Abstract
Intestinal epithelial barrier dysfunction is a well-known sequela of HIV/SIV infection that persists despite anti-retroviral therapy. Although, inflammation is a triggering factor, the underlying molecular mechanisms remain unknown. Emerging evidence suggests that epithelial barrier function is epigenetically regulated by inflammation induced microRNAs. Accordingly, we profiled and characterized miRNA/mRNA expression exclusively in colonic epithelium (CE) and identified 46 differentially expressed (DE) miRNAs (20-up and 26-down) in chronically SIV-infected rhesus macaques (RMs) (Macaca mulatta). We bioinformatically crossed the predicted miRNA targets to transcriptomic data and characterized miR-130a and miR-212 as both were predicted to interact with critical epithelial barrier associated genes. Next, we characterized peroxisome proliferator activator receptor gamma (PPARγ) and occludin (OCLN), predicted targets of miR-130a and miR-212 respectively, as their downregulation has been strongly linked to epithelial barrier disruption and dysbiosis. Immunofluorescence, luciferase-reporter and overexpression studies confirmed the ability of miR-130a and miR-212 to decrease protein expression of OCLN and PPARγ, respectively and reduce transepithelial electrical resistance (TEER). Since delta-9-tetrahydrocannabinol (Δ9-THC) exerted protective effects in the intestine in our previous studies, we successfully used it to reverse miR-130a and miR-212 mediated reduction in TEER. Finally, ex-vivo Δ9-THC treatment of colon tissue from chronically SIV-infected RMs significantly increased PPARγ expression. Our findings suggest that dysregulated miR-130a and miR-212 expression in CE during chronic HIV/SIV infection can facilitate epithelial barrier disruption by downregulating OCLN and PPARγ expression. Most importantly, our results highlight the beneficial effects of cannabinoids on epithelial barrier function in not just HIV/SIV but potentially other chronic intestinal inflammatory diseases.
Keywords: SIV, rhesus macaque, colon, epithelial cell, miR-130a/212, Occludin, PPARγ, THC
INTRODUCTION
HIV/SIV infection of the gastrointestinal tract (GI) / immune system results in rapid and sustained virus induced CD4+ T cell depletion leading to persistent intestinal inflammation, epithelial damage, immune activation, and establishment and replenishment of the viral reservoir (1–2). The gut-associated lymphoid tissue (GALT) accommodates up to 70–80% of the activated memory CD4+ T cells, which makes it a preferential target organ of HIV/SIV (1). A common sequela to this continued CD4+ T cell depletion is chronic GI disease / dysfunction manifested by anorexia, weight loss, nausea, vomiting, dysphagia, abdominal pain and frequent diarrhea that persists despite anti-retroviral therapy (3–4). More strikingly, these symptoms occur in the absence of a detectable enteric pathogen and persist in patients on combination anti-retroviral therapy (cART) (6–7). Damage and impaired regeneration of the intestinal epithelium are critical contributors to the loss of barrier function and subsequent development of gastrointestinal disease in AIDS, which is also seen in SIV-infected rhesus macaques (RMs) (5–6).
The intestinal mucosal barrier is comprised of a layer of mucus, a monolayer of polarized columnar epithelial cells composed of absorptive enterocytes, stem cells, goblet cells, neuroendocrine cells, paneth cells and enteroendocrine cells and a network of tight junction (TJ) proteins that function to protect the sterile subepithelial tissue from luminal antigens and microbes. The persistent proinflammatory environment in the intestinal lamina propria driven by viral replication can damage TJ protein structure and function leading to disruption of the intestinal epithelial barrier. This can facilitate increased penetration of microbes and other antigens resulting in local and systemic immune activation and AIDS progression (7). While the occurrence of epithelial barrier disruption has been well documented in HIV/SIV infection (8), the underlying molecular mechanisms remain unknown. Accordingly, a deeper understanding of the molecular mechanisms responsible for the regulation and alteration of the intestinal barrier function may assist in the identification and development of better therapeutic strategies for the prevention of GI dysfunction in HIV/SIV and other intestinal diseases.
Mounting evidence suggests that the intestinal epithelial barrier is a dynamic structure that is regulated by RNA binding proteins and at the epigenetic level by non-coding RNAs (9–10). At least, 16 microRNAs are predicted to regulate the expression of the different TJ proteins that constitute the epithelial barrier (9–10). While, recent studies have demonstrated that miR-212, miR-122 and miR-29a directly target and downregulate ZO1, OCLN and CLDN1 expression in alcoholic liver disease (ALD) (11), inflammatory bowel disease (IBD) (12) and irritable bowel syndrome (IBS) (13), respectively, mechanistic studies to determine epigenetic regulation of the barrier in HIV/SIV infection remain to be done. Further and more importantly, feasible pharmacological / therapeutic strategies to reverse miRNA-mediated epithelial barrier disruption are yet to be explored. We previously described miRNA dysregulation in the intestine associated with key pathogenic events, namely, viral replication (14), immune cell activation (15) and double-stranded DNA damage response (16). To better understand their critical role in epithelial barrier disruption, we performed an integrative microRNA-mRNA functional analysis exclusively in the colonic epithelium (CE) of chronically SIV-infected rhesus RMs. These studies identified specific miRNAs associated with inflammation, epithelial barrier disruption and double stranded DNA damage response. Among the different miRNA-mRNA interaction networks generated, we identified miR-212 and miR-130a to have the most interactions with several downregulated genes, particularly those associated with epithelial barrier function. Accordingly, we decided to further investigate PPARγ and OCLN and their post-transcriptional regulation by miR-212 and miR-130a, respectively, owing to their well-established role in epithelial barrier function. Using multiple approaches, we confirmed PPARγ and OCLN to be direct targets of miR-130a and miR-212, respectively. Most notably, we demonstrated that moderate overexpression of miR-212 and miR-130a decreased trans epithelial electrical resistance (TEER), a measure of epithelial barrier permeability. Interestingly, in vitro treatment of miRNA transfected cells with delta-9-tetrahydrocannabinol (Δ9-THC), an anti-inflammatory cannabinoid that we had previously demonstrated to be protective in the GI tract (17), fully restored TEER levels to that observed with control miRNA mimics. Finally, ex vivo treatment of colon tissue from chronically SIV-infected RMs with Δ9-THC increased PPARγ mRNA expression. These results strongly identify a role for miRNAs in intestinal inflammation, epithelial barrier function and microbial translocation, all of which are characteristic features of chronic HIV/SIV infection and demonstrate the immense therapeutic potential of cannabinoids in restoration of intestinal epithelial barrier integrity.
MATERIALS AND METHODS
Animal ethics statement
All experiments using rhesus macaques were approved by the Tulane Institutional Animal Care and Use Committee (Protocol No-3574). The Tulane National Primate Research Center (TNPRC) is an Association for Assessment and Accreditation of Laboratory Animal Care International accredited facility (AAALAC #000594). The NIH Office of Laboratory Animal Welfare assurance number for the TNPRC is A3071-01. All clinical procedures, including administration of anesthesia and analgesics, were carried out under the direction of a laboratory animal veterinarian. Animals were anesthetized with ketamine hydrochloride for blood collection procedures. Intestinal resections were performed by laboratory animal veterinarians. Animals were pre-anesthetized with ketamine hydrochloride, acepromazine, and glycopyrolate, intubated and maintained on a mixture of isoflurane and oxygen. Buprenorphine was given intra-operatively and post-operatively for analgesia. All possible measures are taken to minimize discomfort of all the animals used in this study. Tulane University complies with NIH policy on animal welfare, the Animal Welfare Act, and all other applicable federal, state and local laws.
Animals and experimental design
Colon tissues were collected from 36 Indian-origin RMs including 24 animals chronically infected with SIVmac251 (n=22) or SIVmac239 (n=1) or SIVE660 (n=1) and 12 uninfected macaques. Animal IDs, SIV inoculum, duration of infection, plasma and intestinal viral loads and colon histopathology in all chronically SIV-infected animals are provided in Table I. Colon resection segments (~5 cm long) were collected from four macaques (HD08, HH07, HF54, HR42) before SIV infection to be used as controls (Table I) for OpenArray microRNA profiling and RT-qPCR studies. Chronically SIV-infected RMs used for RT-qPCR and ex-vivo Δ9-THC stimulation studies are indicated in Table I.
Table I.
Animal IDs, SIV inoculum, duration of infection, viral loads and colon histopathology in chronically SIV-infected RMs
| Animal ID | SIV Inoculum | Duration of Infection (Days) | Plasma viral loads 106/mL | Colon viral loads 106/mg RNA | Colon Histopathology | Opportunistic Infections |
|---|---|---|---|---|---|---|
|
Chronic SIV-Infected | ||||||
| FT11 | SIVmac251 | 145 | 500 | 2075 | Moderate colitis | ND |
| HN15 | SIVmac251 | 177 | 8 | 21 | Granulomatous colitis | Mycobacterium avium intracellulare |
| FE24 | SIVmac251 | 271 | 3.1 | 0.3 | Mild colitis | ND |
| HB31 | SIVmac251 | 180 | 3000 | 200 | Lymphoid hyperplasia | ND |
| HI68 | SIVmac251 | 158 | 8 | 3 | Mild colitis | ND |
| GA19 | SIVmac251 | 180 | 100 | 600 | Lymphoid hyperplasia | ND |
| HG45 | SIVmac251 | 281 | 0.9 | 0.3 | ND | ND |
| HD08-90D | SIVmac251 | 90 | 37 | 3000 | Moderate colitis/cryptitis | ND |
| HH07-180D* | SIVmac251 | 180 | 9 | 59 | ND | ND |
| HF54-180D* | SIVmac251 | 180 | 0.3 | 56 | ND | ND |
| HR42-180D* | SIVmac251 | 180 | 0.2 | 210 | ND | ND |
| HV48 | SIVmac251 | 180 | 4 | 230 | ND | ND |
| DV42# | SIVmac251 | 226 | 0.1 | 1 | Mild colitis | ND |
| HF27*# | SIVmac251 | 180 | 40 | 0.04 | Lymphoid hyperplasia | ND |
| GK31*# | SIVmac251 | 180 | 30 | 3 | Lymphoid hyperplasia | ND |
| JN82*# | SIVE660 | 141 | NAV | NAV | Lymphoid hyperplasia | ND |
| HL01# | SIVmac251 | 180 | 7 | 0.8 | ND | ND |
| FE53# | SIVmac251 | 140 | 0.3 | 5 | Mild colitis | ND |
|
Chronic SIV-infected macaques used for ex-vivo THC studies | ||||||
| DT09* | SIVmac251 | 186 | NAV | 6 | NAV | NAV |
| IT40* | SIVmac251 | 188 | NAV | 6 | NAV | NAV |
| JR80* | SIVmac251 | 185 | NAV | 3 | NAV | NAV |
| JA32* | SIVmac251 | 187 | 3 | 30 | NAV | NAV |
| KM86 | SIVmac251 | 112 | NAV | 2 | NAV | NAV |
| GA60* | SIVmac239 | 735 | 2 | 400 | NAV | NAV |
|
Uninfected Controls | ||||||
| HT74 | NA | NA | NA | NA | NA | NA |
| IT62 | NA | NA | NA | NA | NA | NA |
| IT16 | NA | NA | NA | NA | NA | NA |
| IT05 | NA | NA | NA | NA | NA | NA |
| IH95 | NA | NA | NA | NA | NA | NA |
| HT73 | NA | NA | NA | NA | NA | NA |
| EJ34 | NA | NA | NA | NA | NA | NA |
| IT13 | NA | NA | NA | NA | NA | NA |
| HD08-Pre | NA | NA | NA | NA | NA | NA |
| HH07-Pre | NA | NA | NA | NA | NA | NA |
| HF54-Pre | NA | NA | NA | NA | NA | NA |
| HR42-Pre | NA | NA | NA | NA | NA | NA |
NAV- Not available, NA- Not applicable, ND- None detected,
Denotes timed necropsy,
used for RT-qPCR only
At necropsy, all tissues were collected in RNAlater® (Ambion, TX) for total RNA extraction and RT-qPCR. For histopathologic evaluation, intestinal tissues were fixed in 10% neutral buffered formalin, embedded in paraffin, sectioned at 7 μm and stained with hematoxylin and eosin for analysis.
Intestinal Epithelial cell isolation
The protocol and purity of epithelial cells isolated from surgical resection segments collected at necropsy were described previously (14,18). Briefly, colon tissue collected at necropsy or as surgical resection segments (~5 cm long) for miRNA/mRNA profiling studies were first incubated with vigorous shaking in Ca++Mg++ free-HBSS containing 1 mM EDTA for two 30-min incubations at 37°C to separate the intestinal epithelial cells. Following incubation, the epithelial cells in the supernatant were harvested by centrifugation at 500 g for 10 min followed by subjecting the cells to percoll density gradient centrifugation to separate intra-epithelial lymphocytes (IELs). Our colleagues at the TNPRC have shown this protocol to consistently yield epithelial cells with >85% purity with minimal contamination with IELs (18).
Global microRNA and mRNA profiling
About 100 ng of total RNA from CE was first reverse transcribed, preamplified and loaded onto human TaqMan® OpenArray® microRNA plates and processed as described previously (17). Transcriptome (mRNA) profiling was performed by Arraystar Inc (Rockville, MD) using whole rhesus macaque 4 x 44K Gene Expression Microarray V2 (Agilent Technologies).
Quantitative Real-Time TaqMan Stem loop microRNA and SYBR Green RT-qPCR
Expression of miR-130a and miR-212 was further confirmed in CE using the TaqMan microRNA predesigned and preoptimized assays (Life Technologies Corp) (14–17). Expression of both miRNAs was normalized to a combination of two endogenous controls, snoU6 and RNU48. PPARγ (For-5′-ATCGCCCAGGTTTGCTGAAT-3′, Rev-5′-GACTCAGGGTGGTTCAGCTTC-3′) and OCLN (For-5′-CATTGCCATCTTTGCATGTGT-3′, Rev-5′- GGTAGCCTACACTACCTCCTATAA-3′) RT-qPCR was performed using the Power SYBR Green RNA to CT kit. PPARγ and OCLN expression in Caco-2 cells was normalized to a combination of GAPDH (For-5′-CAAGAGAGGCATTCTCACCCTGAA-3′, Rev-5′-TGGTGCCAGATCTTCTCCATGTC-3′) and β-Actin (For-5′-CAACAGCCTCAAGATCGTCAGCAA-3′, Rev-5′-GAGTCCTTCCACGATACCAAAGTTGTC-3′). PPARγ expression in ex-vivo colon tissue was normalized to GAPDH as it was found to be more stable than β-Actin. PCR efficiency analysis was performed using serial 10 fold RNA dilutions (500, 50, 5 and 0.5 ng for all target miRNAs and mRNAs) (40, 4, 0.4 and 0.04 ng for RNU48 and snoU6). The amplification curves for all assays were linear and all assays had 100-103% efficiency.
Immunofluorescence for cellular localization of PPARγ and OCLN protein in colon
Immunofluorescence studies for the detection of PPARγ (1 in 100 dilution) (Abcam, Cat No: ab118521) and OCLN (1 in 100 dilution) (Life Span Biosciences, Cat No: LS-B2437) was performed as described previously (16–17). Epithelial expression PPARγ and OCLN positive cells was confirmed using cytokeratin (1 in 500) (Biocare, Denmark) and appropriate Alexa fluor conjugated secondary antibodies (Thermo-Fisher).
Quantitative image analysis
Quantitation of cells and regions of interest (ROI) labeled by PPARγ and OCLN was performed using Volocity 5.5 software (PerkinElmer Inc, MA, USA) after capturing images on a Leica confocal microscope. Several ROI were hand drawn on the epithelial regions in the images captured from colon of chronically SIV-infected and uninfected macaques and processed utilizing the same brightness, density and black level settings. The data was first graphed and then analyzed using Mann Whitney U test employing the Prism v5 software (GraphPad software). p< 0.05 was considered statistically significant.
Cloning of 3′-UTR of PPARγ and OCLN mRNA and Dual-Glo luciferase reporter gene assay
The 3′ UTR of the rhesus PPARγ mRNA contains single predicted miR-130a binding site (TargetScan 7.1) (19) [Site: UUCCCUUCUUCCAGUUGCACUAU (nt 272-279 - Ensembl ID: ENSMMUG00000007191)]. Similarly, the 3′ UTR of the rhesus OCLN mRNA contains two predicted miR-212 binding sites (TargetScan 7.1) (19) [Site 1: ACCACCCUUCAGAUAACUGUUAU (nt 380-386)] and [Site 2: AUCUUGGUCUCUUGGACUGUUAG (nt 2424-2431 - Ensembl ID: ENSMMUG00000006216)]. Accordingly, two separate G-block double stranded DNA sequences containing the miR-130a (~53 bases long) and miR-212 (~117 bases long) binding sites along with few bases flanking the 5′ and 3′ ends of each site was synthesized (IDTDNA Technologies Inc, IA) for cloning into the pmirGLO dual luciferase vector (Promega Corp, Madison, WI). A second G-block sequence with the exact same flanking UTR sequences but with the miR-130a binding sites (8 nucleotides) deleted was also synthesized to serve as a negative control. Cloning of PPARγ and OCLN mRNA 3′ UTR sequences and the dual luciferase reporter assay to confirm direct targeting by miR-130a and miR-212 was performed as described previously (14–17). miRVana miR-130a, miR-212 and negative control (cel-miR-67-3p) mimics were purchased from Thermo Fisher Scientific (Waltham, MA). As recommended by the manufacturer, a dose titration (10, 20, 30 and 50nM) experiment was performed and a final mimic concentration of 30nM was found to cause statistically significant target repression in luciferase reporter assays. This concentration also fell within the best working concentration (3nM-30nM) recommended by the manufacturer.
The ability of Δ9-THC and cannabidiol (CBD) to transactivate PPARγ was determined using the Cignal Reporter assays (Qiagen Inc, CA). The Cignal PPARγ reporter is a mixture of a PPAR-responsive luciferase construct [contains up to three PPAR response elements (3xPPRE)] and a constitutively expressing Renilla construct. Dual reporter assays were performed in six replicate wells and repeated twice as described previously (14–17).
Trans epithelial electrical resistance (TEER) measurements
The effect of miR-130a and miR-212 induced downregulation of PPARγ and OCLN on Caco-2 colon carcinoma cell permeability was determined by monitoring TEER (20). The TEER studies were performed using the xCELLigence cell adherence (CIM) plates (ACEA Biosciences, Inc, San Diego CA) that was connected to the xCELLigence DP system to produce a time zero control for each plate and well. Following this, Caco-2 cells seeded at a density of 1x105 cells in a total volume of 100 μL. When the traces reached a plateau (~72 h) after plating (100% confluency), cells were transfected with miRVana miR-130a and miR-212 or negative control (cel-miR-67-3p) mimics. Δ9-THC (15 μM) treatments were performed at 120 h after miRNA transfection. The plates were returned to the xCELLigence DP system and traces were followed automatically using the installed software to monitor the effects of miRNA overexpression and cannabinoid treatment on electrical resistance. MiRNA transfections for TEER experiments were performed in quadruplicate wells and repeated three times.
Immunoprecipitation and Western Blotting
HEK293 cells were used for reporter and miR-212 overexpression studies as these cells abundantly express OCLN and >95-98% of these cells can be successfully transfected with exogenous nucleic acids using Lipojet® transfection reagent (Signagen Labs, MD). However, to examine the effects of miR-130a on PPARγ, we used Caco-2 cells, as basal PPARγ expression was found to be very low in HEK293 cells. Further, Caco-2 cells continue to be widely used for miRNA mechanistic studies (21). At 96 h post transfection, total protein from transfected HEK293 (cel-miR-67-3p or miR-212) and Caco-2 [pEP-Null or pEP-miR-130a plasmid (Cell Biolabs, San Diego, CA, USA)] cells was extracted using a lysis buffer (Cell Signaling Technology, Inc, Beverly, MA) containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1mM Na2EDTA, 1 mM EGTA, 1% Triton, 2.5 mM sodium pyrophosphate, 1 mM beta-glycerophosphate, 1 mM Na3VO4, 1 μg/ml leupeptin, protease inhibitor cocktail and phosphatase inhibitor cocktail (Sigma Chemical Company, MO). Total protein was quantified using the Pierce BCA protein assay kit (ThermoFisher Scientific, Waltham, MA). Unlike ready to transfect miRNA mimics, the pEP-miR-130a plasmid first generates long primary miR-130a transcripts, which are then processed to precursor and mature miR-130a by the miRNA processing machinery. Approximately, 1500 μg of total protein extract was immunoprecipitated with ~5 μl of a goat polyclonal antibody against OCLN and PPARγ (Santa Cruz Biotechnologies, CA), overnight at 4 °C followed by incubation with 30 μl (50% w/v) of protein G agarose beads (Invitrogen Corp, CA) at 4 °C for 4-5 h. The supernatant was removed and transferred to a new 1.5 mL microcentrifuge tube and immunoprecipitated using goat polyclonal antibody (~5 μl) (Santa Cruz Biotechnologies, CA) to either β-actin or ribosomal protein L5 (RPL5) at 4 °C overnight on a shaker. Immunoprecipitated OCLN, PPARγ and β-actin proteins were heat denatured for 5 min at 100 °C in sample loading buffer containing 62.5 mM Tris-HCl, 5% 2-mercaptoethanol, 10% glycerol, 2% SDS and bromophenol blue, resolved on 8-12% SDS-PAGE gels and transferred to 0.2 μm PVDF membranes (Biorad Laboratories, CA). The membranes were probed with a rabbit polyclonal primary antibody against OCLN and PPARγ (Abcam), and β-actin (Santa Cruz) followed by the appropriate horseradish peroxidase (HRP) conjugated secondary antibody (Santa Cruz, CA). Membranes were treated with West-Dura chemiluminescent substrate (Pierce Biotechnology Inc, Rockford, IL) for 5 min and the signal was captured using the FluorChem-R imaging system (ProteinSimple) and quantified using ImageJ software (NIH).
Quantitation of Plasma and Mucosal Viral Loads
Total RNA samples from all SIV-infected animals were subjected to a quantitative real-time TaqMan two-step RT-qPCR analyses to determine the viral load as described earlier (14–16).
Data Analysis
OpenArray and individual miRNA RT-qPCR data analysis was performed as described before (14–17). QuantStudio™ run files from both groups were first analyzed using ExpressionSuite software v1.0.2 (Thermo Fisher). The output file from the ExpressionSuite software analysis containing five columns (well, sample, detector, task and CT values) was saved as a tab-delimited text file, imported and analyzed using the Omics Office StatMiner qPCR analysis software (TIBCO Spotfire, Perkin Elmer, Waltham, MA) using the global normalization method. Multiple comparisons correction was performed using Benjamini-Hochberg method for false discovery rate. The CT upper limit was set to 28 meaning that all miRNA detectors with a CT value greater than or equal to 28 were excluded. Individual microRNA RT-qPCR data were normalized to a combination of two endogenous controls (snoU6 and RNU48). OpenArray miRNA (Accession number- GSE104767, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE104767) and Agilent gene expression (Accession number- GSE104771, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE104771) data have been deposited with GEO.
Transcriptome profiling and data analysis was performed by Arraystar Inc (Baltimore, USA). Acquired array images were analyzed using Rhesus Agilent Feature Extraction software (version 11.0.1.1). Quantile normalization and subsequent data processing were performed using the GeneSpring GX v12.1 software (Agilent Technologies). After quantile normalization of the raw data, genes that did not have flags were chosen for further data analysis. Differentially expressed (DE) genes with statistical significance were identified through volcano plot filtering.
Individual miRNA RT-qPCR data were analyzed using Wilcoxon’s rank sum test using Omics Office StatMiner qPCR analysis software (Perkin Elmer). For quantification of miR-130a, miR-212 in CE of SIV-infected macaques and PPARγ and OCLN mRNA in intact colon and Caco-2 cells, one uninfected control macaque or vehicle (VEH) treated Caco-2 cell sample with the largest ΔCT value served as the calibrator/reference and assigned a value of 1. MiRNA and mRNA fold change in all other macaques, THC and VEH treated colon and Caco-2 cell samples is shown as an n-fold difference relative to this sample. Quantitative immunoprecipitation/western blotting and immunofluorescence image analysis data were analyzed using Mann-Whitney U test (Prism v5 software; GraphPad software). Firefly/Renila ratios were analyzed using an unpaired “t” test. Ex vivo effects of Δ9-THC on colonic expression of PPARγ were analyzed using matched-pairs Wilcoxon’s signed rank test (Prism v5 software; GraphPad software, La Jolla, CA). p values<0.05 were considered significant.
RESULTS
Mucosal immunophenotyping, viral loads and intestinal histopathology
All chronically SIV-infected macaques used for TaqMan® OpenArray® miRNA profiling had substantial plasma and intestinal viral loads (Table I) and significant intestinal CD4+ T cell depletion (16). Colon histopathology for all six and plasma viral load data for 4 out of 6 chronically SIV-infected RMs used for ex-vivo Δ9-THC stimulation studies (Table 1) were not available. Among the chronically SIV-infected RMs (Table I), all with the exception of 11 (HH07-180D, HF54-180D, HR42-180D, HF27, GK31, JN82, DT09, IT40, JR80, JA32 and GA60) progressed to AIDS. Histologic evaluation of colon sections (Table I) from the majority (n=12) with the exception of 8 chronically SIV-infected macaques revealed the presence of mild to severe colitis including lesions such as crypt abscess / cryptitis and lymphoid hyperplasia (Table I). Mycobacterium avium intracellulare, a major opportunistic pathogen was detected in one chronically SIV-infected RM.
MiRNAs linked to chronic inflammation and epithelial barrier disruption are markedly dysregulated in CE during chronic SIV infection
To better understand the post-transcriptional mechanisms regulating aberrant gene expression associated with epithelial dysfunction, we profiled miRNA expression exclusively in the CE from 12 chronically SIV-infected and 8 uninfected control RMs. After applying Benjamini-Hochberg false discovery rate (FDR), 46 miRNAs (20 up and 26 downregulated) were found to be statistically significant (p<0.05) and differentially expressed (DE) (Figs 1A&B) following analysis using Omics Office StatMiner qPCR analysis software (TIBCO Spotfire, Perkin Elmer). The DE up and downregulated miRNAs with IDs that passed the false discovery rate cutoff are shown in the volcano plot (Fig 1B) as red and green dots, respectively. Supplemental Table 1 shows raw CT, fold change and P values for all DE miRNAs in CE of chronically SIV-infected RMs. Approximately, 60% of the detectors on the human TaqMan OpenArray microRNA panel generated a CT value of 28 or less and thus were considered to cross react with rhesus macaque miRNAs. Detectors with CT values greater than or equal to 29 were considered noise and excluded. Since the TaqMan OpenArray platform is designed for human cells rather than rhesus cells, there is a strong possibility that the species difference could have impacted our ability to detect some of the rhesus miRNA species that exhibit sequence differences from their human orthologs. These findings show that chronic SIV infection is characterized by marked changes in the expression of multiple inflammation related miRNAs in CE, the majority of which (26/46) are downregulated. Red (Fig 1A) and black (Fig 1B) arrows point to miR-130a and miR-212, two miRNAs that were previously shown to be upregulated in intestinal epithelium of IBD (22) and ALD (11) patients, respectively and directly target autophagy and TJ proteins. Accordingly, we decided to further investigate the impact of these two miRNAs on epithelial barrier disruption.
Figure 1.
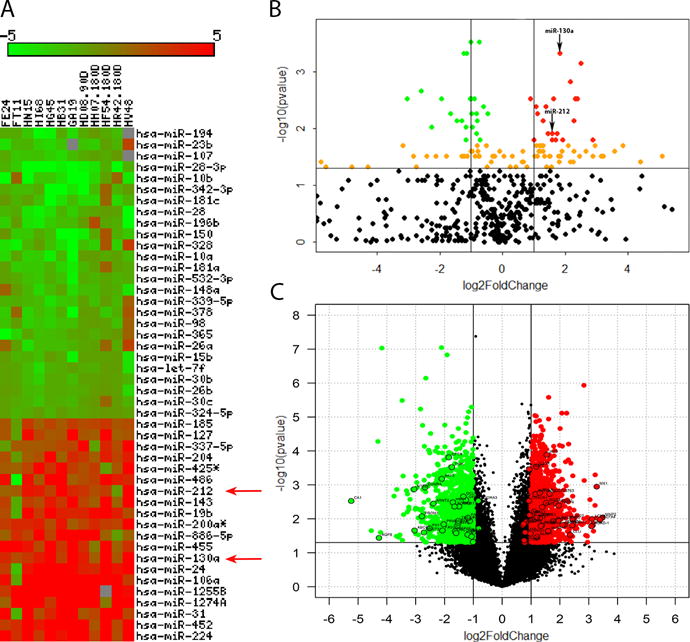
Changes in miRNA expression in colonic epithelium of chronically SIV-infected rhesus macaques. Heat map shows all differentially expressed (adjusted p<0.05) miRNAs in colonic epithelium of chronically SIV-infected rhesus macaques after applying Benjamini-Hochberg multiple comparisons correction (A). MiRNA species originating from the opposite arm of the precursor are denoted with an asterisk (*). Volcano plot shows the relationship between fold-change (X-axis) and statistical significance (Y-axis) of DE miRNAs (B) and mRNAs (C). The vertical lines in (B) correspond to 2.0-fold up and down, respectively, and the horizontal line represents p<0.05. Up and downregulated miRNAs with statistical significance (adjusted p<0.05) are shown in red and green circles, respectively. Orange circles represent up and downregulated miRNAs with adjusted p>0.05. Red (A) and black (B) arrows denote inflammation associated miR-130a and miR-212. The vertical lines in (C) correspond to 2.0-fold up and down, respectively, and the horizontal line represents p<0.05. Up and downregulated mRNAs with statistical significance (p<0.05) are shown in red and green circles, respectively. All DE mRNAs listed in Supplemental tables 2 and 3 are highlighted with a black outer border (C). The negative log of statistical significance (p value) (base 10) is plotted on the Y-axis, and the log of the fold change base (base 2) is plotted on the X-axis in B and C.
Genes associated with epithelial barrier integrity/function, protection against oxidative injury, ion transport, double stranded DNA damage repair and autophagy are significantly downregulated in colonic epithelium of chronically SIV-infected macaques
Since miRNAs have been proposed to function predominantly by destabilizing and degrading mRNAs (22), we next performed genome wide transcriptome analysis on a subset of the samples to determine the functional significance of the DE miRNAs. Supplemental Tables 2 and 3 show a select list of genes with strong relevance to epithelial function that exhibited downregulation or upregulation, respectively. The volcano plot (Fig 1C) shows all DE genes and those listed in Supplemental Tables 2 and 3 are highlighted with a black outer border. More importantly, these tables show which of these rhesus macaque specific genes are predicted targets of any of the 46 miRNAs we found to be DE in CE. Supplemental Table 2 lists significantly downregulated (p<0.05) genes critical to epithelial barrier integrity/structure, epithelial proliferation/differentiation, resistance to oxidative stress, ion transport and double stranded DNA damage repair. Downregulated genes regulating formation and maintenance of epithelial barrier function included 5′-nucleotidase ecto (NT5E, also known as CD73) (24), ATP-binding cassette, sub-family B (MDR/TAP), member 1 (ABCB1) (25), occludin (OCLN) (26), peroxisome proliferator activator receptor gamma (PPARγ) (27), peroxisome proliferator-activated receptor gamma, coactivator 1 α (PPARGC1A) (28), par-3 partitioning defective 3 homolog (PARD3) (29), cystic fibrosis transmembrane regulator (CFTR) (30) and adenosine A3 receptor (ADORA3) (31), endothelin receptor type A and B (EDNRA/EDNRB), trefoil factor-2 (TFF2) (32), aryl hydrocarbon receptor nuclear translocator 2 (ARNT2), hepatocyte nuclear factor 4 gamma (HNF4G) (33), high mobility group-box transcription factor 1 (HBP1) (34), fibronectin 1 (FN1) and integrin alpha 8 (ITGA8). A noteworthy candidate among the epithelial barrier associated genes was CD73, also known as 5′-ectonucleotidase, that was downregulated ~8 fold. Additionally, genes associated with protection against oxidative injury / inflammation (oxidation resistance 1, myocyte enhancer factor 2C) (35–36), intestinal stem cell maintenance [prominin 1 (PROM1) (37), RPTOR independent companion of MTOR, complex 2 (RICTOR) (38), SRY (sex determining region Y)-box 6 (SOX6)], double stranded DNA damage repair (RAD17 and RAD50), intestinal ion/water transport/homeostasis [carbonic anhydrase 1 (CA1) (39), aquaporin 8 (AQP8) (40), bestropin 2 (BEST2)] (41), Wnt signaling [adenomatous polyposis coli (APC), TRAF2 and NCK interacting kinase (TNIK), dickkopf-3 (DKK3)], cell survival [Fas ligand (TNF superfamily, member 6) (FASL), v-akt murine thymoma viral oncogene homolog 3 (AKT3)] (42) and negative regulator of NFκB signaling [(tumor necrosis factor, alpha-induced protein 3 (TNFAIP3, also known as A20)] (43) showed significant downregulation. As evident from the pie chart (Fig 2A), majority of the select list of downregulated genes in the CE during chronic SIV infection were associated with epithelial barrier function.
Figure 2.
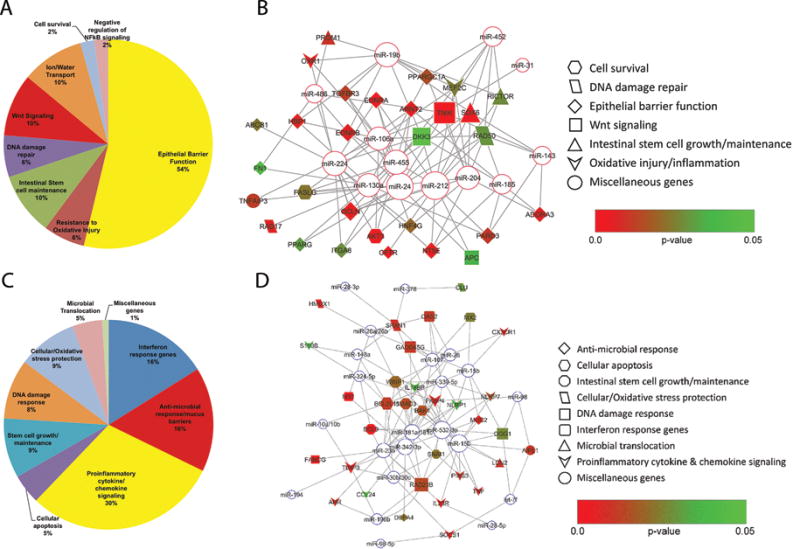
The pie chart displays functional categories of genes that showed decreased (A) or increased expression (C) in the CE during chronic SIV infection. The relative size of each sector in the pie chart is determined by the number of genes in that functional category. Genes with unknown function are not included in the pie chart. The identities of all DE transcripts shown in the pie charts are provided in Supplemental Table 2 and 3. Rhesus macaque specific miRNA-mRNA target pairs in colonic epithelium identified using TargetScan 7.1 (19). Networks linking a select list of differentially expressed miRNAs and their targeting mRNAs were generated using Cytoscape (44): (B) upregulated miRNAs and downregulated mRNAs; and (D) downregulated miRNAs and upregulated mRNAs. MicroRNAs and their interacting genes are connected with lines, and are indicated via various symbols: Open circles with a red or blue border for microRNA and differently shaped symbols for genes associated with colonic epithelial function. The filling colors for gene symbols indicate p-values when gene expression was compared between chronic SIV and uninfected controls. The size of a symbol is proportional to the degree of connectivity of the corresponding miRNA/mRNA, which is the number of interacting mRNAs/miRNAs in the networks. EB- Epithelial barrier, Wnt- Wingless type MMTV integration site.
Based on predicted targets categorized from transcriptomic data, we identified several upregulated (miR-106a, miR-130a, miR-143, miR-185, miR-19b, miR-204, miR-212, miR-24, miR-224 and miR-455) and downregulated (miR-107, miR-15b, miR-150, miR-181a/181c, miR-23b, miR-324-5p, miR-342-3p, miR-532-3p, miR-30b/30c, miR-378 and miR-98) miRNAs that may potentially play significant roles in HIV/SIV induced intestinal epithelial dysfunction. Next, we used TargetScan 7.1, (the only algorithm that provides miRNA target information for the rhesus macaque) to determine which genes are predicted to be targeted and potentially regulated by the 20 upregulated miRNAs (identify miRNA-mRNA pairs). To maintain focus, we restricted this analysis to the genes listed in Supplemental Tables 2 and 3 as these have been previously demonstrated to play well established roles in intestinal epithelial function and homeostasis. Based on this supervised analysis, we identified 8 upregulated miRNAs [miR-212 (n=13), miR-24 (n=11), miR-455 (n=10), miR-19b (n=10), miR-130a (n=9), miR-204 (n=9), miR-224 (n=9) and miR-106a (n-9) that were predicted to directly bind and potentially regulate most of the downregulated genes. Additionally, each of these 8 miRNAs was bioinformatically predicted to regulate, at least, nine rhesus macaque specific target genes listed in Supplemental Table 2 and is shown as an interactive network using Cytoscape (44) in figure 2B. We would like to emphasize that the miRNA-mRNA pairs / interactive networks shown in Fig 2 are only predicted and yet to be validated. Taken together, the significant downregulation of genes linked to formation and maintenance of epithelial tight junctions, resistance to oxidative stress, and DNA damage repair can facilitate epithelial barrier disruption and epithelial dysfunction. The decreased expression of CD73 and ADORA3 suggests dysfunction of the intestinal adenosine signaling pathway that may lead to accumulation of ATP, a key “danger” signal known to trigger persistent proinflammatory responses (24). Finally, miRNA-mRNA target pair analysis identified a potential post-transcriptional mechanism that may partially explain their decreased expression.
MRNA expression of genes associated with interferon response, anti-microbial defense, apoptosis, oxidative DNA damage, endoplasmic reticulum stress and inflammasome signaling are markedly upregulated in colonic epithelium during chronic SIV infection
Consistent with chronic inflammatory signaling, the upregulated gene repertoire was dominated by genes linked to interferon signaling (interferon stimulated gene 12c protein (IFI27), myxovirus resistance 1 (MX1), myxovirus resistance 2, interferon alpha inducible protein 6 (IFI6), 2′-5′-oligoadenylate synthetase 2 (OAS2) and interferon regulatory factor 9 (IRF9), anti-microbial response / mucus barriers [oral alpha defensin 1 (ROAD-1), alpha-defensin 2 (MNP2), defensin alpha 4 (DEFA4), defensin-7-like, beta defensin 122 and alpha-defensin 6 precursor and mucin 2 (MUC2)], inflammasome signaling [NLR family, pyrin domain containing 7, (NLRP7), NLR family, pyrin domain containing 1 (NLRP1) (45), interleukin 18 binding protein (IL18BP)], proinflammatory cytokine and chemokine signaling [tumor necrosis factor (TNF), interleukin 23 receptor (IL23R), chemokine (C-C motif) ligand 24 (CCL24), C-X3-C Motif Chemokine Receptor 1 (CX3CR1)], negative regulators of cytokine signaling [suppressor of cytokine signaling 1 (SOCS1), protein inhibitor of activated STAT3 (PIAS3), and FK506-binding protein 4 (FKBP4), aryl hydrocarbon receptor (AHR), TIMP metallopeptidase inhibitor 3 (TIMP3), S100 calcium binding protein B (S100B)], cellular apoptosis [(BAK1) and (BCL2L11)] and intestinal stem cell growth and maintenance [B-cell lymphoma 9 (BCL9) (46), WNT1 inducible signaling pathway protein 1 (WISP1)] (47), SMAD family member 3 (SMAD3) and snail homolog 1 (Snai1) (50). Narrowing our analysis to the CE also yielded deeper insights into previously unreported molecular changes during chronic infection. These included enhanced expression of genes encoding proteins associated with DNA damage response [8-oxoguanine DNA glycoxylase (OGG1), fanconi anemia, complementation group G (FANCG), RAD23 homolog B (RAD23B) and Growth arrest and DNA-damage-inducible, gamma (GADD45G)], cellular / oxidative stress protection [Clusterin (CLU) (48), (heme oxygenase-1 (HMOX1)] (49), endoplasmic stress response [DNA-damage-inducible transcript 3 (DDIT3), also known as CEBP homologous protein (CHOP)] (51), and microbial translocation [dual oxidase 1 (DUOX1) and lipocalin 2 (LCN2)] (52). It is clear from the pie chart (Fig 2C) that majority of the select list of upregulated genes belonged to the proinflammatory and interferon signaling and anti-microbial defense category.
With the exception of ten genes, each of the 44 upregulated genes shown in Supplemental Table 3 were predicted to be directly targeted by, at least, one and in some cases up to five downregulated miRNAs (19) (Fig 2D). Among these, we previously characterized miR-150 in detail and validated IRAK1 as a direct target of miR-150 in both colonic / jejunal epithelium and lamina propria immune cells (LPLs) of chronically SIV-infected RMs (15). Overall, the data suggests that elevated expression of interferon-induced genes, defensins and cytokine genes may represent a normal host response to SIV replication, and the markedly increased DDIT3 expression may contribute to loss of epithelial barrier function. Further, the significant downregulation of negative regulators like miR-150 and miR-532-3p may assist the intestinal epithelium in mounting an immune response through elevated expression of proinflammatory genes.
PPARγ and OCLN are direct targets of miR-130a and miR-212, respectively, and their protein expression is significantly decreased in colonic epithelium of chronically SIV infected rhesus macaques
We next selected and validated miR-130a and miR-212 as both miRNAs based on the transcriptome analysis described above are predicted to directly target proteins regulating (TargetScan 7.1) (19) intestinal epithelial barrier function and inflammation. Further, both miR-130a and miR-212 have been previously shown to be upregulated in intestinal epithelium of IBD (23) and ALD (11) patients, respectively. Therefore, these and our data suggest that upregulation of both miRNAs is common to intestinal inflammatory diseases. Consistent with the OpenArray data (Fig 1A), RT-qPCR analysis confirmed statistically significant upregulation of both miR-130a and miR-212 (Fig 3A&E) in CE of chronically SIV-infected RMs.
Figure 3.
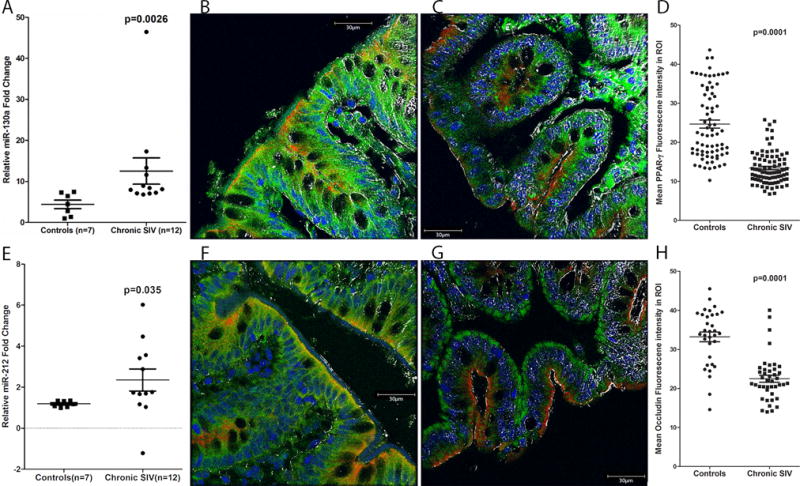
PPARγ and OCLN protein expression is significantly decreased in colonic epithelium of chronically SIV-infected rhesus macaques. RT-qPCR confirmation of miR-130a (A) and miR-212 (E) in colonic epithelium of chronically SIV-infected rhesus macaques (n=12) relative to control samples (n=7). RT-qPCR data were analyzed using the non-parametric Wilcoxon’s rank sum test for independent samples. The error bars represent standard error of mean fold change within each group. Panels B, C, F and G involve triple labels with PPARγ and OCLN (green), cytokeratin (red) and Topro3 for nuclear staining (blue). The grey channel represents differential interference contrast (DIC) to reveal tissue architecture. Note the significantly decreased PPARγ (C) and OCLN (G) staining in the colonic epithelium of the chronically SIV-infected macaque. In contrast, PPARγ (B) and OCLN (F) staining is intense in the colonic epithelium of the control macaque. All panels are 40X magnification. Quantification of cells and regions of interest (ROI) labeled by PPARγ (D) and OCLN (H) was performed using Volocity 5.5 software after capturing images on a Leica confocal microscope. Several ROI were hand drawn on the epithelial regions in the images from three chronic SIV and two uninfected controls. Image analysis data were analyzed using non-parametric Mann-Whitney U test.
Since we had extensively characterized miR-150 (downregulated miRNA in Fig 1A) previously (15), here we decided to focus on two upregulated miRNAs, miR-130a and miR-212 and their target genes PPARγ and OCLN, respectively, as both are central to maintenance of epithelial barrier function (26–28). More importantly, PPARγ has been shown to play a critical role in preventing dysbiosis by decreasing the bio-availability of host derived nitrates and oxygen in colonic epithelial cells (53). The 3′ UTR of rhesus macaque PPARγ mRNA contains a single miR-130a binding site (Position 272-279 - Ensembl ID: ENSMMUG00000007191) that is highly conserved across several mammalian species (19). Similarly, miR-212 was found to have two predicted binding sites (Site 1: Position 380-386 and Site 2: 2424-2431, Ensembl ID: ENSMMUG00000006216) on the 3′ UTR of rhesus macaque OCLN mRNA (19). While site 1 is conserved across multiple mammalian species, site 2 is conserved only in the human, chimpanzee and rhesus macaque. Consistent with changes in mRNA expression (Supplemental Table 2), protein expression of PPARγ (Fig 3C) and OCLN (Fig 3G) was significantly decreased in CE of chronically SIV-infected compared to control RMs (Fig 3B&F). Both PPARγ and OCLN protein localized to epithelial cell membranes and its expression, as evident in figure 3B&F, was intense in both apical border and crypts. Marked loss of PPARγ (Fig 3C) and OCLN (Fig 3G) protein expression from the apical border and reduced expression in crypts was observed in the colons of chronically SIV-infected RMs. Quantitative image analysis further confirmed the reduced PPARγ (Fig 3D) and OCLN (Fig 3H) protein expression in the CE of chronically SIV-infected RMs.
To confirm that both PPARγ and OCLN are strong targets of miR-130a and miR-212, respectively, we further performed an in silico miRNA-mRNA duplex analysis using RNAhybrid (54) to determine the minimum free energy of hybridization for both pairings. Interestingly, both miR-130a-PPARγ and miR-212-OCLN duplexes had significantly low minimum free energy (mfe< −20 kcal/mol) (Fig 4A&E). Consequently, luciferase reporter assays demonstrated that miR-130a and miR-212 at 30nM concentration had the ability to directly bind the 3′ UTRs of PPARγ and OCLN, respectively, and potentially downregulate their protein expression (Fig 4B&F). The 30nM mimic concentration is within the best workable range (3nM-30nM) recommended by the manufacturer (Thermo Fisher Scientific). Note the significant reduction in Firefly/Renila ratios following transfection of HEK293 cells with PPARγ (45%) (Fig 4B) and OCLN (34%) (Fig 4F) wild type pmirGLO vector with miR-130a and miR-212 mimics, respectively. Similar reductions in luciferase activities have been described for miR-215/192 and miR-199b, wherein both miRNAs were shown to play significant roles in the pathogenesis of IBD (55) and IBS (56). Note the absence of reduction in Firefly/Renila ratios following co-transfection of wild type pmirGLO vector and negative control mimic (Fig 4B&F). To confirm functional significance of miRNA function, we moderately overexpressed (30nM) miR-130a (Caco-2 epithelial cells) and miR-212 (HEK293 cells) and detected significant reduction in PPARγ (Fig 4C) and OCLN (Fig 4G) protein levels, respectively. Image quantification (PPARγ and OCLN band intensity relative to β-Actin ratios) confirmed significant reduction in PPARγ (Fig 4D) and OCLN (Fig 4H) target protein levels 96 h following miRNA transfection. These findings provide strong evidence that dysregulated miR-130a and miR-212 expression in the CE can potentially disrupt the integrity of the intestinal epithelial barrier by directly targeting and downregulating tight junction and other proteins like PPARγ critical to maintenance of epithelial barrier function.
Figure 4.
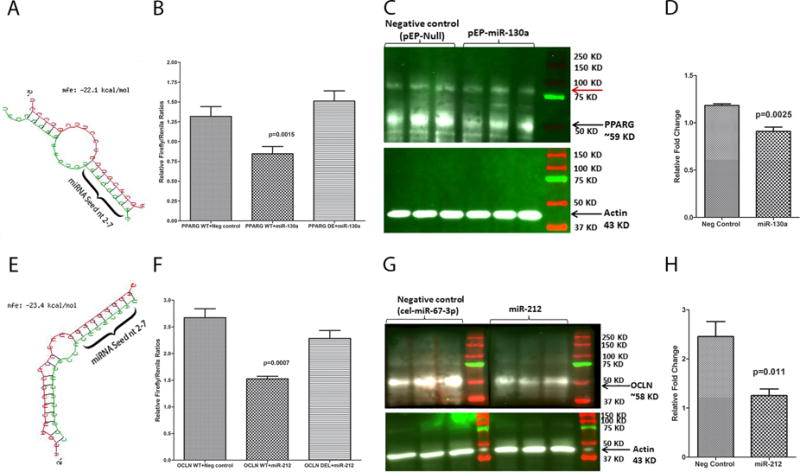
PPARγ and OCLN are direct targets of miR-130a (A-D) and miR-212 (E-H), respectively. Luciferase reporter vectors containing a single highly conserved miR-130a (A) and miR-212 (E) binding site (seed nucleotide region) on the rhesus macaque PPARγ and OCLN mRNA 3′ UTR, respectively, or the corresponding construct with the binding sites deleted were co-transfected into HEK293 cells with 30 nM miRNA mimics (miR-130a or miR-212 or Negative control). Firefly and Renila luciferase activities were detected using the Dual-Glo luciferase assay system 96 h after transfection. Note significantly reduced Firefly/Renilla ratios following co-transfection of miR-130a (B) and miR-212 (F) mimics with pmirGLO vector containing wild type (WT) miRNA binding sties. Representative Western blot (Triplicate wells) and quantification showing reduction in protein expression of PPARγ (C-D) and OCLN (G-H) (both~58 kD) 96 h post transfection of Caco-2 cells with pEP-Null or pEP-miR-130a expressing plasmid and HEK293 cells with negative control (cel-miR-67-3p) or miR-212 mimics. Red arrow (C) indicates a non-specific band detected in all lanes. Firefly/Renila ratios were analyzed using unpaired “t” test. Western blot densitometry data was analyzed using Mann-Whitney “U” test.
MiR-130a and miR-212 mediated reduction in trans epithelial electrical resistance (TEER) is reversed by cannabinoid (Δ9 -THC) treatment in vitro
Since moderate overexpression of miR-130a and miR-212 decreased PPARγ and OCLN protein levels, respectively, we then sought to determine whether this resulted in increased epithelial barrier permeability (biological significance) by measuring trans epithelial electrical resistance. Interestingly, relative to negative control mimic (cel-miR-67-3p) transfected wells (blue lines in figure 5A), moderate overexpression of both miR-130a (green lines in figure 5A) and miR-212 (red lines in figure 5A) in quadruplicate wells markedly reduced TEER at ~72 h post transfection of Caco-2 cells that continued to remain low even at 120 h post transfection. At 120 h post transfection, reductions in TEER induced by miR-212 (p=0.002) but not miR-130a (p=0.0762) showed statistical significance (Fig 5A). While these findings represented an important inflammation induced epigenomic mechanism triggering epithelial barrier disruption, it was equally intriguing to investigate if the miRNA mediated epithelial hyper permeability could be reversed using feasible pharmacological strategies (Δ9-THC) that we had previously used to inhibit gastrointestinal inflammation (17). Interestingly, daily treatment of miR-130a and miR-212 transfected cells with 15 μM Δ9-THC (15, 56–57) (downward facing black arrows in Fig 5A) restored TEER to the levels seen in cells transfected with the negative control miRNA mimic. Nevertheless, some differences were noticed in the ability of Δ9-THC to restore TEER to control levels between miR-130a and miR-212 transfected wells. While TEER restoration to control levels was achieved after two THC-containing media changes for miR-130a, five such changes were required to reverse miR-212 mediated reduction in TEER. Initial studies using both mimics confirmed failure of TEER restoration up to 265 h post transfection (data not shown) in the absence of Δ9-THC treatment despite media changes at 48 h intervals suggesting that the TEER restoration was specifically in response to Δ9-THC treatment. Further, treatment of Caco-2 cells with 15 μM Δ9-THC significantly increased PPARγ but not OCLN gene expression suggesting that Δ9-THC could override the inhibitory effects of miR-130a on PPARγ expression (Fig 5B). Experiments using a PPRE containing luciferase reporter vector confirmed that Δ9-THC (15 μM) could directly trans activate PPARγ transcription in Caco-2 cells and potentially primary intestinal epithelial cells (Fig 5C). These findings also suggested that the PPARγ protein induced by Δ9-THC could then bind PPRE binding elements in the promoters of PPARγ responsive genes and regulate their expression (Fig 5C). Collectively, these results depict a potential molecular mechanism underlying disruption of the intestinal epithelial barrier in chronic HIV/SIV infection, wherein dysregulated miR-130a and miR-212 can increase epithelial paracellular permeability by directly targeting and downregulating the expression of PPARγ and OCLN, respectively. Finally, these findings also demonstrate the ability of cannabinoids, (Δ9-THC) to reverse and restore miRNA-mediated increases in epithelial permeability by directly inducing PPARγ expression.
Figure 5.
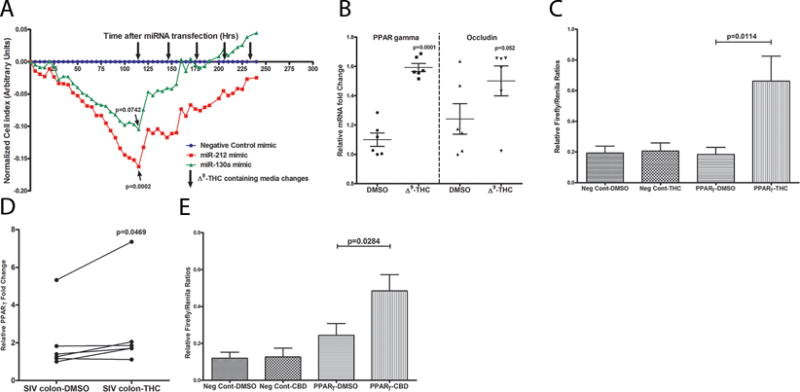
Transfection of Caco-2 cells with 30nM PPARγ targeting miR-130a (green) and OCLN targeting miR-212 (red) mimics markedly decreased TEER at 120h post transfection compared to negative control miRNA mimic (cel-miR-67-3p) transfected cells (blue) (A). MiRNA transfections were performed in quadruplicate wells and repeated three times. Note TEER restoration to control levels after two and five Δ9-THC-containing media changes (down-facing arrows) for miR-130a & miR-212, respectively. Δ9-THC significantly increased PPARγ mRNA expression (B) in Caco-2 cells by directly trans activating (C) PPARγ gene expression. To confirm that Δ9-THC stimulates PPARγ expression, Caco-2 cells were transfected with a luciferase reporter gene containing the PPARγ response element (3×PPRE-TK-Luc) and a negative control vector. Note significant increase in luciferase activity (firefly/renilla ratios) 8 h after 15 μM Δ9-THC treatment of cells transfected with 3xPPRE-TK-Luc (PPARγ) but not negative control vector (C). DMSO treatment of cells transfected with negative control vector or 3xPPRE-TK-Luc (PPARγ) vector did not increase firefly/renilla ratios. Treatment of colon tissue from chronically SIV-infected rhesus macaques with 15 μM Δ9-THC for 16 h significantly increased PPARγ mRNA expression (D). Like Δ9-THC (C), luciferase reporter assays confirmed the ability of cannabidiol, a non-psychotropic/anti-inflammatory cannabinoid to directly trans activate PPARγ expression (E). Data (B, C & E) are shown as means with error bars representing SEM.
Ex vivo Δ9-THC treatment of colon tissue from chronically SIV-infected rhesus macaques significantly upregulated PPARγ but not OCLN mRNA expression
Since Δ9-THC directly trans activated PPARγ expression in Caco-2 cells, we next harvested intact colon tissue from chronically SIV-infected RMs and stimulated them ex vivo with 15 μM Δ9-THC to determine its effect on PPARγ and OCLN expression. This concentration of THC has been previously shown to inhibit B-cell activation in vitro without reducing cell viability (57). The physiological relevance of this concentration was previously described by Ngaotepprutaram et al. (58). Interestingly, similar to the findings in Caco-2 cells, Δ9-THC significantly (p=0.0469) upregulated PPARγ mRNA expression following 16 h of culture (Fig 5E). As observed in Caco-2 cells, Δ9-THC treatment did not impact OCLN mRNA expression (data not shown). Interestingly, like Δ9-THC, the non-pyschotropic CBD, which is a strong inhibitor of intestinal inflammation (59) also trans activated PPARγ expression in Caco-2 cells (Fig 5E). Given that PPARγ has been identified as a therapeutic target for restoring epithelial barrier function (26,60–62) and critical for the prevention of intestinal dysbiosis (53), the positive effects of cannabinoids (both Δ9-THC and CBD) on PPARγ expression has strong translational relevance in not just HIV/SIV but potentially other chronic inflammatory diseases of the intestine.
DISCUSSION
Intestinal epithelial dysfunction, a central feature of HIV/SIV infection, characterized by increased apoptotic cell death, decreased maturation and differentiation of enterocytes and a dysregulated gene expression program is now well known to significantly contribute to AIDS pathogenesis (8,63–64). The clinical consequences of epithelial barrier disruption include chronic malabsorption diarrhea, interference with uptake of nutrients and anti-retroviral medications and most notably, microbial translocation, an important pathogenic event proposed to induce systemic immune activation and drive AIDS progression. A key contributor to epithelial dysfunction is the chronic persistent inflammatory milieu in the underlying lamina propria that potentially drives pathological gene expression in epithelial cells. While we and others have reported dysregulated gene expression (63,65–66), the mechanisms that regulate pathological intestinal epithelial gene expression remains unknown and unexplored. Recently, miRNAs, the well-studied class of non-coding RNAs were demonstrated to regulate epithelial paracellular permeability and several aspects of the inflammatory response through post-transcriptional regulation of epithelial tight junction proteins (11–13) and pro-inflammatory mediators (67). To gain a better understanding of the epigenetic mechanisms underlying epithelial barrier disruption, we first profiled and identified significant dysregulation of epithelial miRNA expression during chronic SIV infection. Next, using a parallel transcriptome analysis, we identified miRNA-mRNA target pairs and regulatory networks linked to epithelial barrier disruption and chronic persistent inflammation. Finally, using in vitro cell line studies, we demonstrated the ability of cannabinoids (THC) to reverse and restore miRNA mediated barrier disruption.
To the best of our knowledge, the present study provided a global view of changes in the miRNA landscape exclusively in CE during chronic HIV/SIV infection. Although, the significantly elevated expression of inflammation associated miR-130a, miR-212, miR-204, miR-24, miR-224 and miR-455 likely represents a normal host response to injury, paradoxically, their persistent upregulation can have deleterious effects on epithelial homeostasis and barrier function. The finding that miRNAs regulate gene expression by destabilizing and degrading mRNAs (23) prompted us to perform a parallel transcriptome analysis on a subset of samples to better understand miRNA function. Consistent with the microbial translocation theory, we identified significant downregulation of genes that not only form TJ structure (OCLN) but also regulate and maintain its integrity (PPARγ, CFTR, TFF2, PARD3, HNF4G, HBP1, ABCB1, ADORA3 and NT5E or CD73). Among these, disruption of the tight junction protein OCLN is well known to cause epithelial barrier defects in chronic inflammatory diseases of the intestine (26,12). At least, six upregulated miRNAs, miR-204, miR-212, miR-224, miR-24, miR-455 and miR-486 were predicted to directly target OCLN with miR-212 and miR-455 having more than one predicted binding site on the 3′ UTR of OCLN mRNA (19). Similarly, PPARγ, is known to regulate expression of genes critical to lipid metabolism, adipocyte differentiation, insulin signaling (27–28) and dysbiosis (53). Although, PPARγ is expressed at high levels in normal CE (60–62), its expression is markedly reduced in CE of IBD (ulcerative colitis) patients and negatively correlated with endoscopic disease activity (26). Further, while LPS was found to downregulate PPARγ expression (68), the underlying regulatory mechanisms remained unknown. Understanding the mechanisms that regulate its expression is necessary to develop novel ways of restoring PPARγ activation, a strategy proposed for the treatment of IBD (26,61). The identification of PPARγ as a predicted target of miR-130a and miR-24 suggested a possible post transcriptional mechanism mediating its downregulation. Similarly, two enzymes apyrase (CD39) and 5′-nucleotidase, ecto (NT5E or CD73) sequentially degrade the high levels of extracellular ATP pool released by dead cells and commensal / dysbiotic bacteria into immunosuppressive adenosine, which then negatively regulates inflammatory responses by binding to adenosine receptors on the epithelium (23). Equally important finding was the markedly reduced expression of CD73 and ADORA3 suggesting major impairment in the generation of anti-inflammatory adenosine and persistence of high levels of pro-inflammatory ATP, thereby resulting in sustained inflammatory signaling. Gaulke et al. (69) had previously performed simultaneous miRNA and gene expression profiling in intact jejunum tissue during chronic SIV infection and identified a DE miRNA profile that showed decreased expression in similar proportions to our study. Using RT-qPCR, decreased expression of miR-16, miR-194 and miR-200c was further confirmed in jejunal crypt epithelial cells. Bioinformatics analysis identified several DE genes known to regulate epithelial cell cycle, differentiation and cell death to be either validated or predicted targets of the three downregulated miRNAs. In contrast, we used a different approach by doing our initial profiling in purified CE cells. This ensured that the differential miRNA and gene expression reported here occurred in the same cell type, which greatly simplified downstream miRNA-mRNA target pair analysis.
In general, immune activation results in enhanced gene expression so that immune cells become well equipped to attack and eliminate invading pathogens. However, successful expression of key immune response genes is contingent on the silencing of negative regulators like miRNAs (70–71). In immune cells, this is accomplished by rapid ubiquitination and degradation of the miRNA processing machinery resulting in attenuation of miRNA expression (71). The fact that epithelial cells, like their immune cell counterparts actively elicit immune responses (72) might partially explain why the majority (~56%) of DE miRNAs showed decreased expression. In accordance with this model, microarray analysis detected significant upregulation of interferon-stimulated genes, anti-microbial peptides (defensins), cytokines (TNF), negative regulators of cytokine signaling, inflammasome activation genes, double stranded DNA damage response genes and endoplasmic stress response (DDIT3, CLU) genes. Interestingly, the multifunctional transcription factor, DDIT3, has been shown to activate the expression of BCL2L11 and GADD34/45 and at the same time downregulate PPARγ expression (73). In perfect agreement with this finding, the expression of BCL2L11 and GADD34/45 increased and that of PPARγ decreased significantly in CE of chronically SIV-infected RMs. More recently, elevated expression of DUOX2, a major producer of hydrogen peroxide (H2O2) in the intestinal epithelium, was correlated with expansion of Proteobacteria in IBD patients (74). This finding is important as DUOX1, a paralog of DUOX2, which also catalyzes H2O2 production was significantly upregulated in CE of chronically SIV-infected RMs. Expansion of proteobacteria was recently reported in colon of chronically SIV-infected RMs (75). Similar to IBD patients, DUOX2 (probe ID: A_01_P1747461) expression was upregulated by ~1.9 fold in CE of SIV-infected RMs but did not reach statistical significance (p=0.14). LCN2 was another important gene linked to bacterial translocation that showed significantly heightened expression. LCN2 limits iron availability by binding to enterobactin, an iron chelator produced by Enterobacteriaceae. Using the TargetScan algorithm, at least, 5 downregulated miRNAs (miR-150, miR-532-3p, miR-342-3p, 181a/c and miR-23b) were predicted to directly target the majority of these downregulated genes (rhesus macaque specific) and their decreased expression is predicted to facilitate an enhanced epithelial immune response to viral replication.
Owing to their functional importance in the maintenance of epithelial barrier integrity, we further characterized PPARγ and OCLN and their targeting miRNAs, miR-130a and miR-212, respectively. Although, it is now clear that decreased PPARγ expression is common to both IBD and HIV/SIV infection, the molecular mechanisms driving its decreased expression in CE remains unknown and undetermined. Moreover, studies performed in mouse models of colitis and ulcerative colitis patients have revealed clinically important roles of PPARγ activation using thiazolidinediones in the regulation of inflammation and immune response, specifically through colonic epithelial cells (60–62). The presence of a miR-130a binding site in the 3′ UTR of RM PPARγ mRNA that is highly conserved across multiple vertebrate species (19) suggested a potential post-transcriptional regulatory mechanism in CE. Similarly, TNFα-induced miR-122 was previously demonstrated to regulate OCLN protein expression in colon of IBD patients (12). As miR-122 expression was undetectable in CE of SIV-infected RMs, we scanned and identified, at least, six different upregulated miRNAs including miR-212, which is predicted to have two binding sites on the 3′ UTR of OCLN and previously shown to downregulate zonula occludens-1 (ZO1) expression in intestinal epithelium of chronic ALD patients (11). Results from the reporter, overexpression and TEER experiments for the first time demonstrated the potential of miR-130a and miR-212 to decrease PPARγ and OCLN expression levels, respectively, in CE and potentially increase intestinal epithelial permeability in chronically SIV infected RMs. Although our findings illustrate an important role for miRNAs in post-transcriptional gene regulation, the reader should be aware that there are other epigenetic mechanisms like chromatin remodeling, histone modifications, DNA methylation., etc that could play critical roles in regulating gene expression.
Healing the intestinal mucosa and restoring epithelial barrier integrity is a top priority of the GI research community. Unlike previously published work by us (14–17) and others (67), in the current study, we not only performed mechanistic studies but also explored the potential of using feasible pharmacological strategies to reverse miRNA mediated epithelial barrier disruption. As PPARγ was shown to be the major functional receptor mediating the effects of commonly prescribed anti-inflammatory amino salicylates in IBD patients (28), strategies to enhance PPARγ expression have gained widespread attention for the clinical management of intestinal inflammation (61). While, miRNA modulation using nanoparticle based delivery systems is being investigated as a viable therapeutic strategy in many diseases (76), there are currently no reliable, safe and cost-effective in vivo delivery systems in place. Therefore, we turned our focus on cannabinoids that have been historically used to treat various GI disorders and accumulating new evidence suggests that they could have long-lasting therapeutic effects in numerous GI disorders (77–78). Moreover, in this context, we recently demonstrated its ability to exert protective effects on the gastrointestinal tract through miRNA modulation in SIV-infected RMs (21). Our findings on the ability of cannabinoids (THC) to reverse miRNA mediated increase in epithelial permeability in vitro and directly trans activate the anti-inflammatory and anti-dysbiotic PPARγ (53) expression in ex vivo colon explant cultures are exciting as it identifies a promising yet feasible strategy to restore epithelial barrier integrity and prevent dysbiosis in not only HIV/SIV but also other chronic inflammatory diseases of the intestine such as IBD, IBS, celiac disease. etc.
In summary, the integrative miRNA-mRNA approach employed in the present study not only identified changes in the epithelial transcriptomic landscape but also provided deeper insights into the regulation of pathological epithelial gene expression during chronic HIV/SIV infection. The ability of miR-130a and miR-212 to directly target and downregulate PPARγ and OCLN for the first time provides a novel molecular mechanism mediating epithelial barrier disruption in HIV/SIV infection. Apart from miR-130a and miR-212, the identification of numerous miRNA-mRNA pairings with potential to regulate epithelial homeostasis including other miRNAs predicted to directly target PPARγ and OCLN underscores the enormous plasticity of miRNA function. Lastly, from a purely clinical perspective, we also show that miRNA mediated increase in intestinal epithelial permeability can be reversed and restored by cannabinoids. With increasing legalization of medical marijuana and given that epithelial barrier dysfunction is a central feature of HIV/SIV infection, future studies are definitely needed to determine whether in vivo cannabinoid administration in combination with anti-retroviral therapy can inhibit residual intestinal inflammation, microbial translocation and restore epithelial barrier integrity.
Supplementary Material
Acknowledgments
The authors would like to thank Xavier Alvarez, Maurice Duplantis, Faith R. Schiro, Cecily C. Midkiff, Christopher Monjure and Coty Tatum for their technical assistance in the study.
Research reported in this publication was supported by the National Institutes of Health Award Numbers R01DK083929, R01DA042524 and R56DE026930 to MM, R01NS104016 to AM and OD011104 (formerly RR00164). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Veazey RS, DeMaria M, Chalifoux LV, Shvetz DE, Pauley DR, Knight HL, Rosenzweig M, Johnson RP, Desrosiers RC, Lackner AA. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science. 1998;280:427–431. doi: 10.1126/science.280.5362.427. [DOI] [PubMed] [Google Scholar]
- 2.Mudd JC, Brenchley JM. Gut Mucosal Barrier Dysfunction, Microbial Dysbiosis, and Their Role in HIV-1 Disease Progression. J Infect Dis. 2016;214(Suppl 2):S58–66. doi: 10.1093/infdis/jiw258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacArthur RD, DuPont HL. Etiology and pharmacologic management of noninfectious diarrhea in HIV-infected individuals in the highly active antiretroviral therapy era. Clin Infect Dis. 2012;55:860–867. doi: 10.1093/cid/cis544. [DOI] [PubMed] [Google Scholar]
- 4.Wang H, Kotler DP. HIV enteropathy and aging: gastrointestinal immunity, mucosal epithelial barrier, and microbial translocation. Curr Opin HIV AIDS. 2014;9:309–16. doi: 10.1097/COH.0000000000000066. [DOI] [PubMed] [Google Scholar]
- 5.Lackner AA, Vogel P, Ramos RA, Kluge JD, Marthas M. Early events in tissues during infection with pathogenic (SIVmac239) and nonpathogenic (SIVmac1A11) molecular clones of simian immunodeficiency virus. Am J Pathol. 1994;145:428–439. [PMC free article] [PubMed] [Google Scholar]
- 6.Kewenig S, Schneider T, Hohloch K, Lampe-Dreyer K, Ullrich R, Stolte N, Stahl-Hennig C, Kaup FJ, Stallmach A, Zeitz M. Rapid mucosal CD4(+) T-cell depletion and enteropathy in simian immunodeficiency virus-infected rhesus macaques. Gastroenterology. 1999;116:1115–1123. doi: 10.1016/s0016-5085(99)70014-4. [DOI] [PubMed] [Google Scholar]
- 7.Tincati C, Douek DC, Marchetti G. Gut barrier structure, mucosal immunity and intestinal microbiota in the pathogenesis and treatment of HIV infection. AIDS Res Ther. 2016;13:19. doi: 10.1186/s12981-016-0103-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Estes JD, Harris LD, Klatt NR, Tabb B, Pittaluga S, Paiardini M, Barclay GR, Smedley J, Pung R, Oliveira KM, Hirsch VM, Silvestri G, Douek DC, Miller CJ, Haase AT, Lifson J, Brenchley JM. Damaged intestinal epithelial integrity linked to microbial translocation in pathogenic simian immunodeficiency virus infections. PLoS Pathog. 2010;6:e1001052. doi: 10.1371/journal.ppat.1001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cichon C, Sabharwal H, Rüter C, Schmidt MA. MicroRNAs regulate tight junction proteins and modulate epithelial/endothelial barrier functions. Tissue Barriers. 2014;2:e944446. doi: 10.4161/21688362.2014.944446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang H, Rao JN, Wang JY. Posttranscriptional Regulation of Intestinal Epithelial Tight Junction Barrier by RNA-binding Proteins and microRNAs. Tissue Barriers. 2014;2:e28320. doi: 10.4161/tisb.28320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang Y, Banan A, Forsyth CB, Fields JZ, Lau CK, Zhang LJ, Keshavarzian A. Effect of alcohol on miR-212 expression in intestinal epithelial cells and its potential role in alcoholic liver disease. Alcohol Clin Exp Res. 2008;32:355–364. doi: 10.1111/j.1530-0277.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- 12.Ye D, Guo S, Al-Sadi R, Ma TY. MicroRNA regulation of intestinal epithelial tight junction permeability. Gastroenterology. 2011;141:1323–1333. doi: 10.1053/j.gastro.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Q, Costinean S, Croce CM, Brasier AR, Merwat S, Larson SA, Basra S, Verne GN. MicroRNA 29 targets nuclear factor-κB-repressing factor and Claudin 1 to increase intestinal permeability. Gastroenterology. 2015;148:158–169. doi: 10.1053/j.gastro.2014.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohan M, Chandra LC, Workineh T, Aye PP, Alvarez X, Lackner AA. miR-190b is markedly upregulated in the gastrointestinal tract in response to SIV infection and partly regulates myotubularin related protein-6 expression. J Immunol. 2014;193:1301–1313. doi: 10.4049/jimmunol.1303479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar V, Workineh T, Fenway C, Schiro F, Mohan M. Longitudinal examination of the intestinal lamina propria cellular compartment of SIV-infected rhesus macaques provides broader and deeper insights into the link between aberrant microRNA expression and persistent immune activation. J Virol. 2016;90:5003–5019. doi: 10.1128/JVI.00189-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohan M, Kumar V, Lackner AA, Alvarez X. Dysregulated miR-34a-SIRT1-Acetyl p65 Axis Is a Potential Mediator of Immune Activation in the Colon during Chronic Simian Immunodeficiency Virus Infection of Rhesus Macaques. J Immunol. 2015;194:291–306. doi: 10.4049/jimmunol.1401447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chandra LC, Kumar V, Amedee A, Vande Stowe Curtis, Winsauer P, Molina PE, Mohan M. Chronic THC administration induces an intestinal anti-inflammatory microRNA expression during acute SIV infection of rhesus macaques. J Virol. 2015;89:1168–1181. doi: 10.1128/JVI.01754-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan D, Das A, Liu D, Veazey RS, Pahar B. Isolation and characterization of intestinal epithelial cells from normal and SIV-infected rhesus macaques. PLoS One. 2012;7:e30247. doi: 10.1371/journal.pone.0030247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee KM, Chiu KB, Renner NA, Sansing HA, Didier PJ, MacLean AG. Form Follows Function: Astrocyte Morphology and Immune Dysfunction in SIV neuroAIDS. J Neurovirol. 2014;20:474–484. doi: 10.1007/s13365-014-0267-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshikawa TJ, Wu M, Otsuka M, Kishikawa T, Suzuki N, Takata A, Ohno M, Ishibashi R, Yamagami M, Nakagawa R, Kato N, Miyazawa M, Han J, Koike K. Repression of MicroRNA Function Mediates Inflammation-associated Colon Tumorigenesis. Gastroenterology. 2017;152:631–643. doi: 10.1053/j.gastro.2016.10.043. [DOI] [PubMed] [Google Scholar]
- 22.Nguyen HT, Dalmasso G, Müller S, Carrière J, Seibold F, Darfeuille-Michaud A. Crohn’s disease-associated adherent invasive Escherichia coli modulate levels of microRNAs in intestinal epithelial cells to reduce autophagy. Gastroenterology. 2014;146:508–519. doi: 10.1053/j.gastro.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 23.Eichhorn SW, Guo H, McGeary SE, Rodriguez-Mias RA, Shin C, Baek D, Hsu SH, Ghoshal K, Villén J, Bartel DP. mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol Cell. 2014;56:104–115. doi: 10.1016/j.molcel.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Francois V, Shehade H, Acolty V, Preyat N, Delrée P, Moser M, Oldenhove G. Intestinal immunopathology is associated with decreased CD73-generated adenosine during lethal infection. Mucosal Immunol. 2015;8:773–784. doi: 10.1038/mi.2014.108. [DOI] [PubMed] [Google Scholar]
- 25.Cario E. P-glycoprotein multidrug transporter in inflammatory bowel diseases: More questions than answers. World J Gastroenterol. 2017;23:1513–1520. doi: 10.3748/wjg.v23.i9.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landy J, Ronde E, English N, Clark SK, Hart AL, Knight SC, Ciclitira PJ, Al-Hassi HO. Tight junctions in inflammatory bowel diseases and inflammatory bowel disease associated colorectal cancer. World J Gastroenterol. 2016;22:3117–3126. doi: 10.3748/wjg.v22.i11.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Torres J, Danese S, Colombel JF. New therapeutic avenues in ulcerative colitis: thinking out of the box. Gut. 2013;62:1642–1652. doi: 10.1136/gutjnl-2012-303959. [DOI] [PubMed] [Google Scholar]
- 28.Cunningham KE, Vincent G, Sodhi CP, Novak EA, Ranganathan S, Egan CE, Stolz DB, Rogers MB, Firek B, Morowitz MJ, Gittes GK, Zuckerbraun BS, Hackam DJ, Mollen KP. Peroxisome Proliferator-activated Receptor-γ Coactivator 1-α (PGC1α) Protects against Experimental Murine Colitis. J Biol Chem. 2016;291:10184–10200. doi: 10.1074/jbc.M115.688812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cunliffe HE, Jiang Y, Fornace KM, Yang F, Meltzer PS. PAR6B is required for tight junction formation and activated PKCζ localization in breast cancer. Am J Cancer Res. 2012;2:478–491. [PMC free article] [PubMed] [Google Scholar]
- 30.McCole DF, Barrett KE. Epithelial transport and gut barrier function in colitis. Curr Opin Gastroenterol. 2003;19:578–582. doi: 10.1097/00001574-200311000-00011. [DOI] [PubMed] [Google Scholar]
- 31.Ren T, Tian T, Feng X, Ye S, Wang H, Wu W, Qiu Y, Yu C, He Y, Zeng J, Cen J, Zhou Y. An adenosine A3 receptor agonist inhibits DSS-induced colitis in mice through modulation of the NF-κB signaling pathway. Sci Rep. 2015;5:9047. doi: 10.1038/srep09047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Judd LM, Chalinor HV, Walduck A, Pavlic DI, Däbritz J, Dubeykovskaya Z, Wang TC, Menheniott TR, Giraud AS. TFF2 deficiency exacerbates weight loss and alters immune cell and cytokine profiles in DSS colitis, and this cannot be rescued by wild-type bone marrow. Am J Physiol Gastrointest Liver Physiol. 2015;308:G12–24. doi: 10.1152/ajpgi.00172.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu F, Dassopoulos T, Cope L, Maitra A, Brant SR, Harris ML, Bayless TM, Parmigiani G, Chakravarti S. Genome-wide gene expression differences in Crohn's disease and ulcerative colitis from endoscopic pinch biopsies: insights into distinctive pathogenesis. Inflamm Bowel Dis. 2007;13:807–821. doi: 10.1002/ibd.20110. [DOI] [PubMed] [Google Scholar]
- 34.Yang R, Miki K, Oksala N, Nakao A, Lindgren L, Killeen ME, Mennander A, Fink MP, Tenhunen J. Bile high-mobility group box 1 contributes to gut barrier dysfunction in experimental endotoxemia. Am J Physiol Regul Integr Comp Physiol. 2009;297:R362–369. doi: 10.1152/ajpregu.00184.2009. [DOI] [PubMed] [Google Scholar]
- 35.Liu KX, Edwards B, Lee S, Finelli MJ, Davies B, Davies KE, Oliver PL. Neuron-specific antioxidant OXR1 extends survival of a mouse model of amyotrophic lateral sclerosis. Brain. 2015;138:1167–1181. doi: 10.1093/brain/awv039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Z, Yoshida T, Wu L, Maiti D, Cebotaru L, Duh EJ. Transcription factor MEF2C suppresses endothelial cell inflammation via regulation of NF-κB and KLF2. J Cell Physiol. 2015;230:1310–1320. doi: 10.1002/jcp.24870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karim BO, Rhee KJ, Liu G, Yun K, Brant SR. Prom1 function in development, intestinal inflammation, and intestinal tumorigenesis. Front Oncol. 2014;4:323. doi: 10.3389/fonc.2014.00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sampson LL, Davis AK, Grogg MW, Zheng Y. mTOR disruption causes intestinal epithelial cell defects and intestinal atrophy postinjury in mice. FASEB J. 2016;30:1263–1275. doi: 10.1096/fj.15-278606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Borenshtein D, Schlieper KA, Rickman BH, Chapman JM, Schweinfest CW, Fox JG, Schauer DB. Decreased expression of colonic Slc26a3 and carbonic anhydrase iv as a cause of fatal infectious diarrhea in mice. Infect Immun. 2009;77:3639–3650. doi: 10.1128/IAI.00225-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Planell N, Lozano JJ, Mora-Buch R, Masamunt MC, Jimeno M, Ordás I, Esteller M, Ricart E, Piqué JM, Panés J, Salas A. Transcriptional analysis of the intestinal mucosa of patients with ulcerative colitis in remission reveals lasting epithelial cell alterations. Gut. 2013;62:967–976. doi: 10.1136/gutjnl-2012-303333. [DOI] [PubMed] [Google Scholar]
- 41.Yu K, Lujan R, Marmorstein A, Gabriel S, Hartzell HC. Bestrophin-2 mediates bicarbonate transport by goblet cells in mouse colon. J Clin Invest. 2010;120:1722–1735. doi: 10.1172/JCI41129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsiperson V, Gruber RC, Goldberg MF, Jordan A, Weinger JG, Macian F, Shafit-Zagardo B. Suppression of inflammatory responses during myelin oligodendrocyte glycoprotein-induced experimental autoimmune encephalomyelitis is regulated by AKT3 signaling. J Immunol. 2013;190:1528–1539. doi: 10.4049/jimmunol.1201387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rhee L, Murphy SF, Kolodziej LE, Grimm WA, Weber CR, Lodolce JP, Chang JE, Bartulis SJ, Messer JS, Schneider JR, Paski S, Nero TM, Boone DL. Expression of TNFAIP3 in intestinal epithelial cells protects from DSS- but not TNBS-induced colitis. Am J Physiol Gastrointest Liver Physiol. 2012;303:G220–227. doi: 10.1152/ajpgi.00077.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams TM, Leeth RA, Rothschild DE, Coutermarsh-Ott SL, McDaniel DK, Simmons AE, Heid B, Cecere TE, Allen IC. The NLRP1 inflammasome attenuates colitis and colitis-associated tumorigenesis. J Immunol. 2015;194:3369–3380. doi: 10.4049/jimmunol.1402098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deka J, Wiedemann N, Anderle P, Murphy-Seiler F, Bultinck J, Eyckerman S, Stehle JC, André S, Vilain N, Zilian O, Robine S, Delorenzi M, Basler K, Aguet M. Bcl9/Bcl9l are critical for Wnt-mediated regulation of stem cell traits in colon epithelium and adenocarcinomas. Cancer Res. 2010;70:6619–28. doi: 10.1158/0008-5472.CAN-10-0148. [DOI] [PubMed] [Google Scholar]
- 47.Wang H, Zhang R, Wen S, McCafferty DM, Beck PL, MacNaughton WK. Nitric oxide increases Wnt-induced secreted protein-1 (WISP-1/CCN4) expression and function in colitis. J Mol Med (Berl) 2009;87:435–445. doi: 10.1007/s00109-009-0445-4. [DOI] [PubMed] [Google Scholar]
- 48.Gassler N, Autschbach F, Heuschen G, Witzgall R, Otto HF, Obermüller N. Expression of clusterin in Crohn’s disease of the terminal ileum. Histol Histopathol. 2001;16:755–762. doi: 10.14670/HH-16.755. [DOI] [PubMed] [Google Scholar]
- 49.Shen NY, Bi JB, Zhang JY, Zhang SM, Gu JX, Qu K, Liu C. Hydrogen-rich water protects against inflammatory bowel disease in mice by inhibiting endoplasmic reticulum stress and promoting heme oxygenase-1 expression. World J Gastroenterol. 2017;23:1375–1386. doi: 10.3748/wjg.v23.i8.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scharl M, Weber A, Fürst A, Farkas S, Jehle E, Pesch T, Kellermeier S, Fried M, Rogler G. Potential role for SNAIL family transcription factors in the etiology of Crohn's disease-associated fistulae. Inflamm Bowel Dis. 2011;17:1907–1916. doi: 10.1002/ibd.21555. [DOI] [PubMed] [Google Scholar]
- 51.Waldschmitt N, Berger E, Rath E, Sartor RB, Weigmann B, Heikenwalder M, Gerhard M, Janssen KP, Haller D. C/EBP homologous protein inhibits tissue repair in response to gut injury and is inversely regulated with chronic inflammation. Mucosal Immunol. 2014;7:1452–1466. doi: 10.1038/mi.2014.34. [DOI] [PubMed] [Google Scholar]
- 52.Bäumler AJ, Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature. 2016;535:85–93. doi: 10.1038/nature18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Byndloss MX, Olsan EE, Rivera-Chávez F, Tiffany CR, Cevallos SA, Lokken KL, Torres TP, Byndloss AJ, Faber F, Gao Y, Litvak Y, Lopez CA, Xu G, Napoli E, Giulivi C, Tsolis RM, Revzin A, Lebrilla CB, Bäumler AJ. Microbiota-activated PPAR-γ signaling inhibits dysbiotic Enterobacteriaceae expansion. Science. 2017;357:570–575. doi: 10.1126/science.aam9949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krüger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451–454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu F, Zikusoka M, Trindade A, Dassopoulos T, Harris ML, Bayless TM, Brant SR, Chakravarti S, Kwon JH. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology. 2008;135:1624–1635. doi: 10.1053/j.gastro.2008.07.068. [DOI] [PubMed] [Google Scholar]
- 56.Zhou Q, Yang L, Larson S, Basra S, Merwat S, Tan A, Croce C, Verne GN. Decreased miR-199 augments visceral pain in patients with IBS through translational upregulation of TRPV1. Gut. 2016;65:797–805. doi: 10.1136/gutjnl-2013-306464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ngaotepprutaram T, Kaplan BL, Carney S, Crawford R, Kaminski NE. Suppression by Δ(9)-tetrahydrocannabinol of the primary immunoglobulin M response by human peripheral blood B cells is associated with impaired STAT3 activation. Toxicology. 2013;310:84–91. doi: 10.1016/j.tox.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ngaotepprutaram T, Kaplan BL, Kaminski NE. Impaired NFAT and NFκB activation are involved in suppression of CD40 ligand expression by Δ(9)-tetrahydrocannabinol in human CD4(+) T cells. Toxicol Appl Pharmacol. 2013;273:209–218. doi: 10.1016/j.taap.2013.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De Filippis D, Esposito G, Cirillo C, Cipriano M, De Winter BY, Scuderi C, Sarnelli G, Cuomo R, Steardo L, De Man JG, Iuvone T. Cannabidiol reduces intestinal inflammation through the control of neuroimmune axis. PLoS One. 2011;6:e28159. doi: 10.1371/journal.pone.0028159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peyrin-Biroulet L, Beisner J, Wang G, Nuding S, Oommen ST, Kelly D, Parmentier-Decrucq E, Dessein R, Merour E, Chavatte P, Grandjean T, Bressenot A, Desreumaux P, Colombel JF, Desvergne B, Stange EF, Wehkamp J, Chamaillard M. Peroxisome proliferator-activated receptor gamma activation is required for maintenance of innate antimicrobial immunity in the colon. Proc Natl Acad Sci U S A. 2010;107:8772–8777. doi: 10.1073/pnas.0905745107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Speca S, Rousseaux C, Dubuquoy C, Rieder F, Vetuschi A, Sferra R, Giusti I, Bertin B, Dubuquoy L, Gaudio E, Desreumaux P, Latella G. Novel PPARγ Modulator GED-0507-34 Levo Ameliorates Inflammation-driven Intestinal Fibrosis. Inflamm Bowel Dis. 2016;22:279–292. doi: 10.1097/MIB.0000000000000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pedersen G, Brynskov J. Topical rosiglitazone treatment improves ulcerative colitis by restoring peroxisome proliferator-activated receptor-gamma activity. Am J Gastroenterol. 2010;105:1595–1603. doi: 10.1038/ajg.2009.749. [DOI] [PubMed] [Google Scholar]
- 63.Mohan M, Kaushal D, Aye PP, Alvarez X, Veazey RS, Lackner AA. Focused examination of the intestinal epithelium reveals transcriptional signatures consistent with disturbances in enterocyte maturation and differentiation during the course of SIV infection. PLoS One. 2013;8:e60122. doi: 10.1371/journal.pone.0060122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Somsouk M, Estes JD, Deleage C, Dunham RM, Albright R, Inadomi JM, Martin JN, Deeks SG, McCune JM, Hunt PW. Gut epithelial barrier and systemic inflammation during chronic HIV infection. AIDS. 2015;29:43–51. doi: 10.1097/QAD.0000000000000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sankaran S, George MD, Reay E, Guadalupe M, Flamm J, Prindiville T, Dandekar S. Rapid onset of intestinal epithelial barrier dysfunction in primary human immunodeficiency virus infection is driven by an imbalance between immune response and mucosal repair and regeneration. J Virol. 2008;82:538–545. doi: 10.1128/JVI.01449-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mohan M, Kaushal D, Aye PP, Alvarez X, Veazey RS, Lackner AA. Focused examination of the intestinal lamina propria yields greater molecular insight into mechanisms underlying SIV induced immune dysfunction. PLoS One. 2012;7:e34561. doi: 10.1371/journal.pone.0034561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kalla R, Ventham NT, Kennedy NA, Quintana JF, Nimmo ER, Buck AH, Satsangi J. MicroRNAs: new players in IBD. Gut. 2015;64:504–517. doi: 10.1136/gutjnl-2014-307891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou M, Wu R, Dong W, Jacob A, Wang P. Endotoxin downregulates peroxisome proliferator-activated receptor-gamma via the increase in TNF-alpha release. Am J Physiol Regul Integr Comp Physiol. 2008;294:R84–92. doi: 10.1152/ajpregu.00340.2007. [DOI] [PubMed] [Google Scholar]
- 69.Gaulke CA, Porter M, Han YH, Sankaran-Walters S, Grishina I, George MD, Dang AT, Ding SW, Jiang G, Korf I, Dandekar S. Intestinal epithelial barrier disruption through altered mucosal microRNA expression in human immunodeficiency virus and simian immunodeficiency virus infections. J Virol. 2014;88:6268–6280. doi: 10.1128/JVI.00097-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, Sharp PA, Manjunath N. miRNA profiling of naïve, effector and memory CD8 T cells. PLoS One. 2007;2:e1020. doi: 10.1371/journal.pone.0001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bronevetsky Y, Villarino AV, Eisley CJ, Barbeau R, Barczak AJ, Heinz GA, Kremmer E, Heissmeyer V, McManus MT, Erle DJ, Rao A, Ansel KM. T cell activation induces proteasomal degradation of Argonaute and rapid remodeling of the microRNA repertoire. J Exp Med. 2013;210:417–432. doi: 10.1084/jem.20111717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pott J, Hornef M. Innate immune signaling at the intestinal epithelium in homeostasis and disease. EMBO Rep. 2012;13:684–698. doi: 10.1038/embor.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Park SH, Choi HJ, Yang H, Do KH, Kim J, Lee DW, Moon Y. Endoplasmic reticulum stress-activated C/EBP homologous protein enhances nuclear factor-kappaB signals via repression of peroxisome proliferator-activated receptor gamma. J Biol Chem. 2010;285:35330–35339. doi: 10.1074/jbc.M110.136259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Haberman Y, Tickle TL, Dexheimer PJ, Kim MO, Tang D, Karns R, Baldassano RN, Noe JD, Rosh J, Markowitz J, Heyman MB, Griffiths AM, Crandall WV, Mack DR, Baker SS, Huttenhower C, Keljo DJ, Hyams JS, Kugathasan S, Walters TD, Aronow B, Xavier RJ, Gevers D, Denson LA. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Invest. 2014;124:3617–3633. doi: 10.1172/JCI75436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Klase Z, Ortiz A, Deleage C, Mudd JC, Quiñones M, Schwartzman E, Klatt NR, Canary L, Estes JD, Brenchley JM. Dysbiotic bacteria translocate in progressive SIV infection. Mucosal Immunol. 2015;8:1009–1020. doi: 10.1038/mi.2014.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kheirolomoom A, Kim CW, Seo JW, Kumar S, Son DJ, Gagnon MK, Ingham ES, Ferrara KW, Jo H. Multifunctional Nanoparticles Facilitate Molecular Targeting and miRNA Delivery to Inhibit Atherosclerosis in ApoE(−/−) Mice. ACS Nano. 2015;9:8885–8897. doi: 10.1021/acsnano.5b02611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gerich ME, Isfort RW, Brimhall B, Siegel CA. Medical marijuana for digestive disorders: high time to prescribe? Am J Gastroenterol. 2015;110:208–214. doi: 10.1038/ajg.2014.245. [DOI] [PubMed] [Google Scholar]
- 78.Goyal H, Singla U, Gupta U, May E. Role of cannabis in digestive disorders. Eur J Gastroenterol Hepatol. 2017;29:135–143. doi: 10.1097/MEG.0000000000000779. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


