Abstract
Background
Blood-based protein biomarkers can be a useful tool as pre-treatment prognostic markers, as they can reflect both variations in the tumor microenvironment and the host immune response. We investigated the influence of a panel of plasma proteins for the development of any failure defined as recurrent disease in the T-, N-, or M-site in HNSCC.
Methods
We used a multiplex bead-based approach to analyze 19 proteins in 86 HNSCC patients and 15 healthy controls. We evaluated the associations between the biomarkers, loco-regional failure, failure in the T-, N-, or M-site, overall survival (OS), p16 status, and hypoxia.
Results
In 41 p16 positive oropharynx cancer patients we identified a profile of biomarkers consisting of upregulation of IL-2, IL-4, IL-6, IL-8, eotaxin, GRO-a, and VEGF and downregulation of VEGFR-1 and VEGFR-2 with a significantly reduced risk of failure (p < 0.01). None of the individual proteins were associated with outcome.
Conclusion
The identified plasma profile potentially reflects an activated immune response in a subgroup of the p16 positive patients.
Keywords: Biomarkers, Circulating proteins, Prognostic, Immune response, Hypoxia, Head and neck cancer
Introduction
Traditionally, the predominant etiological factors for head and neck squamous cell carcinoma (HNSCC) have been tobacco smoking and alcohol consumption [1]. In the past 20 years, it has become increasingly clear that there is an etiological linkage to human papilloma virus (HPV) infections, and a subgroup of HPV-positive HNSCC has been established [2]. Overall, HPV-positive HNSCC constitute an entity of patients with a different molecular biology [3], a different clinical profile, and a more favorable prognosis [4].
Besides HPV status, a number of other prognostic factors are relevant for HNSCC, including tumor stage, nodal stage, a history of tobacco smoking [5], as well as hypoxia [6]. Furthermore, biopsy-based biologically distinct subtypes that are independent of HPV status have been introduced, and the subtype with the most advantageous prognosis shows a prominent immune and mesenchymal phenotype [7].
Blood-based biomarkers can be useful as pre-treatment prognostic markers, as they can reflect variations in tumor microenvironment and host immune response and can complement biopsy-based biomarkers that evaluate tumor cells directly. Although several studies have investigated the prognostic value of various circulating proteins in HNSCC [8], [9], [10], [11], [12], [13], [14], [15], [16], there is no consensus as to which are the most promising prognostic biomarkers, or whether biomarkers should be analyzed individually or combined into profiles. We hypothesize that a panel of circulating endogenous markers in HNSCC is associated with outcome after primary radiotherapy and that these markers are influenced by HPV-status and tumor hypoxia.
In this study, we aimed to investigate the influence of a panel of proteins in the blood for the development of failure defined as recurrent disease in the T-, N-, or M-site in HNSCC. We used a multiplex bead-based approach to analyze 19 previously described proteins (cytokines, chemokines, angiogenic factors, and receptors) [8], [9], [10], [11], [12], [13], [14], [15], [16]. We evaluated the associations between the circulating biomarkers, HPV status, smoking history, and hypoxia evaluated by a 15-gene hypoxia profile [17].
Materials and methods
Patients and samples
The prospectively collected study-population comprised of 86 previously untreated HNSCC patients treated at the Department of Oncology at Aarhus University Hospital, Denmark, between July 2005 and September 2011 (treated according to the DAHANCA 18 protocol) [18], [19]. Additionally, 15 healthy controls from the Danish blood bank at Aarhus University Hospital were enrolled between March 2012 and November 2014. The patients received primary radiotherapy (RT) according to the DAHANCA guidelines (http://www.dahanca.dk). The prescribed dose was 66–68 Gy, two Gy/fraction, six fractions per week. All patients were prescribed orally administered hypoxic radiosensitizer, with Nimorazole 1200 mg/m2, 90 min before each fraction of RT. Patients with locally advanced disease were given Cisplatin intravenously concomitant with RT once a week for a maximum of six cycles (40 mg/m2, maximum dose 70 mg). The patient, tumor and control characteristics are presented in Table 1.
Table 1.
Patient, tumor and control characteristics.
| All patients (n = 86) |
Blood bank controls (n = 15) |
|||
|---|---|---|---|---|
| n | (%) | n | (%) | |
| Age (years) | ||||
| Median | 58 | 55 | ||
| Range | (34–77) | (51–63) | ||
| ⩽60 years | 51 | 59 | 13 | 87 |
| >60 years | 35 | 41 | 2 | 13 |
| Sex | ||||
| Female | 16 | 19 | 7 | 47 |
| Male | 70 | 81 | 8 | 53 |
| Smoking status | ||||
| >10 pack years | 56 | 65 | ||
| ⩽10 pack years | 30 | 35 | ||
| Tumor site | ||||
| Sinonasal carcinoma | 3 | 3 | ||
| Rhinopharynx | 5 | 6 | ||
| Oral cavity | 5 | 6 | ||
| Oropharynx | 56 | 65 | ||
| Hypopharynx | 5 | 6 | ||
| Supraglottic larynx | 8 | 9 | ||
| Glottis | 2 | 2 | ||
| Subglottis | 2 | 2 | ||
| Tumor stage | ||||
| T1-2 | 58 | 67 | ||
| T3-4 | 28 | 33 | ||
| Nodal stage | ||||
| N0 | 8 | 9 | ||
| N1-3 | 78 | 91 | ||
| Disease stage | ||||
| I-II | 4 | 5 | ||
| III-VI | 81 | 94 | ||
| Unknown | 1 | 1 | ||
| HPV/p16 status | ||||
| Positive and oropharynx | 41 | 48 | ||
| Negative or non-oropharynx | 42 | 49 | ||
| Unknown | 3 | 3 | ||
| Hypoxia by gene classifier | ||||
| More hypoxic | 25 | 29 | ||
| Less hypoxic | 57 | 66 | ||
| Unknown | 4 | 5 | ||
| Chemotherapy | ||||
| Yes | 79 | 92 | ||
| No | 7 | 8 | ||
Blood sample processing and multiplex analysis of circulating proteins
Plasma samples were obtained by venipuncture and taken in lithium heparin vials, kept on ice until separation within three hours of collection, and stored at −80 °C until further processing. Procedures on sample processing and analyses have previously been described in detail [20]. Briefly, multiplex analysis was performed of 19 proteins (Table 2), in three different pre-mixed bead-based antibody assays (Bio-Plex Pro™ human Reagent Kit, Bio-Rad), according to the manufacturer’s protocol using the Luminex 100 (BIO-PLEX 200 SYSTEM) and Bio-Plex manager software (version 6.1). For measured values out of range (OOR) the values above the upper limit of quantification were replaced by the highest recorded value of the standard curve. For values below the lower limit of quantification the OOR values were replaced by the lowest recorded value of the standard curve divided by two. For two patients the amount of available plasma was insufficient to perform the analysis in 14 of the investigated proteins.
Table 2.
Associations between baseline levels of proteins and any failure and hazard ratios for any failure from univariate and multivariate Cox analysis. All expression values are log2 transformations of absolute levels in ng/L.
| Protein/Cat.# | Any failure |
Hazard ratio (HR) univariate |
Hazard ratio (HR) multivariate1 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Yes (n = 23) Median (min, max) |
No (n = 63) Median (min, max) |
p-Value | HR | 95% CI | p-Value | HR | 95% CI | p-Value | |||
| EGFR/171BC501M | 15.7 | (14.0; 19.1) | 15.8 | (4.2; 19.1) | 0.78 | 1.12 | (0.89–1.41) | 0.34 | 1.17 | (0.89–1.53) | 0.26 |
| Leptin/171BC508M | 12.7 | (8.1; 16.0) | 13.5 | (8.3; 16.0) | 0.23 | 0.89 | (0.70–1.13) | 0.34 | 1.05 | (0.78–1.41) | 0.74 |
| OPN/171BC509M | 19.6 | (14.8; 19.6) | 17.4 | (7.8; 19.6) | 0.25 | 1.20 | (0.88–1.65) | 0.25 | 1.12 | (0.79–1.59) | 0.51 |
| VEGFR-1/171BC515M | 8.3 | (5.3; 9.9) | 8.2 | (1.3; 10.7) | 0.81 | 1.11 | (0.87–1.43) | 0.40 | 1.04 | (0.81–1.34) | 0.74 |
| VEGFR-2/171BC516 | 15.2 | (13.5; 16.6) | 14.8 | (6.0; 16.7) | 0.22 | 1.44 | (0.91–2.16) | 0.13 | 1.51 | (0.89–2.54) | 0.12 |
| IL-2/171B5003M | 1.8 | (−1.7; 7.0) | 1.7 | (−1.7; 6.1) | 0.88 | 1.02 | (0.85–1.22) | 0.86 | 1.03 | (0.86–1.24) | 0.76 |
| IL-13/171B5012M | 1.2 | (−2.4; 2.7) | 0.6 | (−2.4; 4.0) | 0.11 | 1.24 | (0.94–1.64) | 0.12 | 1.13 | (0.85–1.51) | 0.40 |
| PDGF-bb/171B5024M | 8.1 | (5.2; 10.5) | 7.8 | (4.9; 13.2) | 0.60 | 1.03 | (0.80–1.35) | 0.82 | 0.84 | (0.61–1.16) | 0.29 |
| TNF/171B5026M | 1.1 | (−1.2; 6.4) | 1.1 | (−1.2; 8.4) | 0.37 | 1.05 | (0.86–1.27) | 0.64 | 1.01 | (0.84–1.22) | 0.92 |
| PAI-1/171B7010M | 14 | (12.2; 15.4) | 14 | (11.8; 16.1) | 0.78 | 1.02 | (0.62–1.68) | 0.94 | 0.73 | (0.41–1.30) | 0.29 |
| SDF-1a/171B6019M | 7.3 | (4.8; 9.7) | 7.4 | (4.8; 11.0) | 0.94 | 0.96 | (0.58–1.59) | 0.88 | 0.73 | (0.44–1.22) | 0.23 |
| IL-4/Z50005SADE | −1.7 | (−4.1; 1.8) | −1.7 | (−4.1; 4.8) | 0.93 | 0.86 | (0.61–1.20) | 0.37 | 0.83 | (0.59–1.18) | 0.31 |
| IL-6/Z50005SADE | 2.9 | (−1.3; 6.7) | 3.2 | (−1.3; 6.9) | 0.34 | 0.91 | (0.76–1.10) | 0.33 | 0.91 | (0.75–1.10) | 0.34 |
| IL-8/Z50005SADE | 3.2 | (0.9; 6.3) | 3.3 | (0.7; 9.0) | 0.74 | 0.94 | (0.74–1.20) | 0.63 | 0.95 | (0.74–1.24) | 0.73 |
| Eotaxin/Z50005SADE | 5.6 | (2.7; 8.2) | 5.7 | (0.5; 7.8) | 0.37 | 0.95 | (0.73–1.23) | 0.69 | 0.92 | (0.71–1.18) | 0.52 |
| G-CSF/Z50005SADE | 5.9 | (3.0; 8.4) | 6.1 | (3.4; 9.9) | 0.31 | 0.86 | (0.63–1.18) | 0.35 | 0.91 | (0.63–1.30) | 0.60 |
| VEGF/Z50005SADE | 5.3 | (−0.3; 6.9) | 5.5 | (2.6; 9.3) | 0.08 | 0.78 | (0.62–0.99) | 0.04* | 0.71 | (0.55–0.91) | 0.01 |
| GRO-a/171B6007M | 3.7 | (3.6; 8.3) | 5.3 | (3.6; 10.0) | 0.32 | 0.86 | (0.65–1.14) | 0.29 | 0.82 | (0.60–1.11) | 0.19 |
| HGF/171B6008M | 8.1 | (4.7; 9.2) | 8.1 | (4.0; 9.8) | 0.90 | 0.95 | (0.59: 1.53) | 0.84 | 0.73 | (0.47–1.12) | 0.15 |
Significant.
Adjusted for smoking history and HPV/p16 status.
Tumor characteristics
Previously published data on tumor hypoxia were obtained from a study on a 15-gene hypoxia profile [21]. HPV status was assessed by immunohistochemical detection of p16 as a surrogate marker for HPV infection in FFPE tumor biopsies [4], [22], [23].
Evaluation of treatment response and follow-up
According to the DAHANCA guidelines the follow-up program comprised of a clinical examination two months after completion of RT followed by examination every 3 months during the first 2 years and every 6 months for the next 3 years. Suspected recurrences were evaluated by clinical examination, CT or MRI scans, and eventually confirming biopsies. The median follow-up time was 50 months (range 6–98). Time was calculated from the first date the patient was seen in the center to the last recorded status visit, the date of a confirmed recurrence or to the date of death. The primary endpoint was any failure defined as time from the first visit to the center to the first event i.e. recurrence in the T-, N-, or M-site. Secondary endpoints were loco-regional failure defined as recurrent or residual tumor in the T- and N-site, and overall survival (OS) defined as death from any cause.
Cluster analysis
For outcome driven analysis, the Cutoff Finder web application (molpath.charite.de) was used to find the optimal cut point for each protein by survival analysis using the log-rank test [24]. The patient cohort was divided into a test (n = 56) and a validation (n = 28) cohort by randomization and stratified for HPV/p16, DAHANCA protocol and sex. In the test cohort, proteins for which an optimal cut point could be defined resulting in a log-rank test with a p-value <0.05 were identified. The identified cut points were used as cut off values for dichotomizing each marker, and a combined profile which most significantly separated the groups was tested in the validation cohort. For the non-outcome driven analysis, unsupervised hierarchical clustering was done on log2-transformed baseline protein levels using the SPSS statistics software version 22 implementation (two-step function) of the Balanced Iterative Reducing and Clustering using Hierarchies method (BIRCH) [25], [26] to identify subsets of patients with similar baseline protein profiles. The proteins with the least influence on the cluster formation were removed systematically until no further changes in the cluster formation was observed. OPN and leptin were left out of the analysis due to a large number of missing values. Expression data were visualised as heat-maps using TreeView (Version 1.1.6r2, jtreeview.sourceforge.net). Data were centred on the mean of the cluster means for patients. As no clusters could be identified for the controls, these were median centred. Both patients and controls were randomly ordered.
Statistical analysis
The difference between the log2 transformed protein expression in regards to failure, p16 status, the hypoxia gene profile, and patients versus controls was evaluated by Wilcoxon rank sum test. Correlations between groups of patients were evaluated by chi-squared test. Failure rates were estimated based on cumulative incidence rates [27]. Significance was evaluated by the risk difference at 60 months based on the pseudo-values approach [28]. Overall survival was estimated with the Kaplan–Meier method. Hazard ratios were estimated using the Cox proportional hazards model and the proportional hazard assumption was tested using log-minus-log plots and corroborated with Schoenfeld’s residuals.
The protein levels were treated as continuous variables. Smoking history was dichotomized in more than ten pack years and less than or equal to ten pack years. Smoking history, chemotherapy, and p16 status were included as covariates in multivariate Cox analysis. Bonferroni correction was performed in case of multiple comparisons. All reported p-values were two-sided with a 0.05 significance level. All analyses, excluding the unsupervised hierarchical clustering analysis, were performed using Stata version 12 (StataCorp, College Station, TX, USA).
Ethics
Blood samples were used with permission from the Danish Research Ethics Committee (case number 1-10-72-519-12). The procedures were in accordance with the Helsinki Declaration of 1975 (revised in 1983). Data handling procedures were approved by the Danish Data Protection Agency (case number 2014-41-3510). The Danish registry for use of tissue was consulted before the use of patient material.
Results
Clinical outcome
84 of the 86 patients had complete response to therapy. Two patients had persistent disease by the end of treatment. Twenty-five patients died during the follow-up period; nineteen patients died of their primary cancer, two died of primary cancer of the airways, three died of other cancer, and one died of other disease than cancer.
The overall 5-year mortality rate was 27% (95% CI: 19–38%). Twenty-three patients (27%) experienced a failure defined as recurrence in the T-, N-, or M-site (eleven in T-site, seven in the N-site, three in the M-site, one in both the T- and M-site and one in both the T-, N-, and M-site). The cumulative incidence of any failure was 27% (18–36%) and the 5-year loco-regional failure rate was 24% (15–33%). HPV/p16 status was available for 83/86 patients. Of the 44 HPV/p16 positive patients 41 were oropharynx cancers. Six out of 41 HPV/p16 positive oropharynx cancer patients and 17/42 of the remaining patients experienced an event in the T-, N-, or M-site (p = 0.01). The cumulative incidence of any failure for the HPV/p16 positive oropharynx cancer patients was 15% (4–25%) and 42% (27–57%) for the remaining patients (p = 0.005).
Baseline plasma levels and recurrent disease
The baseline log2 transformed protein levels were compared between patients who experienced a failure (n = 23) and patients who did not (n = 63) (Table 2). None of the baseline protein levels were associated with experiencing a failure. In univariate Cox analysis, only VEGF was associated with risk of failure, as higher baseline VEGF levels were significantly associated with lower risk of failure (HR = 0.78, 95% CI: 0.62–0.99; p = 0.04). After adjusting for smoking history, chemotherapy, and HPV/p16 status in a multivariate Cox analysis, high baseline VEGF remains significantly associated with lower risk of failure (HR = 0.71, 95% CI: 0.55–0.91). VEGF was not associated with failure when using Bonferroni correction for multiple testing. None of the other proteins were associated with failure in uni- and multivariate analysis (Table 2). High levels of VEGF was significantly associated with prolonged OS in multi- but not in univariate Cox analysis (HR = 0.73, 95% CI: 0.57–0.93). None of the baseline protein levels were associated with loco-regional failure.
Baseline plasma levels and HPV/p16 status, hypoxia, and smoking
The baseline protein levels were compared with HPV/p16 status, hypoxia, and smoking history (data not shown). HGF, SDF-1a, PAI-1, VEGF, and PDGF-bb were significantly higher in the HPV/p16 negative patients. When patients were classified as ‘more’ or ‘less’ hypoxic based on a 15-gene hypoxia classifier [17], [29], IL-4 levels were slightly higher in the more hypoxic group. When patients were divided by a smoking history of more or less than or equal 10 pack years, the levels of PAI-1, SDF-1a, and Il-6 were significantly higher in patients having smoked more than 10 pack years. None of the associations were significant when using Bonferroni correction for multiple testing.
Baseline plasma levels in patients versus controls
For eight of the proteins there was a significant difference between the baseline levels of the patient group and the control group (Table 3). EGFR, OPN, VEGFR-1, VEGFR-2, and VEGF levels were higher in the patient group, and IL-2, IL-4, and GRO were lower. For each of the eight proteins, patients were identified as having abnormal levels if they were above the 90% percentile of the control samples for the first 5 proteins or below 10% percentile for the last three. There was no association between the groups with abnormal levels and any failure for any of the proteins. We have previously estimated a measurement uncertainty for evaluating the expression levels of proteins [20]. Based on this, intermediate groups of patients centred on the 90% percentiles and the 10% percentiles of the control samples were identified. After exclusion of the intermediate groups of samples the groups with abnormal protein levels remain un-associated with failure.
Table 3.
Associations between baseline levels of proteins in patients and controls and hazard ratios for any failure from univariate Cox analysis. All expression values are log2 transformations of absolute levels in ng/L.
| Protein | Patients vs controls |
Hazard ratio Abnormal vs normal levels |
Hazard ratio Abnormal vs normal levels Excluding intermediates |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients (n = 86) Median (min, max) |
Controls (n = 15) Median (min, max) |
p-Value | Group size (n/n) | HR | 95% CI | p-Value | Group size (n/n) | HR | 95% CI | p-Value | |||
| EGFR | 15.7 | (4.2; 19.1) | 15.1 | (13.8; 15.9) | 0.03* | 41/45 | 0.81 | (0.35;1.84) | 0.61 | 28/31 | 1.03 | (0.62; 1.71) | 0.92 |
| Leptin | 13.4 | (8.1; 16.0) | 13.4 | (10.7; 16.0) | 0.49 | – | – | – | – | – | – | – | – |
| OPN | 18.5 | (7.8; 19.6) | 17.1 | (14.6; 17.8) | 0.04* | 26/24 | 1.43 | (0.53; 3.84) | 0.48 | 26/17 | 0.75 | (0.41; 1.38) | 0.35 |
| VEGFR-1 | 8.2 | (1.3; 10.7) | 6.9 | (6.2; 9.3) | <0.01* | 28/58 | 0.84 | (0.35; 2.05) | 0.71 | 10/41 | 0.97 | (0.51; 1.84) | 0.93 |
| VEGFR-2 | 14.9 | (6.0; 16.7) | 13.7 | (12.4; 14.8) | <0.001* | 58/28 | 0.91 | (0.39; 2.16) | 0.84 | 36/14 | 0.60 | (0.28; 1.27) | 0.18 |
| IL-2 | 1.8 | (−1.7; 7.0) | 3.8 | (2.6; 6.7) | <0.001* | 28/56 | 0.83 | (0.36; 1.93) | 0.67 | 19/45 | 0.82 | (0.53; 1.29) | 0.39 |
| IL-13 | 0.8 | (−2.4; 4.0) | 0.5 | (−3.6; 2.9) | 0.94 | – | – | – | – | – | – | – | – |
| PDGF-bb | 8.0 | (4.9; 13.2) | 7.6 | (6.1; 9.7) | 0.56 | – | – | – | – | – | – | – | – |
| TNF | 1.1 | (−1.2; 8.4) | 0.3 | (0.3; 5.0) | 0.17 | – | – | – | – | – | – | – | – |
| PAI-1 | 14 | (11.8; 16.1) | 13.8 | (12.7; 15.1) | 0.37 | – | – | – | – | – | – | – | – |
| SDF-1a | 7.3 | (4.8; 11.0) | 7.4 | (6.5; 7.7) | 0.80 | – | – | – | – | – | – | – | – |
| IL-4 | −1.7 | (−4.1; 4.8) | −1.7 | (−1.7; 0.8) | <0.001* | 17/67 | 0.83 | (0.33; 2.11) | 0.70 | 14/14 | 1.13 | (0.57; 2.27) | 0.72 |
| IL-6 | 3.1 | (−1.3; 6.9) | 2.9 | (0.0; 4.7) | 0.67 | – | – | – | – | – | – | – | – |
| IL-8 | 3.3 | (0.7; 9.0) | 3.3 | (1.9; 5.4) | 0.97 | – | – | – | – | – | – | – | – |
| Eotaxin | 5.7 | (0.5; 8.2) | 5.2 | (3.8; 6.1) | 0.14 | – | – | – | – | – | – | – | – |
| G-CSF | 6.0 | (3.0; 9.9) | 5.6 | (3.2; 7.5) | 0.08 | – | – | – | – | – | – | – | – |
| VEGF | 5.4 | (−0.3; 9.3) | 3.1 | (−0.7; 5.6) | <0.001* | 45/39 | 0.87 | (0.38; 1.97) | 0.74 | 31/39 | 0.59 | (0.33; 1.07) | 0.08 |
| GRO-a | 5.3 | (3.6; 10.0) | 6.8 | (5.5; 7.9) | <0.001* | 40/44 | 1.42 | (0.62; 3.24) | 0.41 | 29/37 | 1.19 | (0.78; 1.82) | 0.42 |
| HGF | 8.1 | (4.0; 9.8) | 8.0 | (7.2; 8.5) | 0.60 | – | – | – | – | ||||
Significant.
Outcome driven high risk signature
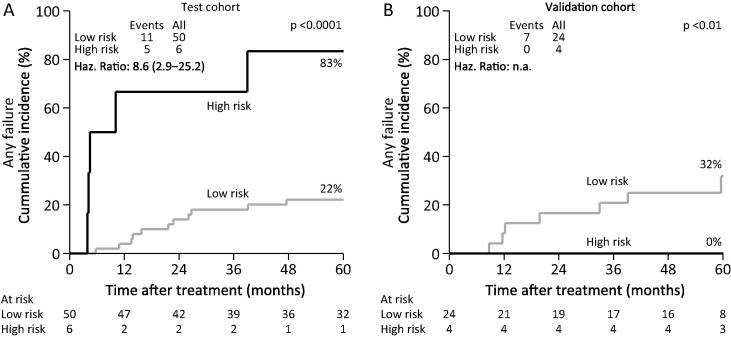
After randomly dividing patients into a test (n = 56) and a validation (n = 28) cohort (stratified by HPV/p16, DAHANCA protocol and sex), optimal cut points were identified for each protein by comparing the survivor functions. Two patients were not included due to a large fraction of missing values. Seven proteins were identified as ‘high risk’ proteins. For IL-2, IL-4, IL-6, eotaxin, G-CSF and GRO, values below the cut point were associated with an increased risk of failure, and for HGF values above the cut point were associated with an increased risk of failure. In the test cohort, the best separation of patients (p < 0.001) was obtained by identifying a group with values of 5–6 of the proteins within the ‘high risk’ expression levels (Fig.1A). The identification of ‘high risk’ patients could not be confirmed in the validation cohort, as the ‘high risk’ patients in the validation cohort had a significantly lower risk of failure than the proposed ‘low risk’ patients (Fig.1B).
Fig. 1.
Associations with any failures for patients with a ‘high risk’ or ‘low risk’ profile of proteins in a subgroup of patients (test cohort, A) and replication of the profile in the remaining subgroup of patients (validation cohort, B). ‘High risk’ patients are identified by having expression of at least 5 of 6 proteins in ‘high risk’ expression ranges.
Cluster analysis, non-outcome driven
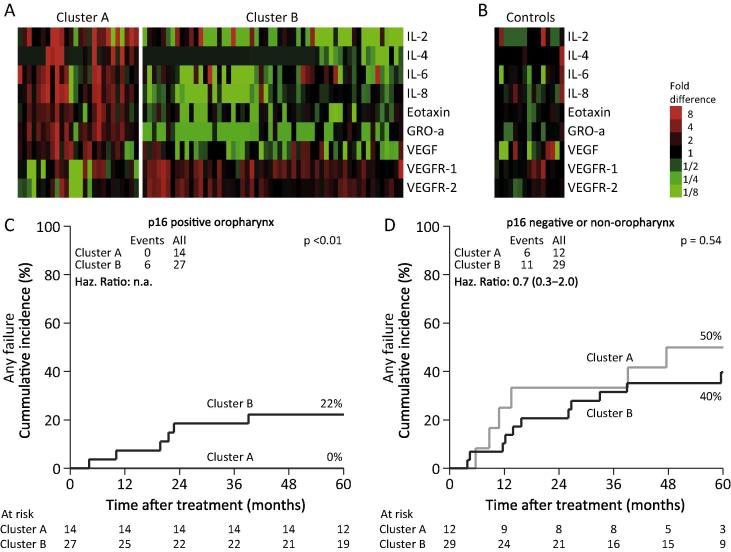
Unsupervised hierarchical clustering analysis of patients resulted in two clusters (cluster A: n = 27 and cluster B: n = 57). Nine proteins were included in the optimal cluster formation: IL-2, IL-4, IL-6 IL-8, eotaxin, GRO-a, VEGF, VEGFR-1, and VEGFR-2 (Fig.2A). The same method of cluster analysis failed to identify any clusters in the control samples (Fig.2B). Overall, univariate Cox analysis showed a non-significant increased risk of any failures in cluster B versus cluster A (HR: 1.45, 95% CI: 0.57–3.67). When correcting for smoking, chemotherapy, and HPV/p16 status HR was 1.31 (95% CI: 0.51–3.38). Comparing the cumulative incidence of any failure, there was a significant increased risk in cluster B for the p16 positive oropharynx cancer patients (p < 0.01) (Fig.2C). There was no increased risk in cluster B in HPV/p16 negative patients or non-oropharynx cancer patients (p = 0.54) (Fig.2D). Cluster B was characterized by lower levels of IL-2, IL-4, IL-6 IL-8, eotaxin, GRO-a, and VEGF and higher levels of VEGFR-1 and VEGFR-2. There was no difference in the sex, smoking, site, tumor stage, nodal stage, disease stage, hypoxia, and HPV/p16 distribution in the two clusters. The patients in cluster A were slightly younger (median age 54.7 versus 58.8 years).
Fig. 2.
Unsupervised clustering of proteins in patients (A) and in controls (B). Associations between clusters and any failures in p16-positive oropharynx patients (C) and other patients (D).
Discussion
Several studies have reported associations between circulating biomarkers and outcome for HNSCC. However, there is no consensus on how the biomarkers should be analyzed; individually or combined in profiles, which cut points should be used or which proteins are of importance [8], [9], [30], [31], [32]. In this study, we approached the analysis with three different strategies. Firstly, we evaluated the relationship between the individual proteins and failure. Secondly, we used two different cut point approaches using a healthy control cohort and an outcome driven method for cut point determination. Finally, we applied a data driven approach using unsupervised hierarchical clustering analysis. Although limited by small numbers in some of the subgroups, we did not find any associations with the first two methods. The data driven approach using unsupervised hierarchical clustering analysis, identified a profile that potentially reflects an activated immune response. In the HPV/p16 positive patients, none of the recurrences were found in patients with this profile.
In the outcome driven cut point analysis, we used the training cohort to identify optimal cut points for the proteins based on a minimum p-value approach [24]. The optimal profile identified in the training cohort could not be confirmed in the validation cohort. This highlights the potential for inflation in the type I error rate when using the minimum p-value approach [33] and the necessity of a validation cohort.
Finally, we performed unsupervised hierarchical clustering analysis using BIRCH. Two clusters based on 9 proteins were identified. Overall, the clusters were not associated with patient outcome. However, in the HPV/p16 positive oropharynx cancer patients one cluster contained 14 patients with no recurrences and the other contained 27 patients with all six recurrences. In the cluster with no recurrences, IL-2, IL-4, IL-6, IL-8, eotaxin, GRO-a, and VEGF levels were higher and VEGFR-1 and VEGFR-2 levels were lower. As this method is not outcome driven, we did not divide the dataset into a test and a validation cohort, in order to maintain more power for the cluster analysis. The finding that the HPV/p16 positive patients could comprise two distinct entities is in accordance with a recent study suggesting two HPV-positive subtypes, based on gene expression clustering and copy number profiling [7]. The two HPV-positive subgroups were classified as a classical subtype and an inflamed mesenchymal subtype. A distinct feature of the second subtype was reported to be expression of immune response genes related to the infiltration of CD8+ T cells in tumors and this subtype had a trend towards a more favorable prognosis [7]. As IL-2/IL-2 receptor interaction is a co-stimulatory signal for activating CD8 cells into cytotoxic killer T-cells it could be hypothesized that IL-2 levels are of importance in the immune elimination of cancer cells. However, the regulatory mechanisms are complex and further investigations into the regulation and importance of the immune system in HPV/p16 positive and HPV/p16 negative head and neck cancer are warranted.
In a previous study on circulating biomarkers in HNSCC, patients were also separated in two clusters by unsupervised hierarchical clustering analysis [9]. Some of the proteins used to identify the patient clusters were described as a ‘hypoxia’ signature and these were the proteins that were upregulated in one of the patient clusters. Several of these proteins were also found in our profile. In the previous study, no recurrences were observed in any of the 12 HPV/p16 positive patients, and could therefore not be linked to any of the clusters. In the HPV/p16 negative patients, recurrences were associated with high expression of the described ‘hypoxia’ signature. No association with outcome in our cohort of HPV/p16 negative patients was found. In our cohort all patients were administered treatment with the hypoxic radiosensitizer nimorazole which has been demonstrated to improve loco-regional control in HNSCC [34]. Thus, the lack of association with outcome in our cohort might be expected if the signature is a marker of hypoxia.
In conclusion, while none of the biomarkers showed strong associations individually, we have identified a profile of circulating pre-treatment biomarkers, which is associated with improved outcome in HPV/p16 positive HNSCC. This profile potentially reflects an activated immune response in a subgroup of the HPV/p16 positive patients. Future studies are needed to validate this profile in an independent cohort, as well as further studies are needed to clarify if and how this profile reflects an activated immune response, or whether it also might be associated with hypoxia.
Conflict of interest
The authors have no conflict of interest to declare.
Acknowledgements
The authors thank Birthe Hermansen for technical assistance. The study was financially supported by The Danish Cancer Society, Lundbeck Foundation Center for Interventional Research in Radiation Oncology (CIRRO), the Danish Council for Strategic Research, Max and Inger Wørzners Mindelegat, and Frits, Georg and Marie Gluds legat.
References
- 1.Decker J., Goldstein J.C. Risk factors in head and neck cancer. N Engl J Med. 1982;306:1151–1155. doi: 10.1056/NEJM198205133061905. [DOI] [PubMed] [Google Scholar]
- 2.Gillison M.L., Koch W.M., Capone R.B. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst. 2000;92:709–720. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- 3.Weinberger P.M., Yu Z., Haffty B.G. Molecular classification identifies a subset of human papillomavirus–associated oropharyngeal cancers with favorable prognosis. J Clin Oncol. 2006;24:736–747. doi: 10.1200/JCO.2004.00.3335. [DOI] [PubMed] [Google Scholar]
- 4.Lassen P. The role of Human papillomavirus in head and neck cancer and the impact on radiotherapy outcome. Radiother Oncol. 2010;95:371–380. doi: 10.1016/j.radonc.2010.04.022. [DOI] [PubMed] [Google Scholar]
- 5.Ang K.K., Harris J., Wheeler R. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Overgaard J. Hypoxic modification of radiotherapy in squamous cell carcinoma of the head and neck–a systematic review and meta-analysis. Radiother Oncol. 2011;100:22–32. doi: 10.1016/j.radonc.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Keck M.K., Zuo Z., Khattri A. Integrative analysis of head and neck cancer identifies two biologically distinct HPV and three non-HPV subtypes. Clin Cancer Res. 2015;21:870–881. doi: 10.1158/1078-0432.CCR-14-2481. [DOI] [PubMed] [Google Scholar]
- 8.Allen C., Duffy S., Teknos T. Nuclear factor-kappaB-related serum factors as longitudinal biomarkers of response and survival in advanced oropharyngeal carcinoma. Clin Cancer Res. 2007;13:3182–3190. doi: 10.1158/1078-0432.CCR-06-3047. [DOI] [PubMed] [Google Scholar]
- 9.Byers L.A., Holsinger F.C., Kies M.S. Serum signature of hypoxia-regulated factors is associated with progression after induction therapy in head and neck squamous cell cancer. Mol Cancer Ther. 2010;9:1755–1763. doi: 10.1158/1535-7163.MCT-09-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ninck S., Reisser C., Dyckhoff G., Helmke B., Bauer H., Herold-Mende C. Expression profiles of angiogenic growth factors in squamous cell carcinomas of the head and neck. Int J Cancer. 2003;106:34–44. doi: 10.1002/ijc.11188. [DOI] [PubMed] [Google Scholar]
- 11.Kaskas N.M., Moore-Medlin T., McClure G.B., Ekshyyan O., Vanchiere J.A., Nathan C.A. Serum biomarkers in head and neck squamous cell cancer. JAMA Otolaryngol Head Neck Surg. 2014;140:5–11. doi: 10.1001/jamaoto.2013.5688. [DOI] [PubMed] [Google Scholar]
- 12.Christopoulos A., Ahn S.M., Klein J.D., Kim S. Biology of vascular endothelial growth factor and its receptors in head and neck cancer: beyond angiogenesis. Head Neck. 2011;33:1220–1229. doi: 10.1002/hed.21588. [DOI] [PubMed] [Google Scholar]
- 13.Aderhold C., Grobschmidt G.M., Sauter A., Faber A., Hormann K., Schultz J.D. Interleukin 4, interleukin 6 and osteopontin-serological markers of head and neck malignancy in primary diagnostics: a pilot study. Oncol Lett. 2014;8:1112–1118. doi: 10.3892/ol.2014.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andersson B.A., Lewin F., Lundgren J. Plasma tumor necrosis factor-alpha and C-reactive protein as biomarker for survival in head and neck squamous cell carcinoma. J Cancer Res Clin Oncol. 2014;140:515–519. doi: 10.1007/s00432-014-1592-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim C.H., Lee J.S., Kang S.O., Bae J.H., Hong S.P., Kahng H. Serum hepatocyte growth factor as a marker of tumor activity in head and neck squamous cell carcinoma. Oral Oncol. 2007;43:1021–1025. doi: 10.1016/j.oraloncology.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 16.Lee Y.C., Yu C.C., Lan C. Plasminogen activator inhibitor-1 as regulator of tumor-initiating cell properties in head and neck cancers. Head Neck. 2015 doi: 10.1002/hed.24124. [DOI] [PubMed] [Google Scholar]
- 17.Toustrup K., Sorensen B.S., Nordsmark M. Development of a hypoxia gene expression classifier with predictive impact for hypoxic modification of radiotherapy in head and neck cancer. Cancer Res. 2011;71:5923–5931. doi: 10.1158/0008-5472.CAN-11-1182. [DOI] [PubMed] [Google Scholar]
- 18.Bentzen J., Toustrup K., Eriksen J.G., Primdahl H., Andersen L.J., Overgaard J. Locally advanced head and neck cancer treated with accelerated radiotherapy, the hypoxic modifier nimorazole and weekly cisplatin. Results from the DAHANCA 18 phase II study. Acta Oncol. 2015;54:1001–1007. doi: 10.3109/0284186X.2014.992547. [DOI] [PubMed] [Google Scholar]
- 19.Mortensen L.S., Johansen J., Kallehauge J. FAZA PET/CT hypoxia imaging in patients with squamous cell carcinoma of the head and neck treated with radiotherapy: results from the DAHANCA 24 trial. Radiother Oncol. 2012;105:14–20. doi: 10.1016/j.radonc.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 20.Brondum L., Sorensen B.S., Eriksen J.G. An evaluation of multiplex bead-based analysis of cytokines and soluble proteins in archived lithium heparin plasma, EDTA plasma and serum samples. Scand J Clin Lab Invest. 2016:1–11. doi: 10.1080/00365513.2016.1230882. [DOI] [PubMed] [Google Scholar]
- 21.Toustrup K., Sorensen B.S., Metwally M.A. Validation of a 15-gene hypoxia classifier in head and neck cancer for prospective use in clinical trials. Acta Oncol. 2016:1–8. doi: 10.3109/0284186X.2016.1167959. [DOI] [PubMed] [Google Scholar]
- 22.Lassen P., Overgaard J. Scoring and classification of oropharyngeal carcinoma based on HPV-related p16-expression. Radiother Oncol. 2012;105:269–270. doi: 10.1016/j.radonc.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 23.Lassen P., Eriksen J.G., Hamilton-Dutoit S., Tramm T., Alsner J., Overgaard J. Effect of HPV-associated p16INK4A expression on response to radiotherapy and survival in squamous cell carcinoma of the head and neck. J Clin Oncol. 2009;27:1992–1998. doi: 10.1200/JCO.2008.20.2853. [DOI] [PubMed] [Google Scholar]
- 24.Budczies J., Klauschen F., Sinn B.V. Cutoff Finder: a comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS One. 2012;7:e51862. doi: 10.1371/journal.pone.0051862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eriksen J.G., Buffa F.M., Alsner J., Steiniche T., Bentzen S.M., Overgaard J. Molecular profiles as predictive marker for the effect of overall treatment time of radiotherapy in supraglottic larynx squamous cell carcinomas. Radiother Oncol. 2004;72:275–282. doi: 10.1016/j.radonc.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 26.Buffa F.M., Bentzen S.M., Daley F.M. Molecular marker profiles predict locoregional control of head and neck squamous cell carcinoma in a randomized trial of continuous hyperfractionated accelerated radiotherapy. Clin Cancer Res. 2004;10:3745–3754. doi: 10.1158/1078-0432.CCR-03-0248. [DOI] [PubMed] [Google Scholar]
- 27.Fine J.P., Gray R.J. A proportional hazards model for the subdistribution of a competing risk. ASA. 1999;94:496. [Google Scholar]
- 28.Andersen P.K., Perme M.P. Pseudo-observations in survival analysis. Stat Methods Med Res. 2010;19:71–99. doi: 10.1177/0962280209105020. [DOI] [PubMed] [Google Scholar]
- 29.Toustrup K., Singers Sørensen B., Hassan Metwally M.A. Validation of a 15-gene hypoxia classifier in head and neck cancer for prospective use in clinical trials. Acta Oncol. 2016 doi: 10.3109/0284186X.2016.1167959. [in press] [DOI] [PubMed] [Google Scholar]
- 30.Meyer F., Samson E., Douville P., Duchesne T., Liu G., Bairati I. Serum prognostic markers in head and neck cancer. Clin Cancer Res. 2010;16:1008–1015. doi: 10.1158/1078-0432.CCR-09-2014. [DOI] [PubMed] [Google Scholar]
- 31.Le Q.T., Fisher R., Oliner K.S. Prognostic and predictive significance of plasma HGF and IL-8 in a phase III trial of chemoradiation with or without tirapazamine in locoregionally advanced head and neck cancer. Clin Cancer Res. 2012;18:1798–1807. doi: 10.1158/1078-0432.CCR-11-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Argiris A., Lee S.C., Feinstein T. Serum biomarkers as potential predictors of antitumor activity of cetuximab-containing therapy for locally advanced head and neck cancer. Oral Oncol. 2011;47:961–966. doi: 10.1016/j.oraloncology.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altman D.G., Lausen B., Sauerbrei W., Schumacher M. Dangers of using “optimal” cutpoints in the evaluation of prognostic factors. J Natl Cancer Inst. 1994;86:829–835. doi: 10.1093/jnci/86.11.829. [DOI] [PubMed] [Google Scholar]
- 34.Overgaard J., Hansen H.S., Overgaard M. A randomized double-blind phase III study of nimorazole as a hypoxic radiosensitizer of primary radiotherapy in supraglottic larynx and pharynx carcinoma. Results of the Danish Head and Neck Cancer Study (DAHANCA) Protocol 5–85. Radiother Oncol. 1998;46:135–146. doi: 10.1016/s0167-8140(97)00220-x. [DOI] [PubMed] [Google Scholar]