wound-induced cDNA from Cucumis meloL. encoding a protein with homology to the ethylene-forming enzyme. Eur J Biochem. 1993;212:27–34. doi: 10.1111/j.1432-1033.1993.tb17628.x. [DOI] [PubMed] [Google Scholar]
- Clendennen SK, May GD. Differential gene expression in ripening banana fruit. Plant Physiol. 1997;115:463–469. doi: 10.1104/pp.115.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies KM, Grierson D. Identification of cDNA clones for tomato (Lycopersicon esculentum, Mill.) mRNAs that accumulate during ripening and leaf senescence. Planta. 1989;179:73–80. doi: 10.1007/BF00395773. [DOI] [PubMed] [Google Scholar]
- Dopico B, Lowe AL, Wilson ID, Merodio C, Grierson D. Cloning and characterization of avocado fruit mRNAs and their expression during ripening and low-temperature storage. Plant Mol Biol. 1993;21:437–449. doi: 10.1007/BF00028802. [DOI] [PubMed] [Google Scholar]
- Fils-Lycaon BR, Wiersma PA, Eastwell KC, Sautiere P. A cherry protein and its gene, abundantly expressed in ripening fruit have been identified as thaumatin-like. Plant Physiol. 1996;111:269–273. doi: 10.1104/pp.111.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fray RG, Grierson D. Identification and genetic analysis of normal and mutant phytoene synthase genes of tomato by sequencing, complementation and co-suppression. Plant Mol Biol. 1993;22:589–602. doi: 10.1007/BF00047400. [DOI] [PubMed] [Google Scholar]
- Giovannoni JJ, DellaPenna D, Bennett AB, Fischer RL. Expression of a chimeric polygalacturonase gene in transgenic rin(ripening inhibitor) tomato fruit results in polyuronide degradation but not fruit softening. Plant Cell. 1989;1:53–63. doi: 10.1105/tpc.1.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gish W, States DJ. Identification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- Guis M, Botondi R, Ben Amor M, Ayub R, Bouyazen M, Pech JC, Latché A. Ripening-associated biochemical traits of cantaloupe charentais melons expressing an antisense ACC oxidase transgene. J Am Soc Hortic Sci. 1997;122:748–751. [Google Scholar]
- Hadfield KA, Bennett AB. Programmed senescence of plant organs. Cell Death Differen. 1997;4:662–670. doi: 10.1038/sj.cdd.4400308. [DOI] [PubMed] [Google Scholar]
- Hadfield KA, Rose JKC, Bennett AB. The respiratory climacteric is present in Charentais (Cucumis melo cv. Reticulatus F1Alpha) melons ripened on or off the plant. J Exp Bot. 1995;46:1923–1925. [Google Scholar]
- Hadfield KA, Rose JKC, Yaver DS, Berka RA, Bennett AB. Polygalacturonase gene expression in ripe melon fruit supports a role for PG in ripening-associated pectin disassembly. Plant Physiol. 1998;117:363–373. doi: 10.1104/pp.117.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AJ, Lycett GW, Grierson D. Antisense gene that inhibits synthesis of the hormone ethylene in transgenic plants. Nature. 1990;346:284–287. [Google Scholar]
- Holdsworth MJ, Schuch W, Grierson D. Organization and expression of a wound/ripening-related small multigene family from tomato. Plant Mol Biol. 1988;11:81–88. doi: 10.1007/BF00015661. [DOI] [PubMed] [Google Scholar]
- Kim J, Leustek T. Cloning and analysis of the gene for cystathionine γ-synthase from Arabidopsis thaliana. Plant Mol Biol. 1996;32:1117–1124. doi: 10.1007/BF00041395. [DOI] [PubMed] [Google Scholar]
- Klann EM, Hall B, Bennett AB. Antisense acid invertase (TIV1) gene alters soluble sugar composition and size in transgenic tomato fruit. Plant Physiol. 1996;112:1321–1330. doi: 10.1104/pp.112.3.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasserre E, Godard F, Bouquin T, Hernandez JA, Pech JC, Roby D, Balaguè D. Differential activation of ACC oxidase gene promoters during development and in response to pathogen attack. Mol Gen Genet. 1997;256:2111–2222. doi: 10.1007/s004380050563. [DOI] [PubMed] [Google Scholar]
- Ledger SE, Gardner RC. Cloning and characterization of five cDNAs for genes differentially expressed during fruit development of kiwifruit (Actinidia deliciosa var. deliciosa) Plant Mol Biol. 1994;25:877–886. doi: 10.1007/BF00028882. [DOI] [PubMed] [Google Scholar]
- Lelièvre J-M, Latché A, Jones B, Bouzayen M, Pech J-C. Ethylene and fruit ripening. Physiol Plant. 1997;101:727–739. [Google Scholar]
- Lincoln JE, Cordes S, Read E, Fischer RL. Regulation of gene expression by ethylene during Lycopersicon esculentum(tomato) fruit development. Proc Natl Acad Sci USA. 1987;84:2793–2797. doi: 10.1073/pnas.84.9.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Suarez R, Manning K, Fletcher J, Aked J, Bird CR, Seymour GB. Gene expression in the pulp of ripening bananas. Plant Physiol. 1997;115:453–461. doi: 10.1104/pp.115.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B, Houlne G, Pozueta-Romero J, Shantz ML, Schantz R. Fruit-specific expression of a defensin-type gene family in bell pepper. Plant Physiol. 1996;112:615–622. doi: 10.1104/pp.112.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nessler CL, Burnett RJ. Organization of the major latex protein gene family in opium poppy. Plant Mol Biol. 1992;20:749–752. doi: 10.1007/BF00046460. [DOI] [PubMed] [Google Scholar]
- Oeller PW, Min-Wong L, Taylor LP, Pike DA, Theologis A. Reversible inhibition of tomato fruit senescence by antisense RNA. Science. 1991;254:437–439. doi: 10.1126/science.1925603. [DOI] [PubMed] [Google Scholar]
- Pear JR, Ridge N, Rasmussen R, Rose RE, Houck CM. Isolation and characterization of a fruit specific cDNA and the corresponding genomic clone from tomato. Plant Mol Biol. 1989;13:639–651. doi: 10.1007/BF00016019. [DOI] [PubMed] [Google Scholar]
- Pozueta-Romero J, Klein M, Houlne G, Schantz M, Meyer B, Schantz R. Characterization of a family of genes encoding a fruit-specific wound-stimulated protein of bell pepper (Capsicum annuum): identification of a new family of transposable elements. Plant Mol Biol. 1995;28:1011–1025. doi: 10.1007/BF00032663. [DOI] [PubMed] [Google Scholar]
- Rose JKC, Hadfield KA, Labavitch JM, Bennett AB. Temporal sequence of cell wall disassembly in rapidly ripening melon fruit. Plant Physiol. 1998;117:345–361. doi: 10.1104/pp.117.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder G, Waitz A, Hotze M, Schroder J. cDNA for S-adenosyl-l-homocysteine hydrolase from Catharanthus roseus. Plant Physiol. 1994;104:1099–1100. doi: 10.1104/pp.104.3.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater A, Maunders MJ, Edwards K, Schuch W, Grierson D. Isolation and characterization of cDNA clones for tomato polygalacturonase and other ripening related proteins. Plant Mol Biol. 1985;5:137–147. doi: 10.1007/BF00015677. [DOI] [PubMed] [Google Scholar]
- Spanu P, Reinhardt D, Boller T. Analysis and cloning of the ethylene-forming enzyme from tomato by functional expression of its mRNA in Xenopus laevisoocytes. EMBO J. 1991;10:2007–2013. doi: 10.1002/j.1460-2075.1991.tb07730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaroop A, Yang-Feng TL, Liu W, Geiser L, Barrow LL, Chen KC, Agarwal N, Meisler MH, Smith DI. Molecular characterization of a novel human gene, SEC13R, related to the yeast secretory pathway gene SEC13, and mapping to a conserved linkage group on human chromosome 3p24–p25 and mouse chromosome 6. Hum Mol Genet. 1994;3:1281–1286. doi: 10.1093/hmg/3.8.1281. [DOI] [PubMed] [Google Scholar]
- Tattersall DB, van-Heeswijck R, Hoj PB. Identification and characterization of a fruit-specific thaumatin-like protein that accumulates at very high levels in conjunction with the onset of sugar accumulation and berry softening in grapes. Plant Physiol. 1997;114:759–769. doi: 10.1104/pp.114.3.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters EJ, Shirley NJ, Williams PJ. Nuisance proteins of wine are grape pathogenesis-related proteins. J Agric Food Chem. 1996;44:3–5. [Google Scholar]