Abstract
Carbapenem-resistant Enterobacteriaceae (CRE) has been considered a serious global threat, but carbapenem resistance remains relatively uncommon in E. cloacae, especially in China. The aim of this study was to characterize carbapenem-resistant E. cloacae (CR-ECL) isolates from 2012 to 2016 in Southwest China. Our study revealed that 20 (15.2%) of the 132 CR-ECL isolates obtained from patients were identified as NDM-1, with most isolates carrying the IncFIIA plasmids. Notably, we initially observed that the E. cloacae strain co-harbored NDM-1 and IMP-8 carbapenemases simultaneously. Analysis of the genetic environment of these two genes has revealed that the highly conserved regions (blaNDM-1-bleMBL-trpF-tat) are associated with the dissemination of NDM-1, while IS26, intI1, and tniC could be involved in the spread of IMP-8. Molecular epidemiology studies showed the nosocomial outbreak caused by NDM-1-producing E. cloacae ST88. Transferring from another hospital and previous carbapenem exposure were identified as independent risk factors for the acquisition of NDM-1-producing E. cloacae. These findings emphasize the need for intensive surveillance and precautions to monitor the further spread of NDM-1 in China.
Keywords: carbapenems, resistance, E. cloacae, outbreak investigation, risk factor
Introduction
As the utility of carbapenem has increased worldwide over the last decade, the emergence and dissemination of carbapenem-resistant Enterobacteriaceae (CRE) have become worsening situations (Cornaglia et al., 2011). Recent surveillance data from the United States, Europe, and South Asia has observed a high resistance rate in CRE, among which Klebsiella pneumoniae and Escherichia coli were the most prevalent (Nordmann et al., 2009; Pollett et al., 2014; Hsu et al., 2017). Similar results were obtained from the CHINET surveillance system, which showed that carbapenem-resistant K. pneumoniae (CRKP) appeared to be most common in China and increased rapidly from 9.4% in 2011 to 15.6% in 2015 (Tian et al., 2016). Unexpectedly, however, surveys released by the Chongqing Resistance Monitoring Network (CQRMN) found that the rate of carbapenem resistance showed the highest in E. cloacae among CRE isolates during the period 2011–2015, while resistance rates for K. pneumoniae and E. coli remained relatively stable and had an average of 2.4 and 1.4%, respectively, compared to 7.8% for E. cloacae (Figure 1). Therefore, investigating carbapenem-resistant E. cloacae is of utmost importance for therapy and control in our region.
FIGURE 1.
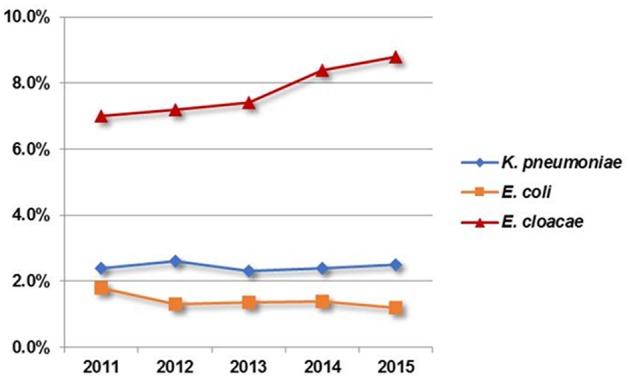
Distribution of carbapenem resistant Enterobacteriaceae from 2011 to 2015 according to the Chongqing Resistance Monitoring Network.
In China, the first CR-ECL was a KPC-producing strain isolated from Shanghai in 2010 (Wu et al., 2010). Later, carbapenemases IMP, VIM, and, recently, NDM have been reported in clinical E. cloacae isolates from different geographical regions (Dai et al., 2013; Yang et al., 2014; Liu et al., 2015), among which NDM-1 should be especially worrisome as the gene encoding this enzyme is often located on mobile genetic elements that can be easily transferred between different species (Rasheed et al., 2013). To date, there have been few studies of outbreak due to NDM-1 producing E. cloacae (Stoesser et al., 2015; Mahida et al., 2017); however, to the best of our knowledge, there are no previous studies of nosocomial outbreaks with E. cloacae that produce NDM-1 in China. More importantly, strains co-producing various carbapenemases are worth being concerned about, since these bacteria may confer a higher-level resistance to carbapenem that further reduces the therapeutic choices. To date, there has been no report of clinical E. cloacae strains simultaneously producing NDM-1 and IMP-8 carbapenemases, and knowledge gaps regarding the genetic context and plasmid characterization of these isolates remain. Additionally, the risk factors and clinical outcomes of nosocomial patients with NDM-1 producing E. cloacae have not been systematically evaluated.
Therefore, the objectives of this study were the following: (i) to describe the prevalence of clinical CR-ECL isolates collected successively for approximately 4 years, (ii) to identify the mechanisms and clonal relatedness among these CR-ECL strains, (iii) to report the first outbreak of NDM-1 producing E. cloacae in China, (iv) to examine the genetic context of NDM-1 and IMP-8, and (v) to evaluate the risk factors and clinical outcomes of NDM-1 positive E. cloacae infections in hospitalized patients.
Materials and Methods
Study Setting and Bacterial Strains
This retrospective study was performed in a 3,200-bed tertiary university-affiliated hospital and associated two branch hospitals located in Southwest China. Between March 2012 and 2016, 1,146 clinical E. cloacae strains were collected in these hospitals. We chose the first isolate from the patient and excluded duplicate isolates from the same patient. All isolates were identified at the species level and routine antimicrobial susceptibility testing was performed by using the VITEK2 compact or VITEK MS (bioMerieux, Hazelwood, MO, United States) automated system. The isolates were collected by the rapid freezing method and stored at -80°C for further analysis. Isolates were included in the study if they were resistant to at least one of the carbapenems by the broth microdilution method, with the criteria of MICs of ≥ 2 μg/mL for ertapenem, ≥ 4 μg/mL for imipenem, or ≥ 4 μg/mL for meropenem. For research involving biohazards, the standard biosecurity or institutional safety procedures have been carried out.
Antimicrobial Susceptibility Testing
All isolates were tested for antibiotic susceptibility to ceftazidime (CAZ), ceftriaxone (CRO), cefepime (FEP), gentamicin (GM), tobramycin (TOB), ciprofloxacin (CIP), and levofloxacin (LEV) by using AST GN13 cards on the VITEK2 compact system. MICs of ertapenem (ETP), imipenem (IPM), meropenem (MEM), colistin (CS), and tigecycline (TGC) were determined using the broth microdilution method, and results were interpreted according to the CLSI M100-S25 interpretive criteria [Clinical and Laboratory Standards Institute [CLSI], 2015]. E. cloacae ATCC 13847 was used as the quality control strain for susceptibility testing. MIC50, MIC90, and the MIC range of each tested agent were also analyzed in our study.
Detection of Antibiotic-Resistant Genes, Expression of Outer Membrane Proteins, and Activity of the Efflux Pump
The PCR was performed to detect for the presence of carbapenemase-related genes, including blaKPC, blaNDM, blaV IM, blaIMP, blaSME, and blaOXA-48, and sequencing was used to confirm the variants of these carbapenemase genes. Moreover, ESBLs, AmpC, resistant genes for aminoglycosides and fluoroquinolones, ompF and ompC genes were also determined by using primers as described previously (Doumith et al., 2009; Zhang et al., 2014). Additionally, the levels of expression of porin-encoding genes were determined from mRNA levels by real-time reverse transcription (RT)-PCR according to the previously described protocols (Bustin et al., 2009). The activity of the efflux pump was examined by comparing the MICs to ertapenem among resistant isolates in the presence and absence of carbonyl cyanide m-chlorophenylhydrazone (CCCP) as an efflux pump inhibitor (Chollet et al., 2004). A twofold decrease in MIC after the addition of CCCP was considered positive results. Plates containing CCCP but no carbapenem were used as controls.
Conjugation, Transformation, and Plasmid Analysis
To assess whether the carbapenemase-producing genes were located on the plasmids and to assess the transferability of these genes, the co-producing blaNDM-1 and blaIMP-8 strains (ECL-86) were conducted by conjugation with E. coli EC600 and by transformation with E. coli DH5α. For conjugative assays, transconjugants were selected on Mueller-Hinton agar plates supplemented with a combination of 8 mg/L ampicillin and 256 mg/L rifampicin. For transformative assays, transformants were selected on Mueller-Hinton agar plates containing 64 mg/L ampicillin. The transconjugants and transformants were tested for antimicrobial susceptibility by the VITEK2 compact system, and they confirmed the presence of resistance determinants by PCR. In addition, all NDM-1-encoding plasmids were characterized for the incompatibility groups by using the PCR-based replicon typing method described previously (Carattoli et al., 2005).
Genetic Environments of NDM-1-Carrying Plasmids and IMP-8-Carrying Plasmids
To investigate the genetic contests in the NDM-1 and IMP-8, we selected 14 representative strains producing single NDM-1 (according to the DiversiLab’s patterns) and one strain co-producing NDM-1and IMP-8 to perform PCR mapping and sequencing in our study. The genetic environment surrounding NDM-1 was established by PCR mapping and subsequent sequencing of the flanking regions. Moreover, we also performed PCR mapping to elucidate the genetic structures of NDM-1 and IMP-8 in E. cloacae ECL-86. PCR amplicons were sequenced and the DNA sequences obtained were compared to those available in the NCBI GenBank database.
Molecular Epidemiological Study
The clonal relationships of isolates harboring the carbapenemase-related genes were further determined by repetitive sequence-based PCR (rep-PCR) typing using the DiversiLab system. Isolates with ≥ 95% similarity were considered of the same rep-PCR type. Multilocus sequence typing (MLST) was performed using amplification of internal fragments of the seven housekeeping genes of carbapenemase-producing E. cloacae isolates according to the MLST1 website.
Risk Factors and Clinical Outcomes of NDM-1-Producing E. cloacae Isolates
We conducted a retrospective case–control study to evaluate the risk factors and clinical outcomes for the isolation of NDM-1-producing E. cloacae. All patients with clinically obtained NDM-1-positive CR-ECL isolates were included as cases. Controls were identified as patients without NDM-1 producers and excluded if isolates harbored other carbapenemase genes, such as KPC-2, IMP-4, and IMP-8. Clinical and epidemiological data were retrieved from the medical record and clinical microbiology laboratory databases included the following parameters: demographics, underlying diseases and primary admission diagnosis, invasive devices prior to the isolation of NDM-1-producing E. cloacae, previous antibiotic exposure within 3 months, and clinical outcomes.
Statistical Analysis
All analyses were performed using SPSS v.22.0 software (SPSS Inc., Chicago, IL, United States). Univariate analyses were performed separately for each of the variables. Categorical variables were calculated using a chi-square test or Fisher’s exact test as appropriate. Continuous variables were calculated using Student’s t-test (normally distributed variables) and Wilcoxon rank-sum test (non-normally distributed variables) as appropriate. The odds ratio (OR) and 95% confidence interval (CI) were calculated to evaluate the strength of any association. All variables with a P-value of ≤ 0.10 in the univariate analyses were considered for inclusion in the multivariate logistic regression model. For all calculations, statistical significance was defined at P < 0.05 for the two-tailed tests.
Ethical Considerations
The data and the samples analyzed in the present study were obtained in accordance with the standards and approved by the Chongqing Medical University Institutional Review Board and Biomedical Ethics Committee. For this study, samples were collected at the Microbiology Laboratory of our hospital, with no contact with the patient. This study was retrospective and there was no patient identification performed during data collection. Therefore, the Ethics Committee determined that informed consent was not required.
Results
General Characteristics and Antimicrobial Susceptibility of CR-ECL Isolates
During the investigation period, a total of 138 strains were identified to be resistant to at least one of the carbapenems and met the study criteria for CR-ECL. These non-duplicated isolates were mainly cultured from urine (n = 55), followed by the respiratory tract (n = 32), wound secretion (n = 28), and blood (n = 21). As shown in Table 1, all CR-ECL isolates were observed in ertapenem resistance, with 60.3% of the strains still performing susceptible to meropenem and imipenem, respectively. In addition, the resistance rate to cephalosporins was relatively high in general. Of these isolates, over half of the CR-ECL strains exhibited resistance to the aminoglycoside and fluoroquinolone agents tested. However, the highest susceptibility rates to the tested antibiotics were to tigecycline (87.7%) and colistin (76.8%). Compared to carbapenemase-negative isolates, our results showed that resistance was a significantly greater proportion of E. cloacae isolates that were carbapenemase-positive for most tested agents, including imipenem, meropenem, cefepime, ciprofloxacin, levofloxacin, gentamycin, and tobramycin. Notably, 65.9% (91/138) of CR-ECL isolates were identified to be MDR as they were resistant to three or more classes of antimicrobial agents.
Table 1.
Antimicrobial susceptibility of CR-ECL isolates with or without carbapenemase.
| Antimicrobial agents | Total (N = 138) |
Carbapenemase positive (N = 27) |
Carbapenemase negative (N = 111) |
P-value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| R (%) | R | MIC50 | MIC90 | Range | R | MIC50 | MIC90 | Range | ||
| Ertapenem | 138 (100.0) | 27 (100.0) | 32 | 128 | 4–128 | 111 (100.0) | 8 | 32 | 2–64 | – |
| Imipenem | 31 (39.7) | 22 (81.5) | 8 | 32 | 0.25–128 | 9 (8.1) | 0.5 | 2 | 0.25–32 | < 0.001 |
| Meropenem | 31 (39.7) | 21 (77.8) | 16 | 128 | 0.5–128 | 10 (9.0) | 0.5 | 2 | 0.25–128 | < 0.001 |
| Ceftriaxone | 136 (98.6) | 27 (100.0) | 128 | 256 | 64–512 | 109 (98.2) | 64 | 128 | 1–512 | 0.482 |
| Ceftazidime | 134 (97.1) | 27 (100.0) | 128 | 512 | 64–512 | 107 (96.4) | 128 | 128 | 1–512 | 0.317 |
| Cefepime | 105 (76.1) | 27 (100.0) | 128 | 512 | 64–512 | 78 (70.3) | 64 | 256 | 1–512 | 0.001 |
| Ciprofloxacin | 74 (53.6) | 22 (81.5) | 8 | 32 | 0.25–32 | 52 (46.8) | 0.5 | 16 | 0.25–32 | 0.001 |
| Levofloxacin | 62 (44.9) | 20 (74.1) | 16 | 32 | 0.25–64 | 42 (37.8) | 4 | 32 | 0.25–128 | 0.001 |
| Gentamycin | 71 (51.4) | 24 (88.9) | 32 | 128 | 1–128 | 47 (42.3) | 1 | 32 | 1–256 | < 0.001 |
| Tobramycin | 73 (52.9) | 23 (85.2) | 32 | 128 | 1–256 | 50 (45.0) | 2 | 128 | 1–256 | < 0.001 |
| Colistin | 32 (23.2) | 7 (25.9) | 1 | 16 | 1–32 | 25 (22.5) | 1 | 8 | 1–32 | 0.707 |
| Tigecycline | 17 (12.3) | 4 (14.8) | 1 | 4 | 0.25–8 | 13 (11.7) | 0.5 | 2 | 0.25–8 | 0.660 |
Data are number resistant (% of resistance rates). P-value for comparisons of the resistance rates of carbapenemase-positive and carbapenemase-negative groups. Bold face indicates values that are significant (P < 0.05). R, resistance.
Genotypic Distribution in CR-ECL Isolates
Among the 138 CR-ECL isolates, 27 (19.6%) were detected as carbapenemase producers: 20 isolates possessed blaNDM-1, five isolates contained blaIMP-8, two isolates carried blaIMP-4, and one isolate had blaKPC-2. Notably, there was one isolate co-harboring blaNDM-1 and blaIMP-8 simultaneously. In addition to the production of carbapenemase, 76.8% (106/138) and 85.5% (118/138) of the CR-ECL isolates were positive for ESBL and AmpC genes, respectively, with 57.9% of the strains positive for both. TEM-type (57.5%, 61/106) was the most prevalent among CR-ECL isolates carrying ESBL, while EBC-type was detected among 93.2% (110/118) of AmpC-producing isolates. Additionally, fluoroquinolone and aminoglycoside genes were detected in 65.9% (91/138) and 44.9% (62/138) of all isolates, with qnrB (39/91) and aac(6′)-Ib (60/62) being the most common, respectively. All of these isolates, except the 11 CP-ECL strains, lost or had a lower expression of at least one major porin with 112 isolates for only one porin and 15 isolates for both porins. Sequencing analysis of the ompF and ompC genes of these CR-ECL isolates ruled out the occurrence of mutations or insertions. Moreover, the MICs of ertapenem were observed to have at least a twofold decrease in the presence of CCCP in 76.8% (106/138) of the CR-ECL isolates (Table 2).
Table 2.
Distribution and corresponding carbapenem MIC ranges for strains with different resistance determinants.
| Carbapenem resistance mechanisms | No. of isolates | MIC range (mg/L) |
||
|---|---|---|---|---|
| ETP | IMP | MEM | ||
| Carbapenemase positive | ||||
| NDM-1, loss or decreased OMPs, efflux pump | 19 | 16–256 | 4–64 | 4–128 |
| NDM-1+IMP-8, loss or decreased OMPs, efflux pump | 1 | 128 | 128 | 32 |
| IMP-8, loss or decreased OMPs, efflux pump | 4 | 8–64 | 0.25–8 | 2–8 |
| IMP-4, no loss or decreased OMPs, efflux pump | 2 | 16 | 0.5–2 | 0.5–1 |
| KPC-2, loss or decreased OMPs | 1 | 64 | 8 | 16 |
| Carbapenemase negative | ||||
| AmpC, ESBLs, loss or decreased OMPs, efflux pump | 42 | 2–64 | 0.5–32 | 0.5–32 |
| AmpC, ESBLs, loss or decreased OMPs | 29 | 2–64 | 0.25–16 | 0.25–32 |
| AmpC, loss or decreased OMPs, efflux pump | 22 | 2–64 | 0.25–16 | 0.25–8 |
| ESBLs, loss or decreased OMPs, efflux pump | 11 | 2–32 | 0.25–2 | 0.25–8 |
| ESBLs, loss or decreased OMPs | 2 | 8 | 0.25–2 | 0.25–4 |
| Loss or decreased OMPs, efflux pump | 5 | 2–8 | 0.25–2 | 0.25–2 |
ETP, ertapenem; IPM, imipenem; MEM, meropenem.
Molecular Analysis of Resistance Mechanisms
As shown in Table 2, all NDM-1-producing strains were resistant to three carbapenem agents, and the MIC range of these isolates was much higher than that of the isolates that carried other carbapenemases. Moreover, approximately half of the carbapenemase-positive isolates with a combination of NDM-1 and porin loss along with an active efflux pump showed the highest MICs of ertapenem, imipenem, and meropenem, while IMP-4, IMP-8, or KPC-2 producers exhibited varied carbapenem MICs. Compared to carbapenemase-positive strains, the remaining 111 isolates showed lost or decreased expression of outer membrane porins, and these isolates were often accompanied by a high prevalence of ESBL (84/111), AmpC (93/111), and efflux pump (80/111). Thus, the resistance mechanism of carbapenemase-negative isolates could be attributed to the productions of ESBL or AmpC enzymes coupled with the efflux pump and porin loss.
Plasmid Analysis
As shown in Table 3, all of the transconjugants and transformants exhibited multidrug resistance phenotypes similar to those of the donor strain with reduced susceptibility to the tested carbapenems and cephalosporins, but remained susceptible to colistin and tigecycline. PCR assays confirmed that blaNDM-1 along with blaCTX-M-9, aac(6′)-Ib-cr and aac(6′)-Ib were successfully transferred by both conjugation and transformation, however, blaIMP-8, TEM, and qnrS were only obtained via the transformation assay. Compared to transconjugants, transformants containing two carbapenemase genes showed a twofold increase in the MIC values for ertapenem and imipenem. Although both transformants and transconjugants exhibited susceptibility to quinolones and aminoglycosides, the MICs of ciprofloxacin and levofloxacin for the transformants were four- and eightfold higher than those for the transconjugants, possibly due to the transformants containing qnrS. PCR-based replicon typing revealed that NDM-1 plasmids belonged to the incompatibility groups IncFIIA (n = 7), IncFIA (n = 3), IncN (n = 3), IncA/C (n = 2), and IncP (n = 1).
Table 3.
Antibiotic resistance genes and susceptibility profiles in donor, E. coli EC600, transconjugant, E.coli DH5α, and transformant.
| Donor strain |
Recipient strains |
Transformant | Transconjugant | ||
|---|---|---|---|---|---|
| EC86 | DH5α | EC600 | |||
| Resistance genes | NDM-1, IMP-8, SHV, TEM, CTX-M-9, aac(6′)-Ib-cr, qnrS, aac(6′)-Ib | NA | NA | NDM-1, IMP-8, TEM, CTX-M-9, aac(6′)-Ib-cr, qnrS, aac(6′)-Ib | NDM-1, CTX-M-9 aac(6′)-Ib-cr, aac(6′)-Ib |
| Sequence type | ST 93 | NA | NA | NA | NA |
| Plasmid replicon type | IncP | NA | NA | IncP | IncP |
| Minimum inhibitory concentration (μg/mL) | |||||
| Ertapenem | 128 | ≤ 0.5 | ≤ 0.5 | 64 | 32 |
| Imipenem | 128 | ≤ 1 | ≤ 1 | 64 | 32 |
| Meropenem | 32 | ≤ 1 | ≤ 1 | 16 | 16 |
| Ceftriaxone | ≥ 64 | ≤ 1 | ≤ 1 | ≥ 64 | ≥ 64 |
| Ceftazidime | ≥ 64 | ≤ 1 | ≤ 1 | ≥ 64 | ≥ 64 |
| Cefepime | ≥ 64 | ≤ 1 | ≤1 | 16 | 8 |
| Ciprofloxacin | ≥ 4 | ≤ 0.25 | ≤ 0.25 | 1 | 0.25 |
| Levofloxacin | ≥ 8 | ≤ 0.25 | 0.5 | 2 | 0.25 |
| Gentamycin | ≥ 16 | ≤ 1 | ≤ 1 | ≤ 1 | ≤1 |
| Tobramycin | ≥ 16 | ≤ 1 | ≤ 1 | ≤ 1 | ≤1 |
| Colistin | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 |
| Tigecycline | ≤ 1 | ≤1 | ≤ 1 | ≤ 1 | ≤ 1 |
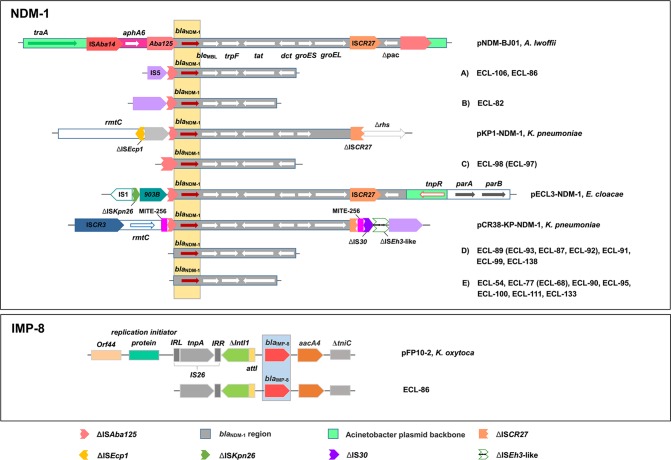
Characterization of the Genetic Environment of NDM-1 and IMP-8
For NDM-1, 15 NDM-1-producing strains could be divided into five different types (A–E) based on the analysis of genetic structures, among which type E was the most predominant (n = 7), followed by type D (n = 4), type A (n = 2), type B (n = 1), and type C (n = 1). Type A had a partial insertion sequence, IS5, truncating the ISAba125 upstream of the blaNDM-1 gene and followed by the bleMBL (bleomycin resistance gene), the trpF, the tat, and the truncated dct genes. Type B harbored similar structures as type A, but it accompanied an intact IS5 element and a complete deletion of dct gene. Type C presented as a truncated ISAba125 upstream of the blaNDM-1 gene compared with Type A, but without the IS5 element. Type D carried four genetic elements downstream of the blaNDM-1 gene, including bleMBL, trpF, tat, and dct. Type E possessed similar genetic structures as type D, but it carried a complete deletion of dct gene.
For IMP-8, the genetic structure identified from CR-ECL86 was similar to the one previously reported in Klebsiella oxytoca plasmids pFP10-2 (GenBank Accession No. HQ651093) from China. The blaIMP-8 gene in CR-ECL86 was preceded by a recombination site (attI1) and followed by an aminoglycoside acetyltransferase gene (aacA4) and a truncated transposase gene (ΔtniC). A class 1 integron (Intl1), located upstream of the blaIMP-8 gene in this isolate, was truncated due to the insertion of IS26. Compared to pFP10-2, the IS26 insertion in CR-ECL86 shared identical gene cassettes except for the missing left-inverted repeat sequence (LRR) in the 5′-conserved region (Figure 2).
FIGURE 2.
Comparison of the genetic elements surrounding the blaNDM-1 and blaIMP-8 genes identified in this study. Reference sequences: A. lwoffii (pNDM-BJ01, GenBank accession no. JQ001791), K. pneumoniae (pKP1-NDM-1, GenBank accession no. KF992018 and pCR38-KP-NDM-1, GenBank accession no. KP826710), and E. cloacae (pECL3-NDM-1, GenBank accession no. KC887917).
Risk Factors and Clinical Outcomes of NDM-1-Producing E. cloacae
To further clarify the risk factors and clinical outcomes for patients with NDM-1-producing E. cloacae infection, 20 NDM-1-positive E. cloacae cases were matched to 111 NDM-1-negative controls (Table 4). In the univariate analysis, patients with acquisition of NDM-1-positive E. cloacae were significantly more frequent in elderly, transferring from another hospital, some underlying diseases, and previous cephalosporins and carbapenem exposure (P < 0.05). In the multivariate analysis, transferring from another hospital (OR: 4.82, 95% CI: 1.14–20.35, P = 0.033) and previous carbapenem exposure (OR: 7.00, 95% CI: 1.86–26.33, P = 0.004) were identified as independent risk factors. For clinical outcomes, we found that the total length of hospital stay and functional status deterioration were significantly more frequent in patients with NDM-1 isolates. For in-hospital mortality, 4 (20.0%) patients infected with NDM-1 and 8 (7.2%) patients infected without NDM-1 died. However, there was no statistical significance between the cases and the controls with respect to in-hospital mortality.
Table 4.
Univariable and multivariable analysis of risk factors and outcomes for isolation of NDM-1-positive CR-ECL isolates compared with negative controls.
| Variable | NDM-1-negative E. cloacae (N = 111) | NDM-1-positive E. cloacae (N = 20) | Univariate analysis |
Multivariate analysis |
||
|---|---|---|---|---|---|---|
| OR (95% CI) | P-value | OR (95% CI) | P-value | |||
| Demographics, n (%) | ||||||
| Elderly (≥ 60 years) | 21 (18.9%) | 8 (40.0%) | 2.86 (1.04-7.87) | 0.045 | ||
| Male gender | 52 (46.8%) | 9 (45.0%) | 0.93 (0.36-2.42) | 1.000 | ||
| Surgical units | 32 (28.8%) | 9 (45.0%) | 2.02 (0.76-5.34) | 0.191 | ||
| Transfer from another hospital | 18 (16.2%) | 10 (50.0%) | 5.17 (1.88-14.21) | 0.001 | 4.82 (1.14-20.35) | 0.033 |
| Admission to ICU | 22 (19.8%) | 7 (35.0%) | 2.18 (0.78-6.11) | 0.132 | ||
| Comorbid conditions, n (%) | ||||||
| Hypertension | 23 (20.7%) | 4 (20.0%) | 0.96 (0.29-3.14) | 1.000 | ||
| Malignant disease | 13 (11.7%) | 5 (25.0%) | 2.51 (0.78-8.06) | 0.152 | ||
| Cardiovascular disease | 12 (10.8%) | 2 (10.0%) | 0.92 (0.19-4.45) | 1.000 | ||
| Chronic kidney disease | 16 (14.4%) | 4 (20.0%) | 1.48 (0.44-5.01) | 0.508 | ||
| Hepatobiliary disease | 38 (34.2%) | 6 (30.0%) | 0.82 (0.29-2.31) | 0.712 | ||
| Gastrointestinal disease | 25 (22.5%) | 5 (25.0%) | 1.15 (0.38-3.47) | 0.778 | ||
| Respiratory diseases | 21 (18.9%) | 9 (45.0%) | 3.51 (1.29-9.54) | 0.018 | ||
| Neurological disease | 16 (14.4%) | 2 (10.0%) | 0.66 (0.14-3.12) | 1.000 | ||
| Endocrine, metabolic disease | 3 (2.7%) | 1 (5.0%) | 1.89 (0.19-19.19) | 0.489 | ||
| Vascular, hematological disease | 15 (13.5%) | 4 (20.0%) | 1.60 (0.47-5.44) | 0.491 | ||
| Hypoalbuminaemia | 24 (21.6%) | 9 (45.0%) | 2.97 (1.10-7.98) | 0.047 | ||
| Neutropenia | 24 (21.6%) | 6 (30.0%) | 1.55 (0.54-4.47) | 0.412 | ||
| Urinary tract infection | 8 (7.2%) | 1 (5.0%) | 0.68 (0.08-5.74) | 1.000 | ||
| Respiratory infection | 27 (24.3%) | 8 (40.0%) | 2.07 (0.77-5.61) | 0.172 | ||
| Invasive procedures within prior 4 weeks, n (%) | ||||||
| Surgery in the past 6 months | 29 (26.1%) | 9 (45.0%) | 2.31 (0.87-6.15) | 0.109 | ||
| Receipt of total parenteral nutrition | 8 (7.2%) | 3 (15.0%) | 2.27 (0.55-9.43) | 0.372 | ||
| Bronchoscopes | 9 (8.1%) | 3 (15.0%) | 2.00 (0.49-8.14) | 0.325 | ||
| Mechanical ventilation | 30 (27.0%) | 11 (55.0%) | 3.30 (1.24-8.75) | 0.013 | ||
| Bladder irrigation | 7 (6.3%) | 2 (10.0%) | 1.65 (0.32-8.59) | 0.626 | ||
| Drainage tube | 42 (37.8%) | 9 (45.0%) | 1.34 (0.51-3.51) | 0.621 | ||
| Urinary catheter | 33 (29.7%) | 8 (40.0%) | 1.58 (0.59-4.21) | 0.433 | ||
| Tracheal cannula | 25 (22.5%) | 2 (10.0%) | 0.38 (0.08-1.76) | 0.247 | ||
| Nasal catheter | 14 (12.6%) | 4 (20.0%) | 1.73 (0.51-5.93) | 0.477 | ||
| Central venous catheter | 19 (17.1%) | 3 (15.0%) | 0.85 (0.23-3.21) | 1.000 | ||
| Antimicrobial exposure within 3 months, n (%) | ||||||
| Penicillin | 2 (1.8%) | 0 (0.0%) | 0.98 (0.96-1.01) | 1.000 | ||
| Cephalosporins | 37 (33.3%) | 12 (60.0%) | 3.00 (1.13-7.98) | 0.042 | ||
| Carbapenem | 22 (19.8%) | 13 (65.0%) | 7.51 (2.68-21.01) | < 0.001 | 7.00 (1.86-26.33) | 0.004 |
| Fluoroquinolone | 17 (15.3%) | 4 (20.0%) | 1.38 (0.41-4.64) | 0.599 | ||
| Aminoglycosides | 13 (11.7%) | 4 (20.0%) | 1.89 (0.55-6.51) | 0.294 | ||
| Glycopeptide | 16 (14.4%) | 5 (25.0%) | 1.98 (0.63-6.20) | 0.316 | ||
| Combined use of antibiotics | 20 (18.0%) | 4 (20.0%) | 1.14 (0.34-3.77) | 0.762 | ||
| Clinical outcomes | ||||||
| In-hospital mortality, n (%) | 8 (7.2%) | 4 (20.0%) | 3.22 (0.87-11.94) | 0.087 | ||
| Functional status deterioration | 13 (11.7%) | 6 (30.0%) | 3.23 (1.06-9.88) | 0.044 | ||
| Postculture length of stay, median (IQR), days | 9 (5-16) | 13.5 (7.25-18.75) | NA | 0.300 | ||
| Total length of hospital stay, median (IQR), days | 29 (22-38) | 39 (27.25-64.75) | NA | 0.015 | ||
Bold face indicates values that are significant (P < 0.05). ICU, intensive care unit.
Molecular Epidemiology of CP-ECL Isolates
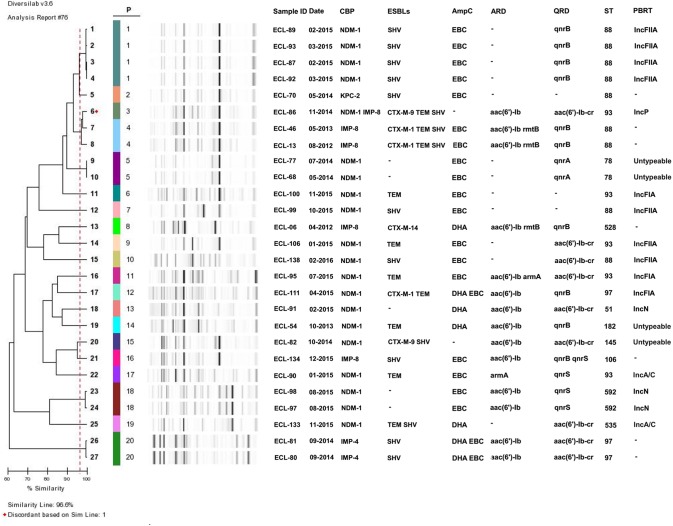
The 27 carbapenemase-producing E. cloacae isolates belonging to 20 different clusters are shown in Figure 3. Of these, the main cluster (cluster 1), showing four NDM-1-carrying E. cloacae strains, was isolated from the respiratory department between February and March in 2015. Four smaller clusters (cluster 4, 5, 18, and 20) represented two IMP-8 isolates, two IMP-4 isolates, and four NDM-1 isolates, respectively, but these strains were collected in different wards. MLST typing revealed that ST88 was the most common among CP-ECL isolates (9/27, 33.3%), followed by ST93 (5/27, 18.5%), ST97 (3/27, 11.1%), ST78 (2/27, 7.4%), and ST592 (2/27, 7.4%).
FIGURE 3.
Dendrogramanalysis of DiversiLab Rep-PCR fingerprint of carbapenemase-producing E. cloacae isolates. A genetic similarity index scale is shown in the left of the dendrogram. Isolate number, collection data, resistance determinants, MLST, and plasmid type. CBP, carbapenemase; ESBLs, extended-spectrum β-lactamase; PBRT, PCR-based replicon typing.
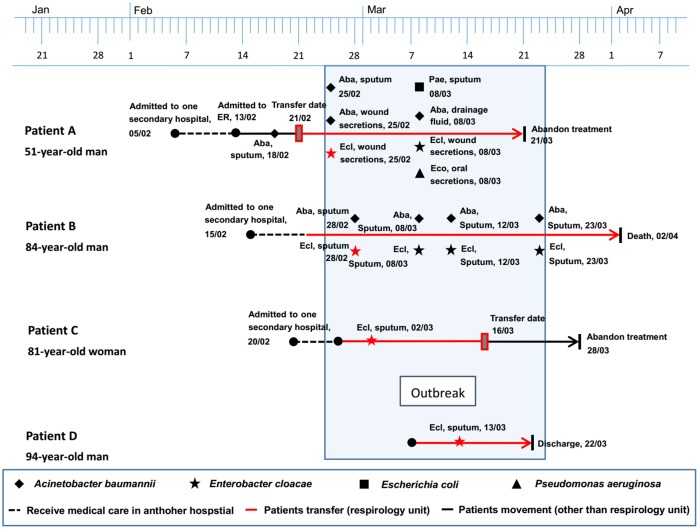
Description of the Outbreak Caused by NDM-1-Producing E. cloacae Strains
This outbreak involved four E. cloacae isolates retrieved from four hospitalized patients aged 51–94 years between February and March in 2015. Three of these four patients, who developed complications of respiratory failure and severe pneumonia, had been transferred from a secondary hospital to the respiratory unit in our setting. Strain ECL-87 was isolated from patient A on February 25, 2015, which could be the first of the outbreak NDM-1-producing isolates. Over the following 17 days, another three isolates (ECL-89, ECL-92, and ECL-93) were retrieved from three patients (patient B, C, and D) in the same department, having an average of a new NDM-1-producing E. cloacae infection emerging approximately every 5.6 days. In addition to the isolation of E. cloacae, multiple species of bacteria (including Acinetobacter baumannii, E. coli, and Pseudomonas aeruginosa) were recovered from the wound secretion, sputum, and drainage fluid of patients A and B. Additionally, all of the patients underwent a series of antimicrobial treatment and some invasive procedures. During the outbreak, patients A and C stopped further treatment with unimproved symptoms, and patient B died because of multiple organ dysfunction. The timeline of the outbreak investigation is illustrated in Figure 4.
FIGURE 4.
Timeline of the NDM-1-producing E. cloacae outbreak cases.
Nucleotide Sequence Accession Number
The sequence described in this paper has been submitted to GenBank under the Accession No. MH087224 (E. cloacae isolate CQECL-100).
Discussion
In light of carbapenem resistant E. cloacae being widely distributed and showing the highest resistant rate among CRE in Southwest China, which was substantially different from other CRE studies in South Asia, the United States, and Europe (Nordmann et al., 2009; Pollett et al., 2014; Hsu et al., 2017), the issue of carbapenem resistant E. cloacae in our region deserved special concern. In this work, we have found some particularly noteworthy findings. First, our results revealed that the main mechanism of CR-ECL isolates collected in this study could be attributed to the productions of ESBL, AmpC enzymes coupled with the efflux pump and porin loss. Notably, the high prevalence (74%) of NDM-1-producing E. cloacae was observed among the CP-ECL isolates, with the annual increasing tendency during the study period. Previous studies have showed the emergence and spread of NDM-1-producing E. cloacae isolates in China, with most isolates carrying on the IncX3 and IncA/C plasmids (Yang et al., 2015). However, we reported the high prevalence of IncFIIA plasmids carrying NDM-1 (55%) in our region, revealing that this type of plasmids could be further disseminated in China.
Second, of the NDM-1 producers detected in our study, four isolates from the respiratory department showed the identical DiversiLab pattern, belonged to the same clone ST88 and displayed the same IncFIIA plasmids, providing evidence that the outbreak has emerged in our hospital. One possible source of this outbreak might be that patient A was exposed to a patient infected with NDM-1 in another hospital before the admission to our hospital. This alternative possibility may have contributed to the transmission from another unidentified colonized patient who has been discharged in the same department before the identification of these patients. To the best of our knowledge, this is the first report of a nosocomial outbreak of NDM-1-producing E. cloacae ST88. Most previous studies appeared to be restricted to report the sporadic cases of E. cloacae isolates harboring NDM-1, with diverse clones from geographic regions such as ST92 in Croatia, ST265 in Australia, and ST90, ST93, ST120, and ST177 in China (Atalić et al., 2014; Wailan et al., 2015). ST88, a globally distributed resistant clone belonging to clonal complex 23, was the most frequent ST found in E. cloacae isolates possessing NDM-1 in our study. The outbreak of this sequence type may provide a new model for the spread of the blaNDM-1-haboring CRE in China and should be of great concern.
Third, we reported for the first time the emergence of E. cloacae strain co-producing NDM-1 and IMP-8, which was isolated from a patient with urinary tract infection. This isolate showed highly resistance to many antibiotics, but remained susceptible to tigecycline and colistin. Moreover, the transmission assays of these two carbapenemase genes revealed that both NDM-1 and IMP-8 were located on separate IncP plasmids, suggesting two plasmids containing IncP replicons might be compatible in a single strain. Of greater interest was that all NDM-1 producers carried the highly conserved regions (blaNDM-1-bleMBL-trpF-tat) surrounding the blaNDM-1 gene, which were similar to that found in various NDM-1-harboring plasmids, including K. pneumoniae from Australia, Acinetobacter lwoffii from China, and E. cloacae from Australia (Wailan et al., 2015, 2016). This finding suggested that the transmission of NDM-1 could be widely spread among different bacterial species and geographic regions. In addition, although several reports on IMP-producing E. cloacae have emerged in recent years, the genetic elements with respect to blaIMP-8 in this species are still missing. Our results demonstrated that the genetic environment of this species was similar to the previous report found in K. oxytoca from China (Li et al., 2011), with the exception of missing a left-inverted repeat sequence in the 5′-conserved region. It should be noted that the transposon elements IS26, intI1, and tniC could facilitate the mobilization of IMP-8. Considering that the strain coproducing NDM-1 and IMP-8 may confer a higher-level resistance to multiple antibiotics that further reduce the therapeutic choices, surveillance for carbapenemase detection and infection control measures should be implemented to prevent their further spread.
Fourth, we initially performed a retrospective analysis to assess clinical predictors and outcomes for NDM-1-producing E. cloacae. Compared to previous studies, our results demonstrated that transferring from another hospital was an independent risk factor associated with the acquisition of NDM-1-producing E. cloacae. One possible explanation may be that these patients could have poor functional status and severe clinical symptoms which placed them at a greater risk for the infection caused by NDM-1-producing E. cloacae. Furthermore, few laboratories in Chinese secondary hospitals regularly conducted testing for CRE resistance mechanisms, especially for certain novel carbapenemases, such as NDM-1. Therefore, patients infected or colonized with NDM-1 could be the source of the horizontal transmission in different hospital environments. In addition, our study identified carbapenem exposure was a risk factor for case patients, in agreement with previous reports on the assessment of CRE (Fukuda et al., 2008; Hyle et al., 2010). Receipt of these agents could potentially produce a disruption in gastrointestinal flora and accelerate bacterial colonization, emphasizing the important role of antimicrobial stewardship in controlling NDM-1 transmission. For clinical outcomes, patients infected with NDM-1 were at a higher risk, in general, of poor outcomes than those without NDM-1. However, our results unexpectedly revealed that no statistical significance was observed in in-hospital mortality, probably due to the limited number of NDM-1-positive cases in our study.
In summary, this present study revealed the high prevalence of NDM-1 among carbapenem resistant E. cloacae in Southwest China. We initially reported the nosocomial outbreak caused by NDM-1-producing E. cloacae ST88, as well as the co-production of NDM-1 and IMP-8 in a single E. cloacae isolate. We also identified independent risk factors for the acquisition of NDM-1-producing E. cloacae, and our findings highlight an urgent need to develop effective measures to prevent and control the further spread of NDM-1 in China.
Author Contributions
LZ, XJ, and WD designed the study. XJ, WD, WM, JY, JH, and SL performed the experiments. SY, XX, SS, JS, and CL analyzed the data. XJ wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This study was supported in part by the National Natural Science Foundation of China (Grant Nos. 81471992, 81772239, and 31500749) and the Science Foundation of Chongqing (Grant Nos. cstc2016jcyjA0248 and cstc2015jcyjA10102).
References
- Atalić V. Z., Bedenić B., Kocsis E., Mazzariol A., Sardelić S., Barišić M., et al. (2014). Diversity of carbapenemases in clinical isolates of Enterobacteriaceae in Croatia—the results of a multicentre study. Clin. Microbiol. Infect. 20 894–903. 10.1111/1469-0691.12635 [DOI] [PubMed] [Google Scholar]
- Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55 611–622. 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- Carattoli A., Bertini A., Villa L., Falbo V., Hopkins K. L., Threlfall E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63 219–228. [DOI] [PubMed] [Google Scholar]
- Chollet R., Chevalier J., Bollet C., Pages J. M., Davin-Regli A. (2004). RamA is an alternate activator of the multidrug resistance cascade in Enterobacter aerogenes. Antimicrob. Agents Chemother. 48 2518–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute [CLSI] (2015). M100-S25. Performance Standards for Antimicrobial Susceptibility Testing. 25th Informational Supplement. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Cornaglia G., Giamarellou H., Rossolini G. M. (2011). Metallo-beta-lactamases: a last frontier for beta-lactams? Lancet Infect. Dis. 11 381–393. 10.1016/S1473-3099(11)70056-1 [DOI] [PubMed] [Google Scholar]
- Dai W., Sun S., Yang P., Huang S., Zhang X., Zhang L. (2013). Characterization of carbapenemases, extended spectrum beta-lactamases and molecular epidemiology of carbapenem-non-susceptible Enterobacter cloacae in a Chinese hospital in Chongqing. Infect. Genet. Evol. 14 1–7. 10.1016/j.meegid.2012.10.010 [DOI] [PubMed] [Google Scholar]
- Doumith M., Ellington M. J., Livermore D. M., Woodford N. (2009). Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J. Antimicrob. Chemother. 63 659–667. 10.1093/jac/dkp029 [DOI] [PubMed] [Google Scholar]
- Fukuda S., Kuwabara S., Yasuda M., Mizuno K., Kato T., Sugiura T., et al. (2008). Predictors of carbapenem-resistant Klebsiella pneumoniae acquisition among hospitalized adults and effect of acquisition on mortality. Antimicrob. Agents Chemother. 52 1028–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu L. Y., Apisarnthanarak A., Khan E., Suwantarat N. (2017). Carbapenem-resistant Acinetobacter baumannii and Enterobacteriaceae in South and Southeast Asia. Clin. Microbiol. Rev. 30 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyle E. P., Ferraro M. J., Silver M., Lee H., Hooper D. C. (2010). Ertapenem-resistant Enterobacteriaceae: risk factors for acquisition and outcomes. Infect. Control Hosp. Epidemiol. 31 1242–1249. [DOI] [PubMed] [Google Scholar]
- Li B., Sun J. Y., Liu Q. Z., Han L. Z., Huang X. H., Ni Y. X. (2011). First report of Klebsiella oxytoca strain coproducing KPC-2 and IMP-8 carbapenemases. Antimicrob. Agents Chemother. 55 2937–2941. 10.1128/AAC.01670-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Qin S., Xu H., Xu L., Zhao D., Liu X., et al. (2015). New Delhi metallo-beta-lactamase 1(NDM-1), the dominant carbapenemase detected in carbapenem-resistant Enterobacter cloacae from Henan Province, China. PLoS One 10:e0135044. 10.1371/journal.pone.0135044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahida N., Clarke M., White G., Vaughan N., Boswell T. (2017). Outbreak of Enterobacter cloacae with New Delhi metallo-beta-lactamase (NDM)-1: challenges in epidemiological investigation and environmental decontamination. J. Hosp. Infect. 97 64–65. [DOI] [PubMed] [Google Scholar]
- Nordmann P., Cuzon G., Naas T. (2009). The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect. Dis. 9 228–236. 10.1016/S1473-3099(09)70054-4 [DOI] [PubMed] [Google Scholar]
- Pollett S., Miller S., Hindler J., Uslan D., Carvalho M., Humphries R. M. (2014). Phenotypic and molecular characteristics of carbapenem-resistant Enterobacteriaceae in a health care system in Los Angeles, California, from 2011 to 2013. J. Clin. Microbiol. 52 4003–4009. 10.1128/JCM.01397-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasheed J. K., Kitchel B., Zhu W., Anderson K. F., Clark N. C., Ferraro M. J., et al. (2013). New Delhi metallo-beta-lactamase-producing Enterobacteriaceae, United States. Emerg. Infect. Dis. 19 870–878. 10.3201/eid1906.121515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoesser N., Sheppard A. E., Shakya M., Sthapit B., Thorson S., Giess A., et al. (2015). Dynamics of MDR Enterobacter cloacae outbreaks in a neonatal unit in Nepal: insights using wider sampling frames and next-generation sequencing. J. Antimicrob. Chemother. 70 1008–1015. 10.1093/jac/dku521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L., Tan R., Chen Y., Sun J., Liu J., Qu H., et al. (2016). Epidemiology of Klebsiella pneumoniae bloodstream infections in a teaching hospital: factors related to the carbapenem resistance and patient mortality. Antimicrob. Resist. Infect. Control 5:48. 10.1186/s13756-016-0145-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wailan A. M., Paterson D. L., Kennedy K., Ingram P. R., Bursle E., Sidjabat H. E. (2015). Genomic characteristics of NDM-producing Enterobacteriaceae isolates in Australia and their blaNDM genetic contexts. Antimicrob. Agents Chemother. 60 136–141. 10.1128/AAC.01243-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wailan A. M., Sidjabat H. E., Yam W. K., Alikhan N. F., Petty N. K., Sartor A. L., et al. (2016). Mechanisms involved in acquisition of blaNDM genes by IncA/C2 and IncFIIY plasmids. Antimicrob. Agents Chemother. 60 4082–4088. 10.1128/AAC.00368-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Liu Q., Han L., Sun J., Ni Y. (2010). Plasmid-mediated carbapenem-hydrolyzing enzyme KPC-2 and ArmA 16S rRNA methylase conferring high-level aminoglycoside resistance in carbapenem-resistant Enterobacter cloacae in China. Diagn. Microbiol. Infect. Dis. 66 326–328. 10.1016/j.diagmicrobio.2009.10.003 [DOI] [PubMed] [Google Scholar]
- Yang L., Wu A. W., Su D. H., Lin Y. P., Chen D. Q., Qiu Y. R. (2014). Resistome analysis of Enterobacter cloacae CY01, an extensively drug-resistant strain producing VIM-1 metallo-beta-lactamase from China. Antimicrob. Agents Chemother. 58 6328–6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Fang L., Fu Y., Du X., Shen Y., Yu Y. (2015). Dissemination of NDM-1-producing Enterobacteriaceae mediated by the IncX3-type plasmid. PLoS One 10:e0129454. 10.1371/journal.pone.0129454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Xu X., Pu S., Huang S., Sun J., Yang S., et al. (2014). Characterization of carbapenemases, extended spectrum beta-lactamases, quinolone resistance and aminoglycoside resistance determinants in carbapenem-non-susceptible Escherichia coli from a teaching hospital in Chongqing, Southwest China. Infect. Genet. Evol. 27 271–276. 10.1016/j.meegid.2014.07.031 [DOI] [PubMed] [Google Scholar]