Abstract
BACKGROUND
In immunosurveillance, bone-derived immune cells infiltrate the tumor and secrete inflammatory cytokines to destroy cancer cells. However, cancer cells have evolved mechanisms to usurp inflammatory cytokines to promote tumor progression. In particular, the inflammatory cytokine, interleukin-1 (IL-1), is elevated in prostate cancer (PCa) patient tissue and serum and promotes PCa bone metastasis. IL-1 also represses androgen receptor (AR) accumulation and activity in PCa cells, yet the cells remain viable and tumorigenic; suggesting that IL-1 may also contribute to AR-targeted therapy resistance. Furthermore, IL-1 and AR protein levels negatively correlate in PCa tumor cells. Taken together, we hypothesize that IL-1 reprograms AR positive (AR+) PCa cells into AR negative (AR−) PCa cells that co-opt IL-1 signaling to ensure AR-independent survival and tumor progression in the inflammatory tumor microenvironment.
METHODS
LNCaP and PC3 PCa cells were treated with IL-1β or HS-5 bone marrow stromal cell (BMSC) conditioned medium and analyzed by RNA sequencing and RT-QPCR. To verify genes identified by RNA sequencing, LNCaP, MDA-PCa-2b, PC3 and DU145 PCa cell lines were treated with the IL-1 family members, IL-1α or IL-1β, or exposed to HS-5 BMSC in the presence or absence of Interleukin-1 Receptor Antagonist (IL-1RA). Treated cells were analyzed by western blot and/or RT-QPCR.
RESULTS
Comparative analysis of sequencing data from the AR+ LNCaP PCa cell line versus the AR− PC3 PCa cell line reveals an IL-1-conferred gene suite in LNCaP cells that is constitutive in PC3 cells. Bioinformatics analysis of the IL-1 regulated gene suite revealed that inflammatory and immune response pathways are primarily elicited; likely facilitating PCa cell survival and tumorigenicity in an inflammatory tumor microenvironment.
CONCLUSIONS
Our data supports that IL-1 reprograms AR+ PCa cells to mimic AR− PCa gene expression patterns that favor AR-targeted treatment resistance and cell survival.
Keywords: interleukin-1, prostate cancer, gene expression pattern, androgen receptor, treatment resistance
INTRODUCTION
Inflammatory cytokines are present in the tumor microenvironment due to infiltrating immune cell paracrine secretion and tumor cell autocrine signaling1. The normal function of inflammatory cytokines includes signaling the destruction and removal of damaged and foreign cells in wound healing1. Tumor cells, however, have evolved mechanisms that use inflammatory cytokines to promote tumor cell survival and disease progression1. For example, in prostate cancer (PCa), the interleukin-1 (IL-1) inflammatory cytokine family member, IL-1 beta (IL-1β), has been shown to promote PCa tumor angiogenesis2 and PCa metastasis and bone colonization3, thereby contributing to PCa progression.
We and others have shown that IL-1β and the other major IL-1 family member, IL-1 alpha (IL-1α), can repress androgen receptor (AR) accumulation and activity4–7. AR is a nuclear receptor transcription factor8. AR and its hormone ligand, androgen, are required for prostate cell growth8. As such, AR drives PCa initiation and progression and is a PCa therapeutic target8. Therapies that target and block AR signaling include chemical or surgical androgen deprivation (hormone therapy) or treatment with anti-androgen antagonists which directly bind AR8. AR-targeted therapies are initially effective, but PCa patients can relapse within a few years and develop bone metastatic tumors that acquire androgen independence and treatment resistance9. Treatment at the bone metastatic stage is palliative9, highlighting the need to investigate and rigorously dissect mechanisms of PCa progression and acquired resistance to AR-targeted therapy.
The intended target of androgen deprivation and anti-androgens are the AR+ luminal cells which comprise the bulk of the prostate adenocarcinoma tumor10. However, the AR+ luminal PCa cells can evolve androgen independence and AR-targeting drug resistance due to multiple different mechanisms, including AR overexpression, AR gene mutation, or androgen-independent AR activation10. Another mechanism of androgen independence and treatment resistance is the accumulation of the AR− prostate gland cell types – basal, neuroendocrine, or stem cells – which are not susceptible to AR-targeting therapy and thus can repopulate the tumor and contribute to relapse10. Importantly, it has been found that high grade, metastatic, and castration-resistant PCa tumors are enriched in cells with low or no accumulation of AR11,12 or the AR target gene, Prostate Specific Antigen (PSA)13, suggesting that the loss of AR activity is associated with PCa progression. It is thus becoming increasingly apparent that PCa tumors with low or no AR accumulation or activity, either through the enrichment of AR− prostate gland cell types or the de novo reduction or loss of AR in AR+ luminal cells, represent an inherently treatment resistant cell phenotype that should not be overlooked and warrants further study.
We propose that a possible mechanism of AR-targeted treatment resistance and PCa tumor progression is IL-1-mediated AR repression in luminal PCa cells. Clinical support for such a mechanism is the observation that PCa patient bone metastases show an inverse correlation between IL-1β and AR protein accumulation in tumor cells3,14. Bone-derived immune cells present in the tumor microenvironment can also produce high levels of IL-11; and we previously discovered that the secretory bone marrow stromal cell line, HS-5, which secretes IL-1α and IL-1β15, causes paracrine repression of AR mRNA and protein in PCa cell lines4. Concomitant with AR repression, we found that HS-5 bone marrow stromal cells (BMSCs) also induce prosurvival proteins, one of which, Sequestome-1 (p62), is also induced by IL-1β4,16,17. p62 is an adaptor protein that functions in inflammatory, antioxidant, and autophagy pathways18 and is cytoprotective for AR− PCa cell lines16. Therefore, we hypothesize that IL-1 promotes PCa progression and treatment resistance by reprograming AR+ PCa cells into AR− PCa cells that acquire AR-independent survival and tumor promoting mechanisms. To begin to address our hypothesis, we set out to identify the IL-1-confered gene expression profile in AR+ PCa cells that mimics AR− PCa cell gene expression patterns to uncover the elicited signaling pathways that might promote AR-independent PCa cell survival and tumorigenesis.
MATERIALS AND METHODS
Cell Culture
LNCaP, MDA-PCa-2b, PC3, and DU145 prostate cancer (PCa) cell lines, and HS-5 and HS-27a bone marrow stromal (BMSC) cell lines were grown in a 37°C, 5.0% (v/v) CO2 growth chamber. All cell lines, except for MDA-PCa-2b, were cultured in DMEM (Gibco/Thermo Scientific; 1185-092) supplemented with 10% (v/v) fetal bovine serum essence (FBS; Seradigm; 3100-500), 0.4mM L-glutamine (L-glut; Gibco/Invitrogen; 25030-081), and 10U/ml penicillin G sodium and 10 mg/ml streptomycin sulfate (pen-strep; Gibco/ Invitrogen; 15140-122). MDA-PCa-2b cells were cultured in BRFF-HPC1 (AthenaES; 403) supplemented with 20% (v/v) fetal bovine serum essence (FBS; Seradigm; 3100-500), 0.4mM L-glutamine (L-glut; Gibco/Invitrogen; 25030-081), and 10U/ml penicillin G sodium and 10 mg/ml streptomycin sulfate (pen-strep; Gibco/ Invitrogen; 15140-122).
Cell Treatments
Conditioned Medium (CM): Conditioned medium was obtained from LNCaP, PC3, or DU145 PCa cell lines or HS-5 or HS-27a BMSC cell lines grown in DMEM/10% FBS for 5 days, the medium collected, and filtered (Milipore; SCGPU05RE, pore size 0.22 μm) to remove cell debris. The MDA-PCa-2b PCa cell line was grown in BRFF-HPC1 medium. Conditioned medium (CM) was stored at -80°C until use. For conditioned medium treatment experiments, PCa cells were grow in their own PCa conditioned medium (control CM) or BMSC conditioned medium (BMSC CM). MDA-PCa-2b cells were grown in 1:1 BRFF-HPC1: DMEM (control CM) or 1:1 BRFF-HPC1: BMSC conditioned medium (BMSC CM). Intereleukin-1 (IL-1): Phosphate buffered saline (PBS) vehicle control, 25 ng/ml IL-1α (R&D Systems; 200-LA/CF), or 25 ng/ml IL-1β (R&D Systems; 201-LB-005) were added to DMEM/10% FBS growth medium. Interleukin-1 Receptor Antagonist (IL-1Ra): Cells were pre-treated for 1 or 2 days with PBS vehicle control or 500 ng/ml human recombinant IL-1Ra (R&D Systems; 280-RA/CF) in the DMEM/10% FBS growth medium. The next day the medium was removed and replaced with fresh DMEM/10% FBS (control CM) or HS-5 conditioned medium (HS-5 CM), plus an additional 500 ng/ml IL-1Ra for 1 or 3 days. Phosphate buffer saline (PBS) served as the IL-Ra vehicle control.
RNA Isolation and Reverse Transcription Quantitative PCR (RT-QPCR)
Total RNA was extracted from the cells using the GeneJET RNA Purification Kit according to the manufacturer’s instructions (Thermo Scientific; K0732). cDNA was generated using iScript Reverse Transcription Supermix (Biorad; 170-8841). The RT-QPCR reaction was prepared using iTaq Universal SYBR Green Supermix (Biorad; 172-5125). Primer sequences for genes of interest are listed below. Gene of interest cycle times (CT) were normalized to the β-actin. Relative mRNA levels were calculated using the 2−ΔΔCT method. Primer sequences, 5′-3′: Androgen Receptor (AR), forward AAGACGCTTCTACCAGCTCACCAA, reverse TCCCAGAAAGGATCTTGGGCACTT; Beta actin (β-actin), forward GATGAGATTGGCATGGCT TT, reverse CACCTTCACCGGTCCAGTTT; CD24 molecule (CD24), forward GCTCCTACCCACGCAGATTTA, reverse TGGTGGTGGCATTAGTTGGA; CD44 molecule (CD44), forward AGGAATGATGTCACAGGTGGA, reverse AGGTCCTGCTTTCCTTCGTG; E74 like ETS transcription factor 3 (ELF3), forward CTGGTCGAAGACGCAGGTT, reverse CGTGAGAAGTCAATGGCGCT; NK3 homeobox 1 (NKX3.1), forward CCCACACTCAGGTGATCGAG, reverse GTCTCCGTGAGCTTGAGGTT; Prostate Specific Antigen (PSA), forward CACCTGCTCGGGTGATTCTG, reverse ACTGCCCCATGACGTGATAC; Sequestosome-1 (p62), forward AAATGGGTCCACCAGGAAACTGGA, reverse TCAACTTCAATGCCCAGAGGGCTA; Superoxide dismutase 2 (SOD2), forward GGCCTACGTGAACAACCTGA, reverse GTTCTCCACCACCGTTAGGG; SRY-box 9 (SOX9), forward GAGACTTCTGAACGAGAGCGA, reverse CGTTCTTCACCGACTTCCTCC; Transmembrane protease serine 2 (TMPRSS2), forward TCAGGGTCACCACCAGCTAT, reverse CTGTGCGGGATAGGGGTTTT.
RNA-Sequencing Analysis
Fastq files were checked for quality using fastqc (v0.11.2)19 and fastq_screen (v0.4.4)20 and were quality trimmed using fastq-mcf (ea-utils/1.1.2-806)21. Trimmed fastq files were mapped to mm10 (UCSC version from igenomes) using TopHat22, duplicates were marked using picard-tools (v1.127 https://broadinstitute.github.io/picard/), read counts were generated using featureCounts23 and differential expression analysis was performed using edgeR24. Pathway analysis was conducted using QIAGEN’s Ingenuity Pathway Analysis tool (http://www.qiagen.com/ingenuity). RNA-seq datasets generated for this study are available at GEO NCBI, accession GSE105088.
Western Blot
Protein was isolated from cells using NP40 lysis buffer (0.5% NP-40 (US Biologicals; NP40S), 50mM Tris (pH 7.5), 150mM NaCl, 3mM MgCl2, 1X protease inhibitors (Roche; 0505489001)). Protein concentration was measured using the Pierce BCA Protein Assay Kit (Thermo Scientific; 23227). For western blot analysis, equal protein concentrations were loaded onto and separated in 12% (w/v) sodium dodecyl sulfate polyacrylamide gel (40% acrylamide/bis-acrylamide solution; Bio-Rad; 161-0146). Proteins were transferred from the gel to 0.45 mm pore size nitrocellulose membrane (Maine Manufacturing; 1215471) and total protein visualized using Ponceau S (Amresco; K793). The membrane was blocked with 2.5% (w/v) bovine serum albumin (BSA; Fisher; BP 1600-1) in 1X TBST (20mM Tris, pH 7.6, 150mM NaCl, 0.002% Tween-20). Primary and secondary antibodies were diluted in 2.5% BSA/1X TBST. Protein blot bands were visualized using SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific; 34095) and imaged using the GE Amersham Imager 600 (General Electric). Primary antibodies: AR (Cell Signaling; D6F11), p62/SQSTM1 (Abnova; H00008878-M01), SOD2 (Abgent; AM7579a), β-actin (Abcam; ab8226), β-actin (Novus; NB600-505), NKX3.1 (Cell Signaling, D2Y1A). Secondary antibodies: Sheep anti-mouse (Jackson ImmunoResearch Laboratories; 515-035-062), goat anti-rabbit (Abnova; PAB10822).
Statistical Analysis
Statistical significance was determined using unpaired student t test. Graphs are shown as the average of a minimum of n = 3 biological replicates +/− standard deviation (STDEV).
RESULTS
IL-1β modulates a suite of genes in LNCaP PCa cells that mimics basal PC3 PCa gene expression patterns
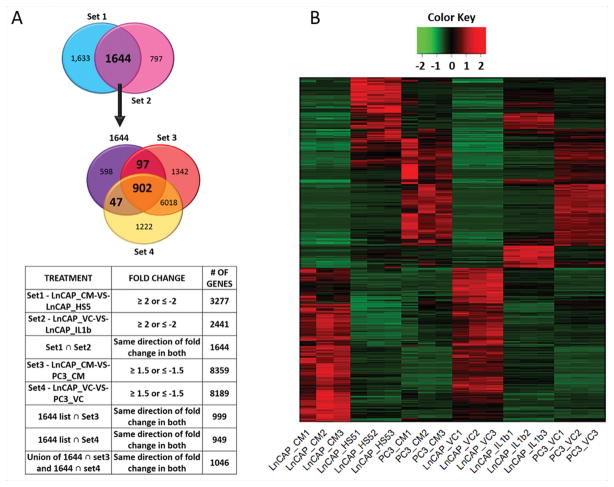
Based on our previous observations that HS-5 BMSC conditioned medium (CM) or IL-1β repress AR mRNA levels in AR+ PCa cell lines concomitant with induced expression of prosurvival p62, a gene constitutively expressed in AR− PCa cell lines4,16, we set out to identify the gene expression profile acquired by IL-1-exposed AR+ PCa cells that mimics intrinsic gene expression in AR− PCa cells. In doing so, we would identify conserved mechanisms that promote PCa cell survival and tumor progression either as a consequence of or despite AR loss or reduction. To obtain a profile of genes that might drive AR+ PCa cells towards an AR− PCa cell molecular phenotype, we performed RNA sequencing (RNA-seq) on the AR+ LNCaP and the AR− PC3 PCa cell line models. Four sets of differential gene expression analysis were performed: (Set 1) LNCaP cells grown in LNCaP conditioned medium (CM) versus HS-5 BMSC conditioned medium (“LnCAP_CM-VS-LnCAP_HS5”); (Set 2) LNCaP cells treated with phosphate buffered saline (PBS) vehicle control (VC) versus IL-1β (“LnCAP_VC-VS-LnCAP_IL1β”); (Set 3) LNCaP cells grown in LNCaP conditioned medium versus PC3 cells grown in PC3 conditioned medium (“LnCAP_CM-VS-PC3_CM”); and (Set 4) LNCaP cells grown in PBS vehicle control versus PC3 cells grown in PBS vehicle control (“LnCAP_VC-VS-PC3_VC”) (Fig. 1A, Supplementary Table T1).
Figure 1. RNA-seq reveals that HS-5 BMSC conditioned medium and IL-1β modulate a suite of 1,046 genes in the AR+ LNCaP PCa cell line that mimics basal gene expression in the AR− PC3 PCa cell line.
(A) Venn diagrams and table illustrating the generation of our gene expression profile. Four sets of differential gene expression analysis were performed: (Set 1) LNCaP cells grown for 3 days in LNCaP conditioned medium (CM) versus HS-5 BMSC conditioned medium (“LnCAP_CM-VS-LnCAP_HS5”); (Set 2) LNCaP cells treated for 3 days with phosphate buffered saline (PBS) vehicle control (VC) versus IL-1β (“LnCAP_VC-VS-LnCAP_IL1β”); (Set 3) LNCaP cells grown in LNCaP conditioned medium versus PC3 cells grown in PC3 conditioned medium for 3 days (“LnCAP_CM-VS-PC3_CM”); and (Set 4) LNCaP cells grown in PBS vehicle control versus PC3 cells grown in PBS vehicle control for 3 days (“LnCAP_VC-VS-PC3_VC”). The intersection (ȩ) of gene sets (1) and (2) generated a 1,644 genes list. The union of gene set (3) and gene set (4) intersected with the 1,644 genes list produced a final list of 1,024 genes (902 + 97 + 47) which we refer to as our “gene expression profile.” (B) The heat map of the induced (red) and repressed (green) genes for our RNA sequencing analysis. Three biological replicates for each treatment are shown: LNCaP cells grown in LNCaP CM (LNCaP_CM1,2,3), LNCaP cells grown in HS-5 BMSC CM (LNCaP_HS51,2,3), PC3 cells grown in PC3 CM (PC3-CM-1,2,3), LNCaP cells grown in PBS vehicle control (LNCaP_VC1,2,3), LNCaP cells treated with 25 ng/ml IL-1β (LNCaP_IL-1β1,2,3), PC3 cells grown in PBS vehicle control (PC3_VC1,2,3).
To identify a gene expression pattern representing IL-1β-dependent HS-5 paracrine signaling in LNCaP cells, we first obtained a consensus list of 1,644 genes that were either induced or repressed at least 2-fold in both gene sets (1) and (2) (log fold change (logFC) ≥ 1 or ≤ -1, false discovery rate (FDR) ≤ 0.05, and log counts per million (logCPM) > 0) (Fig. 1A, Supplementary Table T1). Next, to identify genes that are induced or repressed by IL-1β in LNCaP cells and are, respectively, basally high or low at least 1.5-fold in PC3 cells, we took the union of the intersection between the 1,644 genes list and gene set (3) and the 1,644 genes list and gene set (4) (logFC ≥ 0.6 or ≤ -0.6, FDR ≤ 0.05, logCPM > 0) (Fig. 1A, Supplementary Table T1). The union produced a list of 1,046 genes (Fig. 1A, Supplementary Table T1), which represent an IL-1β-induced gene expression profile in LNCaP cells that mimics basal PC3 gene expression patterns (Fig. 1A & B).
We previously published that IL-1β represses AR mRNA expression in AR+ PCa cell lines, yet cells can remain viable4. Therefore, the 1,046 gene expression profile we generated might provide insight into genes and pathways that confer AR-independent PCa cell survival and treatment resistance.
HS-5 CM, IL-1α, and IL-1β show similar regulation of selected genes from our expression profile in AR+ PCa cell lines
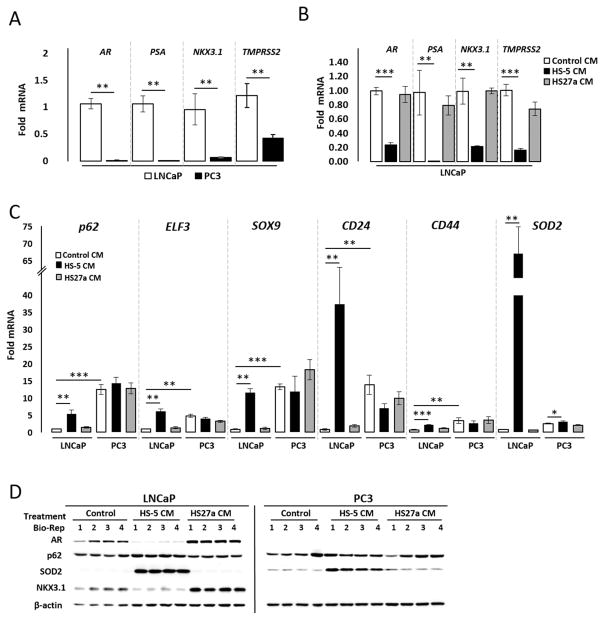
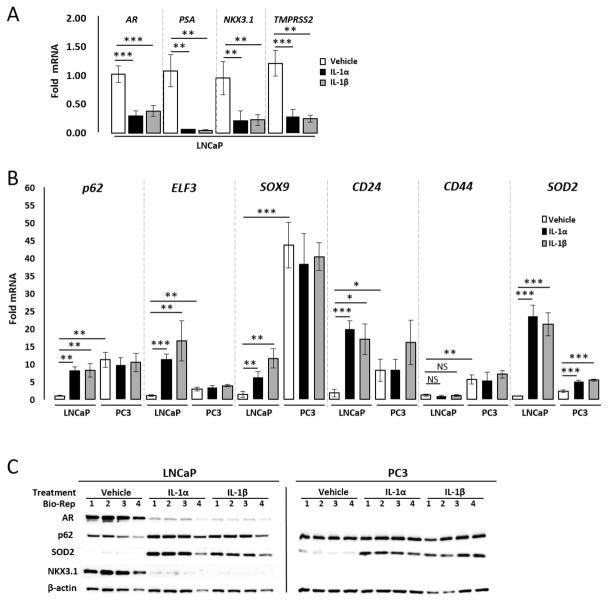
The 1,046 genes list is based on a comparison between HS-5 CM and IL-1β treatments in LNCaP cells. To confirm the RNA-seq data and to determine if IL-1α induces similar gene regulation in LNCaP cells, we performed quantitative RT-PCR (RT-QPCR) and/or western blot analysis for rationally selected genes based on previous studies showing IL-1 regulation4 (p62, AR, and AR target genes25, PSA, NKX3.1, and TMPRSS2) or randomly selected genes (CD24, CD44, ELF3, and SOX9) from the 1,046 genes list (Supplementary Table T1). RT-QPCR and/or western blot confirmed that AR, PSA, NKX3.1, and TMPRSS2 are repressed by HS-5 CM (Fig. 2B & D), IL-1α (Fig. 3A & C), or IL-1β (Fig. 3A & C) in LNCaP cells and are basally low or absent in PC3 cells (Fig. 2A). Likewise, RT-QPCR and/or western blot confirmed that p62, ELF3, SOX9, CD24, and/or CD44 are induced by HS-5 CM (Fig. 2C &D), IL-1α (Fig. 3B & C), or IL-1β (Fig. 3B & C) in LNCaP cells and are basally high in PC3 cells (Figs. 2 & 3). Conditioned medium from the HS-27a bone marrow stromal cell line, which does not express or secrete IL-1 levels comparable to HS-5 cells15,26 and does not induce p62 or repress AR4,16, serves as a negative control and Superoxide Dismutase (SOD2), which is a known IL-1-induced antioxidant gene27, serves as a positive control for experimental efficacy (Figs. 2 &3). The AR+ MDA-PCa-2b PCa cell line shows similar mRNA expression and/or protein accumulation response to HS-5 CM, IL-1α, and/or IL-1β as the LNCaP cells for AR, PSA, NKX3.1, TMPRSS2, p62, ELF3, CD24, and SOD2 (Supplementary Fig. S1). Likewise, the AR− DU145 PCa cell line shows similar high and low basal expression patterns as PC3 cells for all selected genes (Supplementary Fig. S2). Thus, while LNCaP, MDA-PCa-2b, PC3, and DU145 have many underlying genetic differences, such as differences in p53 or PTEN status28, that could influence basal or IL-1-induced gene expression profiles, we were yet able to show conserved intrinsic (e.g. PC3, DU145) or IL-1-confered (e.g. LNCaP, MDA-PCa-2b) expression patterns for our selected genes. Thus, the IL-1-confered gene expression pattern might be a common signature in AR+ PCa cell lines that reflects a functionally relevant AR− PCa cell molecular phenotype.
Figure 2. Confirmation that HS-5 BMSC conditioned medium modulates select genes identified in the 1,046 gene expression profile.
(A, B, C) RT-QPCR and (C) western blot analysis was performed for the AR+ LNCaP PCa cell line or the AR− PC3 PCa cell line treated for 3 days with PCa control conditioned medium (Control CM), HS-5 BMSC CM, or HS-27a BMSC CM. (A) PC3 cells have little or no detectable AR, PSA, NKX3.1, or TMPRSS2 mRNA relative to LNCaP cells. (B) HS-5 CM can repress AR, PSA, NKX3.1, and TMPRSS2 mRNA in LNCaP cells. (C) HS-5 CM can induce p62, ELF3, SOX9, CD24 and CD44 mRNA LNCaP cells. PC3 cells have high basal p62, ELF3, SOX9, CD24, and CD44 mRNA relative to LNCaP cells and show no significant induction response to HS-5 CM for these genes. (D) HS-5 CM can induce p62 protein and downregulate AR and NKX3.1 protein in LNCaP cells. PC3 cells do not accumulate AR or NKX3.1 protein and show no detectable p62 induction response to HS-5 CM. HS-5 CM induces SOD2 mRNA and protein in both LNCaP and PC3 cells and is used as a treatment efficacy control. n = 4 biological replicates (Bio-Rep); error bars = +/−STDEV; p-value = * ≤ 0.05, ** ≤ 0.005, ***≤ 0.0005. mRNA fold change is normalized to LNCaP vehicle control. β-actin is a western blot loading control.
Figure 3. Confirmation that IL-1α and IL1-β modulate select genes identified in the 1,046 gene expression profile.
(A, B) RT-QPCR and (C) western blot analysis was performed for the AR+ LNCaP PCa cell line or the AR− PC3 PCa cell line treated for 3 days with vehicle control, IL-1α (25 ng/ml), or IL-1β (25 ng/ml). (A) IL-1α or IL-1β can repress AR, PSA, NKX3.1, and TMPRSS2 mRNA in LNCaP cells. (B) IL-1α or IL-1β can induce p62, ELF3, SOX9, and CD24 mRNA LNCaP cells; no change was detected for CD44. PC3 cells have high basal p62, ELF3, SOX9, CD24, and CD44 mRNA relative to LNCaP cells and show no significant induction response to IL-1 for these genes. (C) IL-1α or IL-1β can induce p62 protein and downregulate AR and NKX3.1 protein in LNCaP cells. PC3 cells do not accumulate AR or NKX3.1 protein and show no detectable p62 induction response to IL-1. IL-1 induces SOD2 mRNA and protein in both LNCaP and PC3 cells and is used as a treatment efficacy control. n = 4 biological replicates (Bio-Rep); error bars = +/−STDEV; p-value = * ≤ 0.05, ** ≤ 0.005, ***≤ 0.0005. mRNA fold change is normalized to LNCaP vehicle control. β-actin is a western blot loading control.
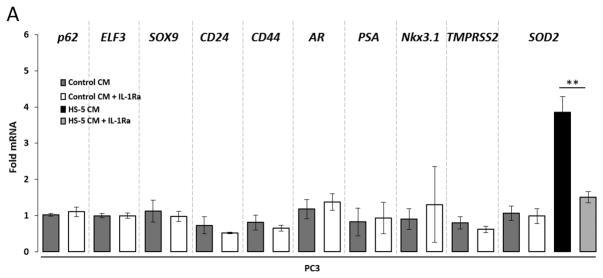
PC3 cells show IL-1-independent expression levels of selected genes from the 1,046 genes list
The 1,046 genes list is comprised of IL-1-induced or -repressed genes in LNCaP cells that are basally high or low, respectively, in PC3 cells. However, this does not preclude IL-1 from modulating expression of these genes in PC3 cells. Interestingly, while HS-5 CM (Fig. 2) or IL-1 (Fig. 3) induce SOD2 mRNA and protein in PC3 cells, HS-5 CM or IL-1 do not significantly modulate mRNA expression and/or protein accumulation of the selected genes (AR, PSA, NKX3.1, TMPRSS2, p62, ELF3, SOX9, CD24, or CD44) in PC3 cells. To determine if these genes are induced or repressed independent of IL-1 in PC3 cells, we grew PC3 cells in PC3 conditioned medium in the presence of the IL-1 Receptor Antagonist (IL-1Ra) to block IL-1 receptor activity. As an indication of IL-1Ra treatment efficacy, we found that IL-1Ra is sufficient to attenuate HS-5 CM-induced SOD2 mRNA (Fig. 4) in PC3 cells. IL-1Ra, however, does not induce AR, PSA, NKX3.1, or TMPRSS2 nor repress p62, ELF3, SOX9, CD24, or CD44 mRNA in PC3 cells (Fig. 4). Thus, AR, PSA, NKX3.1, and TMPRSS2 are basally low or absent and p62, ELF3, SOX9, CD24, and CD44 are basally high independent of IL-1 receptor activation in PC3 cells.
Figure 4. The AR− PC3 PCa cell line shows IL-1-independent regulation of basal AR, PSA, NKX3.1, TMPRSS2, p62, ELF3, SOX9, CD24, and CD44 levels.
PC3 cells were pretreated with 500 ng/ml Interleukin-1 Receptor Antagonist (IL-1Ra) for 1 day (to analyze basally high genes) or 2 days (to analyze basally low genes). Following pretreatment, the growth medium was replaced with PC3 conditioned medium (Control CM) + 500 ng/ml IL-1Ra or with HS-5 CM + 500 ng/ml IL-1Ra for an additional 1 day (to analyze basally low genes) or 3 days (to analyze basally high genes). PBS is the IL-1Ra vehicle control. RT-QPCR was performed for PC3 cells. IL-1Ra attenuates HS-5 CM-induced SOD2 mRNA, indicating IL-1Ra treatment efficacy. IL-1Ra does not modulate basal AR, PSA, NKX3.1, TMPRSS2, p62, ELF3, SOX9, CD24, or CD44 mRNA. n = 3 biological replicates (Bio-Rep); error bars = +/−STDEV; p-value = * ≤ 0.05. mRNA fold change is normalized to the respective Control CM for each gene.
IL-1 signaling is sufficient to mediate HS-5 BMSC paracrine modulation in AR+ PCa cell lines of selected genes from the 1,046 genes list
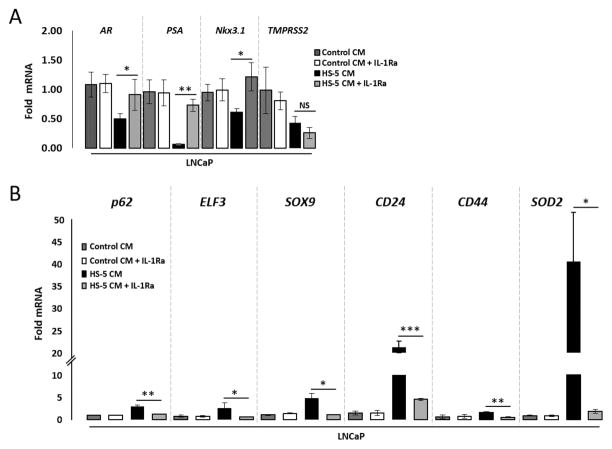
HS-5 CM contains a milieu of cytokines15; therefore, we reasoned that the intersection between the HS-5 CM-modulated gene set and IL-1β-modulated gene set in LNCaP cells (Fig. 1A, Supplementary Table 1) are those genes regulated by HS-5 BMSCs through IL-1 paracrine signaling. To provide support for our reasoning, we incubated LNCaP cells with IL-1Ra in the presence of HS-5 CM and used RT-QPCR to determine if IL-1Ra could attenuate HS-5 CM regulation of selected genes. As an indication of IL-1Ra treatment efficacy, we found that IL-1Ra is sufficient to attenuate HS-5 CM-induced SOD2 mRNA (Fig. 5B) in LNCaP cells. IL-1Ra also attenuates HS-5 CM-induction of p62, ELF3, SOX9, CD24, and CD44 mRNA (Fig. 5B) and attenuates HS-5 CM repression of AR, PSA, and NKx3.1 mRNA (Fig. 5A), but not TMPRSS2 mRNA levels. MDA-PCA-2b cells show similar responses to IL-1Ra as LNCaP cells (Supplementary Fig. S3). Thus, the gene expression patterns induced by IL-1-mediated BMSC paracrine signaling in AR+ PCa cell lines is likely conserved in PCa.
Figure 5. IL-1 signaling is sufficient to mediate HS-5 BMSC conditioned medium modulation of AR, PSA, Nkx3.1, p62, ELF3, SOX9, CD24, and CD44 mRNA in the AR+ LNCaP PCa cell line.
LNCaP cells were pretreated with 500 ng/ml Interleukin-1 Receptor Antagonist (IL-1Ra) for 1 day (to analyze induced genes) or 2 days (to analyze repressed genes). Following pretreatment, the growth medium was replaced with LNCaP conditioned medium (Control CM) with 500 ng/ml IL-1Ra or with HS-5 CM with 500 ng/ml IL-1Ra for an additional 1 day (to analyze repressed genes) or 3 days (to analyze induced genes). PBS is the IL-1Ra vehicle control. Treated cells were analyzed by RT-QPCR. IL-1Ra attenuates HS-5 CM-induced SOD2 mRNA (B), indicating IL-1Ra treatment efficacy. (A) IL-1Ra attenuates HS-5 CM repression of AR, PSA and Nkx3.1 mRNA; no statistical change was detected for TMPRSS2 mRNA. (B) IL-1Ra attenuates HS-5 CM induction of p62, ELF3, SOX9, CD24, and CD44 mRNA. n = 3 biological replicates (Bio-Rep); error bars = +/−STDEV; p-value = * ≤ 0.05, ** ≤ 0.005, ***≤ 0.0005. mRNA fold change is normalized to the respective Control CM for each gene.
The 1,046 genes list expression profile is predicted to regulate inflammatory, immune, and AR signaling pathways
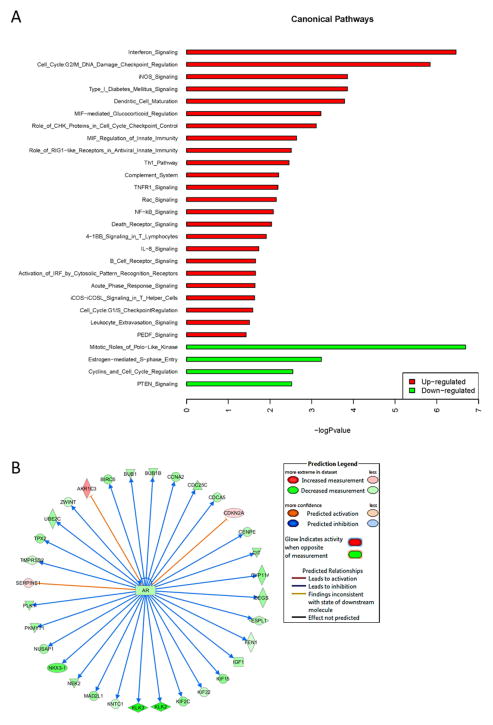
Ingenuity Pathway Analysis (IPA; Qiagen) was used to perform pathway and network analysis on the 1046 genes. The canonical pathway analysis module predicted that, primarily, canonical IL-1 inflammatory and immune response pathways are activated, including NFκB29–31, IL-832, and iNOS29 signaling cascades (z-score ≥ 1.5 or ≤ -1.5; -log (p-value) ≥ 1.3) (Fig. 6A). The canonical pathway analysis module did not predict the repression of AR signaling; however, the upstream analysis module which predicts pathway activation or repression based on the gene expression changes of known protein interactors and their functional interactions, predicts that the 1,046 gene expression profile reflects inhibition of AR signaling (Fig. 6B). This is in line with our directed look at AR and the AR target genes, PSA, NKX3.1, and TMPRSS2 by RT-QPCR and/or western blot, which we find are repressed by IL-1 in LNCaP and/or MDA-PCa-2b cells (Fig. 3, Supplementary Fig. S1) and are basally low or not expressed in PC3 and DU145 cells (Fig. 2, Supplementary Fig. S2). Taken together, concomitant with the repression of AR signaling, IL-1-confered gene expression patterns elicit inflammatory and immune response pathways that are basally high in AR− PCa cells and likely help promote AR-independent PCa cell survival and tumor progression in the inflammatory tumor microenvironment.
Figure 6. The 1,046 gene expression profile is predicted to regulate inflammatory, immune, and AR signaling pathways.
Bioinformatic enrichment and upstream analyses of the 1,046 gene expression profile was performed to identify (A) canonical pathways and (B) interactomes. (A) Enrichment analysis predicts that, primarily, canonical IL-1 inflammatory and immune pathways are elicited, including NFκB, IL-8, and iNOS signaling cascades (z-score ≥ 1.5 or ≤ 1.5; -log (p-value) ≥ 1.3). (B) Upstream analysis predicts that AR signaling is repressed.
DISCUSSION
IL-1 signaling may promote the de novo accumulation of ARlow/− PCa cells
In addition to tumor autocrine signaling, infiltrating bone-derived immune cells and tumor stromal cells secrete inflammatory cytokines and these cytokines have various pro- and anti-tumorigenic functions33. Thus, dissecting the paracrine interactions in the inflammatory tumor microenvironment better defines tumor biology and reveals cell survival mechanisms that could be therapeutically targeted.
Inflammation is implicated in PCa initiation, progression, and treatment resistance34, as such, PCa cells are not only exposed to a rich inflammatory cytokine environment in the primary tumor, but in the bone microenvironment, as well, where PCa cells preferentially metastasize. Thus, to explore how bone cell paracrine signaling effects PCa cell survival, we had previously shown that the macrophage-like, secretory bone marrow stromal cell line, HS-5, secrete factors that induce apoptosis in multiple different AR+ PCa cell lines35; however a subpopulation of the HS-5 CM-treated cells survive apoptosis, elicit cytoprotective autophagy and transdifferentiate into neuroendocrine-like cells that lose AR and have elevated p62 accumulation4,16,17,35. We found that HS-5-secreted interleukin-6 (IL-6) promotes PCa autophagy and neuroendocrine differentiation17 and that HS-5-secreted IL-1 mediates AR loss and p62 accumulation4 (Fig. 5, Supplementary Fig. S3). Interestingly, AR− PCa cell lines show reduced sensitivity to HS-5-induced apoptosis and have high basal autophagy, neuroendocrine makers, and cytoprotective p62 accumulation16,17,35,36. Therefore, a pattern emerges wherein HS-5 BMSCs cause AR+ PCa cells to acquire an AR− PCa prosurvival phenotype.
AR overexpression, AR gene mutation, androgen-independent AR activation, and the less well studied loss of AR accumulation, are each mechanisms that can drive resistance to AR-targeted therapy. In this paper, we focus on the potential role of IL-1 in transforming AR+ PCa cells into a viable AR− PCa cell type by identifying the IL-1-confered expression pattern in an AR+ PCa cell line that mimics intrinsic gene expression patterns of a viable AR− PCa cell line. IL-1 promotes PCa metastasis and skeletal colonization and inversely correlates with AR activity in bone metastases3. Based on these published and present data, we propose a hypothetical model wherein IL-1 in the primary or metastatic PCa tumor microenvironment causes the de novo accumulation of ARlow/− PCa cells that evade AR-targeted therapy and re-emerge in treatment resistant tumors. Thus, IL-1-targeted therapies, such as the FDA approved anti-inflammatory IL-1Ra drug, anakinra37, could be used to prevent the de novo accumulation of ARlow/− PCa cells. Chronic exposure to IL-1, however, is likely to produce ARlow/− PCa cell subpopulations that develop the ability to maintain constitutive activation of IL-1-confered pathways independent of IL-1 receptor activity, similar to what we observe for PC3 cells (Fig. 4). Thus, targeting such downstream pathways would predictably be effective against both IL-1-sensitive AR+ PCa cells and ARlow/− PCa cells that have evolved IL-1-independence.
The IL-1-confered 1,046 expression pattern is enriched in prosurvival genes and pathways that promote PCa progression
To identify the prosurvival proteins and pathways that could be promising therapeutic targets against both IL-1-senstive AR+ PCa cells that downregulate or lose AR and AR− PCa cells that have evolved IL-1-independent constitutive pathways, we identified an IL-1-confered gene expression pattern in the AR+ LNCaP PCa cell line that is intrinsic to the AR− PC3 PCa cell line (Fig. 1, Supplementary Table 1). The gene expression pattern is enriched in inflammatory and immune response signaling pathways (Fig. 6A) that mediate inflammation-induced tumorigenicity and provide cellular defense against the inflammatory microenvironment. For example, NFκB38 and two of its many downstream inflammatory signaling molecules, IL-839,40 and iNOS31,41, have been shown to promote androgen receptor-independent PCa cell survival.
NFκB is an inflammatory, immune and antioxidant response transcription factor that inhibits PCa cell apoptosis and promotes PCa cell metastasis and skeletal colonization30,39. NFκB protein levels and activity are elevated in PCa patients and AR− PCa cell lines30. p62 and ELF3, two of the selected genes that we confirmed by RT-QPCR and/or western blot analysis to be induced by IL-1 in AR+ PCa cell lines and basally high in AR− PCa cell lines (Fig. 3, Supplementary Figs. 1 & 2), encode proteins that promote IL-1-induced NFκB signaling. In response to IL-1, p62 binds and adds activating ubiquitin to TRAF6, leading to downstream NFκB activation18 and IL-1β induces expression of the ELF3 transcription factor through an ELF3-NFκB positive feedback loop42. Both p6243 and ELF342 are elevated in PCa patient tissue; and constitutive ELF3 overexpression has been shown to enhance PCa xenograft tumor growth 42, while p62 downregulation reduces cell viability in AR− PCa cell lines16. Thus, the IL-1-activated or constitutive p62-NFκB and/or ELF3-NFκB axes may promote AR-independent survival and tumorigenic behavior for ARlow/− PCa cells.
In addition to p62 and ELF3, the other selected IL-1-induced genes, SOX9, CD44, and CD24 (Fig. 3, Supplementary Figs. 1 & 2), also encode proteins that are important in PCa initiation and progression. SOX9 is a cell fate determinant transcription factor that regulates normal prostate cell development44–46. SOX9 is elevated in PCa tissue, correlates with tumor progression, represses AR activity in PCa cells, shows inverse accumulation with AR in prostate glands, and promotes PCa tumor growth in xenograft and genetic mouse models44–46. CD24 and CD44 are glycoproteins that are used to distinguish prostate cell subpopulations at different cell fate stages. CD44 is expressed in AR− neuroendocrine and stem cells36,47 and CD24+ cells represent transient amplifying cells48. CD24 is associated with poor prognosis49 and CD44+ enriched stem cell populations promote PCa tumorigenicity and androgen-resistant tumor growth47,50,51. Thus, IL-1 may regulate cell fate programs that provide ARlow/− PCa cells with a survival and tumor-promoting advantage.
The repressed genes in our gene expression profile are equally as important in PCa progression as the elevated genes. For example, the AR target gene, NKX3.1, encodes a PCa tumor suppressor protein that is reduced or lost in PCa patient tissue and cooperates with PTEN loss to promote PCa initiation52. Interestingly, our enrichment analysis suggests that PTEN signaling is downregulated in our gene expression profile (Fig. 6). Thus, IL-1-repressed NKX3.1 expression would be expected to facilitate the survival and promote tumorigenicity of ARlow/− PCa cells.
CONCLUSIONS
In closing, the IL-1-conferred gene expression profile that we identified in AR+ LNCaP cells that mimics AR− PC3 gene expression patterns reveals potential prosurvival, tumorigenic pathways and proteins that could be effective targets to prevent IL-1-induced ARlow/− PCa de novo accumulation and to eliminate existing AR− PCa cells, thereby, thwarting PCa hormone therapy resistance and AR-independent survival and tumor progression. Of note, as we have previously shown for IL-1 regulation of p62 versus AR4, our expression profile includes genes that are regulated both independently (e.g. p62) and dependently (e.g. AR target genes such as PSA) of IL-1-induced AR repression. It will be important to delineate the independent versus dependent genes, as genes that encode for signaling pathways conferring survival or tumor-promoting advantages independent of AR repression would be particularly advantageous in the face of AR-targeting therapies. For example, a cell exposed to AR-targeting therapy in an IL-1-rich microenvironment would, concomitant with losing target AR, still be able to elicit p62 pathways independent of AR repression and, thus, would be expected to still survive and promote tumor progression. As a next step, it will also be important to prioritize the genes in our expression profile and to characterize their IL-1-mediated mechanistic regulation and their functional significance in PCa treatment resistance and AR-independent survival and tumor progression in order to identify novel therapeutic targets and identify PCa patients that would benefit from novel therapies.
Supplementary Material
(A, B) RT-QPCR and (C) western blot analysis was performed for the AR+ MDA-PCa-2b PCa cell line treated for 3 days with MDA-PCa-2b conditioned medium (Control CM), HS-5 CM, or HS-27a CM or treated for 3 days with vehicle control, IL-1α (25 ng/ml), or IL-1β (25 ng/ml). (A) HS-5 CM can repress AR, PSA, NXK3.1, and TMPRSS2 and induce p62 and ELF3 mRNA; no change was detected for SOX9, CD24, or CD44 mRNA. (B) IL-1α or IL-1β can repress NKX3.1 and induce p62, ELF3, and CD24; no change was detected for AR, PSA, TMPRSS2, SOX9, or CD44 mRNA. (C) HS-5 CM, IL-1α, or IL-1β can downregulate AR and NKX3.1 protein accumulation and upregulate p62 protein. HS-5 CM, IL-1α, or IL-1β induce SOD2 mRNA and protein in MDA-PCa-2b cells and is used as a treatment efficacy control. n = 4 biological replicates (Bio-Rep); error bars = +/−STDEV; p-value = * ≤ 0.05, ** ≤ 0.005, ***≤ 0.0005. mRNA fold change is normalized to Control CM or MDA-PCa-2b vehicle control. β-actin or Ponceau are western blot loading controls.
(A, B) RT-QPCR and (C) western blot analysis was performed for the AR+ LNCaP PCa cell line or the AR− DU145 PCa cell line treated for 3 days with PCa control conditioned medium (Control CM), HS-5 BMSC CM, or HS-27a BMSC CM or treated for 3 days with vehicle control, IL-1α (25 ng/ml), or IL-1β (25 ng/ml). (A) DU145 cells have little or no detectable AR, PSA, NKX3.1, or TMPRSS2 mRNA relative to LNCaP cells. (B) DU145 cells have high basal p62, ELF3, SOX9, CD24, and CD44 mRNA relative to LNCaP cells and show no significant induction response to HS-5 CM, IL-1α, or IL-1β for these genes. (C) DU145 show no detectable p62 induction response to HS-5 CM, IL-1α, or IL-1β. HS-5 CM, IL-1α, or IL-1β induce SOD2 mRNA and protein in both DU145 cells and is used as a treatment efficacy control. n = 4 biological replicates (Bio-Rep); error bars = +/−STDEV; p-value = * ≤ 0.05, ** ≤ 0.005, ***≤ 0.0005. mRNA fold change is normalized to LNCaP Control CM or LNCaP vehicle control. β-actin is a western blot loading control.
MDA-PCa-2b cells were pretreated with 500 ng/ml IL-1Ra for 1 day (to analyzed induced genes) or 2 days (to analyze repressed genes). Following pretreatment, the growth medium was replaced with 1:1 BRFF-HPC1:DMEM (Control CM) + 500 ng/ml IL-1Ra or 1:1 BRFF-HPC1:HS-5 CM + 500 ng/ml IL-1Ra for an additional 1 day (to analyze repressed genes) or 3 days (to analyze induced genes). PBS is the IL-1Ra vehicle control. IL-1Ra attenuates HS-5 CM-induced SOD2 mRNA (B), indicating IL-1Ra treatment efficacy. (A) IL-1Ra attenuates HS-5 CM repression of AR, PSA, NKX3.1, and TMPRSS2 mRNA. (B) IL-1Ra attenuates HS-5 CM induction of p62 and ELF3 mRNA; no change was detected for SOX9, CD24, or CD44 mRNA. n = 2 biological replicates (Bio-Rep); p-value = * ≤ 0.05, ** ≤ 0.005. mRNA fold change is normalized to the respective Control CM for each gene.
Acknowledgments
Financial support: NIH/NCI R21CA175798 (Delk); NIH/NCI K01CA160602 (Delk), University of Texas at Dallas (Delk), NIH UL1TR001105 (Xing)
For their advice and support throughout this process, we would like to thank all the members of the labs of Drs. Nikki Delk, Chao Xing, and Jung-whan Kim (UT Dallas), including Michael Nugent, as well as Vanessa Schmid (Next Generation Sequencing Core, UTSW) and Drs. Cindy Farach-Carson (UT Health Science Center) and Pamela Constantinou (Rice University). We would also like to acknowledge financial support for the Delk lab from the University of Texas at Dallas and financial support from the National Institutes of Health (NIH/NCI R21CA175798 (Delk); NIH/NCI K01CA160602 (Delk); NIH UL1TR001105 (Xing)).
Footnotes
Conflict of Interest: the authors declare no potential conflicts of interest
References
- 1.Multhoff G, Molls M, Radons J. Chronic inflammation in cancer development. Front Immunol. 2012;2(JAN):1–17. doi: 10.3389/fimmu.2011.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voronov E, Shouval DS, Krelin Y, et al. IL-1 is required for tumor invasiveness and angiogenesis. Proc Natl Acad Sci U S A. 2003;100(5):2645–2650. doi: 10.1073/pnas.0437939100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Q, Russell MR, Shahriari K, et al. Interleukin-1β promotes skeletal colonization and progression of metastatic prostate cancer cells with neuroendocrine features. Cancer Res. 2013;73(11):3297–3305. doi: 10.1158/0008-5472.CAN-12-3970. [DOI] [PubMed] [Google Scholar]
- 4.Chang MA, Patel V, Gwede M, et al. IL-1β Induces p62/SQSTM1 and Represses Androgen Receptor Expression in Prostate Cancer Cells. J Cell Biochem. 2014;115(12):2188–2197. doi: 10.1002/jcb.24897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Culig Z, Hobisch a, Herold M, et al. Interleukin 1beta mediates the modulatory effects of monocytes on LNCaP human prostate cancer cells. Br J Cancer. 1998;78(8):1004–1011. doi: 10.1038/bjc.1998.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Staverosky JA, Zhu X, Ha S, Logan SK. Anti-androgen resistance in prostate cancer cells chronically induced by interleukin-1β. 2013;1(1):53–65. [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang B, Kwon OJ, Henry G, et al. Non-Cell-Autonomous Regulation of Prostate Epithelial Homeostasis by Androgen Receptor. Mol Cell. 2016;63(6):976–989. doi: 10.1016/j.molcel.2016.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katzenwadel A, Wolf P. Androgen deprivation of prostate cancer: Leading to a therapeutic dead end. Cancer Lett. 2015;367(1):12–17. doi: 10.1016/j.canlet.2015.06.021. [DOI] [PubMed] [Google Scholar]
- 9.Frieling JS, Basanta D, Lynch CC, et al. Current and emerging therapies for bone metastatic castration-resistant prostate cancer. Cancer Control. 2015;22(1):109–120. doi: 10.1177/107327481502200114. http://www.ncbi.nlm.nih.gov/pubmed/25504285%5Cnhttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC4673894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santer FR, Erb HHH, McNeill RV. Therapy escape mechanisms in the malignant prostate. Semin Cancer Biol. 2015;35:133–144. doi: 10.1016/j.semcancer.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 11.Tomlins SA, Mehra R, Rhodes DR, et al. Integrative molecular concept modeling of prostate cancer progression. Nat Genet. 2007;39(1):41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- 12.Davis JN, Wojno KJ, Daignault S, et al. Elevated E2F1 inhibits transcription of the androgen receptor in metastatic hormone-resistant prostate cancer. Cancer Res. 2006;66(24):11897–11906. doi: 10.1158/0008-5472.CAN-06-2497. [DOI] [PubMed] [Google Scholar]
- 13.Qin J, Liu X, Laffin B, et al. The PSA-/loprostate cancer cell population harbors self-renewing long-term tumor-propagating cells that resist castration. Cell Stem Cell. 2012;10(5):556–569. doi: 10.1016/j.stem.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shahriari KS. Interleukin-1β drives prostate cancer cell cooperation in the bone metastatic niche. 2016 [Google Scholar]
- 15.Roecklein BA, Torok-Storb B. Functionally distinct human marrow stromal cell lines immortalized by transduction with the human papilloma virus E6/E7 genes. Blood. 1995;85(4):997–1005. [PubMed] [Google Scholar]
- 16.Chang MA, Morgado M, Warren CR, Hinton CV, Farach-Carson MC, Delk NA. p62/SQSTM1 is required for cell survival of apoptosis-resistant bone metastatic prostate cancer cell lines. Prostate. 2014;74(2):149–163. doi: 10.1002/pros.22737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delk NA, Farach-Carson MC. Interleukin-6: A bone marrow stromal cell paracrine signal that induces neuroendocrine differentiation and modulates autophagy in bone metastatic PCa cells. Autophagy. 2012;8(4):650–663. doi: 10.4161/auto.19226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bitto A, Lerner CA, Nacarelli T, Crowe E, Torres C, Sell C. P62/SQSTM1 at the interface of aging, autophagy, and disease. Age (Dordr) 2014;36(3):9626. doi: 10.1007/s11357-014-9626-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.SA . FastQC: a quality control tool for high throughput sequence data. 2010. [Google Scholar]
- 20.SW FastQ Screen: quality control tool to screen a library of sequences in FastQ format against a set of sequence databases. 2011 [Google Scholar]
- 21.Aronesty E. Comparison of Sequencing Utility Programs. Open Bioinforma J. 2013;7:1–8. doi: 10.2174/1875036201307010001. [DOI] [Google Scholar]
- 22.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao Y, Smyth GK, Shi W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 24.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waltering KK, Helenius MA, Sahu B, et al. Increased expression of androgen receptor sensitizes prostate cancer cells to low levels of androgens. Cancer Res. 2009;69(20):8141–8149. doi: 10.1158/0008-5472.CAN-09-0919. [DOI] [PubMed] [Google Scholar]
- 26.Graf L, Iwata M, Torok-storb B, Mackinnon S. Gene expression profiling of the functionally distinct human bone marrow To the editor: Gene expression profiling of the functionally distinct human bone marrow. 2008;100(4):1509–1511. doi: 10.1182/blood-2002-03-0844. [DOI] [PubMed] [Google Scholar]
- 27.Miao L, StClair DK. Regulation of superoxide dismutase genes: implcations in diseases. Free Radic Biol Med. 2010;47(4):344–356. doi: 10.1016/j.freeradbiomed.2009.05.018.Regulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cunningham D, You Z. In vitro and in vivo model systems used in prostate cancer research. J Biol Methods. 2015;2(1):1–28. doi: 10.14440/jbm.2015.63.In. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vanini F, Kashfi K, Nath N. The dual role of iNOS in cancer. Redox Biol. 2015;6:334–343. doi: 10.1016/j.redox.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen DP, Li J, Yadav SS, Tewari AK. Recent insights into NF-κB signalling pathways and the link between inflammation and prostate cancer. BJU Int. 2014;114(2):168–176. doi: 10.1111/bju.12488. [DOI] [PubMed] [Google Scholar]
- 31.Morgan MJ, Liu Z. Crosstalk of reactive oxygen species and NF-κB signaling. Cell Res. 2011;21(1):103–115. doi: 10.1038/cr.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baggiolini M, Clark-Lewis I. Interleukin 8, a chemotactic and inflammatory. FEBS Lett. 1992;307(I):97–101. doi: 10.1016/0014-5793(92)80909-z. [DOI] [PubMed] [Google Scholar]
- 33.Grivennikov SI, Greten FR, Karin M. Immunity, Inflammation, and Cancer. Cell. 2011;140(6):883–899. doi: 10.1016/j.cell.2010.01.025.Immunity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sciarra A, Gentilucci A, Salciccia S, et al. Prognostic value of inflammation in prostate cancer progression and response to therapeutic: a critical review. J Inflamm. 2016;13(1):35. doi: 10.1186/s12950-016-0143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chu Z, Mehrnoosh S, Fayth LM, et al. Paracrine factors produced by bone marrow stromal cells induce apoptosis and neuroendocrine differentiation in prostate cancer cells. Prostate. 2011;71(2):157–167. doi: 10.1002/pros.21231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palapattu GS, Wu C, Silvers CR, et al. Selective expression of CD44, a putative prostate cancer stem cell marker, in neuroendocrine tumor cells of human prostate cancer. Prostate. 2009;69(7):787–798. doi: 10.1002/pros.20928. [DOI] [PubMed] [Google Scholar]
- 37.Dinarello Ca. Immunological and inflammatory functions of the interleukin-1 family. Annu Rev Immunol. 2009;27:519–550. doi: 10.1146/annurev.immunol.021908.132612. [DOI] [PubMed] [Google Scholar]
- 38.Nelius T, Filleur S, Yemelyanov A, et al. Androgen receptor targets NFkappaB and TSP1 to suppress prostate tumor growth in vivo. Int J cancer. 2007;121(5):999–1008. doi: 10.1002/ijc.22802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang S, Pettaway Ca, Uehara H, Bucana CD, Fidler IJ. Blockade of NF-κB activity in human prostate cancer cells is associated with suppression of angiogenesis, invasion, and metastasis. Oncogene. 2001;20(31):4188–4197. doi: 10.1038/sj.onc.1204535. [DOI] [PubMed] [Google Scholar]
- 40.Araki S, Omori Y, Lyn D, et al. Interleukin-8 is a molecular determinant of androgen independence and progression in prostate cancer. Cancer Res. 2007;67(14):6854–6862. doi: 10.1158/0008-5472.CAN-07-1162. [DOI] [PubMed] [Google Scholar]
- 41.Cronauer MV, Ince Y, Engers R, et al. Nitric oxide-mediated inhibition of androgen receptor activity: possible implications for prostate cancer progression. Oncogene. 2007;26(13):1875–1884. doi: 10.1038/sj.onc.1209984. [DOI] [PubMed] [Google Scholar]
- 42.Longoni N, Sarti M, Albino D, et al. ETS transcription factor ESE1/ELF3 orchestrates a positive feedback loop that constitutively activates NF-κB and drives prostate cancer progression. Cancer Res. 2013;73(14):4533–4547. doi: 10.1158/0008-5472.CAN-12-4537. [DOI] [PubMed] [Google Scholar]
- 43.Burdelski C, Reiswich V, Hubemagg C, et al. Cytoplasmic Accumulation of Sequestosome 1 ( p62 ) Is a Predictor of Biochemical Recurrence, Rapid Tumor Cell Proliferation, and Genomic Instability in Prostate Cancer. 1:3471–3479. doi: 10.1158/1078-0432.CCR-14-0620. [DOI] [PubMed] [Google Scholar]
- 44.Wang H, McKnight NC, Zhang T, Lu ML, Balk SP, Yuan X. SOX9 is expressed in normal prostate basal cells and regulates androgen receptor expression in prostate cancer cells. Cancer Res. 2007;67(2):528–536. doi: 10.1158/0008-5472.CAN-06-1672. [DOI] [PubMed] [Google Scholar]
- 45.Wang H, Leav I, Ibaragi S, et al. SOX9 is expressed in human fetal prostate epithelium and enhances prostate cancer invasion. Cancer Res. 2008;68(6):1625–1630. doi: 10.1158/0008-5472.CAN-07-5915. [DOI] [PubMed] [Google Scholar]
- 46.Thomsen MK, Ambroisine L, Wynn S, et al. SOX9 elevation in the prostate promotes proliferation and cooperates with PTEN loss to drive tumor formation. Cancer Res. 2010;70(3):979–987. doi: 10.1158/0008-5472.CAN-09-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patrawala L, Calhoun T, Schneider-Broussard R, et al. Highly purified CD44+ prostate cancer cells from xenograft human tumors are enriched in tumorigenic and metastatic progenitor cells. Oncogene. 2006;25(12):1696–1708. doi: 10.1038/sj.onc.1209327. [DOI] [PubMed] [Google Scholar]
- 48.Petkova N, Hennenlotter J, Sobiesiak M, et al. Surface CD24 distinguishes between low differentiated and transit-amplifying cells in the basal layer of human prostate. Prostate. 2013;73(14):1576–1590. doi: 10.1002/pros.22708. [DOI] [PubMed] [Google Scholar]
- 49.Kristiansen G, Pilarsky C, Pervan J, et al. CD24 expression is a significant predictor of PSA relapse and poor prognosis in low grade or organ confined prostate cancer. Prostate. 2004;58(2):183–192. doi: 10.1002/pros.10324. [DOI] [PubMed] [Google Scholar]
- 50.Seiler D, Zheng J, Liu G, et al. Enrichment of putative prostate cancer stem cells after androgen deprivation: upregulation of pluripotency transactivators concurs with resistance to androgen deprivation in LNCaP cell lines. Prostate. 2013;73(13):1378–1390. doi: 10.1002/pros.22685. [DOI] [PubMed] [Google Scholar]
- 51.Salvatori L, Caporuscio F, Verdina A, et al. Cell-to-cell signaling influences the fate of prostate cancer stem cells and their potential to generate more aggressive tumors. PLoS One. 2012;7(2):1–14. doi: 10.1371/journal.pone.0031467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim MJ, Cardiff RD, Desai N, et al. Cooperativity of Nkx3.1 and Pten loss of function in a mouse model of prostate carcinogenesis. Proc Natl Acad Sci U S A. 2002;99(5):2884–2889. doi: 10.1073/pnas.042688999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A, B) RT-QPCR and (C) western blot analysis was performed for the AR+ MDA-PCa-2b PCa cell line treated for 3 days with MDA-PCa-2b conditioned medium (Control CM), HS-5 CM, or HS-27a CM or treated for 3 days with vehicle control, IL-1α (25 ng/ml), or IL-1β (25 ng/ml). (A) HS-5 CM can repress AR, PSA, NXK3.1, and TMPRSS2 and induce p62 and ELF3 mRNA; no change was detected for SOX9, CD24, or CD44 mRNA. (B) IL-1α or IL-1β can repress NKX3.1 and induce p62, ELF3, and CD24; no change was detected for AR, PSA, TMPRSS2, SOX9, or CD44 mRNA. (C) HS-5 CM, IL-1α, or IL-1β can downregulate AR and NKX3.1 protein accumulation and upregulate p62 protein. HS-5 CM, IL-1α, or IL-1β induce SOD2 mRNA and protein in MDA-PCa-2b cells and is used as a treatment efficacy control. n = 4 biological replicates (Bio-Rep); error bars = +/−STDEV; p-value = * ≤ 0.05, ** ≤ 0.005, ***≤ 0.0005. mRNA fold change is normalized to Control CM or MDA-PCa-2b vehicle control. β-actin or Ponceau are western blot loading controls.
(A, B) RT-QPCR and (C) western blot analysis was performed for the AR+ LNCaP PCa cell line or the AR− DU145 PCa cell line treated for 3 days with PCa control conditioned medium (Control CM), HS-5 BMSC CM, or HS-27a BMSC CM or treated for 3 days with vehicle control, IL-1α (25 ng/ml), or IL-1β (25 ng/ml). (A) DU145 cells have little or no detectable AR, PSA, NKX3.1, or TMPRSS2 mRNA relative to LNCaP cells. (B) DU145 cells have high basal p62, ELF3, SOX9, CD24, and CD44 mRNA relative to LNCaP cells and show no significant induction response to HS-5 CM, IL-1α, or IL-1β for these genes. (C) DU145 show no detectable p62 induction response to HS-5 CM, IL-1α, or IL-1β. HS-5 CM, IL-1α, or IL-1β induce SOD2 mRNA and protein in both DU145 cells and is used as a treatment efficacy control. n = 4 biological replicates (Bio-Rep); error bars = +/−STDEV; p-value = * ≤ 0.05, ** ≤ 0.005, ***≤ 0.0005. mRNA fold change is normalized to LNCaP Control CM or LNCaP vehicle control. β-actin is a western blot loading control.
MDA-PCa-2b cells were pretreated with 500 ng/ml IL-1Ra for 1 day (to analyzed induced genes) or 2 days (to analyze repressed genes). Following pretreatment, the growth medium was replaced with 1:1 BRFF-HPC1:DMEM (Control CM) + 500 ng/ml IL-1Ra or 1:1 BRFF-HPC1:HS-5 CM + 500 ng/ml IL-1Ra for an additional 1 day (to analyze repressed genes) or 3 days (to analyze induced genes). PBS is the IL-1Ra vehicle control. IL-1Ra attenuates HS-5 CM-induced SOD2 mRNA (B), indicating IL-1Ra treatment efficacy. (A) IL-1Ra attenuates HS-5 CM repression of AR, PSA, NKX3.1, and TMPRSS2 mRNA. (B) IL-1Ra attenuates HS-5 CM induction of p62 and ELF3 mRNA; no change was detected for SOX9, CD24, or CD44 mRNA. n = 2 biological replicates (Bio-Rep); p-value = * ≤ 0.05, ** ≤ 0.005. mRNA fold change is normalized to the respective Control CM for each gene.