Abstract
Integrins are α/β heterodimeric transmembrane adhesion receptors. Evidence exists that their transmembrane domain (TMD) separates upon activation. Subunit-specific differences in activation sensitivity of integrins were reported. However, whether sequence variations in the TMD lead to differential TMD association has remained elusive. Here, we show by molecular dynamics simulations and association free energy calculations on TMDs of integrin αIIbβ3, αvβ3, and α5β1 that αIIbβ3 TMD is most stably associated; this difference is related to interaction differences across the TMDs. The order of TMD association stability is paralleled by the basal activity of these integrins, which suggests that TMD differences can have a decisive effect on integrin conformational free energies. We also identified a specific order of clasp disintegration upon TMD dissociation, which suggests that the closed state of integrins may comprise several microstates. Our results provide unprecedented insights into a possibly contributing role of TMD towards subunit-specific sensitivity of integrin activation.
Introduction
Integrins are a major class of heterodimeric adhesion receptors consisting of α and β subunits1 and are involved in the regulation of many biological events2. Each subunit is formed by a large extracellular domain (ectodomain) connected to a short cytoplasmic tail through a single transmembrane domain (TMD)3. In providing a physical link between the exterior and the interior of the cell, the TMD serves as a translator of mechanical and biochemical signals in both directions across the plasma membrane, leading to inside-out and outside-in signaling4–6. TMDs are directly involved in the mechanism of integrin activation7 in that the two transmembrane (TM) segments associate in the resting state8–10 and dissociate upon activation11–14. Structural features of the integrin TMD were revealed by nuclear magnetic resonance (NMR) structures15–17, biochemical data18,19, and electron microscopy20–22. Two structural elements, the inner and outer membrane clasps (IMC and OMC)23, were recognized as principal mediators of TMD assembly24, together with electrostatic interactions in the membrane-proximal region. However, despite these unifying principles, there is also evidence for specific ways of helix-helix association among different isoforms25, and it has remained unclear if and how these differences are linked to the subunit-specific sensitivity of integrin activation26,27: Integrin αIIbβ3 has been described as basally inactive, in contrast to αvβ3, which was found to be in the active state by default in certain cell types28,29, and β1 integrins, which are considered to be basally active26, with integrin α5β1 being among the most conformationally flexible integrins containing β127. In addition, contradicting findings of NMR24,30 and in vitro31 versus in vivo32 studies with respect to a salt bridge interaction between αIIb-R995 and β3-D723 might also be related to specific TMD associations (see below for details).
The TMD of a subunit is formed by a short α-helix33, and two such TMDs are arranged in a right-handed coiled-coil conformation16 in the resting state of the integrin9,10,34. The structure of the TMD of αIIbβ3 integrin15 revealed that the TM and membrane-proximal regions of the β3 subunit form a continuous helical structure that is tilted by ~25° with respect to the membrane normal16,17. In contrast, the αIIb subunit is oriented in parallel to the membrane normal, breaks at G991, and bents towards the β3 TMD. This allows the dimer to be stabilized by the OMC and IMC23. The former is a GXXXG-like motif 35, while the latter is a highly conserved GFFKR motif 36, with the two Phe residues found in all α subunits36. Mutational studies13, disulphide scanning11 and Leu scanning11 experiments confirmed the importance of the OMC, whose alteration prevents correct helix packing and abolishes helix association36. However, different compositions of the GXXXG motif are found in α subunits, which makes it reasonable to hypothesize that the different OMC interfaces contribute to differential integrin activation. Likewise, the importance of the IMC, and, in particular, the two conserved Phe residues, in maintaining correct TMD packing and restraining integrin in the resting state has been shown31,37. In contrast, it has remained controversial whether αIIb-F992 engages β3-K716 in hydrogen bond formation to support IMC formation17, or whether K716 “snorkels” towards the lipid head groups38. Moreover, NMR spectroscopy24 revealed the presence of a salt bridge between αIIb-R995 and β3-D723 at the membrane-proximal region, in immediate vicinity of the IMC, whose functional role in restraining integrin in the inactive state has been demonstrated24,30,39–41. However, NMR structures in which both αIIb-F992 and αIIb-R995 point away from the β3 subunit, thus, not making any interactions with the β3 subunit, have also been determined30. Likewise, mutational studies in which the breaking of the R995-D723 salt bridge did not cause immediate integrin activation were reported14,39.
In order to provide insights at the atomistic level as to a potential influence of the TMD on the subunit-specific sensitivity of integrin activation, including the role of interactions across the IMC and OMC, here, we performed equilibrium molecular dynamics (MD) simulations in an explicit membrane environment of in total 9 μs length on the associated TMDs of integrins αIIbβ3, αvβ3, and α5β1, respectively, and potential of mean force (PMF) computations of TMD association of in total 3.5 μs sampling time, from which we derived association free energies. Our results show that the TMD of integrin αIIbβ3 is most stably associated compared to αvβ3 and α5β1. We relate these differences to particular interactions across the TMDs, with a focus on the different OMC compositions and an “OMC before IMC” order of clasp disintegration found for TMD dissociation. Our results provide unprecedentedly detailed and comparative insights into a possibly contributing role of the TMD towards subunit-specific sensitivity of integrin activation.
Results
Structural dynamics of the αIIbβ3, αvβ3, and α5β1 TMDs
In order to investigate at an atomistic level possible subunit-specific differences in the association of TMDs of integrins αIIbβ3, αvβ3, and α5β1, the three TMDs were subjected to all-atom MD simulations in an explicit membrane and solvent environment (Fig. 1a). Prior to that, we generated homology models of αvβ3 and α5β1 TMDs using the NMR structure of αIIbβ3 TMD (PDB ID 2K9J)15 as a template. To estimate the quality of the models, we used the QMEANBrane version42 of the QMEAN scoring function implemented in the QMEAN server43. QMEANBrane employs specifically trained potentials for three different segments (membrane, interface and soluble) in a transmembrane protein model to determine local (i.e., per residue) absolute quality estimates on the basis of a single model. With 1.0 as the optimal score, we found local scores of ~0.8 for the residues embedded in the membrane and ~0.6–0.8 when considering the overall structures (Fig. S1). The NMR structure showed very similar QMEANBrane scores, suggesting a sufficient local structural quality of the models. Sequence identities of 40% to 65% between respective template and target sequences furthermore suggest that the global folds are conserved. Note that we generally refer to the simulated systems as “TMD” here, although the actual TM region is prepended by 9–11 (8) residues of the linker to the calf-2 (β-tail) domain at the N-terminal end of the α (β) subunit, and appended by 5 (6) residues of the respective cytosolic domains at the C-terminal ends (Fig. 1b). For clarity, we furthermore only refer to the sequence numbering of integrin αIIbβ3 below (Fig. 1b).
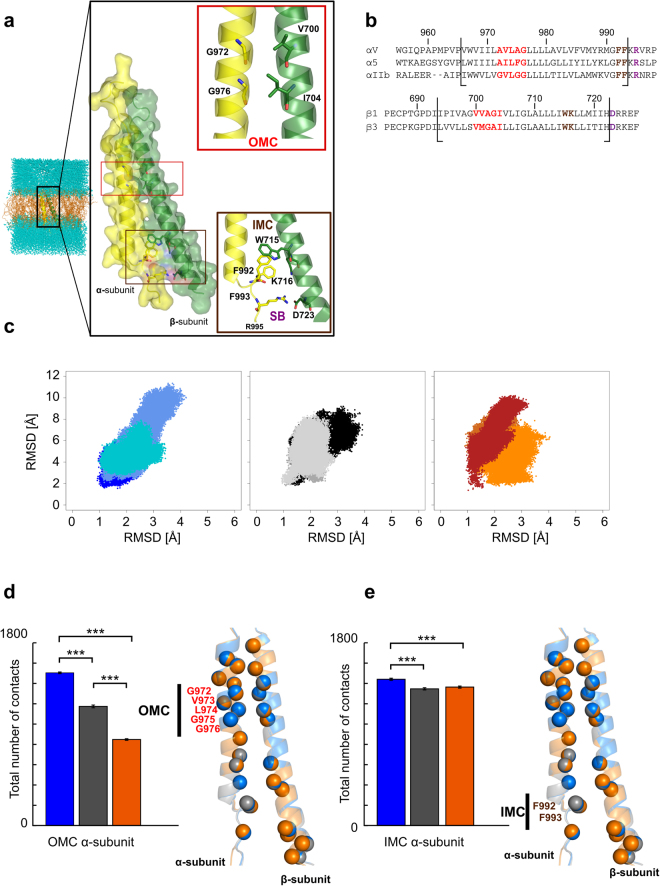
Figure 1.
Structural integrity of the membrane-embedded helices throughout the MD simulations and contacts across the interface. (a) Exemplary simulation box generated to perform MD simulations including the NMR structure of αIIbβ3 TMD (PDB ID 2K9J; cartoon and surface representation) with the position of the OMC (red box) and IMC (brown box) indicated; phospholipids are shown as orange sticks, and water layers as blue spheres. Close-up views of the box contents show essential residues mediating the clasps: αIIb-G972/G976 and β3-V700/I704 for the OMC, αIIb-F992/F993/R995 and β3-W715/K716/D723 for the IMC plus the putative salt bridge. (b) Sequence alignment of the αIIbβ3, αvβ3, α5β1 TMD sequences used to generate the homology models. GXXXG and GFFKR motifs are highlighted in red and brown, respectively, and R995/D723 residues in purple. Black bars indicate the TMD borders as reported in ref.45. (c) Two dimensional histograms of the RMSD values of all Cα atoms (ordinate values) and only those that are embedded in the membrane (abscissa values) (range of residues considered: P996-V1015 and D718-I747) calculated over three MD simulations. Blue, cornflowerblue, turquoiseblue colored dots represent αIIbβ3 (MD simulations 1, 2 and 3); darkgrey, black, lightgrey αvβ3; chocolate, orange, firebrick α5β1. (d,e) Histograms of the total number of overall contacts per residue at the OMC and IMC averaged over three MD simulations, with error bars showing the standard error of the mean (SEM; eq. 6) and stars indicating the statistical significance (see Methods section for definition). Here, only residues of the α subunit are shown (numbering refers to the αIIb subunit). On the right of the plots, a superimposition of the αIIbβ3, αvβ3, and α5β1 TMDs in blue, grey, and orange, respectively, is shown. The Cα atoms of residues considered in the contact analysis are indicated as spheres, and the residues of the α subunit considered in the analyses are labeled.
Using the three TMD structures as starting structures, we performed three independent MD simulations of 1 μs length for each system, yielding in total 9 μs of simulation time. The structural variability was assessed in terms of root mean square deviations (RMSD) after a mass-weighted superimpositioning onto the respective starting structure. The RMSD of all Cα atoms, including those not embedded in the membrane, raises to values of ~7–10 Å. However, if only the TM region is considered, the RMSD amounts to ~2–4 Å (Fig. 1c). This data indicates that the structural integrity of the two TM helices remains intact throughout the MD simulations, whereas the TMD ends fray at about nine residues at either terminus of either helix. The finding that the overall configuration of the TMD remains intact during our simulations is congruent with a slow dissociation rate found for αIIbβ3 TMD44. In agreement with our findings, NMR spectroscopy revealed a dynamically unstructured αIIb linker45. The convergence of the internal motions between independent MD simulations was assessed following ref.46. In short, the overlap of histograms of principal component (PC) projections obtained in a pair-wise manner from each simulation for a given TMD system are looked at as a function of time (Fig. S2). The results reveal that the first three PCs are relatively well-converged after ~400 ns for αIIbβ3 TMD, whereas for αvβ3 and α5β1 TMDs it takes ~800 ns to reach a comparable level of convergence.
To conclude, RMSD values of membrane-embedded TMD parts generally below 4 Å reveal that both the secondary structure and overall configuration of the two TM helices remain intact throughout the MD simulations of all three TMDs. The convergence behavior of PC projections suggests larger and/or slower internal motions in αvβ3 and α5β1 TMDs than αIIbβ3 TMD.
Residue-wise analysis of contacts and mobility in the TMD interfaces
Next, we examined differences in the TMD topology of each system in terms of changes in the number of contacts present in the starting structure (“native contacts”) and those formed over the MD simulation time (“non-native contacts”). A contact is considered formed between the α and β subunits if any two atoms of two residues come closer than 7 Å47. First, for globally comparing the three systems, we computed the average number of overall contacts. The total number of native contacts already present in the NMR structure of αIIbβ3 TMD (PDB ID 2K9J) is almost equal to that of the initial structure of αvβ3 TMD (6217 versus 6244), and only ~2% smaller than that of the initial structure of α5β1 TMD (6344). In contrast, compared to αIIbβ3, both αvβ3 and α5β1 reveal a highly significant reduction of overall contacts by ~10% (p < 0.0001) (Fig. S3) during the MD simulations. Second, from a list of residues conserved across the three α subunits and the contact map information, we extracted those residues accounting for the native and non-native contacts at the OMC and IMC interface between α and β subunit (Table S1). As to the former, compared to αIIbβ3 TMD, the overall average number of contacts is ~20% smaller in the αvβ3 TMD and ~40% smaller in the α5β1 TMD (Fig. 1d). Both differences are highly significant (p < 0.0001). As to the latter, compared to αIIbβ3 TMD, both αvβ3 and α5β1 TMDs reveal a significant reduction of overall contacts by ~10% (Fig. 1e) (p ≈ 0.003 and 0.006, respectively).
Furthermore, we computed residue-wise root mean square fluctuations (RMSF) as a measure of atomic mobility to identify local differences in the structural dynamics of the three TMD complexes, averaged over the respective three simulations. For both the α and β subunit, the αIIbβ3 TMD shows less pronounced residue motions, with the RMSF values of 60 out of 89 residues (~67%) being lower than those of αvβ3 and α5β1 (Table S2). To conclude, these results demonstrate on a per-residue level that the αIIbβ3 TMD forms generally more contacts across the TMD interface and shows less mobile residues in the TMD.
Subunit-specific differences in OMC and IMC distances
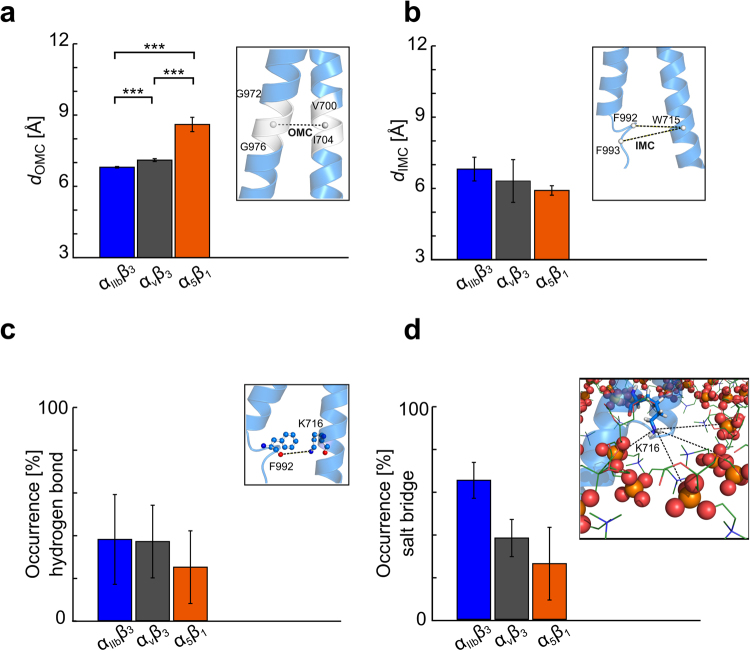
Both the OMC and IMC are considered necessary to maintain integrin in the low affinity state2 (Fig. 1a). However, particularly in the OMC, each TMD has subunit-specific amino acids (Fig. 1b) that may be responsible for the above observed differences. The dOMC (distance computed between the centers of mass (COM) of the Cα atoms of the GXXXG motif (G972-G976) on the αIIb subunit and V700-I704 on the β3 subunit) is 9.3 Å in the NMR structure of αIIbβ3 TMD (PDB ID 2K9J) and 7.3 Å in a structure based on Cys cross-linking results48. The distance dIMC computed as the minimal distance between the COMs of the aromatic rings of F992 or F993 on the αIIb subunit and the aromatic ring of W715 on the β3 subunit is 4.0 Å in the NMR structure and 5.4 Å in the structure based on Cys cross-linking results.
As to the MD simulations, first, the dOMC averaged over three simulations is smaller by ~0.3 Å in the αIIbβ3 TMD (~6.8 Å, SEM ≈ 0.03 Å) than in the αvβ3 TMD (~7.1 Å, SEM ≈ 0.06 Å), and smaller by ~1.5 Å than in the α5β1 TMD (~8.4 Å, SEM ≈ 0.3 Å). All values are well in the range of inter-helical distances found for TM heterodimers containing an OMC-like structural motif49. The differences are highly significant in all cases (p < 0.0001) (Fig. 2a, Table S3). Hence, the OMC interface is most compact in αIIbβ3 TMD, followed by αvβ3 and α5β1 TMDs. Second, the dIMC averaged over three simulations remains below 7 Å in all cases: αIIbβ3 (~6.8 Å, SEM ≈ 0.5 Å), αvβ3 (~6.3 Å, SEM ≈ 0.9 Å) and α5β1 TMDs (~5.9 Å, SEM ≈ 0.2 Å) (Fig. 2b, Table S3). The differences are not significant (p ≈ 0.6 for αIIbβ3/αvβ3, ≈ 0.05 for αIIbβ3/α5β1, ≈ 0.5 for αvβ3/α5β1), however. Hence, we conclude that the IMC interface is equally maintained over the course of the MD simulations.
Figure 2.
Differences between interdomain interactions in αIIbβ3, αvβ3, and α5β1 TMDs. (a,b) Histograms of the distances dOMC and dIMC (see main text for definition) averaged over three MD simulations. From left to right, the αIIbβ3, αvβ3, and α5β1 TMDs are displayed in blue, grey, and orange, respectively. Within each plot, a close-up view of the NMR structure of αIIbβ3 TMD (PDB ID 2K9J) is colored in blue and indicates the analyzed distances (black dashed lines). White spheres indicate Cα atoms of the labeled amino acids. (c) Histogram of the mean relative occurrence of the hydrogen bond between K716Nε and F992O using a distance cutoff of 3.5 Å and an angle cutoff of 120°. Within the plot, a close-up view of the respective distance measured is shown (color code as in panel a). (d) Histogram of the mean relative occurrence of the salt bridge between K716Nε and the oxygens of phospholipids head groups of the lower lipid leaflet applying a distance cutoff of 4 Å. Within the plot, a close-up view of the respective distance measured is shown, with the phospholipid head groups depicted as spheres. (a–d) Error bars show the SEM (eq. 5) and stars indicate the statistical difference (see Methods section for definition).
Subunit-specific interactions formed across the TMD interfaces
The IMC interface is adjacent to the membrane-proximal region, which is believed to be mainly stabilized by a salt bridge formed between αIIb-R995 and β3-D7232. First, we computed the minimal distance between the atoms R995Nη1/Nη2 and D723Oδ1/Oδ2 for each system (Table 1). As next to R995 and D723, respectively, R997 is present on the α subunit and E726 on the β subunit, we also analyzed the minimal distances between atoms R995Nη1/Nη2 and E726Oε1/Oε2, R997Nη1/Nη2 and D723Oδ1/Oδ2, and R997Nη1/Nη2 and E726Oε1/Oε2 (Table 1; α5β1 integrin contains a Leu at position 997 such that the last two distances were not evaluated there). Then, to assess the frequency of occurrence of formed salt bridges, we applied a cutoff of 4 Å to the computed minimal distances, according to a previous study50 (Table 1): 1) the R995-D723 salt bridge has the highest occupancy in the αIIbβ3 TMD (~58%), followed by the αvβ3 (~37%) and α5β1 TMDs (~39%). The differences between αIIbβ3 TMD and either αvβ3 or α5β1 TMDs are not statistically significant (p ≈ 0.3 for αIIbβ3/αvβ3, αIIbβ3/α5β1, and ≈ 0.9 for αvβ3/α5β1); 2) the R995-E726 salt bridge (the only alternative interaction that can be formed in α5β1) has the highest occupancy in the α5β1 TMD (~42%), followed by the αIIbβ3 (~33%) and αvβ3 TMDs (~39%). The differences are not significant (p ≈ 0.8 for αIIbβ3/αvβ3, αIIbβ3/α5β1, and ≈ 0.9 for αvβ3/α5β1); 3) the R997-D723 salt bridge is formed in both the αIIbβ3 (~47%) and αvβ3 TMDs (~48%) to a very similar extent (the difference is not significant (p ≈ 0.9)); 4) the R997-E726 salt bridge is formed to a lower extent in the αIIbβ3 (~17%) and αvβ3 TMDs (~31%), and the difference is not significant (p ≈ 0.4). To conclude, our results, in agreement with ref.24, indicate that R995-D723 is not the only salt bridge that is formed across the TMD interface. In agreement with ref.51, the R995-D723 salt bridge dissociates intermittently, as indicated by occupancies ≪100%. However, the MD simulations reveal that, among all four possible salt bridges that can form in the membrane-proximal region, R995-D723 is the most prevalent interaction, followed by R995-E726, and R997-E726 is the least prevalent one.
Table 1.
Frequency of occurrence of the up to four salt bridges in the membrane-proximal region for each TMD system[a].
| Salt bridge | αIIbβ3 TMD | αvβ3 TMD | α5β1 TMD | ||||||
|---|---|---|---|---|---|---|---|---|---|
| SimI | SimII | SimIII | SimI | SimII | SimIII | SimI | SimII | SimIII | |
| R995-D723 | 47 | 92 | 35 | 57 | 8 | 46 | 14 | 66 | 38 |
| 58 ± 6.7[b] | 37 ± 14.8[b] | 39 ± 15.0[b] | |||||||
| R995-E726 | 47 | 50 | 2 | 70 | 29 | 18 | 24 | 78 | 24 |
| 33 ± 15.5[b] | 39 ± 18.8[b] | 42 ± 18.0[b] | |||||||
| R997-D723 | 23 | 95 | 23 | 84 | 52 | 7 | —[c] | —[c] | —[c] |
| 47 ± 24.0[b] | 48 ± 22.3[b] | —[c] | |||||||
| R997-E726 | 10 | 39 | 2 | 10 | 31 | 51 | —[c] | —[c] | —[c] |
| 17 ± 11.2[b] | 31 ± 11.8[b] | —[c] | |||||||
[a]In %.
[b]Mean values and SEM (eq. 6), calculated over three MD simulations.
[c]Interactions that cannot be formed in the TMD of integrin α5β1.
Subunit-specific contribution of K716 to the TMD stability
Finally, we investigated the behavior of K716, monitoring where the residue preferentially places its side chain, i.e., either as part of a hydrogen bond with F992 or by orienting the positive charge of Nε towards the negatively charged head groups of the lipid bilayer38. In the NMR structure of αIIbβ3 TMD (PDB ID 2K9J), the distance between Nε of K716 and the carbonyl oxygen of F992 is 6.2 Å; in the structure based on Cys cross-linking results, it is 2.9 Å. During the three simulations of each system, the occupancy of the K716-F992 hydrogen bond was investigated applying a distance cutoff of 3.5 Å and an angle cutoff of 120°. On average, the hydrogen bond is formed in a similar manner in the αIIbβ3 (~38%) and αvβ3 TMDs (~37%), and to a lower extent in the α5β1 TMD (~25%) (Fig. 2c, Table S4). The differences between αIIbβ3 or αvβ3 TMDs with respect to α5β1 TMD result in p ≈ 0.6, respectively.
To evaluate the presence of electrostatic interactions between the K716 side chain and the head groups of lipids, first, we computed the minimal distance between Nε of K716 and the nearest O atom of the phospholipid head groups from the lower lipid leaflet (dsnorkeling). Then, we applied a 4 Å cutoff to dsnorkeling and calculated the frequency of occurrence of a salt bridge. On average, the salt bridge is mainly formed in the αIIbβ3 TMD (~65%), followed by αvβ3 TMD (~38%), and to a lower extent in the α5β1 TMD (~26%) (Fig. 2d, Table S5). The differences between αIIbβ3 TMD with respect to αvβ3 or α5β1 TMDs result in p ≈ 0.08 and ≈ 0.1, while the difference between αvβ3 and α5β1 TMDs results in p ≈ 0.5. To conclude, the K716 sidechain is engaged in either a hydrogen bond across the TMD or a salt bridge with phospholipid head groups in the αIIbβ3 TMD (~100%), followed by αvβ3 (~75%) and α5β1 (~50%) TMDs, with the salt bridge being the more prevalent interaction in the case of αIIbβ3 TMD.
Configurational free energies of TM helix association
The above analyses strongly suggest that the TMDs of the integrin isoforms differ in their potential to associate. To corroborate these findings in an independent manner, we computed the configurational free energy (potential of mean force, PMF) of TM helix association using the distance between COMs of the sections of the α and β subunits embedded in the membrane as a reaction coordinate (referred to as dCOM-COM). The PMFs were computed using umbrella sampling52 along a pathway from the bound subunits to subunits where any pair of atoms between the two subunits is at least 10 Å apart, and WHAM53 post-processing. The PMF profiles were obtained employing 16 biased MD simulations of 200 ns length for reaction coordinate values of dCOM-COM = 8 Å to 20 Å, and four biased MD simulations of 70 ns length for reaction coordinate values of dCOM-COM = 21 Å to 24 Å. Together, this sums up to a total of ~ 3.5 μs simulation time. Approximately Gaussian-shaped frequency distributions were obtained for each reference point along the reaction coordinates, with all such distributions well overlapping (Fig. S4). These are prerequisites for the successful application of WHAM to extract a PMF from these distributions53. Repeating the computations of the PMFs for the range of dCOM-COM = 8 Å to 20 Å for parts of the simulation time demonstrates that, for all three systems, the PMFs are converged after at most 160 ns of simulation time per window (maximal difference between two PMFs: 0.2 kcal mol−1) (Fig. S5). The same procedure was repeated for the PMFs from dCOM-COM = 20 Å to 24 Å (fully dissociated state), demonstrating that these PMFs are converged after at most 50 ns of simulation time per window (Fig. S5). For comparison, the PMF values at dCOM-COM = 20 Å were set to zero in all three cases (Fig. 3a).
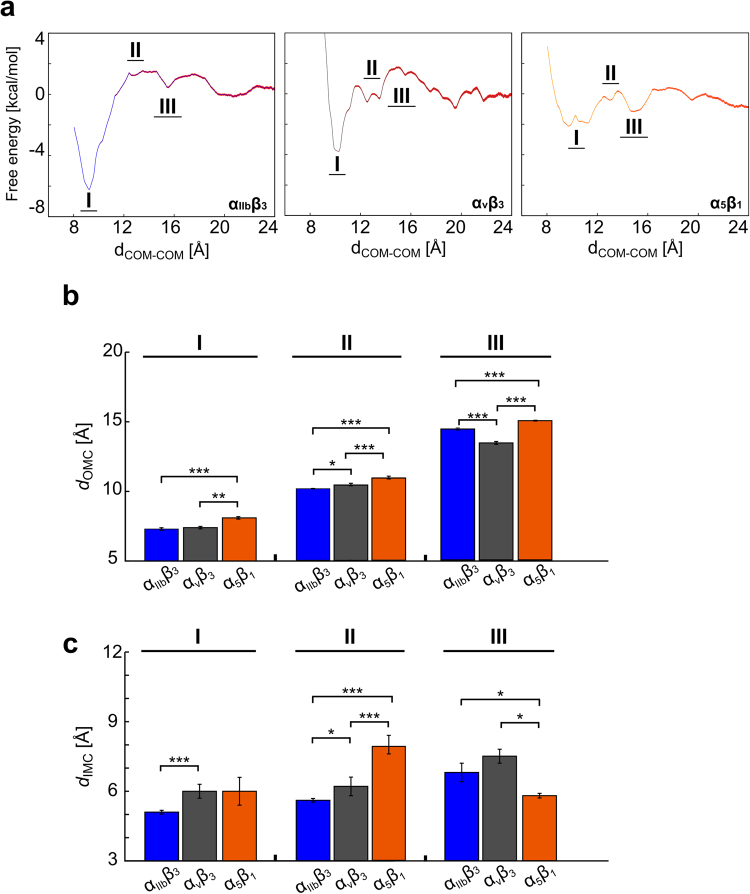
Figure 3.
Potential of mean force of TM helix association of αIIbβ3, αvβ3, and α5β1 TMDs and differences at the OMC/IMC interface with increasing dCOM-COM. (a) Configurational free energies as a function of the dCOM-COM used as a reaction coordinate for αIIbβ3 (left panel), αvβ3 (middle panel), and α5β1 (right panel) TMDs. Roman numbers indicate free energy minima. Statistical errors, calculated using bootstrap analysis, are displayed as red shaded curves added to the PMF profiles. The PMF values at dCOM-COM = 20 Å were set to zero. (b,c) Histograms of the averaged dOMC (B) and dIMC (C) across umbrella sampling windows linked to free energy minima I–III observed in panel (A) (see also Table S7), using reweighted (“unbiased”) TMD configurations. Error bars denote the SEM (eq. 5) and stars indicate the statistical difference (see Methods section for definition).
The global minima (dCOM-COM ≈ 9 Å) are in all three cases close to the initial distance (10 Å resp. 12.6 Å) calculated from the NMR structure of the αIIbβ3 TMD (PDB ID 2K9J) or the structure based on Cys cross-linking results, and the general shapes of the PMFs are comparable, with a rising free energy with increasing reaction coordinate values and rather flat PMFs beyond dCOM-COM ≈ 20 Å. Furthermore, in going from the global minima to dCOM-COM ≈ 20 Å, two local minima are passed (marked by roman numbers II and III in Fig. 3a). These minima are located at dCOM-COM ≈ 12 to 13 Å and dCOM-COM ≈ 15 Å in all three systems.
However, the PMFs also show pronounced differences. First, the global minima show values of −6.5, −3.8, and −2.1 kcal mol−1 for the αIIbβ3, αvβ3, and α5β1 TMDs, respectively, demonstrating the largest tendency for the TM helices to associate in the case of αIIbβ3, and the lowest in the case of α5β1. For computing association free energies ΔG from the PMFs (eqs 1–3), we, first, assessed to what extent the dissociated TMD helices of αIIb and β3 sample the accessible configuration space on the simulated time scales. This yielded values for ||Ω||, a factor describing the restriction of the configurational space of the monomers upon dimer formation (eq. 4), of 0.05 to 0.10 (Table 2). Compared to the value of the overall accessible space of (2π)2, these values indicate that the dissociated TMD helices sample only a fraction of the overall accessible space. Hence, we followed a procedure by Johnston et al.54 to compute dimerization and mole fraction dimerization constants Ka and KX, respectively, (eqs 1,2) and from there ΔG (eq. 3) (Table 2). The results indicate that αIIbβ3 TMD is the most stable system (ΔG = −3.8 kcal mol−1), followed by αvβ3 and α5β1 TMDs (ΔG = −0.8 kcal mol−1 and 0.5 kcal mol−1, respectively). Note that the exact value of the dimerization distance D (eq. 1; here evaluated between 8 and 12 Å) has a low impact on the results (see Table S6 for ΔG values evaluated between 8 and 14 Å), because the integrals reach plateaus after ~10 Å55. Second, the TM association requires to pass configurational free energy barriers that are similar for αIIbβ3 and α5β1 TMDs (~1.5 kcal mol−1, respectively), whereas a larger barrier occurs in the case of αvβ3 TMD (~3 kcal mol−1).
Table 2.
Thermodynamic quantities for each TMD system[a].
| System | αIIbβ3 TMD | αvβ3 TMD | α5β1 TMD |
|---|---|---|---|
| ||Ω||[b] | 0.07 | 0.05 | 0.1 |
| K a [c] | 38009.8 | 280.9 | 27.7 |
| K X | 543.0 | 4.0 | 0.4 |
| ΔG[d] | −3.8 | −0.8 | 0.5 |
To conclude, the PMFs of TM helix association and computed association free energies show overall similar shapes but reveal that the αIIbβ3 TMD has the strongest tendency to associate, and the PMF of the α5β1 TMD shows the smallest configurational free energy changes.
Distinct order of TMD clasp formation and differences in the persistence of IMC and OMC with increasing helix-helix distance
To further investigate differences between the integrin isoforms in more detail, averaged dOMC and dIMC were computed on the reweighted (“unbiased”) configurations from umbrella sampling (reweighting done according to ref.56) (Fig. S6) for free energy minima I – II, respectively (Fig. 3b,c; Table S7).
At the global minimum I, the dOMC, averaged over windows 3–5, is very similar in the αIIbβ3 and αvβ3 TMDs (~7.3 Å, ~7.4 Å, SEM ≈ 0.09 Å), and smaller by ~0.8 Å than in the α5β1 TMD (~8.1 Å, SEM ≈ 0.09 Å). The differences between αIIbβ3 or αvβ3 TMDs versus α5β1 TMD are (highly) significant (p < 0.0001, and p ≈ 0.0003). The dIMC is smaller by ~0.9 Å in the αIIbβ3 TMD (~5.1 Å, SEM ≈ 0.08 Å) than in the αvβ3 and α5β1 TMDs (~6.0 Å, SEM ≈0.3 Å and ≈ 0.6 Å, respectively). The difference between αIIbβ3 and αvβ3 TMDs is highly significant (p < 0.0001), while for that between αIIbβ3 and α5β1 TMDs, p ≈ 0.06 results. Hence, both the OMC and IMC are conserved, with the clasps being more compact in the αIIbβ3 TMD, followed by αvβ3 and α5β1 TMDs.
At minimum II, the dOMC, averaged over windows 8–9, is slightly smaller in the αIIbβ3TMD (~10.1 Å, SEM ≈ 0.03 Å) than in the αvβ3 TMD (~10.4 Å, SEM ≈ 0.09 Å), and smaller by ~0.8 Å than in the α5β1 TMD (~10.9 Å, SEM ≈ 0.1 Å). The difference between αIIbβ3 and αvβ3 TMDs is significant (p ≈ 0.04) and highly significant in the other cases (p < 0.0001). The dIMC is smaller by ~0.6 Å in the αIIbβ3 TMD (~5.6 Å, SEM ≈ 0.08 Å) than in the αvβ3 TMD (~6.2 Å, SEM ≈ 0.4 Å), and by ~2.4 Å than in the α5β1 TMD (~8.0 Å, SEM ≈ 0.4 Å). The difference between αIIbβ3 and αvβ3 TMDs is significant (p ≈ 0.03 Å), and highly significant in the other cases (p < 0.0001). Hence, the interface at the OMC is less compact than before and starts to disintegrate (dOMC > 10 Å). In contrast, the IMC packing is conserved, similar to as before, but the interface is tighter in αIIbβ3 TMD than in αvβ3 and α5β1 TMDs.
Finally, at minimum III, dOMC and dIMC, averaged over windows 11–12, reveal that the OMC packing is largely lost (distances > 13 Å), while the IMC packing is still conserved. The dIMC is ~0.8 Å smaller in the αIIbβ3 TMD (~6.8 Å, SEM ≈ 0.4 Å) than in the αvβ3 TMD (~7.5 Å, SEM ≈ 0.3 Å), and ~1 Å larger than in the α5β1 TMD (~5.8 Å, SEM ≈ 0.1 Å). The difference between αIIbβ3 and αvβ3 TMDs is not significant (p ≈ 0.2), but both differences versus α5β1 TMD are significant (p ≈ 0.005).
To conclude, our results suggest for all investigated integrin isoforms that helix association in the TMDs proceeds first via IMC formation, and that OMC formation then reinforces the coiled-coil conformation. The reverse order is suggested to occur upon helix dissociation. However, pronounced differences among the three TMDs as to the conservation of OMC/IMC packing with increasing dCOM-COM became obvious, with αIIbβ3 TMD showing the most persistent clasps.
Discussion
In this study, we have shown by molecular simulations at the atomistic level that the TMD of integrin αIIbβ3 is most stably associated compared to that of αvβ3 and α5β1, and that this difference is related to differences in particular interactions across the TMDs, notably in the OMC. We furthermore identified an “OMC before IMC” order of clasp disintegration upon TMD dissociation as a uniform property of all three investigated TMDs. Our study was motivated by the considerable evidence that the TMD separate upon integrin activation7,11,13,14,17,57–60 and so are expected to influence conformational properties of integrins, which, in turn, have been correlated to integrin adhesiveness and affinity26,61–66. Furthermore, while the overall structural organization and activation mechanism of integrins appears largely conserved48,62,66–68, subunit-specific differences in the activation sensitivity had been reported26,27,29. Accordingly, here, we comparatively assessed the TMD of α5β1, a physiologically relevant69,70 member of the β1 integrin subfamily that is considered basally active26 and TMDs of αIIbβ3 and αvβ3, physiologically relevant71,72 members of the β3 integrin subfamily that is considered basally inactive26, although a cell-specific influence on the activation sensitivity has been reported for integrin αvβ329.
That αIIbβ3 TMD is most stably associated compared to that of αvβ3 and α5β1 has been demonstrated in two independent ways. First, we performed unbiased microsecond-long MD simulations at the atomistic level in explicit solvent and an explicit lipid bilayer on the three TMDs, likely the currently most accurate way to explore structure and dynamics of transmembrane proteins73. The length of our MD simulations surpasses comparable previous ones on integrin TMDs by at least one order of magnitude51,74,75. We performed triplicate MD simulations for each system, which allows probing for the influence of the starting conditions and determining the significance of the computed results by statistical testing and rigorous error estimation46. As to the latter, we paid close attention to only consider uncorrelated instances for SEM calculations (eqs 5 and 6). The assessment of convergence of internal motions between independent MD simulations revealed for motions of the TMDs described by the first three PCs that they are relatively well-converged on the timescale of the simulations. For the MD simulations, we used established parameterizations for the solvent76, lipids77, and proteins78; the latter, we had also applied successfully in other integrin simulations79–82, although we note that more recent protein force fields have become available83,84. Yet, the impact of force field deficiencies on our results is expected to be small due to cancellation of errors when comparatively assessing the TMDs. While for αIIbβ3 TMD an experimental structure was available for system setup15, structures generated by homology modeling were used for αvβ3 and α5β1 TMDs. Still, with sequence identities of 40 to 65%, Cα-RMSD values to the native structure below 1 Å and, hence, close to experimental uncertainty can be expected for transmembrane regions85. The quality of the modeled starting structures is indirectly supported by the fact that for all three systems, very similar magnitudes of structural deviations along the MD trajectories were found (Fig. 1c). The simulated protein sequences contain a linker region at the N-terminal ends and up to six residues of the cytoplasmic domains at the C-terminal ends, in addition to the TM helices. For β1 and β3, the linker region is almost conserved (Fig. 1b), and the linker of αIIb has been shown to be dynamically unstructured45. Together with crystallographic studies of inactive ectodomains that were unable to obtain structural information on these linkers, indicating their high flexibility48,86,87, we thus do not expect these linker regions to influence the TMD association differentially. In contrast, for the cytoplasmic membrane-proximal region, an influence on maintaining integrin inactivity has been suggested36 (see also below). Of particular relevance is the choice of lipid type, as it has been shown that annular anionic lipids can stabilize αIIbβ3 TMD51. Therefore, our lipid bilayer consisted of zwitterionic 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) lipids, which were shown to interfere much less with inter-TM helix interactions51.
As a second, independent means to investigate the energetics of TMD association and because we cannot expect to observe a dissociation of the TMDs on the time scale of our MD simulations44, we performed biased MD simulations at the atomistic level followed by PMF computations, using established protocols successfully applied previously by us50,88,89 and dCOM-COM as an intuitive reaction coordinate previously applied on similar systems6,90. To our knowledge, so far, the association energetics of integrin TMDs, by computational means, has only been investigated by coarse-grained MD simulations followed by PMF computations6,90. By repeating the PMF computations for parts of the biased simulations, we demonstrated that the PMFs are converged with respect to the overall simulation time per sampling window (Fig. S5). Still, even with sampling times of up to 200 ns per sampling window, pronounced helix tilting or even helix rotation around an axis perpendicular to the helix axis, once the helices are separated by a large enough distance, cannot be expected. Likewise, such sampling times may not be sufficient to yield sampled helix-helix configurations that are completely unbiased from the respective starting structures. While these potential issues may be expected to disfavor separated helix-helix configurations with respect to associated TMD, their impact on our results is expected to be small due to cancellation of errors when comparatively assessing the respective TMD. Comparing quantitative results from the PMFs (Fig. 3a) and subsequent association free energy calculations54 (Table 2) to experimental data lends remarkable support to the quality of the setup, parameterization, and execution of our simulations: (I) For αIIbβ3 TMD, association free energies of −4.33 and −4.84 kcal mol−1 have been determined in 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) lipids by NMR spectroscopy and calorimetry44,51, and our computed ΔG of −3.8 kcal mol−1 (Table 2) is within chemical accuracy of these results; (II) for α5β1, to our knowledge, the energetics of TMD association has not been explicitly probed experimentally. However, from comparing conformational equilibria between the extended-closed and extended-open states for full-length integrin α5β1 versus the α5β1 ectodomain, one may infer that the associated and dissociated states of the TMDs and cytoplasmic domains are almost isoenergetic26, which is in very good agreement with a computed ΔG of 0.5 kcal mol−1 (Table 2); (III) obtaining a barrier height pertinent to kinetics via a PMF has been debated91. Still, when inserting the configurational free energy barrier for αIIbβ3 TMD association of ~1.5 kcal mol−1 (Fig. 3a) as ΔG‡ in the transition state theory equation k = k‡ exp(−ΔG‡/RT)92 and approximating k‡ with a collision frequency of a transmembrane protein of 105 to 106 s−1 93, a rate k of ~8 * 103 to ~8 * 104 s−1 is obtained as a coarse upper bound94, in good to fair agreement with an association rate of αIIbβ3 TMD in phospholipid bicelles of 4.5 * 103 s−1 found by NMR spectroscopy44. As to probing the internal consistency of our results, the shallow contact minimum found in the PMF for α5β1 TMD and the apparent lack of pronounced barriers towards the dissociated state suggest that in MD simulations that are long enough, the two helices should (start to) come apart. In fact, such a tendency is found as dCOM-COM in the unbiased MD simulations of α5β1 TMD is ~1 and 0.6 Å larger than for αIIbβ3 and αVβ3 TMDs (Fig. S7, Table S8). Likewise, internal consistency can be probed by analyzing structural parameters of configurations from unbiased MD simulations and reweighted (“unbiased”) configurations from umbrella sampling simulations. This reveals qualitatively similar results for subtype-specific differences in the distances characterizing OMC formation (dOMC; Fig. 2a versus Fig. 3b, part I), and likewise similar results for αvβ3 and α5β1 TMDs concerning the distance characterizing IMC formation (dIMC; Fig. 2b versus Fig. 3c, part I), although the results for αIIbβ3 deviate in this case. The latter discrepancy may be explained in that distances in Fig. 2b where directly computed from the (tightly associated) αIIbβ3 TMD configurations in the unbiased MD simulations, while three umbrella sampling windows were evaluated for distances in Fig. 3c.
The PMF and association free energy calculations clearly revealed that αIIbβ3 TMD is most stably associated, followed by αvβ3 TMD and α5β1 TMD (Table 2). It had been previously suggested by experiment13,39 and computations49,95 that the two structural motifs OMC and IMC are responsible for the correct TMD packing but if, and how, sequence variations there (Fig. 1b), especially in the OMC, lead to differential TMD association has remained largely elusive. Our unbiased MD simulations reveal that the most stable association of αIIbβ3 TMD is paralleled by this TMD forming more contacts in general across the whole TMD interface (Fig. S3) as well as particularly in the OMC interface (Fig. 1d); also, this TMD shows the most compact OMC (Fig. 2a; Fig. 3b in part I). Although differences in the prevalence of single hydrogen bonds or salt bridges among the three TMDs were not significant, our unbiased MD simulations still suggest a trend, according to which the importance of the membrane-proximal region for TMD association is confirmed, particularly of the R995-D723 salt bridge24,39,51, as this interaction is also most prevalent in αIIbβ3 TMD (>20%, Table 1). Our results may also provide an explanation as to why, in in vivo experiments on transgenic mice carrying a point mutation of the respective D723 of the β1 subunit, a normal integrin function was found32: In α5β1 TMD, the R995-E726 salt bridge is more prevalent than the R995-D723 one. Other questions relate to the role of β3-K716 as a key determinant for the stability at the IMC interface96 and whether the proposed stabilization arises from an engagement of K716 with the surroundings lipid molecules96 or by forming a hydrogen bond with F99217. Our MD simulations revealed that K716 occurs most frequently in either a hydrogen bond across the TMD or a salt bridge with phospholipid head groups in αIIbβ3 TMD, followed by αvβ3 and α5β1 TMDs, consistent with the notion that K716 is important for integrin function in αIIbβ3, but not in α5β125.
Although more work would be required to establish unequivocally a link between differences in K716/phospholipid head group interactions and differences in TMD association free energies, we do not find it unlikely that this link exists. This link would then stress that (differences in) TMD association may be governed by additional factors besides sequence differences, including interactions to annular lipids, as previously demonstrated for the αIIbβ3 TMD51, or membrane tension97. Along these lines, recent work on integrin α5β1 suggested the hypothesis that the non-ligand binding leg domains and N-glycans may have previously unappreciated roles in regulating integrin conformations26. Hence, while considering a subsystem such as the TMD in this study has the benefit of yielding detailed answers under well-defined conditions, at the same time, it leads to the limitation that effects from other parts of the system are not accounted for.
To our knowledge, a novel aspect resulting from our study with respect to the question what governs integrin adhesiveness and affinity in relation to conformational changes is the finding that the two clasps disintegrate in the order “OMC before IMC” upon TMD dissociation. This finding leads to the relevant prediction that the closed state of integrins might not be single but rather comprising several microstates that vary in the extent of TMD association, e.g., with the TMD associated at both OMC and IMC, or with the TMD associated only at the IMC. A similar proposition was made for the extended state(s) of integrins based on the flexibility of integrin legs26,67. Our finding may also be related to, not yet fully understood, results on conformational free energies for intact integrin α5β1 that revealed that the presence of the TMD and cytoplasmic domains favors considerably the bent-closed over the extended-closed conformation26, when one considers that the bent-closed → extended-closed transition may already contain energetic contributions from a partial TMD dissociation in the OMC region. In that respect, computations as performed here may support valuable efforts of gaining affinity information for specific integrin conformational states26 providing access to the energetic contributions of defined subsystems. Finally, we find it striking to note that the order of association free energies of the TMDs (αIIbβ3 ≪ αvβ3 < α5β1 (Table 2)) parallels reports on the basal activity of these integrins (αIIbβ3 ≪ αvβ3 < α5β1)26,27. While it is not possible to establish a direct link between these two series from the comparative simulation studies on TMDs alone, our finding suggests, to our knowledge for the first time, that the sequence composition of the TM helices can have a decisive effect on free energies associated with distinct conformational states of different integrins. A likewise suggestion has been made regarding the strength of interactions between leg domains for integrins αIIbβ3 and αvβ329. Notably, the magnitude of differences in association free energies across the investigated TMDs (~2.7 kcal mol−1; Table 2) is clearly in the range of observed changes in conformational free energies upon activation of full-length integrin α5β1 (~3.7 kcal mol−1)26, suggesting that TMD differences can indeed significantly impact overall conformational integrin energetics.
In summary, αIIbβ3 TMD is most stably associated compared to that of αvβ3 and α5β1, which is related to differences in particular interactions across the TMDs, notably in the OMC. The order of TMD association stability is paralleled by the basal activity of these integrins, which suggests that TMD differences can have a decisive effect on conformational free energies of integrin states. The “OMC before IMC” order of clasp disintegration upon TMD dissociation uniformly identified for all three investigated TMDs suggests that the closed state of integrins might not be single but rather comprising several microstates that vary in the extent of TMD association.
Methods
Generation of starting structures
The starting structure for MD simulations of the αIIbβ3 TMD was obtained from the coordinates of the NMR structure (PDB ID 2K9J) available in the RCSB Protein Data Bank98. The starting structures for MD simulations of the αvβ3 and α5β1 TMDs were generated by homology modeling. The homology models were generated using MODELLER v9.999, and the NMR structure of the αIIbβ3 TMD was used as a template. In the case of αvβ3 integrin, only the αv sequence was modeled based on an alignment with the αIIb TMD with a sequence identity of 45%. In the case of the α5β1 TMD, the α5 and β1 sequences are 40% and 65% identical to those of αIIbβ3 TMD, respectively. The quality of the models was assessed by the QMEANBrane scoring function available in QMEAN server42,43. A global QMEAN score of 0.60, 0.64, and 0.67 was computed for the αIIbβ3, αvβ3, and α5β1 TMDs, respectively.
Setup of simulation systems
The membrane builder tool available on the CHARMM-GUI website100 was used for embedding the TMDs in a pre-equilibrated bilayer of DOPC lipids101 using the replacement method102. The PPM web server103 was used to assess the correct orientation of the αIIbβ3 TMD relative to the hydrocarbon core of the lipid bilayer. The required rectangular simulation box was generated by defining the number of lipids (88 and 85 for the upper and lower leaflet, respectively) and setting a value of 17 Å for the water layer above and below the protein. The total system size is ~60,000 atoms, including TIP3P water molecules76 and Cl- counter ions.
Molecular dynamics simulations
All MD simulations were performed using the AMBER 14 suite of programs47, the ff99SB force field for the proteins78, the Lipid14 force field77 for the lipids, and the TIP3P water model76. The particle mesh Ewald method104 was used to treat long-range electrostatic interactions, and bond lengths involving bonds to hydrogen atoms were constrained using the SHAKE algorithm105. The time step for integrating Newton’s equations of motion was 2 fs with a direct space, nonbonded cutoff of 8 Å. Initially, harmonic restraints with a force constant of 500 kcal mol−1 Å−2 were applied to all solute atoms during the first 250 steps of steepest descent and then reduced to 25.0 kcal mol−1 Å−2 for the second 2500 steps of conjugate gradient minimization and 10 kcal mol−1 Å−2 for the last 2500 steps of conjugate gradient minimization. MD simulations in the NVT (constant number of particles, volume, and temperature) ensemble were carried out for 50 ps, during which the system was heated from 100 to 300 K. Subsequent MD simulations in the NPT (constant number of particles, pressure, and temperature) ensemble were used for 150 ps to adjust the solvent density. In both steps, a force constant of 10 kcal mol−1 Å−2 was applied to all solute and lipid atoms. The production MD simulations of 9 μs length were performed with the GPU version of the program pmemd106 in the tensionless NPT ensemble using the anisotropic Berendsen barostat107 to control the pressure (coupling constant = 1 ps) and the Langevin thermostat107 to control the temperature (coupling constant = 1 ps), as suggested in ref.77.
Potential of mean force computations
Profiles of the free energy of association of αIIbβ3, αvβ3, and α5β1 TMDs were constructed from umbrella sampling MD simulations108,109 in combination with the WHAM method53. As reaction coordination, the distance between the COMs of the TM segments embedded in the membrane was considered (Cα atoms of residues P996-V1015 and D718-I747, for the αIIb and β3 subunits, respectively (equivalent ranges of residues were used for αvβ3 and α5β1)). The initial distance computed from the NMR structure of the TMD of αIIbβ3 integrin is 10.0 Å. To generate starting structures for umbrella sampling, the initial distance was reduced to 8 Å and increased to 11 Å in 0.5 Å steps, and from 11 Å increased to 24 Å in 1 Å steps. Each TMD configuration was inserted in a pre-equilibrated bilayer of DOPC lipids as described above. This resulted in a total of 20 initial systems per TMD. Each of the 20 windows of the three integrin systems was subjected to umbrella sampling simulations, carried out in the NPT ensemble for 200 ns each for dCOM-COM = 8 Å to 20 Å and 70 ns each for dCOM-COM = 21 Å to 24 Å. This resulted in a total of ~3.5 μs of MD simulation time.
Within each umbrella sampling window, a harmonic potential with a force constant of 4 kcal mol−1 Å−2 was applied to restrain the conformations close to the reference point. Force constants of 20 kcal mol−1 Å−2 were also used to restrain conformations whose initial dCOM-COM ranged from 8.0 Å to 10 Å to generate approximately Gaussian-shaped frequency distributions. Otherwise, the parameters described above were used for thermalization and production runs. Finally, to compute the errors at the reference points of the PMF profiles, the Monte Carlo bootstrapping analysis implemented in WHAM using 200 resampling trials was applied.
Estimation of association free energy
An association free energy was estimated from the obtained PMF following the membrane two-body derivation from Johnston et al.54. In brief, the PMF is integrated along the reaction coordinate to calculate an association constant (Ka), transformed to the mole fraction scale (Kx) taking into account the number of lipids NL per surface area A, and this value is used to calculate the difference in free energy between dimer and monomers (ΔG), according to eqs 1–3
| 1 |
| 2 |
| 3 |
where r is the value of the reaction coordinate, w(r) is the PMF at value r, D is the maximum distance at which the protein is still considered a dimer, kB is the Boltzmann constant, and T is the temperature at which the simulations were performed. Additionally, a factor that considers the restriction of the configurational space of the monomers upon dimer formation is included in terms of the sampled angle between the two chains in the dimeric state (eq. 4)
| 4 |
and the accessible space for the monomers, (2π)2. In eq. 4, the angle θa is defined as the angle formed between the vectors connecting the COM of helix 1 with the COM of helix 2 and with the COM of residues V971 to V973 of the latter helix; θb is defined analogously starting from the COM of helix 2 and using the COM of residues L698 to V700 in helix 1.
Analysis of trajectories
For the analysis of the trajectories, ptraj/cpptraj110 of the AmberTools 14 suite of programs was applied. For the unbiased MD simulations, the first 200 ns were not considered for analysis. To evaluate the helix-helix interface (indicated as dCOM-COM-), a maximal distance of 3.5 Å and a minimal angle of 120° were used as exclusion criteria to identify hydrogen bond formation, as was a maximal distance of 4 Å to identify salt bridge formation. To examine the OMC interface, the distance between the Cα atoms of G972/G976 (αIIb subunit) and V700/I704 (β3 subunit) was computed (indicated as dOMC). To evaluate the IMC interface, the distance between the centers of mass of the phenyl rings of F992 or F993 (αIIb subunit) and W715 (β3 subunit) (indicated as dIMC1 and dIMC2, respectively) was computed, and the smaller distance of the two was considered further as dIMC. To evaluate the TMD association, we calculated the total number of native and non-native contacts formed between the two helices. A native contact was defined as a contact satisfying the distance cutoff of 7 Å in the first frame (used as reference frame), a non-native contact as a contact satisfying the distance cutoff of 7 Å without being already found in the reference frame. This definition is according to ref.47. The same set of analyses was also carried out to analyze the configurations sampled during umbrella sampling. For all the configurations t generated, we estimated a weight wt according to eqs 7 and 8 from ref.56. The reweighting is performed over the entire ensemble of each system and, then, normalized by dividing each wt by the sum of all wt for each integrin system.
Statistical analysis
Results from three independent MD simulations are expressed as arithmetic means ± SEM calculated over the time. The overall SEM for each simulated system was calculated according to the law of error propagation (eq. 5):
| 5 |
where the subscripts i = {1, 2, 3} indicate the three trajectories. SEMi was computed following ref.111 (see particularly eqs 11–13 there) by, first, detecting the decorrelation time of an investigated variable along each MD simulation and, second, establishing the effective sample size from that time, which is then used to compute SEMi. In the case of hydrogen bond, salt bridge, and contact analyses, SEM is calculated from the standard deviation (SD) of the three means of the three MD simulations according to eq. 6, assuming that the three MD simulations are statistically independent:
| 6 |
p values related to eq. 5 are calculated according to the Student’s t-test for parametric testing, with , where is the total number of uncorrelated frames within the trajectories based on the statistical inefficiency gt0 and the simulation time range [t0, T] following ref.111; the same test is applied for p values related to eq. 6, with Neff = 3. Differences between mean values are considered to be statistically significant if p < 0.05 and p < 0.001 (indicated as “*” and “**” in figures, respectively) and highly statistically significant if p < 0.0001 (indicated as “***” in figures). The statistical analysis was performed using the R software112 and the pymbar module for MBAR113.
Data availability
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).
Electronic supplementary material
Acknowledgements
We are grateful for computational support and infrastructure provided by the “Zentrum für Informations- und Medientechnologie” (ZIM) at the Heinrich Heine University Düsseldorf and the computing time provided by the John von Neumann Institute for Computing (NIC) to H.G. on the supercomputer JURECA at Jülich Supercomputing Centre (JSC) (project IDs: 6710, 8348, 10307; user ID: HDD11). We are grateful to Stephan Schott Verdugo for the calculation of the thermodynamic quantities.
Author Contributions
H.G. designed research, G.P. performed research, G.P. and H.G. analyzed data and wrote the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-23778-5.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Luo BH, Springer TA. Integrin structures and conformational signaling. Curr. Opin. Cell. Biol. 2006;18:579–586. doi: 10.1016/j.ceb.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ye F, Kim C, Ginsberg MH. Reconstruction of integrin activation. Blood. 2012;119:26–33. doi: 10.1182/blood-2011-04-292128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/S0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 4.Luo BH, Carman CV, Springer TA. Structural basis of integrin regulation and signaling. Annu. Rev. Immunol. 2007;25:619–647. doi: 10.1146/annurev.immunol.25.022106.141618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calderwood DA. Integrin activation. J. Cell. Sci. 2004;117:657–666. doi: 10.1242/jcs.01014. [DOI] [PubMed] [Google Scholar]
- 6.Chng CP, Tan SM. Leukocyte integrin alpha(L)beta(2) transmembrane association dynamics revealed by coarse-grained molecular dynamics simulations. Proteins. 2011;79:2203–2213. doi: 10.1002/prot.23044. [DOI] [PubMed] [Google Scholar]
- 7.Peterson JA, Visentin GP, Newman PJ, Aster RH. A recombinant soluble form of the integrin alpha(IIb)beta(3) (GPIIb-IIIa) assumes an active, ligand-binding conformation and is recognized by GPIIb-IIIa-specific monoclonal, allo-, auto-, and drug-dependent platelet antibodies. Blood. 1998;92:2053–2063. [PubMed] [Google Scholar]
- 8.Wegener KL, Campbell ID. Transmembrane and cytoplasmic domains in integrin activation and protein-protein interactions (Review) Mol. Membr. Biol. 2008;25:376–387. doi: 10.1080/09687680802269886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adair BD, Yeager M. Three-dimensional model of the human platelet integrin alpha(IIb)beta(3) based on electron cryomicroscopy and X-ray crystallography. Proc. Natl. Acad. Sci. USA. 2002;99:14059–14064. doi: 10.1073/pnas.212498199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gottschalk KE. A coiled-coil structure of the alpha(IIb)beta(3) integrin transmembrane and cytoplasmic domains in its resting state. Structure. 2005;13:703–712. doi: 10.1016/j.str.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 11.Luo BH, Springer TA, Takagi J. A specific interface between integrin transmembrane helices and affinity for ligand. PLOS Biol. 2004;2:e153. doi: 10.1371/journal.pbio.0020153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W, et al. A push-pull mechanism for regulating integrin function. Proc. Natl. Acad. Sci. USA. 2005;102:1424–1429. doi: 10.1073/pnas.0409334102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo BH, Carman CV, Takagi J, Springer TA. Disrupting integrin transmembrane domain heterodimerization increases ligand binding affinity, not valency or clustering. Proc. Natl. Acad. Sci. USA. 2005;102:3679–3684. doi: 10.1073/pnas.0409440102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Partridge AW, Liu S, Kim S, Bowie JU, Ginsberg MH. Transmembrane domain helix packing stabilizes integrin alpha(IIb)beta(3) in the low affinity state. J. Biol. Chem. 2005;280:7294–7300. doi: 10.1074/jbc.M412701200. [DOI] [PubMed] [Google Scholar]
- 15.Lau TL, Kim C, Ginsberg MH, Ulmer TS. The structure of the integrin alpha(IIb)beta(3) transmembrane complex explains integrin transmembrane signalling. EMBO J. 2009;28:1351–1361. doi: 10.1038/emboj.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang J, et al. Structure of an integrin alpha(IIb)beta(3) transmembrane-cytoplasmic heterocomplex provides insight into integrin activation. Proc. Natl. Acad. Sci. USA. 2009;106:17729–17734. doi: 10.1073/pnas.0909589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu J, et al. The structure of a receptor with two associating transmembrane domains on the cell surface: integrin alpha(IIb)beta(3) Mol. Cell. Biol. 2009;34:234–249. doi: 10.1016/j.molcel.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo BH, Springer TA, Takagi J. Stabilizing the open conformation of the integrin headpiece with a glycan wedge increases affinity for ligand. Proc. Natl. Acad. Sci. USA. 2003;100:2403–2408. doi: 10.1073/pnas.0438060100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tng E, Tan SM, Ranganathan S, Cheng M, Law SK. The integrin alpha(L)beta(2) hybrid domain serves as a link for the propagation of activation signal from its stalk regions to the I-like domain. J. Biol. Chem. 2004;279:54334–54339. doi: 10.1074/jbc.M407818200. [DOI] [PubMed] [Google Scholar]
- 20.Hantgan RR, Paumi C, Rocco M, Weisel JW. Effects of ligand-mimetic peptides Arg-Gly-Asp-X (X = Phe, Trp, Ser) on alpha(IIb)beta(3) integrin conformation and oligomerization. Biochemistry. 1999;38:14461–14474. doi: 10.1021/bi9907680. [DOI] [PubMed] [Google Scholar]
- 21.Plow EF, Haas TK, Zhang L, Loftus J, Smith JW. Ligand binding to integrins. J. Biol. Chem. 2000;275:21785–21788. doi: 10.1074/jbc.R000003200. [DOI] [PubMed] [Google Scholar]
- 22.Takagi J, Erickson HP, Springer TA. C-terminal opening mimics ‘inside-out’ activation of integrin alpha(5)beta(1) Nat. Struct. Biol. 2001;8:412–416. doi: 10.1038/87569. [DOI] [PubMed] [Google Scholar]
- 23.Shattil SJ, Kim C, Ginsberg MH. The final steps of integrin activation: the end game. Nat. Rev. Mol. Cell. Biol. 2010;11:288–300. doi: 10.1038/nrm2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vinogradova O, et al. A structural mechanism of integrin alpha(IIb)beta(3) “inside-out” activation as regulated by its cytoplasmic face. Cell. 2002;110:587–597. doi: 10.1016/S0092-8674(02)00906-6. [DOI] [PubMed] [Google Scholar]
- 25.Lu Z, et al. Implications of the differing roles of the beta(1) and beta(3) transmembrane and cytosplasmic domains for integrin function. eLife. 2016;5:e18633. doi: 10.7554/eLife.18633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, et al. Conformational equilibria and intrinsic affinities define integrin activation. EMBO J. 2017;36:629–645. doi: 10.15252/embj.201695803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bazzoni G, Ma L, Blue ML, Hemler ME. Divalent cations and ligands induce conformational changes that are highly divergent among beta(1) integrins. J. Biol. Chem. 1998;273:6670–6678. doi: 10.1074/jbc.273.12.6670. [DOI] [PubMed] [Google Scholar]
- 28.Askari JA, Buckley PA, Mould AP, Humphries MJ. Linking integrin conformation to function. J. Cell Sci. 2009;122:165–170. doi: 10.1242/jcs.018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamata T, Handa M, Sato Y, Ikeda Y, Aiso S. Membrane-proximal alpha/beta stalk interactions differentially regulate integrin activation. J. Biol. Chem. 2005;280:24775–24783. doi: 10.1074/jbc.M409548200. [DOI] [PubMed] [Google Scholar]
- 30.Weljie AM, Hwang PM, Vogel HJ. Solution structures of the cytoplasmic tail complex from platelet integrin alpha(IIb) and beta(3) subunits. Proc. Natl. Acad. Sci. USA. 2002;99:5878–5883. doi: 10.1073/pnas.092515799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Toole TE, et al. Integrin cytoplasmic domains mediate inside-out signal transduction. J. Cell. Biol. 1994;124:1047–1059. doi: 10.1083/jcb.124.6.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Czuchra A, Meyer H, Legate KR, Brakebusch C, Fassler R. Genetic analysis of beta(1) integrin “activation motifs” in mice. J. Cell. Biol. 2006;174:889–899. doi: 10.1083/jcb.200604060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Campbell ID, Humphries MJ. Integrin structure, activation, and interactions. Cold Spring Harb. Perspect. Biol. 2011;3:a004994. doi: 10.1101/cshperspect.a004994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma YQ, Qin J, Plow EF. Platelet integrin alpha(IIb)beta(3): activation mechanisms. J. Thromb. Haemost. 2007;5:1345–1352. doi: 10.1111/j.1538-7836.2007.02537.x. [DOI] [PubMed] [Google Scholar]
- 35.Schneider D, Engelman DM. Involvement of transmembrane domain interactions in signal transduction by alpha/beta integrins. J. Biol. Chem. 2004;279:9840–9846. doi: 10.1074/jbc.M312749200. [DOI] [PubMed] [Google Scholar]
- 36.Kim C, Ye F, Ginsberg MH. Regulation of integrin activation. Annu. Rev. Cell. Dev. Biol. 2011;27:321–345. doi: 10.1146/annurev-cellbio-100109-104104. [DOI] [PubMed] [Google Scholar]
- 37.Hughes PE, O’Toole TE, Ylanne J, Shattil SJ, Ginsberg MH. The conserved membrane-proximal region of an integrin cytoplasmic domain specifies ligand binding affinity. J. Biol. Chem. 1995;270:12411–12417. doi: 10.1074/jbc.270.21.12411. [DOI] [PubMed] [Google Scholar]
- 38.Strandberg E, Killian JA. Snorkeling of lysine side chains in transmembrane helices: how easy can it get? FEBS Lett. 2003;544:69–73. doi: 10.1016/S0014-5793(03)00475-7. [DOI] [PubMed] [Google Scholar]
- 39.Hughes PE, et al. Breaking the integrin hinge. A defined structural constraint regulates integrin signaling. J. Biol. Chem. 1996;271:6571–6574. doi: 10.1074/jbc.271.12.6571. [DOI] [PubMed] [Google Scholar]
- 40.Haas TA, Plow EF. Development of a structural model for the cytoplasmic domain of an integrin. Protein Eng. 1997;10:1395–1405. doi: 10.1093/protein/10.12.1395. [DOI] [PubMed] [Google Scholar]
- 41.Vallar L, et al. Divalent cations differentially regulate integrin alpha(IIb) cytoplasmic tail binding to beta(3) and to calcium- and integrin-binding protein. J. Biol. Chem. 1999;274:17257–17266. doi: 10.1074/jbc.274.24.17257. [DOI] [PubMed] [Google Scholar]
- 42.Studer G, Biasini M, Schwede T. Assessing the local structural quality of transmembrane protein models using statistical potentials (QMEANBrane) Bioinformatics. 2014;30:I505–I511. doi: 10.1093/bioinformatics/btu457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benkert P, Tosatto SCE, Schomburg D. QMEAN: A comprehensive scoring function for model quality assessment. Proteins: Struct., Funct., Bioinf. 2008;71:261–277. doi: 10.1002/prot.21715. [DOI] [PubMed] [Google Scholar]
- 44.Situ AJ, Schmidt T, Mazumder P, Ulmer TS. Characterization of membrane protein interactions by isothermal titration calorimetry. J. Mol. Biol. 2014;426:3670–3680. doi: 10.1016/j.jmb.2014.08.020. [DOI] [PubMed] [Google Scholar]
- 45.Schmidt T, et al. A Conserved Ectodomain-Transmembrane Domain Linker Motif Tunes the Allosteric Regulation of Cell Surface Receptors. J. Biol. Chem. 2016;291:17536–17546. doi: 10.1074/jbc.M116.733683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Galindo-Murillo R, Roe DR, Cheatham TE., 3rd Convergence and reproducibility in molecular dynamics simulations of the DNA duplex d(GCACGAACGAACGAACGC) Biochim. Biophys. Acta. 2015;1850:1041–1058. doi: 10.1016/j.bbagen.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Case DA, et al. The Amber biomolecular simulation programs. J. Comp. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu J, et al. Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces. Mol. Cell. Biol. 2008;32:849–861. doi: 10.1016/j.molcel.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berger BW, et al. Consensus motif for integrin transmembrane helix association. Proc. Natl. Acad. Sci. USA. 2010;107:703–708. doi: 10.1073/pnas.0910873107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ciupka D, Gohlke H. On the potential alternate binding change mechanism in a dimeric structure of Pyruvate Phosphate Dikinase. Sci. Rep. 2017;7:8020. doi: 10.1038/s41598-017-08521-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmidt T, et al. Annular anionic lipids stabilize the integrin alpha(IIb)beta(3) transmembrane complex. J. Biol. Chem. 2015;290:8283–8293. doi: 10.1074/jbc.M114.623504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kastner J. Umbrella sampling. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2011;1:932–942. doi: 10.1002/wcms.66. [DOI] [Google Scholar]
- 53.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. The Weighted Histogram Analysis Method for Free-Energy Calculations onBiomolecules .1. The Method. J. Comp. Chem. 1992;13:1011–1021. doi: 10.1002/jcc.540130812. [DOI] [Google Scholar]
- 54.Johnston JM, Wang H, Provasi D, Filizola M. Assessing the Relative Stability of Dimer Interfaces in G Protein-Coupled Receptors. PLOS Comp. Biol. 2012;8:e1002649. doi: 10.1371/journal.pcbi.1002649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hénin J, Pohorille A, Chipot C. Insights into the Recognition and Association of Transmembrane α-Helices. The Free Energy of α-Helix Dimerization in Glycophorin A. J. Am. Chem. Soc. 2005;127:8478–8484. doi: 10.1021/ja050581y. [DOI] [PubMed] [Google Scholar]
- 56.Moradi M, Enkavi G, Tajkhorshid E. Atomic-level characterization of transport cycle thermodynamics in the glycerol-3-phosphate: phosphate antiporter. Nat. Commun. 2015;6:8393. doi: 10.1038/ncomms9393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim M, Carman CV, Springer TA. Bidirectional transmembrane signaling by cytoplasmic domain separation in integrins. Science. 2003;301:1720–1725. doi: 10.1126/science.1084174. [DOI] [PubMed] [Google Scholar]
- 58.Yin H, et al. Activation of platelet alpha(IIb)beta(3) by an exogenous peptide corresponding to the transmembrane domain of alpha(IIb) J. Biol. Chem. 2006;281:36732–36741. doi: 10.1074/jbc.M605877200. [DOI] [PubMed] [Google Scholar]
- 59.Dana N, Fathallah DM, Arnaout MA. Expression of a soluble and functional form of the human beta(2) integrin CD11b/CD18. Proc. Natl. Acad. Sci. USA. 1991;88:3106–3110. doi: 10.1073/pnas.88.8.3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mehta RJ, et al. Transmembrane-truncated alpha(V)beta(3) integrin retains high affinity for ligand binding: evidence for an ‘inside-out’ suppressor? Biochem. J. 1998;330(Pt 2):861–869. doi: 10.1042/bj3300861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takagi J, Petre BM, Walz T, Springer TA. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell. 2002;110:599–511. doi: 10.1016/S0092-8674(02)00935-2. [DOI] [PubMed] [Google Scholar]
- 62.Takagi J, Strokovich K, Springer TA, Walz T. Structure of integrin alpha(5)beta(1) in complex with fibronectin. EMBO J. 2003;22:4607–4615. doi: 10.1093/emboj/cdg445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiao T, Takagi J, Coller BS, Wang JH, Springer TA. Structural basis for allostery in integrins and binding to fibrinogen-mimetic therapeutics. Nature. 2004;432:59–67. doi: 10.1038/nature02976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schurpf T, Springer TA. Regulation of integrin affinity on cell surfaces. EMBO J. 2011;30:4712–4727. doi: 10.1038/emboj.2011.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu J, Zhu J, Springer TA. Complete integrin headpiece opening in eight steps. J. Cell Biol. 2013;201:1053–1068. doi: 10.1083/jcb.201212037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen X, et al. Requirement of open headpiece conformation for activation of leukocyte integrin alpha(X)beta(2) Proc. Natl. Acad. Sci. USA. 2010;107:14727–14732. doi: 10.1073/pnas.1008663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takagi J, Springer TA. Integrin activation and structural rearrangement. Immunol. Rev. 2002;186:141–163. doi: 10.1034/j.1600-065X.2002.18613.x. [DOI] [PubMed] [Google Scholar]
- 68.Su Y, et al. Relating conformation to function in integrin alpha(5)beta(1) Proc. Natl. Acad. Sci. USA. 2016;113:E3872–E3881. doi: 10.1073/pnas.1605074113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gohlke H, Schmitz B, Sommerfeld A, Reinehr R, Häussinger D. alpha(5)beta(1)-integrins are sensors for tauroursodeoxycholic acid in hepatocytes. Hepatology. 2013;57:1117–1129. doi: 10.1002/hep.25992. [DOI] [PubMed] [Google Scholar]
- 70.Schwarzbauer JE, DeSimone DW. Fibronectins, their fibrillogenesis, and in vivo functions. Cold Spring Harb. Perspect. Biol. 2011;3:a005041. doi: 10.1101/cshperspect.a005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gottschalk KE, Adams PD, Brunger AT, Kessler H. Transmembrane signal transduction of the alpha(IIb)beta(3) integrin. Protein Sci. 2002;11:1800–1812. doi: 10.1110/ps.4120102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Müller MA, et al. Cytoplasmic salt bridge formation in integrin alpha(V)beta(3) stabilizes its inactive state affecting integrin-mediated cell biological effects. Cell Signal. 2014;26:2493–2503. doi: 10.1016/j.cellsig.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 73.Chavent M, Duncan AL, Sansom MS. Molecular dynamics simulations of membrane proteins and their interactions: from nanoscale to mesoscale. Curr. Opin. Struct. Biol. 2016;40:8–16. doi: 10.1016/j.sbi.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kalli AC, Campbell ID, Sansom MS. Multiscale simulations suggest a mechanism for integrin inside-out activation. Proc. Natl. Acad. Sci. USA. 2011;108:11890–11895. doi: 10.1073/pnas.1104505108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vararattanavech A, et al. A transmembrane polar interaction is involved in the functional regulation of integrin alpha(L)beta(2) J. Mol. Biol. 2010;398:569–583. doi: 10.1016/j.jmb.2010.03.027. [DOI] [PubMed] [Google Scholar]
- 76.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 77.Dickson CJ, et al. Lipid14: The Amber Lipid Force Field. J. Chem. Theory Comput. 2014;10:865–879. doi: 10.1021/ct4010307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hornak V, et al. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reinehr R, Gohlke H, Sommerfeld A. vom Dahl, S. & Häussinger, D. Activation of integrins by urea in perfused rat liver. J. Biol. Chem. 2010;285:29348–29356. doi: 10.1074/jbc.M110.155135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bopp B, et al. Design and biological testing of peptidic dimerization inhibitors of human Hsp90 that target the C-terminal domain. Biochim. Biophys. Acta. 2016;1860:1043–1055. doi: 10.1016/j.bbagen.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 81.Ciglia E, et al. Resolving hot spots in the C-terminal dimerization domain that determine the stability of the molecular chaperone Hsp90. PLOS one. 2014;9:e96031. doi: 10.1371/journal.pone.0096031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Metz A, et al. Hot spots and transient pockets: predicting the determinants of small-molecule binding to a protein-protein interface. J. Chem. Inf. Model. 2012;52:120–133. doi: 10.1021/ci200322s. [DOI] [PubMed] [Google Scholar]
- 83.Perez A, MacCallum JL, Brini E, Simmerling C, Dill KA. Grid-based backbone correction to the ff12SB protein force field for implicit-solvent simulations. J. Chem. Theory. Comput. 2015;11:4770–4779. doi: 10.1021/acs.jctc.5b00662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Maier JA, et al. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Forrest LR, Tang CL, Honig B. On the accuracy of homology modeling and sequence alignment methods applied to membrane proteins. Biophys. J. 2006;91:508–517. doi: 10.1529/biophysj.106.082313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xiong JP, et al. Crystal structure of the complete integrin alpha(V)beta(3) ectodomain plus an alpha/beta transmembrane fragment. J. Cell. Biol. 2009;186:589–600. doi: 10.1083/jcb.200905085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dong X, et al. alpha(V)beta(3) integrin crystal structures and their functional implications. Biochemistry. 2012;51:8814–8828. doi: 10.1021/bi300734n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Minges A, et al. Structural intermediates and directionality of the swiveling motion of Pyruvate Phosphate Dikinase. Sci. Rep. 2017;7:45389. doi: 10.1038/srep45389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Koller AN, Bozilovic J, Engels JW, Gohlke H. Aromatic N versus aromatic F: bioisosterism discovered in RNA base pairing interactions leads to a novel class of universal base analogs. Nucleic Acids Res. 2010;38:3133–3146. doi: 10.1093/nar/gkp1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sengupta D, Marrink SJ. Lipid-mediated interactions tune the association of glycophorin A helix and its disruptive mutants in membranes. Phys. Chem. Chem. Phys. 2010;12:12987–12996. doi: 10.1039/c0cp00101e. [DOI] [PubMed] [Google Scholar]
- 91.Dellago C, Bolhuis PG, Csajka FS, Chandler D. Transition path sampling and the calculation of rate constants. J. Phys. Chem. 1998;108:1964–1977. doi: 10.1063/1.475562. [DOI] [Google Scholar]
- 92.Eyring, H. The Activated Complex in Chemical Reactions. J. Chem. Phys. 3 (1935).
- 93.Poo M, Cone RA. Lateral diffusion of rhodopsin in the photoreceptor membrane. Nature. 1974;247:438–441. doi: 10.1038/247438a0. [DOI] [PubMed] [Google Scholar]
- 94.Rhee YM, Pande VS. One-dimensional reaction coordinate and the corresponding potential of mean force from commitment probability distribution. J. Phys. Chem. B. 2005;109:6780–6786. doi: 10.1021/jp045544s. [DOI] [PubMed] [Google Scholar]
- 95.Kalli A, Campbell ID, Sansom SP. Multiscale simulations suggest a mechanism for integrin inside-out activation. PNAS. 2011;108:11890–11895. doi: 10.1073/pnas.1104505108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kim C, et al. Basic amino-acid side chains regulate transmembrane integrin signalling. Nature. 2012;481:209–213. doi: 10.1038/nature10697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ferraris GM, et al. The interaction between uPAR and vitronectin triggers ligand-independent adhesion signalling by integrins. EMBO J. 2014;33:2458–2472. doi: 10.15252/embj.201387611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bernstein, F. C., et al. The Protein Data Bank: A Computer-based Archival File For Macromolecular Structures. J. Mol. Biol. 112, (1977). [DOI] [PubMed]
- 99.Enswar N, et al. Comparative Protein Structure Modeling using MODELLER. Curr. Protoc. Bioinformatics. 2006;15:5.6.1–5.6.30. doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jo S, Kim T, Lyer VG, Lm W. CHARMM-GUI: A Web-Based Graphical User Interface for CHARMM. J. Comput. Chem. 2008;29:1860–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 101.Domański J, Stansfeld P, Sansom MSP, Beckstein O. Lipidbook: A Public Repository for Force Field Parameters Used in Membrane Simulations. J. Memb. Biol. 2010;236:255–258. doi: 10.1007/s00232-010-9296-8. [DOI] [PubMed] [Google Scholar]
- 102.Jo S, Kim T, Im W. Automated Builder and Database of Protein/Membrane Complexes for Molecular Dynamics Simulations. PLOS one. 2007;2:e880. doi: 10.1371/journal.pone.0000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 2012;40:D370–D376. doi: 10.1093/nar/gkr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cheatham TE, Miller JL, Fox T, Darden TA, Kollman PA. Molecular-Dynamics simulations on solvated biomolecular systems - the particle Mesh Ewald method leads to stable trajectories of DNA, RNA, and proteins. J. Am. Chem. Soc. 1995;117:4193–4194. doi: 10.1021/ja00119a045. [DOI] [Google Scholar]
- 105.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical-Integration of Cartesian Equations of Motion of a System with Constraints - Molecular-Dynamics of N-Alkanes. J. Comput. Phys. 1977;23:327–341. doi: 10.1016/0021-9991(77)90098-5. [DOI] [Google Scholar]
- 106.Salomon-Ferrer R, Gotz AW, Poole D, Le Grand S, Walker RC. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle Mesh Ewald. J. Chem. Theory Comput. 2013;9:3878–3888. doi: 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- 107.Berendsen HJC, Postma JPM, van Gunsteren WF, Dinola A, Haak JR. Molecular-Dynamics with Coupling to an External Bath. J. Chem. Phys. 1984;81:3684–3690. doi: 10.1063/1.448118. [DOI] [Google Scholar]
- 108.Torrie GM, Valleau JP. Monte-Carlo Free-Energy Estimates Using Non-Boltzmann Sampling - Application to Subcritical Lennard-Jones Fluid. Chem. Phys. Lett. 1974;28:578–581. doi: 10.1016/0009-2614(74)80109-0. [DOI] [Google Scholar]
- 109.Torrie GM, Valleau JP. Non-Physical Sampling Distributions in Monte-Carlo Free-Energy Estimation - Umbrella Sampling. J. Comp. Phys. 1977;23:187–199. doi: 10.1016/0021-9991(77)90121-8. [DOI] [Google Scholar]
- 110.Roe DR, Cheatham TE. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 111.Chodera JD. A Simple Method for Automated Equilibration Detection in Molecular Simulations. J. Chem. Theory Comput. 2016;12:1799–1805. doi: 10.1021/acs.jctc.5b00784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Team, R. D. C. R: a language and environment for statistical computing, http://www.R-project.org (2010).
- 113.Shirts MR, Chodera JD. Statistically optimal analysis of samples from multiple equilibrium states. J. Chem. Phys. 2008;129:124105. doi: 10.1063/1.2978177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).