Abstract
Background
Long term survival of lung transplant recipients (LTRs) is limited by the occurrence of bronchiolitis obliterans syndrome (BOS). Recent evidence suggests a role for microbiome alterations in the occurrence of BOS, although the precise mechanisms are unclear. In this study, we evaluated the relationship between the airway microbiome and distinct subsets of immune regulatory myeloid derived suppressor cells (MDSCs) in LTRs.
Methods
Bronchoalveolar lavage (BAL) and simultaneous oral wash and nasal swab samples were collected from adult LTRs. Microbial genomic DNA was isolated, 16S rRNA genes amplified using V4 primers, PCR products sequenced and analyzed. BAL MDSC subsets were enumerated using flow cytometry.
Results
The oral microbiome signature differs from that of the nasal, proximal and distal airway microbiomes, while the nasal microbiome is closer to the airway microbiome. Proximal and distal airway microbiome signatures of individual subjects are distinct. We identified phenotypic subsets of MDSCs in BAL, with a higher proportion of immunosuppressive MDSCs in the proximal airways in contrast to a preponderance of pro-inflammatory MDSCs in distal airways. Relative abundance of distinct bacterial phyla in proximal and distal airways correlated with particular airway MDSCs. Expression of C/EBP-homologous protein (CHOP), an endoplasmic (ER) stress sensor was increased in immunosuppressive MDSCs as compared to pro-inflammatory MDSCs.
Conclusions
The nasal microbiome closely resembles the microbiome of the proximal and distal airways in LTRs. The association of distinct microbial communities with airway MDSCs suggests a functional relationship between the local microbiome and MDSC phenotype, which may contribute to the pathogenesis of BOS.
Keywords: Bronchiolitis Obliterans Syndrome, lung microbiome, Myeloid-derived suppressor cells, Immune regulation, lung transplantation
Introduction
Over the last decade, most solid organ transplantations have been associated with improved survival 1. In contrast to heart, liver and kidney transplantation, 5-year survival for lung transplant recipients (LTR) remains low 1. Long-term survival is primarily limited by bronchiolitis obliterans syndrome (BOS), which is the leading cause of chronic allograft failure 2,3. Many risk factors have been implicated in the development of BOS though the exact pathogenesis remains unclear 4-7. There is emerging evidence of a role for microbiome in the development and/or progression of acute/chronic lung diseases, including BOS 8-13. Previous studies have shown association of Pseudomonas infection 14,15 and dysbiosis in microbiome after transplantation with development of BOS 11,12; however, mechanisms linking alterations in the lung microbiome with BOS have not been clearly defined.
Dysregulation of immune mechanisms underlying chronic allograft failure has not been well defined. Myeloid-derived suppressor cells (MDSCs), a heterogeneous group of immature myeloid cells, function as immune regulators by various mechanisms 16. We and others have characterized phenotypic subsets of MDSCs with differential functions, including a pro-inflammatory phenotype in asthma and sepsis in addition to the immunosuppressive subsets reported in cancer and other chronic inflammatory diseases 17-19. It has been shown that the differentiation of MDSCs is enhanced during Pseudomonas and Staphylococcus infections resulting in suppression of host immunity 20,21.
We hypothesized that changes in the diversity and/or abundance of the lung microbiome modulate host immune responses in transplanted lung allografts by the presence and activity of recruited MDSCs. An imbalance in the ratio of anti-inflammatory and pro-inflammatory MDSC subsets may contribute to the pathogenesis of BOS and other chronic inflammatory lung disorders.
In this study, we compared the oral, nasal and airway microbiomes of lung transplant recipients. Additionally, we report that phenotypic subsets of airway MDSCs correlate with the local lung microbiota.
Methods
Baseline patient characteristics
Samples were collected from consecutive adult (>18 years) LTRs presenting for routine surveillance bronchoscopy at our center (Table 1). A 20 ml saline oral wash (O) and nasal swab (N) was performed (reconstituted in 10ml saline) prior to bronchoscopy. Controls from bronchoscope used were also collected prior to the procedure (25 ml of saline flushed in bronchoscope and collected). For standardization, all bronchoscopies were conducted via the oral route. Bronchoalveolar lavage (BAL) was performed in the right middle lobe or lingula based on clinician preference. A total of 200 ml of saline was used for the BAL in 6 fractions of 33.5 ml each. Serial fractions of return were collected in separate tubes and these samples are defined as BAL #1 (B1) thru BAL #6 (B6). BAL #1 represented the proximal airways and BAL# 6 the distal airways (alveolar region). Baseline characteristics of the subjects are detailed in Table 1. Bronchiolitis obliterans syndrome was adjudicated based on published guidelines 22. Written consents were obtained for sample collection under the IRB approved protocol (University of Alabama at Birmingham, IRB number X120606006).
Table 1. Baseline characteristics of lung transplant subjects.
| S.No | Age | Gender | Bronchoscopy Indication |
Months from Transplant |
Transplant Type |
Pre- transplant diagnosis |
Transplant Medications |
Prophylactic Antibiotic |
Culture Data | BOS(Y/N)/ Stage/ Duration (months) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 37 | M | PSB | 15 | Double | ES | Tac, Pred | Atova, Vori | Neg | N |
| 2 | 46 | F | DLF | 12 | Single | COPD | Tac, Pred, MMF | TMP-SMX, Valcyte, Itra | Neg | Y-0P/10 |
| 3 | 52 | F | DLF | 17 | Double | CF | Tac, Pred, MMF | Azithro, TMP-SMX, Vori, | Malbranchia species | Y/1/7 |
| 4 | 24 | M | DLF | 31 | Double | CF | Tac, Pred, MMF | Dapsone, Azithro, Vori, | Negative | Y/1/9 |
| 5 | 31 | M | PSB | 32 | Double | CF | Tac, Pred, MMF | Azithro, Pentam | MRSA | N |
| 6 | 67 | F | DLF | 31 | Single | COPD | Tac, Pred, Siro | Azithro, TMP-SMX, | Negative | Y/1/17 |
| 7 | 72 | M | AI | 83 | Single | IPF | Tac, Pred, Siro | TMP-SMX, Azithro | Negative | N |
| 8 | 63 | M | PSB | 12 | Single | IPF | Tac, Pred | Dapsone, Itra, Valcyte | Negative | N |
| 9 | 50 | F | AI | 156 | Double | Sarcoid | Tac, Pred, MMF | None | Pseudomonas aeruginosa | Y/2/48 |
| 10 | 59 | M | PSB | 4 | Double | IPF | Tac, Pred, MMF | Vori, Valcyte, TMP-SMX | Negative | N |
Definitions of abbreviations: ES- Eisenmenger's syndrome, A1AT- Alpha 1 antitrypsin deficiency, CF-Cystic Fibrosis, COPD- Chronic Obstructive Pulmonary Disease, IPF- Idiopathic Pulmonary Fibrosis, Sarcoid- Sarcoidosis, Tac- Tacrolimus, Pred- Prednisone, MMF-Mycophenolate Mofetil; Siro- Sirolimus, Vori- Voriconazole, Valcyte- Valganciclovir, Itra-Itraconazole, Azithro- Azithromycin, BOS- Bronchiolitis Obliterans Syndrome, Atova-Atovaquone, TMP-SMX- Trimethoprim/Sulfamethoxazole, Pentam-Pentamidine, PSB- Protocol driven surveillance bronchoscopy (center specific), AI- abnormal imaging, DLF- Declining lung function.
Processing of samples
4 aliquots (1 ml each) of BAL fraction 1 and 6 samples were centrifuged at 1000 rpm for 5 minutes to separate the cellular fraction. The supernatants were centrifuged again at 15000 rpm for 10 minute to pellet the bacterial component. Similarly, 3 aliquots (1 ml each) of samples O, N, C1, C2 fractions (oral, nasal and control bronchoscope washes respectively) were collected and centrifuged at 15000 rpm for 10 minutes. All pellets were then stored at -80°C. DNA was extracted from the cell pellets. PCR amplification of V4 region of the 16S rRNA gene was performed and products used for microbial DNA sequencing. BAL supernatant 1 and 6 was used to measure SP-D and RAGE levels. Flowcytometry was performed on fresh BAL 1 and 6 samples to enumerate MDSC subsets. Details provided in the supplementary text.
Statistical Analysis
To compare beta diversities between individual patient proximal and distal airway samples, Weighted UniFrac distances were calculated between all pairs of samples and then each sample type was plotted separately in 3D space by principal coordinate analysis. The plots were then transformed by Procrustes analysis to achieve maximum alignment. Within the 3D plots, blue color represents one sample and the red color represents the other sample and the two points from individual subjects are connected by a bar. If both plots are similar, then the relative distance will be small. The overall similarity is summarized by the M2 value, and statistical goodness of fit is measured by a Monte Carlo label permutation approach. To identify individual OTUs at the phylum and/or genus levels that were distinctive between the two airway compartments, Wilcoxon signed rank test was used to perform the primary analysis on the microbiome signatures t. Additionally, linear mixed models were used where airway compartment was the fixed effect and both technical replicate ID and individual ID were random effects. To compare cell fractions across different cells, ANOVA was used as an omnibus test followed by paired t tests to compare all pairs of groups. To test if correlations between proportion of a cell population and a phylum differ across B1 and B6, a multiple regression with phylum OTU abundance as dependent variable and cell proportion, B1/B6 fraction, and their interaction as independent variables was performed, and the p-value for the interaction was reported. For main effects, multiple tests were adjusted by Bonferroni correction or false discovery rate as applicable. P-values not reaching Bonferroni-corrected threshold but smaller than 0.05 were noted to be ‘nominally’ significant. For interaction, only nominal significance was considered. To identify unique OTUs in BOS subjects, Wilcoxon signed rank test was performed separately for samples collected from both proximal and distal airways for the BOS and non-BOS Patients. Graphpad prism version 7.0, R version 3.1.0 and SAS version 9.4 were used for statistical analysis and generation of figures.
Results
SP-D and RAGE levels were higher in distal airway compared to proximal airway fluid
We first tested whether B1 and B6 samples were representative of two distinct compartments namely the proximal and distal airways. Previous studies have shown that distal airway (alveolar) samples have higher concentration of SP-D and RAGE 23,24. The total amounts of both RAGE (p=0.01) and SP-D (p=0.07) were higher in the B6 compared to the B1 (Figure 1A and 1B). Levels of RAGE were significantly different while SP-D did not reach statistical significance. These data demonstrate that B1 and B6 represent sampling of two different regions of the airway.
Figure 1. Levels of SP-D, RAGE and alpha diversity of bacterial communities in compartments of the respiratory tract.
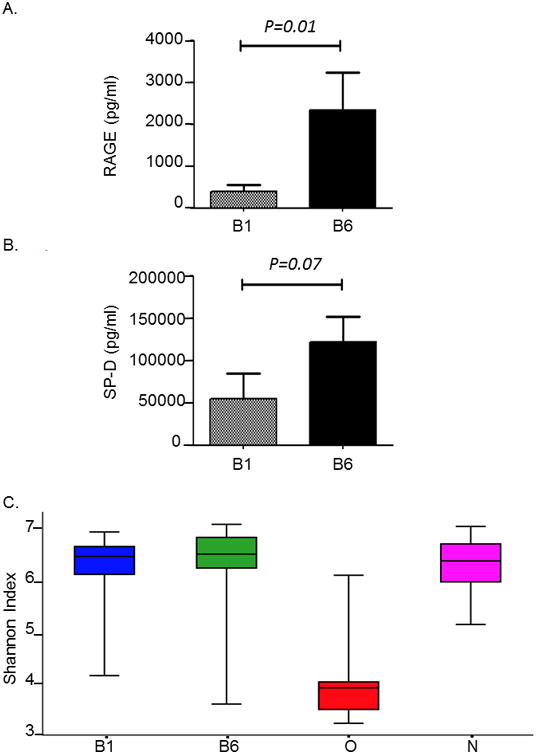
A. Bar plot showing the level of RAGE (ng/ml) found in the proximal (B1) and distal airway (B6). Levels of RAGE were significantly higher in the distal as compared to the proximal airway (*P<0.01)
B. Bar plot showing the level of SP-D (ng/ml) found in the proximal (B1) and distal airway (B6). Levels of SP-D were higher in the distal as compared to the proximal airway, but it did not reach statistical significance (P=0.07)
C. Box plots showing alpha diversity of bacterial communities in transplant subjects. Alpha diversity was calculated using Shannon Index for oro-pharyngeal wash (O),nasal (N) and bronchoalveolar lavage samples represented by B1 (proximal airways)and B6 (distal airways). One way ANOVA analysis of B1 versus O, *P<0.01; B6versus O, P<0.01; N versus O; P<0.01.
Oral microbiome is less diverse than nasal or airway microbiome
To determine if O, N and airway (B1 and B6) samples differed in species richness, we first calculated the number of observed OTUs at various sequencing depths for bacteria in O, N, B1, and B6. Rarefaction analyses indicated that the species richness was significantly reduced (P <0.001) in the O compared to N, B1 and B6 samples (Supplementary Figure E1). We then evaluated diversity of bacterial communities between samples, using Shannon diversity index, a measure of alpha diversity within a sample that represents both species richness and evenness (Figure 1C). Shannon diversity index of 3.5 and above indicate highly diverse bacterial communities. While all samples including airway, nasal and oral samples had high bacterial community diversity, B1 (6.6), B6 (6.8) and N (6.3) had a significantly higher Shannon diversity index when compared to O (3.6) (P<0.001); differences in diversity between airway and nasal samples were not significantly different (P=0.8).
Nasal and airway microbiomes are distinct from the oral microbiome
Previous studies in healthy subjects have shown that the oral microbiome contributes to the composition of the lung microbiome 25. We compared the bacterial communities of O and B1 microbiome, using the beta diversity metric, the weighted UniFrac distance. The weighted UniFrac distances are inclusive of abundance, the presence or absence of OTUs between samples and their phylogenetic distances and were visualized using a principal coordinate analysis (PCoA) plot. PCoA plot between the individual subject's O and B1 microbiome showed wide separation with the first two principle components explaining 75% variability (Figure 2A). Additionally, combined analysis of all subject samples also showed that the O and B1 microbiomes clustered separately with the first two principal components explaining 74% variability (Figure 2B). When comparing O and N microbiomes with B1, the Unifrac distance between the O and B1 was greater as compared to N and B1 microbiome (P<0.0006, paired t test) (Figure 2C). The phyla Acidobacteria, Proteobacteria, Firmicutes and Chloroflexi contributed significantly to the differences found between the O and B1 microbiome (Supplementary Table S1A). Similarly, weighted Unifrac distance between O and B6 of individual subjects showed a greater separation with the first two principal components accounting for 74% variability (Figure 2D). Likewise, combined analysis of all subjects showed distinct clustering of O and B6 microbiome with the first two principal components accounting for 64% and the first three components accounting for 73% variability (Figure 2E). In addition, the Unifrac distances were also greater between O and B6 as compared to N and B6 microbiome (P<0.0001, paired t test) (Figure 2F). Significant differences in the O and B6 microbiome were contributed by phyla Tenericutes, Fusobacteria, Deferibacteres and Proteobacteria (Supplementary Table S1B).
Figure 2. Principal component analysis showing Weighted Unifrac distance between oral and airway microbiome in lung transplant recipients.
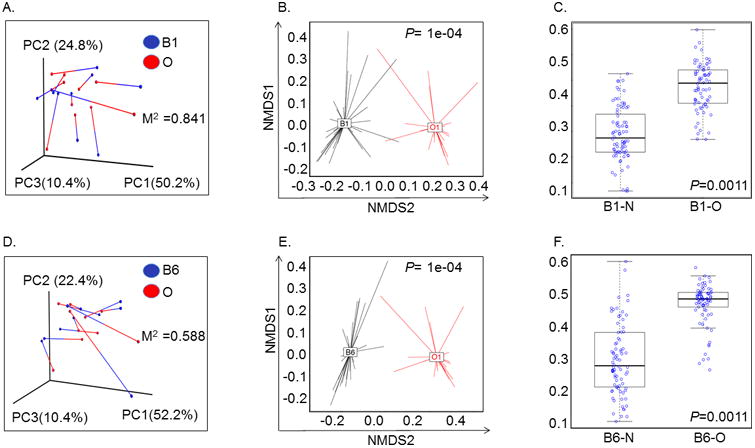
A. Relationship between proximal airway (B1) and oropharyngeal (O) bacterial communities within individual subjects. Weighted UniFrac distances were calculated between all pairs of samples within B1 or O, and then each sample type was plotted separately in 3D space by principal coordinate analysis. The two plots (B1 and O) were then transformed by Procrustes analysis to achieve maximum alignment. Each point corresponds to a bacterial community, with B1 communities shown in blue, O communities shown in red, and the two communities from each subject connected by a bar. The red end of each bar connects to the O sample data; the blue end connects to the B1 sample data from the same individual. If B1 and O plots are similar, then the relative distance between connected points (residuals) will be small. The overall similarity is summarized by the M2 value, and statistical goodness of fit is measured by a Monte Carlo label permutation approach (10,000 iterations). The M2 value ranges from 0-1, with 0 suggesting complete overlap (i.e. similarity) and 1 suggesting maximum variation.
B. Principal coordinate analyses (PCoA) plots with centroids showing weighted Unifrac distances between oral and proximal airway microbiomes. The p-values were calculated by Permanova (adonis) test over the weighed Unifrac distance to test the differences between a pair of groups. * P=1e-04
C. Box Plot analysis showing significant difference in Unifrac-weighted distance between proximal airways (B1) – nasal (N) and proximal airways (B1) – oral (O) microbiome. Y-axis represents Unifrac-weighted distance and samples are represented on the X-axis. *P=0.001 (paired t-test).
D. Relationship between distal airway (B6) and oropharyngeal (O) bacterial communities within individual subjects. Weighted UniFrac distances were calculated between all pairs of samples within B6 or O, and then each sample type was plotted separately in 3D space by principal coordinate analysis. The two plots (B6 and O) were then transformed by Procrustes analysis to achieve maximum alignment. Each point corresponds to a bacterial community, with B6 communities shown in blue, O communities shown in red, and the two communities from each subject connected by a bar. The red end of each bar connects to the O sample data; the blue end connects to the B6 sample data from the same individual. If B6 and O plots are similar, then the relative distance between connected points (residuals) will be small. The overall similarity is summarized by the M2 value, and statistical goodness of fit is measured by a Monte Carlo label permutation approach (10,000 iterations). The M2 value ranges from 0-1, with 0 suggesting complete overlap i.e. similarity and 1 suggesting maximum variation.
E. Principal coordinate analyses (PCoA) plots with centroids showing weighted Unifrac distances between oral (O) and distal airway (B6) microbiomes. The p-values were calculated by Permanova (adonis) test over the weighed Unifrac distance to test the differences between a pair of groups. * P=1e-04
F. Box Plot analysis showing significant difference in Unifrac-weighted distance between distal airways (B6) – nasal (N) and distal airways (B6) – oral (O) microbiome. Y-axis represents Unifrac-weighted distance and samples are represented on the X-axis. *P=0.001 (paired t-test).
Pooled analysis of microbiomes of N and B1 and N and B6 from all subject samples did not show discrete clustering (Supplementary Figure E2A and E2B). No significant differences were found between the Unifrac distances between B1 and N as compared to B6 and N (P=0.5) (Supplementary Figure E2C). Common and distinct OTU signatures present in nasal and airway microbiome have been detailed in Supplementary Table S2 & S3 and Supplementary Figure E3A & E3B.
Proximal and distal airways microbiomes are distinct
Previous data suggests that proximal and distal airways are significantly different from each other in their cellular components, cytokine/chemokine profiles and reactive species 26. It is not known if differences in microbiomes of the airway compartments contribute to these or result from these variations. We evaluated the differences in microbiome compositions between B1 and B6 of each individual patient. PCoA analysis using weighted Unifrac distances showed wide separation between B1 and B6 microbiome of each patient (Figure 3A). At the level of phyla, Proteobacteria, Firmicutes, Bacteroidetes and Actinobacteria contributed the most to the differences in B1 and B6 microbiome signatures (Supplementary Table S4).
Figure 3. Principal component analysis showing Weighted Unifrac distance between proximal and distal airway microbiome lung transplant recipients.
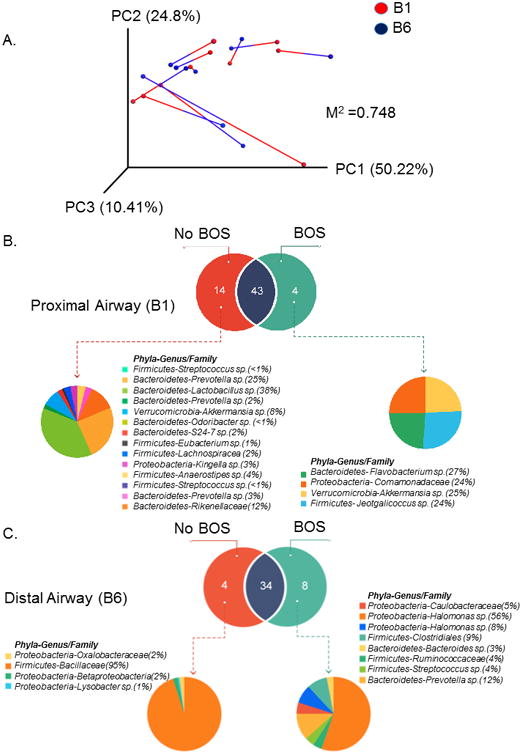
A. Beta-diversity metric comparing distances between the proximal (B1) and distal airway (B6) microbiomes within individual subjects. Weighted UniFrac distances were calculated between all pairs of samples within B1 or B6, and then each sample type was plotted separately in 3D space by principal coordinate analysis. The two plots (B1 and B6) were then transformed by Procrustes analysis to achieve maximum alignment. Each point corresponds to a bacterial community, with B1 communities shown in blue, B6 communities shown in red, and the two communities from each subject connected by a bar. The red end of each bar connects to the B1 sample data; the blue end connects to the B6 sample data from the same individual. If B1 and B6 plots are similar, then the relative distance between connected points (residuals) will be small. The overall similarity is summarized by the M2 value, and statistical goodness of fit is measured by a Monte Carlo label permutation approach (10,000 iterations). The M2 value ranges from 0-1, with 0 suggesting complete overlap i.e. similarity and 1 suggesting maximum variation.
B. Venn diagram showing the distinct and common OTUs present in the proximal airways (B1) of subjects with bronchiolitis obliterans syndrome (BOS) and those without BOS. The subsequent pie chart details the Phyla-genus level description of the unique OTUs found in the B1 sample of BOS and Non-BOS subjects.
C. Venn diagram showing the distinct and common OTUs present in the distal airways (B6) of subjects with bronchiolitis obliterans syndrome (BOS) and those without BOS. The subsequent pie chart details the Phyla-genus level description of the unique OTUs found in the B6 sample of BOS and Non-BOS subjects.
Next, we sought to identify the distinct airway microbial signatures that may be present in subjects with BOS. We found several unique OTUs in both the B1 and B6 samples of BOS subjects as compared to those without BOS (Figure 3B and 3C). The proximal airway (B1) of BOS patients has 4 unique OTUs corresponding to phyla Proteobacteria, Firmicutes, Verrucomicrobia and Bacteroidetes, whereas the distal airway (B6) of BOS patients had 8 unique OTUs corresponding to phyla Proteobacteria, Firmicutes and Bacteroidetes (Figure 3 B, 3C; Supplementary Table S5A & S5B). These unique OTU signatures in the proximal and distal airways of BOS subjects did not overlap with each other.
To confirm the validity of our findings, we compared the microbial signatures present in the bronchoscope washes (C1) collected prior to each procedure with the airway samples. PCoA analysis shows significant Unifrac distances between the microbiome of individual subjects present in the C1 as compared to B1 and B6. (Supplementary Figure E4A & E4B, Supplementary Table S6 & S7). We also compared the O microbiome with the C1 microbiome and found significant differences in relative abundance of various OTUs (Supplementary Table S8).
Pro-inflammatory MDSCs predominate in the distal airways of LTRs
Previous studies have shown that adoptive transfer of MDSCs was able to induce tolerance to autoimmune T cells against islet antigen, islet allograft and prevent graft versus host disease in mice 27-29. Studies to date have not addressed the involvement of microbial communities in influencing their function in causing allograft rejection. We first identified three distinct phenotypes of MDSCs in the BAL of transplanted subjects: Subset (A), CD14+CD11b+CD16-HLA-DR- (monocytic) subset (B) CD66+CD16+CD14-HLA-DR-CD11b+ (neutrophilic) and subset (C) CD163+HLA-DR+CD11b+CD66b-CD16- (macrophage like) MDSCs in both B1 and B6 samples (Figure 4A). Previous studies have shown that A and B are immunosuppressive 30,31, while C has pro-inflammatory functions 32. The gating strategy for characterizing these subsets is described in detail in Supplementary Figure E5. While subset A has been reported in asthmatics and cancer subjects, subset B has only been noted in cancer and not in asthma. Subset C has been reported in asthma and sepsis. In the B1 samples, both A and C were not significantly different, but proportion of B was significantly lower (P<0.05, paired t test). In B6, C was significantly higher than A and B (P<0.001). When comparing B1 and B6, while the proportion of A was significantly higher in B1 (P<0.01), the proportion of C was significantly higher in B6 (P<0.05) suggesting a more pro-inflammatory immune cell profile in the lower airways. No differences were noted in the proportion of B in both B1 and B6 (P=0.8).
Figure 4. Relative proportions of myeloid-derived suppressor cells in the proximal and distal airways and its correlation with phylum level-OTUs.
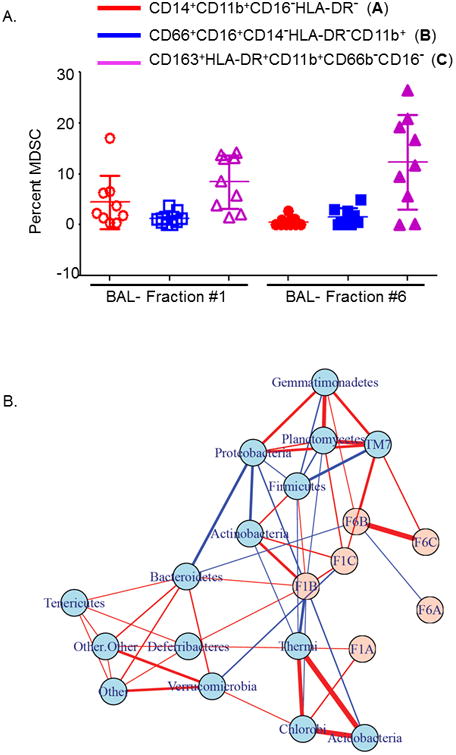
A Plot showing different phenotypes of myeloid-derived suppressor cells (MDSCs) in the proximal (B1) and distal airway (B6) bronchoalveolar lavages of lung transplant recipients. Y-axis represents the proportion of MDSCs and X-axis the B1 and B6 fractions. Circles in red represent monocytic MDSCs with a known immunosuppressive function, squares in blue are the neutrophilic MDSCs also with immunosuppressive function and triangles in purple show macrophage like MDSCs with a known pro-inflammatory function. In B1, P<0.05 for comparisons of B versus A & C. In B6, C was significantly higher than A and B (P<0.001). P<0.01 when comparing proportion of subset A in B1 and B6, and P<0.05 when comparing proportion of C in B1 and B6.
B. Correlation networks of bacterial phyla and proportions of the different phenotypic subsets of MDSCs. MDSCs are shown in pink and microbiome measurements of phylum-level OTUs in light blue. Links indicate pairwise Pearson's correlations |r|>0.30. Links with red color indicate a positive correlation while blue indicates a negative correlation. Thickness of the link indicates the strength of the pairwise correlations.
Phyla Bacteroidetes, Proteobacteria and TM7 are differentially correlated with neutrophilic MDSCs in proximal versus distal airways
We performed correlation analyses between the airway (B1 and B6) microbiomes and the proportions of MDSC subsets. First, we correlated MDSC phenotypes with OTUs in either B1 or B6 (Supplementary Figure E6). We found that in B1, the phyla Actinobacteria had a significant positive correlation with MDSC subsets B (r= 0.56, P= 0.002) and C (r=0.45, P=0.02) (Supplementary Figures E6 & E7). In addition, the phyla Chlorobi positively correlated with subset A (r=0.40, P=0.03) and phyla Planctomycetes (r=0.44, P= 0.02) and TM7 (r=0.40, P=0.04) positively correlated with subset C (Supplementary Figures E6 & E7). In contrast, significant negative correlation with subset B was observed with the phyla Acidobacteria (r=-0.41, P=0.04), Proteobacteria (r=-0.40, P=0.04) and Thermi (r=-0.54, P=0.003), and phyla Verrucomicrobia (r=-0.40, P=0.04) had a negative correlation with subset C (Supplementary Figures E6 & E7). In comparison, in B6 only phyla TM7 had a significant positive correlation with subset B (r=0.53, P=0.003) and C (r=0.44, p=0.01) (Supplementary Figures E6 & E7). These positive and negative correlations between MDSC subsets and bacterial phyla are represented in the network diagram shown in Figure 4B.
Next, we analyzed if MDSC-OTU correlations differed between B1 and B6 fractions using regression analysis. We observed differential interaction of various bacterial phyla with MDSCs in the proximal and distal airway compartments. Phyla Bacteroidetes with subset B was significantly different, with a positive correlation with subset B in B1 (r=0.35) and negative correlation in B6 (r=-0.3) (P=0.01) (Supplementary Figures E6, E8, E9A). However, phyla Proteobacteria was negatively correlated with subset B in B1 (r=-0.4) and positively correlated in B6 (r=0.27) (P=0.01) (Supplementary Figures E6, E8, E9B). Similarly, phyla TM7 negatively correlated with subset B in B1 (r=-0.2) and positively correlated in B6 (r=0.53) (P=0.01) (Supplementary Figures E6, E8, E9C). Phyla Firmicutes and Thermi had differential interactions with subset B in B1 and B6, with marginal significance (P=0.05) (Supplementary Figure E6, E9D and E9E). Previously, it has been shown that neutrophilic MDSCs are preferentially activated in gram-positive sepsis in humans33. We performed correlation analysis to evaluate interactions of the gram-positive bacteria (GPB) with neutrophilic MDSC (subset B) in the airways of LTRs. The interaction between proportions of subset B and airway microbiome (B1/B6) was significant (P=0.046) (Figure E9F). The total gram+ abundance and subset B cells were significantly correlated in B1 (P=0.036), but not in B6 (P=0.684).
The endoplasmic reticulum (ER) stress response is an evolutionary conserved process that is important in cellular quality control and host defense. Bacterial and viral infections modulate ER stress responses 34,35. C/EBP-homologous protein (CHOP), a cellular stress sensor, regulates the function and accumulation of suppressive MDSCs 36,37. We hypothesized that the associations between bacterial phyla and MDSC subsets may account for stress responses leading to regulation of differential functions of the pro-inflammatory and anti-inflammatory MDSC subsets in the airways of LTRs. We noted significant increase in gene expression of stress response sensor CHOP and XBP1, a transcription factor which functions as an additional sensor of ER stress response (Supplementary Figure E10). These increases were noted specifically in suppressive HLA-DR−MDSCs, compared to the pro-inflammatory HLA-DR+ MDSCs. Additionally, Nox2 expression that generates ROS production in MDSCs was significantly elevated in HLA-DR+ MDSCs.
Discussion
BOS is the major cause of reduced long-term survival in lung transplantation 2,3. At five years after transplant, almost 50% of LTRs develop BOS and median survival thereafter is 3 years 38. Previous studies have suggested a correlation between changes in the lung microbiome and occurrence of BOS 11,12,39. In this study, we characterized the relationships between the composition and diversity of airway microbiomes with the presence of immune regulatory cells (MDSCs) that may contribute to BOS.
The lung microbiomes of healthy subjects have been shown to be similar to that of the oral microbiome; whereas, the nasal microbiome is distinct from both the oral and lung microbiomes 25,40. In this study, we found that the oral microbiome was distinct from both proximal and distal airway microbiomes in LTRs. Additionally, the nasal microbiome was also dissimilar from the oral microbiome but more closely aligned with both proximal and distal airway microbiomes. In contrast to findings in healthy, non-transplant subjects, our studies in LTRs show a marked divergence between the BAL and oral microbiota. Other studies in lung transplant and cystic fibrosis have demonstrated the greater dissimilarity between oral and lung microbiome 41-43. In addition, HIV individuals on antiretroviral therapy (ART) harbor similar oral and airway microbiota compared to those not on ART 44,45. It is known that various treatments affecting mucosal immunity play a role in defining the microbiome 46. It is possible that immunosuppressive agents in LTRs or immune deficiency in HIV subjects not on ART may contribute to these differences. We performed all bronchoscopies via the oral route and thus the potential for bronchoscopic contamination of microbiota from the nasal tract was eliminated. The microbiome differences identified in our study was validated using multiple replicates.
Our group previously defined the MDSC subsets in proximal and distal airways of asthmatics 26. We used sequential BAL sampling and found distinct microbiome and MDSC signatures in the proximal and distal airways of LTRs of individual subjects. Importantly, the distal airways had a higher proportion of pro-inflammatory MDSCs (C subset), whereas the proximal airways harbored more immuno-suppressive MDSCs (subsets A and B). Nakajima et al. have hypothesized that certain microbial populations in the lungs may trigger inflammatory pathways, resulting in allograft rejection 10. Although our study was not powered to detect statistical significance between microbial signatures in BOS and non-BOS subjects, the BOS cohort had increased relative abundance of Proteobacteria, Firmicutes, and Bacteroidetes in their distal airways as compared to non-BOS subjects (Figure 3C). While the phyla Firmicutes dominated the microbiome signature in the distal airways of non-BOS subjects, this shifted to a Proteobacteria dominant signature in the BOS cohort. It is possible that the higher abundance of the distinct bacterial populations that we identified in the distal airways may contribute to the activation of MDSCs in the distal airways, thus leading to increased airway inflammation and risk of allograft rejection.
Interestingly, two separate species of genus Prevotella were found to differentially associate with non-BOS subjects in the proximal, and BOS subjects in the distal airways. Prevotella intermedia was associated with non-BOS in B1, while Prevotella melaninogenica was associated with BOS group in B6. Although different Prevotella species are known to be upper respiratory tract commensals, some of those may differentially act as commensals in the proximal airways and pathobionts in the distal airways based on the local microenvironment and bacterial biomass present. Future studies designed to study the virulence potential of bacteria are required to delineate this further.
We also found significant correlations between bacterial OTUs and subsets of MDSCs (Figure 4B). Phyla Actinomycetes, Planctomycetes and TM7 positively correlated with the pro-inflammatory subset C. Interestingly, phyla Proteobacteria, Bacteroidetes and TM7 were differentially correlated with neutrophilic MDSCs (subset B) in the proximal and distal airways. Positive or negative correlation of these bacteria to the subset B in proximal and distal airways was determined by the relative abundance of the bacteria in that compartment. A higher relative abundance led to a negative correlation, and lower abundance of bacteria led to a positive correlation with neutrophilic MDSCs. It remains to be determined whether the correlations of MDSCs with these bacteria may reflect direct involvement in the activation of the pro-inflammatory MDSCs or the primary activation of immunosuppressive neutrophilic MDSCs by these bacteria contributing to a secondary proinflammatory state of activation of non-neutrophilic MDSCs. Hence, it is plausible that presence of specific bacterial communities may be stimulatory or inhibitory in the activation of MDSCs; the imbalance in the ratio of immunosuppressive and pro-inflammatory MDSCs may trigger inflammatory pathways leading to allograft rejection. Significant correlation between gram-positive bacteria and neutrophilic MDSCs were also found. This is similar to previous studies that reported preferential activation of neutrophilic MDSCs in sepsis by Staphylococcus spp. and gram-positive bacteria 33,47. Neutrophilic MDSCs are immunosuppressive in function and their activation by gram-positive bacteria in LTR may reduce host immunity and trigger further bacterial growth in the airways. Bacterial phyla are known to utilize unique metabolic pathways to maintain homeostasis (Supplementary Table S9). It is possible that these unique metabolic pathways determine the immune response generated by different bacterial phyla leading to the differential function of MDSCs. Additionally, our preliminary studies have indicated that ER stress response sensors are differentially expressed in MDSC subsets in LTRs, with higher expression in immunosuppressive HLA-DR−MDSCs. This increased ER stress response can lead to apoptosis of the immunosuppressive MDSCs and may account for the imbalance in the proportions of the pro-inflammatory HLA-DR+MDSCs. Bacterial and viral infections have also been shown to modulate stress responses 34,35. Future studies are warranted to investigate if differential correlations of identified bacterial phyla and MDSC subsets may account for stress responses leading to regulation of differential functions by MDSC subsets in LTRs.
One of the challenges in the field of lung microbiome is presence of relatively lower biomass in BAL samples. Interpretation of results obtained from a single low biomass sample without appropriate controls can be misleading 40,48-50. To circumvent this potential problem, we collected control samples from the bronchoscope prior to each procedure. These were found to be dissimilar to the airway microbiome (Figure E4A and E4B). Additionally, we processed quadruplicates of each biological sample to evaluate variability in relative abundance (Supplementary Figure E11). While we observed that the beta diversity between B1 and B6 samples were greater than that within B1 and B6, respectively, the large within-sample variability for B1 and B6 suggest that technical replicates are critical. Our study has several limitations. The sample size and cross sectional nature of the study limits our ability to establish a causal relationship between microbiome-immune cell interaction and BOS. However, this was a pilot study to evaluate the microbiome-immune cell interactions in LTRs. The use of quadruplet samples for microbiome analysis and inclusion of appropriate controls enhances the validity of our observations. Further studies are currently underway to validate our observations in a longitudinal cohort. Even though we found an association between the airway microbiome and MDSCs in LTRs, it is possible that several other variables such as previous acute rejection episodes, infections, immunosuppressive drugs, and antimicrobial therapies, may influence the microbiome and immune cell profiles in these subjects. Additionally, there may be an inverse effect and/or a bidirectional interplay between microbiome and the immune cells. Although, the gene expression data did show increased NOX2 expression in the HLADR+ MDSCs suggesting increased potential for reactive oxygen species formation, it does not demonstrate actual MDSC effector response. Future studies evaluating the response of MDSCs on T cell effector response will have to be conducted to confirm our observations.
Our study identifies, for the first time, the association of distinct MDSC sub-populations with the lung microbiome in LTRs. Future studies with gnotobiotic and humanized murine models will facilitate further investigations regarding the crosstalk between the lung microbiome and immune cell responses in LTR; this will also aid in defining the role of extra-pulmonary microbiome in contributing to the lung microbiota.
Supplementary Material
Acknowledgments
Authors gratefully acknowledge the staff at the bronchoscopy lab, Kirklin Clinic, University of Alabama at Birmingham, AL for their support in collection of BAL specimens. We thank the Microbiome Resource at the University of Alabama at Birmingham: School of Medicine, Comprehensive Cancer Center, Center for AIDS Research, Center for Clinical Translational Science and Heflin Center. We acknowledge the Center for Aids Research Flow Cytometry Core facility for support in acquisition of Flow cytometry data.
Funding Support: Supported by Flight Attendant Medical Research Institute (Young Clinical Scientist faculty award [YCSA2010] and NIH 1R01HL128502 to J.S.D, Angus Cooper Award to N.S.S., Parker B Francis fellowship award to J.S.D, NIH P01 HL114470 award to V.J.T. Microbiome analysis was supported by the Microbiome Resource at the University of Alabama at Birmingham: School of Medicine, Comprehensive Cancer Center (P30AR050948), Center for AIDS Research (5P30AI027767), Center for Clinical Translational Science (UL1TR000165) and Heflin Center.
Footnotes
Author contributions: N.S.S, K.M.W., E.D.G., S.R., V.J.T., D.D.C., C.M. and J.S.D. designed the research; N.S.S., E.D.G., and K.M.W., C.W. collected the samples; N.S.S., Y.W., P.E., J.V.R.D., C.M., R.K. and J.S.D. processed samples and performed research; N.S.S., S.A., M.A., R.S. D.Z., K.Z. K.P.H. and J.S.D. analyzed data; N.S.S. and J.D. wrote the manuscript and N.S.S., D.Z., S.R., K.M.W., D.D.C., C.W.H., K.P.H, C.M., V.J.T. and J.S.D. performed critical review of the manuscript.
Disclosures: None
References
- 1.Annual Data Report of the US Organ Procurement and Transplantation Network Preface. Am J Transplant. 2014;14(1):5–7. doi: 10.1111/ajt.12626. [DOI] [PubMed] [Google Scholar]
- 2.Burton CM, Carlsen J, Mortensen J, et al. Long-term survival after lung transplantation depends on development and severity of bronchiolitis obliterans syndrome. J Heart Lung Transplant. 2007;26:681–686. doi: 10.1016/j.healun.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 3.Finlen Copeland CA, Snyder LD, Zaas DW, et al. Survival after bronchiolitis obliterans syndrome among bilateral lung transplant recipients. Am J Respir Crit Care Med. 2010;182:784–789. doi: 10.1164/rccm.201002-0211OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bharat A, Kuo E, Steward N, et al. Immunological link between primary graft dysfunction and chronic lung allograft rejection. Ann Thorac Surg. 2008;86:189–195. doi: 10.1016/j.athoracsur.2008.03.073. discussion 196-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bobadilla JL, Jankowska-Gan E, Xu Q, et al. Reflux-induced collagen type v sensitization: potential mediator of bronchiolitis obliterans syndrome. Chest. 2010;138:363–370. doi: 10.1378/chest.09-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daud SA, Yusen RD, Meyers BF, et al. Impact of immediate primary lung allograft dysfunction on bronchiolitis obliterans syndrome. Am J Respir Crit Care Med. 2007;175:507–513. doi: 10.1164/rccm.200608-1079OC. [DOI] [PubMed] [Google Scholar]
- 7.Snyder LD, Finlen-Copeland CA, Turbyfill WJ, et al. Cytomegalovirus pneumonitis is a risk for bronchiolitis obliterans syndrome in lung transplantation. Am J Respir Crit Care Med. 2010;181:1391–1396. doi: 10.1164/rccm.200911-1786OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gottlieb J, Mattner F, Weissbrodt H, et al. Impact of graft colonization with gram-negative bacteria after lung transplantation on the development of bronchiolitis obliterans syndrome in recipients with cystic fibrosis. Respir Med. 2009;103:743–749. doi: 10.1016/j.rmed.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 9.Husain S, Singh N. Bronchiolitis obliterans and lung transplantation: evidence for an infectious etiology. Semin Respir Infect. 2002;17:310–314. doi: 10.1053/srin.2002.36442. [DOI] [PubMed] [Google Scholar]
- 10.Nakajima T, Palchevsky V, Perkins DL, et al. Lung transplantation: infection, inflammation, and the microbiome. Semin Immunopathol. 2011;33:135–156. doi: 10.1007/s00281-011-0249-9. [DOI] [PubMed] [Google Scholar]
- 11.Willner DL, Hugenholtz P, Yerkovich ST, et al. Reestablishment of recipient-associated microbiota in the lung allograft is linked to reduced risk of bronchiolitis obliterans syndrome. Am J Respir Crit Care Med. 2013;187:640–647. doi: 10.1164/rccm.201209-1680OC. [DOI] [PubMed] [Google Scholar]
- 12.Bernasconi E, Pattaroni C, Koutsokera A, et al. Airway Microbiota Determines Innate Cell Inflammatory or Tissue Remodeling Profiles in Lung Transplantation. Am J Respir Crit Care Med. 2016;194:1252–1263. doi: 10.1164/rccm.201512-2424OC. [DOI] [PubMed] [Google Scholar]
- 13.Syed SA, Whelan FJ, Waddell B, et al. Reemergence of Lower-Airway Microbiota in Lung Transplant Patients with Cystic Fibrosis. Ann Am Thorac Soc. 2016;13:2132–2142. doi: 10.1513/AnnalsATS.201606-431OC. [DOI] [PubMed] [Google Scholar]
- 14.Botha P, Archer L, Anderson RL, et al. Pseudomonas aeruginosa colonization of the allograft after lung transplantation and the risk of bronchiolitis obliterans syndrome. Transplantation. 2008;85:771–774. doi: 10.1097/TP.0b013e31816651de. [DOI] [PubMed] [Google Scholar]
- 15.Vos R, Vanaudenaerde BM, Geudens N, et al. Pseudomonal airway colonisation: risk factor for bronchiolitis obliterans syndrome after lung transplantation? Eur Respir J. 2008;31:1037–1045. doi: 10.1183/09031936.00128607. [DOI] [PubMed] [Google Scholar]
- 16.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuenca AG, Delano MJ, Kelly-Scumpia KM, et al. A paradoxical role for myeloid-derived suppressor cells in sepsis and trauma. Mol Med. 2011;17:281–292. doi: 10.2119/molmed.2010.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brudecki L, Ferguson DA, McCall CE, et al. Myeloid-derived suppressor cells evolve during sepsis and can enhance or attenuate the systemic inflammatory response. Infect Immun. 2012;80:2026–2034. doi: 10.1128/IAI.00239-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deshane J, Zmijewski JW, Luther R, et al. Free radical-producing myeloid-derived regulatory cells: potent activators and suppressors of lung inflammation and airway hyperresponsiveness. Mucosal Immunol. 2011;4:503–518. doi: 10.1038/mi.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heim CE, Vidlak D, Scherr TD, et al. Myeloid-derived suppressor cells contribute to Staphylococcus aureus orthopedic biofilm infection. J Immunol. 2014;192:3778–3792. doi: 10.4049/jimmunol.1303408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rieber N, Brand A, Hector A, et al. Flagellin induces myeloid-derived suppressor cells: implications for Pseudomonas aeruginosa infection in cystic fibrosis lung disease. J Immunol. 2013;190:1276–1284. doi: 10.4049/jimmunol.1202144. [DOI] [PubMed] [Google Scholar]
- 22.Estenne M, Maurer JR, Boehler A, et al. Bronchiolitis obliterans syndrome 2001: an update of the diagnostic criteria. J Heart Lung Transplant. 2002;21:297–310. doi: 10.1016/s1053-2498(02)00398-4. [DOI] [PubMed] [Google Scholar]
- 23.Jambo KC, French N, Zijlstra E, et al. AIDS patients have increased surfactant protein D but normal mannose binding lectin levels in lung fluid. Respir Res. 2007;8:42. doi: 10.1186/1465-9921-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu J, Kobayashi M, Sousa EA, et al. Differential proteomic analysis of bronchoalveolar lavage fluid in asthmatics following segmental antigen challenge. Mol Cell Proteomics. 2005;4:1251–1264. doi: 10.1074/mcp.M500041-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Charlson ES, Bittinger K, Haas AR, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. 2011;184:957–963. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson JT, Zeng M, Li Q, et al. Elevated levels of NO are localized to distal airways in asthma. Free Radic Biol Med. 2011;50:1679–1688. doi: 10.1016/j.freeradbiomed.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin B, Ma G, Yen CY, et al. Myeloid-derived suppressor cells prevent type 1 diabetes in murine models. J Immunol. 2010;185:5828–5834. doi: 10.4049/jimmunol.0903636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marigo I, Bosio E, Solito S, et al. Tumor-induced tolerance and immune suppression depend on the C/EBPbeta transcription factor. Immunity. 2010;32:790–802. doi: 10.1016/j.immuni.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 29.Zhou Z, French DL, Ma G, et al. Development and function of myeloid-derived suppressor cells generated from mouse embryonic and hematopoietic stem cells. Stem Cells. 2010;28:620–632. doi: 10.1002/stem.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vasquez-Dunddel D, Pan F, Zeng Q, et al. STAT3 regulates arginase-I in myeloid-derived suppressor cells from cancer patients. J Clin Invest. 2013;123:1580–1589. doi: 10.1172/JCI60083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piskin G, Bos JD, Teunissen MB. Neutrophils infiltrating ultraviolet B-irradiated normal human skin display high IL-10 expression. Arch Dermatol Res. 2005;296:339–342. doi: 10.1007/s00403-004-0522-z. [DOI] [PubMed] [Google Scholar]
- 32.Deshane JS, Redden DT, Zeng M, et al. Subsets of airway myeloid-derived regulatory cells distinguish mild asthma from chronic obstructive pulmonary disease. J Allergy Clin Immunol. 2015;135:413–424 e415. doi: 10.1016/j.jaci.2014.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janols H, Bergenfelz C, Allaoui R, et al. A high frequency of MDSCs in sepsis patients, with the granulocytic subtype dominating in gram-positive cases. J Leukoc Biol. 2014;96:685–693. doi: 10.1189/jlb.5HI0214-074R. [DOI] [PubMed] [Google Scholar]
- 34.Lim YJ, Choi JA, Choi HH, et al. Endoplasmic reticulum stress pathway-mediated apoptosis in macrophages contributes to the survival of Mycobacterium tuberculosis. PLoS One. 2011;6:e28531. doi: 10.1371/journal.pone.0028531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van 't Wout EF, van Schadewijk A, van Boxtel R, et al. Virulence Factors of Pseudomonas aeruginosa Induce Both the Unfolded Protein and Integrated Stress Responses in Airway Epithelial Cells. PLoS Pathog. 2015;11:e1004946. doi: 10.1371/journal.ppat.1004946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Condamine T, Kumar V, Ramachandran IR, et al. ER stress regulates myeloid-derived suppressor cell fate through TRAIL-R-mediated apoptosis. J Clin Invest. 2014;124:2626–2639. doi: 10.1172/JCI74056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thevenot PT, Sierra RA, Raber PL, et al. The stress-response sensor chop regulates the function and accumulation of myeloid-derived suppressor cells in tumors. Immunity. 2014;41:389–401. doi: 10.1016/j.immuni.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyer KC, Raghu G, Verleden GM, et al. An international ISHLT/ATS/ERS clinical practice guideline: diagnosis and management of bronchiolitis obliterans syndrome. Eur Respir J. 2014;44:1479–1503. doi: 10.1183/09031936.00107514. [DOI] [PubMed] [Google Scholar]
- 39.Jaramillo A, Fernandez FG, Kuo EY, et al. Immune mechanisms in the pathogenesis of bronchiolitis obliterans syndrome after lung transplantation. Pediatr Transplant. 2005;9:84–93. doi: 10.1111/j.1399-3046.2004.00270.x. [DOI] [PubMed] [Google Scholar]
- 40.Bassis CM, Erb-Downward JR, Dickson RP, et al. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio. 2015;6:e00037. doi: 10.1128/mBio.00037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Charlson ES, Diamond JM, Bittinger K, et al. Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant. Am J Respir Crit Care Med. 2012;186:536–545. doi: 10.1164/rccm.201204-0693OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown PS, Pope CE, Marsh RL, et al. Directly sampling the lung of a young child with cystic fibrosis reveals diverse microbiota. Ann Am Thorac Soc. 2014;11:1049–1055. doi: 10.1513/AnnalsATS.201311-383OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goddard AF, Staudinger BJ, Dowd SE, et al. Direct sampling of cystic fibrosis lungs indicates that DNA-based analyses of upper-airway specimens can misrepresent lung microbiota. Proc Natl Acad Sci U S A. 2012;109:13769–13774. doi: 10.1073/pnas.1107435109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iwai S, Fei M, Huang D, et al. Oral and airway microbiota in HIV-infected pneumonia patients. J Clin Microbiol. 2012;50:2995–3002. doi: 10.1128/JCM.00278-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cook CHF, Thompson RL. A meta-analysis of response rates in web- or internet-based surveys. Educ Psychol Meas. 2000;606:821–836. [Google Scholar]
- 46.Ivanov II, Frutos Rde L, Manel N, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tebartz C, Horst SA, Sparwasser T, et al. A major role for myeloid-derived suppressor cells and a minor role for regulatory T cells in immunosuppression during Staphylococcus aureus infection. J Immunol. 2015;194:1100–1111. doi: 10.4049/jimmunol.1400196. [DOI] [PubMed] [Google Scholar]
- 48.Venkataraman A, Bassis CM, Beck JM, et al. Application of a neutral community model to assess structuring of the human lung microbiome. MBio. 2015;6 doi: 10.1128/mBio.02284-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salter SJ, Cox MJ, Turek EM, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lusk RW. Diverse and widespread contamination evident in the unmapped depths of high throughput sequencing data. PLoS One. 2014;9:e110808. doi: 10.1371/journal.pone.0110808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


