Abstract
MicroRNAs are implicated in regulating cancer progression and metastasis. Here we show that miR-720 is positively associated with renal cell carcinoma (RCC). Elevated levels of miR-720 were observed in a panel of RCC cell lines and clinical tissues compared to non-malignant cell line and normal samples. Loss of miR-720 function inhibited proliferation, migration and invasion and induced apoptosis in RCC cell lines in vitro and repressed tumor growth in xenograft mouse model. Conversely, gain of miR-720 function in non-malignant HK-2 cells induced pro-cancerous characteristics. Silencing of miR-720 caused a marked induction in the levels of endogenous αE-catenin and E-cadherin protein levels in anti720 transfected cells compared to control. Whereas, miR-720 overexpression in RCC cell lines reduced activity of a luciferase reporter gene fused to the wild-type αE-catenin or E-cadherin 3′ UTR compared to non-specific 3′ UTR control indicating that αE-catenin-E-cadherin complex is a direct and functional target of miR-720 in RCC. We also observed attenuation of β-Catenin, CD44 and Akt expression in RCC cells transfected with miR-720 inhibitor compared to control. Further, miR-720 exhibited clinical significance in RCC. Expression of miR-720 significantly distinguished malignant from normal samples. Elevated miR-720 levels positively correlated with higher Fuhrman grade, pathological stage and poor overall survival of RCC patients. These findings uncover a new regulatory network in RCC involving metastasis-promoting miR-720 that directly targets expression of key metastasis-suppressing proteins E-cadherin and αE-catenin complex. These results suggest that therapeutic regulation of miR-720 may provide an opportunity to regulate EMT and metastasis in RCC.
Keywords: MicroRNA, renal cancer, E-cadherin, αE-catenin, miR-720
Introduction
Renal cell carcinoma (RCC) is among the ten leading cancer types in United States with an estimated 62,700 new cases and 14,240 deaths in 2016(1). RCC can be histologically classified into several subtypes, with clear cell renal cell carcinoma (ccRCC) being the most common (2). Approximately 25–30% of ccRCC patients have metastatic disease at the time of diagnosis, while in 30% recurrence develops after complete resection of the primary tumor(3). The 5 year relative survival rate of regional or localized RCC is 65–92%, while that of metastatic RCC is only 12%(1). These patients have very poor prognosis because of the refractory nature of RCC to current treatment regimens. Therapeutic options for RCC are limited due to its lack of sensitivity to both chemo-, radio- and immunotherapy(4–6). RCC is characterized by considerable molecular heterogeneity in tumors, making prediction of disease progression and therapeutic response difficult.
Tumor recurrence and metastasis represent two major obstacles in the successful treatment of cancer. Emerging lines of evidence suggest that cancer aggressiveness is associated with epithelial-mesenchymal transition (EMT)(7). EMT is reactivated to promote tumorigenic progression of epithelial cells with increased cell migration and invasion, ‘stemness,’ and inhibition of apoptosis and senescence(8–10). A hallmark of EMT is the functional loss of cell-cell adheren junctions (AJs)(10). The transmembrane core of AJs is composed of E-cadherin, whose cytoplasmic domain interacts with β-catenin, which in turn binds to α-catenin(11, 12). E-cadherin is an established tumor suppressor(13) and its loss induces cancer invasion and metastasis(14–16), correlating with the aggressiveness of numerous carcinomas and worsening prognosis(13, 17). In ccRCC, microarray analysis of primary tumor samples showed a diminution of E-cadherin expression in the majority of patient samples(16) and E-cadherin loss was reported to be an early pathogenic event(16, 18). Whereas, forced expression of E-cadherin suppressed tumor development and invasion in various in vitro and in vivo tumor model systems(17). αE-catenin, a member of the α-catenin family has been reported to be a tumor suppressor in many cancers(19–22). The proportion of tumors that fail to express either E-cadherin, αE-catenin or both has been reported to be as high as 80%(11), and their loss often correlates with the degree of tumor differentiation and metastasis(12, 23–25). These findings indicate that perturbation of the E-cadherin-αE-catenin complex is an important molecular event in the progression of several cancers. Functional loss of E-cadherin-αE-catenin junctions activates EMT, contributes to metastasis and more aggressive behavior of these human cancers(23). Therefore, it is of critical importance to regulate EMT and to develop effective therapeutic strategies for the treatment of recurrent and metastatic cancer. Critical regulators of EMT include transcription repressors that suppress E-cadherin expression (7, 26, 27) and microRNAs that target key proteins involved in EMT (28, 29).
MicroRNAs (miRNA) are small evolutionarily conserved non-coding RNA molecules that negatively regulate transcript levels through sequence-dependent recognition mechanisms(30). In a previous study, we performed an initial screen for differentially expressed miRNAs in RCC and identified that miR-205 is significantly downregulated in cancer compared to normal cell line(31). A second miRNA, miR-720, was significantly over-expressed in RCC cells compared to normal cell line. Thus the present study was undertaken to define the role of miR-720 in RCC. Here we report that (i) miR-720 is overexpressed in RCC cell lines and clinical samples, (ii) loss of miR-720 induces tumor suppressor effects in vitro in RCC cell lines and in-vivo in nude mouse xenografts, (iii) overexpression of miR-720 in normal kidney cells leads to tumorigenic effects, iv) miR-720 directly targets E-cadherin and αE-catenin, core proteins of AJs that are established tumor suppressors, (iv) attenuation of miR-720 rescues expression of E-cadherin and E-catenin protein levels while inhibiting the expression of CD44, CTNNB1 and Akt that are known to have oncogenic function in RCC, (v) finally we show that miR-720 has diagnostic and prognostic potential in RCC. Therefore, targeting of miR-720 in RCC may be an important strategy to regulate RCC growth and metastasis.
Materials and Methods
Cell culture, plasmids and transfection
Human renal cell carcinoma cell lines 786-O, ACHN, A498 and 769-P and a non-malignant renal cell line HK-2 were obtained from the American Type Culture Collection (ATCC) (Manassas, VA) in the year 2016 and grown according to ATCC protocol. These human-derived cell lines were authenticated by DNA short-tandem repeat analysis by ATCC. Cell line experiments were performed within 6 months of their procurement/resuscitation. Plasmids for nonspecific miRNA control vector for pEZX-MT01 (CmiT000001-MT01) was purchased fromGeneCopoeia (GeneCopoeia, Rockville, MD). For luciferase reporter assays, pMIR-REPORT dual luciferase vector was purchased from Ambion, Cambridge MA. TaqMan probes for hsa-miR-720 (miR-720), anti-miR-720 and negative controls pre-miR and anti-miR-Control (cont-miR) were purchased from Applied Biosystems (Life Technologies, CA). Lipofectamine 2000 and 3000 reagents were used for transfection.
Tissue samples and laser capture microdissection (LCM)
Tissue samples from radical nephrectomy were obtained from the Veterans Affairs Medical Center, San Francisco, CA, USA in accordance with the institutional guidelines (IRB approval no. 16-18555). Written informed consent was obtained from patients for tissue collection. LCM was performed as described previously(32). Briefly 8μm sections were placed on glass slides, deparaffinized, stained with hematoxylin, dehydrated, and placed in the AutoPix instrument for microdissection (AutoPix System; Arcturus). Areas of interest were captured with infrared laser pulses onto CapSure Macro LCM Caps.
Quantitative real-time PCR and expression analysis in TCGA data cohort
Total RNA was extracted and assayed for mature miRNAs and mRNAs using the TaqMan MicroRNA Assays and Gene Expression Assays, respectively, in accordance with the manufacturer’s instructions (Applied Biosystems). All RT reactions were run in a 7500 Fast Real Time PCR System (Applied Biosystems). Relative expression was calculated using comparative Ct. Expression of miR-720 was also analyzed in a publicly available data cohort of renal carcinoma from the Cancer Genome Atlas (TCGA) now available as genomic data commons (GDC) data portal (https://gdc-portal.nci.nih.gov/).
Flow cytometry, cell viability, migration and invasion assays
FACS analysis for cell cycle and apoptosis was done 72 hours post-transfection. The cells were harvested, washed with cold PBS, and resuspended in the nuclear stain DAPI for cell cycle analysis or stained with 7-AAD and Annexin-V-FITC using ANNEXIN V-FITC/7-AAD KIT (BD Biosciences) for apoptosis analysis according to the manufacturer’s protocol. Stained cells were immediately analyzed by FACS (BD FACSVerse; BD Biosciences). Cell viability was determined at 24, 48 and 72 h by using the CellTiter 96 AQueous One Solution Cell Proliferation Assay kit (Promega, Madison, WI) according to the manufacturer’s protocol. Also a cytoselect 24-well cell migration and invasion assay kit (Cell Biolabs, Inc) was used for migration and invasion assays according to the manufacturer’s protocol. For migration and invasion assays two replicates were used and the experiment was repeated three times.
Immunoblotting
Immunoblotting was performed as described previously(31). Briefly RCC cell lines 786-O and A498 were transfected atleast three times at different time points with anti-miR control or antimiR-720 (miR-720 inhibitors). Protein was isolated from 70–80% confluent cultured cells using the M-PER Mammalian Protein Extraction Reagent (Pierce Biotechnology, Rockfield, IL) following the manufacturer’s directions. Equal amounts of protein from all the transfections were resolved on the same 4–20% SDS polyacrylamide gels to keep the setting same for all the transfections. Protein was then transferred to nitrocellulose membrane. The resulting blots were blocked with 5% non-fat dry milk and probed with antibodies. Antibodies were obtained from Cell Signaling Technology Inc. Denver, MA for alpha-E-catenin (cat. #. 36611), Santa Cruz Biotechnology, Inc, CA for GAPDH (cat. #. sc-32233) and from ThermoFisher Scientific, CA for E-cadherin (cat. #. MA5-11496). Blots were visualized using enhanced chemiluminescence (Pierce Biotechnology, Rockford, IL) or Western Blotting Luminol reagent (Santa Cruz Biotechnology, Inc, CA).
Luciferase Assays
The wild type 3′UTR complimentary sequences of αE-catenin and E-cadherin for miR-720 are given in Figure 5C. These sequences were cloned downstream of the luciferase gene in the pMIR-REPORT luciferase vector (Ambion, Cambridge MA). For non-specific 3′ UTR control, we used control plasmid (CmiT000001-MT01) from GeneCopoeia (Rockville, MD). The complete sequence of vector is available at http://www.genecopoeia.com/. For reporter assays, cells were transiently transfected with wild-type or control plasmids and miR-720 or control-miR. Firefly luciferase activities were measured using the Dual Luciferase Assay (Promega, Madison, WI) 24 hr after transfection and the results were normalized with Renilla luciferase.
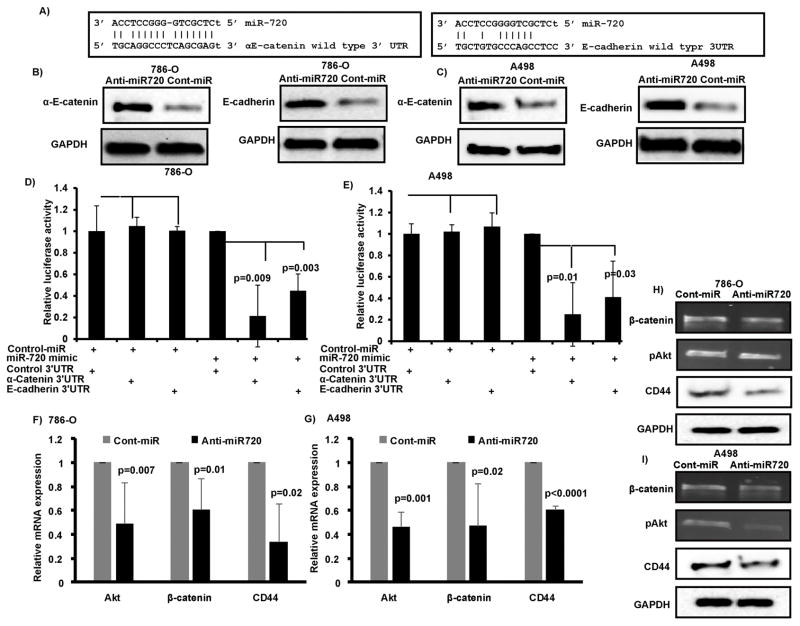
Figure 5. αE-catenin-E-cadherin complex is a direct target of miR-720 and miR-720 silencing attenuates EMT and metastasis associated genes.
A–B) Transient transfection of anti720 caused a significant increase in αE-catenin and E-cadherin protein levels compared to control in both cell lines. C) Complimentary binding sites for miR-720 in 3′UTR of αE-catenin and E-cadherin. D–E) Luciferase assays showing decreased reporter activity after co-transfection of either wild type 3′UTR sequences compared to non-specific control 3′UTR with miR-720 in 786-O and A498 cells. F–I) Decreased Akt, CTNNB1 (β-catenin) and CD44 expression observed at mRNA (F–G) and protein (H–I) levels after miR-720 silencing in both A498 and 786-O RCC cell lines.
In vivo intratumoral delivery of anti720
The antitumor effect of silencing miR-720 was determined by local administration of miR-720 miRNA inhibitor (anti720) in established tumors in athymic nude mice and compared to an antimiR-negative control mice group. Each mouse was injected sub-cutaneously with 5.0×106 A498 cells. Once palpable tumors developed, 6.25 μg of synthetic anti720 or antimiR-negative control (control) complexed with 1.6 μl siPORT Amine transfection reagent (Ambion, Austin, TX) in 50 μl PBS was delivered eleven times intratumorally at 3-day intervals over the course of experiment. In total 6 mice received anti720 and 6 mice received control miR. Tumor growth was followed for 6 weeks from the first injection. All animal care was in accordance with institutional guidelines (IACUC approval no. 16-004).
Statistical analysis
Statistical analyses were performed with StatView for Windows (SAS Institute Inc. NC, USA), GraphPad-Pris-5 and MedCalc. All quantified data represents the average of at least triplicate samples and three experiments performed at different times or as indicated. For migration and invasion assays, two replicates were used and the experiment was repeated three times. Error bars represent standard deviation of the mean or as indicated. All tests were performed two tailed and p-values <0.05 were considered statistically significant. TCGA data cohort was analyzed using Mann-Whitney test (independent samples) from Rank sum tests to compare reads per million between normal and tumor samples. Receiver operating curves (ROC) were calculated to determine potential of miR-720 to discriminate between malignant and non-malignant tissues and for overall survival Kaplan-Meier analyses (log-rank tests) were also performed. Chi-square tests were performed to determine correlation between miR-720 expression and clinicopathological characteristics.
Results
miR-720 is highly expressed in RCC
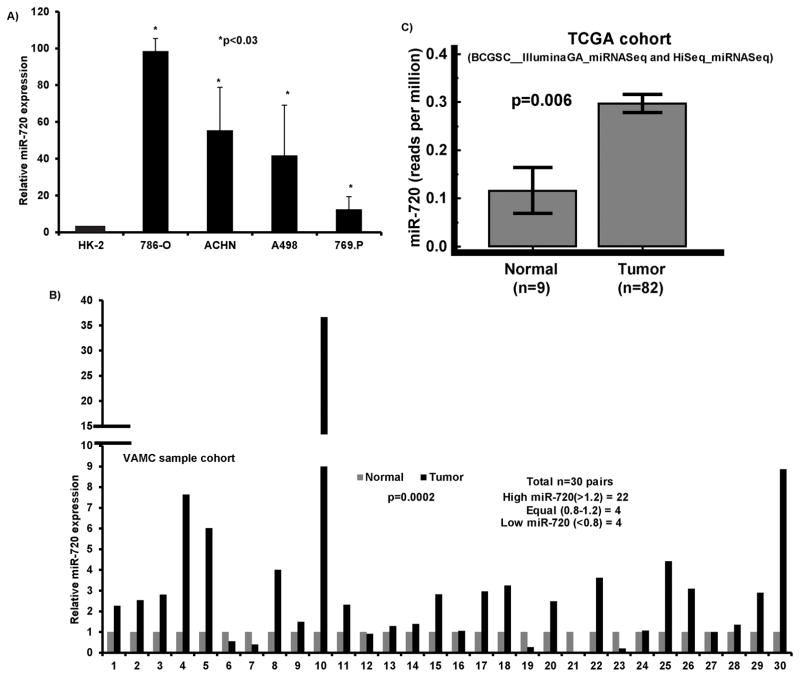
We previously found that miR-720 was significantly upregulated in RCC cell lines compared to non-malignant cell line HK2(31) and validated this data by miRNA-quantitative real time PCR (miR qRT-PCR) analysis. The results confirmed that miR-720 is overexpressed in several RCC cell lines compared to normal HK-2 cells (Figure 1A). We investigated the expression of miR-720 in 60 (30 pairs) laser-captured micro-dissected (LCM) matched clinical samples (Figure 1B). miR-720 expression was significantly elevated in cancer samples compared to matched normal controls (Figure 1B). Further, we analyzed miR-720 expression in The Cancer Genome Atlas (TCGA) sample cohort using Mann-Whitney test (independent samples) from Rank sum tests to compare normal and tumor samples. miR-720 expression levels were significantly high in tumors compared to normal samples (p=0.006) (Figure 1C). Thus the data from clinical tissues is in agreement with cell line data showing that miR-720 is highly expressed and may have an oncogenic function in RCC.
Figure 1. miR-720 expression is elevated in RCC.
A) Quantitative RT-PCR analysis of miR-720 in a panel of cell lines. B) Quantitative real time PCR analysis of miR-720 expression in 30 pairs of matched Laser-Captured Microdissected tissue samples. C) miR-720 expression in a renal carcinoma sample cohort publicly available at the TCGA data base.
miR-720 loss of function exerts tumor suppressor effects in RCC
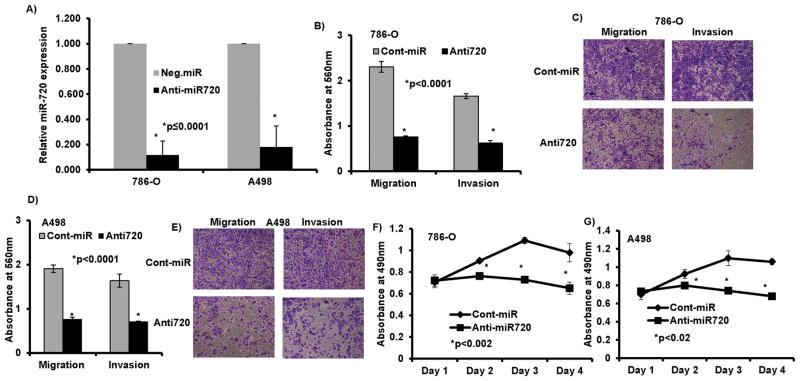
We performed a series of in vitro loss-of-function analyses by silencing miR-720 with antisense oligonucleotides (anti720) in two RCC cell lines, 786-O and A498 (Figure 2A). To discern whether miR-720 has any role in RCC metastasis, we performed in vitro chemotactic transwell invasion and migration assays. Silencing of miR-720 caused a marked reduction in migration (67%, p<0.0001) and invasion (62%, p<0.0001) of 786-O cancer cells post 72h transfection (Figure 2B–C). Similarly there was a decrease in migration (60%, p<0.0001) and invasion (57%, p<0.0001) with A498 cells (Figure 2D–E) compared to controls. These results suggest that loss of miR-720 expression impairs RCC cell migration and invasion.
Figure 2. miR-720 loss of function exerts tumor suppressor effects in RCC.
A) Expression of miR-720 post transient transfection of 50nM miR-720 inhibitor (anti720) in kidney cancer cells compared to anti-miR-control inhibitor (control). B–E) Migration and invasion assays in 786-O cells (B–C) and A498 cells (D–E) transfected with anti720 compared to anti-negative control miR (cont-miR). F–G) Proliferation of 786-O and A498 cells after anti720 transfection was significantly reduced compared to control.
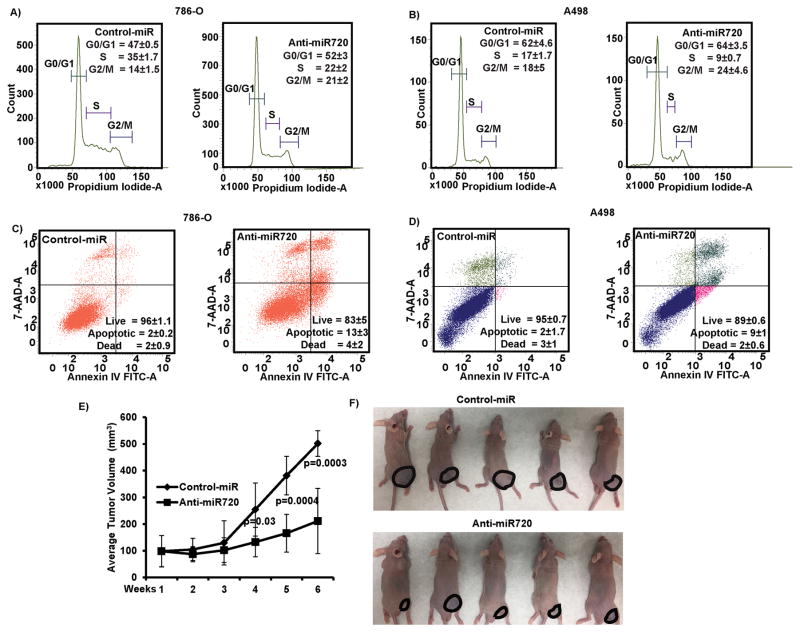
Unregulated cell proliferation and survival are common features of cancer cells that favor metastatic dissemination. Silencing of miR-720 impaired cell viability compared to control (Cont-miR) in both 786-O and A498 cell lines (Figure 2F–G). Flow cytometry cell cycle analysis revealed a significant decrease in S-phase (8–13%) with a concomitant but modest increase in G2/M cell population (6–7%) in cells transfected with anti720 compared to control (Figure 3A–B). Depletion of miR-720 also induced apoptosis in both cancer cell lines as compared to control (7–11%) (Figure 3C–D). Taken together, these observations suggest that miR-720 function is required for tumorigenic attributes of RCC cell lines.
Figure 3. Silencing of miR-720 induces apoptosis and G2/M cell cycle arrest in vitro and tumor growth suppression effects in vivo in an intratumoral xenograft mouse model.
A–B) Cell cycle analysis showing a significant decrease in S-phase along with a modest increase in the G2/M phase of 786-O and A498 cells after transfection with anti720. C–D) Apoptosis assay showing induction of apoptosis by anti720. Numbers on panels are mean populations±SEM. E) Tumor volume following intratumoral injection of control or anti720 into established tumors. Data represents the mean of each group (6 mice) and error bars are SD. Week 1 is the start of injection and tumor growth was followed for 6 weeks and each mice received a total of 11 injections. F) Representative pictures of mice injected with either control or miR-720 at the termination of experiment.
In vivo tumor suppressor effect of loss of function of miR-720 in RCC
To recapitulate the in vitro tumor suppressor effects of miR-720 loss of function in vivo, we performed growth suppression experiment in nude mice by intra-tumoral delivery of anti720 (miR-720 inhibitor) or control (non-specific inhibitor control) in established tumors. Tumor growth was significantly suppressed in the mouse group that received anti720 compared to controls (Figure 3E–F). Average tumor volume in controls was 245mm3 compared to 133mm3 in mice that received anti720 at the termination of the experiment. These results confirm the in vitro tumor suppressive effect of silencing miR-720 in RCC xenograft model.
miR-720 gain of function exerts tumorigenic attributes in non-malignant HK-2 cells
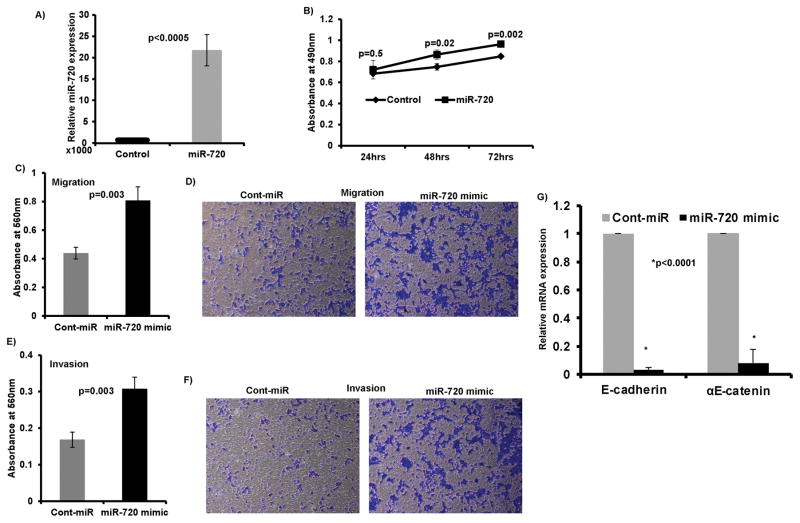
We next ascertained whether miR-720 gain of function induces tumorigenic characteristics in non-malignant HK-2 cells. miR-720 was overexpressed in HK-2 cells by transient transfection of 50 nM miR-720 mimic along with a miRNA-control (control) (Figure 4A). Indeed, miR-720 overexpression mediated pro-cancerous phenotypes as indicated by increased cell proliferation (Figure 4B) migration (Figure 4C–D) and invasion (Figure 4 E–F) compared to controls. In addition, a significant decrease was observed in E-cadherin and αE-catenin expression in HK-2 cells overexpressing miR-720 compared to the control (Figure 4G). These results attest to the oncogenic role of miR-720 in RCC.
Figure 4. miR-720 gain of function exerts tumorigenic attributes in non-malignant cell line HK-2.
A) miR-720 expression after transient transfection of miR-720 mimic compared to pre-miR-negative control (cont-miR). B) miR-720 overexpression in non-malignant HK-2 cells induced cell proliferation. C–F) An increase in the migration (C–D) and invasion (E–F) of HK-2 cells. G) A significant decrease in the expression of E-cadherin and αE-catenin in HK-2 cells overexpressing miR-720 compared to control.
αE-catenin-E-cadherin complex is a direct and functional target of miR-720
To understand the mechanism by which miR-720 induces tumor suppressor effects in RCC, we used computational methods to identify potential miR-720 targets in humans. We performed an initial screening of several targets such as cMyc, N-cadherin, vimentin, VEGFA, Cyclin-A2, Cyclin-B2, Cyclin-E2, MAPK7, Pax2, CA9, BNIP2, KLF10, TCF7L2, ZBTB17, p27, p16, IGFBP1, TP73, Caspase 8 and Caspase 1. There was either no or a modest effect of miR-720 silencing observed on these targets in Western or qRT-PCR analysis. We focused on αE-catenin-E-cadherin complex because they are key negative regulators of EMT and metastasis and have complimentary binding sites in their 3′UTR for miR-720 (Figure 5A). In addition, our functional assays showed a robust reduction in migration and invasion when miR-720 was silenced in RCC cell lines. In support of these results, we observed a marked induction in the levels of endogenous αE-catenin and E-cadherin protein levels in miR-720 silenced cells compared to control (Figure 5 B–C). Further, miR-720 overexpression in 786-O and A498 RCC cell lines reduced activity of a luciferase reporter gene fused to the wild-type αE-catenin (75–78% reduction, p=0.009, p=0.01) or E-cadherin (55–60% reduction, p=0.003, p=0.03) 3′ UTR compared to non-specific 3′ UTR control (Figure 5D–E). These results indicate that miR-720 directly targets these proteins and regulates their expression through translational inhibition.
Silencing of miR-720 attenuates EMT and metastasis associated genes
We further demonstrated the effect of miR-720 silencing on EMT and metastasis associated such as CD44 that plays pivotal role in EMT(7), β-catenin (CTNNB1), another core component of cell-cell adheren junctions (AJs) and Akt that are associated with tumorigenesis, EMT and metastasis in multiple cancers including RCC(33, 34). A significant decrease in the expression of all these genes was observed at both mRNA (Figure 5F–G) and protein levels (Figure 5H–I) in both A498 and 786-O RCC cell lines. These results indicate that attenuation of miR-720 in RCC negatively regulates EMT and metastatic genes in RCC.
Clinical relevance of miR-720 in RCC
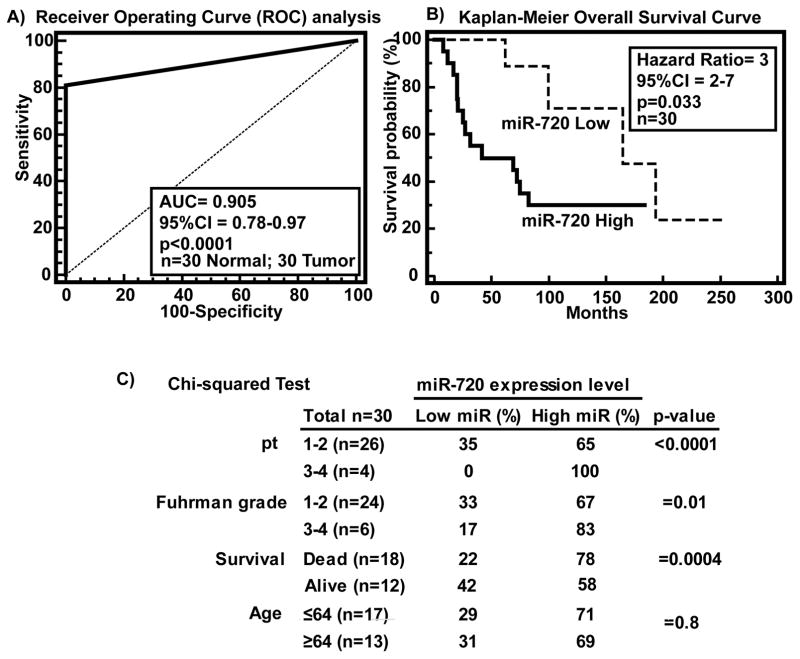
Clinical demographics of the study cohort are summarized in supplementary Table 1. Receiver operating curve (ROC) analysis was performed to evaluate the ability of miR-720 expression to discriminate between normal (n=30) and tumor cases (n=30). An area under the ROC curve (AUC) of 0.905 (P<0.0001; 95% CI=0.78 to 0.97; Specificity=100%; Sensitivity=80%) (Figure 6A) was obtained suggesting that miR-720 expression can discriminate between malignant and non-malignant tissues and hence has potential to be used as a diagnostic marker for RCC. To determine whether miR-720 has prognostic significance, we divided patients into low miR-720 (expression T/N<0.8 fold) and high miR-720 (expression T/N>0.8 fold) groups and performed Kaplan-Meier survival analysis. The results show that the low miR-720 group displayed significantly higher overall survival probability compared to the high miR-720 group (Logrank Test p<0.03, HR=3, 95% CI=2–7) (Figure 6B). We also determined the correlation of miR-720 expression with clinicopathological variables such as Fuhrman grade, pathological stage (pT), age and survival (Figure 6C). Chi-squared test revealed that cases with high miR-720 expression increased from low grade, low pathological stage to high grade and high pathological stage (Figure 6C). Among deceased patients, 78% had high miR-720 expression (p<0.0001) compared to 22% with low expression. Collectively, these results suggest that miR-720 has potential to be a marker for diagnosis and predicting survival of RCC patients though it needs to be validated in a large sample cohort.
Figure 6. Clinical relevance of miR-720 in RCC.
A) ROC curve analysis showing ability of miR-720 expression to discriminate between malignant and non-malignant tissue samples. B) Kaplan-Meier analysis for overall survival based on miR-720 expression. C) Chi-square test showing correlation of clinicopathological characteristics with miR-720 expression.
Discussion
In this study, we identified miR-720 as a pro-metastatic miRNA and as a negative regulator of the key metastasis suppressors E-cadherin and αE-catenin in RCC. We observed miR-720 to be significantly elevated in a panel of RCC cell lines and tumor samples when compared to normal cell line or matched normal tissue samples. This is in agreement with a previous study of microRNA profiling in RCC tissue samples where miR-720 expression was reported to be almost 3 fold higher in tumors compared to adjacent non-tumorous tissues(35). This suggests an oncogenic role for miR-720 in RCC, however neither the functional relevance nor the clinical significance of miR-720 expression in RCC has been previously defined.
EMT is a conserved embryologic genetic program that is essential in cancer cell metastasis. Functional loss of E-cadherin-αE-catenin junctions activates EMT, contributes to metastasis and more aggressive behavior of various human cancers(23). Our study indicates that E-cadherin and αE-catenin are the direct functional targets of miR-720 and attenuation of miR-720 in RCC cell lines rescued expression of both proteins. A study in breast cancer has reported a pivotal role for the CD44 in accelerating EMT(7). Expression of CD44s led to the activation of Akt and attenuation of E-cadherin(7). In RCC, CD44 has been identified as a prognostic marker with high expression correlating with higher Fuhrman grade, recurrence and poor prognosis(36). Our results indicated that silencing of miR-720 suppressed CD44 and Akt expression in RCC cell lines. We also observed a decrease in β-catenin (CTNNB1), another core component of cell-cell adheren junctions, mRNA expression in anti-miR720 transfected cells compared to control. CTNNB1 and Akt are associated with tumorigenesis, EMT and metastasis in multiple cancers including RCC(33, 34). EMT promotes tumorigenic progression and metastasis by increased migration and invasion while inhibiting apoptosis and senescence(8–10). Our results revealed that silencing of miR-720 by anti-agomiRs induced tumor suppressor effects as indicated by impaired proliferation, migration, invasion and induction of apoptosis in RCC cells in vitro. We also confirmed the tumor growth suppression effects of miR-720 silencing in an in vivo intra-tumoral xenograft mouse model. In non-malignant HK-2 cells over-expression of miR-720 induced pro-cancerous characteristics such as increased proliferation, migration and invasion. Collectively, these results indicate that miR-720 is positively associated with RCC. Our findings show that elevated expression of the miR-720 contributes to EMT and metastasis in RCC. Expression of miR-720 is also high in several other cancers and studies have implicated this miRNA in transformed phenotypes and have suggested its use in cancer diagnosis or as a prognostic marker(37–40).
MiRNAs have generated great interest as potential prognostic biomarkers and powerful tools for early cancer diagnosis, in part due to their lineage-specific expression and stability in tissues and body fluids (41–44). MicroRNA expression patterns have been reported to be more accurate in establishing the right diagnosis than the mRNA criteria, in poorly differentiated metastatic cancer cases with cancer of unknown primary origin (45). In ccRCC, miRNA-429 was found to be a potential diagnostic and prognostic biomarker and a potential regulator of EMT(46). miR-429 was significantly associated with cancer metastasis, shorter disease-free and overall survival(46). Our results reveal that miR-720 expression can robustly (p<0.0001) discriminate malignant from non-malignant tissues and hence can be used as a diagnostic marker for RCC. We also observed that high miR-720 expression was significantly associated with poor overall survival. Correlation of miR-720 expression with clinicopathological variables indicated that high miR-720 expression is positively associated with high grade and high pathological stage and interestingly, 78% of deceased patients had high miR-720 expression. Collectively, our study highlights the biomarker potential of miR-720 expression in RCC, though it needs to be validated in a larger sample cohort.
In conclusion, this study shows that miR-720 is significantly upregulated and has a clinical significance in RCC. Silencing of miR-720 induced in vitro and in vivo tumor suppressor effects in RCC. In contrast, overexpression of miR-720 elicited tumorigenic effects in non-malignant cells. In addition, E-cadherin-αE-catenin complex was affirmed to be the direct and functional target of miR-720. Our data suggests that miR-720 is positively associated with RCC and may contribute to EMT and metastasis through negative regulation of E-cadherin-αE-catenin expression. We envisage that therapeutic regulation of miR-720 may provide an opportunity to regulate EMT and metastasis in RCC.
Supplementary Material
Acknowledgments
Financial support: This work was supported by the National Cancer Institute at the National Institutes of Health through grant number RO1CA199694 and U.S. Department of Veterans Affairs through VA Merit Review number BX001604P1 awarded to R. Dahiya.
We thank Dr. Roger Erickson for his support and assistance with the preparation of the manuscript and Dr. Z. Laura Tabatabai for the help with pathology of the clinical tissues.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.van Vlodrop IJ, Joosten SC, de Meyer T, Smits KM, Van Neste L, Melotte V, et al. A four-gene promoter methylation marker panel consisting of GREM1, NEURL, LAD1 and NEFH predicts survival of clear cell renal cell cancer patients. Clin Cancer Res. doi: 10.1158/1078-0432.CCR-16-1236. [DOI] [PubMed] [Google Scholar]
- 4.McDermott DF, Regan MM, Clark JI, Flaherty LE, Weiss GR, Logan TF, et al. Randomized phase III trial of high-dose interleukin-2 versus subcutaneous interleukin-2 and interferon in patients with metastatic renal cell carcinoma. J Clin Oncol. 2005;23:133–41. doi: 10.1200/JCO.2005.03.206. [DOI] [PubMed] [Google Scholar]
- 5.Belldegrun AS, Klatte T, Shuch B, LaRochelle JC, Miller DC, Said JW, et al. Cancer-specific survival outcomes among patients treated during the cytokine era of kidney cancer (1989–2005): a benchmark for emerging targeted cancer therapies. Cancer. 2008;113:2457–63. doi: 10.1002/cncr.23851. [DOI] [PubMed] [Google Scholar]
- 6.Rydzanicz M, Wrzesinski T, Bluyssen HA, Wesoly J. Genomics and epigenomics of clear cell renal cell carcinoma: recent developments and potential applications. Cancer Lett. 2013;341:111–26. doi: 10.1016/j.canlet.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 7.Brown RL, Reinke LM, Damerow MS, Perez D, Chodosh LA, Yang J, et al. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J Clin Invest. 2011;121:1064–74. doi: 10.1172/JCI44540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 9.Sanchez-Tillo E, Liu Y, de Barrios O, Siles L, Fanlo L, Cuatrecasas M, et al. EMT-activating transcription factors in cancer: beyond EMT and tumor invasiveness. Cell Mol Life Sci. 2012;69:3429–56. doi: 10.1007/s00018-012-1122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larsen JE, Nathan V, Osborne JK, Farrow RK, Deb D, Sullivan JP, et al. ZEB1 drives epithelial-to-mesenchymal transition in lung cancer. J Clin Invest. 2016;126:3219–35. doi: 10.1172/JCI76725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobielak A, Fuchs E. Alpha-catenin: at the junction of intercellular adhesion and actin dynamics. Nat Rev Mol Cell Biol. 2004;5:614–25. doi: 10.1038/nrm1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piao HL, Yuan Y, Wang M, Sun Y, Liang H, Ma L. alpha-catenin acts as a tumour suppressor in E-cadherin-negative basal-like breast cancer by inhibiting NF-kappaB signalling. Nat Cell Biol. 2014;16:245–54. doi: 10.1038/ncb2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gervais ML, Henry PC, Saravanan A, Burry TN, Gallie BL, Jewett MA, et al. Nuclear E-cadherin and VHL immunoreactivity are prognostic indicators of clear-cell renal cell carcinoma. Lab Invest. 2007;87:1252–64. doi: 10.1038/labinvest.3700684. [DOI] [PubMed] [Google Scholar]
- 14.Onder TT, Gupta PB, Mani SA, Yang J, Lander ES, Weinberg RA. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008;68:3645–54. doi: 10.1158/0008-5472.CAN-07-2938. [DOI] [PubMed] [Google Scholar]
- 15.Derksen PW, Liu X, Saridin F, van der Gulden H, Zevenhoven J, Evers B, et al. Somatic inactivation of E-cadherin and p53 in mice leads to metastatic lobular mammary carcinoma through induction of anoikis resistance and angiogenesis. Cancer Cell. 2006;10:437–49. doi: 10.1016/j.ccr.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 16.Evans AJ, Russell RC, Roche O, Burry TN, Fish JE, Chow VW, et al. VHL promotes E2 box-dependent E-cadherin transcription by HIF-mediated regulation of SIP1 and snail. Mol Cell Biol. 2007;27:157–69. doi: 10.1128/MCB.00892-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–7. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esteban MA, Tran MG, Harten SK, Hill P, Castellanos MC, Chandra A, et al. Regulation of E-cadherin expression by VHL and hypoxia-inducible factor. Cancer Res. 2006;66:3567–75. doi: 10.1158/0008-5472.CAN-05-2670. [DOI] [PubMed] [Google Scholar]
- 19.Liu TX, Becker MW, Jelinek J, Wu WS, Deng M, Mikhalkevich N, et al. Chromosome 5q deletion and epigenetic suppression of the gene encoding alpha-catenin (CTNNA1) in myeloid cell transformation. Nat Med. 2007;13:78–83. doi: 10.1038/nm1512. [DOI] [PubMed] [Google Scholar]
- 20.Ji H, Wang J, Fang B, Fang X, Lu Z. alpha-Catenin inhibits glioma cell migration, invasion, and proliferation by suppression of beta-catenin transactivation. J Neurooncol. 2011;103:445–51. doi: 10.1007/s11060-010-0413-4. [DOI] [PubMed] [Google Scholar]
- 21.Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, Rodriguez JR, Zhou D, et al. Yap1 acts downstream of alpha-catenin to control epidermal proliferation. Cell. 2011;144:782–95. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silvis MR, Kreger BT, Lien WH, Klezovitch O, Rudakova GM, Camargo FD, et al. alpha-catenin is a tumor suppressor that controls cell accumulation by regulating the localization and activity of the transcriptional coactivator Yap1. Sci Signal. 2011;4:ra33. doi: 10.1126/scisignal.2001823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Papadavid E, Katsambas A. The interactions and role of epithelial cadherin and catenins in tumorigenicity. Int J Dermatol. 2001;40:254–7. doi: 10.1046/j.1365-4362.2001.01179.x. [DOI] [PubMed] [Google Scholar]
- 24.Schipper JH, Frixen UH, Behrens J, Unger A, Jahnke K, Birchmeier W. E-cadherin expression in squamous cell carcinomas of head and neck: inverse correlation with tumor dedifferentiation and lymph node metastasis. Cancer Res. 1991;51:6328–37. [PubMed] [Google Scholar]
- 25.Vermeulen SJ, Bruyneel EA, Bracke ME, De Bruyne GK, Vennekens KM, Vleminckx KL, et al. Transition from the noninvasive to the invasive phenotype and loss of alpha-catenin in human colon cancer cells. Cancer Res. 1995;55:4722–8. [PubMed] [Google Scholar]
- 26.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–39. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 27.Bolos V, Peinado H, Perez-Moreno MA, Fraga MF, Esteller M, Cano A. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J Cell Sci. 2003;116:499–511. doi: 10.1242/jcs.00224. [DOI] [PubMed] [Google Scholar]
- 28.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–56. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 31.Majid S, Saini S, Dar AA, Hirata H, Shahryari V, Tanaka Y, et al. MicroRNA-205 inhibits Src-mediated oncogenic pathways in renal cancer. Cancer Res. 2011;71:2611–21. doi: 10.1158/0008-5472.CAN-10-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suh SO, Chen Y, Zaman MS, Hirata H, Yamamura S, Shahryari V, et al. MicroRNA-145 is regulated by DNA methylation and p53 gene mutation in prostate cancer. Carcinogenesis. 2011;32:772–8. doi: 10.1093/carcin/bgr036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuan H, Meng X, Guo W, Cai P, Li W, Li Q, et al. Transmembrane-Bound IL-15-Promoted Epithelial-Mesenchymal Transition in Renal Cancer Cells Requires the Src-Dependent Akt/GSK-3beta/beta-Catenin Pathway. Neoplasia. 2015;17:410–20. doi: 10.1016/j.neo.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fodde R, Brabletz T. Wnt/beta-catenin signaling in cancer stemness and malignant behavior. Curr Opin Cell Biol. 2007;19:150–8. doi: 10.1016/j.ceb.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 35.Yi Z, Fu Y, Zhao S, Zhang X, Ma C. Differential expression of miRNA patterns in renal cell carcinoma and nontumorous tissues. J Cancer Res Clin Oncol. 2010;136:855–62. doi: 10.1007/s00432-009-0726-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X, Ma X, Chen L, Gu L, Zhang Y, Zhang F, et al. Prognostic value of CD44 expression in renal cell carcinoma: a systematic review and meta-analysis. Sci Rep. 2015;5:13157. doi: 10.1038/srep13157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Das SG, Romagnoli M, Mineva ND, Barille-Nion S, Jezequel P, Campone M, et al. miR-720 is a downstream target of an ADAM8-induced ERK signaling cascade that promotes the migratory and invasive phenotype of triple-negative breast cancer cells. Breast Cancer Res. 2016;18:40. doi: 10.1186/s13058-016-0699-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lerebours F, Cizeron-Clairac G, Susini A, Vacher S, Mouret-Fourme E, Belichard C, et al. miRNA expression profiling of inflammatory breast cancer identifies a 5-miRNA signature predictive of breast tumor aggressiveness. Int J Cancer. 2013;133:1614–23. doi: 10.1002/ijc.28171. [DOI] [PubMed] [Google Scholar]
- 39.Hara ES, Ono M, Eguchi T, Kubota S, Pham HT, Sonoyama W, et al. miRNA-720 controls stem cell phenotype, proliferation and differentiation of human dental pulp cells. PLoS One. 2013;8:e83545. doi: 10.1371/journal.pone.0083545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X, Kuang Y, Shen X, Zhou H, Chen Y, Han Y, et al. Evaluation of miR-720 prognostic significance in patients with colorectal cancer. Tumour Biol. 2015;36:719–27. doi: 10.1007/s13277-014-2697-z. [DOI] [PubMed] [Google Scholar]
- 41.Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101:2087–92. doi: 10.1111/j.1349-7006.2010.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Glud M, Klausen M, Gniadecki R, Rossing M, Hastrup N, Nielsen FC, et al. MicroRNA expression in melanocytic nevi: the usefulness of formalin-fixed, paraffin-embedded material for miRNA microarray profiling. J Invest Dermatol. 2009;129:1219–24. doi: 10.1038/jid.2008.347. [DOI] [PubMed] [Google Scholar]
- 43.Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, Zepeniuk M, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26:462–9. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- 44.Heinzelmann J, Henning B, Sanjmyatav J, Posorski N, Steiner T, Wunderlich H, et al. Specific miRNA signatures are associated with metastasis and poor prognosis in clear cell renal cell carcinoma. World J Urol. 2011;29:367–73. doi: 10.1007/s00345-010-0633-4. [DOI] [PubMed] [Google Scholar]
- 45.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 46.Machackova T, Mlcochova H, Stanik M, Dolezel J, Fedorko M, Pacik D, et al. MiR-429 is linked to metastasis and poor prognosis in renal cell carcinoma by affecting epithelial-mesenchymal transition. Tumour Biol. 2016;37:14653–8. doi: 10.1007/s13277-016-5310-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.